Abstract
Gut microbial communities play vital roles in the modulation of many insects' immunity, including Apis mellifera. However, little is known about the interaction of Apis cerana gut bacteria and A. cerana immune system. Here in this study, we conducted a comparison between germ-free gut microbiota deficient (GD) workers and conventional gut community (CV) workers, to reveal the possible impact of gut microbiota on the expression of A. cerana antimicrobial peptides and immune regulate pathways. We also test whether A. cerana gut microbiota can strengthen host resistance to Nosema ceranae. We find that the expression of apidaecin, abaecin and hymenoptaecin were significantly upregulated with the presence of gut bacteria, and JNK pathway was activated; in the meanwhile, the existence of gut bacteria inhibited the proliferation of Nosema ceranae. These demonstrated the essential role of A. cerana gut microbiota to host health and provided critical insight into the honeybee host–microbiome interaction.
Keywords: Apis cerana, gut microbiota, antimicrobial peptide, immune system, Nosema ceranae
1. Introduction
In animal gastrointestinal tracts live complex assemblages of microorganisms, some of which are emerging as key players in governing host health [1]. In particular, the gut bacteria communities of insects have been found involved in food digestion, nutrient provisioning, host development and intraspecific communication, and they generally can contribute to host health through immune system stimulation as well as conferring resistance against pathogens [2–4].
The honeybee (Apis spp.) gut is colonized by 8 to 10 phylotypes of bacteria, namely, Snodgrassella alvi, Gilliamella apicola, Lactobacillus spp., Bifidobacterium spp., Frischella perrara, Bartonella apis, Parasaccharibacter apium and Commensalibacter spp. [5]. Due to their social behaviour and the ways of acquiring gut microbiota, honeybees consistently harbour a highly conserved gut community, many of which have coevolved with their host for millions of years [6]. Thus, it is not surprising that these gut bacteria are tightly intertwined with the physiology of honeybees. Study has demonstrated that Apis mellifera gut microbiota possesses a large repertoire of metabolic capabilities [7]. For example, Zheng et al. [8,9] have shown that A. mellifera gut microbiota can promote host development and participate in the metabolism of toxic sugars.
Several studies also clearly demonstrated that A. mellifera gut microbiota make a strong contribution to their hosts' health. One way they can do this is by promoting the expression of apidaecin and hymenoptaecin [10], or strongly activating the host immune system as caused by the gut bacteria S. alvi and F. perrara [11]. Further, a recent study of antibiotic-treated A. mellifera revealed that destruction of their gut bacteria could increase the vulnerability of honeybee to Nosema infection [12]. Nosema ceranae is a microsporidia parasite originally found in the eastern honeybee (Apis cerana) in 1996 [13]. Now it is a common pathogen infecting European honeybee species and it is highly pathogenic to its novel host [14]. Nosema ceranae infection impairs midgut integrity and alters the energy demand in A. mellifera, and it can also significantly suppress bee immune response and modify pheromone production in A. mellifera workers and their queens, leading to precocious foraging [15].
However, most studies of honeybee host–microbiome interactions have focused on A. mellifera, leaving our knowledge about the function of A. cerana gut symbionts quite limited. The A. cerana honeybee's native range spans southern, southeastern and eastern Asia. This species is in the same subgenus as the western honeybee (A. mellifera), which is naturally spread though Africa and Europe. Both species share many biological similarities, in terms of their physiology, behaviour and life history, and their guts are both colonized by the same phylotypes of bacteria. Interestingly, studies have revealed a distinctive strain-level diversity between these two species [6,16], which could be a source of functional diversity [17]. Therefore, research on the interaction between A. cerana and its gut microbiota could not only shed light upon the functioning of A. cerana gut microbiota functions but also provide insight into the functional differences between A. cerana and A. mellifera gut microbial communities.
In the current study, we focused on the contribution of A. cerana gut microbiota to host health in two key aspects: (i) the interaction between gut microbiota and immune system of A. cerana, and (ii) the interaction between gut microbiota and parasites of A. cerana. By comparing workers colonized with or without gut bacteria, we compared the gene expression of both antimicrobial peptides (AMPs) and the regulators of immune pathways, and we also evaluated the role of gut microbiota in Nocema resistance. We found that A. cerana gut microbiota can upregulate the expression of genes apidaecin, abaecin, hymenoptaecin and vitellogenin, regulate key components of the JAK/STAT and JNK pathways and demonstrate their contribution to host resistance against N. ceranae.
2. Material and methods
2.1. Rearing of honeybees
Apis cerana colonies for the experiments and A. mellifera colony for Nosema spore were all kept in the apiary maintained at the Honey Bee Research Laboratory in the College of Animal Sciences, Zhejiang University, Hangzhou, China. Experiments were replicated three times using three different A. cerana colonies, and in each replicate, A. cerana workers and A. cerana gut bacteria from the same colony were used.
Gut microbiota deficient (GD) A. cerana workers and conventional gut community (CV) A. cerana workers were obtained using the protocol described by Zheng et al. [8]. Briefly, late stage pupae (dark-eyed) were removed from brood frames and transferred into sterile 48-well cell culture plates. These plates were placed in an incubator at 34°C ± 1°C with 80% ± 5% relative humidity until bees emerged. Workers emerged within the 48 h after transformation were collected for the experiments.
