Graphical abstract
Abbreviations: ATR-FTIR, attenuated total reflection Fourier transform infrared; dDMPC, 1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; MCMC, Markov chain Monte Carlo; MP, membrane protein; MSP, membrane scaffold protein; MWCO, molecular weight cut-off; NaOAc, sodium acetate; NR, neutron reflectometry; RAFT, reversible addition-fragmentation chain transfer; RAFT-SMA, RAFT-synthesised SMA; SEC, size exclusion chromatography; SiMW, silicon-matched water; SLD, scattering length density; SMA, poly(styrene-co-maleic acid); SMALP, SMA lipid particle; SMAnh, poly(styrene-co-maleic anhydride); SMI, poly(styrene-co-maleimide); SMILP, SMI lipid particle
Keywords: Supported lipid bilayer, Polymer-stabilized phospholipid nanodisc, styrene-maleic acid lipid particle (SMALP), styrene maleic acid (SMA), Neutron reflectometry, Lipid exchange, Adsorption
Abstract
Over recent years, there has been a rapid development of membrane-mimetic systems to encapsulate and stabilize planar segments of phospholipid bilayers in solution. One such system has been the use of amphipathic copolymers to solubilize lipid bilayers into nanodiscs. The attractiveness of this system, in part, stems from the capability of these polymers to solubilize membrane proteins directly from the host cell membrane. The assumption has been that the native lipid annulus remains intact, with nanodiscs providing a snapshot of the lipid environment. Recent studies have provided evidence that phospholipids can exchange from the nanodiscs with either lipids at interfaces, or with other nanodiscs in bulk solution. Here we investigate kinetics of lipid exchange between three recently studied polymer-stabilized nanodiscs and supported lipid bilayers at the silicon-water interface. We show that lipid and polymer exchange occurs in all nanodiscs tested, although the rate and extent differs between different nanodisc types. Furthermore, we observe adsorption of nanodiscs to the supported lipid bilayer for one nanodisc system which used a polymer made using reversible addition-fragmentation chain transfer polymerization. These results have important implications in applications of polymer-stabilized nanodiscs, such as in the fabrication of solid-supported films containing membrane proteins.
1. Introduction
The inherent amphiphilic nature of biological membranes has been a persistent challenge when attempting to study membrane proteins (MPs). These molecules represent an important class of biological macromolecules, as exemplified by representing approximately 30% of the protein-coding regions of the human genome and 60% of therapeutic drug targets [1], [2], [3]. MPs require stabilization of a hydrophobic, transmembrane core for solubility in aqueous media. This has traditionally been achieved by the use of ‘head and tail’ surfactants which act by assembling a micelle around the MP of interest, with hydrophobic tails stabilizing the transmembrane regions of the protein, and hydrophilic head groups allowing interactions with the aqueous solvent [4], [5], [6]. However, this leads to issues with stability, denaturation and suppression of conformational flexibility which is frequently a bottleneck when attempting in vitro studies of isolated MPs [7], [8].
Over the past 20 years there has been a rapid development of alternative solubilization strategies attempting to overcome the limitations of classical surfactant mediated MP solubilization [9], [10], [11], [12]. One such solution has been nanodiscs: nanoscale discoidal colloidal particles which consist of a central core of a planar phospholipid bilayer which are stabilized by an amphiphilic ‘belt’ consisting of either peptides, membrane scaffold proteins (MSPs) or synthetic copolymers [12], [13], [14]. An amphipathic copolymer, poly(styrene-co-maleic acid) (SMA), has been shown to self-assemble in the presence of phospholipids to form nanodiscs [15]. These polymer-stabilized nanodiscs, where the lipid core is stabilized in aqueous solution by a polymer belt [16], have been termed SMA lipid particles (SMALPs). SMA has the advantage of being able to solubilize MPs directly from the cell membrane, without the need for classical surfactants at any stage [17]. SMALPs have been reported to both maintain protein function and improve stability over classical surfactant solubilized MPs and also MPs in native membranes [18], [19]. Furthermore, the capability of SMA to extract MPs directly from the cell membrane maintains a more native-like lipid environment, facilitating functional [20], [19], [21] and structural [22], [23], [24], [25] studies of SMALP-solubilized MPs.
There have now been a number of studies detailing the self-assembly and structure of SMALP nanodiscs from a range of commercially available SMA copolymers [26], [27], [28], [29], [30]. This has spawned a drive to develop new nanodisc-forming polymers which are able to form nanodiscs with increased stability, enhanced buffer compatibility, modified size and improved properties for a larger range of downstream applications [14]. One example is the synthesis of SMA polymers by reversible addition-fragmentation chain transfer (RAFT) polymerization. These polymers have a narrower size distribution than commercial SMA polymers and have a substantially different monomer architecture along the polymer chain, whereby the styrene and maleic acid monomers are initially alternating, tending towards longer poly(styrene) stretches along the length of the chain as maleic acid is consumed throughout the synthesis reaction [31]. RAFT-SMA has been shown to form nanodiscs in a similar manner to commercial SMAs but forming nanodiscs of increased size and with greater thermodynamic efficiency [30], [32].
Another polymer, poly(styrene-co-maleimide) (SMI) has recently been shown to form nanodiscs termed SMI lipid particles (SMILPs) [33]. In contrast to SMA, SMI contains positively charged dimethylaminopropylamine maleimide in place of maleic acid as the hydrophilic component. This has a number of advantages over SMA, including solubility under acidic conditions and a high tolerance to divalent cations. While SMI is capable of efficiently solubilizing phospholipids to form nanodiscs, SMILPs are somewhat smaller than SMALPs and SMI is less efficient at MP solubilization from biological membranes.
Due to the therapeutic importance of MPs, nanodiscs have gained significant interest in application to characterizing MP-ligand interactions and drug discovery. An increasingly adopted strategy involves adsorption of the MP of interest to interfaces allowing interaction screens against large compound libraries to be performed using techniques such as surface plasmon resonance (SPR) [34]. MSP-nanodiscs have been shown to adsorb to the air-water and Si-water interfaces where the plane of the membrane lies parallel to the surface under investigation [35]. Further studies have shown that MSP-nanodiscs containing cytochrome P450 reductase align at the Si-water interface in a similar manner whilst retaining dynamic conformational flexibility and solvent accessibility of globular, soluble domains [36], [37].
Inspired by this work, the interaction of SMALP nanodiscs containing a phospholipid bilayer with a net-negative charge on synthetic lipid monolayers containing positively charged surfactants was studied using neutron reflectometry. It was hoped that an electrostatic interaction between nanodiscs and lipid interfaces would aid the adsorption process. However, the data could not be fitted assuming a meaningful coverage of adsorbed nanodiscs, but instead lipid exchange between the lipids in the monolayers and nanodiscs was found to be the only explanation, providing the first evidence of the phenomenon of lipid exchange occurring with SMALPs [38].
Lipid exchange between SMALP nanodiscs in solution has since been investigated in more detail. These studies revealed a combination of a rapid collisional and slower monomer diffusion mechanism underlying the transfer of lipids between nanodiscs in solution where the rate of lipid exchange is increased further with increasing ionic strength [39], [40]. Furthermore, lipid exchange is not unique to an individual SMA polymer, where RAFT-SMA [38], SMA(2:1) [40] (containing a 2:1 ratio of styrene:maleic acid monomers, tradename Xiran SZ30010) and SMA(3:1) [39] (tradename Xiran SL25010 S25) have now been shown to exhibit rapid lipid exchange between nanodiscs.
While much research has been conducted characterizing lipid exchange between nanodiscs in solution, the interaction with different nanodisc types and model membranes remains relatively unknown. Here, we have investigated the interactions between nanodiscs and lipid bilayers at the Si-water interface as mimics of biological membranes. We have use nanodiscs formed of three polymers: SMA2000 (SMA), SMA2000I (SMI) and a RAFT-synthesized SMA (RAFT-SMA), each containing a 2:1 ratio of styrene:maleic acid (or maleimide in the case of SMI) [30], [33]. While lipid exchange at interfaces has been reported for nanodiscs bound by RAFT-SMA [38], there have been no such reports for SMA2000 or SMI. We determine the kinetics of lipid exchange using attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy and show that each nanodisc type exhibits different kinetics for deposition and removal of lipids. We proceed to structurally characterize lipid bilayers before and after interaction with nanodiscs using neutron reflectometry (NR). Contrary to earlier studies, in the process of carrying out these measurements we have observed an adsorption of SMALPs formed by RAFT-SMA to phospholipid bilayers at the Si-water interface in addition to lipid exchange, an unexpected result. The presence of polymer in the membrane can also be identified with each nanodisc system. The results presented here have important implications in membrane and membrane protein research, challenging the viewpoint of polymer-bound nanodiscs representing a static and kinetically trapped snapshot of a native cell-membrane environment.
2. Experimenal section
2.1. Materials
Fully hydrogenated 1,2-dimyristoyl-sn-glycero-3-phosphocholine (hDMPC) and tail-deuterated 1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine (dDMPC) were purchased from Avanti Polar Lipids. SMA2000 and SMA2000I resin was purchased from Cray Valley. All other chemicals were purchased from Sigma-Aldrich at >98% purity and used without further purification.
