Abstract
Objective
Clinical diagnostics often requires the detection of multiple bacterial species in limited clinical samples with a single DNA extraction method. This study aimed to compare the bacterial DNA extraction efficiency of two lysis methods automated with the MagNA-Pure LC instrument. The samples included five oral bacterial species (three Gram-positive and two Gram-negative) with or without human saliva background.
Methods
Genomic DNA (gDNA) was extracted from bacterial cultures by bead-beating lysis (BMP) or chemical lysis (MP), followed by automated purification and measurement by quantitative PCR.
Results
For pure bacterial cultures, the MP method yielded higher quantities of extracted DNA and a lower detection limit than the BMP method, except where the samples contained high numbers of Gram-positive bacteria. For bacterial cultures with a saliva background, no difference in gDNA extraction efficacy was observed between the two methods.
Conclusions
The efficiency of a bacterial DNA extraction method is not only affected by the bacterial cell wall structure but also by the sample milieu. The MP method provided superior gDNA extraction efficiency when the samples contained a single bacterial species, whereas either of the BMP and MP methods could be applied with similar efficiencies to samples containing multiple species of bacteria.
Keywords: Genomic DNA, extraction methods, Gram-negative bacteria, Gram-positive bacteria, saliva, in vitro, molecular diagnostics
Introduction
The oral cavity harbors a rich, diverse, and complex microbial community. Depending on the detection methods used, 700 to more than 10,000 phylotypes of microbial species have been found in this community.1,2 Increasing evidence has indicated that most oral infections, including caries, root canal infections, periodontitis, and peri-implantitis, are polymicrobial infections.3 The oral pathogens and commensal bacterial species in a microbial community interact and compete with each other, thereby modulating the progress of infections.4 Therefore, reliable quantification of various bacterial species in an oral microbial community is crucial.
Modern molecular techniques have allowed the examination of multiple bacterial species, including unculturable species, in clinical samples.5,6 Genomic DNA (gDNA) extraction is a prerequisite step for most of these techniques. To capture the true microbial composition, it is imperative that no bias is introduced in the gDNA extraction step. In principle, every gDNA extraction method includes cell lysis and the subsequent recovery of gDNA, free of amplification inhibitors. Cell lysis is often the “bottleneck” of the whole process because its efficiency is influenced by many factors such as bacterial cell wall characteristics, bacterial cell physical status, and the presence of matrix components, including polysaccharides, in the samples.7,8
In general, it is more difficult to lyse Gram-positive microorganisms than Gram-negative microorganisms because of the thick, rigid cell walls of Gram-positive species.9 To address this problem, researchers have developed specialized procedures, such as bead-beating or the use of specific lytic enzymes, to facilitate routine gDNA extraction from resistant species. Therefore, separate extraction protocols are commonly used for these two types of bacteria.10,11 This approach works well for a single microorganism target. However, it is not optimal for the examination of clinical oral microbial samples, which contain numerous Gram-positive and Gram-negative bacterial species. Clinical samples are typically too limited to facilitate two separate gDNA extractions. Therefore, a single cell lysis procedure, which overcomes resistant cell walls without degrading gDNA from easily lysed cells, would be preferable for clinical studies.
MagNA Pure DNA extraction is an automated method that ensures a high degree of reproducibility, minimal hands-on time, and the parallel processing of a large number of samples.12 The original protocol includes the chemical lysis of bacterial cells and subsequent DNA purification with magnetic bead technology. A recent study demonstrated that physical disruption, by beating with 0.1-mm beads, in combination with MagNA Pure DNA extraction (i.e., bead-beating + MagNA Pure, termed the “BMP” method) enhances the lysis of diverse Gram-positive microorganisms without compromising the DNA isolated from Gram-negative microorganisms.7 However, this study tested a limited number of pure microbial species over a limited range of concentrations. It did not evaluate the influence of the microbial community on gDNA extraction. Therefore, it is necessary to further evaluate the proposed BMP method using oral clinical samples under clinically relevant conditions.
The aim of this study was to evaluate the efficiency of gDNA extraction from Gram-positive and Gram-negative oral bacteria via two lysis procedures in combination with automated MagNA Pure DNA purification. Five common oral bacterial species (three Gram-positive and two Gram-negative) were subjected to gDNA extraction using either the bead-based BMP method or the original chemical lysis extraction protocol (termed the “MP” method) as the control. These five bacterial species are frequently reported to be associated with two major oral infectious diseases, namely dental caries and periodontitis.13–15 We examined the extraction efficiencies by quantitative polymerase chain reaction (qPCR) with species-specific 16S rDNA probes. To examine the influence of the microbial community on gDNA extraction, we also tested saliva samples spiked with the five oral bacterial species investigated here.