The newly emerged germ-free bees were randomly assigned to GD or CV groups (50 workers per group). GD workers were supplied with sterile pollen and sterile sugar water (50% sucrose solution, w/v), while CV workers were supplied with food containing gut bacteria for 5 days and then switched to sterile pollen and sterile sugar water. For the quantification of bacterial loads of GD and CV bees, six workers from each cage were sampled on day 5 after emergence. In addition, we have also sampled 10 forager workers from the field to serve as a positive control. For the gene expression profiling, six workers per cage were collected from each of the GD and CV groups on day 14. Experiments were replicated three times, with a total of three cages used for GD and three cages for CV.
To analyse the interaction between A. cerana gut bacteria and Nosema, newly emerged germ-free workers were randomly assigned to four different groups (50 bees per group). GDT: germ-free workers were inoculated individually with 2 µl of a syrup-spore suspension containing 1 × 105 Nosema spores [18] and supplied with sterile pollen and sterile sugar water; GDC: germ-free workers were inoculated with PBS and supplied with sterile pollen and sterile sugar water; CVT: germ-free workers were inoculated individually with 2 µl syrup-spore suspension containing 1 × 105 Nosema spores and colonized with gut homogenates; CVC: germ-free workers were inoculated with PBS and colonized with gut homogenates. The number of dead bees were recorded daily and removed. Six live bees from each cage were randomly sampled to monitor Nosema spore numbers on 7th and 14th day post infection (dpi), respectively. Experiments were replicated three times, with a total of 12 cages: three cages each for GDT, GDC, CVT and CVC treatment groups.
2.2. Preparation of Apis cerana gut bacteria
Ten A. cerana forager workers were sampled from the entrance of bee hives; their hindguts were dissected immediately and homogenized together in 1 ml PBS. This gut homogenate was centrifuged at 10 000g for 10 min and the supernatant were removed to eliminate the possible viral contamination. Then 1 ml PBS was used to resuspend the bacteria, from which a 100 µl suspension was added to and mixed with sterilized pollen.
2.3. Quantification of bacterial loads in the gut of honeybees
Bacterial loads of GD and CV workers were determined by qPCR [19], using the universal bacterial 16S rRNA primers as listed in table 1. Workers were immobilized at 4°C and their whole guts were immediately dissected. From these, their DNA was extracted using the TIANamp Stool DNA Kit (Tiangen Biotech Co., Ltd, Beijing, China) according to the manufacturer's protocol and then used for the qPCR. The absolute quantification of 16S rRNA copy numbers was quantified using the StepOne Plus real-time PCR system for which the thermal cycling conditions were as follow: initial denaturing step of 95°C for 30 s, 40 amplification cycles of 95°C for 5 s and 60°C annealing for 30 s, with a melt curve analysis done from 60°C to 95°C at 0.5°C/5 s increments to confirm expected dissociation curves. The qPCR reaction mixtures were set up with 1 µl of DNA, 0.2 µl of forward and reverse primers (10 µM), 5 µl of TB Green™ Premix Ex Taq (Takara Biomedical Technology Co., Ltd, Beijing, China) and 3.6 µl of distilled water. All DNA samples were replicated in three wells.
Table 1.
The primers used in this study.
| target gene name and accession no. | sequence (5′ to 3′) |
gene classification | amplicon size (bp) | reference | |
|---|---|---|---|---|---|
| universal bacterial 16S rRNA | F: | AGAGTTTGATCCTGGCTCAG | bacteria quantification | 328 | [19] |
| R: | CTGCTGCCTCCCGTAGGAGT | ||||
| Arp1 (XM_017059067.2) | F: | CTCACAGTGTTCGCAACTCG | housekeeping gene | 206 | this study |
| R: | CGAAACCGGCTTTGCACATA | ||||
| abaecin (XM_017063755.2) | F: | ATCTTCGCACTACTCGCCAC | antimicrobial peptide | 103 | this study |
| R: | CCTGACCAGGAAACGTTGGA | ||||
| apidaecin (XM_017060818.2) | F: | CCAGATCCGCCTACTCAACC | antimicrobial peptide | 131 | this study |
| R: | GGTTTAGCTTCACGGCGTAG | ||||
| defensin 1 (XM_017050425.2) | F: | AGCCACTTGAGCATCCTGAG | antimicrobial peptide | 151 | this study |
| R: | CCGTTCTTGCAATGACCTCC | ||||
| defensin 2 (XM_017060723.2) | F: | TTTCGCGATTCTCGTCGCTA | antimicrobial peptide | 156 | this study |
| R: | TGTCGTAGCAGTAGCGGTTC | ||||
| hymenoptaecin (XM_017049926.1) | F: | CGTGTTGGTTGTCTTCTGCG | antimicrobial peptide | 209 | this study |
| R: | CACCATAGGCATCTCCCGTC | ||||
| Vg NM_001328484.1) | F: | ACCAACGACTTCATGGGACC | immune marker | 181 | this study |
| R: | CGCTGTCGCTGATCACATTG | ||||
| relish (XM_017053040.2) | F: | TGAAGCTGGTGCATGTGTTG | IMD pathway | 105 | this study |
| R: | CCTGCTTTTGCTGCAAGATGT | ||||
| dorsal (XM_017054012.2) | F: | TTTATCACGATTGTAGATGCTGC | toll pathway | 149 | this study |
| R: | GGAGAAGTTGTTGCCATCGG | ||||
| basket (XM_028666319.1) | F: | AGGAGAACGTGGACATTTGG | JNK pathway | 243 | [20] |
| R: | AATCCGATGGAAACAGAACG | ||||
| Imd (XM_017057615.2) | F: | TGTTAACGACCGATGCAAAA | IMD pathway | 153 | [20] |
| R: | CATCGCTCTTTTCGGATGTT | ||||
| toll (XM_017053307.2) | F: | TCGATGTCCAACGGAGCAAA | toll pathway | 102 | this study |
| R: | ACTTTCACAACGAAGGCCGA | ||||
| domeless (XM_028666710.1) | F: | TTGTGCTCCTGAAAATGCTG | JAK/STAT pathway | 180 | [20] |
| R: | AACCTCCAAATCGCTCTGTG | ||||
| kayak (XM_028666620.1) | F: | CGACAGATCCGCAGAGAAAG | JNK pathway | 148 | [21] |
| R: | CCTGTTGCAGCTGTTGTATC | ||||
| hopscotch (XM_028664411.1) | F: | ATTCATGGCATCGTGAACAA | JAK/STAT pathway | 141 | [21] |
| R: | CTGTGGTGGAGTTGTTGGTG | ||||
2.4. Profiling the gene expression levels of antimicrobial peptides and immune pathways
Workers were immobilized at 4°C and their whole abdomens were immediately dissected and used for RNA extractions. We assayed the transcript levels of the following genes: apidaecin, abaecin, defensin 1, defensin 2, hymenoptaecin, relish, dorsal, basket, Imd, toll, domeless, kayak, hopscotch and vitellogenin (Vg), with the housekeeping gene actin related protein 1 (Arp1) chosen as the reference control. The primers used are listed in table 1.
