Abstract
mTOR complex 1 (mTORC1) senses nutrients to mediate anabolic processes within the cell. Exactly how mTORC1 promotes cell growth remains unclear. Here, we identified a novel mTORC1-interacting protein called protein kinase A anchoring protein 8L (AKAP8L). Using biochemical assays, we found that the N-terminal region of AKAP8L binds to mTORC1 in the cytoplasm. Importantly, loss of AKAP8L decreased mTORC1-mediated processes such as translation, cell growth, and cell proliferation. AKAPs anchor protein kinase A (PKA) through PKA regulatory subunits, and we show that AKAP8L can anchor PKA through regulatory subunit Iα. Reintroducing full-length AKAP8L into cells restored mTORC1-regulated processes, whereas reintroduction of AKAP8L missing the N-terminal region that confers the interaction with mTORC1 did not. Our results suggest a multifaceted role for AKAPs in the cell. We conclude that mTORC1 appears to regulate cell growth, perhaps in part through AKAP8L.
Keywords: mTOR complex (mTORC), mTORC1, protein synthesis, cell biology, cell proliferation, protein kinase A (PKA), A-kinase anchoring protein (AKAP), cell size, mRNA translation, nutrient sensing, scaffolding protein, AKAP8L, anabolic pathway, cAMP signaling
Introduction
Nutrients promote anabolic pathways like protein synthesis to increase cell size and proliferation. Alternatively, when nutrients are limited, catabolic processes such as autophagy are initiated to produce energy for the cell. mTORC1 controls these events by sensing nutrient availability and is often referred to as a “master regulator” of cell growth (1–4). mTORC1 is comprised of three main components: the catalytic subunit, mTOR,2 Raptor (the substrate-recognizing component), and mLST8 (a positive regulator of mTORC1). In the current model, increased nutrients such as amino acids promote mTORC1 lysosomal localization and subsequent activation (5, 6). When mTORC1 is at the lysosome, it binds to and is activated by the small G-protein, Rheb (Ras homolog enriched in brain), downstream of growth factor signaling. Once activated, mTORC1 can control multiple downstream processes such as protein translation (3, 4, 7–9).
mTORC1 is known to control protein translation through the phosphorylation of two well-characterized substrates, ribosomal S6 kinases (S6Ks) and the eIF4E-binding proteins (4EBPs) (1, 10, 11). S6K modulates translation initiation factors and ribosome biogenesis through eIF4B and S6 (12). In studies using mTORC1 active-site inhibitors (PP242 and Torin1), 4EBPs were found to play a major role in regulating cell proliferation (13). Specifically, 4EBPs inhibit protein translation through the eIF4F (eukaryotic translation initiation factor 4F) complex assembly by associating with eIF4E (14, 15). 4EBPs have also been implicated in the regulation of specific mTORC1-regulated transcripts, known as pyrimidine-rich 5′ TOP or “TOP-like” motifs (16–18). These TOP mRNAs are responsible for encoding ribosomal proteins and elongation factors. La-related protein 1 (LARP1), also modulates TOP mRNAs, and a recent study revealed that LARP1 is a direct substrate of mTORC1 (19–22). The phosphorylation of LARP1 by mTORC1 results in LARP1 dissociation from the 5′-UTR of ribosomal protein mRNAs. LARP1 acts as a molecular switch for the activation or repression of specific mTORC1-regulated transcripts. Thus, S6K, 4EBP1, and LARP1 are identified as mTORC1 substrates that regulate protein translation. Additional mTORC1 substrates, like Unc-51 like autophagy activating kinase 1 (ULK1), are involved in other biological processes like autophagy (23).