Materials and methods
Cultivation of bacterial cells
The bacterial species tested in this study were three Gram-positive bacteria (Streptococcus mutans UA159, Actinomyces naeslundii DSM 43013, and Lactobacillus casei ATCC 334) and two Gram-negative bacteria (Aggregatibacter actinomycetemcomitans Y4 and Porphyromonas gingivalis W83). S. mutans, A. naeslundii, and A. actinomycetemcomitans were grown in Brain Heart Infusion (BHI) broth or BHI agar (BD Diagnostics, Sparks, MD, USA); L. casei was grown in de Man, Rogosa, and Sharpe (MRS) broth or MRS agar (Oxoid, Basingstoke, UK); and P. gingivalis was grown in BHI broth supplemented with hemin (5 µg/mL) and menadione (1 µg/mL) or 5% horse blood agar supplemented with hemin (5 µg/mL) and menadione (1 µg/mL). S. mutans, L. casei, and P. gingivalis were grown anaerobically (80% N2, 10% H2, and 10% CO2) at 37°C. A. naeslundii and A. actinomycetemcomitans were grown at 37°C in static culture in ambient air supplemented with 10% CO2.
Saliva collection
This study was carried out between 2015 and 2017. It was approved by the Medical Ethical Committee of the VU University Medical Center Amsterdam (approval number 2011/236). At the beginning of the study, unstimulated saliva was collected on ice from a donor. The saliva donor was informed of the study design and provided verbal informed consent. The donor refrained from food and drink for 2 hours before saliva collection. The collected saliva was divided into 450 µL aliquots and stored at −80°C for further analysis or bacterial spiking.
Preparation of bacterial cells
All five bacterial species were grown to mid-log phase. Bacterial cells (1 mL cell suspensions) were then washed once by centrifugation for 2 minutes at 16,060 ×g. The pellets were resuspended in 1 mL sterile TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0). The resuspensions were then serially diluted (10-fold up to a 107-fold dilution) in TE buffer. Aliquots of each dilution were used for viable cell counts and saliva sample spiking. The remainder of each dilution was divided into 100-µL aliquots and stored at −80°C until gDNA extraction.
To obtain comparable saliva samples spiked with the five oral bacterial species, 50 µL of each bacterial dilution was added into 450 µL saliva, and this mixture was then aliquoted in 100-µL portions and stored at −80°C. These aliquots were also subjected to gDNA extraction.
To count the viable cell numbers in the bacterial suspensions, 100 µL of the 105-, 106-, and 107-fold dilutions of each species were plated on suitable agar plates. The plates were incubated for 3 to 7 days before the colonies were counted.
We repeated all experiments using three batches of all five bacterial species. For each gDNA extraction method, we tested at least two aliquots per batch of bacterial cells.
gDNA extraction methods
We used the MagNA Pure LC DNA Isolation Kit III and the MagNA Pure LC workstation (Roche Diagnostics Corp., Indianapolis, IN, USA) for gDNA extraction. The extraction method included two steps performed on the workstation: cell lysis and DNA purification.
The BMP cell lysis protocol, which uses physical disruption, was a modification of the method described by de Boer et al.7 Each of the bacterial dilutions (100 µL) was mixed with 0.25 µL glass beads (0.1 mm diameter), 250 µL lysis buffer (LGC Genomics, Berlin, Germany), and 250 µL Roti Phenol (Carl Roth, Karlsruhe, Germany). The mixture was beaten for 2 minutes at 2,100 oscillations/minute in a bead-beater (Biospec Products, Bartlesville, OK, USA). This step was repeated four times at 3-minute intervals to prevent over-heating of the samples. The samples were centrifuged at 16,060 ×g for 15 minutes at 4°C, then 250 µL of the aqueous phase were transferred into the workstation for DNA purification.
The MP cell lysis protocol was based on the manufacturer’s instructions for the MagNA Pure LC DNA Isolation Kit III. Each of the bacterial dilutions (100 µL) was mixed with 150 µL bacterial lysis buffer (Roche Diagnostics Corp.) containing proteinase K (20 mg/mL). Before being transferred into the workstation, the mixtures were incubated at 55°C for 1 hour and then 95°C for 10 minutes. After isolation, the gDNA was eluted in 100 µL elution buffer and stored at −20°C for further analysis.