Total RNA was extracted with the RNApure Total RNA Kit (Aidlab Biotechnologies Co. Ltd, Beijing, China) according to the manufacturer's protocol. The cDNA synthesis reaction was performed using 0.5 µg of total RNA with the PrimeScript™ RT Master Mix (Takara Biomedical Technology Co., Ltd, Beijing, China). The concentration and quality of this RNA was determined using the Nanodrop 2000 (Thermo Fisher Scientific, MA, US). The qPCR reaction mixtures were set up with 1 µl of cDNA (10X diluted) as described above. The relative expression levels of target genes were calculated by applying the 2−ΔΔCt method [22].
2.5. Isolation and counting of Nosema spore
Nosema spores for inoculation were freshly isolated from a heavily infected A. mellifera colony kept at the experimental apiary; hence, all the N. ceranae spores used in this study were isolated from a single colony. A total of 30 midguts were homogenized in 30 ml of distilled water. To sediment the Nosema spores, this homogenate was centrifuged at 3000 r.p.m. for 5 min and the supernatant was discarded. The centrifugation process was repeated twice to obtain a crude Nosema spore suspension. To obtain the pure Nosema spores, the suspension was then purified by the method of Percoll gradient centrifugation [14]. Nosema spore concentrations were determined by counting spore numbers within a haemocytometer chamber (Hausser Scientific 3100). Nosema spore concentration was calculated by the following equation: ((total numbers of spores counted in the haemocytometer) × 4 × 106)/square numbers. The spore-syrup suspension was freshly prepared by mixing the pure Nosema spores with 30% sucrose syrup before use. The species identity of Nosema spores was confirmed by PCR [23]. To count the Nosema spore numbers in individual worker, each bee's abdomen was smashed and macerated in 1 ml of distilled water. From this, a 10 µl tissue solution was placed in a haemocytometer to quantify Nosema spore numbers [24].
2.6. Data analysis
Statistical analysis was carried out using SPSS software version 22.0. For the gene expression profiling and Nosema spore numbers, results from three replicates were pooled together and statistical significance was calculated using the independent sample t-test with Bonferroni-corrected p-values (p = 0.05/3 = 0.016). The Kaplan–Meier survival curve and Cox regression analyses were used for the survivorship data. All the figures were drawn in GraphPad Prism 7.
3. Results
3.1. Apis cerana gut microbiota stimulates host antimicrobial peptides and vitellogenin expression
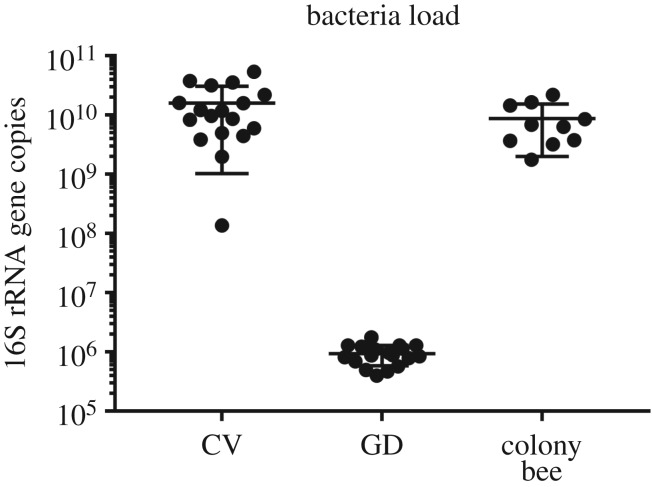
The copy numbers of the 16S rRNA gene in the gut of A. cerana workers was significantly increased when inoculated with normal gut microbiota (Student's t-test, t = 4.538, d.f. = 34, p < 0.001), with a total of approximately 1010 bacterial cells found per gut of CV bees versus a total bacterial load of around 106 cells per gut of GD bees (figure 1). Furthermore, the comparison between CV workers and colony workers revealed that they had similar bacteria loads in their gut (Student's t-test, t = 1.438 d.f. = 26, p = 0.163). These results verified that bees fed with homogenized guts were successfully colonized by honeybee gut bacteria.