2.2. Methods
2.2.1. Polymer solubilization
Both RAFT-SMAnh (synthesized as previously described) [30], [41] and SMAnh require hydrolysis from the anhydride to the acid forms to become soluble in aqueous solution, and for functionality in SMALP nanodisc self-assembly. Copolymer hydrolysis was performed as previously described. In brief, a 10% w/v suspension of either SMAnh or RAFT-SMAnh in 1 M NaOH was heated under reflux for 2 hrs. Once the clarified solution had cooled, soluble SMA was precipitated by the dropwise addition of concentrated HCl until pH < 5.0. Precipitated SMA was pelleted by centrifugation at 10,000 × g and washed 3 times in water. After washing, precipitated SMA was dissolved in 0.6 M NaOH and incubated at 37 °C for 16 h before repeating the precipitation and washing procedure. The final hydrolyzed SMAs were again dissolved in a minimal volume of 0.6 M NaOH, adjusted to pH 8.0 by dropwise addition of concentrated HCl, lyophilized and used without further purification.
Similarly SMI was solubilized as previously described [33]. This was essentially performed in the same manner as for both SMA copolymers, except 10% w/v SMA2000I resin in 1 M HCl was heated under reflux for 2 h. The precipitation and washing procedure was identical as for SMA, except the polymer was precipitated by the dropwise addition of 5 M NaOH until pH > 9.0 prior to redissolution in 0.6 M HCl. After washing, precipitated, solubilized SMI was dissolved in a minimal volume of 0.6 M HCl, adjusted to pH 5.0 by dropwise addition of 5 M NaOH, lyophilized and used without further purification.
2.2.2. Nanodisc preparation
hDMPC powder was dissolved in CHCl3 and dried under a stream of N2 to create a multilamellar lipid film around a glass vial. Vials were desiccated for at least 2 h to remove residual solvent. For experiments using SMA, lipids were rehydrated in 50 mM sodium phosphate, 200 mM NaCl, pH 8.0. For experiments using SMI, lipids were rehydrated in 50 mM sodium acetate, 200 mM NaCl, pH 5.0. In all cases, the buffer was warmed to 30 °C and lipids hydrated to 10 mg/mL. Vesicles were formed by sonication at 30 °C for 30 min. 3% w/v polymer was prepared in the appropriate buffer (SMA − 50 mM sodium phosphate, 200 mM NaCl, pH 8.0; SMI − 50 mM sodium acetate (NaOAc), 200 mM NaCl, pH 5.0) and added to vesicles at a 1:1 v/v ratio to a final volume of 10 mL. The nanodisc solutions were allowed to equilibrate at room temperature for 24 h. Nanodiscs were concentrated in 10,000 MWCO centrifugal concentrator tubes to a volume of 3 mL. Concentrated nanodiscs were purified by size exclusion chromatography (SEC) using a HiLoad Superdex 200 26/600 column, equilibrated in the same buffer, attached to an Äkta purification system (GE Healthcare Life Sciences, Buckinghamshire, UK) monitoring absorbance at 254 nm (Fig. S1). Fractions containing nanodiscs were pooled, flash frozen in liquid nitrogen and stored at −80 °C.
2.2.3. Attenuated total reflection Fourier transform infrared spectroscopy
Attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR) describes the chemical species present at the solid-liquid interface through the identification of discreet IR active functional groups. An infrared (IR) beam is totally internally reflected through a substrate in contact with bulk solvent, where the refractive index of the substrate, n1, must be higher than that of the solvent, n2, to allow for total internal reflection. As the beam is reflected from the substrate-solvent interface, the beam penetrates into the solvent, with a depth, d, given by:
where is the wavelength of the IR light and is the angle of incidence of the beam at the interface [42]. Beam penetration depth is on the order of a few microns but its intensity decays exponentially with increasing distance from the surface, resulting in increased sensitivity to material close to or at the bulk interface. The absorption of IR light within the penetration depth occurs when the characteristic frequency of vibrational modes of a given chemical bond matches the frequency of the incident light. As deuterium has a higher mass than hydrogen, the vibrational frequency of deuterium-containing bonds is lower than equivalent hydrogen-containing bonds, resulting in absorption of lower frequency IR light.
Our experiment involved the deposition of a dDMPC bilayer on a Si ATR crystal before injection of hDMPC nanodiscs, whereafter spectra were periodically taken. Due to the increased mass of deuterium compared to hydrogen, the frequency of C-D bond vibrations is decreased relative to C—H bonds. This allows direct observation of the relative decreases and increases in C-Dx and C-Hx, respectively from the lipid tails upon the introduction of nanodiscs into the superphase. Importantly, ATR-FTIR allows direct observation of lipid exchange, separate to polymer exchange due to the relatively low C-H2 content of the polymers, and the presence of aromatic C C bonds and carbonyl bonds in different chemical environments to the glycerol-ester present in the phospholipid head groups.
ATR-FTIR spectra were collected using a ThermoNicolet Nexus instrument fitted with an ATR flow cell accessory (Specac) attached to a calibrated syringe pump, a cryo-cooled mercury cadmium telluride detector and a dry-air purge operating at a flow rate of 40 L/min in order to minimize absorbance from residual water vapor in the beam path. All spectra were collected with a resolution of 4 cm−1 and 128 interferograms collected for each spectra. Throughout the measurements, temperature was maintained at 25 °C by a temperature controlled water cooling loop passing through hollow aluminium blocks in direct contact with the Si substrate and the aqueous superphase.
Silicon ATR substrates were cleaned by sonication in 2% (w/v) SDS for 30 min before rinsing extensively with ultrapure water and drying under a stream of nitrogen. Substrates were then UV-ozone cleaned for 10 min, washed with ultrapure water and then UV-ozone cleaned a final time for 10 min. The substrate was mounted in a dry flow cell and the volume filled with either 50 mM Na2HPO4, 200 mM NaCl, pD 8.0 in D2O for measurements using SMALPs and RAFT-SMALPs, or 50 mM NaOAc, 200 mM NaCl, pD 5.0 in D2O for measurements using SMILPs. A background spectra was taken of the bare Si substrate in the appropriate buffer and subtraction spectra collected for later removal of water vapor absorbance. 20 µL dDMPC as a 10 mg/mL stock in CHCl3 (Avanti) was transferred to a clean glass vial and dried under a stream of nitrogen. Lipid films were rehydrated in either Na2HPO4 or NaOAc buffer as described above to a concentration of 0.1 mg/mL. The lipid suspension was sonicated in a sonic bath for 30 min until optically transparent, suggesting small unilamellar vesicle (SUV) formation, consistent with our previous studies [30], [33]. Vesicle suspensions were subsequently centrifuged at 14,000 × g, 25 °C for 10 min to pellet large aggregates. A 2 mL dDMPC SUV suspension was manually injected into the flow cell and incubated for 20 min with continuous spectra collection to allow for deposition of the dDMPC bilayer until no further spectral changes were observed. The flow cell was then flushed with 2 mL buffer at a flow rate of 0.5 mL/min and a spectrum collected of the supported dDMPC bilayer at the Si-water interface. Nanodiscs were diluted to 50 µM polymer concentration and 2 mL manually injected into the flow cell over the dDMPC bilayer. The deuterated bilayer was incubated with the hDMPC nanodiscs for 300 min. Spectra were continuously recorded with a time resolution of 80 s.
All spectra were corrected for removal of water vapor absorbance bands by scaling and subtraction of spectra collected before the deposition of the dDMPC bilayer from the bilayer spectra. No further processing was performed.
ATR-FTIR peak integrations were preformed over the aliphatic C—H stretching and aliphatic C—D stretching regions from 2990 to 2810 cm−1 and 2230 – 2050 cm−1, respectively. This includes contributions from the symmetric and asymmetric CH2/CD2 stretching vibrations and the CH3/CD3 stretching vibrations arising primarily from the aliphatic phospholipid tails. Due to the different frequencies and amplitudes of CH and CD bond vibrations, this allows the relative changes in hydrogenated and deuterated material at the Si-water interface to be compared as a function of time.
2.2.4. Analysis of lipid exchange kinetics
The exchange of dDMPC from bilayers at the Si-water interface to nanodiscs in bulk solution was modelled by a two-phase first order process which was fit to the experimental data by non-linear regression:
where the Integrated area at time t, It, can be described by two apparent first order rate constants for a fast and slow process, kFast and kSlow, respectively and the proportion of the decrease of the initial integrated area, I0, to that at equilibrium, Ieq, for which each process is responsible, and .
The exchange of hDMPC from the nanodiscs to the bilayer showed more variation between different nanodisc systems. Therefore, each process was modelled by both a single-phase first order process:
and two-phase first order process:
which were fit to the experimental data by nonlinear regression. The model with the highest probability of being correct was then selected by performing the Akaike’s Information Criteria (AIC) test implemented within GraphPad Prism.
