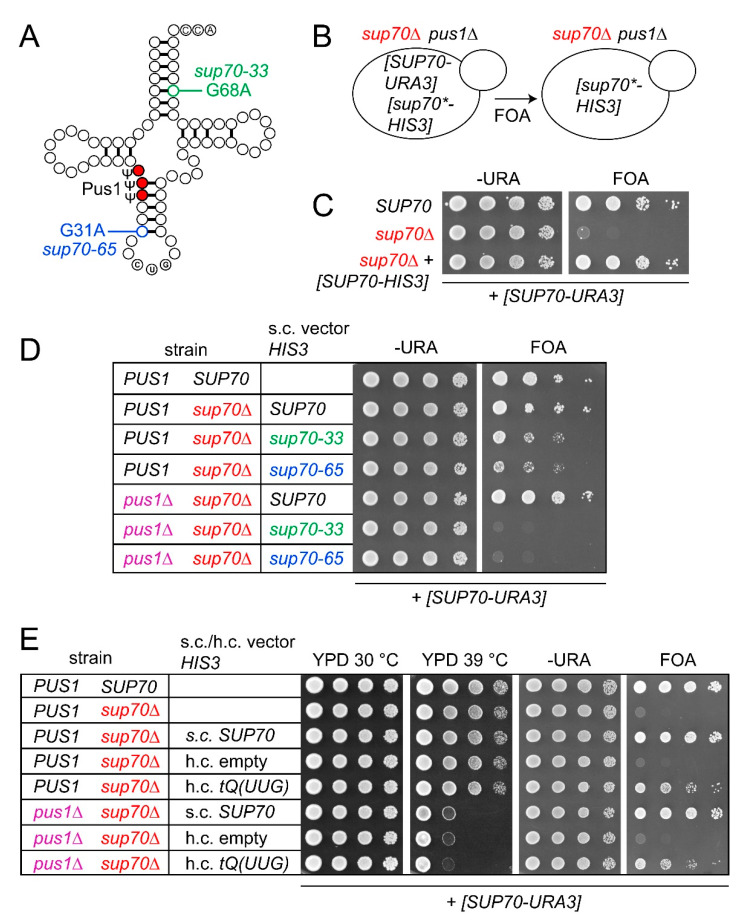
Figure 1.
Complementation of sup70 mutation by sup70-33, sup70-65, and tQ(UUG) in presence and absence of PUS1. (A) Scheme depicting tRNAGlnCUG with base exchanges present in sup70-33 and sup70-65 alleles, as well as Pus1-dependent pseudouridylation sites highlighted. (B) Outline of plasmid shuffle approach. Yeast strains carrying a deletion of SUP70 and 5-fluoro-orotate (5-FOA) counter-selectable SUP70 plasmids [SUP70-URA3] were used. In addition, HIS3-selected plasmids were introduced, which carry either native SUP70 or sup70-33/sup70-65 alleles [sup70*-HIS3]. The complementation ability of the HIS3-selected plasmids was determined in the presence or absence of a genomic PUS1 deletion. (C) Proof of principle assay demonstrating inviability of the reporter strain (sup70 [SUP70-URA3]) on 5-FOA medium (FOA) in the absence of native SUP70 ([SUP70-HIS3]). (D) Complementation of sup70 by SUP70, sup70-33, and sup70-65 in the presence and absence of PUS1. Relevant strain genotype is indicated (strain). s.c.: single copy. (E) Complementation of sup70 by high-copy (h.c.) tQ(UUG) or single copy (s.c.) SUP70 in the presence and absence of PUS1. h.c. empty: empty vector.