Abstract
Acinetobacter baumannii is a common cause of serious nosocomial infections. Although community-acquired infections are observed, the vast majority occur in people with preexisting comorbidities. A. baumannii emerged as a problematic pathogen in the 1980s when an increase in virulence, difficulty in treatment due to drug resistance, and opportunities for infection turned it into one of the most important threats to human health. Some of the clinical manifestations of A. baumannii nosocomial infection are pneumonia; bloodstream infections; lower respiratory tract, urinary tract, and wound infections; burn infections; skin and soft tissue infections (including necrotizing fasciitis); meningitis; osteomyelitis; and endocarditis. A. baumannii has an extraordinary genetic plasticity that results in a high capacity to acquire antimicrobial resistance traits. In particular, acquisition of resistance to carbapenems, which are among the antimicrobials of last resort for treatment of multidrug infections, is increasing among A. baumannii strains compounding the problem of nosocomial infections caused by this pathogen. It is not uncommon to find multidrug-resistant (MDR, resistance to at least three classes of antimicrobials), extensively drug-resistant (XDR, MDR plus resistance to carbapenems), and pan-drug-resistant (PDR, XDR plus resistance to polymyxins) nosocomial isolates that are hard to treat with the currently available drugs. In this article we review the acquired resistance to carbapenems by A. baumannii. We describe the enzymes within the OXA, NDM, VIM, IMP, and KPC groups of carbapenemases and the coding genes found in A. baumannii clinical isolates.
Keywords: β-lactam, β-lactamase, antibiotic resistance, plasmid, ESKAPE, Acinetobacter
1. A Brief Summary of Acinetobacter baumannii as a Pathogen
Acinetobacter spp., a Gram-negative coccobacillus found in virtually all environments [1,2], used to be viewed as a “low-virulence” opportunistic pathogen of negligible significance. Despite signals about the potential this group of bacteria had as a nosocomial pathogen [3,4,5,6], its importance remained unappreciated until the mid-1990s [7]. Later, a better appreciation of the impact of Acinetobacter occurred after an increase in the understanding of its epidemiology identified it as the etiology of numerous hospital infections. In addition, an increase in virulence, difficulty of treatment due to drug resistance, and opportunities for infection, made this pathogen one of the most important threats to human health [8,9]. The spread and prevalence of A. baumannii in health care institutions was helped by its ability to withstand dry as well as humid environments, its resistance to disinfectants and antibiotics, and its biofilm-forming property that leads to colonization of inert surfaces and medical devices [10,11,12,13]. Taxonomy of the genus Acinetobacter has been complex, in part, due to the high genetic variability found among its members [1,11].
A. baumannii, the most common cause of nosocomial infections caused by Acinetobacter, is part of what is known as the Acinetobacter calcoaceticus-baumannii complex, a group of bacteria that also includes Acinetobacter pittii, Acinetobacter nosocomialis, and Acinetobacter calcoaceticus [14]. A. baumannii is characterized by being catalase-positive, oxidase-negative, nonfermenting, and nonpigmented (although a pigmented strain has been recently described [15]). Although it was thought to be nonmotile, that property has been disputed by newer reports [10]. A. baumannii infections are almost exclusively nosocomial [10,11], but community-acquired cases have been reported [10,11,16,17,18,19]. However, the vast majority of community-acquired infections occur in people with preexisting comorbidities [10,20,21]. The most common clinical manifestation of A. baumannii nosocomial infection is pneumonia, which has been widely reported to increase patient mortality [2,22,23]. However, some reports still dispute this fact [24]. A vast majority of these infections occur in patients undergoing mechanical ventilation in intensive care units [25]. A. baumannii is also responsible for bloodstream infections mainly originating from intravascular devices [10,26,27]. The mortality rates of bloodstream infections caused by this bacterium range between 30% and 52% [26,27]. Other sources of A. baumannii bloodstream infections are lower respiratory tract, urinary tract, and wound infections [10]. Other manifestations caused by this bacterium include, but are not limited to, burn infections, skin and soft tissue infections (including necrotizing fasciitis), meningitis, osteomyelitis, and endocarditis [28,29,30,31,32,33,34,35,36].
A. baumannii has an extraordinary capacity to attach and survive on abiotic surfaces from non-medical objects like linen or door handles to medical equipment like catheters or respirators [37,38,39,40]. This property permits this bacterium to survive in health care environments despite the highly desiccated and starvation conditions that would kill other Gram-negatives [41,42,43,44]. As a consequence, A. baumannii is transmitted through contact with inanimate objects, making it a constant threat to immunosuppressed and weakened patients. The ability to attach to abiotic surfaces and to resist desiccation, together with the usual multidrug and disinfectant resistance exhibited by A. baumannii, are the major factors behind the success of this bacterium as a nosocomial pathogen [21,45,46,47,48,49,50].
In the past couple of decades, A. baumannii was studied intensely and, as a result, some virulence factors were identified and characterized [10,20,51,52,53]. Resistance to complement-mediated killing in the vast majority of A. baumannii clinical isolates is due to a capsular polysaccharide, of which numerous types are identified [54,55,56,57,58]. As it is the case for other bacteria [59,60,61,62], the synthesis of the capsular polysaccharide occurs through the undecaprenol-linked glycan pathway (the Wzx/Wzy-dependent pathway) [63,64,65,66]. An additional exopolysaccharide, poly-β-(1-6)-N-acetylglucosamine, is biosynthesized through the synthase-dependent pathway, i.e., a single synthase protein is responsible for the polymerization and the translocation process [65,67]. This polymer seems to play a role in efficient A. baumannii biofilm development [68].
Another carbohydrate-containing macromolecule, the lipooligosaccharide, so named because the antigen-O characteristic of lipopolysaccharides is absent in this bacterium [69,70], plays a role in resistance to colistin, an antibiotic that despite its toxicity is used as a last recourse against infection [70]. Resistance occurs through modifications in the chemistry or total loss of the lipooligosaccharide lipid A [71,72,73,74]. Membrane-associated protein O-glycosylation has been associated to virulence and biofilm formation in A. baumannii [64,75]. O-linked glycosylation of membrane proteins shares the initial steps of the capsular polysaccharide biosynthesis [64]. The role in A. baumannii virulence played by pili is less clear. Evidence that type IV pili play a fundamental role in the virulence of this pathogen [70] is not available. Its synthesis has been recently associated to the presence of thioredoxin-A [76].
Another kind of pili, the chaperone-usher pili may play a role in biofilm formation and infection [49,70]. The Type 2 secretion systems are associated to pathogenesis or antibiotic resistance [77,78]. Information on the involvement of the Type 6 secretion system is controversial [79,80,81].
Another group of potential A. baumannii virulence factors are those systems that facilitate efficient uptake of micronutrients [82,83,84,85,86,87,88,89,90]. A paradigmatic example of these virulence factors are the iron uptake systems [91,92] that bacteria evolved in response to the host’s sequestration of iron, a group of non-specific systems of defense against bacterial infection. Despite its abundance, iron is poorly available in the host organism because it is complexed to ferritin in intracellular compartments or tightly bound to high-affinity iron-binding proteins like serum transferrin and lactoferrin [3,91,92,93,94,95,96]. To cause infection, bacteria must overcome this hurdle. Numerous iron acquisition systems were found in A. baumannii, and each strain possesses more than one of them. One of the systems facilitates transport of ferrous iron, known as Feo, consisting of three genes, feoABC, coding for a transmembrane (FeoB) and two hydrophilic cytoplasmic (FeoA and FeoC) proteins [88,97,98]. Two systems mediate the use of heme as source of iron, the hemO and hemT clusters [85,88,99,100,101]. Finally, three siderophore-mediated iron uptake systems have been identified to date. The acinetobactin system was the first to be discovered; it has been thoroughly studied, and it was shown to be critical for virulence [87,102,103]. Acinetobactin is closely related to anguibactin, another siderophore that was associated to virulence in Vibrio anguillarum [104,105,106]. The other two systems are those that utilize the siderophores fimsbactins (A–F) or baumannoferrins (A and B) [89,107,108]. Exhaustive and detailed descriptions of A. baumannii virulence factors can be found in recent excellent articles [10,20,51,53,70,78,85,109,110,111]. Other confirmed or proposed virulence factors include the phospholipases and elastase [112,113], the surface autotransporter Ata [114], the formation of outer membrane vesicles [115,116], survival factors such as serum resistance [42,117,118,119,120,121], and others [57,122,123,124,125,126].
Virulence of A. baumannii is enhanced in patients with predisposing factors such as diabetes, cancer, obstructive pulmonary disorders, immunocompromising diseases or treatment, and others [10,20,21]. Chronic alcoholism is also a risk factor for A. baumannii infection. Despite the rarity of community-acquired A. baumannii infection in the healthy population, this bacterium is a common etiologic agent of community-acquired pneumonia in individuals with a history of alcoholism [18]. Alcohol abuse causes a series of disturbances in the immune response, such as alteration of the monocytes function in presenting antigens to T-cells, alteration of the levels of cytokines and natural killer cells, as well as impairment of B-cells [127,128,129,130,131]. The detrimental effects of alcohol abuse on the immune system were shown to be a factor in the increased morbidity and mortality of A. baumannii [132].
Further studies on the alcohol-related A. baumannii-enhanced virulence showed elevated expression of 49 genes involved in multiple cellular functions after exposure to ethanol [125]. This study concluded that the effect of ethanol on virulence of A. baumannii involves numerous factors including the stress response [125]. Another study concluded that ethanol not only induces expression of certain genes, but also results in repression of others [126]. Some of the effects of ethanol described in this article include increased lipid and carbohydrate anabolism, enhanced biofilm formation, and decreased motility on semi-solid surfaces [126]. The presence of ethanol also induced the acidification of bacterial cultures and the production of indole-3-acetic acid. In summary, alcohol causes changes in the human body predisposing it for infection and induces changes that enhance or decrease expression of multiple bacterial functions that result in higher pathogenicity [125,126,132,133,134].
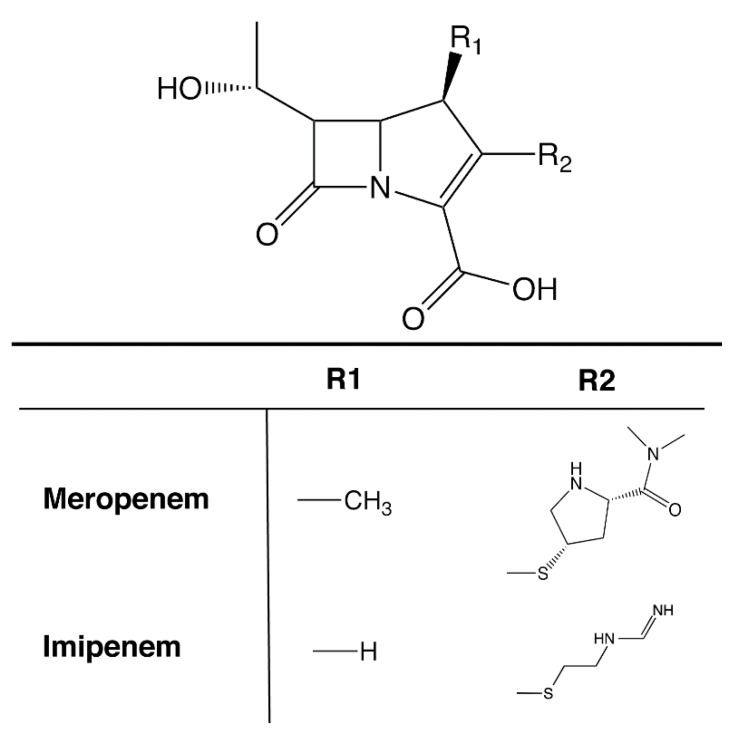
A property that turned A. baumannii into the human health threat that it has become is the resistance to multiple antibiotics exhibited by most clinical strains that complicate treatment [8]. In particular, resistance to carbapenems (Figure 1), which are among the antimicrobials of last resort for treatment of multidrug-resistant infections, is increasing among A. baumannii strains compounding the problem of nosocomial infections caused by this pathogen [9,135]. This is the reason why in the recent CDC’s 2019 Antibiotic Resistance Threats Report, Carbapenem-resistant A. baumannii (CRAB) is listed as an “Urgent” threat [8]. Considering the importance of this urgent threat, in this review, we focus on carbapenemase-mediated resistance to carbapenems in A. baumannii.
Figure 1.
Structure of carbapenems. Generic chemical structure of carbapenems. The R1 and R2 groups for imipenem and meropenem, the most used in the clinics, are shown.
2. Mechanisms of Resistance to Carbapenems in A. baumannii
A. baumannii would not be the problematic pathogen it is without the multiple mechanisms of drug resistance the strains usually possess and its versatility to acquire new ones [9,51,136,137,138,139,140]. Multiple intrinsic and acquired mechanisms of resistance have been detected in A. baumannii isolates [51]. These mechanisms include enzymatic modification of the antibiotic, changes in permeability, efflux pumps, or modifications in the target sites [51,138,141,142,143]. Multidrug-resistant (MDR; resistance to at least three classes of antimicrobials), extensively drug-resistant (XDR; MDR plus resistance to carbapenems), and pan-drug-resistant (PDR; XDR plus resistance to polymixins) [144] A. baumannii strains are currently being isolated in clinics making treatment increasingly harder [9,139,145,146,147]. In particular, a “quantum leap” in the difficulty to treat A. baumannii infections occurred with the appearance of CRAB strains. Identified mechanisms of resistance to carbapenems include efflux pumps [51,148,149,150,151,152], reduction or inactivation of expression of porins [51,153,154,155], modification in expression or synthesis of new penicillin binding proteins [156,157], and presence of carbapenemases. Since the latter is the most frequently observed and worrisome mechanism present in CRAB strains, in this article, we review the carbapenemases detected in this bacterium.
3. Carbapenemases in A. baumannii: OXA β-Lactamases
One of the most common, if not the most common, mechanisms of resistance to β-lactam antibiotics is their enzymatic hydrolysis mediated by β-lactamases [158,159,160]. There is more than one classification scheme for β-lactamases, which number in the hundreds, but one of the most frequently used is that proposed by Ambler (amino acid sequence-based) that divides these enzymes into four molecular classes: A, B, C, and D [160,161,162]. Enzymes belonging to classes A, C, and D catalyze the hydrolysis of the β-lactam substrate forming an intermediate covalent acyl–enzyme complex with a serine residue within the active site. In the case of class B β-lactamases, the hydrolysis reaction is mediated by one or more zinc ions [160,161]. OXA β-lactamases are class D enzymes that were originally differentiated by TEM enzymes by their ability to hydrolyze oxacillin [163]. The first OXA enzymes showed relatively narrow spectrum and were plasmid-mediated [164,165,166,167]. Later, numerous cases of proteins, encoded by chromosome- or plasmid-located genes, with characteristics of extended spectrum β-lactamase (ESBL) or carbapenemase OXA enzymes were described [160,163,168,169].
3.1. OXA-23-Like Group
The first OXA enzyme with carbapenemase activity to be identified in A. baumannii was the OXA-23 (first named ARI-1), found in an isolate from Scotland [170]. This enzyme gave the name to the first group of OXA enzymes with the capability to confer resistance to carbapenems (Table 1). Besides OXA-23, some enzymes within this group, such as OXA-27 or OXA-146, were subjected to more detailed analysis [171,172,173].
Table 1.
OXA enzymes identified in A. baumannii *.
| Enzyme Group | Genetic Location | Predominant Isolation Countries * | Isolation Source | Other Reported Species | Total Reported |
|---|---|---|---|---|---|
| OXA-23-like | Plasmid, chromosome | USA (564), India (125), South Korea (122) | Clinical (2,830) Environmental/other (1128) | Providencia alcalifaciens, Proteus mirabilis, Klebsiella pneumoniae, Citrobacter freundii, E. coli/Shigella, Serratia marcescens, Acinetobacter non-baumannii, Pseudomonas aeruginosa | 4048 |
| OXA-24/40-like | USA (100), Spain (5) | Clinical (124) Environmental/other (21) | Acinetobacter non-baumannii, Klebsiella pneumoniae, Providencia rettgeri, Staphylococcus aureus | 162 | |
| OXA-51-like | Germany (8), Brazil (8), Japan (6) | Clinical (39) Environmental/other (20) | Acinetobacter non-baumannii, Klebsiella pneumoniae | 88 | |
| OXA-58-like | Plasmid, Chromosome |
USA (84), Spain (12), Thailand (8) | Clinical (90) Environmental/other (177) | Providencia alcalifaciens, Klebsiella pneumoniae, E. coli/Shigella, Proteus mirabilis, Enterobacter sp., Acinetobacter non-baumannii. | 284 |
| OXA-143-like | Brazil (3) | Clinical (3) | Acinetobacter non-baumannii | 15 |
* The number of isolates was calculated using the US National Library of Medicine Pathogen Detection tool (https://www.ncbi.nlm.nih.gov/pathogens) and complemented using Blast [174]. Enzymes in each group have 90% identity and 96% coverage.