2.2.5. Neutron reflectometry
Detailed descriptions of the theoretical basis of neutron reflectometry (NR), instrumental details and applications to soft-matter have been described in depth elsewhere [43], [44], [45], so here we will only include a brief description of the technique. NR measures the specular reflection of neutrons as a function of the scattering vector perpendicular to the surface normal (), defined by:
where represents the angle of reflection and represents the wavelength of the reflected neutrons. The reflected intensity is dependent on the structure and scattering length density (SLD) of the material at the interface, as well as the scattering length density of the bulk phases through an inverse Fourier transform. The SLD of a molecule is defined as:
where SLD, , is given by the sum of the coherent scattering lengths, , for each atomic nuclei, n, within a given molecular volume, . Due to the distinctly different neutron scattering lengths of deuterium and hydrogen(−3.74 × 10−5 Å and 6.67 × 10−5 Å, respectively), NR is very sensitive to the incorporation of hydrogenated material from nanodiscs into a deuterated monolayer, or vice versa.
Single crystals of silicon with dimensions of 50 × 80 × 15 mm with a single 80 × 50 mm 111 face polished to ~5 Å RMS roughness was cleaned by immersion in piranha solution (5:3:1 H2O:H2SO4:H2O2 heated to 90 °C for ten minutes) followed by two cycles of extensive washing in ultrapure water and ozone cleaning. Clean Si blocks were then mounted in sealed PEEK lamellar flow cells underwater to avoid introduction of air bubbles during cell assembly.
Data for the interaction of SMALPs and RAFT-SMALPs with d54DMPC bilayers at the Si-water interface were collected using the Polref reflectometer (ISIS neutron and muon source, UK) in non-polarized TOF-NR mode. NR was measured at three angles: 0.5, 1.2 and 2.5° with a neutron wavelength range of 1–14 Å covering an effective -range of 0.01–0.3 Å−1 where δQ/Q is 2.88%. Data for the interaction of SMILPs with d54DMPC bilayers at the Si-water interface were measured using the Surf reflectometer (ISIS neutron and muon source, UK). NR was measured at three incident angles: 0.35, 0.65 and 1.5° with a neutron wavelength range of 0.55–6.8 Å covering a similar -range of 0.012–0.3 Å−1.
In both instruments, Si crystal flow cells were mounted on a horizontal geometry translation stage, at an ambient temperature of 22 °C, and connected to a HPLC pump (Knauer) in order to change solution isotopic contrasts. The measured reflected neutron intensity was normalized to the incident neutron flux measured in transmission through each substrate.
Si crystal substrates were initially characterized by NR in H2O and D2O buffers (50 mM Na2HPO4, 200 mM NaCl, pH 8 for substrates to be injected with SMALP and RAFT-SMALP nanodiscs and 50 mM NaOAc, 200 mM NaCl pH 5 for substrates to be injected with SMILP nanodiscs). dDMPC in chloroform was transferred to a clean glass vial and dried under a stream of nitrogen. Lipids were rehydrated to 0.3 mg/mL in 20 mM HEPES pH 7 and small unilamellar vesicles (SUVs) formed by sonication in a water bath for 30 min. 10 mL of d54DMPC was manually injected into each flow cell and incubated for 40 min to allow for vesicular rupture and bilayer deposition on the Si substrates. Resultant bilayers were characterized by NR in H2O, D2O and silicon-matched water (SiMW, 38% v/v D2O) buffers as described above. SEC-purified Nanodiscs were diluted to 50 µM polymer concentration in the appropriate deuterated buffer and 15 mL flowed over the supported bilayers at a flow rate of 1.2 mL/min. Excess nanodiscs in bulk solution were removed by flushing the flow cells with deuterated buffer and the final interfacial structure characterized in H2O, D2O and SiMW.
2.2.6. Analysis of lipid bilayer structure
NR data corresponding to lipid bilayers at the Si-water interface were analyzed using RasCAL [46]. This uses a “slab model” to construct an SLD profile for the system, from which the reflectivity can be calculated and compared to the data. For each layer we therefore obtain an SLD, a thickness, a hydration and a roughness, which can then be constrained by our pre-existing knowledge of the sample.
Data for each sample consisted of up to eight measurements. The first two datasets were from the silicon substrate before bilayer deposition, measured twice using two solution isotopic contrasts: H2O and D2O. These data were modelled using a single layer corresponding to SiO2. The SLD of Si and SiO2 were fixed based on literature values and the only parameter which varied between the individual contrasts is the SLD of the bulk solution to account for different isotopic contrasts measured and the possibility of imperfect exchange of the water within a sample.
The next three datasets correspond to d54DMPC bilayers, prior to the injection of nanodiscs, in three solution contrasts (H2O, SiMW and D2O). Five layers were used to model this data corresponding to SiO2 (constrained to be equal to the measurement prior to bilayer deposition, Fig. S5), an inner leaflet of phosphatidylcholine headgroups, two identical layers corresponding to dimyristoyl tails and an outer leaflet of phosphatidylcholine headgroups. We assumed that the bilayer was always symmetrical across each leaflet and applied the constraint that the mean molecular area is consistent for both headgroup and tail layers ensuring that the resulting model represents a physically realistic bilayer structure. This was achieved by calculating a mean molecular area from the fitted thickness of the phospholipid tail layer, where
Here, represents the mean molecular area associated with the phospholipid tails, represents the partial specific molecular volume of the phospholipid tails and corresponds to the fitted thickness of the tail layer.
Similarly, the apparent hydrated area of the lipid headgroups,, can be calculated as
where represents the partial specific molecular volume of headgroups as represents the fitted thickness of the headgroup layer.
It is likely that the bilayer surface coverage is incomplete, so we have allowed for an additional parameter: the solvent volume fraction within the tail layer, . Since the mean molecular area of the headgroups must equal that of the phospholipid tails, the volume fraction of ‘dry’ lipid headgroups,, within the layer can be calculated as
allowing calculation of the volume fraction of solvent, , in the headgroup layer from
An additional constraint was applied such that and could not exceed the maximum thickness for a fully extended (all-trans) tail conformation based on the Tanford equation.
SLD values for the headgroups were calculated based on literature values for partial specific molecular volumes [47] and held as a constant. Surface roughness was fitted to be equal for all layers within the lipid bilayer, since these layers are all physically coupled.
The final three datasets correspond to measurements made after incubation with the nanodiscs, again in same three solution contrasts. To fit this data, we required two different approaches. For the case of SMALPs and SMILPs we were able to use the simplest model possible, where the only parameter allowed to differ between the 6 datasets modelling the bilayer was the mole fraction of hydrogenated tails (effectively the SLD of the two phospholipid tail layers). All other parameters describing the bilayer structure were held constant. This very simple but highly constrained approach allows us to account for lipid exchange between the bilayer and nanodiscs in solution (but explicitly not adsorbed to the surface).
In contrast we found that we could not use the same highly constrained model for the RAFT-SMALP system. This is demonstrated in the supporting information (see Fig. S5 and Table S1). Thus, in order to model the data, we required some additional layers. Since the introduction of new layers considerably increases the complexity of our model we have considered a number of different possible models, discussed in some detail in the results section and in the supporting information.
In all cases the fitting process co-refined across all eight data sets. The error associated with the parameters in each model was estimated by MCMC analysis implemented within RasCAL to account for the covariance between parameters. We assumed a Gaussian prior distribution for each parameter. The posterior distribution was determined by performing 50,000 iterations with 5,000 burn-in points to allow for location of the global minima. The 95% confidence interval was then calculated from the posterior distribution obtained after three independent repeat runs. For any individual contrast, the smallest real space structures which can be resolved by defined features in the data is given by , though structures smaller than this limit can still contribute partial features to the experimental reflectivity profiles if there is suitable scattering contrast between the structure and its surrounding medium. By utilizing multiple isotopic contrasts, either interfacial or bulk, and some a priori information about the sample (most notable the substrate SLD) the structural contribution of these components to the sample can be resolved with a useful range of uncertainty. Throughout this work, we have quoted fit parameters and errors to the nearest Ångström to account for both the limited contrasts and finite range.
3. Results
3.1. Kinetics of nanodisc interactions with phospholipid bilayers at the Si-water interface
As discussed above, there is currently limited information regarding the interaction of nanodiscs stabilized by different polymers with interfacial lipid membranes. In order to determine the role that the stabilizing polymer belt has in determining the nature of these interactions, we first probed the kinetics of nanodisc-bilayer interactions using time-resolved ATR-FTIR at the Si-water interface.