Figure 1.
The bacterial colonization levels in the guts of GD worker (n = 5 bees × 3 replicates), CV workers (n = 5 bees × 3 replicates) and colony bees (n = 10). For each boxplot, the centre line displays the median, + indicates expression mean, the boxes correspond to the 25th and 75th percentiles and whiskers span the 10th–90th percentile.
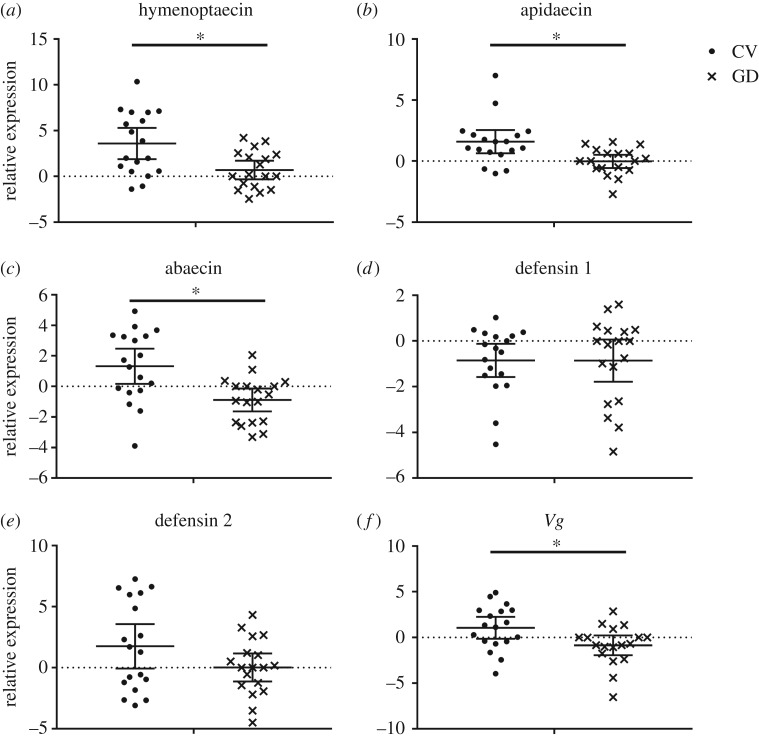
We found that A. cerana gut microbiota had a significant impact on the expression of three AMP coding genes: apidaecin (Student's t-test, t = 3.108, d.f. = 34, p = 0.004), abaecin (Student's t-test, t = 3.381, d.f. = 34, p = 0.002) and hymenoptaecin (Student's t-test, t = 3.065, d.f. = 34, p = 0.004), which were increased 3.1-fold, 4.6-fold and 7.4-fold, respectively. The upregulation of Vg (Student's t-test, t = 2.159, d.f. = 34, p = 0.015), a common marker for overall honeybee health, was also detected in CV workers. By contrast, the transcripts for defensin 1 (Student's t-test, t = 0.015, d.f. = 34, p = 0.988) and defensin 2 (Student's t-test, t = 1.696, d.f. = 34, p = 0.099) did not show any significant differences between the CV and GD workers (figure 2).
Figure 2.
The expression level of AMPs and Vg in the abdomen of GD worker (n = 6 bees × 3 replicates) and CV workers (n = 6 bees × 3 replicates). The relative expression was log2 transformed. Means ± 95 CI are shown by the black bars and whiskers. *represents the significant difference (p < 0.016).
3.2. Apis cerana gut microbiota actives JNK pathways
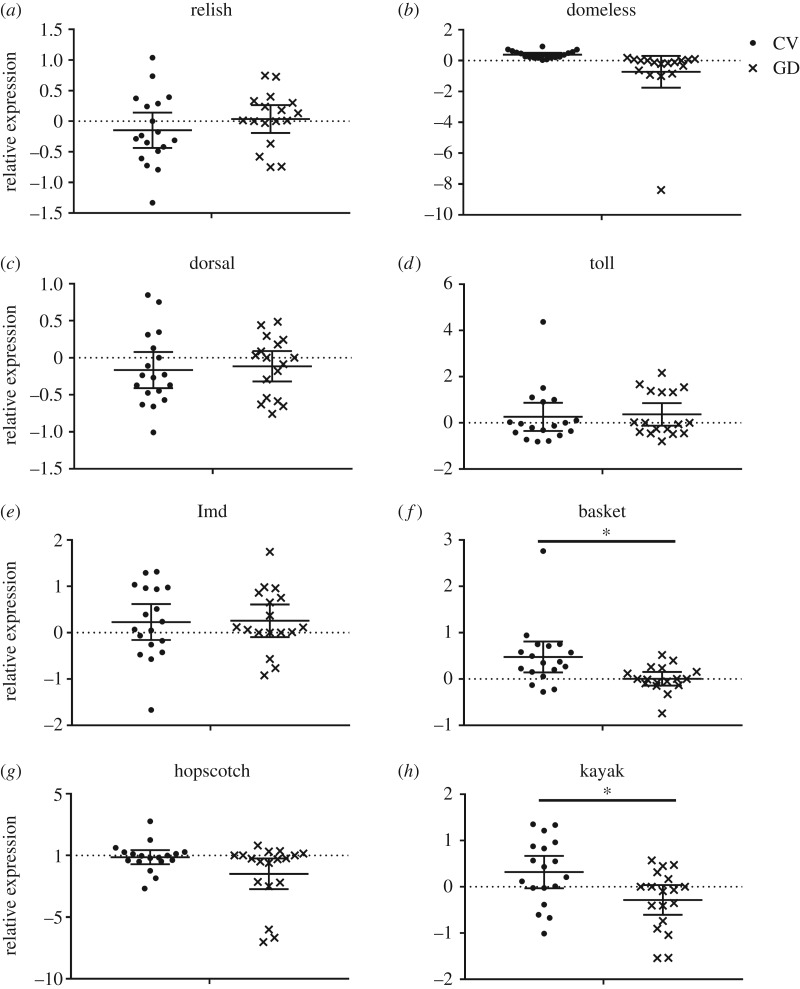
As shown in figure 3, gut bacteria colonization dramatically increased the expression of basket (Student's t-test, t = 2.847, d.f. = 34, p = 0.012) and kayak (Student's t-test, t = 2.671, d.f. = 34, p = 0.012) genes, which are major components of the JNK pathway. Interestingly, the expression levels of domeless (Student's t-test, t = 2.309, d.f. = 34, p = 0.026) and hopscotch (Student's t-test, t = 2.073, d.f. = 34, p = 0.046), both of which are immune regulators of JAK/STAT pathway, in the CV workers were not significantly higher than those of GD workers. Also, negligible interactions were found between A. cerana gut microbiota and toll as well as the IMD pathways, in that the expression level of key components of these two pathways, namely relish (Student's t-test, t = 1.127, d.f. = 34, p = 0.304), dorsal (Student's t-test, t = 0.457, d.f. = 34, p = 0.742), toll (Student's t-test, t = 0.286, d.f. = 34, p = 0.777) and Imd (Student's t-test, t = 0.076, d.f. = 34, p = 0.913), did not undergo significant regulation with gut bacteria present in the host (figure 3).
Figure 3.
The expression level of key immune components in the abdomen of GD worker (n = 6 bees × 3 replicates) and CV workers (n = 6 bees × 3 replicates). The relative expression was log2 transformed. Means ± 95 CI are shown by the black bars and whiskers. *represents the significant difference (p < 0.016).