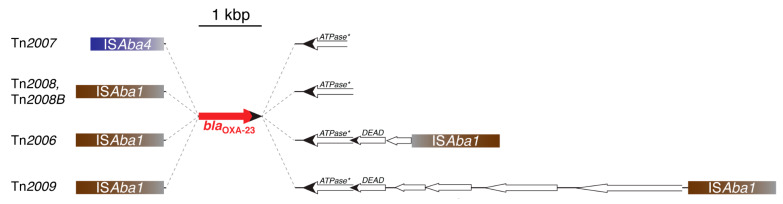
The blaOXA-23 gene is thought to have originated in A. radioresistens where close relatives are found located within the chromosome contiguous to a gene potentially coding for an ATPase [175,176]. After the first identification, the blaOXA-23 gene was detected in A. baumannii isolates from several countries, usually as part of Tn2006, Tn2007, Tn2009, Tn2008, and Tn2008B (Figure 2). In all these elements, the blaOXA-23 is preceded by an IS element, ISAba1 or ISAba4, and contiguous to a truncated version of the ATPase [175,177]. Tn2006 has the typical structure of a composite transposon where the blaOXA-23 is part of a DNA stretch also containing the truncated ATPase gene and two genes, one of them coding for a helicase [177,178]. Tn2008 and Tn2008B consist of a copy of ISAba1 associated to the fragment including blaOXA-23 and the incomplete ATPase gene (Figure 2). The difference between these two elements is the number of nucleotides between the ISAba1 and the OXA-23 initiation of translation codon, 27 and 34, respectively [177]. The ISAba1- blaOXA-23-helicase region, i.e., the complete Tn2008 sequence, is contained within Tn2006. Furthermore, Tn2009 is a larger derivative of Tn2006 that acquired a DNA fragment between both ISAba1 copies [177]. Tn2006, Tn2008, and Tn2009 have been found within plasmids in addition to the chromosome [175,177,179,180]. Tn2007, detected in plasmids, consists of a DNA fragment including blaOXA-23 and a truncated version of the helicase gene preceded by a copy of ISAb4 (Figure 2) [177]. Table 1 summarizes the number of OXA-23-like enzymes identified to date.
Figure 2.
Genetic structures of blaOXA-23-containing elements. The structure of Tn2007, which has not been proven to transpose, is shown on top. Tn2008 and Tn2008B differ in the number of nucleotides (27 and 34, respectively) that separate blaOXA-23 from ISAba1. Tn2006, the only structure experimentally shown to transpose [178], includes copies of ISAba1 in opposite orientations, while Tn2009 carries these insertion sequences in the same orientation. These two transposons are the only ones with the typical composite transposon structure. An extensive and detailed description of these elements was recently published [177]. ATPase*, gene coding for a putative ATPase truncated at the N-terminus. DADE, gene coding for a putative DEAD box family helicase [181]. Figure interpreted from Nigro and Hall (2016) [177].
3.2. OXA-24/40-Like Group
The number of beta-lactamases belonging to the OXA-24/40 group is less than that of the OXA-23 group and others to be discussed below. The first representative of this group to be isolated from A. baumannii was OXA-24, now known as OXA-40 after sequencing revision showed that they were identical enzymes [150,182]. The number of genes in this group, species that were found, and genetic location are shown in Table 1. Some plasmid-mediated genes belonging to the blaOXA24/40-like were found flanked by Xer site-specific recombination target sites, which led researchers to propose that this recombination mechanism plays a role in mobilization of these genes at the molecular level [183,184,185]. Xer site-specific recombination has been associated with plasmid evolution [186,187,188]. Structural studies showed that OXA-24/40 specifically utilizes carbapenems as substrate through a hydrophobic barrier for which the side chains of amino acids Y112 and M223 are essential [189]. This structure forms a tunnel-like entrance (hydrophobic bridge) to the active site. OXA-40 was also crystallized in complex to doripenem and inhibitors [190,191]. These studies were extended and details about the mechanism by which inhibitors inactivate OXA-24/40, such as decarboxylation of a K residue within, were clarified [189,191,192]. A clinical variant of OXA-24/40 identified in a clinical A. baumannii isolate has the mutation P227S, which results in an expanded substrate range that includes cephalosporins and aztreonam [193].
3.3. OXA-51-Like Group
OXA-51 was initially found as a chromosomally-mediated β-lactamase in a clinical A. baumannii isolate from Argentina [194]. Further studies showed the existence of numerous members of what is called the OXA-51-like group of enzymes [163]. The number of genes in this group, species that were found, and genetic location are shown in Table 1. Studies on the OXA-51 enzyme also showed that blaOXA-51-like genes are specific to the A. baumannii chromosome. As a consequence, blaOXA-51-like genes are used as a landmark in the identification of Acinetobacter isolates as A. baumannii [195,196]. A review describing available methodologies and their effectivity to identify A. baumannii has been recently published [197]. In contrast, identification of A. baumannii using blaOXA-51-like as a marker can present difficulties. Although these genes are mostly located within the chromosome, a few cases of plasmid location have been detected [198]. Plasmid-mediated blaOXA-51-like was the cause of misidentification of three non-baumannii Acinetobacter isolates that potentially acquired the gene from a plasmid originated in A. baumannii [199]. A. baumannii isolates from an outbreak in a hospital in Iran were hard to identify because the blaOXA-51-like was interrupted by an insertion of ISAba19 [200].
Early biophysical and biochemical studies on OXA-51 showed that the protein did not significantly denature when exposed to broad pH (4–10) and temperature (30–60 °C) ranges, and up to 75% of the enzymatic activity was retained in these conditions [201]. Enzymatic studies showed that OXA-51-like is a weak carbapenemase and the question of whether this enzyme or others within this group are responsible for carbapenem-resistance was the focus of numerous studies [152,163,198,202,203,204]. Various blaOXA51-like genes were found associated to ISAbA1, which, when in the proper orientation, enhances expression and alone or in association with expression of other mechanisms can lead to phenotypic resistance to carbapenems [204,205,206]. However, levels of expression may not be the only way OXA-51-like enzyme confers diminished susceptibility to carbapenems. Variants with one or more amino acid substitutions with respect to OXA-51 exhibit enhanced hydrolytic activity [205,207,208,209,210,211,212]. The molecular bases of several of these changes in activity associated to amino acids substitution were investigated by comparison of the three-dimensional structure of OXA-51 and the variant OXA-51 I129L in complex with doripenem [207,213]. This substitution increases the affinity of the protein for carbapenem molecules such as doripenem and imipenem. Variants including the substitution I129L show a significant increase in hydrolytic activity [208]. The results of the comparative analysis of these structures indicate that the presence of a Leu residue instead of Ile at position 129 causes the protein to better accommodate the substrate, increasing affinity and concomitantly enzymatic activity [207,213]. In conclusion, different OXA-51-like enzymes confer different degrees of or no clinical resistance to carbapenems, and they serve as a tool for molecular epidemiology of A. baumannii.
3.4. OXA-58-Like Group
This is a group of enzymes with a reduced number of variants (Table 1). The first blaOXA-58 gene was present in a non-self-transmissible 30-kbp plasmid residing in a multiresistant A. baumannii isolated in the Rangueil University hospital, Toulouse, France [214]. The gene was flanked by ISAba3-like insertion element copies in opposite orientations. The same gene was detected in seven carbapenem-resistant isolates, six from patients, and one from the environment, in the burns unit of the same hospital [215], and rapidly spread to other countries [216]. A recent study showed that an A. seifertii plasmid is 99% homologous to one from A. baumannii, but the ISAba3-like insertion element located upstream of blaOXA-58 is interrupted by a copy of ISAba825 [217]. These studies argue in favor of horizontal transmission as the main mechanism of dissemination of blaOXA-58-like genes. Due to the difference in GC percent found between the gene and the A. baumannii chromosome, it is currently thought that blaOXA-58-like genes originated in a different bacterium [163].
The blaOXA-58 gene was carried by plasmids harbored by six clonally related A. baumannii strains isolated in 2005 from a hospital in Rome, Italy. The strains showed different levels of resistance to carbapenems, and the plasmids were nearly identical but differed at the regions carrying the blaOXA-58 gene. A structure consisting of IS26-ISAba2- blaOXA-58-ISAba3 was identified that can induce amplification of the gene, producing an increase in MIC by gene dosage [218]. Gene amplification can occur through a wide variety of mechanisms [218], and its role in increasing levels of resistance has been shown multiple times [219,220,221,222,223].
3.5. OXA-143-Like Group
OXA-143 was first identified in a ~30-kbp plasmid present in an A. baumannii blood culture isolate from a patient in an intensive care unit in Brazil in 2004 (Table 1) [224]. OXA-143 hydrolyzed penicillin and carbapenems but did not significantly hydrolyze expanded-spectrum cephalosporins. The blaOXA-143 gene was neither associated with insertion sequences nor located within an integron, but later, at least a blaOXA-143-like gene, blaOXA-253, was located in a plasmid, downstream of ISAba47 [225]. On the other hand, blaOXA-143 and a downstream gene were flanked by copies of a gene coding for a replication protein, suggesting that its incorporation in the plasmid could have occurred by homologous recombination [224]. The OXA-143 amino acid sequence shows 88%, 63%, and 52% identity with OXA-40, OXA-23, and OXA-58, respectively. Later, other OXA-143-like enzymes were isolated in Brazil, Korea, Peru, and Honduras [225,226,227,228,229,230]. On the basis of the GC percent, these genes, which are located in plasmids, seem to have originated in a different Acinetobacter species [163].
3.6. OXA-235-Like Group
The OXA-235 and two variants, OXA-236 and OXA-237, which have one amino acid substitution (OXA-236 G173V and OXA-237 D208G), were identified in nine A. baumannii strains from the US and Mexico [231]. Location of the genes coding for these enzymes was chromosome or plasmid, with some strains showing positive signal within a plasmid and colocalization with blaOXA-51. In all nine isolates having the blaOXA-235, the gene was flanked by two copies of ISAba1. In a surveillance study performed between 2010 and 2016 in Canadian acute care hospitals, 94 carbapenem-resistant Acinetobacter sp. strains, of which 90 were A. baumannii, were selected for further analysis [232]. OXA-235-like proteins were detected in 48% of the strains. Furthermore, ISAba1 was associated to the blaOXA-235-like gene in all A. baumannii strains.
An outbreak that occurred across five healthcare facilities in Oregon, US, and lasted from June 2012 to October 2014, affected 16 patients and was caused by extensively drug-resistant A. baumannii [233]. The resistance to carbapenem was caused by the presence of the blaOXA-235-like gene blaOXA-237, which was flanked by ISAba1 elements in opposite orientations in a 15.198-kbp plasmid present in all 16 isolates [233,234]. It was of special concern that blaOXA-237 is plasmid-mediated, a characteristic that enhances the potential for dissemination, and was found in a strain belonging to clonal group IC2, the most prominent worldwide [234].
4. Carbapenemases in A. baumannii: Metallo-β-Lactamases
The mechanism used by β-lactamases to catalyze hydrolysis of the antibiotic molecule can involve a two-step cycle consisting of acylation by formation of an acylenzyme intermediate by establishing a covalent bond between the β-lactam and a serine residue within the enzyme’s active site followed by a deacylation step or a cation-facilitated hydrolytic reaction in which one or two essential zinc ions locate at the active site of the enzyme [235]. These enzymes, known as metallo-β-lactamases or Class B β-lactamases, are subdivided into three subclasses, B1–B3 [235].
Enzymes belonging to subclasses B1 and B3 utilize two Zn2+ ions, while those within subclass B2 need just one zinc atom to catalyze inactivation. A distinctive characteristic of metallo-β-lactamases is that they are active against carbapenems [158,235,236,237]. There are still no inhibitors available for clinical use for metallo-β-lactamases. However, recently described compounds such as QPX7728 and VNRX-5133 (Taniborbactam) show inhibition activity against metallo-β-lactamases. Both compounds are active at nanomolar concentrations and could be developed as part of β-lactam/β-lactamase inhibitor formulations against metallo-β-lactamases [238,239,240].
4.1. NDM Group
NDM-1 is a metallo-β-lactamase (New Delhi metallo-β-lactamase) first isolated in India in 2008 from a patient with a urinary tract infection caused by a carbapenem-resistant Klebsiella pneumoniae [241]. The blaNDM-1 gene, located adjacent to a truncated IS26 element, was part of a 180-kbp plasmid that includes genetic determinants that confer resistance to all antibiotics except fluoroquinolones and colistin. It was also troublesome that an E. coli strain isolated form the same patient harbored a plasmid containing blaNDM-1, suggesting that transfer by conjugation occurs at high frequency. This possibility was supported in mating assays and the authors predicted a worrisome scenario where the gene would spread to other bacteria [241]. Unfortunately, the predictions were correct, and bacteria harboring a plasmid containing the blaNDM-1 gene were soon isolated in infections across the world, [8,242,243,244,245,246].
A search at the National Center for Biotechnology Information Pathogen Detection Browser (https://www.ncbi.nlm.nih.gov/pathogens) performed at the time this article was being written produced 2407 isolates carrying NDM-1. The number of NDM-1-possesing A. baumannii strains was 240, only surpassed by K. pneumoniae (1527) and Escherichia coli/Shigella (265 combined).
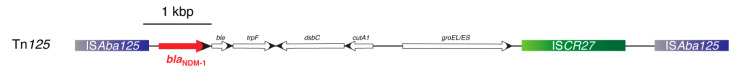
Except for monobactams, NDM-1 confers resistance to all other β-lactams [247]. Twenty-seven variants were described in addition to NDM-1 that present amino acid substitutions, and in one case there is a 5-amino acid repeat. A detailed description of the NDM-1 variants has been recently published [243]. According to this publication and newer data, NDM-1, NDM-2, NDM-3, NDM-5, and NDM-7 have been detected in A. baumannii [243,248,249]. Unfortunately, blaNDM-1 is usually associated to other genetic determinants that specify resistance to numerous antibiotics leaving only last-line antimicrobials, usually used in combination therapies, as options for treatment [250,251,252]. While the blaNDM-1 gene was found in more than one genetic context, in all cases described to date it is located downstream of ISAb125 sequences that provide the -35 region of the promoter [243]. In some instances, there is complete copy of the insertion sequence and in others only a fragment (identified as ISAba125*) [253]. This structure is associated to other genes, in many cases flanked by insertion sequences to form a composite transposon [243]. In most, but not all, cases, downstream of blaNDM-1, there is a bleMBL gene (resistance to bleomycin) followed by trpF (phosphoribosylanthranilate isomerase), and dsbC (tat twin-arginine translocation pathway signal sequence domain protein). Other genes that usually follow these genes are cutA1 (periplasmic divalent cation tolerance protein) and groES-groEL (chaperonin) [243,254,255]. In many instances, this structure is followed by a copy of ISCR27 like in the A. baumannii transposon Tn125 and other elements (Figure 3) [256,257,258,259]. These structures are found flanked by different elements that imply the existence of a wide variety of mechanisms of dissemination [243,253,255,260,261,262,263]. In those cases where the structure is part of a composite transposon, this cluster of genes is flanked by ISAba125 (forming Tn125) [257], IS903, IS26, or IS3000, in direct or opposite orientations [243,252,253,256,264]. The structure ISAba125*-blaNDM-1-bleMBL-trpF-dsbC-cutA1-groES-groEL has been also found flanked by short repeats with features characteristic of miniature inverted repeat elements [254].
Figure 3.
Genetic map of A. baumannii transposon Tn125. Figure interpreted from Poirel et al. [257].
As it is the case with other resistance proteins like the pJHCMW1-coded AAC(6′)-Ib, which has a fusion at the N-terminus where the first six amino acids are identical to those of the TEM β-lactamase [141,142,166], analysis of the nucleotide sequence of blaNDM-1 and the upstream region showed that the first 6 amino acids of the protein are identical to those of the aminoglycoside phosphotransferase APH(3′)-Via [256], a protein associated to A. baumannii strains [265]. Furthermore, the GC content of the gene changes abruptly at the point of divergence. These facts, together with the known association of ISAba125 to A. baumannii, led to propose that blaNDM-1 originated through a fusion that took place in this bacterium [256]. With a few exceptions in which blaNDM-1 was found located within the chromosome, the gene is harbored in plasmids. Upwards of 350 different plasmids and 20 replicons harboring blaNDM genes were described [243]. The rich number of plasmid varieties and transposable elements that carry blaNDM genes explain their fast dissemination at the molecular and cellular levels [243]. The structure and biochemistry of NDM-1 has been thoroughly studied [252,266,267,268,269,270,271].