Quantitative PCR
The sequences and concentrations of the primer/probe sets used in this study are shown in Table 1. Quantitative PCR (qPCR) amplification was performed in a total reaction volume of 20 µL. The reaction mixtures contained 16 µL of 2× LightCycler 480 Probes Master (Roche Diagnostics Corp.), bacterial species-specific primers and probes, and 4 µL gDNA sample. The gDNA samples included a serial dilution of the purified gDNA of each species (for standard curve generation) and the gDNA extracted by the BMP or MP methods. All qPCR amplifications were carried out in the LightCycler 480 System (Roche Diagnostics Corp.). The PCR conditions comprised initial pre-incubation at 95°C for 5 minutes, followed by 45 cycles at 95°C for 10 s and 60°C for 20 s. The data were analyzed with the LightCycler 480 SW1.5 software. Only cycle threshold (cycle number, CT) values ≤40 were considered as indicating a positive result. A standard curve for each species was generated by plotting the log of the gDNA against the CT value determined by qPCR. The gDNA concentration of each sample could then be calculated based on the relevant standard curve. The linear relationship between the log of the colony-forming units (CFU) and the corresponding gDNA concentration for each 10-fold dilution series was established, allowing the slope and correlation coefficient to be calculated for each series. The DNA extraction efficiency (%) was determined from the slope value with the following formula: [10(−1/slope)−1] × 100.
Table 1.
PCR primers and probes.
| Organism | Primer/Probe | Sequence (5′–3′) | Concentration (nM) | Reference |
|---|---|---|---|---|
| A. naeslundii | Forward | GAGCACGCCGCTCTGTA | 450 | Ciric et al.16 |
| Reverse | ACCTTGCCGCCTCCGAA | 450 | ||
| Probe | 6FAM-CCTCGTCGCCACGGTGGGTCA-BHQ1 | 150 | ||
| L. casei | Forward | TGGATGCCTTGGCACTAGGA | 900 | Haarman and Knol17 |
| Reverse | AAATCTCCGGATCAAAGCTTACTTAT | 900 | ||
| Probe | 6FAM-TATTAGTTCCGGTCCTTCATC-BHQ1 | 200 | ||
| S. mutans | Forward | CCAGGTCTTGACATCCCGAT | 450 | This study |
| Reverse | CACCTGTCTCCGATGTACCGA | 450 | ||
| Probe | 6FAM-ATTCTTAGAGATAGGAAGTTAC-BHQ1 | 150 | ||
| A. actinomycetemcomitans | Forward | GAACCTTACCTACTCTTGACATCCGAA | 90 | Boutaga et al.18 |
| Reverse | TGCAGCACCTGTCTCAAAGC | 90 | ||
| Probe | 6FAM-AGAACTCAGAGATGGGTTTGTGCCTTAGGG-BBQ | 20 | ||
| P. gingivalis | Forward | GCGCTCAACGTTCAGCC | 90 | Boutaga et al.18 |
| Reverse | CACGAATTCCGCCTGC | 90 | ||
| Probe | Cyan500-CACTGAACTCAAGCCCGGCAGTTTCAA-BBQ | 20 |
Statistical analysis
Data were analyzed with SPSS Statistics for Windows, Version 17.0 (SPSS Inc., Chicago, IL, USA). Analysis of variance with repeated measures was used to evaluate the effects of the extraction method and the presence of saliva on the DNA extraction efficiency. Student’s paired samples t-test was used to compare the DNA extraction efficiencies of the BMP method with those of the MP method. The limit of detection (LOD) for each bacterial species was determined based on the lowest concentration at which all four replicates produced a positive result. The LODs of the two extraction methods were compared with the Student’s paired samples t-test.
To understand the performance of each extraction method when the initial bacterial cell count was either low or high, CT values for 105 CFU/mL and 107 CFU/mL of each bacterial species, representing the quantity of extracted gDNA, were analyzed. These data were separated into Gram-positive and Gram-negative groups, based on their bacterial cell wall characteristics. The effects of the extraction method and the bacterial cell wall group on the CT values were analyzed with a non-parametric test (Wilcoxon signed-ranks test). Similarly, the CT values of the spiked saliva samples and the pure culture samples were also compared at 105 CFU/mL and 107 CFU/mL (CFU counts based on the pure cultures). The influence of the extraction method on the CT values was analyzed with a non-parametric test (Wilcoxon signed-ranks test).
Results
Before comparing the BMP and MP methods, we first performed a pilot experiment to determine the optimal bead-beating protocol for DNA extraction from a Gram-positive bacterial culture (A. naeslundii). In brief, the mid-log phase A. naeslundii cells were resuspended and serially diluted in TE buffer. Each dilution was split into two portions (100 µL/tube). One dilution series was subjected to bead beating four times followed by MagNA Pure purification, while the other dilution series was subjected to bead beating once before purification. This experiment was repeated in triplicate. The LOD when bead beating four times was significantly lower (∼104 CFU/mL) than that when bead beating once only (∼106 CFU/mL; data not shown). Therefore, the BMP method used in this study adopted the scheme of bead beating four times.