G protein–coupled receptors (GPCRs) are members of one of the most widely therapeutically targeted protein families, consisting of about 34% of all Food and Drug Administration–approved drugs (24). GPCRs mediate specific downstream signaling cascades depending on the specific G-protein coupled to the receptor (24–27). GPCRs coupled to Gαs proteins can regulate downstream targets through the secondary messenger cAMP (28). Elevated levels of cAMP activate the Ser/Thr protein kinase A (PKA). PKA is a holoenzyme and contains two catalytic subunits and two regulatory subunits (29). The regulatory subunits RI and RII isoforms exist as I/IIα and I/IIβ. RI subunits are mostly in the cytoplasm, whereas RII subunits are frequently at membrane organelles (30–37). RI/IIα are ubiquitously expressed, and RI/IIβ are enriched in specific tissues such as spinal cord, brain, endocrine, fat, liver, and reproductive tissues (33, 38). cAMP activates PKA via binding to the regulatory subunits to release the PKA catalytic subunits. The PKA catalytic subunits can then phosphorylate downstream targets such as cAMP-response element–binding protein (CREB) on Ser-133 (39). Protein kinase A anchoring proteins (AKAPs) provide compartmentalization to cAMP signaling, by tethering the regulatory subunits of PKA to various cellular localizations (25, 40). At least 40 members of the AKAP family exist (41). Because of the specificity of AKAPs, they have been considered practical targets for cancer therapeutics (42). Additionally, there has been precedent for a relationship between AKAP family members and mTORC1 (43, 44). Expression levels of AKAP1, for example, have been correlated with high mTORC1 activity and have been implicated in supporting tumor growth (44). Further research into AKAP family members might provide more targets for therapeutic treatment of mTORC1-mediated diseases. Importantly, we recently discovered that GPCRs paired to Gαs proteins increase cAMP to activate PKA, which then inhibits mTORC1 (45). PKA directly phosphorylates the mTORC1 component Raptor on Ser-791, leading to inhibition of mTORC1 activity in multiple cell lines and mouse tissue. Thus, we are actively searching for other machinery involved in the cross-talk between mTORC1 and PKA.
Here, we identify AKAP8L as a new mTORC1-binding protein in the cytoplasm. Although AKAP8L does not alter the phosphorylation of Raptor at Ser-791 leading to mTORC1 inhibition, AKAP8L appears to play a crucial role in mTORC1-mediated processes. AKAP8L promotes protein translation, cell size, and cell proliferation. Moreover, we found that the N-terminal region of AKAP8L, which is required for the interaction with mTORC1, is critical in mediating these events.
Results
AKAP8L interacts with mTORC1
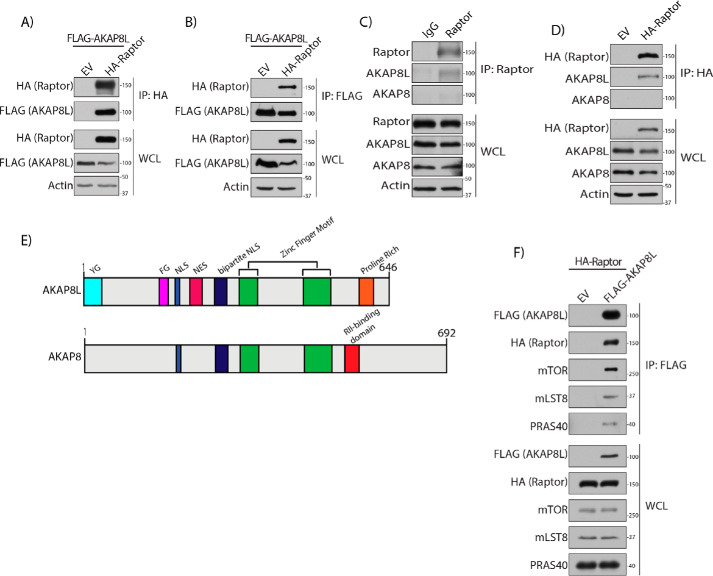
To identify potential mTORC1-interacting proteins, we expressed FLAG-tagged Raptor in human embryonic kidney 293A (HEK293A) cells and analyzed anti-FLAG immunoprecipitates by MS. We identified AKAP8L as one of the hits. To confirm that AKAP8L-mTORC1 could interact in cells, HA-tagged Raptor and FLAG-tagged AKAP8L were overexpressed in HEK293A cells (Fig. 1A). Immunoprecipitation of HA-tagged Raptor conferred interaction with FLAG-tagged AKAP8L under normal cell culturing conditions. Likewise, a reverse immunoprecipitation of FLAG-tagged AKAP8L could co-immunoprecipitate HA-tagged Raptor (Fig. 1B). Interestingly, under amino acid starvation, the interaction between FLAG-tagged AKAP8L and HA-tagged Raptor increases compared with normal conditions (Fig. S1). Moreover, endogenous AKAP8L was able to interact with both endogenous Raptor (Fig. 1C) and an overexpressed HA-tagged Raptor (Fig. 1D). AKAP8L and homologous relative AKAP8 contain 61% sequence identity (Fig. 1E) (46). Both AKAP8L and AKAP8 have zinc finger motifs and mono- and bipartite nuclear localization sequences. AKAP8L also contains a nuclear export sequence (NES), a YG domain of unknown function, an FG domain that resembles nuclear pore-like repeats (47), and a proline-rich region similar to an SRC homology 3–binding domain (48, 49). The proline-rich domain on AKAP8L exists in a similar area as the AKAP8 RII-binding region. Although AKAP8L interacts with Raptor, AKAP8 does not (Fig. 1C). FLAG-tagged AKAP8L interacted with the mTORC1 components mTOR, mLST8, and PRAS40 (Fig. 1F). Thus, AKAP8L is an mTORC1-interacting protein.