3.3. Apis cerana gut microbiota promote host resistance against Nosema ceranae infection
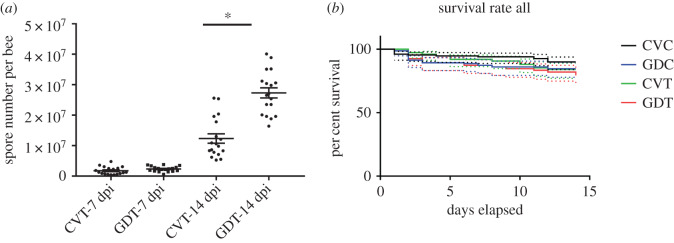
The numbers of Nosema spores in honeybee workers' gut were significantly affected by the presence of gut bacteria. An average of 2.73 × 107 spore per bee were counted in the gut of GDT workers, whereas only 1.23 × 107 spores were found in CVT workers at 14 dpi (Student's t-test, t = 6.561, d.f. = 34, p < 0.001); however, no significant difference were observed at 7 dpi (Student's t-test, t = 1.58, d.f. = 34, p = 0.123, figure 4a). Additionally, no spores were identified in the GDC and CVC bees, which demonstrated that workers used in the experiments were Nosema free.
Figure 4.
The Nosema ceranae spore number (a) in CVT and GDT worker (n = 6 bees × 3 replicates) after 7 dpi and 14 dpi and survival rate of A. cerana workers in three replicates (b). Means ± SEM are shown by the black bars and whiskers. * represents the significant difference (p < 0.05).
We also noticed that although the spore numbers were significantly different between CVT and GDT workers, the survival rate of Nosema-treated workers did not differ greatly during the experiment period (figure 4b). Furthermore, no increase in mortality rates was observed in workers lacking their microbiota, which agrees with similar findings on A. mellifera [25].
4. Discussion
Many studies have shown that gut bacteria are key players in immune modulation and are essential for a healthy immune system [26,27]. In all stages of their life cycle, insects are threatened by a multitude of predators, parasites, parasitoids and pathogens. To counteract these threats, insects have evolved mechanical, chemical and behavioural defences as well as a complex immune system, and in addition to the host's own defences, whereby some insects are associated with protective symbionts [28]. Our study provides the first evidence of a close relation in A. cerana between its gut microbiota and the host immune system. The results of our study strongly emphasized the importance of commensal gut bacteria in stimulating the immune system of A. cerana and for strengthening the resistance of A. cerana to N. ceranae infection.
Vg, the precursor of yolk proteins, was traditionally seen as being the energy reserve for nourishment of the developing embryos, yet, its role extends beyond this nutrient function [29]. In the honeybee A. mellifera, it was found to participate in the regulation of social organization and individual physiology [29], and it has been linked to host immune defence as an immune-relevant molecule [30]. Thus, Vg has become widely accepted as a marker of honeybees' overall health. Accordingly, the upregulation of Vg expression by gut microbiota we found here clearly shows that these A. cerana gut inhabitants have a positive impact on host health.