An interesting feature of NDM-1 is related to its dependence on zinc ions, a property shared with other metallo-β-lactamases. It is well known that one of the mammals’ innate immune responses to bacterial infections is the chelation of metal ions by high-affinity binding proteins [272,273]. Calprotectin, an important component of the cytosolic protein pool, was first identified by its ability to interfere with the growth of fungal and bacterial pathogens [274]. This property seems to be related to its ability to bind Zn2+ with high affinity. Calprotectin is released at the infection foci in the body, and one of its effects is to reduce the action of metallo-β-lactamases and induce degradation because the apo-enzyme is degraded in the periplasm [275]. Many metallo-β-lactamases elude this defense mechanism by being very efficient at binding the scarce zinc ions present in the periplasmic space [275]. NDM-1 differs from most metallo-β-lactamases in that limitation of zinc ions does not result in degradation because it is anchored to the outer membrane, a property that makes it refractory to destabilization [276]. Furthermore, this feature results in secretion of the protein with the release of outer membrane vesicles that shield infecting bacterial cells from high levels of β-lactams in their environment [276,277].
4.2. VIM Group
This type of metallo-β-lactamases were first identified in Pseudomonas aeruginosa isolates in Italy and France [278,279]. The variants were denominated VIM-1 and VIM-2, respectively, and were part of gene cassettes as part of class 1 integrons. However, while VIM-1-carrying integron was located within the chromosome and included a second aac(6′)-Ib-containing gene cassette in its variable region, the VIM-2-carrying integron was part of a ~45-kbp non conjugative plasmid and possessed a unique gene cassette. The geographical location of the first detection of these enzymes and their genetic environment are represented in the VIM name of the group (Verona integron-related metallo-β-lactamase). After the first identification, the number of VIM variants increased rapidly, mainly found in Enterobacteriaceae [160,280,281]. Forty-six VIM variants were identified in a search at the National Center for Biotechnology Information Pathogen Detection Browser (https://www.ncbi.nlm.nih.gov/pathogens) but 70 are presently uploaded in GenBank. Early studies to identify metallo-β-lactamases in a collection of isolates from Korean hospitals showed that in one study an A. baumannii isolate and in the other 27 out of 38 tested Acinetobacter spp. samples carried VIM-2 [282,283]. Five unrelated A. baumannii isolates obtained in two Greek hospitals in the years 2004 and 2005 included a class 1 integron that carried blaVIM-1 in the variable portion [284]. The nontransferable characteristic of blaVIM-1 in these isolates suggests chromosomal or a nonconjugative plasmid. The gene was also detected in eight out of 13 strains tested in a study in Saudi Arabia and Iran where blaVIM-2 was also present [285,286,287]. Both blaVIM-1 and blaVIM-2 were also detected in a study of 100 A. baumannii isolates from three teaching hospitals in Iran [288]. Other VIM-type enzymes identified in A. baumannii were VIM-3 and VIM-11 in strains from a university hospital in Taiwan [289]. Several studies identified unspecified VIM-type enzymes in A. baumannii clinical isolates [290,291,292,293,294,295,296]. Blast analysis, carried out at the time of writing of this article, demonstrated that the genes coding for VIM-6, VIM-11, and VIM-25 were also detected in A. baumannii.
4.3. IMP Group
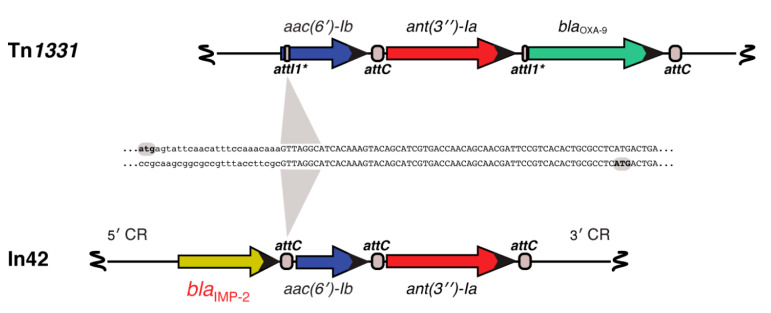
The first metallo-β-lactamase in the IMP (active on imipenem) group was identified in an imipenem-resistant P. aeruginosa clinical strain collected in 1988 in Japan [297]. The blaIMP gene resided in a conjugative 47.7-kbp plasmid, pMS350, belonging to the P-9 incompatibility group that also conferred resistance to gentamicin and sulfonamide [297]. The blaIMP gene was then found in the chromosome and as part of an integron in a transferable plasmid of urinary tract infection Serratia marcescens isolates from Japan [298,299,300]. Rapid dissemination was soon described, an expected outcome due to the location in transferable plasmids. P. aeruginosa and K. pneumoniae clinical strains carrying blaIMP were identified, and a six-years study at the Nagasaki University Hospital, Japan, identified blaIMP in 80 clinical strains belonging to the species P. aeruginosa, P. putida, P. stutzeri, and Citrobacter freundii [301,302,303]. The first report of an IMP-type enzyme in Europe occurred in a multidrug-resistant A. baumannii strain isolated from the respiratory tract of an intensive care unit patient in Italy. Further analysis showed that the gene coded for IMP-2 and was carried in a gene cassette as part of a class I integron located in the chromosome (Figure 4) [304,305]. The blaIMP-2 gene cassette was located downstream of intI1, and the variable region also included aac(6′)-Ib and ant(3′′)-Ia [304]. A Blast comparison between the aac(6′)-Ib- ant(3′′)-Ia region from this transposon and that one from the Tn1331 [306,307,308] showed 99% homology with divergence at the point of action of IntI1 at the N-terminus coding sequences of aac(6′)-Ib, a well-known characteristic of this gene that exhibits heterogeneous N-termini [141,309,310,311] (Figure 4). The second identification of an IMP-type enzyme in Europe was also on a nosocomial A. baumannii isolate in Portugal. Analysis of this gene concluded that it was a new member, named blaIMP-5 because blaIMP-3 and blaIMP-4 had been identified earlier in Asia [312,313,314,315]. IMP-4 was later found within a class 1 integron in A. baumannii isolates [316] and in similar integrons in A. pitti, K. pneumoniae, E. coli, and Enterobacter cloacae strains. In the cases of the E. coli and two E. cloacae strains, it was confirmed that the gene resided in a plasmid [317,318]. The genetic context of blaIMP-4 present in A. junii was not determined [319]. To date, A. baumannii strains carrying blaIMP-1, blaIMP-2, blaIMP-4, blaIMP-5, blaIMP-6, blaIMP-8, blaIMP-11, blaIMP-14a, blaIMP-19, and blaIMP-55, have been reported. The blaIMP-55 was identified in a study of 65 multidrug-resistant A. baumannii strains isolated in two intensive care units in Iran which showed that 6 of them contained integrons including blaIMP-55 genes [320].
Figure 4.
Comparison of the In42 variable region and a Tn1331 resistance genes fragment. The genetic maps of the variable region of In42 and a resistance genes fragment of Tn1331 are aligned. All attC loci are shown equally in the figure, but they are not identical at the nucleotide sequence level. The attI1* structure and functionality have been described [306]. The aac(6′)-Ib genes code for proteins that show differences at the N-termini, a known characteristic of this gene [141,309,310].
A search at the National Center for Biotechnology Information Pathogen Detection Browser (https://www.ncbi.nlm.nih.gov/pathogens) found 85 variants of IMP metallo-β-lactamases and 865 IMP-carrying isolates, the vast majority of which were P. aeruginosa (240 strains), Enterobacter (215 strains), and K. pneumoniae (184 strains). Forty-one IMP-carrying A. baumannii isolates were found in this search, the most prevalent variants in this bacterium were IMP-1 (21 strains) and IMP-4 (3 strains).
5. Carbapenemases in A. baumannii: KPC β-Lactamases
Enzymes within the KPC (K. pneumoniae carbapenemase) group are class A serine carbapenem-hydrolyzing β-lactamases that were initially reported in 2001, when KPC-1 was identified in a K. pneumoniae strain collected in 1996 from a hospital in North Carolina [321]. The gene quickly disseminated among Klebsiella strains and other bacteria and a high percentage of multidrug-resistant isolates were found to carry a blaKPC gene [322,323,324,325,326,327,328,329,330]. Unfortunately, bacteria carrying KPC enzymes are usually multidrug resistant, which seriously limits the antibiotic options for treatment; as a consequence, mortality among patients infected with KPC-harboring bacteria is high [328]. Interestingly, in some locations, there has been a decline in the isolates harboring these genes [325,331]. The quick and efficient dissemination of blaKPC genes is due to their usual location within transposable elements, mainly Tn4401 or close relatives [332] in conjugative plasmids [333,334,335,336]. Tn4401 is also commonly inserted within other transposon or insertion sequences generating multidrug resistance transposable elements [337,338]. It is worth noting that although unusual, blaKPC genes are found associated to non-Tn4401 genetic environments [339,340,341]. A search at the National Center for Biotechnology Information Pathogen Detection Browser (https://www.ncbi.nlm.nih.gov/pathogens) found 47 variants of KPC and 8716 records. There are numerous reports of A. baumannii carrying undetermined blaKPC genes [294,342,343]. Confirmed variants of blaKPC found in A. baumannii include blaKPC-2 and blaKPC-3, first identified in a study to characterize the resistance to carbapenem in A. baumannii (CRAB) isolates from patients with burn injury in Brazil [344]. Moreover, a study of Acinetobacter isolates from 17 hospitals in Puerto Rico identified blaKPC-4 and blaKPC-10 [345].
Enzymes belonging to the KPC group are the only ones that recognize as substrates all FDA-approved β-lactams. KPCs are inhibited by avibactam, relebactam, and vaborbactam [160,326,346,347,348,349,350,351]. Inhibition of KPC enzymes by clavulanate, tazobactam, and sulbactam is minimal [352].
6. Final Remarks
A. baumannii emerged as a problematic nosocomial pathogen in the mid-1980s, when acquisition of resistance traits became significant, making treatment more challenging [11,353]. The genetic plasticity of Acinetobacter spp. permitted this bacterium to evolve rapidly, turning it into one the most serious threats to hospitalized patients [52,53,354,355,356]. Currently, multi- and pan-drug-resistant A. baumannii are ubiquitous, and options for treatment are shrinking. The rise of carbapenem-resistant A. baumannii strains (CRABs) further compounds the problem, which requires urgent attention to avoid expanding the number of deaths due to nosocomial infection. Among the many mechanisms causing resistance, the acquisition of carbapenemase-coding genes are the most relevant. The majority of carbapenem-resistant A. baumannii isolates owes this property to the presence of OXA-23. The prevalence of blaOXA-23 is, at least in part, attributed to the spread of successful global clones such us GC1 and GC2. However, the number of isolates containing NDM-1 is rapidly growing. Research efforts geared to developing new antimicrobials must be complemented with strategies to overcome the presence of these enzymes such as the introduction of new inhibitors to be used in combination with antibacterial drugs. Despite the grim perspective of witnessing the rise of strains resilient to all available treatments, investigative attempts to introduce new inhibitors and drugs give hope that options to control these infections will continue to be available.
Author Contributions
Conceptualization, M.S.R., R.A.B., and M.E.T.; writing—original draft preparation, M.E.T.; writing—review and editing, M.S.R., R.A.B., and M.E.T. All authors have read and agreed to the published version of the manuscript.
Funding
Authors’ work cited in this review article was funded by Public Health Service Grants SC3GM125556 (MSR), 2R15AI047115 (MET), R01AI100560 (RAB), R01AI063517 (to RAB), and R01AI072219 (to RAB) from the National Institutes of Health and VA 1I01BX001974 (to RAB) form the Cleveland Department of Veterans Affairs. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Giamarellou H., Antoniadou A., Kanellakopoulou K. Acinetobacter baumannii: A universal threat to public health? Int. J. Antimicrob. Agents. 2008;32:106–119. doi: 10.1016/j.ijantimicag.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Fournier P.E., Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 2006;42:692–699. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 3.Actis L.A., Tolmasky M.E., Crosa L.M., Crosa J.H. Effect of iron-limiting conditions on growth of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 1993;31:2812–2815. doi: 10.1128/JCM.31.10.2812-2815.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartstein A.I., Rashad A.L., Liebler J.M., Actis L.A., Freeman J., Rourke J.W., Jr., Stibolt T.B., Tolmasky M.E., Ellis G.R., Crosa J.H. Multiple intensive care unit outbreak of Acinetobacter calcoaceticus subspecies anitratus respiratory infection and colonization associated with contaminated, reusable ventilator circuits and resuscitation bags. Am. J. Med. 1988;85:624–631. doi: 10.1016/S0002-9343(88)80233-X. [DOI] [PubMed] [Google Scholar]
- 5.Hartstein A.I., Morthland V.H., Rourke J.W., Jr., Freeman J., Garber S., Sykes R., Rashad A.L. Plasmid DNA fingerprinting of Acinetobacter calcoaceticus subspecies anitratus from intubated and mechanically ventilated patients. Infect. Control. Hosp. Epidemiol. 1990;11:531–538. doi: 10.2307/30151321. [DOI] [PubMed] [Google Scholar]
- 6.Larson E. A decade of nosocomial Acinetobacter. Am. J. Infect. Control. 1984;12:14–18. doi: 10.1016/0196-6553(84)90067-1. [DOI] [PubMed] [Google Scholar]
- 7.Bergogne-Berezin E., Towner K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996;9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC . Antibiotic resistance threats in the United States. Centers for Disease Control; Atlanta, GA, USA: 2019. [Google Scholar]
- 9.Isler B., Doi Y., Bonomo R.A., Paterson D.L. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 2019;63:e01110-18. doi: 10.1128/AAC.01110-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McConnell M.J., Actis L., Pachon J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013;37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 11.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomaras A.P., Dorsey C.W., Edelmann R.E., Actis L.A. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: Involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 13.Tomaras A.P., Flagler M.J., Dorsey C.W., Gaddy J.A., Actis L.A. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology. 2008;154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 14.Nemec A., Krizova L., Maixnerova M., van der Reijden T.J., Deschaght P., Passet V., Vaneechoutte M., Brisse S., Dijkshoorn L. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU) Res. Microbiol. 2011;162:393–404. doi: 10.1016/j.resmic.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Traglia G., Chiem K., Quinn B., Fernandez J.S., Montana S., Almuzara M., Mussi M.A., Tolmasky M.E., Iriarte A., Centron D., et al. Genome sequence analysis of an extensively drug-resistant Acinetobacter baumannii indigo-pigmented strain depicts evidence of increase genome plasticity. Sci. Rep. 2018;8:16961. doi: 10.1038/s41598-018-35377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu A., Zhu H., Gao B., Weng H., Ding Z., Li M., Weng X., He G. Diagnosis of severe community-acquired pneumonia caused by Acinetobacter baumannii through next-generation sequencing: A case report. BMC Infect. Dis. 2020;20:45. doi: 10.1186/s12879-019-4733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C.T., Wang Y.C., Kuo S.C., Shih F.H., Chen T.L., How C.K., Yang Y.S., Lee Y.T. Community-acquired bloodstream infections caused by Acinetobacter baumannii: A matched case-control study. J. Microbiol. Immunol. Infect. 2018;51:629–635. doi: 10.1016/j.jmii.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Leung W.S., Chu C.M., Tsang K.Y., Lo F.H., Lo K.F., Ho P.L. Fulminant community-acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest. 2006;129:102–109. doi: 10.1378/chest.129.1.102. [DOI] [PubMed] [Google Scholar]
- 19.Chen M.Z., Hsueh P.R., Lee L.N., Yu C.J., Yang P.C., Luh K.T. Severe community-acquired pneumonia due to Acinetobacter baumannii. Chest. 2001;120:1072–1077. doi: 10.1378/chest.120.4.1072. [DOI] [PubMed] [Google Scholar]
- 20.Harding C.M., Hennon S.W., Feldman M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018;16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong D., Nielsen T.B., Bonomo R.A., Pantapalangkoor P., Luna B., Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: A century of challenges. Clin. Microbiol. Rev. 2017;30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbo A., Carmeli Y., Navon-Venezia S., Siegman-Igra Y., Schwaber M.J. Impact of multi-drug-resistant Acinetobacter baumannii on clinical outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2007;26:793–800. doi: 10.1007/s10096-007-0371-8. [DOI] [PubMed] [Google Scholar]
- 23.Falagas M.E., Rafailidis P.I. Attributable mortality of Acinetobacter baumannii: No longer a controversial issue. Crit. Care. 2007;11:134. doi: 10.1186/cc5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garnacho J., Sole-Violan J., Sa-Borges M., Diaz E., Rello J. Clinical impact of pneumonia caused by Acinetobacter baumannii in intubated patients: A matched cohort study. Crit. Care Med. 2003;31:2478–2482. doi: 10.1097/01.CCM.0000089936.09573.F3. [DOI] [PubMed] [Google Scholar]
- 25.Ciginskiene A., Dambrauskiene A., Rello J., Adukauskiene D. Ventilator-associated pneumonia due to drug-resistant Acinetobacter baumannii: Risk factors and mortality relation with resistance profiles, and independent predictors of in-hospital mortality. Medicina (Kaunas) 2019;55:49. doi: 10.3390/medicina55020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dijkshoorn L., Nemec A., Seifert H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 27.Wisplinghoff H., Paulus T., Lugenheim M., Stefanik D., Higgins P.G., Edmond M.B., Wenzel R.P., Seifert H. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Infect. 2012;64:282–290. doi: 10.1016/j.jinf.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Zurawski D.V., Banerjee J., Alamneh Y.A., Shearer J.P., Demons S.T. Skin and soft tissue models for Acinetobacter baumannii infection. Methods Mol. Biol. 2019;1946:271–287. doi: 10.1007/978-1-4939-9118-1_25. [DOI] [PubMed] [Google Scholar]