Upon incubation of the dDMPC bilayer with hDMPC SMALPs (Fig. 1a–c) an increase in the intensity of absorbances arising from the symmetric and asymmetric C-H2 and C-H3 stretching vibrations is observed as hDMPC from the nanodiscs become incorporated at the interface (Fig. 1a). A concurrent decrease in the intensity of absorbances arising from the symmetric and asymmetric C-D2 and C-D3 stretching vibrations is also observed as lipids are exchanged from the bilayer to the nanodiscs in bulk solution (Fig. 1b). By inspecting the carbonyl and aromatic peaks (Fig. 1c), we observed a decrease and then apparent plateau of the ester C O stretch absorbance from the DMPC tails. Importantly, we also observe a gradual increase in the carboxylic acid C O stretch absorbance and the aromatic C C stretching absorbance, suggesting polymer is interacting with the bilayer. In comparison, spectra collected during the incubation of hDMPC SMILPs (Fig. 1d–f) and RAFT-SMALPs (Fig. 1h–j) show similar trends, although with smaller decreases in C-Dx and increases in C-Hx bonds. This suggests that the bilayer experiences a much decreased lipid exchange with SMILPs and RAFT-SMALPs compared to SMALPs. However, when inspecting the polymer-specific absorbances, we can observe a larger increase in the imide or carboxylic acid carbonyl bonds present within SMI and RAFT-SMA, respectively along with increases in the aromatic C C bonds. This suggests that both SMI and RAFT-SMA have an increased propensity to interact with planar bilayers than SMA. Due to peak overlap, noise from imperfect water vapor subtraction and differing dipole moments between different bond vibrations, it is difficult for us to quantify the total amount of polymer incorporation into the bilayer.
Fig. 1.
Excerpts of ATR-FTIR spectra measuring the interaction of hDMPC containing nanodiscs with dDMPC bilayers at the Si-water interface. Spectral regions showing aliphatic C-Hx peaks, aliphatic C-Dx peaks and carbonyl/aromatic peaks are shown for incubations of hDMPC-containing SMALP (a–c), SMILP (d–f) and RAFT-SMALP (g–i) nanodiscs with dDMPC supported bilayers at the Si-water interface, respectively. A spectrum was collected prior to the injection of nanodiscs (0 min, red spectrum) and then at various time points during nanodisc incubation (15, 30, 60, 150 and 300 min, colored orange through indigo, respectively). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
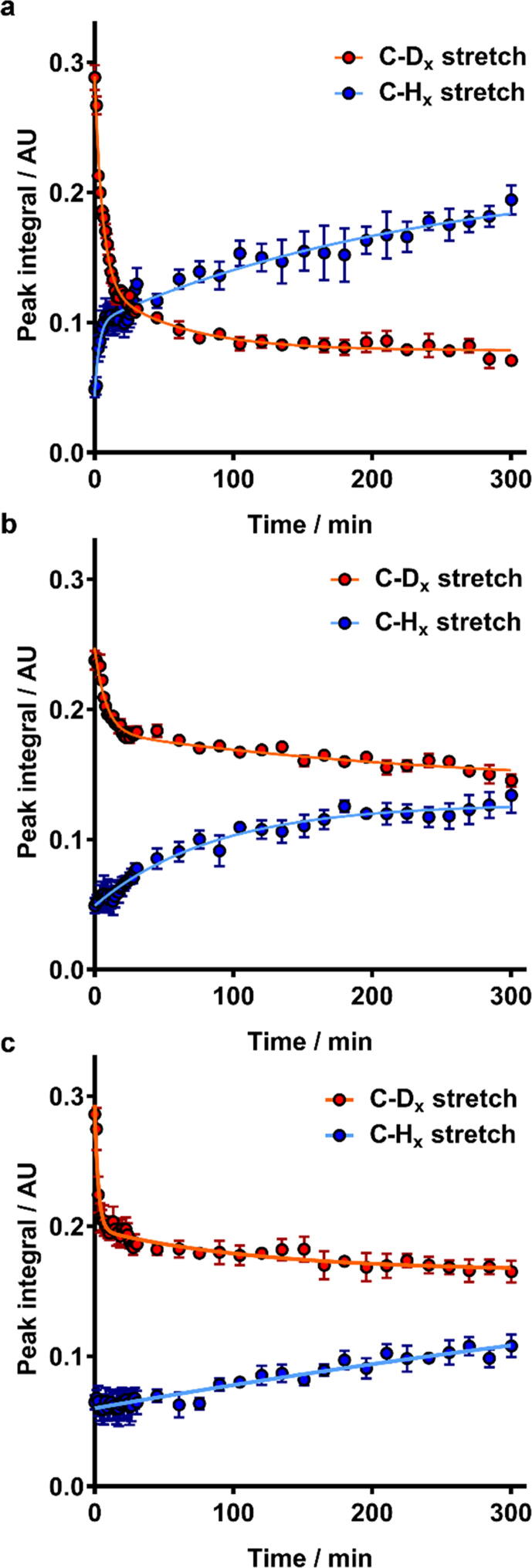
By integrating spectra collected at different time points during incubation with nanodiscs over the C–Hx stretch regions (2990–2810 cm−1) and C-Dx stretch regions (2230–2050 cm−1), we were able to obtain kinetic plots of lipid exchange from the bilayer to the nanodiscs and from the nanodiscs to the bilayer with minimal contribution from polymer incorporation (Fig. 2, Fig. S4). In contrast to exchange kinetics at the air-water interface, each system shows a similar two-phase removal of dDMPC from the bilayer. These data were analyzed using a two-phase first-order rate equation, similar to the analysis for lipid/polymer addition to monolayers at the air-water interface. This analysis yields two rate constants, kFast, B→N and kSlow, B→N, describing the fast and slow phases of lipid exchange from the bilayer to the nanodiscs, respectively (Table 1). The initial fast phase of lipid exchange from the bilayer to the nanodiscs is responsible for a similar proportion of the total removal of lipid from the bilayer for both SMALP and RAFT-SMALP systems, with approximately 75% of the total lipid removal occurring during this phase. SMILPs also show an initial rapid exchange of phospholipids from the bilayer to the nanodiscs, although this is responsible for less of the total lipid removal, at approximately 60%. Interestingly, the rate of this phase is significantly higher for the RAFT-SMALP system compared to SMALPs and SMILPs. The rate of the second, slower phase of lipid exchange from the bilayer to the nanodiscs show a different trend, whereby SMALPs exhibit a higher value for kSlow, B→N than either SMILPs or RAFT-SMALPs.
Fig. 2.
Kinetics of lipid exchange between hDMPC-containing SMALPs (a), SMILPs (b) and RAFT-SMALPs (c) and supported dDMPC bilayers at the Si-water interface. The integral of regions corresponding to C-Dx stretching vibrations are shown as red points and C-Hx stretching vibrations are shown as blue points. Points are the mean of three separate experiments with the error bars representing ± 1 standard error. Kinetic fits to the experimental data are shown as solid lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Lipid exchange parameters obtained for kinetics of lipid exchange occurring between dDMPC bilayers at the Si-water interface and hDMPC nanodiscs.
| Exchange with SMALPs | Exchange with SMILPs | Exchange with RAFT-SMALPs | |
|---|---|---|---|
| Kinetic parameters of lipid exchange from the bilayer to nanodiscs | |||
| /min−1 | 0.18 ± 0.02 | 0.12 ± 0.02 | 0.39 ± 0.06 |
| /% | 75.37 ± 2.83 | 60.17 ± 8.60 | 74.81 ± 3.56 |
| /×10−3min−1 | 17.70 ± 4.56 | 3.94 ± 3.08 | 8.66 ± 5.29 |
| /% | 24.63 ± 2.83 | 39.83 ± 8.60 | 25.19 ± 3.56 |
| Kinetic parameters of lipid exchange from nanodiscs to the bilayer | |||
| /×10−3min−1 | 321.40 ± 110.60 | 11.69 ± 1.90 | 1.08 ± 2.59 |
| /% | 32.96 ± 6.30 | – | – |
| /×10−3min−1 | 4.30 ± 2.16 | – | – |
| /% | 67.04 ± 6.30 | – | – |
Analysis of C-Hx stretch peak integrals yields information describing lipid exchange from the nanodiscs to the bilayer. In the case of SMILPs and RAFT-SMALPs, the best fits to the experimental data were obtained by using a single-phase, first-order rate equation (Table 1). Here the rate of lipid exchange from the nanodiscs to the bilayer is over ten-fold higher for SMILPs than RAFT-SMALPs. SMALPs, however, show two phase kinetics, with a fast and slow phase, described by kFast, N→B and kSlow, N→B. kFast, N→B is significantly higher than the rate constants obtained for lipid exchange from either SMILPs or RAFT-SMALPs to the bilayer, but of a similar order of magnitude as kFast, B→N, discussed above. In contrast, kSlow, N→B has a value more akin to the rate of lipid deposition at the bilayer by both SMILPs and RAFT-SMALPs. This suggests that there may be also be a biphasic process occurring for SMILPs and RAFT-SMALPs, though the rate of the initial fast phase is faster than can be measured within the achievable time-resolution. In addition, the majority of lipid deposition at the bilayer by SMALPs occurs during the slow phase, in contrast to lipid removal discussed above. We therefore suggest that in all cases, lipid exchange from the nanodiscs to the bilayer is dominated by a slow, diffusion limited, collisional lipid exchange.