AMPs are crucial effectors of insects’ innate immune system, providing the first line of defence against a variety of pathogens [31]. Due to their importance for maintaining honeybee health, AMPs have increasingly become a focus of research investigations. Only a reduced number of AMPs were detected in the honeybee haemolymph [32], including apidaecin, abaecin, defensin 1, defensin 2 and hymenoptaecin and their respective genes' expression is generally regulated by four intracellular signalling pathways (Toll, IMD, JNK and JAK/STAT). Many studies have shown that symbiotic bacteria can activate the immune system of their insect host and thereby increase the efficiency of pathogen defence [33,34]. Of all the AMPs expressed in honeybee haemolymph, we found that gut bacteria colonization led to significant upregulation in the expression levels of apidaecin, abaecin and hymenoptaecin in workers’ abdomens, while the bacteria inoculation had no apparent influence on defensin 1 and defensin 2. Intriguingly, Kwong et al. [10] found that A. mellifera gut microbiota can increase the expression of apidaecin and hymenoptaecin, but they had no impact on abaecin, suggesting that A. cerana and A. mellifera gut microbiota have similar albeit not identical functioning in host immune modulation, even though they both harbour a small, recurring set of bacterial phylotypes [6]. Experiments on mono-colonized A. mellifera bees have reported that S. alvi colonization can only upregulate apidaecin expression [10], whereas F. perrara colonization can upregulate the expression of apidaecin, abaecin and defensin 1 [11]. Hence, host immune modulation is the outcome of complex host–microbiota interactions involving multiple phylotypes, as differing bacteria phylotypes/strains generate different microbe-associated molecular patterns for the pathway leading to AMP production. Therefore, we speculated that the difference in the regulation of AMP expression between our study and previous studies is likely to be caused by the presence of host-specific strains or the variation in the relative abundance of bacterial species in honeybees' gut microbial communities.
Recent studies have led to important advances in our understanding of the regulation of honeybee AMPs; however, there are still wide gaps between our current knowledge and the full understanding of the underlying molecular mechanisms [32]. Four non-autonomous pathways are implicated in inducible host defence, the Toll, IMD, JAK/STAT and JNK pathways, and are considered as the major directors of this process [35]. In our study, the expression of key components of these pathways was analysed. We found that key regulators of JNK pathways (basket and kayak) were significantly regulated by the colonization of gut bacteria, while the JAK/STAT, IMD and Toll pathways remained mostly unchanged. Functional study of the interaction between social bee AMP and JNK pathways is quite limited; however, these relationships have been well studied on other insects like the model Drosophila melanogaster. The JNK pathway has been identified as a regulator of AMP gene expression in Drosophila S2 cells [36] and the expression of Drosophila AMPs was found blocked by an inhibitor of JNK signalling and also in JNK mutant clones [37]. Given those findings, our results indicated that A. cerana gut symbionts enhanced host immunity though regulating JNK pathways. A study of gut microbiota dysbiosis in A. mellifera workers revealed that antibiotic treatment decreased the expression of relish [38], a transcription factor in the IMD pathway, leading to the downregulation of AMPs [12]. However, considering that gut microbiota dysbiosis and being gut microbiota deficient are different situations, it is premature to say that A. cerana and A. mellifera gut microbiota regulate different immune pathways. Therefore, studies on honeybee immune–microbiota interactions are urgently needed, especially those that use the mono-colonized worker model or studies which combine germ-free workers and RNAi knockdown.
Colonization of the gut with commensal or mutualistic microbial communities can increase the resistance of the host against parasite invasion. For example, Hamiltonella defensa in pea aphids (Acyrthosiphon pisum) and black bean aphids (Aphis fabae) confer protection against parasitic wasps [39]. In addition, a variety of studies on the bumblebee showed that its intestinal symbiont of bumblebee (Bombus terrestris) influences the infection of the parasite Crithidia bombi [40,41]. Currently, N. ceranae, which jumped from the Asian to the western honeybee some decades ago, is the most widespread and deadly gut parasite of A. mellifera [15,42]. Rubanov et al. [43] have found that two specific strains of Gilliamella were significantly more abundant in bees from colonies with high Nosema loads versus those with low Nosema loads, and that eliminating their gut bacteria using antibiotics made them more susceptible to Nosema infections [12]. That work suggested a clear association between A. mellifera gut microbiota and honeybee resistance to Nosema spores. As its original host, A. cerana has coevolved with N. ceranae for millions of years, so it is perhaps not surprising that N. ceranae proliferation was inhibited by A. cerana gut bacteria, as significantly higher spore loads were noticed in GD workers than CV workers in our study. This result demonstrated that A. cerana gut microbiota contributes greatly to how the host resists this parasite. The interactions between gut microbiota, gut parasites and host are complicated; the inhibition of parasites could be accomplished through their direct interaction with microbes, or due to changes to the physical gastrointestinal landscape or the immune landscape of the gut [44]. Thus, the mechanism underlying this inhibition of a potent honeybee parasite merits further study. In addition, we have noticed that Nosema infection had little or no impact on honeybee longevity, in contrast with a previous study of A. cerana and A. mellifera [45]. This discrepancy could be due to the modulation of the N. ceranae virulence that is related to a polymorphism between N. ceranae isolates from different geographic origins [15]. Hence, the interspecies variants of N. ceranae should be explicitly considered in future studies of honeybee gut microbiota–N. ceranae interactions.
Taken together, our experimental work demonstrated the contribution of A. cerana gut microbiota to host health, pointing out the beneficial role of a balanced gut microbiome in honeybee A. cerana and providing new insights into the honeybee host–microbiome interactions. Also, our findings further suggest the potential use of A. cerana gut bacteria as probiotics for promoting honeybee resistance to Nosema in apiculture.
Supplementary Material
Ethics
Our study does not present research with ethical considerations.