- 29.Matthews L., Goodrich J.S., Weber D.J., Bergman N.H., Miller M.B. The brief case: A fatal case of necrotizing fasciitis due to multidrug-resistant Acinetobacter baumannii. J. Clin. Microbiol. 2019;57:e01751-18. doi: 10.1128/JCM.01751-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerrero D.M., Perez F., Conger N.G., Solomkin J.S., Adams M.D., Rather P.N., Bonomo R.A. Acinetobacter baumannii-associated skin and soft tissue infections: Recognizing a broadening spectrum of disease. Surg. Infect. (Larchmt.) 2010;11:49–57. doi: 10.1089/sur.2009.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X., Wang L., Ye Y.Z., Yu H. Postoperative multidrug-resistant Acinetobacter baumannii meningitis successfully treated with intravenous doxycycline and intraventricular gentamicin: A case report. World J. Clin Cases. 2019;7:4342–4348. doi: 10.12998/wjcc.v7.i24.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zurawski D.V., Thompson M.G., McQueary C.N., Matalka M.N., Sahl J.W., Craft D.W., Rasko D.A. Genome sequences of four divergent multidrug-resistant Acinetobacter baumannii strains isolated from patients with sepsis or osteomyelitis. J. Bacteriol. 2012;194:1619–1620. doi: 10.1128/JB.06749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charnot-Katsikas A., Dorafshar A.H., Aycock J.K., David M.Z., Weber S.G., Frank K.M. Two cases of necrotizing fasciitis due to Acinetobacter baumannii. J. Clin. Microbiol. 2009;47:258–263. doi: 10.1128/JCM.01250-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturiale M., Corpina C., Sturiale L. Endocarditis due to Acinetobacter baumannii. Int. J. Cardiol. 2016;209:161–163. doi: 10.1016/j.ijcard.2014.10.098. [DOI] [PubMed] [Google Scholar]
- 35.Kunhi M., Sanagar S., Jagadeesh N., Shankar B., Abraham A. Emergency cardiac double valve surgery in active infective endocarditis due to Acinetobacter baumannii with aortic root abscess in a patient with dialysis-dependent end-stage renal failure: A rare case report. J. Surg Case Rep. 2016;2016:rjw168. doi: 10.1093/jscr/rjw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel G., Perez F., Hujer A.M., Rudin S.D., Augustine J.J., Jacobs G.H., Jacobs M.R., Bonomo R.A. Fulminant endocarditis and disseminated infection caused by carbapenem-resistant Acinetobacter baumannii in a renal-pancreas transplant recipient. Transpl. Infect. Dis. 2015;17:289–296. doi: 10.1111/tid.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borer A., Gilad J., Smolyakov R., Eskira S., Peled N., Porat N., Hyam E., Trefler R., Riesenberg K., Schlaeffer F. Cell phones and Acinetobacter transmission. Emerg. Infect. Dis. 2005;11:1160–1161. doi: 10.3201/eid1107.050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen R., Shimoni Z., Ghara R., Ram R., Ben-Ami R. Effect of a ventilator-focused intervention on the rate of Acinetobacter baumannii infection among ventilated patients. Am. J. Infect. Control. 2014;42:996–1001. doi: 10.1016/j.ajic.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Villegas M.V., Hartstein A.I. Acinetobacter outbreaks, 1977-2000. Infect. Control. Hosp. Epidemiol. 2003;24:284–295. doi: 10.1086/502205. [DOI] [PubMed] [Google Scholar]
- 40.Weernink A., Severin W.P., Tjernberg I., Dijkshoorn L. Pillows, an unexpected source of Acinetobacter. J. Hosp. Infect. 1995;29:189–199. doi: 10.1016/0195-6701(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 41.Farrow J.M., 3rd, Wells G., Pesci E.C. Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PLoS ONE. 2018;13:e0205638. doi: 10.1371/journal.pone.0205638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aranda J., Bardina C., Beceiro A., Rumbo S., Cabral M.P., Barbe J., Bou G. Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J. Bacteriol. 2011;193:3740–3747. doi: 10.1128/JB.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapartegui-Gonzalez I., Lazaro-Diez M., Bravo Z., Navas J., Icardo J.M., Ramos-Vivas J. Acinetobacter baumannii maintains its virulence after long-time starvation. PLoS ONE. 2018;13:e0201961. doi: 10.1371/journal.pone.0201961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boll J.M., Tucker A.T., Klein D.R., Beltran A.M., Brodbelt J.S., Davies B.W., Trent M.S. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. mBio. 2015;6:e00478-15. doi: 10.1128/mBio.00478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roca I., Espinal P., Vila-Farres X., Vila J. The Acinetobacter baumannii oxymoron: Commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 2012;3:148. doi: 10.3389/fmicb.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaddy J.A., Actis L.A. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 2009;4:273–278. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson M.G., Black C.C., Pavlicek R.L., Honnold C.L., Wise M.C., Alamneh Y.A., Moon J.K., Kessler J.L., Si Y., Williams R., et al. Validation of a novel murine wound model of Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 2014;58:1332–1342. doi: 10.1128/AAC.01944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greene C., Vadlamudi G., Newton D., Foxman B., Xi C. The influence of biofilm formation and multidrug resistance on environmental survival of clinical and environmental isolates of Acinetobacter baumannii. Am. J. Infect. Control. 2016;44:e65–e71. doi: 10.1016/j.ajic.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood C.R., Ohneck E.J., Edelmann R.E., Actis L.A. A light-regulated type I pilus contributes to Acinetobacter baumannii biofilm, motility, and virulence functions. Infect. Immun. 2018;86:e00442-18. doi: 10.1128/IAI.00442-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hassan K.A., Jackson S.M., Penesyan A., Patching S.G., Tetu S.G., Eijkelkamp B.A., Brown M.H., Henderson P.J., Paulsen I.T. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. USA. 2013;110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee C.R., Lee J.H., Park M., Park K.S., Bae I.K., Kim Y.B., Cha C.J., Jeong B.C., Lee S.H. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell Infect. Microbiol. 2017;7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antunes L.C., Visca P., Towner K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 53.Weber B.S., Harding C.M., Feldman M.F. Pathogenic Acinetobacter: From the cell surface to infinity and beyond. J. Bacteriol. 2015;198:880–887. doi: 10.1128/JB.00906-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russo T.A., Luke N.R., Beanan J.M., Olson R., Sauberan S.L., MacDonald U., Schultz L.W., Umland T.C., Campagnari A.A. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun. 2010;78:3993–4000. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh J.K., Adams F.G., Brown M.H. Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front. Microbiol. 2018;9:3301. doi: 10.3389/fmicb.2018.03301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niu T., Guo L., Luo Q., Zhou K., Yu W., Chen Y., Huang C., Xiao Y. Wza gene knockout decreases Acinetobacter baumannii virulence and affects Wzy-dependent capsular polysaccharide synthesis. Virulence. 2020;11:1–13. doi: 10.1080/21505594.2019.1700659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chin C.Y., Tipton K.A., Farokhyfar M., Burd E.M., Weiss D.S., Rather P.N. A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat. Microbiol. 2018;3:563–569. doi: 10.1038/s41564-018-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cress B.F., Englaender J.A., He W., Kasper D., Linhardt R.J., Koffas M.A. Masquerading microbial pathogens: Capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol. Rev. 2014;38:660–697. doi: 10.1111/1574-6976.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tolmasky M.E., Staneloni R.J., Leloir L.F. Lipid-bound saccharides in Rhizobium meliloti. J. Biol. Chem. 1982;257:6751–6757. [PubMed] [Google Scholar]
- 60.Tolmasky M.E., Staneloni R.J., Ugalde R.A., Leloir L.F. Lipid-bound sugars in Rhizobium meliloti. Arch. Biochem. Biophys. 1980;203:358–364. doi: 10.1016/0003-9861(80)90187-3. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez J.E., Semino C.E., Wang L.X., Castellano-Torres L.E., Walker G.C. Biosynthetic control of molecular weight in the polymerization of the octasaccharide subunits of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA. 1998;95:13477–13482. doi: 10.1073/pnas.95.23.13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ielpi L., Couso R.O., Dankert M.A. Sequential assembly and polymerization of the polyprenol-linked pentasaccharide repeating unit of the xanthan polysaccharide in Xanthomonas campestris. J. Bacteriol. 1993;175:2490–2500. doi: 10.1128/JB.175.9.2490-2500.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hug I., Feldman M.F. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 2011;21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- 64.Lees-Miller R.G., Iwashkiw J.A., Scott N.E., Seper A., Vinogradov E., Schild S., Feldman M.F. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 2013;89:816–830. doi: 10.1111/mmi.12300. [DOI] [PubMed] [Google Scholar]
- 65.Schmid J., Sieber V., Rehm B. Bacterial exopolysaccharides: Biosynthesis pathways and engineering strategies. Front. Microbiol. 2015;6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staneloni R.J., Tolmasky M.E., Leloir L.F. Lipid-bound saccharides containing glucose and galactose in Agrobacterium tumefaciens. J. Gen. Microbiol. 1984;130:869–879. doi: 10.1099/00221287-130-4-869. [DOI] [Google Scholar]
- 67.Low K.E., Howell P.L. Gram-negative synthase-dependent exopolysaccharide biosynthetic machines. Curr. Opin. Struct. Biol. 2018;53:32–44. doi: 10.1016/j.sbi.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 68.Choi A.H., Slamti L., Avci F.Y., Pier G.B., Maira-Litran T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 2009;191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kenyon J.J., Nigro S.J., Hall R.M. Variation in the OC locus of Acinetobacter baumannii genomes predicts extensive structural diversity in the lipooligosaccharide. PLoS ONE. 2014;9:e107833. doi: 10.1371/journal.pone.0107833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geisinger E., Huo W., Hernandez-Bird J., Isberg R.R. Acinetobacter baumannii: Envelope determinants that control drug resistance, virulence, and surface variability. Annu. Rev. Microbiol. 2019;73:481–506. doi: 10.1146/annurev-micro-020518-115714. [DOI] [PubMed] [Google Scholar]
- 71.Pelletier M.R., Casella L.G., Jones J.W., Adams M.D., Zurawski D.V., Hazlett K.R., Doi Y., Ernst R.K. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013;57:4831–4840. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arroyo L.A., Herrera C.M., Fernandez L., Hankins J.V., Trent M.S., Hancock R.E. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 2011;55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Snitkin E.S., Zelazny A.M., Gupta J., Program N.C.S., Palmore T.N., Murray P.R., Segre J.A. Genomic insights into the fate of colistin resistance and Acinetobacter baumannii during patient treatment. Genome Res. 2013;23:1155–1162. doi: 10.1101/gr.154328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beceiro A., Llobet E., Aranda J., Bengoechea J.A., Doumith M., Hornsey M., Dhanji H., Chart H., Bou G., Livermore D.M., et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 2011;55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iwashkiw J.A., Seper A., Weber B.S., Scott N.E., Vinogradov E., Stratilo C., Reiz B., Cordwell S.J., Whittal R., Schild S., et al. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 2012;8:e1002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.May H.C., Yu J.J., Zhang H., Wang Y., Cap A.P., Chambers J.P., Guentzel M.N., Arulanandam B.P. Thioredoxin-A is a virulence factor and mediator of the type IV pilus system in Acinetobacter baumannii. PLoS ONE. 2019;14:e0218505. doi: 10.1371/journal.pone.0218505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elhosseiny N.M., Elhezawy N.B., Attia A.S. Comparative proteomics analyses of Acinetobacter baumannii strains ATCC 17978 and AB5075 reveal the differential role of type II secretion system secretomes in lung colonization and ciprofloxacin resistance. Microb. Pathog. 2019;128:20–27. doi: 10.1016/j.micpath.2018.12.039. [DOI] [PubMed] [Google Scholar]
- 78.Harding C.M., Kinsella R.L., Palmer L.D., Skaar E.P., Feldman M.F. Medically relevant Acinetobacter species require a type II secretion system and specific membrane-associated chaperones for the export of multiple substrates and full virulence. PLoS Pathog. 2016;12:e1005391. doi: 10.1371/journal.ppat.1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopez J., Ly P.M., Feldman M.F. The tip of the VgrG spike Is essential to functional type VI secretion system assembly in Acinetobacter baumannii. mBio. 2020;11:e02761-19. doi: 10.1128/mBio.02761-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Repizo G.D., Gagne S., Foucault-Grunenwald M.L., Borges V., Charpentier X., Limansky A.S., Gomes J.P., Viale A.M., Salcedo S.P. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS ONE. 2015;10:e0138265. doi: 10.1371/journal.pone.0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang J., Zhou Z., He F., Ruan Z., Jiang Y., Hua X., Yu Y. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS ONE. 2018;13:e0192288. doi: 10.1371/journal.pone.0192288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hesse L.E., Lonergan Z.R., Beavers W.N., Skaar E.P. The Acinetobacter baumannii Znu system overcomes host-imposed nutrient zinc limitation. Infect. Immun. 2019;87:e00746-19. doi: 10.1128/IAI.00746-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mortensen B.L., Skaar E.P. The contribution of nutrient metal acquisition and metabolism to Acinetobacter baumannii survival within the host. Front. Cell Infect. Microbiol. 2013;3:95. doi: 10.3389/fcimb.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Green E.R., Juttukonda L.J., Skaar E.P. The manganese-responsive transcriptional regulator MumR protects Acinetobacter baumannii from oxidative stress. Infect. Immun. 2019;88:e00762-19. doi: 10.1128/IAI.00762-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramirez M.S., Penwell W.F., Traglia G.M., Zimbler D.L., Gaddy J.A., Nikolaidis N., Arivett B.A., Adams M.D., Bonomo R.A., Actis L.A., et al. Identification of potential virulence factors in the model strain Acinetobacter baumannii A118. Front. Microbiol. 2019;10:1599. doi: 10.3389/fmicb.2019.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Penwell W.F., Actis L.A. Isolation and characterization of the acinetobactin and baumannoferrin siderophores produced by Acinetobacter baumannii. Methods Mol. Biol. 2019;1946:259–270. doi: 10.1007/978-1-4939-9118-1_24. [DOI] [PubMed] [Google Scholar]
- 87.Penwell W.F., Arivett B.A., Actis L.A. The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS ONE. 2012;7:e36493. doi: 10.1371/journal.pone.0036493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Antunes L.C., Imperi F., Towner K.J., Visca P. Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 2011;162:279–284. doi: 10.1016/j.resmic.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 89.Proschak A., Lubuta P., Grun P., Lohr F., Wilharm G., De Berardinis V., Bode H.B. Structure and biosynthesis of fimsbactins A-F, siderophores from Acinetobacter baumannii and Acinetobacter baylyi. Chembiochem. 2013;14:633–638. doi: 10.1002/cbic.201200764. [DOI] [PubMed] [Google Scholar]
- 90.Juttukonda L.J., Chazin W.J., Skaar E.P. Acinetobacter baumannii coordinates urea metabolism with metal import to resist host-mediated metal limitation. mBio. 2016;7:e01475–e01486. doi: 10.1128/mBio.01475-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crosa J.H., Walsh C.T. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wooldridge K.G., Williams P.H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 93.Tolmasky M.E., Crosa J.H. Regulation of plasmid-mediated iron transport and virulence in Vibrio anguillarum. Biol. Met. 1991;4:33–35. doi: 10.1007/BF01135554. [DOI] [PubMed] [Google Scholar]
- 94.Di Lorenzo M., Stork M. Plasmid-encoded iron uptake systems. Microbiol. Spectr. 2014;2:PLAS–0030–2014. doi: 10.1128/microbiolspec.PLAS-0030-2014. [DOI] [PubMed] [Google Scholar]
- 95.Parrow N.L., Fleming R.E., Minnick M.F. Sequestration and scavenging of iron in infection. Infect. Immun. 2013;81:3503–3514. doi: 10.1128/IAI.00602-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Porcheron G., Garenaux A., Proulx J., Sabri M., Dozois C.M. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: Correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell Infect. Microbiol. 2013;3:90. doi: 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lau C.K., Krewulak K.D., Vogel H.J. Bacterial ferrous iron transport: The Feo system. FEMS Microbiol. Rev. 2016;40:273–298. doi: 10.1093/femsre/fuv049. [DOI] [PubMed] [Google Scholar]
- 98.Alvarez-Fraga L., Vazquez-Ucha J.C., Martinez-Guitian M., Vallejo J.A., Bou G., Beceiro A., Poza M. Pneumonia infection in mice reveals the involvement of the feoA gene in the pathogenesis of Acinetobacter baumannii. Virulence. 2018;9:496–509. doi: 10.1080/21505594.2017.1420451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Leseleuc L., Harris G., KuoLee R., Xu H.H., Chen W. Serum resistance, gallium nitrate tolerance and extrapulmonary dissemination are linked to heme consumption in a bacteremic strain of Acinetobacter baumannii. Int. J. Med. Microbiol. 2014;304:360–369. doi: 10.1016/j.ijmm.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 100.Zimbler D.L., Penwell W.F., Gaddy J.A., Menke S.M., Tomaras A.P., Connerly P.L., Actis L.A. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals. 2009;22:23–32. doi: 10.1007/s10534-008-9202-3. [DOI] [PubMed] [Google Scholar]
- 101.Ramirez M.S., Adams M.D., Bonomo R.A., Centron D., Tolmasky M.E. Genomic analysis of Acinetobacter baumannii A118 by comparison of optical maps: Identification of structures related to its susceptibility phenotype. Antimicrob. Agents Chemother. 2011;55:1520–1526. doi: 10.1128/AAC.01595-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dorsey C.W., Tomaras A.P., Connerly P.L., Tolmasky M.E., Crosa J.H., Actis L.A. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology. 2004;150:3657–3667. doi: 10.1099/mic.0.27371-0. [DOI] [PubMed] [Google Scholar]