3.2. Structural changes to phospholipid bilayer structure upon interaction with phospholipid nanodiscs at the Si-water interface
From ATR-FTIR measurements, we were able to determine the kinetics of lipid exchange between phospholipid nanodiscs and supported phospholipid bilayers at the Si-water interface. However, we were also able to identify the presence chemical bonds arising from each of the polymers at the interface. ATR-FTIR does not give any structural information on nanodisc-bilayer interactions. We therefore used NR at the Si-water interface to determine the large-scale structure of DMPC bilayers which have been subject to incubation with nanodiscs. There are two main possibilities which could account for the observed spectral changes, which are not necessarily mutually exclusive. Firstly, a similar scenario to that observed previously with lipid monolayers at the Si-water interface where no nanodisc adsorption can be identified [38], but where polymers embed into the bilayer along with lipid exchange. The second possibility is that nanodiscs adsorb to the surface of the bilayer. Both of these scenarios would account for the presence of C-Hx absorbances and polymer-specific absorbances in the ATR-FTIR spectra.
After characterization by NR of bare Si substrates, dDMPC bilayers were deposited on the substrates and measured in three contrasts (Fig. 3a, 4a, S4). This data was modelled with 5 layers, constrained as described earlier, with corresponding SLD profiles shown in Fig. 3b, 4b and S4. The model provided good fits for the data for the SMALP and SMILP samples and the structural parameters (Table 2, Table 3) are consistent with a high-coverage dDMPC bilayer that is consistent with previous studies. [48], [49], [50] The structural parameters for the RAFT-SMALP sample (Table S1-S7 in the supporting info) is also consistent with a high coverage DMPC bilayer, but the silicon substrate used was not as good as for the other two samples. This is seen in a rougher more solvated oxide layer which results in a less well defined bilayer and means we have more uncertainty in the structural parameters extracted from this sample.
Fig. 3.
Neutron reflectometry data (points) with overlaid fits (solid lines) plotted as RQ4 (a, c) and corresponding model SLD profiles (b, d) of a dDMPC supported bilayer deposited on a Si substrate before (a, b) and after (c, d) incubation with dDMPC SMALPs. Colored shaded regions represent the 95% confidence interval associated with the fit and model SLD profiles determined by Bayesian MCMC error estimation routines. Grey shaded regions denote layer boundaries in the model SLD profiles. In all cases, red, green and blue points/lines correspond to data collected in D2O, SiMW and H2O solution contrasts, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Structural parameters obtained by fitting NR data of dDMPC bilayers at the Si-water interface before and after incubation with hDMPC SMALPs. Values marked * were held as a constant throughout the fitting procedure. Values in parentheses indicate the 95% confidence intervals estimated from Bayesian MCMC error estimation.
| Layer | Parameter | dDMPC bilayer | dDMPC bilayer after hDMPC SMALP incubation |
|---|---|---|---|
| Si | SLD/×10−6 Å−2 | 2.07* | |
| Roughness/Å | 6 (5, 6) | ||
| SiO2 | SLD/×10−6 Å−2 | 3.47* | |
| Thickness/Å | 17 (14, 20) | ||
| Hydration/% | 8 (6, 10) | ||
| Roughness/Å | 5 (4, 6) | ||
| PC Headgroups | SLD/×10−6 Å−2 | 2.14* | |
| Thickness/Å | 11 (11, 12) | ||
| Hydration/% | 58 (57, 58) | 71 (70, 71) | |
| Roughness/Å | 4 (4, 5) | ||
| DM Tails | SLD/×10−6 Å−2 | 6.5* | 3.41 (3.36, 3.48) |
| Thickness/Å | 14 (13, 14) | ||
| Hydration/% | 6 (4, 8) | 35 (34, 37) | |
| Roughness/Å | 4 (4, 5) | ||
| Area Per Molecule/Å2 | 60 (57, 62) | 87 (83, 91) | |
| Bulk Solvent | SLD (D2O)/×10−6 Å−2 | 6.28 (6.27, 6.29) | 6.27 (6.26, 6.28) |
| SLD (SiMW)/×10−6 Å−2 | 3.33 (3.30, 3.37) | 1.52 (1.45, 1.59) | |
| SLD (H2O)/×10−6 Å−2 | −0.46 (−0.55, −0.36) | −0.44 (−0.50, −0.38) |
Table 3.
Structural parameters obtained by fitting NR data of dDMPC bilayers at the Si-water interface before and after incubation with hDMPC SMILPs. Values marked * were held as a constant throughout the fitting procedure. Values in parentheses indicate the 95% confidence intervals estimated from Bayesian MCMC error estimation.
| Layer | Parameter | dDMPC bilayer | dDMPC bilayer after hDMPC SMALP incubation |
|---|---|---|---|
| Si | SLD/×10−6 Å−2 | 2.07* | |
| Roughness/Å | 4, (3, 6) | ||
| SiO2 | SLD/×10−6 Å−2 | 3.47* | |
| Thickness/Å | 13 (12, 14) | ||
| Hydration/% | 6 (2, 12) | ||
| Roughness//Å | 3 (3, 4) | ||
| PC Headgroups | SLD/×10−6 Å−2 | 2.14* | |
| Thickness/Å | 8 (7, 9) | ||
| Hydration//% | 39 (37, 42) | 39 (37, 42) | |
| Roughness/Å | 4 (3, 4) | ||
| DM Tails | SLD/×10−6 Å−2 | 6.5* | 4.98 (4.88, 5.07) |
| Thickness/Å | 14 (13, 14) | ||
| Hydration/% | 2 (1, 4) | 2 (1, 4) | |
| Roughness/Å | 4 (3, 4) | ||
| Area Per Molecule/Å2 | 57 (55, 60) | 57 (55, 60) | |
| Bulk Solvent | SLD (D2O)/×10−6 Å−2 | 6.13 (6.12, 6.14) | 6.13 (6.13, 6.14) |
| SLD (SiMW)/×10−6 Å−2 | 2.11 (1.94, 2.29) | 2.00 (1.84, 2.18) | |
| SLD (H2O)/×10−6 Å−2 | −0.45 (-0.55, −0.31) | −0.42 (-0.54, −0.30) |
After initial characterization of the bilayers, hDMPC nanodiscs were then injected into the sample cells and incubated for 5 h, after which the cells were flushed with buffer to remove excess, non-adsorbed polymer/nanodiscs and the resultant structures analyzed by NR in three solution contrasts.
In the case of dDMPC bilayers with hDMPC SMALPs, we were able to obtain satisfactory fits to the experimental data by allowing for lipid exchange of hDMPC lipids into the bilayer. The same 5 layer model as used before nanodisc interaction (Fig. 3c, d), was sufficient to fit the data indicating that SMALP nanodiscs do not adsorb. Structural parameters (Table 2) indicate that incubation with hDMPC SMALPs leads to a net loss of material from the bilayer, as supported by FTIR data (Fig. S4). Hydration of the DMPC tails region increases from approximately 6 to 35% and the area per molecule of the bilayer lipids increases from 60 Å2 to 87 Å2. We also observe a decrease in SLD of the tails, from 6.5 to 3.41 × 10−6 Å−2. If this change is solely down to the introduction of hDMPC, then we can estimate that 45% of the remaining lipid is hDMPC from the nanodiscs. However, since the hydrogenous SMA happens to have a very similar SLD to the lipid headgroups (1.89 × 10−6 Å−2 compared to 2.13 × 10−6 Å−2), we are unable to distinguish between exchange of hDMPC and the introduction of polymer into the bilayer. Given the ATR-FTIR results above, is likely that some polymer is incorporated into the bilayer.
Using the same approach, we were also able to obtain satisfactory fits to the experimental data for a dDMPC bilayer interacting with hDMPC SMILPs (see Fig. 4c, d and Table 3). In this case, the structural parameters suggest that, within error, there is no addition or removal of phospholipids from the bilayer, as shown by the similarity in both the hydration of the lipid tails and the area per molecule of the bilayer lipids before and after SMILP interaction, which is again consistent with FTIR data (Fig. S4). We also observed a smaller decrease in SLD of the lipid tails from 6.5 to 4.98 × 10−6 Å−2. Again, if this decrease in SLD is solely due to lipid exchange, we can estimate that 22% of the lipid in the resultant bilayer is hDMPC from SMILPs, although we are still unable to distinguish the polymer in this system.
Fig. 4.