Figure 1.
AKAP8L is an mTORC1-interacting protein. A, Raptor interacts with AKAP8L. Shown is co-immunoprecipitation of FLAG-tagged AKAP8L with HA-tagged Raptor. B, co-immunoprecipitation of HA-tagged Raptor with FLAG-tagged AKAP8L. C, co-immunoprecipitation of endogenous AKAP8L with endogenous Raptor. D, co-immunoprecipitation of HA-tagged Raptor with endogenous AKAP8L. E, schematic of the domains of AKAP8L and AKAP8. YG, YG-rich domain; FG, FG repeat region; NLS, nuclear localization sequence. F, mTORC1 interacts with AKAP8L. Shown is co-immunoprecipitation of HA-tagged Raptor and endogenous mTORC1 components with FLAG-tagged AKAP8L. IP, immunoprecipitation; EV, empty vector.
AKAP8L interacts with mTORC1 through the N terminus
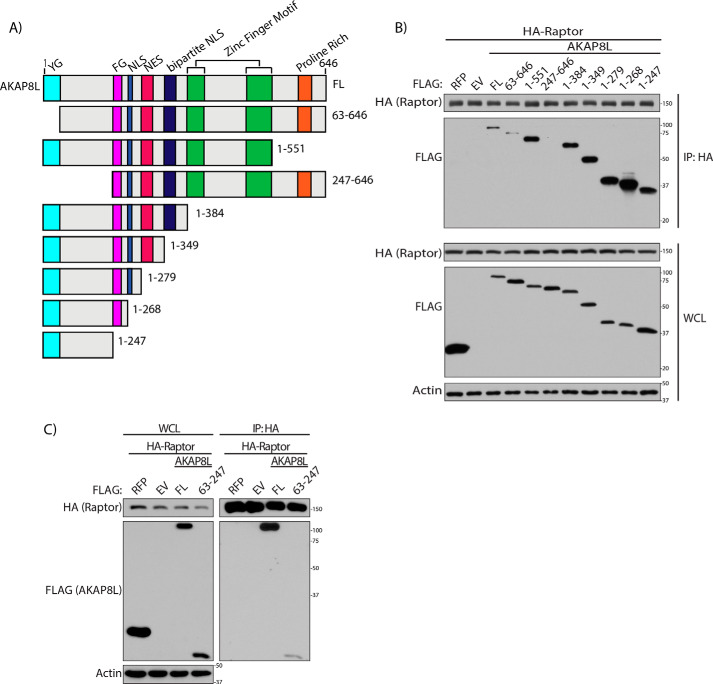
To determine the region on AKAP8L that could interact with Raptor, we generated several AKAP8L truncations. We designed and generated eight different AKAP8L truncations similar to that of another study (Fig. 2A) (49). HA-tagged Raptor was co-expressed with either full-length FLAG-tagged AKAP8L (residues 1–646), FLAG-tagged N-terminal truncations of AKAP8L (residues 63–646 or 247–646), or C-terminal truncations of AKAP8L (residues 1–551, 1–384, 1–349, 1–279, 1–268, or 1–247) (Fig. 2B). FLAG-tagged AKAP8L and the truncations of FLAG-tagged AKAP8L were expressed at similar levels in the whole-cell lysate (WCL) (Fig. 2B, bottom). Immunoprecipitation experiments of HA-tagged Raptor showed that full-length FLAG-tagged AKAP8L and most of the FLAG-tagged AKAP8L truncations could still interact with HA-tagged Raptor (Fig. 2B, top). However, FLAG-tagged AKAP8L 247–646 was unable to bind to Raptor, indicating that the N terminus of AKAP8L is critical for the interaction with mTORC1. Because FLAG-tagged AKAP8L 63–646 could still bind to mTORC1, we narrowed down amino acids 63–247 of AKAP8L to be important for the AKAP8L-mTORC1 interaction. To test whether amino acids 63–247 on AKAP8L alone are capable of binding to Raptor, we overexpressed FLAG-tagged AKAP8L 63–246 with HA-tagged Raptor (Fig. 2C). Amino acids 63–247 of AKAP8L were sufficient to bind to Raptor. Taken together, these results suggest that amino acids 63–247 of AKAP8L are necessary and sufficient for interaction with mTORC1.