Data accessibility
All the data generated and used in our study are included in the electronic supplementary material: table S1 includes all the gene expression data, table S2 includes the bacteria load, table S3 includes the Nosema spore number and table S4 includes the survival rate of Nosema treated CV and GD workers.
Authors' contributions
Y.W., Y.Z., H.Z. and F.H. conceived and designed the study. Y.W., Y.Z., Y.C and G.C. performed experiments. Y.W and Y.Z. analysed data and drafted the manuscript. All authors edited and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Funding
The work was supported by National Natural Science Foundation of China (31902222, Y.W., 31672498, H.Z. and 31872431, F.H.), Science and Technology Department of Zhejiang Province, China (2016C02054-11, F.H.) and the Modern Agroindustry Technology Research System (CARS-44, F.H. and H.Z.).
References
- 1.Heintz-Buschart A, Wilmes P. 2018. Human gut microbiome: function matters. Trends Microbiol. 26, 563–574. ( 10.1016/j.tim.2017.11.002) [DOI] [PubMed] [Google Scholar]
- 2.Engel P, Moran NA. 2013. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. ( 10.1111/1574-6976.12025) [DOI] [PubMed] [Google Scholar]
- 3.Dong Y, Manfredini F, Dimopoulos G. 2009. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 5, e1000423 ( 10.1371/journal.ppat.1000423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch H, Schmid-Hempel P. 2011. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl Acad. Sci. USA 108, 19 288–19 292. ( 10.1073/pnas.1110474108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellegaard KM, Engel P. 2019. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 10, 446 ( 10.1038/s41467-019-08303-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, Moran NA. 2017. Dynamic microbiome evolution in social bees. Sci. Adv. 3, e1600513 ( 10.1126/sciadv.1600513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kesnerova L, Mars RAT, Ellegaard KM, Troilo M, Sauer U, Engel P. 2017. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 15, e2003467 ( 10.1371/journal.pbio.2003467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. 2017. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl Acad. Sci. USA 114, 4775–4780. ( 10.1073/pnas.1701819114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng H, Nishida A, Kwong WK, Koch H, Engel P, Steele MI, Moran NA. 2016. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio. 7, e01326-16 ( 10.1128/mBio.01326-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong WK, Mancenido AL, Moran NA. 2017. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 4, 170003 ( 10.1098/rsos.170003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emery O, Schmidt K, Engel P. 2017. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 26, 2576–2590. ( 10.1111/mec.14058) [DOI] [PubMed] [Google Scholar]
- 12.Li JH, Evans JD, Li WF, Zhao YZ, DeGrandi-Hoffman G, Huang SK, Li ZG, Hamilton M, Chen YP. 2017. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee's vulnerability to Nosema infection. PLoS ONE. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fries I, Feng F, daSilva A, Slemenda SB, Pieniazek NJ. 1996. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 32, 356–365. ( 10.1016/S0932-4739(96)80059-9) [DOI] [Google Scholar]
- 14.Chen YP, Evans JD, Murphy C, Gutell R, Zuker M, Gundensen-Rindal D, Pettis J. 2009. Morphological, molecular, and phylogenetic characterization of Nosema ceranae, a microsporidian parasite isolated from the European honey bee, Apis mellifera. J. Eukaryot. Microbiol. 56, 142–147. ( 10.1111/j.1550-7408.2008.00374.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paris L, El Alaoui H, Delbac F, Diogon M. 2018. Effects of the gut parasite Nosema ceranae on honey bee physiology and behavior. Curr. Opin. Insect Sci. 26, 149–154. ( 10.1016/j.cois.2018.02.017) [DOI] [PubMed] [Google Scholar]
- 16.Koch H, Abrol DP, Li J, Schmid-Hempel P. 2013. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 22, 2028–2044. ( 10.1111/mec.12209) [DOI] [PubMed] [Google Scholar]
- 17.Greenblum S, Carr R, Borenstein E. 2015. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 160, 583–594. ( 10.1016/j.cell.2014.12.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Wang S, Xu Y, Gong H, Wu Y, Chen Y, Hu F, Zheng H. et al. In press. Protective potential of Chinese herbal extracts against microsporidian Nosema ceranae, an emergent pathogen of western honey bees, Apis mellifera L. J. Asia Pac. Entomol. [Google Scholar]
- 19.Powell JE, Martinson VG, Urban-Mead K, Moran NA. 2014. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 80, 7378–7387. ( 10.1128/AEM.01861-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Huang X, Xu Z, Han R, Chen J. 2013. Differential gene transcription in honeybee (Apis cerana) larvae challenged by Chinese sacbrood virus (CSBV) Sociobiology 60, 413–420. [Google Scholar]
- 21.Tesovnik T, Cizelj I, Zorc M, Citar M, Bozic J, Glavan G, Narat M. 2017. Immune related gene expression in worker honey bee (Apis mellifera carnica) pupae exposed to neonicotinoid thiamethoxam and Varroa mites (Varroa destructor). PLoS ONE. 12, e0187079 ( 10.1371/journal.pone.0187079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Evans JD, Zhou L, Boncristiani H, Kimura K, Xiao T, Litkowski AM, Pettis JS. 2009. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 101, 204–209. ( 10.1016/j.jip.2009.05.012) [DOI] [PubMed] [Google Scholar]