Neutron reflectometry data (points) with overlaid fits (solid lines) plotted as RQ4 (a, c) and corresponding model SLD profiles (b, d) of a dDMPC supported bilayer deposited on a Si substrate before (a, b) and after (c, d) incubation with hDMPC SMILPs. Colored shaded regions represent the 95% confidence interval associated with the fit and model SLD profiles determined by Bayesian MCMC error estimation routines. Grey shaded regions denote layer boundaries in the model SLD profiles. In all cases, red, green and blue points/lines correspond to data collected in D2O, SiMW and H2O solution contrasts, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
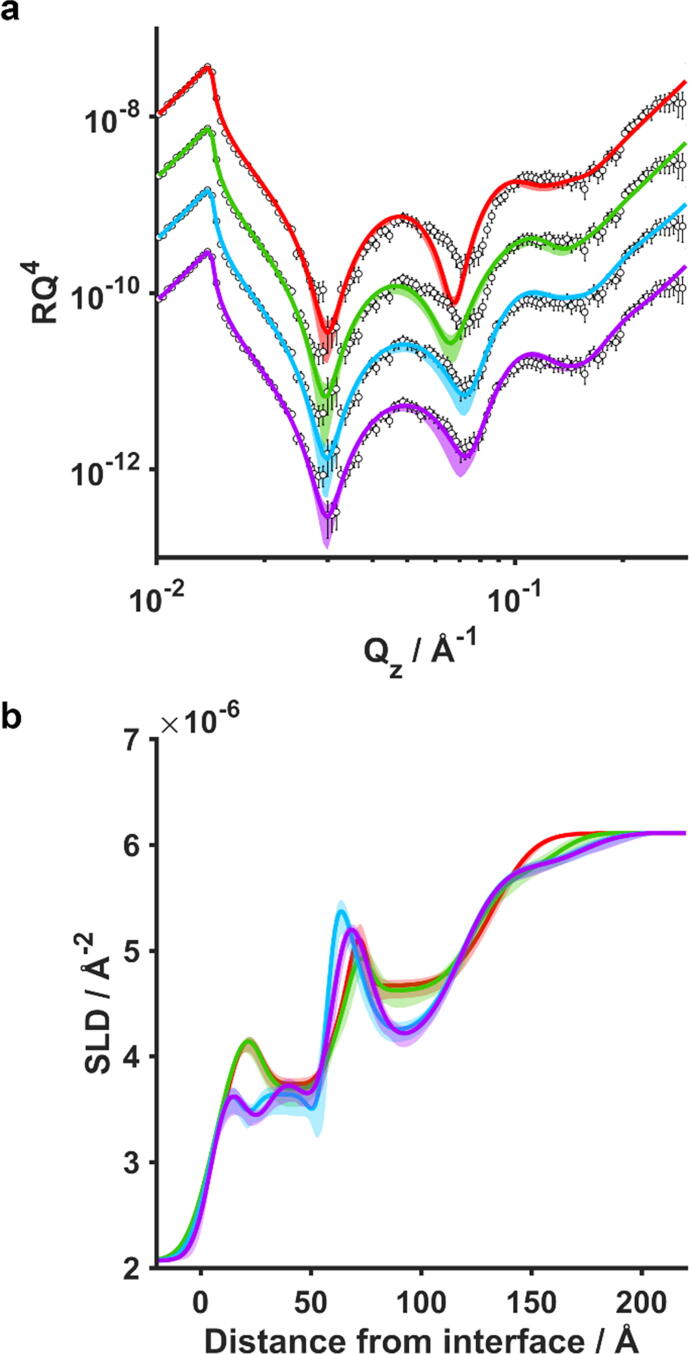
In contrast to SMALPs and SMILPs, the NR data collected after the incubation of a dDMPC bilayer with hDMPC RAFT-SMALPs shows the appearance of strong Keissig fringes which are indicative of a significantly thicker interfacial layer (Fig. 5). This was an unexpected result and we were unable to use the same approach to fitting for this data as in the other two systems. As mentioned earlier, in order to fit the data some additional layers were required. As a result, we have cautiously examined several models of increasing complexity in an effort to extract physically meaningful data without over-analyzing. The details of these models are in the supporting information. Briefly, we started with a similar model as utilized for SMALP and SMILP interactions except with the addition of a floating layer corresponding to adsorbed nanodiscs (Model 2). We subsequently increased the model complexity by relaxing structural constraints, allowing the bilayer structure to be influenced by nanodisc interaction (Model 4), including an additional layer representing adsorbed nanodiscs (Model 5), and finally removing the requirement for the bilayer structure to be constrained by area-per-molecule calculations (Model 6).
Fig. 5.
(a) Neutron reflectometry data (points) with overlaid fits (solid lines) plotted as RQ4 for the D2O contrast of a dDMPC supported bilayer deposited on a Si substrate after incubation with hDMPC RAFT-SMALPs. The data is shown four times with different fits to models of increasing complexity (Models 2, 4, 5 & 6, corresponding to red, green, blue and purple, respectively) discussed in the supporting information. The SLD profiles for the D2O contrast for the different models are compared in (b). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
From our analysis we can draw some general conclusions. Most unsurprisingly, if we increase the number of variables we can get towards a very good fit to the data. Unfortunately, this does not necessarily mean that we are getting ever more detailed structural information about this system since we inherently lack the additional contrasts to be certain of those details. We can, however, compare the SLD profiles for these models, shown in Fig. 5 for Models 2, 4, 5 and 6. This figure clearly shows that despite the difference in detail between these models, the overall fit always tends towards a similar outcome. It is this SLD profile that represents the truly meaningful structural information for this sample. Specifically, it shows that there is significant adsorption of nanodiscs as well as the lipid exchange seen for the other bilayer /nanodisc systems. The lipid exchange is of the order of 35%, similar to the level of exchange seen for the other SMALP system. The adsorbed nanodiscs do not form a well-defined bilayer-like layer but are significantly more diffuse with most of the density within about 50 Å of the bilayer but with some extending up to 100 Å. This thickness is similar to the size of 1–2 nanodiscs. If we assume the SLD of the adsorbed nanodiscs within this layer is similar to the calculated value of 2.63 × 10−6 Å−2, then the coverage of this layer is around 50% in the first 50 Å.
4. Discussion and conclusion
By using a combination of ATR-FTIR spectroscopy and NR, we have been able to quantify lipid exchange between nanodiscs in bulk solution and phospholipid bilayers at the Si-water interface. Our ATR-FTIR results show an interesting trend; RAFT-SMALPs exhibit the least exchange and SMALPs the most. Previous studies investigating the thermodynamics of self-assembly of these nanodiscs show that the Gibbs energy of transition for the polymers associating with the lipids is most negative for RAFT-SMALPs and least negative for SMALPs [30], [33]. It may therefore be that the thermodynamic stability associated with each of the polymers when in a nanodisc dictates the propensity of said nanodisc to exchange material with planar lipid bilayers.
When comparing the kinetics of lipid exchange for each nanodisc system, biphasic kinetics can be seen in all cases. This suggests a two-step process for this interaction. The initial lipid exchange on the introduction of nanodiscs in solution is rapid but subsequently slows. The initial rapid exchange occurs when the bilayer first interacts with the nanodiscs and thereafter the rate slows probably limited by diffusion.
The ATR-FTIR also suggest the presence of polymer in each of the bilayers after incubation with nanodiscs. As briefly discussed above this observation can be explained by either adsorption of nanodiscs to the bilayer or by the incorporation of polymer within it, or both. Our NR is not directly able to see the polymer but we can infer its presence.
Our proposed explanation for these observations is as follows. In each case the nanodiscs rapidly approach the interface and briefly adsorb. This adsorption results in quick exchange of lipids with the bilayer as well as some interaction of the polymer with the surface. The relative extent of the exchange is likely to be governed by the particular nanodisc stability in each case. In the case of the SMALP and SMILP nanodiscs the adsorption is not strong enough to have a substantial residence time that would allow for the formation of a stable adsorbed nanodisc layer, but the process does leave a small amount of polymer on the surface. For the RAFT-SMALP nanodiscs the adsorption is stronger and this results in an adsorbed nanodisc layer that survives the solvent rinses. We believe that the increased interaction strength in this case is due to the hydrophobic polystyrene tail present in this polymer that is not present in the other polymers. Once this tail interacts with the bilayer it effectively acts as a tether and prevents rapid desorption. After this initial interaction, subsequent lipid exchange is determined by the rate at which nanodiscs diffuse away from the interface region and are replaced by fresh nanodiscs containing h-lipids.
In all of these measurements we cannot quantify the extent of polymer interaction with the surface. However, the results do show that the image of discreet nanodisc particles interacting with a surface bilayer is not quite right. There is clearly extensive exchange of material, both lipids and polymer, in all of these samples. Exchange of polymers in addition to lipids is not unexpected. Recent studies have shown by fluorescently labelling SMA polymers, that in addition to the now-accepted lipid exchange between nanodiscs in solution [39], [40], there is also exchange of polymers between nanodiscs [51]. Furthermore, Langmuir balance and molecular dynamics studies of nanodisc self-assembly have demonstrated that nanodiscs form via an initial adsorption of polymers to the bilayer surface, prior to embedding into the hydrophobic core and subsequent solubilization [27], [52]. This is not dissimilar to what we observe here.
Previous studies investigating the interaction of RAFT-SMALPs with DOPC monolayers deposited on a hydrophobic surface at the Si-water interface saw little to no nanodisc adsorption [38]. The reasons as to why, in this case, clear increases in thickness were observed is unclear. It is important to note that the sample used for the RAFT-SMALPs was rougher with a less well-defined bilayer and it is possible that this is the reason why the nanodiscs are able to strongly adsorb in this case. It is also possible that monolayers on a hydrophobic surface, rather than bilayers on a hydrophilic surface, are less amenable to adsorption. Alternatively, the difference in the gel to liquid transition temperatures of DOPC (−17 °C) relative to DMPC (24 °C) could affect the adsorption mechanism and thermodynamics. Only further experiments to investigate the link between polymer architecture and adsorption will be able to determine whether reliable and robust assembly of adsorbed nanodisc films will be possible.