Figure 2.
The N terminus of AKAP8L is required to interact with mTORC1. A, schematic of AKAP8L truncations generated based on relevant domains. B, AKAP8L amino acid region 247–646 does not interact with Raptor. Shown is co-immunoprecipitation of HA-tagged Raptor with the indicated FLAG-tagged AKAP8L truncations. C, AKAP8L amino acid region 63–247 interacts with Raptor. Shown is co-immunoprecipitation of FLAG-tagged AKAP8L amino acid region 63–247 with HA-tagged Raptor. IP, immunoprecipitation; FL, full-length; RFP, red fluorescent protein.
AKAP8L-mTORC1 interaction occurs in the cytoplasm
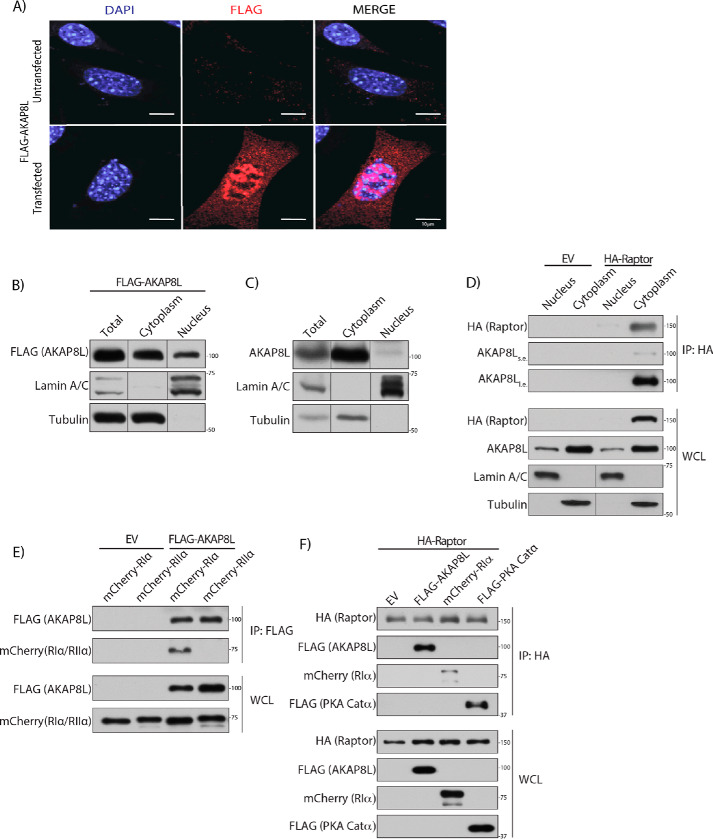
AKAP8L has been shown to localize to the nucleus (46, 50, 51). However, another study has reported that AKAP8L can also reside in the cytoplasm (49, 52). AKAP8 localizes to the nucleus and has no known NES or role in the cytoplasm (53, 54). We confirmed that AKAP8L localizes both to the nucleus and cytoplasm by overexpressing FLAG-tagged AKAP8L (red) in mouse embryonic fibroblasts, followed by immunofluorescence (Fig. 3A). AKAP8L appears not to co-localize with the tested organelle markers calreticulin (endoplasmic reticulum), Cox IV (mitochondria), LAMP2 (lysosome), and GM130 (Golgi apparatus) (Fig. S2). Complementary experiments using subcellular fractionation confirmed that FLAG-tagged AKAP8L and endogenous AKAP8L localized to both the nucleus and cytoplasm (Fig. 3, B and C). To determine where the Raptor-AKAP8L interaction occurred, we performed nuclear and cytoplasmic isolation experiments followed by immunoprecipitation of HA-tagged Raptor (55) (Fig. 3D). HA-tagged Raptor bound to endogenous AKAP8L in the cytoplasm. Thus, AKAP8L interacts with mTORC1 in the cytoplasm and not the nucleus.