DNA extraction efficiency
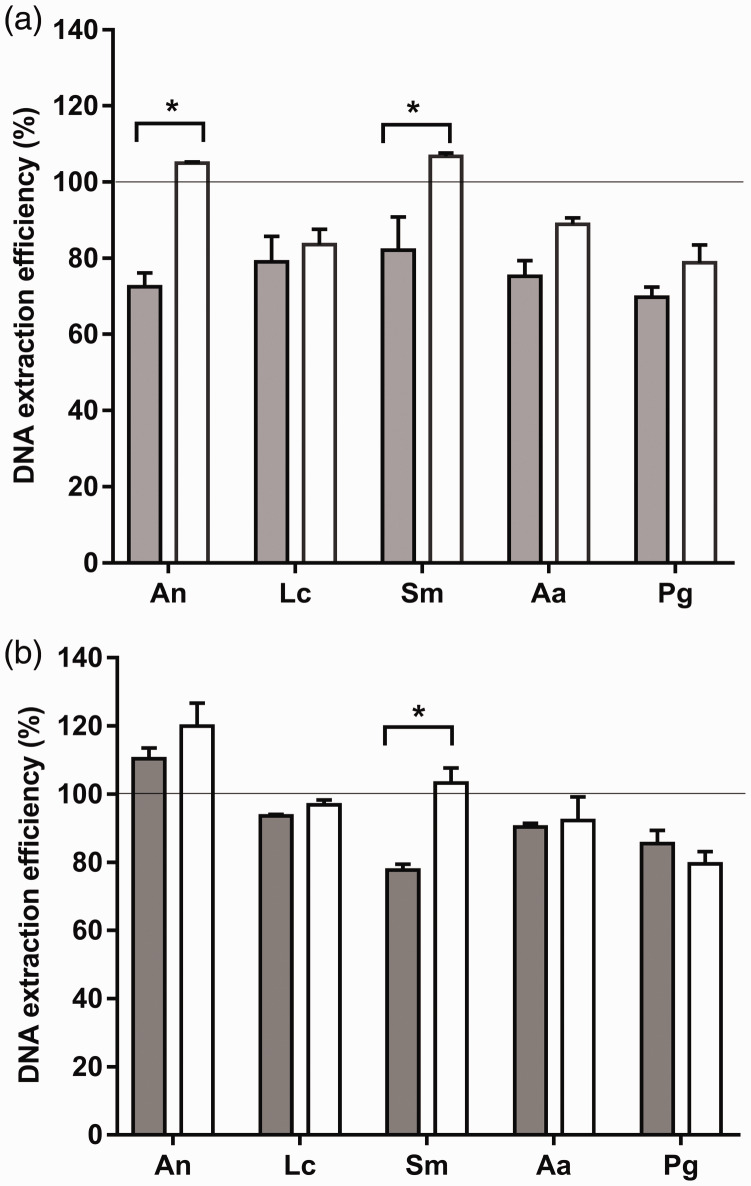
For single-species (pure) cultures, the MP method generally resulted in higher gDNA extraction efficiencies than the BMP method for all tested bacterial species (Figure 1a), although the difference between the two methods reached significance only in samples of the Gram-positive bacteria A naeslundii and S. mutans (P < 0.05). When the bacterial cultures were spiked into saliva samples, the differences in gDNA extraction efficiency between the two methods were generally less obvious. Only the S. mutans gDNA extraction efficiency was significantly higher with the MP method than with the BMP method (P < 0.05, Figure 1b). The saliva used in all experiments was supplied by a single donor.
Figure 1.
The DNA extraction efficiencies of two methods were compared using serial dilutions of five oral bacterial species. (a) Pure bacterial cultures and (b) spiked saliva samples were subjected to the BMP (gray bars) and MP (white bars) DNA extraction methods. The extracted DNA was quantified by qPCR. DNA extraction efficiency (%) was determined based on a standard curve of the log of the colony-forming units vs the corresponding gDNA concentration of a 10-fold dilution series for each species. Data represent the mean ± standard deviation (n = 3 per group), *P < 0.05. An, A. naeslundii; Lc, L. casei; Sm, S. mutans; Aa, A. actinomycetemcomitans; Pg, P. gingivalis.
Assessment of the limit of detection (LOD)
For the pure bacterial cultures, the LODs of both Gram-negative species were significantly lower with the MP method than with the BMP method (Table 2). In contrast, the extraction method used did not affect the LODs when using pure cultures of the Gram-positive species. For the spiked samples, we first quantified the bacterial cell counts for each species in the donor saliva. The donor saliva did not contain detectable levels of A. actinomycetemcomitans, L. casei or S. mutans, but it did contain A. naeslundii (3 × 104 CFU/mL) and P. gingivalis (5 × 102 CFU/mL). Therefore, it was not possible to evaluate the LODs of A. naeslundii and P. gingivalis in the spiked saliva samples. The LODs of the remaining Gram-negative and Gram-positive species (A. actinomycetemcomitans, L. casei, and S. mutans) in the spiked samples did not differ between the two extraction methods.
Table 2.