- 103.Bailey D.C., Bohac T.J., Shapiro J.A., Giblin D.E., Wencewicz T.A., Gulick A.M. Crystal structure of the siderophore binding protein BauB bound to an unusual 2:1 complex between acinetobactin and ferric iron. Biochemistry. 2018;57:6653–6661. doi: 10.1021/acs.biochem.8b00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Di Lorenzo M., Poppelaars S., Stork M., Nagasawa M., Tolmasky M.E., Crosa J.H. A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibrio anguillarum. J. Bacteriol. 2004;186:7327–7336. doi: 10.1128/JB.186.21.7327-7336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Actis L.A., Fish W., Crosa J.H., Kellerman K., Ellenberger S.R., Hauser F.M., Sanders-Loehr J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1) J. Bacteriol. 1986;167:57–65. doi: 10.1128/JB.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Actis L., Tolmasky M.E., Crosa J.H. Vibriosis. In: Woo P.T., Bruno D.W., editors. Fish Diseases and Disorders. Volume 3. CAB International; New York, NY, USA: 1999. pp. 523–557. [Google Scholar]
- 107.Bohac T.J., Fang L., Giblin D.E., Wencewicz T.A. Fimsbactin and acinetobactin compete for the periplasmic siderophore binding protein BauB in pathogenic Acinetobacter baumannii. ACS Chem. Biol. 2019;14:674–687. doi: 10.1021/acschembio.8b01051. [DOI] [PubMed] [Google Scholar]
- 108.Penwell W.F., DeGrace N., Tentarelli S., Gauthier L., Gilbert C.M., Arivett B.A., Miller A.A., Durand-Reville T.F., Joubran C., Actis L.A. Discovery and characterization of new hydroxamate siderophores, baumannoferrin A and B, produced by Acinetobacter baumannii. Chembiochem. 2015;16:1896–1904. doi: 10.1002/cbic.201500147. [DOI] [PubMed] [Google Scholar]
- 109.Morris F.C., Dexter C., Kostoulias X., Uddin M.I., Peleg A.Y. The mechanisms of disease caused by Acinetobacter baumannii. Front. Microbiol. 2019;10:1601. doi: 10.3389/fmicb.2019.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li F.J., Starrs L., Burgio G. Tug of war between Acinetobacter baumannii and host immune responses. Pathog. Dis. 2018;76 doi: 10.1093/femspd/ftz004. [DOI] [PubMed] [Google Scholar]
- 111.Kroger C., Kary S.C., Schauer K., Cameron A.D. Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes (Basel) 2016;8:12. doi: 10.3390/genes8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kareem S.M., Al-Kadmy I.M.S., Al-Kaabi M.H., Aziz S.N., Ahmad M. Acinetobacter baumannii virulence is enhanced by the combined presence of virulence factors genes phospholipase C (plcN) and elastase (lasB) Microb. Pathog. 2017;110:568–572. doi: 10.1016/j.micpath.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 113.Fiester S.E., Arivett B.A., Schmidt R.E., Beckett A.C., Ticak T., Carrier M.V., Ghosh R., Ohneck E.J., Metz M.L., Sellin Jeffries M.K., et al. Iron-regulated phospholipase C activity contributes to the cytolytic activity and virulence of Acinetobacter baumannii. PLoS ONE. 2016;11:e0167068. doi: 10.1371/journal.pone.0167068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bentancor L.V., Routray A., Bozkurt-Guzel C., Camacho-Peiro A., Pier G.B., Maira-Litran T. Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect. Immun. 2012;80:3381–3388. doi: 10.1128/IAI.06096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chatterjee S., Mondal A., Mitra S., Basu S. Acinetobacter baumannii transfers the blaNDM-1 gene via outer membrane vesicles. J. Antimicrob. Chemother. 2017;72:2201–2207. doi: 10.1093/jac/dkx131. [DOI] [PubMed] [Google Scholar]
- 116.Kim S.Y., Kim M.H., Kim S.I., Son J.H., Kim S., Lee Y.C., Shin M., Oh M.H., Lee J.C. The sensor kinase BfmS controls production of outer membrane vesicles in Acinetobacter baumannii. BMC Microbiol. 2019;19:301. doi: 10.1186/s12866-019-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Koenigs A., Stahl J., Averhoff B., Gottig S., Wichelhaus T.A., Wallich R., Zipfel P.F., Kraiczy P. CipA of Acinetobacter baumannii is a novel plasminogen binding and complement inhibitory protein. J. Infect. Dis. 2016;213:1388–1399. doi: 10.1093/infdis/jiv601. [DOI] [PubMed] [Google Scholar]
- 118.Koenigs A., Zipfel P.F., Kraiczy P. Translation elongation factor Tuf of Acinetobacter baumannii is a plasminogen-binding protein. PLoS ONE. 2015;10:e0134418. doi: 10.1371/journal.pone.0134418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gebhardt M.J., Shuman H.A. GigA and GigB are master regulators of antibiotic resistance, stress responses, and virulence in Acinetobacter baumannii. J. Bacteriol. 2017;199:e00066-17. doi: 10.1128/JB.00066-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gebhardt M.J., Gallagher L.A., Jacobson R.K., Usacheva E.A., Peterson L.R., Zurawski D.V., Shuman H.A. Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. mBio. 2015;6:e01660-15. doi: 10.1128/mBio.01660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cerqueira G.M., Kostoulias X., Khoo C., Aibinu I., Qu Y., Traven A., Peleg A.Y. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 2014;210:46–55. doi: 10.1093/infdis/jiu024. [DOI] [PubMed] [Google Scholar]
- 122.Lin F., Xu Y., Chang Y., Liu C., Jia X., Ling B. Molecular characterization of reduced susceptibility to biocides in clinical isolates of Acinetobacter baumannii. Front. Microbiol. 2017;8:1836. doi: 10.3389/fmicb.2017.01836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Darwish Alipour Astaneh S., Rasooli I., Mousavi Gargari S.L. Filamentous hemagglutinin adhesin FhaB limits A.baumannii biofilm formation. Front. Biosci. 2017;9:266–275. doi: 10.2741/e801. [DOI] [PubMed] [Google Scholar]
- 124.Tipton K.A., Rather P.N. An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J. Bacteriol. 2017;199:e00705–e00716. doi: 10.1128/JB.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Camarena L., Bruno V., Euskirchen G., Poggio S., Snyder M. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 2010;6:e1000834. doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nwugo C.C., Arivett B.A., Zimbler D.L., Gaddy J.A., Richards A.M., Actis L.A. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS ONE. 2012;7:e51936. doi: 10.1371/journal.pone.0051936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Szabo G., Verma B., Catalano D. Selective inhibition of antigen-specific T lymphocyte proliferation by acute ethanol exposure: The role of impaired monocyte antigen presentation capacity and mediator production. J. Leukoc. Biol. 1993;54:534–544. doi: 10.1002/jlb.54.6.534. [DOI] [PubMed] [Google Scholar]
- 128.Szabo G., Dolganiuc A., Mandrekar P., White B. Inhibition of antigen-presenting cell functions by alcohol: Implications for hepatitis C virus infection. Alcohol. 2004;33:241–249. doi: 10.1016/j.alcohol.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 129.Achur R.N., Freeman W.M., Vrana K.E. Circulating cytokines as biomarkers of alcohol abuse and alcoholism. J. Neuroimmune Pharmacol. 2010;5:83–91. doi: 10.1007/s11481-009-9185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gallucci R.M., Pfister L.J., Meadows G.G. Effects of ethanol consumption on enriched natural killer cells from C57BL/6 mice. Alcohol. Clin. Exp. Res. 1994;18:625–631. doi: 10.1111/j.1530-0277.1994.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 131.Spinozzi F., Agea E., Bassotti G., Belia S., Rondoni F., Broccucci L., Solinas A., Gerli R., Bertotto A. Ethanol-specific impairment of T-lymphocyte activation is caused by a transitory block in signal-transduction pathways. Gastroenterology. 1993;105:1490–1501. doi: 10.1016/0016-5085(93)90156-7. [DOI] [PubMed] [Google Scholar]
- 132.Gandhi J.A., Ekhar V.V., Asplund M.B., Abdulkareem A.F., Ahmadi M., Coelho C., Martinez L.R. Alcohol enhances Acinetobacter baumannii-associated pneumonia and systemic dissemination by impairing neutrophil antimicrobial activity in a murine model of infection. PLoS ONE. 2014;9:e95707. doi: 10.1371/journal.pone.0095707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Asplund M.B., Coelho C., Cordero R.J., Martinez L.R. Alcohol impairs J774.16 macrophage-like cell antimicrobial functions in Acinetobacter baumannii infection. Virulence. 2013;4:467–472. doi: 10.4161/viru.25641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Smith M.G., Des Etages S.G., Snyder M. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 2004;24:3874–3884. doi: 10.1128/MCB.24.9.3874-3884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Thomson J.M., Bonomo R.A. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: Beta-lactams in peril! Curr. Opin. Microbiol. 2005;8:518–524. doi: 10.1016/j.mib.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 136.Mohd Sazlly Lim S., Zainal Abidin A., Liew S.M., Roberts J.A., Sime F.B. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: A systematic review and meta-analysis. J. Infect. 2019;79:593–600. doi: 10.1016/j.jinf.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 137.Butler D.A., Biagi M., Tan X., Qasmieh S., Bulman Z.P., Wenzler E. Multidrug resistant Acinetobacter baumannii: Resistance by any other name would still be hard to treat. Curr. Infect. Dis. Rep. 2019;21:46. doi: 10.1007/s11908-019-0706-5. [DOI] [PubMed] [Google Scholar]
- 138.Nowak P., Paluchowska P. Acinetobacter baumannii: Biology and drug resistance - role of carbapenemases. Folia Histochem. Cytobiol. 2016;54:61–74. doi: 10.5603/FHC.a2016.0009. [DOI] [PubMed] [Google Scholar]
- 139.Paterson D.L., Bonomo R.A. Multidrug-resistant gram-negative pathogens: The urgent need for ‘old’ polymyxins. Adv. Exp. Med. Biol. 2019;1145:9–13. doi: 10.1007/978-3-030-16373-0_2. [DOI] [PubMed] [Google Scholar]
- 140.Perez F., Hujer A.M., Hujer K.M., Decker B.K., Rather P.N., Bonomo R.A. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ramirez M.S., Nikolaidis N., Tolmasky M.E. Rise and dissemination of aminoglycoside resistance: The aac(6’)-Ib paradigm. Front. Microbiol. 2013;4:121. doi: 10.3389/fmicb.2013.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ramirez M.S., Tolmasky M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bonomo R.A., Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006;43(Suppl. 2):S49–S56. doi: 10.1086/504477. [DOI] [PubMed] [Google Scholar]
- 144.Manchanda V., Sanchaita S., Singh N. Multidrug resistant Acinetobacter. J. Glob. Infect. Dis. 2010;2:291–304. doi: 10.4103/0974-777X.68538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Perez F., El Chakhtoura N.G., Yasmin M., Bonomo R.A. Polymyxins: To combine or not to combine? Antibiotics (Basel) 2019;8:38. doi: 10.3390/antibiotics8020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shin B., Park W. Antibiotic resistance of pathogenic Acinetobacter species and emerging combination therapy. J. Microbiol. 2017;55:837–849. doi: 10.1007/s12275-017-7288-4. [DOI] [PubMed] [Google Scholar]
- 147.Nasr P. Genetics, epidemiology, and clinical manifestations of multidrug-resistant Acinetobacter baumannii. J. Hosp. Infect. 2020;104:4–11. doi: 10.1016/j.jhin.2019.09.021. [DOI] [PubMed] [Google Scholar]
- 148.Hou P.F., Chen X.Y., Yan G.F., Wang Y.P., Ying C.M. Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy. 2012;58:152–158. doi: 10.1159/000335599. [DOI] [PubMed] [Google Scholar]
- 149.Xu C., Bilya S.R., Xu W. adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 2019;30:100549. doi: 10.1016/j.nmni.2019.100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hamidian M., Nigro S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom. 2019;5:e000306. doi: 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang Y., Li Z., He X., Ding F., Wu W., Luo Y., Fan B., Cao H. Overproduction of efflux pumps caused reduced susceptibility to carbapenem under consecutive imipenem-selected stress in Acinetobacter baumannii. Infect Drug Resist. 2017;11:457–467. doi: 10.2147/IDR.S151423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rumbo C., Gato E., Lopez M., Ruiz de Alegria C., Fernandez-Cuenca F., Martinez-Martinez L., Vila J., Pachon J., Cisneros J.M., Rodriguez-Bano J., et al. Contribution of efflux pumps, porins, and beta-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013;57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Poirel L., Nordmann P. Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin. Microbiol. Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 154.Mussi M.A., Limansky A.S., Viale A.M. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: Natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob. Agents Chemother. 2005;49:1432–1440. doi: 10.1128/AAC.49.4.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.del Mar Tomas M., Beceiro A., Perez A., Velasco D., Moure R., Villanueva R., Martinez-Beltran J., Bou G. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005;49:5172–5175. doi: 10.1128/AAC.49.12.5172-5175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vashist J., Tiwari V., Das R., Kapil A., Rajeswari M.R. Analysis of penicillin-binding proteins (PBPs) in carbapenem resistant Acinetobacter baumannii. Indian J. Med. Res. 2011;133:332–338. [PMC free article] [PubMed] [Google Scholar]
- 157.Fernandez-Cuenca F., Martinez-Martinez L., Conejo M.C., Ayala J.A., Perea E.J., Pascual A. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 2003;51:565–574. doi: 10.1093/jac/dkg097. [DOI] [PubMed] [Google Scholar]
- 158.Bush K. Past and present perspectives on beta-lactamases. Antimicrob. Agents Chemother. 2018;62:e01076-18. doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bonomo R.A. beta-lactamases: A focus on current challenges. Cold Spring Harb. Perspect. Med. 2017;7:a025239. doi: 10.1101/cshperspect.a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bush K., Bradford P.A. Epidemiology of beta-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020;33:e00047-19. doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bush K., Jacoby G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ambler R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 163.Evans B.A., Amyes S.G. OXA beta-lactamases. Clin. Microbiol. Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sykes R.B., Matthew M. The beta-lactamases of gram-negative bacteria and their role in resistance to beta-lactam antibiotics. J. Antimicrob. Chemother. 1976;2:115–157. doi: 10.1093/jac/2.2.115. [DOI] [PubMed] [Google Scholar]
- 165.Medeiros A.A., Cohenford M., Jacoby G.A. Five novel plasmid-determined beta-lactamases. Antimicrob. Agents Chemother. 1985;27:715–719. doi: 10.1128/AAC.27.5.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tolmasky M.E. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid. 1990;24:218–226. doi: 10.1016/0147-619X(90)90005-W. [DOI] [PubMed] [Google Scholar]
- 167.Tolmasky M.E., Crosa J.H. Genetic organization of antibiotic resistance genes (aac(6’)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- 168.Hall L.M., Livermore D.M., Gur D., Akova M., Akalin H.E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1993;37:1637–1644. doi: 10.1128/AAC.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Queenan A.M., Bush K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Donald H.M., Scaife W., Amyes S.G., Young H.K. Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 2000;44:196–199. doi: 10.1128/AAC.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Afzal-Shah M., Woodford N., Livermore D.M. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D beta-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2001;45:583–588. doi: 10.1128/AAC.45.2.583-588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kaitany K.C., Klinger N.V., June C.M., Ramey M.E., Bonomo R.A., Powers R.A., Leonard D.A. Structures of the class D carbapenemases OXA-23 and OXA-146: Mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam. Antimicrob. Agents Chemother. 2013;57:4848–4855. doi: 10.1128/AAC.00762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Smith C.A., Antunes N.T., Stewart N.K., Toth M., Kumarasiri M., Chang M., Mobashery S., Vakulenko S.B. Structural basis for carbapenemase activity of the OXA-23 beta-lactamase from Acinetobacter baumannii. Chem. Biol. 2013;20:1107–1115. doi: 10.1016/j.chembiol.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mugnier P.D., Poirel L., Naas T., Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 2010;16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Poirel L., Figueiredo S., Cattoir V., Carattoli A., Nordmann P. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 2008;52:1252–1256. doi: 10.1128/AAC.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nigro S.J., Hall R.M. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 2016;71:1135–1147. doi: 10.1093/jac/dkv440. [DOI] [PubMed] [Google Scholar]
- 178.Mugnier P.D., Poirel L., Nordmann P. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 2009;191:2414–2418. doi: 10.1128/JB.01258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chen Y., Gao J., Zhang H., Ying C. Spread of the blaOXA-23-containing Tn2008 in carbapenem-resistant Acinetobacter baumannii isolates grouped in CC92 from China. Front. Microbiol. 2017;8:163. doi: 10.3389/fmicb.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hua X., Xu Q., Zhou Z., Ji S., Yu Y. Relocation of Tn2009 and characterization of an ABGRI3-2 from re-sequenced genome sequence of Acinetobacter baumannii MDR-ZJ06. J. Antimicrob. Chemother. 2019;74:1153–1155. doi: 10.1093/jac/dky521. [DOI] [PubMed] [Google Scholar]