Figure 3.
AKAP8L interacts with mTORC1 in the cytoplasm. A, AKAP8L localizes to the cytoplasm and nucleus. Expression of FLAG-tagged AKAP8L in mouse embryonic fibroblast cells. Red, FLAG-tagged AKAP8L (1.0 μg). B, expression of FLAG-tagged AKAP8L in the cytoplasmic and nuclear fractions of HEK293A cells. C, expression of endogenous AKAP8L in the cytoplasmic and nuclear fractions of HEK293A cells. D, Raptor interacts with AKAP8L in the cytoplasm. Co-immunoprecipitation of endogenous AKAP8L with HA-tagged Raptor. E, AKAP8L interacts with RIα. Co-immunoprecipitation of mCherry-tagged RIα with FLAG-tagged AKAP8L. F, Raptor interacts with AKAP8L and PKA machinery. Co-immunoprecipitation of FLAG-tagged AKAP8L, mCherry-tagged RIα, and FLAG-tagged PKA Catα with HA-tagged Raptor. IP, immunoprecipitation. s.e., short exposure; l.e., long exposure.
We recently showed that PKA inhibits mTORC1 activity directly through the phosphorylation of Raptor at Ser-791 (45). Because AKAPs typically anchor PKA to distinct regions of the cell through regulatory subunits, we tested whether Raptor could form a complex with AKAP8L and the catalytic and regulatory subunits of PKA. Previously, AKAP8L was shown not to associate with PKA regulatory subunit IIα in vitro (46). Consistently, FLAG-tagged AKAP8L did not interact with mCherry-tagged PKA regulatory subunit IIα in cells (Fig. 3E). Interestingly, FLAG-tagged AKAP8L could bind to PKA regulatory subunit Iα and the PKA catalytic subunit (Fig. 3F). Thus, mTORC1 can form a complex with AKAP8L, PKA regulatory subunit Iα, and PKA catalytic subunit. To determine whether AKAP8L is involved in the phosphorylation of Raptor at Ser-791, we overexpressed HA-tagged Raptor with or without FLAG-tagged AKAP8L (Fig. S3A). Cells were treated with or without forskolin, and HA-tagged Raptor was immunoprecipitated and assessed for Ser-791 phosphorylation via a phospho-PKA substrate antibody. A commercially available antibody recognizes phosphorylated proteins on Ser or Thr residues within the PKA recognition motif RRX(S*/T*). We previously showed that it is specific for Raptor Ser-791 phosphorylation (45). There was no change in Raptor Ser-791 phosphorylation when FLAG-tagged AKAP8L was overexpressed. Overexpression of full-length FLAG-AKAP8L or the region of AKAP8L that binds to mTORC1 (FLAG-AKAP8L 63–247) did not alter the phosphorylation of known mTORC1 substrates (Figs. S3 (B and C) and S4A). Likewise, depletion of AKAP8L did not change the mTORC1 activity via the phosphorylation of known mTORC1 substrates (Figs. S3 (D and E) and S4 (B and C)). Taken together, AKAP8L does not appear to regulate the phosphorylation of Raptor Ser-791 or the phosphorylation of the mTORC1 substrates S6K, 4EBP1, LARP1, or ULK1.