LOD determined by qPCR using DNA extracted from pure and spiked bacterial cultures.
| Sample type |
LOD (CFU/mL) |
||
|---|---|---|---|
| Species | BMP method | MP method | |
| Pure bacterial | A. naeslundii | 6 × 104 (3 × 104–1 × 105) | 6 × 104 (3 × 104–1 × 105) |
| culture | L. casei | 6 × 102 (4 × 101–3 × 103) | 4 × 102 (4 × 101–3 × 103) |
| S. mutans | 7 × 102 (1 × 102–1 × 103) | 1 × 103 (1 × 103) | |
| A. actinomycetemcomitans* | 1 × 104 (7 × 103–7 × 104) | 7 × 102 (7 × 102) | |
| P. gingivalis* | 2.8 × 103 (5 × 102–5 × 103) | 1.6 × 102 (5 × 101–5 × 102) | |
| Spiked saliva | L. casei | 4 × 101 (4 × 101) | 4 × 101 (4 × 101) |
| S. mutans | 1 × 103 (1 × 103) | 1 × 103 (1 × 103) | |
| A. actinomycetemcomitans | 7 × 102 (7 × 102) | 7 × 102 (7 × 102) | |
Data represent the mean CFU of four replicate samples (the CFU range is presented in brackets).
*P < 0.05, when comparing the LOD for samples processed with the BMP and MP methods.
CT value comparison at high and low bacterial cell counts
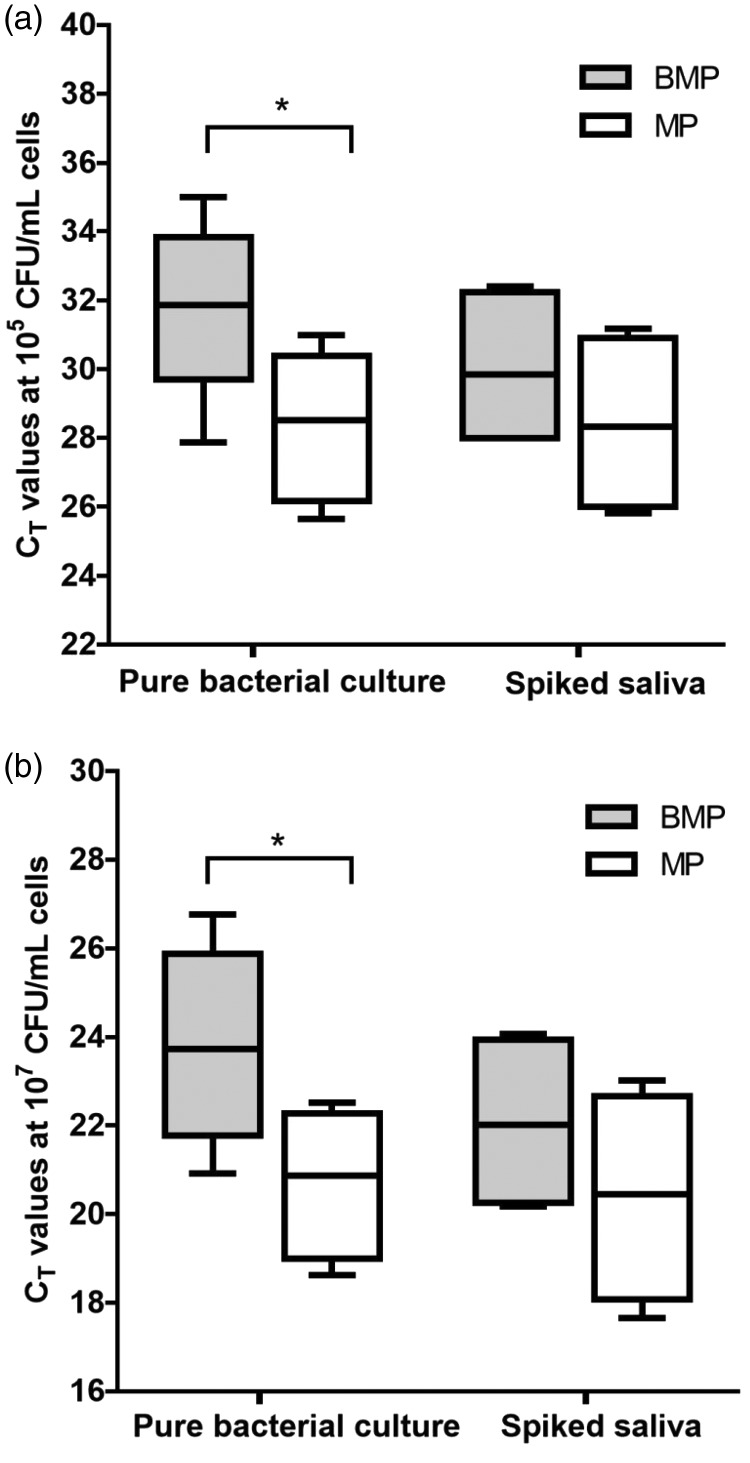
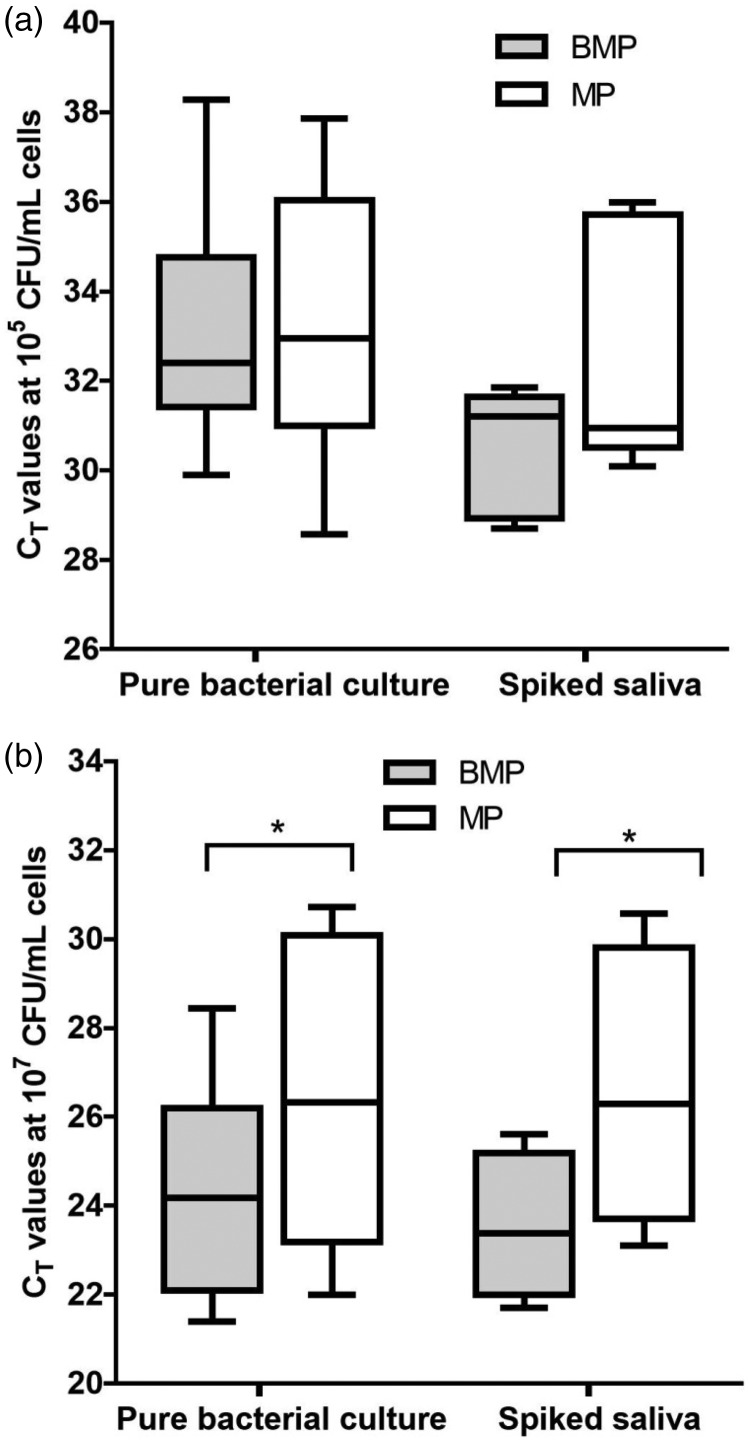
We further evaluated the efficiencies of the BMP and MP methods for samples containing low (105 CFU/mL) or high (107 CFU/mL) cell counts. The CT values of the samples were used as an indicator of the concentrations of the extracted gDNA. The distributions of the CT values of the samples with low or high cell counts in the pure bacterial cultures and the spiked samples are shown in Figure 2 (Gram-negative cultures) and Figure 3 (Gram-positive cultures).
Figure 2.
Comparison of the CT values obtained with the two gDNA extraction methods when targeting Gram-negative bacterial cultures. The samples comprised (a) 105 CFU/mL or (b) 107 CFU/mL of each bacterial species. The CT values were used as a measure of the gDNA yields when using the BMP method (gray boxes) or the MP method (white boxes). The box plots indicate the mean and range of the data (n = 3 per group), *P < 0.05.
Figure 3.
Comparison of the CT values obtained with the two gDNA extraction methods when targeting Gram-positive bacterial cultures. The samples comprised (a) 105 CFU/mL or (b) 107 CFU/mL of each bacterial species. The CT values were used as a measure of the gDNA yields when using the BMP method (gray boxes) or the MP method (white boxes). The box plots indicate the mean and range of the data (n = 3 per group), *P < 0.05.