- 181.Linder P., Lasko P.F., Ashburner M., Leroy P., Nielsen P.J., Nishi K., Schnier J., Slonimski P.P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 182.Bou G., Oliver A., Martinez-Beltran J. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 2000;44:1556–1561. doi: 10.1128/AAC.44.6.1556-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.D’Andrea M.M., Giani T., D’Arezzo S., Capone A., Petrosillo N., Visca P., Luzzaro F., Rossolini G.M. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:3528–3533. doi: 10.1128/AAC.00178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tian G.B., Adams-Haduch J.M., Bogdanovich T., Pasculle A.W., Quinn J.P., Wang H.N., Doi Y. Identification of diverse OXA-40 group carbapenemases, including a novel variant, OXA-160, from Acinetobacter baumannii in Pennsylvania. Antimicrob. Agents Chemother. 2011;55:429–432. doi: 10.1128/AAC.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Merino M., Acosta J., Poza M., Sanz F., Beceiro A., Chaves F., Bou G. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 2010;54:2724–2727. doi: 10.1128/AAC.01674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tran T., Andres P., Petroni A., Soler-Bistue A., Albornoz E., Zorreguieta A., Reyes-Lamothe R., Sherratt D.J., Corso A., Tolmasky M.E. Small plasmids harboring qnrB19: A model for plasmid evolution mediated by site-specific recombination at oriT and Xer sites. Antimicrob. Agents Chemother. 2012;56:1821–1827. doi: 10.1128/AAC.06036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zakharova M.V., Beletskaya I.V., Denjmukhametov M.M., Yurkova T.V., Semenova L.M., Shlyapnikov M.G., Solonin A.S. Characterization of pECL18 and pKPN2: A proposed pathway for the evolution of two plasmids that carry identical genes for a Type II restriction-modification system. Mol Genet. Genomics. 2002;267:171–178. doi: 10.1007/s00438-002-0644-y. [DOI] [PubMed] [Google Scholar]
- 188.Ramirez M.S., Traglia G.M., Lin D.L., Tran T., Tolmasky M.E. Plasmid-mediated antibiotic resistance and virulence in Gram-negatives: The Klebsiella pneumoniae paradigm. Microbiology Spectr. 2014;2:1–15. doi: 10.1128/microbiolspec.PLAS-0016-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Santillana E., Beceiro A., Bou G., Romero A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. USA. 2007;104:5354–5359. doi: 10.1073/pnas.0607557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schneider K.D., Ortega C.J., Renck N.A., Bonomo R.A., Powers R.A., Leonard D.A. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J. Mol. Biol. 2011;406:583–594. doi: 10.1016/j.jmb.2010.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bou G., Santillana E., Sheri A., Beceiro A., Sampson J.M., Kalp M., Bethel C.R., Distler A.M., Drawz S.M., Pagadala S.R., et al. Design, synthesis, and crystal structures of 6-alkylidene-2’-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase. J. Am. Chem. Soc. 2010;132:13320–13331. doi: 10.1021/ja104092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Che T., Bonomo R.A., Shanmugam S., Bethel C.R., Pusztai-Carey M., Buynak J.D., Carey P.R. Carboxylation and decarboxylation of active site Lys 84 controls the activity of OXA-24 beta-lactamase of Acinetobacter baumannii: Raman crystallographic and solution evidence. J. Am. Chem. Soc. 2012;134:11206–11215. doi: 10.1021/ja303168n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Staude M.W., Leonard D.A., Peng J.W. Expanded substrate activity of OXA-24/40 in carbapenem-resistant Acinetobacter baumannii involves enhanced binding loop flexibility. Biochemistry. 2016;55:6535–6544. doi: 10.1021/acs.biochem.6b00806. [DOI] [PubMed] [Google Scholar]
- 194.Brown S., Young H.K., Amyes S.G. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 2005;11:15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- 195.Merkier A.K., Centron D. bla(OXA-51)-type beta-lactamase genes are ubiquitous and vary within a strain in Acinetobacter baumannii. Int. J. Antimicrob. Agents. 2006;28:110–113. doi: 10.1016/j.ijantimicag.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 196.Turton J.F., Woodford N., Glover J., Yarde S., Kaufmann M.E., Pitt T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006;44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Vijayakumar S., Biswas I., Veeraraghavan B. Accurate identification of clinically important Acinetobacter spp.: An update. Future Sci. OA. 2019;5:FSO395. doi: 10.2144/fsoa-2018-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen T.L., Lee Y.T., Kuo S.C., Hsueh P.R., Chang F.Y., Siu L.K., Ko W.C., Fung C.P. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents Chemother. 2010;54:4575–4581. doi: 10.1128/AAC.00764-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lee Y.T., Kuo S.C., Chiang M.C., Yang S.P., Chen C.P., Chen T.L., Fung C.P. Emergence of carbapenem-resistant non-baumannii species of Acinetobacter harboring a blaOXA-51-like gene that is intrinsic to A. baumannii. Antimicrob. Agents Chemother. 2012;56:1124–1127. doi: 10.1128/AAC.00622-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ahmadi A., Salimizand H. Delayed identification of Acinetobacter baumannii during an outbreak owing to disrupted blaOXA-51-like by ISAba19. Int. J. Antimicrob. Agents. 2017;50:119–122. doi: 10.1016/j.ijantimicag.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 201.Tiwari V., Moganty R.R. Conformational stability of OXA-51 beta-lactamase explains its role in carbapenem resistance of Acinetobacter baumannii. J. Biomol. Struct. Dyn. 2014;32:1406–1420. doi: 10.1080/07391102.2013.819789. [DOI] [PubMed] [Google Scholar]
- 202.Wong M.H., Chan B.K., Chan E.W., Chen S. Over-expression of ISAba1-linked intrinsic and exogenously acquired OXA type carbapenem-hydrolyzing-class D-beta-lactamase-encoding genes is key mechanism underlying carbapenem resistance in Acinetobacter baumannii. Front. Microbiol. 2019;10:2809. doi: 10.3389/fmicb.2019.02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Nigro S.J., Hall R.M. Does the intrinsic oxaAb (blaOXA-51-like) gene of Acinetobacter baumannii confer resistance to carbapenems when activated by ISAba1? J. Antimicrob. Chemother. 2018;73:3518–3520. doi: 10.1093/jac/dky334. [DOI] [PubMed] [Google Scholar]
- 204.Figueiredo S., Poirel L., Croize J., Recule C., Nordmann P. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural bla(OXA-66) oxacillinase gene. Antimicrob. Agents Chemother. 2009;53:2657–2659. doi: 10.1128/AAC.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Turton J.F., Ward M.E., Woodford N., Kaufmann M.E., Pike R., Livermore D.M., Pitt T.L. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006;258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 206.Hu W.S., Yao S.M., Fung C.P., Hsieh Y.P., Liu C.P., Lin J.F. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3844–3852. doi: 10.1128/AAC.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.June C.M., Muckenthaler T.J., Schroder E.C., Klamer Z.L., Wawrzak Z., Powers R.A., Szarecka A., Leonard D.A. The structure of a doripenem-bound OXA-51 class D beta-lactamase variant with enhanced carbapenemase activity. Protein Sci. 2016;25:2152–2163. doi: 10.1002/pro.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mitchell J.M., Leonard D.A. Common clinical substitutions enhance the carbapenemase activity of OXA-51-like class D beta-lactamases from Acinetobacter spp. Antimicrob. Agents Chemother. 2014;58:7015–7016. doi: 10.1128/AAC.03651-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Schroder E.C., Klamer Z.L., Saral A., Sugg K.A., June C.M., Wymore T., Szarecka A., Leonard D.A. Clinical variants of the native class D beta-lactamase of Acinetobacter baumannii pose an emerging threat through Increased hydrolytic activity against carbapenems. Antimicrob. Agents Chemother. 2016;60:6155–6164. doi: 10.1128/AAC.01277-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li H., Liu F., Zhang Y., Wang X., Zhao C., Chen H., Zhang F., Zhu B., Hu Y., Wang H. Evolution of carbapenem-resistant Acinetobacter baumannii revealed through whole-genome sequencing and comparative genomic analysis. Antimicrob. Agents Chemother. 2015;59:1168–1176. doi: 10.1128/AAC.04609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Evans B.A., Brown S., Hamouda A., Findlay J., Amyes S.G. Eleven novel OXA-51-like enzymes from clinical isolates of Acinetobacter baumannii. Clin. Microbiol. Infect. 2007;13:1137–1138. doi: 10.1111/j.1469-0691.2007.01828.x. [DOI] [PubMed] [Google Scholar]
- 212.Tsakris A., Ikonomidis A., Spanakis N., Pournaras S., Bethimouti K. Identification of a novel bla(OXA-51) variant, bla(OXA-92), from a clinical isolate of Acinetobacter baumannii. Clin. Microbiol. Infect. 2007;13:348–349. doi: 10.1111/j.1469-0691.2006.01598.x. [DOI] [PubMed] [Google Scholar]
- 213.Smith C.A., Antunes N.T., Stewart N.K., Frase H., Toth M., Kantardjieff K.A., Vakulenko S. Structural basis for enhancement of carbapenemase activity in the OXA-51 family of class D beta-lactamases. ACS Chem. Biol. 2015;10:1791–1796. doi: 10.1021/acschembio.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Poirel L., Marque S., Heritier C., Segonds C., Chabanon G., Nordmann P. OXA-58, a novel class D {beta}-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005;49:202–208. doi: 10.1128/AAC.49.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Heritier C., Dubouix A., Poirel L., Marty N., Nordmann P. A nosocomial outbreak of Acinetobacter baumannii isolates expressing the carbapenem-hydrolysing oxacillinase OXA-58. J. Antimicrob. Chemother. 2005;55:115–118. doi: 10.1093/jac/dkh500. [DOI] [PubMed] [Google Scholar]
- 216.Marque S., Poirel L., Heritier C., Brisse S., Blasco M.D., Filip R., Coman G., Naas T., Nordmann P. Regional occurrence of plasmid-mediated carbapenem-hydrolyzing oxacillinase OXA-58 in Acinetobacter spp. in Europe. J. Clin. Microbiol. 2005;43:4885–4888. doi: 10.1128/JCM.43.9.4885-4888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Matos A.P., Cayo R., Almeida L.G.P., Streling A.P., Nodari C.S., Martins W., Narciso A.C., Silva R.M., Vasconcelos A.T.R., Gales A.C. Genetic characterization of plasmid-borne blaOXA-58 in distinct Acinetobacter species. mSphere. 2019;4:e00376-19. doi: 10.1128/mSphere.00376-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bertini A., Poirel L., Bernabeu S., Fortini D., Villa L., Nordmann P., Carattoli A. Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:2324–2328. doi: 10.1128/AAC.01502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Reams A.B., Roth J.R. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 2015;7:a016592. doi: 10.1101/cshperspect.a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Peterson B.C., Rownd R.H. Drug resistance gene amplification of plasmid NR1 derivatives with various amounts of resistance determinant DNA. J. Bacteriol. 1985;161:1042–1048. doi: 10.1128/JB.161.3.1042-1048.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tolmasky M.E., Chamorro R.M., Crosa J.H., Marini P.M. Transposon-mediated amikacin resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 1988;32:1416–1420. doi: 10.1128/AAC.32.9.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Peterson B.C., Rownd R.H. Recombination sites in plasmid drug resistance gene amplification. J. Bacteriol. 1985;164:1359–1361. doi: 10.1128/JB.164.3.1359-1361.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Schechter L.M., Creely D.P., Garner C.D., Shortridge D., Nguyen H., Chen L., Hanson B.M., Sodergren E., Weinstock G.M., Dunne W.M., Jr., et al. Extensive gene amplification as a mechanism for piperacillin-tazobactam resistance in Escherichia coli. mBio. 2018;9:e00583-18. doi: 10.1128/mBio.00583-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Higgins P.G., Poirel L., Lehmann M., Nordmann P., Seifert H. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:5035–5038. doi: 10.1128/AAC.00856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Levy-Blitchtein S., Roca I., Plasencia-Rebata S., Vicente-Taboada W., Velasquez-Pomar J., Munoz L., Moreno-Morales J., Pons M.J., Del Valle-Mendoza J., Vila J. Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru. Emerg. Microbes Infect. 2018;7:119. doi: 10.1038/s41426-018-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gionco B., Pelayo J.S., Venancio E.J., Cayo R., Gales A.C., Carrara-Marroni F.E. Detection of OXA-231, a new variant of blaOXA-143, in Acinetobacter baumannii from Brazil: A case report. J. Antimicrob. Chemother. 2012;67:2531–2532. doi: 10.1093/jac/dks223. [DOI] [PubMed] [Google Scholar]
- 227.Kim C.K., Lee Y., Lee H., Woo G.J., Song W., Kim M.N., Lee W.G., Jeong S.H., Lee K., Chong Y. Prevalence and diversity of carbapenemases among imipenem-nonsusceptible Acinetobacter isolates in Korea: Emergence of a novel OXA-182. Diagn. Microbiol. Infect. Dis. 2010;68:432–438. doi: 10.1016/j.diagmicrobio.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 228.Mostachio A.K., Levin A.S., Rizek C., Rossi F., Zerbini J., Costa S.F. High prevalence of OXA-143 and alteration of outer membrane proteins in carbapenem-resistant Acinetobacter spp. isolates in Brazil. Int. J. Antimicrob. Agents. 2012;39:396–401. doi: 10.1016/j.ijantimicag.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 229.Zander E., Bonnin R.A., Seifert H., Higgins P.G. Characterization of blaOXA-143 variants in Acinetobacter baumannii and Acinetobacter pittii. Antimicrob. Agents Chemother. 2014;58:2704–2708. doi: 10.1128/AAC.02618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Girlich D., Damaceno Q.S., Oliveira A.C., Nordmann P. OXA-253, a variant of the carbapenem-hydrolyzing class D beta-lactamase OXA-143 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014;58:2976–2978. doi: 10.1128/AAC.02640-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Higgins P.G., Perez-Llarena F.J., Zander E., Fernandez A., Bou G., Seifert H. OXA-235, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013;57:2121–2126. doi: 10.1128/AAC.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Boyd D.A., Mataseje L.F., Pelude L., Mitchell R., Bryce E., Roscoe D., Embree J., Katz K., Kibsey P., Lavallee C., et al. Results from the Canadian Nosocomial Infection Surveillance Program for detection of carbapenemase-producing Acinetobacter spp. in Canadian hospitals, 2010–2016. J. Antimicrob. Chemother. 2019;74:315–320. doi: 10.1093/jac/dky416. [DOI] [PubMed] [Google Scholar]
- 233.Buser G.L., Cassidy P.M., Cunningham M.C., Rudin S., Hujer A.M., Vega R., Furuno J.P., Marshall S.H., Higgins P.G., Jacobs M.R., et al. Failure to communicate: Transmission of extensively drug-resistant blaOXA-237-containing Acinetobacter baumannii - multiple facilities in Oregon, 2012-2014. Infect. Control. Hosp. Epidemiol. 2017;38:1335–1341. doi: 10.1017/ice.2017.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hujer A.M., Higgins P.G., Rudin S.D., Buser G.L., Marshall S.H., Xanthopoulou K., Seifert H., Rojas L.J., Domitrovic T.N., Cassidy P.M., et al. Nosocomial outbreak of extensively drug-resistant Acinetobacter baumannii isolates containing blaOXA-237 carried on a plasmid. Antimicrob. Agents Chemother. 2017;61:e00797-17. doi: 10.1128/AAC.00797-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Palzkill T. Metallo-beta-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mojica M.F., Bonomo R.A., Fast W. B1-Metallo-beta-Lactamases: Where Do We Stand? Curr. Drug Targets. 2016;17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Walsh T.R., Toleman M.A., Poirel L., Nordmann P. Metallo-beta-lactamases: The quiet before the storm? Clin. Microbiol. Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lomovskaya O., Nelson K., Rubio-Aparicio D., Tsivkovski R., Sun D., Dudley M.N. The impact of intrinsic resistance mechanisms on potency of QPX7728, a new ultra-broad-spectrum beta-lactamase inhibitor of serine and metallo beta-lactamases in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Antimicrob. Agents Chemother. 2020:AAC.00552-20. doi: 10.1128/AAC.00552-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hamrick J.C., Docquier J.D., Uehara T., Myers C.L., Six D.A., Chatwin C.L., John K.J., Vernacchio S.F., Cusick S.M., Trout R.E.L., et al. VNRX-5133 (Taniborbactam), a broad-spectrum inhibitor of serine- and metallo-beta-lactamases, restores activity of cefepime in Enterobacterales and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2020;64:e01963-19. doi: 10.1128/AAC.01963-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Krajnc A., Brem J., Hinchliffe P., Calvopina K., Panduwawala T.D., Lang P.A., Kamps J., Tyrrell J.M., Widlake E., Saward B.G., et al. Bicyclic boronate VNRX-5133 inhibits metallo- and serine-beta-lactamases. J. Med. Chem. 2019;62:8544–8556. doi: 10.1021/acs.jmedchem.9b00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yong D., Toleman M.A., Giske C.G., Cho H.S., Sundman K., Lee K., Walsh T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Khan A.U., Maryam L., Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-beta-lactamase (NDM): A threat to public health. BMC Microbiol. 2017;17:101. doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wu W., Feng Y., Tang G., Qiao F., McNally A., Zong Z. NDM metallo-beta-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019;32:e00115–e00118. doi: 10.1128/CMR.00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Karthikeyan K., Thirunarayan M.A., Krishnan P. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 2010;65:2253–2254. doi: 10.1093/jac/dkq273. [DOI] [PubMed] [Google Scholar]
- 245.Adams M.D., Pasteran F., Traglia G.M., Martinez J., Huang F., Liu C., Fernandez J.S., Lopez C., Gonzalez L.J., Albornoz E., et al. Distinct mechanisms of dissemination of NDM-1 metallo-beta-lactamase in Acinetobacter species in Argentina. Antimicrob. Agents Chemother. 2020;64:e00324-20. doi: 10.1128/AAC.00324-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Dortet L., Poirel L., Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed. Res. Int. 2014;2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Groundwater P.W., Xu S., Lai F., Varadi L., Tan J., Perry J.D., Hibbs D.E. New Delhi metallo-beta-lactamase-1: Structure, inhibitors and detection of producers. Future Med. Chem. 2016;8:993–1012. doi: 10.4155/fmc-2016-0015. [DOI] [PubMed] [Google Scholar]
- 248.Khalid S., Ahmad N., Ali S.M., Khan A.U. Outbreak of efficiently transferred carbapenem-resistant blaNDM-producing gram-negative bacilli isolated from neonatal intensive care unit of an indian hospital. Microb Drug Resist. 2020;26:284–289. doi: 10.1089/mdr.2019.0092. [DOI] [PubMed] [Google Scholar]
- 249.Singh A., Singh S., Singh K., Pathak A., Prasad K.N. Acinetobacter baumannii strain ASKPNAB1 Subclass B1 Metallo-Beta-Lactamase NDM-5 (blaNDM) Gene, blaNDM-5 Allele, Complete Cds. [(accessed on 30 April 2020)]; Available online: https://www.ncbi.nlm.nih.gov/nuccore/MK682761.1.