mTORC1 regulates many cellular processes, including protein translation. Additionally, studies in yeast have found that cAMP levels modulate cell size (56) and the rate of protein synthesis for cell division (57) and inhibit translation in mammalian cells (58). Recent work from our laboratory demonstrated that increasing cAMP levels could inhibit global protein translation in mammalian cells (45). Therefore, we were interested in investigating whether AKAP8L and the mTORC1-AKAP8L interaction had a role in translation. AKAP8L has been implicated in transcription (59) and was observed to interact with RNA helicase A (RHA), also known as DHX9 (49, 52). Interestingly, RHA has been suggested to partake in the translation process of highly structured RNAs (60, 61) and was shown to recruit to the 5′ mRNA cap structure under mTORC1-activating conditions (20). Therefore, we investigated whether RHA could be involved in our mTORC1-AKAP8L interaction. RHA has been thought to bind AKAP8L at both the N terminus and C terminus (49). Indeed, we confirmed that HA-tagged RHA could interact with FLAG-tagged AKAP8L (Fig. S5A). Next, we overexpressed increasing amounts of HA-tagged RHA to determine whether RHA could disrupt the Raptor-AKAP8L interaction (Fig. S5B). HA-tagged RHA did not complex with immunoprecipitated Myc-tagged Raptor; nor did it disrupt FLAG-tagged AKAP8L binding. Thus, RHA does not appear to partake in this interaction in this context.
AKAP8L promotes mTORC1-mediated biology
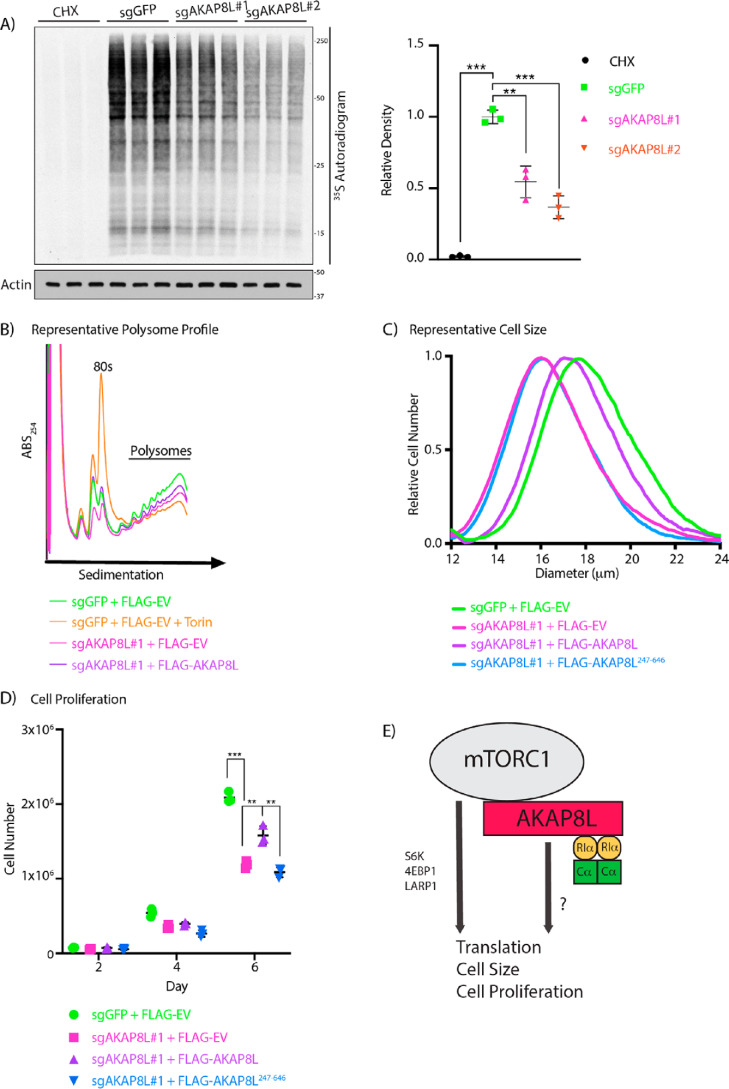
Next, we investigated whether AKAP8L could play a role in translation and mTORC1-mediated biology. To test whether AKAP8L is essential in mTORC1-mediated biology, we generated two different AKAP8L knockout (KO) HEK293A cells using two different guide RNAs via the CRISPR-Cas9 system (Fig. S6, A and B). HEK293A cells contain at least three copies of AKAP8L. All indels in the AKAP8L KO clonal cell lines resulted in frameshift mutations. Loss of AKAP8L protein expression was further confirmed by immunoblotting with an AKAP8L antibody. Interestingly, AKAP8L KO cells have a significant reduction in global protein translation when compared with control cells (Fig. 4A). A widely used technique in the mTORC1 field to measure actively translating mRNAs is polysome profiling (13, 17, 18, 62), which is based on the separation of translated mRNAs associated with polysomes compared with untranslated mRNAs. Similar to global protein translation, a loss of AKAP8L led to a reduction in actively translating mRNAs shown by a decrease in the polysome fraction (Fig. 4B). It has previously been reported that mTORC1 inhibition results in the reduction of nearly all mRNAs to some extent; as expected, inhibition of mTORC1 with the ATP mimetic Torin1 also decreased the polysome fraction (17). To confirm that the loss of protein translation is indeed due to the deletion of AKAP8L, we stably overexpressed FLAG-tagged AKAP8L in AKAP8L KO HEK293A cells and found that it could rescue the polysome fraction (Fig. 4B and Figs. S6C and S7). mTORC1 is a critical regulator of cell size and proliferation (63, 64). Because a loss of AKAP8L led to translation reduction, we examined the role of AKAP8L in cell size and proliferation. Indeed, a loss of AKAP8L significantly reduced cell size (Fig. 4C and Fig. S8A) and cell proliferation (Fig. 4D and Fig. S8B). Importantly, overexpression of FLAG-tagged full-length AKAP8L, but not the mTORC1-defective binding mutant of AKAP8L (FLAG-AKAP8L 247–646), could rescue cell size and proliferation (Fig. 4 (C and D) and Fig. S6D). Thus, AKAP8L and the region of AKAP8L that binds to mTORC1 are important for promoting mTORC1-mediated biology.