For the pure Gram-negative bacterial cultures, the BMP method did not appear to perform well when evaluating it based on the CT values. It led to significantly higher CT values (indicative of lower gDNA concentrations) than the MP method, irrespective of the bacterial cell count in the sample (Figure 2a and Figure 2b). In contrast, we did not observe any statistically significant difference in the CT values for gDNA extracted with the BMP and MP methods in the spiked saliva samples, irrespective of the number of spiked bacterial cells.
For the Gram-positive bacterial species, the BMP method appeared to perform well when the numbers of examined bacterial cells were high. This method resulted in significantly lower CT values (indicative of higher gDNA concentrations) than the MP method in the high-cell count groups of both the pure cultures and the spiked saliva samples (Figure 3b).
Discussion
The polymicrobial nature of oral infections requires the simultaneous detection of various bacterial species in limited clinical samples. However, the differing cell wall properties of different bacterial species often necessitate distinct DNA extraction methods to ensure similar extraction efficiencies.8 Here, we found that the efficiency of the two bacterial DNA extraction methods tested was influenced by several factors, including the cell wall characteristics of the individual bacterial species, their sample milieu, and their abundance in the sample.
For pure bacterial cultures, we found that the MP method resulted in higher quantities of extracted gDNA and a lower detection limit than the BMP method. A previous study comparing the MP and BMP methods recommended the use of the BMP method because it enhances the cell lysis of Gram-positive microorganisms without any negative effects on the gDNA concomitantly isolated from Gram-negative microorganisms.7 However, our results indicated that the bead-beating step in the BMP method did not improve the LODs for Gram-positive oral bacteria and it compromised the LODs for Gram-negative oral bacteria. In the previous study,7 the samples were subjected to bead beating once for 2 minutes, whereas we beat the samples with beads four times for 2 minutes each. Our pilot experiment showed that this enhanced bead-beating protocol is necessary for reasonable gDNA extraction from A. naeslundii. This enhanced bead-beating protocol may account for the poorer performance of the BMP method in our study for the extraction of gDNA from Gram-negative bacteria, given that both the duration and speed of the bead beating influence the quality of the gDNA extracted.19 The conflicting results may also be due to the different bacterial species tested in the two studies.The study by de Boer et al.7 only evaluated methods for the extraction of DNA from pure bacterial cultures. However, samples obtained from the oral cavity always contain multiple bacterial species. Therefore, we analyzed spiked saliva samples in this study to mimic in vivo conditions. We found that the observed differences in gDNA extraction efficiency between the extraction methods when targeting pure bacterial cultures were not always observed for the spiked saliva samples. The two tested extraction methods demonstrated similar DNA extraction efficiencies for the spiked samples (except for S. mutans) and similar LODs. These results indicate that the polymicrobial milieu also influences the efficiency of a gDNA extraction method. The reasons for the contrasting results between the spiked saliva samples and the pure bacterial cultures are not clear. It has been reported that sample matrices can reduce the efficiency of DNA extraction and the sensitivity of subsequent qPCR assays.20 We did not observe reduced qPCR sensitivity in the saliva samples. On the contrary, we found improved DNA extraction efficiencies and LODs for L. casei and A. actinomycetemcomitans in spiked saliva samples. More research is required to better understand how the polymicrobial milieu affects gDNA extraction efficiency.
We also evaluated the effect of the bacterial cell count in the samples on the gDNA extraction efficiency because the efficiency of a good DNA extraction method is independent of the cell counts of the targeted bacterial species in a sample. We found that the efficiencies of the extraction methods were greatly affected by bacterial cell counts in pure cultures. However, this effect was less evident in the saliva samples. When the samples contained high numbers of Gram-positive bacteria, the MP method resulted in lower quantities of extracted gDNA than the BMP method. One possible explanation for this is that the efficiency of an extraction method is related to the total bacterial cell counts, regardless of species, in a sample. On average, a saliva sample contains 108 CFU/mL bacterial cells.21 This concentration far exceeds the concentration of the bacteria spiked into the saliva samples (the most concentrated bacterial dilution contained 107 CFU/mL bacterial cells before spiking). Therefore, the total bacterial cell counts in each spiked saliva sample were essentially constant and were unaffected by the spiked bacterial dilutions. Consequently, the extraction of gDNA from the spiked samples varied less than that from pure bacterial cultures based on the extraction method. Other factors in the saliva samples, such as polysaccharides and enzymes, may also affect the efficiency of gDNA extraction.
One limitation of this study is that saliva from only one donor was used for all experiments. However, the composition of the saliva microbiome appears to be relatively stable among healthy individuals whereas the function of the saliva microbiome is variable.22 Because our study indicates that the gDNA extraction efficiency for a specific bacterial species is correlated with the total bacterial cell count in the sample, the results reported here are unlikely to be influenced by variations in salivary microbial flora among healthy donors. Nevertheless, it is worth examining this issue in future studies to determine whether the efficiency of gDNA extraction of a specific bacterial species is influenced by the use of saliva obtained from different donors.