In summary, we have demonstrated that both lipid and polymer exchange occurs between polymer-stabilized phospholipid nanodiscs of three different types. We were also able to show that RAFT-SMALPs show substantial adsorption to a bilayer at the silicon-water interface. This is the first such observation for polymer stabilized nanodiscs. This result was surprising and it remains to be seen as to whether changing the sample conditions will have an effect on this behavior. Nonetheless, this result demonstrates the potential of polymer stabilized nanodiscs to be used for applications in which adsorption may be important. The fact that the polymer seems to have such a dramatic effect suggests that the adsorption may be controllable by modifying the properties of the polymer. For example, use of an optimized polymer could enable structural and functional characterization of solubilized MPs within a phospholipid environment by techniques such as X-ray and neutron reflectometry, surface plasmon resonance and quartz-crystal microbalance.
Funding sources
This work has been supported by an Engineering and Physical Sciences Research Council (EPSRC: EP/M506461/1) and Diamond Light Source studentship for SCLH and funding from the Biotechnology and Biological Sciences Research Council (BBSRC: BB/M018261/1 [TRD], BB/P009840/1 [TJK] and BB/L00335X/1 [TJK]). CT acknowledges STFC BioMemNet (studentship agreement #2990) and the University of Bath for PhD studentship funding.
CRediT authorship contribution statement
Stephen C.L. Hall: Conceptualization, Methodology, Investigation, Formal Analysis, Writing - Original Draft and Writing - Review & Editing. Luke A. Clifton: Conceptualization, Methodology, Investigation, Formal Analysis and Writing - Review & Editing. Cecilia Tognoloni: Investigation. Kerrie A. Morrison: Investigation. Timothy J. Knowles: Methodology, Investigation and Formal Analysis. Christian J. Kinane: Methodology, Formal Analysis, Investigation. Tim R. Dafforn: Conceptualization, Writing - Review & Editing and Funding acquisition. Karen J. Edler: Conceptualization, Writing - Review & Editing and Funding acquisition. Thomas Arnold: Conceptualization, Methodology, Investigation, Formal Analysis, Writing - Original Draft, Writing - Review & Editing and Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors wish to thank the ISIS Pulsed Neutron Source for beamtime awarded using Polref and Surf (RB1900009), for access to ATR-FTIR instrumentation and for provision of D2O used throughout this study. We particularly thank Marité Cardenas for very useful discussions regarding the NR fitting methodology. We also thank Bert Klumperman and Gareth Price for assistance with polymer synthesis and Chris Nicklin for his support with SCLH's studentship placement at Diamond Light Source.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jcis.2020.04.013.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Fagerberg L., Jonasson K., von Heijne G., Uhlén M., Berglund L. Prediction of the Human Membrane Proteome. Proteomics. 2010;10(6):1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 2.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A.-K., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., Feilitzen K., Von Forsberg M., Persson L., Johansson F., Zwahlen M., Heijne G., Von Nielsen J., Pontén F. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 3.Overington J.P., Al-Lazikani B., Hopkins A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 4.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap Opera. Biochim. Biophys. Acta BBA - Biomembr. 2004;1666(1–2):105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Arnold Thomas, Linke Dirk. The use of detergents to purify membrane proteins. Current Protocols Protein Sci. 2008;53(1) doi: 10.1002/0471140864.2008.53.issue-110.1002/0471140864.ps0408s53. [DOI] [PubMed] [Google Scholar]
- 6.Smith S.M. Strategies for the purification of membrane proteins. Meth. Mol. Biol. Clifton NJ. 2011;681:485–496. doi: 10.1007/978-1-60761-913-0_29. [DOI] [PubMed] [Google Scholar]
- 7.Krishnamani V., Lanyi J.K. Molecular dynamics simulation of the unfolding of individual bacteriorhodopsin helices in sodium dodecyl sulfate micelles. Biochemistry. 2012;51(6):1061–1069. doi: 10.1021/bi201770y. [DOI] [PubMed] [Google Scholar]
- 8.Frey L., Lakomek N.-A., Riek R., Bibow S. Micelles, bicelles, and nanodiscs: comparing the impact of membrane mimetics on membrane protein backbone dynamics. Angew. Chem. Int. Ed. 2016;56(1):380–383. doi: 10.1002/anie.201608246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popot J.-L. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 10.Parmar M.J., Lousa C.D.M., Muench S.P., Goldman A., Postis V.L.G. Artificial membranes for membrane protein purification, functionality and structure studies. Biochem. Soc. Trans. 2016;44(3):877–882. doi: 10.1042/BST20160054. [DOI] [PubMed] [Google Scholar]
- 11.Hardy D., Bill R.M., Jawhari A., Rothnie A.J. Overcoming bottlenecks in the membrane protein structural biology pipeline. Biochem. Soc. Trans. 2016;44(3):838–844. doi: 10.1042/BST20160049. [DOI] [PubMed] [Google Scholar]
- 12.Denisov I.G., Sligar S.G. Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 2017;117(6):4669–4713. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Midtgaard S.R., Pedersen M.C., Kirkensgaard J.J.K., Sørensen K.K., Mortensen K., Jensen K.J., Arleth L. Self-assembling peptides form nanodiscs that stabilize membrane proteins. Soft Matter. 2014;10(5):738–752. doi: 10.1039/C3SM51727F. [DOI] [PubMed] [Google Scholar]
- 14.Stroud Z., Hall S.C.L., Dafforn T.R. Purification of membrane proteins free from conventional detergents: SMA, new polymers new opportunities and new insights. Methods. 2018;147:106–117. doi: 10.1016/j.ymeth.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Knowles T.J., Finka R., Smith C., Lin Y.-P., Dafforn T., Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009;131(22):7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 16.Jamshad M., Grimard V., Idini I., Knowles T.J., Dowle M.R., Schofield N., Sridhar P., Lin Y., Finka R., Wheatley M., Thomas O.R.T., Palmer R.E., Overduin M., Govaerts C., Ruysschaert J.-M., Edler K.J., Dafforn T.R. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 2015;8(3):774–789. doi: 10.1007/s12274-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S.C., Knowles T.J., Postis V.L.G., Jamshad M., Parslow R.A., Lin Y.-P., Goldman A., Sridhar P., Overduin M., Muench S.P., Dafforn T.R. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11(7):1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- 18.Morrison K.A., Akram A., Mathews A., Khan Z.A., Patel J.H., Zhou C., Hardy D.J., Moore-Kelly C., Patel R., Odiba V., Knowles T.J., Javed M.-U.-H., Chmel N.P., Dafforn T.R., Rothnie A.J. Membrane protein extraction and purification using styrene-maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem. J. 2016;473(23):4349–4360. doi: 10.1042/BCJ20160723. [DOI] [PubMed] [Google Scholar]
- 19.Jamshad M., Charlton J., Lin Y.-P., Routledge S.J., Bawa Z., Knowles T.J., Overduin M., Dekker N., Dafforn T.R., Bill R.M., Poyner D.R., Wheatley M. G-Protein coupled receptor solubilization and purification for biophysical analysis and functional studies, in the total absence of detergent. Biosci. Rep. 2015;35(2) doi: 10.1042/BSR20140171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swainsbury D.J.K., Scheidelaar S., van Grondelle R., Killian J.A., Jones M.R. Bacterial reaction centers purified with styrene maleic acid copolymer retain native membrane functional properties and display enhanced stability. Angew. Chem. Int. Ed Engl. 2014;53(44):11803–11807. doi: 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laursen T., Borch J., Knudsen C., Bavishi K., Torta F., Martens H.J., Silvestro D., Hatzakis N.S., Wenk M.R., Dafforn T.R., Olsen C.E., Motawia M.S., Hamberger B., Møller B.L., Bassard J.-E. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science. 2016;354(6314):890–893. doi: 10.1126/science.aag2347. [DOI] [PubMed] [Google Scholar]