- 24.Fries I, et al. 2013. Standard methods for Nosema research. J. Apicult. Res. 52, 1–28. ( 10.3896/IBRA.1.52.1.14) [DOI] [Google Scholar]
- 25.Raymann K, Shaffer Z, Moran NA. 2017. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 15, e2001861 ( 10.1371/journal.pbio.2001861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly D, Conway S, Aminov R. 2005. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 26, 326–333. ( 10.1016/j.it.2005.04.008) [DOI] [PubMed] [Google Scholar]
- 27.Grizotte-Lake M, Zhong G, Duncan K, Kirkwood J, Iyer N, Smolenski I, Isoherranen N, Vaishnava S. 2018. Commensals suppress intestinal epithelial cell retinoic acid synthesis to regulate interleukin-22 activity and prevent microbial dysbiosis. Immunity 49, 1103–1115. ( 10.1016/j.immuni.2018.11.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaltenpoth M, Engl T. 2014. Defensive microbial symbionts in Hymenoptera. Funct. Ecol. 28, 315–327. ( 10.1111/1365-2435.12089) [DOI] [Google Scholar]
- 29.Zhang S, Wang S, Li H, Li L. 2011. Vitellogenin, a multivalent sensor and an antimicrobial effector. Int. J. Biochem. Cell Biol. 43, 303–305. ( 10.1016/j.biocel.2010.11.003) [DOI] [PubMed] [Google Scholar]
- 30.Amdam GV, Simoes ZL, Hagen A, Norberg K, Schroder K, Mikkelsen O, Kirkwood TBL, Omholt SW. 2004. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp. Gerontol. 39, 767–773. ( 10.1016/j.exger.2004.02.010) [DOI] [PubMed] [Google Scholar]
- 31.Wu Q, Patocka J, Kuca K. 2018. Insect antimicrobial peptides, a mini review. Toxins. 10, 461–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danihlik J, Aronstein K, Petrivalsky M. 2015. Antimicrobial peptides: a key component of honey bee innate immunity. J. Apicult. Res. 54, 123–136. ( 10.1080/00218839.2015.1109919) [DOI] [Google Scholar]
- 33.de Souza DJ, Bézier A, Depoix D, Drezen J-M, Lenoir A. 2009. Blochmannia endosymbionts improve colony growth and immune defence in the ant Camponotus fellah. BMC Microbiol. 9, 29 ( 10.1186/1471-2180-9-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans JD, Lopez DL. 2004. Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae). J. Econ. Entomol. 97, 752–756. ( 10.1093/jee/97.3.752) [DOI] [PubMed] [Google Scholar]
- 35.Evans JD, et al. 2006. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 15, 645–656. ( 10.1111/j.1365-2583.2006.00682.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallio J, Leinonen A, Ulvila J, Valanne S, Ezekowitz RA, Rämet M. 2005. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 7, 811–819. ( 10.1016/j.micinf.2005.03.014) [DOI] [PubMed] [Google Scholar]
- 37.Delaney JR, Stöven S, Uvell H, Anderson KV, Engström Y, Mlodzik M. 2006. Cooperative control of Drosophila immune responses by the JNK and NF-κB signaling pathways. EMBO J. 25, 3068–3077. ( 10.1038/sj.emboj.7601182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Heerman MC et al. 2019. Pollen reverses decreased lifespan, altered nutritional metabolism and suppressed immunity in honey bees (Apis mellifera) treated with antibiotics. J. Exp. Biol. 222, 1. [DOI] [PubMed] [Google Scholar]
- 39.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl Acad. Sci. USA 100, 1803–1807. ( 10.1073/pnas.0335320100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer-Young EC, Raffel TR, McFrederick QS. 2019. pH-mediated inhibition of a bumble bee parasite by an intestinal symbiont. Parasitology. 146, 380–388. ( 10.1017/S0031182018001555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Praet J, Parmentier A, Schmid-Hempel R, Meeus I, Smagghe G, Vandamme P. 2018. Large-scale cultivation of the bumblebee gut microbiota reveals an underestimated bacterial species diversity capable of pathogen inhibition. Environ. Microbiol. 20, 214–227. ( 10.1111/1462-2920.13973) [DOI] [PubMed] [Google Scholar]
- 42.Emsen B, Guzman-Novoa E, Hamiduzzaman MM, Eccles L, Lacey B, Ruiz-Pérez RA, Nasr M. 2016. Higher prevalence and levels of Nosema ceranae than Nosema apis infections in Canadian honey bee colonies. Parasitol. Res. 115, 175–181. ( 10.1007/s00436-015-4733-3) [DOI] [PubMed] [Google Scholar]
- 43.Rubanov A, Russell KA, Rothman JA, Nieh JC, McFrederick QS. 2019. Intensity of Nosema ceranae infection is associated with specific honey bee gut bacteria and weakly associated with gut microbiome structure. Sci. Rep. 9, 3820 ( 10.1038/s41598-019-40347-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung JM, Graham AL, Knowles SC. 2018. Parasite-microbiota interactions with the vertebrate gut: synthesis through an ecological lens. Front. Microbiol. 9, 843 ( 10.3389/fmicb.2018.00843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinpoo C, Paxton RJ, Disayathanoowat T, Krongdang S, Chantawannakul P. 2018. Impact of Nosema ceranae and Nosema apis on individual worker bees of the two host species (Apis cerana and Apis mellifera) and regulation of host immune response. J. Insect. Physiol. 105, 1–8. ( 10.1016/j.jinsphys.2017.12.010) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated and used in our study are included in the electronic supplementary material: table S1 includes all the gene expression data, table S2 includes the bacteria load, table S3 includes the Nosema spore number and table S4 includes the survival rate of Nosema treated CV and GD workers.