- 250.Rogers B.A., Sidjabat H.E., Silvey A., Anderson T.L., Perera S., Li J., Paterson D.L. Treatment options for New Delhi metallo-beta-lactamase-harboring enterobacteriaceae. Microb Drug Resist. 2013;19:100–103. doi: 10.1089/mdr.2012.0063. [DOI] [PubMed] [Google Scholar]
- 251.Abdul Rahim N., Cheah S.E., Johnson M.D., Yu H., Sidjabat H.E., Boyce J., Butler M.S., Cooper M.A., Fu J., Paterson D.L., et al. Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two ’old’ antibiotics-polymyxin B and chloramphenicol. J. Antimicrob. Chemother. 2015;70:2589–2597. doi: 10.1093/jac/dkv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Linciano P., Cendron L., Gianquinto E., Spyrakis F., Tondi D. Ten years with New Delhi metallo-beta-lactamase-1 (NDM-1): From structural insights to inhibitor design. ACS Infect. Dis. 2019;5:9–34. doi: 10.1021/acsinfecdis.8b00247. [DOI] [PubMed] [Google Scholar]
- 253.Poirel L., Bonnin R.A., Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob. Agents Chemother. 2011;55:4224–4229. doi: 10.1128/AAC.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Huang T.W., Wang J.T., Lauderdale T.L., Liao T.L., Lai J.F., Tan M.C., Lin A.C., Chen Y.T., Tsai S.F., Chang S.C. Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan. Antimicrob. Agents Chemother. 2013;57:4072–4076. doi: 10.1128/AAC.02266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wailan A.M., Paterson D.L., Kennedy K., Ingram P.R., Bursle E., Sidjabat H.E. Genomic characteristics of NDM-producing Enterobacteriaceae isolates in Australia and their blaNDM genetic contexts. Antimicrob. Agents Chemother. 2016;60:136–141. doi: 10.1128/AAC.01243-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Toleman M.A., Spencer J., Jones L., Walsh T.R. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2012;56:2773–2776. doi: 10.1128/AAC.06297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Poirel L., Bonnin R.A., Boulanger A., Schrenzel J., Kaase M., Nordmann P. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2012;56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bontron S., Nordmann P., Poirel L. Transposition of Tn125 encoding the NDM-1 carbapenemase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016;60:7245–7251. doi: 10.1128/AAC.01755-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Jones L.S., Toleman M.A., Weeks J.L., Howe R.A., Walsh T.R., Kumarasamy K.K. Plasmid carriage of blaNDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob. Agents Chemother. 2014;58:4211–4213. doi: 10.1128/AAC.02500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.McGann P., Hang J., Clifford R.J., Yang Y., Kwak Y.I., Kuschner R.A., Lesho E.P., Waterman P.E. Complete sequence of a novel 178-kilobase plasmid carrying bla(NDM-1) in a Providencia stuartii strain isolated in Afghanistan. Antimicrob. Agents Chemother. 2012;56:1673–1679. doi: 10.1128/AAC.05604-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Sekizuka T., Matsui M., Yamane K., Takeuchi F., Ohnishi M., Hishinuma A., Arakawa Y., Kuroda M. Complete sequencing of the bla(NDM-1)-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS ONE. 2011;6:e25334. doi: 10.1371/journal.pone.0025334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wailan A.M., Sartor A.L., Zowawi H.M., Perry J.D., Paterson D.L., Sidjabat H.E. Genetic contexts of blaNDM-1 in patients carrying multiple NDM-producing Strains. Antimicrob. Agents Chemother. 2015;59:7405–7410. doi: 10.1128/AAC.01319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hudson C.M., Bent Z.W., Meagher R.J., Williams K.P. Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS ONE. 2014;9:e99209. doi: 10.1371/journal.pone.0099209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Partridge S.R., Iredell J.R. Genetic contexts of blaNDM-1. Antimicrob. Agents Chemother. 2012;56:6065–6067. doi: 10.1128/AAC.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Martin P., Jullien E., Courvalin P. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: Evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol. Microbiol. 1988;2:615–625. doi: 10.1111/j.1365-2958.1988.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 266.Zhang H., Hao Q. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J. 2011;25:2574–2582. doi: 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
- 267.King D., Strynadka N. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011;20:1484–1491. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kim Y., Tesar C., Mire J., Jedrzejczak R., Binkowski A., Babnigg G., Sacchettini J., Joachimiak A. Structure of apo- and monometalated forms of NDM-1--a highly potent carbapenem-hydrolyzing metallo-beta-lactamase. PLoS ONE. 2011;6:e24621. doi: 10.1371/journal.pone.0024621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lisa M.N., Palacios A.R., Aitha M., Gonzalez M.M., Moreno D.M., Crowder M.W., Bonomo R.A., Spencer J., Tierney D.L., Llarrull L.I., et al. A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-beta-lactamases. Nat. Commun. 2017;8:538. doi: 10.1038/s41467-017-00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zheng M., Xu D. New Delhi metallo-beta-lactamase I: Substrate binding and catalytic mechanism. J. Phys. Chem. B. 2013;117:11596–11607. doi: 10.1021/jp4065906. [DOI] [PubMed] [Google Scholar]
- 271.Kim Y., Cunningham M.A., Mire J., Tesar C., Sacchettini J., Joachimiak A. NDM-1, the ultimate promiscuous enzyme: Substrate recognition and catalytic mechanism. FASEB J. 2013;27:1917–1927. doi: 10.1096/fj.12-224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kozlyuk N., Monteith A.J., Garcia V., Damo S.M., Skaar E.P., Chazin W.J. S100 Proteins in the Innate Immune Response to Pathogens. Methods Mol. Biol. 2019;1929:275–290. doi: 10.1007/978-1-4939-9030-6_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Diaz-Ochoa V.E., Jellbauer S., Klaus S., Raffatellu M. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front. Cell Infect. Microbiol. 2014;4:2. doi: 10.3389/fcimb.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Corbin B.D., Seeley E.H., Raab A., Feldmann J., Miller M.R., Torres V.J., Anderson K.L., Dattilo B.M., Dunman P.M., Gerads R., et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 275.Gonzalez J.M., Meini M.R., Tomatis P.E., Medrano Martin F.J., Cricco J.A., Vila A.J. Metallo-beta-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat. Chem. Biol. 2012;8:698–700. doi: 10.1038/nchembio.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Gonzalez L.J., Bahr G., Nakashige T.G., Nolan E.M., Bonomo R.A., Vila A.J. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat. Chem. Biol. 2016;12:516–522. doi: 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lopez C., Ayala J.A., Bonomo R.A., Gonzalez L.J., Vila A.J. Protein determinants of dissemination and host specificity of metallo-beta-lactamases. Nat. Commun. 2019;10:3617. doi: 10.1038/s41467-019-11615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lauretti L., Riccio M.L., Mazzariol A., Cornaglia G., Amicosante G., Fontana R., Rossolini G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999;43:1584–1590. doi: 10.1128/AAC.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Poirel L., Naas T., Nicolas D., Collet L., Bellais S., Cavallo J.D., Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 2000;44:891–897. doi: 10.1128/AAC.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Kazmierczak K.M., Rabine S., Hackel M., McLaughlin R.E., Biedenbach D.J., Bouchillon S.K., Sahm D.F., Bradford P.A. Multiyear, multinational survey of the incidence and global distribution of metallo-beta-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016;60:1067–1078. doi: 10.1128/AAC.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Matsumura Y., Peirano G., Devinney R., Bradford P.A., Motyl M.R., Adams M.D., Chen L., Kreiswirth B., Pitout J.D.D. Genomic epidemiology of global VIM-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2017;72:2249–2258. doi: 10.1093/jac/dkx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Lee K., Lee W.G., Uh Y., Ha G.Y., Cho J., Chong Y., Korean Nationwide Surveillance of Antimicrobial Resistance Group VIM- and IMP-type metallo-beta-lactamase-producing Pseudomonas spp. and Acinetobacter spp. in Korean hospitals. Emerg. Infect. Dis. 2003;9:868–871. doi: 10.3201/eid0907.030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yum J.H., Yi K., Lee H., Yong D., Lee K., Kim J.M., Rossolini G.M., Chong Y. Molecular characterization of metallo-beta-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: Identification of two new integrons carrying the bla(VIM-2) gene cassettes. J. Antimicrob. Chemother. 2002;49:837–840. doi: 10.1093/jac/dkf043. [DOI] [PubMed] [Google Scholar]
- 284.Tsakris A., Ikonomidis A., Pournaras S., Tzouvelekis L.S., Sofianou D., Legakis N.J., Maniatis A.N. VIM-1 metallo-beta-lactamase in Acinetobacter baumannii. Emerg. Infect. Dis. 2006;12:981–983. doi: 10.3201/eid1206.051097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.El-Ageery S.M., Al-Hazmi S.S. Microbiological and molecular detection of VIM-1 metallo beta lactamase-producing Acinetobacter baumannii. Eur. Rev. Med. Pharmacol. Sci. 2014;18:965–970. [PubMed] [Google Scholar]
- 286.Beigverdi R., Sattari-Maraji A., Emaneini M., Jabalameli F. Status of carbapenem-resistant Acinetobacter baumannii harboring carbapenemase: First systematic review and meta-analysis from Iran. Infect. Genet. Evol. 2019;73:433–443. doi: 10.1016/j.meegid.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 287.Davoodi S., Boroumand M.A., Sepehriseresht S., Pourgholi L. Detection of VIM- and IMP-type metallo-beta-lactamase genes in Acinetobacter baumannii isolates from patients in two hospitals in Tehran. Iran. J. Biotechnol. 2015;13:63–67. doi: 10.15171/ijb.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Rezaei A., Fazeli H., Halaji M., Moghadampour M., Faghri J. Prevalence of metallo-beta-lactamase producing Acinetobacter baumannii isolated from intensive care unit in tertiary care hospitals. Ann. Ig. 2018;30:330–336. doi: 10.7416/ai.2018.2224. [DOI] [PubMed] [Google Scholar]
- 289.Lee M.F., Peng C.F., Hsu H.J., Chen Y.H. Molecular characterisation of the metallo-beta-lactamase genes in imipenem-resistant Gram-negative bacteria from a university hospital in southern Taiwan. Int. J. Antimicrob. Agents. 2008;32:475–480. doi: 10.1016/j.ijantimicag.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 290.Peymani A., Nahaei M.R., Farajnia S., Hasani A., Mirsalehian A., Sohrabi N., Abbasi L. High prevalence of metallo-beta-lactamase-producing Acinetobacter baumannii in a teaching hospital in Tabriz, Iran. Jpn. J. Infect. Dis. 2011;64:69–71. [PubMed] [Google Scholar]
- 291.El-Badawy M.F., Abdelwahab S.F., Alghamdi S.A., Shohayeb M.M. Characterization of phenotypic and genotypic traits of carbapenem-resistant Acinetobacter baumannii clinical isolates recovered from a tertiary care hospital in Taif, Saudi Arabia. Infect. Drug Resist. 2019;12:3113–3124. doi: 10.2147/IDR.S206691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Benmahmod A.B., Said H.S., Ibrahim R.H. Prevalence and mechanisms of carbapenem resistance among Acinetobacter baumannii clinical isolates in Egypt. Microb. Drug Resist. 2019;25:480–488. doi: 10.1089/mdr.2018.0141. [DOI] [PubMed] [Google Scholar]
- 293.Azimi L., Talebi M., Pourshafie M.R., Owlia P., Rastegar Lari A. Characterization of carbapenemases in extensively drug resistant Acinetobacter baumannii in a burn care center in Iran. Int. J. Mol Cell Med. 2015;4:46–53. [PMC free article] [PubMed] [Google Scholar]
- 294.AlAmri A.M., AlQurayan A.M., Sebastian T., AlNimr A.M. Molecular surveillance of multidrug-resistant Acinetobacter baumannii. Curr. Microbiol. 2020;77:335–342. doi: 10.1007/s00284-019-01836-z. [DOI] [PubMed] [Google Scholar]
- 295.Girija S.A., Jayaseelan V.P., Arumugam P. Prevalence of VIM- and GIM-producing Acinetobacter baumannii from patients with severe urinary tract infection. Acta Microbiol. Immunol. Hung. 2018;65:539–550. doi: 10.1556/030.65.2018.038. [DOI] [PubMed] [Google Scholar]
- 296.Amin M., Navidifar T., Saleh Shooshtari F., Goodarzi H. Association of the genes encoding metallo-beta-lactamase with the presence of integrons among multidrug-resistant clinical isolates of Acinetobacter baumannii. Infect. Drug Resist. 2019;12:1171–1180. doi: 10.2147/IDR.S196575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Watanabe M., Iyobe S., Inoue M., Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1991;35:147–151. doi: 10.1128/AAC.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Osano E., Arakawa Y., Wacharotayankun R., Ohta M., Horii T., Ito H., Yoshimura F., Kato N. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 1994;38:71–78. doi: 10.1128/AAC.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Arakawa Y., Murakami M., Suzuki K., Ito H., Wacharotayankun R., Ohsuka S., Kato N., Ohta M. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 1995;39:1612–1615. doi: 10.1128/AAC.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Ito H., Arakawa Y., Ohsuka S., Wacharotayankun R., Kato N., Ohta M. Plasmid-mediated dissemination of the metallo-beta-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob. Agents Chemother. 1995;39:824–829. doi: 10.1128/AAC.39.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Hirakata Y., Izumikawa K., Yamaguchi T., Takemura H., Tanaka H., Yoshida R., Matsuda J., Nakano M., Tomono K., Maesaki S., et al. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 1998;42:2006–2011. doi: 10.1128/AAC.42.8.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Koh T.H., Babini G.S., Woodford N., Sng L.H., Hall L.M., Livermore D.M. Carbapenem-hydrolysing IMP-1 beta-lactamase in Klebsiella pneumoniae from Singapore. Lancet. 1999;353:2162. doi: 10.1016/S0140-6736(05)75604-X. [DOI] [PubMed] [Google Scholar]