Figure 4.
AKAP8L regulates protein translation, cell size, and proliferation. A, depletion of AKAP8L decreases global protein synthesis. AKAP8L KO cells were incubated in methionine and cysteine-free Dulbecco's modified Eagle's medium for 1 h. Cycloheximide (CHX) treatment for 1 h was used as a positive control. 35S-labeled l-methionine and l-cysteine mix was then added to the medium for 10 min, and newly synthesized proteins were detected by autoradiography. Values are displayed as means ± S.D. (error bars). Significance was analyzed using Student's t test. B, loss of AKAP8L reduces the number of polysomes. AKAP8L KO cells with or without stably expressing full-length FLAG-tagged AKAP8L were subjected to polysome profiling. Negative control WT cells were treated with 100 nm Torin1 for 1 h. A representative image is shown from three biological replicates. C, loss of AKAP8L reduces cell size. The size of AKAP8L KO cells with or without stably expressing full-length FLAG-tagged AKAP8L or FLAG-tagged AKAP8L 247–646 was measured using a Coulter counter. Samples with the closest value to the mean are plotted as a representative image. Significance (p) was as follows: sgGFP versus sgAKAP8L#1 + FLAGEV, <0.01; sgGFP versus FLAGAKAP8L 247–646, <0.01; sgAKAP8L#1 + FLAGAKAP8L versus sgAKAP8L#1 + FLAGEV, <0.001; sgAKAP8L#1 + FLAGAKAP8L versus sgAKAP8L#1 + FLAGAKAP8L 247–646, <0.001. Significance was analyzed using Student's t test. The number of biological repeats is n ≥ 3. The number of technical repeats of each sample is n = 3 per experiment. D, loss of AKAP8L reduces cell proliferation. AKAP8L KO cells with or without stably expressing full-length FLAG-tagged AKAP8L or FLAG-tagged AKAP8L 247–646 were counted on the indicated days using trypan blue and a Bio-Rad automated cell counter. Values are displayed as means ± S.D. Significance was analyzed using Student's t test. The number of biological repeats is n ≥ 3. The number of technical repeats of each sample is n = 3 per experiment. E, working model of AKAP8L regulating translation, cell size, and proliferation. Significance is indicated as follows. **, p < 0.01; ***, p < 0.001.
Discussion
It is well-established that mTORC1 controls cell growth (65). Although the mechanism of this regulation has not been fully elucidated, key substrates of protein translation like 4EBP1 (14, 15, 66), S6K (18, 63, 64, 67), and LARP1 (19–22) have been discovered to regulate this process (1, 9). Here, we report a previously unidentified interaction between mTORC1 and AKAP8L. We demonstrate that amino acids 63–247 of AKAP8L bind to mTORC1. Moreover, the subcellular localization of this AKAP8L-mTORC1 interaction resides in the cytoplasm. We also find that AKAP8L can complex with the PKA holoenzyme, through regulatory subunit Iα. Deletion of AKAP8L in cells led to a reduction in protein synthesis, cell size, and proliferation. Furthermore, rescuing AKAP8L KO cells with the full-length protein, restored mTORC1-mediated processes. AKAP8L protein missing the N-terminal region, crucial to bind mTORC1, did not rescue mTORC1 biology. We propose that the mTORC1-AKAP8L interaction has an important role promoting anabolic processes like translation, through an unknown molecular mechanism (Fig. 4E).