Conclusion
The efficiency of a DNA extraction method is not only affected by bacterial cell wall structure but also by the bacterial sample milieu. The MP method resulted in superior gDNA extraction efficiency when the samples contained a single bacterial species, whereas both of the BMP and MP methods could be applied with similar efficiencies to samples containing multiple species of bacteria.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81400505]; the Medical Scientific Research Foundation of Guangdong Province of China [grant number A2015198]; and the State Scholarship Fund of China Scholarship Council [grant number 201706385079].
ORCID iDs
Xiaolan Li https://orcid.org/0000-0003-3055-8067
References
- 1.Aas JA, Paster BJ, Stoke LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 2005; 43: 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keijser BJ, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 2008; 87: 1016–1020. [DOI] [PubMed] [Google Scholar]
- 3.Larsen T, Fiehn NE. Dental biofilm infections – an update. APMIS 2017; 125: 376–384. [DOI] [PubMed] [Google Scholar]
- 4.Marsh PD. Dental plaque as a biofilm and a microbial community – implications for health and disease. BMC Oral Health 2006; 6: S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki N, Yoshida A, Nakano Y. Quantitative analysis of multi-species oral biofilms by TaqMan Real-Time PCR. Clin Med Res 2005; 3: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liotti FM, Posteraro B, Mannu F, et al. Development of a multiplex PCR platform for the rapid detection of bacteria, antibiotic resistance, and Candida in human blood samples. Front Cell Infect Microbiol 2019; 9: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Boer R, Peters R, Gierveld S, et al. Improved detection of microbial DNA after bead-beating before DNA isolation. J Microbiol Methods 2010; 80: 209–211. [DOI] [PubMed] [Google Scholar]
- 8.Guo F, Zhang T. Biases during DNA extraction of activated sludge samples revealed by high throughput sequencing. Appl Microbiol Biotechnol 2013; 97: 4607–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auer GK, Weibel DB. Bacterial cell mechanics. Biochemistry 2017; 56: 3710–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadkarni MA, Martin FE, Hunter N, et al. Methods for optimizing DNA extraction before quantifying oral bacterial numbers by real-time PCR. FEMS Microbiol Lett 2009; 296: 45–51. [DOI] [PubMed] [Google Scholar]
- 11.Hwang KY, Kwon SH, Jung SO, et al. Miniaturized bead-beating device to automate full DNA sample preparation process for gram-positive bacteria. Lab Chip 2011; 11: 3649–3655. [DOI] [PubMed] [Google Scholar]
- 12.Kessler HH, Mühlbauer G, Stelzl E, et al. Fully automated nucleic acid extraction: MagNA Pure LC. Clin Chem 2001; 47: 1124–1126. [PubMed] [Google Scholar]
- 13.Abranches J, Zeng L, Kajfasz JK, et al. Biology of oral streptococci. Microbiol Spectr 2018; 6: 10.1128/microbiolspec.GPP3-0042-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Göhler A, Samietz S, Schmidt CO, et al. Comparison of oral microbe quantities from tongue samples and subgingival pockets. Int J Dent 2018; 2018: 2048390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brailsford SR, Shah B, Simons D, et al. The predominant aciduric microflora of root-caries lesions. J Dent Res 2001; 80: 1828–1833. [DOI] [PubMed] [Google Scholar]
- 16.Ciric L, Pratten J, Wilson M, et al. Development of a novel multi-triplex qPCR method for the assessment of bacterial community structure in oral populations. Environ Microbiol Rep 2010; 2: 770–774. [DOI] [PubMed] [Google Scholar]
- 17.Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl Environ Microbiol 2006; 72: 2359–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutaga K, van Winkelhoff AJ, Vandenbroucke-Grauls CM, et al. Periodontal pathogens: a quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol Med Microbiol 2005; 45: 191–199. [DOI] [PubMed] [Google Scholar]
- 19.Miller DN, Bryant JE, Madsen EL, et al. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol 1999; 65: 4715–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dauphin LA Moser BD, andBowen MD.. Evaluation of five commercial nucleic acid extraction kits for their ability to inactivate Bacillus anthracis spores and comparison of DNA yields from spores and spiked environmental samples. J Microbiol Methods 2009; 76: 30–37. [DOI] [PubMed] [Google Scholar]
- 21.Mantilla Gómez S, Danser MM, Sipos PM, et al. Tongue coating and salivary bacterial counts in healthy/gingivitis subjects and periodontitis patients. J Clin Periodontol 2001; 28: 970–978. [DOI] [PubMed] [Google Scholar]
- 22.Zaura E, Brandt BW, Prodan A, et al. On the ecosystemic network of saliva in healthy young adults. ISME J 2017; 11: 1218–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]