- 22.Broecker J., Eger B.T., Ernst O.P. Crystallogenesis of membrane proteins mediated by polymer-bounded lipid nanodiscs. Structure. 2017;25(2):384–392. doi: 10.1016/j.str.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Bersch B., Dörr J.M., Hessel A., Killian J.A., Schanda P. Proton-detected Solid-State NMR spectroscopy of a zinc diffusion facilitator protein in native nanodiscs. Angew. Chem. Int. Ed Engl. 2017;56(9):2508–2512. doi: 10.1002/anie.201610441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parmar M., Rawson S., Scarff C.A., Goldman A., Dafforn T.R., Muench S.P., Postis V.L.G. Using a SMALP platform to determine a sub-nm single particle Cryo-EM membrane protein structure. Biochim. Biophys. Acta BBA - Biomembr. 2018;1860(2):378–383. doi: 10.1016/j.bbamem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun C., Benlekbir S., Venkatakrishnan P., Wang Y., Hong S., Hosler J., Tajkhorshid E., Rubinstein J.L., Gennis R.B. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature. 2018;557(7703):123–126. doi: 10.1038/s41586-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vargas C., Arenas R.C., Frotscher E., Keller S. Nanoparticle self-assembly in mixtures of phospholipids with styrene/maleic acid copolymers or fluorinated surfactants. Nanoscale. 2015;7(48):20685–20696. doi: 10.1039/c5nr06353a. [DOI] [PubMed] [Google Scholar]
- 27.Scheidelaar S., Koorengevel M.C., Pardo J.D., Meeldijk J.D., Breukink E., Killian J.A. Molecular model for the solubilization of membranes into nanodisks by styrene maleic acid copolymers. Biophys. J. 2015;108(2):279–290. doi: 10.1016/j.bpj.2014.11.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheidelaar S., Koorengevel M.C., van Walree C.A., Dominguez J.J., Dörr J.M., Killian J.A. Effect of polymer composition and ph on membrane solubilization by styrene-maleic acid copolymers. Biophys. J. 2016;111(9):1974–1986. doi: 10.1016/j.bpj.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grethen A., Oluwole A.O., Danielczak B., Vargas C., Keller S. Thermodynamics of nanodisc formation mediated by styrene/maleic acid (2:1) copolymer. Sci. Rep. 2017;7(1):11517. doi: 10.1038/s41598-017-11616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall S.C.L., Tognoloni C., Price G.J., Klumperman B., Edler K.J., Dafforn T.R., Arnold T. Influence of poly(styrene-Co-maleic acid) copolymer structure on the properties and self-assembly of SMALP nanodiscs. Biomacromolecules. 2018;19(3):761–772. doi: 10.1021/acs.biomac.7b01539. [DOI] [PubMed] [Google Scholar]
- 31.Smith A.A.A., Autzen H.E., Laursen T., Wu V., Yen M., Hall A., Hansen S.D., Cheng Y., Xu T. Controlling styrene maleic acid lipid particles through RAFT. Biomacromolecules. 2017;18(11):3706–3713. doi: 10.1021/acs.biomac.7b01136. [DOI] [PubMed] [Google Scholar]
- 32.Craig A.F., Clark E.E., Sahu I.D., Zhang R., Frantz N.D., Al-Abdul-Wahid M.S., Dabney-Smith C., Konkolewicz D., Lorigan G.A. Tuning the size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for biophysical studies. Biochim. Biophys. Acta BBA - Biomembr. 2016;1858(11):2931–2939. doi: 10.1016/j.bbamem.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Hall S.C.L., Tognoloni C., Charlton J., Bragginton É.C., Rothnie A.J., Sridhar P., Wheatley M., Knowles T.J., Arnold T., Edler K.J., Dafforn T.R. An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles. Nanoscale. 2018;10(22):10609–10619. doi: 10.1039/C8NR01322E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patching Simon G. Surface plasmon resonance spectroscopy for characterisation of membrane protein–ligand interactions and its potential for drug discovery. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2014;1838(1):43–55. doi: 10.1016/j.bbamem.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 35.Wadsäter M., Simonsen J.B., Lauridsen T., Tveten E.G., Naur P., Bjørnholm T., Wacklin H., Mortensen K., Arleth L., Feidenhans’l R., Cárdenas M. Aligning nanodiscs at the air-water interface, a neutron reflectivity study. Langmuir. 2011;27(24):15065–15073. doi: 10.1021/la203100n. [DOI] [PubMed] [Google Scholar]
- 36.Wadsäter M., Laursen T., Singha A., Hatzakis N.S., Stamou D., Barker R., Mortensen K., Feidenhans R., l, Møller B.L., Cárdenas M. Monitoring shifts in the conformation equilibrium of the membrane protein cytochrome P450 reductase (POR) in nanodiscs. J. Biol. Chem. 2012;287(41):34596–34603. doi: 10.1074/jbc.M112.400085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertram N., Laursen T., Barker R., Bavishi K., Møller B.L., Cárdenas M. Nanodisc films for membrane protein studies by neutron reflection: effect of the protein scaffold choice. Langmuir. 2015;31(30):8386–8391. doi: 10.1021/acs.langmuir.5b00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hazell G., Arnold T., Barker R.D., Clifton L.A., Steinke N.-J., Tognoloni C., Edler K.J. Evidence of lipid exchange in styrene maleic acid lipid particle (SMALP) nanodisc systems. Langmuir. 2016;32(45):11845–11853. doi: 10.1021/acs.langmuir.6b02927. [DOI] [PubMed] [Google Scholar]
- 39.Cuevas Arenas R., Danielczak B., Martel A., Porcar L., Breyton C., Ebel C., Keller S. Fast collisional lipid transfer among polymer-bounded nanodiscs. Sci. Rep. 2017;7:45875. doi: 10.1038/srep45875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grethen A., Glueck D., Keller S. Role of coulombic repulsion in collisional lipid transfer among SMA(2:1)-bounded nanodiscs. J. Membr. Biol. 2018;251(3):443–451. doi: 10.1007/s00232-018-0024-0. [DOI] [PubMed] [Google Scholar]
- 41.Harrisson S., Wooley K.L. Shell-crosslinked nanostructures from amphiphilic ab and ABA block copolymers of styrene-alt-(maleic anhydride) and styrene: polymerization, assembly and stabilization in one pot. Chem. Commun. 2005;26:3259–3261. doi: 10.1039/b504313a. [DOI] [PubMed] [Google Scholar]
- 42.Oelichmann J. Surface and depth-profile analysis using FTIR spectroscopy. Fresenius Z. Für Anal. Chem. 1989;333(4–5):353–359. doi: 10.1007/BF00572327. [DOI] [Google Scholar]
- 43.Braun L., Uhlig M., von Klitzing R., Campbell R.A. Polymers and Surfactants at Fluid interfaces studied with specular neutron reflectometry. Adv. Colloid Interf. Sci. 2017;247:130–148. doi: 10.1016/j.cis.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 44.X-Ray and Neutron Reflectivity; Daillant, J., Gibaud, A., Eds.; Beiglböck, W., Ehlers, J., Hepp, K., Weidenmüller, H. A., Beig, R., Domcke, W., Englert, B.-G., Frisch, U., Hänggi, P., Hasinger, G., Hillebrandt, W., Jaffe, R. L., Janke, W., Löhneysen, H. von, Mangano, M., Raimond, J.-M., Sornette, D., Theisen, S., Weise, W., Zittartz, J., Guinea, F., Vollhardt, D., Series Eds.; Lecture Notes in Physics; Springer Berlin Heidelberg: Berlin, Heidelberg, 2009; Vol. 770. https://doi.org/10.1007/978-3-540-88588-7
- 45.Clifton L.A., Hall S.C.L., Mahmoudi N., Knowles T.J., Heinrich F., Lakey J.H. In: Lipid-Protein Interactions: Methods and Protocols. Methods in Molecular Biology. Kleinschmidt J.H., editor. Springer New York; New York, NY: 2019. Structural investigations of protein–lipid complexes using neutron scattering; pp. 201–251. https://doi.org/10.1007/978-1-4939-9512-7_11. [DOI] [PubMed] [Google Scholar]
- 46.RasCAL https://sourceforge.net/projects/rscl/(accessed May 16, 2019).
- 47.Kučerka N., Kiselev M.A., Balgavý P. Determination of bilayer thickness and lipid surface area in unilamellar dimyristoylphosphatidylcholine vesicles from small-angle neutron scattering curves: a comparison of evaluation methods. Eur. Biophys. J. 2004;33(4):328–334. doi: 10.1007/s00249-003-0349-0. [DOI] [PubMed] [Google Scholar]
- 48.Tristram-Nagle S., Liu Y., Legleiter J., Nagle J.F. Structure of gel phase DMPC determined by X-Ray diffraction. Biophys. J. 2002;83(6):3324–3335. doi: 10.1016/S0006-3495(02)75333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollinshead C.M., Hanna M., Barlow D.J., De Biasi V., Bucknall D.G., Camilleri P., Hutt A.J., Lawrence M.J., Lu J.R., Su T.J. Neutron Reflection from a Dimyristoylphosphatidylcholine Monolayer Adsorbed on a Hydrophobised Silicon Support. Biochim. Biophys. Acta BBA - Biomembr. 2001;1511(1):49–59. doi: 10.1016/S0005-2736(00)00380-1. [DOI] [PubMed] [Google Scholar]
- 50.Hughes A.V., Roser S.J., Gerstenberg M., Goldar A., Stidder B., Feidenhans’l R., Bradshaw J. Phase behavior of DMPC free supported bilayers studied by neutron reflectivity. Langmuir. 2002;18(21):8161–8171. doi: 10.1021/la025765u. [DOI] [Google Scholar]
- 51.Schmidt V., Sturgis J.N. Modifying styrene-maleic acid co-polymer for studying lipid nanodiscs. Biochim. Biophys. Acta BBA - Biomembr. 2018;1860(3):777–783. doi: 10.1016/j.bbamem.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Xue M., Cheng L., Faustino I., Guo W., Marrink S.J. Molecular mechanism of lipid nanodisk formation by styrene-maleic acid copolymers. Biophys. J. 2018 doi: 10.1016/j.bpj.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.