- 303.Senda K., Arakawa Y., Nakashima K., Ito H., Ichiyama S., Shimokata K., Kato N., Ohta M. Multifocal outbreaks of metallo-beta-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum beta-lactams, including carbapenems. Antimicrob. Agents Chemother. 1996;40:349–353. doi: 10.1128/AAC.40.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Riccio M.L., Franceschini N., Boschi L., Caravelli B., Cornaglia G., Fontana R., Amicosante G., Rossolini G.M. Characterization of the metallo-beta-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 2000;44:1229–1235. doi: 10.1128/AAC.44.5.1229-1235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Cornaglia G., Riccio M.L., Mazzariol A., Lauretti L., Fontana R., Rossolini G.M. Appearance of IMP-1 metallo-beta-lactamase in Europe. Lancet. 1999;353:899–900. doi: 10.1016/S0140-6736(98)05954-6. [DOI] [PubMed] [Google Scholar]
- 306.Ramirez M.S., Parenteau T.R., Centron D., Tolmasky M.E. Functional characterization of Tn1331 gene cassettes. J. Antimicrob. Chemother. 2008;62:669–673. doi: 10.1093/jac/dkn279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Sarno R., McGillivary G., Sherratt D.J., Actis L.A., Tolmasky M.E. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 2002;46:3422–3427. doi: 10.1128/AAC.46.11.3422-3427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Tolmasky M.E., Crosa J.H. Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 1987;31:1955–1960. doi: 10.1128/AAC.31.12.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Casin I., Hanau-Bercot B., Podglajen I., Vahaboglu H., Collatz E. Salmonella enterica serovar Typhimurium bla(PER-1)-carrying plasmid pSTI1 encodes an extended-spectrum aminoglycoside 6’-N-acetyltransferase of type Ib. Antimicrob. Agents Chemother. 2003;47:697–703. doi: 10.1128/AAC.47.2.697-703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Dery K.J., Soballe B., Witherspoon M.S., Bui D., Koch R., Sherratt D.J., Tolmasky M.E. The aminoglycoside 6’-N-acetyltransferase type Ib encoded by Tn1331 is evenly distributed within the cell’s cytoplasm. Antimicrob. Agents Chemother. 2003;47:2897–2902. doi: 10.1128/AAC.47.9.2897-2902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Nobuta K., Tolmasky M.E., Crosa L.M., Crosa J.H. Sequencing and expression of the 6’-N-acetyltransferase gene of transposon Tn1331 from Klebsiella pneumoniae. J. Bacteriol. 1988;170:3769–3773. doi: 10.1128/JB.170.8.3769-3773.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Da Silva G.J., Correia M., Vital C., Ribeiro G., Sousa J.C., Leitao R., Peixe L., Duarte A. Molecular characterization of bla(IMP-5), a new integron-borne metallo-beta-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol. Lett. 2002;215:33–39. doi: 10.1111/j.1574-6968.2002.tb11366.x. [DOI] [PubMed] [Google Scholar]
- 313.Iyobe S., Kusadokoro H., Ozaki J., Matsumura N., Minami S., Haruta S., Sawai T., O’Hara K. Amino acid substitutions in a variant of IMP-1 metallo-beta-lactamase. Antimicrob. Agents Chemother. 2000;44:2023–2027. doi: 10.1128/AAC.44.8.2023-2027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Chu Y.W., Afzal-Shah M., Houang E.T., Palepou M.I., Lyon D.J., Woodford N., Livermore D.M. IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 2001;45:710–714. doi: 10.1128/AAC.45.3.710-714.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Hawkey P.M., Xiong J., Ye H., Li H., M’Zali F.H. Occurrence of a new metallo-beta-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People’s Republic of China. FEMS Microbiol. Lett. 2001;194:53–57. doi: 10.1111/j.1574-6968.2001.tb09445.x. [DOI] [PubMed] [Google Scholar]
- 316.Koh T.H., Sng L.H., Wang G.C., Hsu L.Y., Zhao Y. IMP-4 and OXA beta-lactamases in Acinetobacter baumannii from Singapore. J. Antimicrob. Chemother. 2007;59:627–632. doi: 10.1093/jac/dkl544. [DOI] [PubMed] [Google Scholar]
- 317.Partridge S.R., Ginn A.N., Paulsen I.T., Iredell J.R. pEl1573 Carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob. Agents Chemother. 2012;56:6029–6032. doi: 10.1128/AAC.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Sidjabat H.E., Heney C., George N.M., Nimmo G.R., Paterson D.L. Interspecies transfer of blaIMP-4 in a patient with prolonged colonization by IMP-4-producing Enterobacteriaceae. J. Clin. Microbiol. 2014;52:3816–3818. doi: 10.1128/JCM.01491-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Peleg A.Y., Franklin C., Walters L.J., Bell J.M., Spelman D.W. OXA-58 and IMP-4 carbapenem-hydrolyzing beta-lactamases in an Acinetobacter junii blood culture isolate from Australia. Antimicrob. Agents Chemother. 2006;50:399–400. doi: 10.1128/AAC.50.1.399-400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Azizi O., Shakibaie M.R., Badmasti F., Modarresi F., Ramazanzadeh R., Mansouri S., Shahcheraghi F. Class 1 integrons in non-clonal multidrug-resistant Acinetobacter baumannii from Iran, description of the new blaIMP-55 allele in In1243. J. Med. Microbiol. 2016;65:928–936. doi: 10.1099/jmm.0.000315. [DOI] [PubMed] [Google Scholar]
- 321.Yigit H., Queenan A.M., Anderson G.J., Domenech-Sanchez A., Biddle J.W., Steward C.D., Alberti S., Bush K., Tenover F.C. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Miriagou V., Tzouvelekis L.S., Rossiter S., Tzelepi E., Angulo F.J., Whichard J.M. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 2003;47:1297–1300. doi: 10.1128/AAC.47.4.1297-1300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Smith Moland E., Hanson N.D., Herrera V.L., Black J.A., Lockhart T.J., Hossain A., Johnson J.A., Goering R.V., Thomson K.S. Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 2003;51:711–714. doi: 10.1093/jac/dkg124. [DOI] [PubMed] [Google Scholar]
- 324.Woodford N., Tierno P.M., Jr., Young K., Tysall L., Palepou M.F., Ward E., Painter R.E., Suber D.F., Shungu D., Silver L.L., et al. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 2004;48:4793–4799. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Abdallah M., Olafisoye O., Cortes C., Urban C., Landman D., Ghitan M., Collins B., Bratu S., Quale J. Rise and fall of KPC-producing Klebsiella pneumoniae in New York City. J. Antimicrob. Chemother. 2016;71:2945–2948. doi: 10.1093/jac/dkw242. [DOI] [PubMed] [Google Scholar]
- 326.Kazmierczak K.M., Biedenbach D.J., Hackel M., Rabine S., de Jonge B.L., Bouchillon S.K., Sahm D.F., Bradford P.A. Global dissemination of blaKPC into bacterial species beyond Klebsiella pneumoniae and in vitro susceptibility to ceftazidime-avibactam and aztreonam-avibactam. Antimicrob. Agents Chemother. 2016;60:4490–4500. doi: 10.1128/AAC.00107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Naas T., Nordmann P., Vedel G., Poyart C. Plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 2005;49:4423–4424. doi: 10.1128/AAC.49.10.4423-4424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Munoz-Price L.S., Poirel L., Bonomo R.A., Schwaber M.J., Daikos G.L., Cormican M., Cornaglia G., Garau J., Gniadkowski M., Hayden M.K., et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ramirez M.S., Xie G., Marshall S.H., Hujer K.M., Chain P.S., Bonomo R.A., Tolmasky M.E. Multidrug-resistant (MDR) Klebsiella pneumoniae clinical isolates: A zone of high heterogeneity (HHZ) as a tool for epidemiological studies. Clin. Microbiol. Infect. 2012;18:E254–E258. doi: 10.1111/j.1469-0691.2012.03886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ramirez M.S., Xie G., Traglia G.M., Johnson S.L., Davenport K.W., van Duin D., Ramazani A., Perez F., Jacobs M.R., Sherratt D.J., et al. Whole-genome comparative analysis of two carbapenem-resistant ST-258 Klebsiella pneumoniae strains Isolated during a North-eastern Ohio outbreak: Differences within the high heterogeneity zones. Genome Biol. Evol. 2016;8:2036–2043. doi: 10.1093/gbe/evw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Park S.O., Liu J., Furuya E.Y., Larson E.L. Carbapenem-resistant Klebsiella pneumoniae infection in three New York City hospitals trended downwards from 2006 to 2014. Open Forum Infect. Dis. 2016;3:ofw222. doi: 10.1093/ofid/ofw222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Naas T., Cuzon G., Villegas M.V., Lartigue M.F., Quinn J.P., Nordmann P. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob. Agents Chemother. 2008;52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Leavitt A., Chmelnitsky I., Ofek I., Carmeli Y., Navon-Venezia S. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J. Antimicrob. Chemother. 2010;65:243–248. doi: 10.1093/jac/dkp417. [DOI] [PubMed] [Google Scholar]
- 334.Gootz T.D., Lescoe M.K., Dib-Hajj F., Dougherty B.A., He W., Della-Latta P., Huard R.C. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 2009;53:1998–2004. doi: 10.1128/AAC.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Chen L., Chavda K.D., Al Laham N., Melano R.G., Jacobs M.R., Bonomo R.A., Kreiswirth B.N. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 2013;57:5019–5025. doi: 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ho P.L., Cheung Y.Y., Lo W.U., Li Z., Chow K.H., Lin C.H., Chan J.F., Cheng V.C. Molecular characterization of an atypical IncX3 plasmid pKPC-NY79 carrying blaKPC-2 in a Klebsiella pneumoniae. Curr. Microbiol. 2013;67:493–498. doi: 10.1007/s00284-013-0398-2. [DOI] [PubMed] [Google Scholar]
- 337.Partridge S.R. Tn4401 carrying blaKPC is inserted within another insertion in pKpQIL and related plasmids. J. Clin. Microbiol. 2014;52:4448–4449. doi: 10.1128/JCM.02426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Rice L.B., Carias L.L., Hutton R.A., Rudin S.D., Endimiani A., Bonomo R.A. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2008;52:3427–3429. doi: 10.1128/AAC.00493-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Souza R.C., Dabul A.N.G., Boralli C., Zuvanov L., Camargo I. Dissemination of blaKPC-2 in an NTEKPC by an IncX5 plasmid. Plasmid. 2019;106:102446. doi: 10.1016/j.plasmid.2019.102446. [DOI] [PubMed] [Google Scholar]
- 340.Chen L., Mathema B., Chavda K.D., DeLeo F.R., Bonomo R.A., Kreiswirth B.N. Carbapenemase-producing Klebsiella pneumoniae: Molecular and genetic decoding. Trends Microbiol. 2014;22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Wang L., Fang H., Feng J., Yin Z., Xie X., Zhu X., Wang J., Chen W., Yang R., Du H., et al. Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front. Microbiol. 2015;6:838. doi: 10.3389/fmicb.2015.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Azimi L., Talebi M., Khodaei F., Najafi M., Lari A.R. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis typing of carbapenemases producing Acinetobacter baumannii isolated from burn patients. Burns. 2016;42:441–445. doi: 10.1016/j.burns.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 343.Lima W.G., Silva Alves G.C., Sanches C., Antunes Fernandes S.O., de Paiva M.C. Carbapenem-resistant Acinetobacter baumannii in patients with burn injury: A systematic review and meta-analysis. Burns. 2019;45:1495–1508. doi: 10.1016/j.burns.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 344.Ribeiro P.C., Monteiro A.S., Marques S.G., Monteiro S.G., Monteiro-Neto V., Coqueiro M.M., Marques A.C., de Jesus Gomes Turri R., Santos S.G., Bomfim M.R. Phenotypic and molecular detection of the blaKPC gene in clinical isolates from inpatients at hospitals in Sao Luis, MA, Brazil. BMC Infect. Dis. 2016;16:737. doi: 10.1186/s12879-016-2072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Robledo I.E., Aquino E.E., Sante M.I., Santana J.L., Otero D.M., Leon C.F., Vazquez G.J. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 2010;54:1354–1357. doi: 10.1128/AAC.00899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Ju L.C., Cheng Z., Fast W., Bonomo R.A., Crowder M.W. The continuing challenge of metallo-beta-lactamase inhibition: Mechanism matters. Trends Pharmacol. Sci. 2018;39:635–647. doi: 10.1016/j.tips.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Tolmasky M.E. Strategies to prolong the useful life of existing antibiotics and help overcoming the antibiotic resistance crisis. In: Atta-ur-Rhaman, editor. Frontiers in Clinical Drug Research-Anti Infectives. Volume 1. Bentham Books; Sharjah, UAE: 2017. pp. 1–27. [Google Scholar]
- 348.Papp-Wallace K.M., Bonomo R.A. New beta-lactamase inhibitors in the clinic. Infect. Dis. Clin. North. Am. 2016;30:441–464. doi: 10.1016/j.idc.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Noval M., Banoub M., Claeys K.C., Heil E. The battle is on: New beta-lactams for the treatment of multidrug-resistant gram-negative organisms. Curr. Infect. Dis. Rep. 2020;22:1. doi: 10.1007/s11908-020-0710-9. [DOI] [PubMed] [Google Scholar]
- 350.Carpenter J., Neidig N., Campbell A., Thornsberry T., Truex T., Fortney T., Zhang Y., Bush K. Activity of imipenem/relebactam against carbapenemase-producing Enterobacteriaceae with high colistin resistance. J. Antimicrob. Chemother. 2019;74:3260–3263. doi: 10.1093/jac/dkz354. [DOI] [PubMed] [Google Scholar]
- 351.Haidar G., Clancy C.J., Shields R.K., Hao B., Cheng S., Nguyen M.H. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 2017;61:e02534-16. doi: 10.1128/AAC.02534-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Porreca A.M., Sullivan K.V., Gallagher J.C. The epidemiology, evolution, and treatment of KPC-producing organisms. Curr. Infect. Dis. Rep. 2018;20:13. doi: 10.1007/s11908-018-0617-x. [DOI] [PubMed] [Google Scholar]
- 353.Doi Y., Murray G.L., Peleg A.Y. Acinetobacter baumannii: Evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 2015;36:85–98. doi: 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Traglia G.M., Chua K., Centron D., Tolmasky M.E., Ramirez M.S. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol. Evol. 2014;6:2235–2239. doi: 10.1093/gbe/evu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Imperi F., Antunes L.C., Blom J., Villa L., Iacono M., Visca P., Carattoli A. The genomics of Acinetobacter baumannii: Insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life. 2011;63:1068–1074. doi: 10.1002/iub.531. [DOI] [PubMed] [Google Scholar]
- 356.Traglia G.M., Place K., Dotto C., Fernandez J.S., Montana S., Bahiense C.D.S., Soler-Bistue A., Iriarte A., Perez F., Tolmasky M.E., et al. Interspecies DNA acquisition by a naturally competent Acinetobacter baumannii strain. Int. J. Antimicrob. Agents. 2019;53:483–490. doi: 10.1016/j.ijantimicag.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]