An RNA-binding protein called S6K1 Aly/REF-like substrate (SKAR) (68, 69) has been reported to serve as a scaffolding protein between S6K1 and newly spliced mRNA (69). SKAR has been shown to regulate cell growth, as siRNA knockdown of SKAR caused a reduction in cell size (68). Similar to SKAR, we found that AKAP8L KO cells experienced a significant decrease in cell size. Like SKAR, it could be possible that AKAP8L serves as a scaffold for components involved in translation and cell growth. This could explain why the phosphorylation of known mTORC1 substrates was unaffected, yet growth was impacted. Alternatively, AKAP8L may be involved in active translation through RHA (Fig. S5A). mRNAs that are highly structured require helicase activity for better translation efficiency. This process may need a scaffolding protein, like AKAP8L, to proceed.
Another interesting similarity between SKAR and AKAP8L is the potential of being a substrate for kinases in the mTOR pathway. It has already been demonstrated that SKAR is a substrate of S6K1 (68). However, AKAP8L has never been implicated as a substrate of S6K or kinases in the mTOR signaling pathway. In efforts to find new mTORC1 substrates and expand the phosphoproteome, two MS studies identified a multitude of potential new targets (70, 71). In those studies, the AKAP8L homolog AKAP8 was among those newly identified proteins with possible mTORC1 phosphorylation sites. Interestingly, when comparing sequences between AKAP8 and AKAP8L, there were conserved potential sites that could be targeted by mTORC1 for phosphorylation. A follow-up study indeed showed possible sites on AKAP8L, similar to our prediction using the AKAP8 sites (72). AKAP8L sites Ser-297, Ser-300, and Ser-302, of note, are located proximal to the NES. It would be interesting if AKAP8L itself was a substrate of mTORC1. For example, AKAP8L phosphorylation near the NES by mTORC1 could regulate the subcellular localization and function of AKAP8L.
AKAP8L and AKAP8 share 61% protein sequence identity (46), prompting the question of how AKAP8L is able to interact with mTORC1 whereas AKAP8 is not (Fig. 1, C and D). Within the AKAP8L region that interacts with mTORC1, amino acids 63–247, there is about 30% protein sequence similarity to AKAP8, possibly indicating that the mTORC1 binding motif is not conserved. Alternatively, the NES motif on AKAP8L allows for the proper subcellular location in order to interact with mTORC1 (Fig. 1E). Another difference between AKAP8L and AKAP8 is the ability to bind RIIα (46). Surprisingly, AKAP8L was observed to bind RIα (Fig. 3, E and F), which has been shown to be more cytoplasmic (30–37). Additionally, RIα has previously been implicated in regulating mTORC1-mediated processes such as autophagy (73). Previously, AKAP8L was not considered a canonical AKAP due to the inability to bind RIIα (46); however, it is now more recognized that AKAP family members can bind RIα/RIIα or both (40). Because AKAP8L is not involved in the phosphorylation of Raptor at Ser-791 by PKA (Fig. S3A), it suggests that AKAP8L has a different role in promoting mTORC1-mediated processes. Like AKAP1, which also associates with mTORC1 (44), AKAP8L positively mediates mTORC1 biology and anchors the PKA holoenzyme. We currently have no evidence that PKA plays a direct role with respect to AKAP8L-mediated mTORC1 biology. Activation of cAMP shows a further decrease in cell proliferation in AKAP8L KO cells (Fig. S9), indicating that cAMP signaling may regulate protein translation through another pathway (74).
Experimental procedures
Cell lines and tissue culture
HEK293A cells were maintained at 37 °C with 5% CO2, cultured in high-glucose Dulbecco's modified Eagle's medium (#D5796 from Sigma) supplemented with 10% FBS (#F2442 from Sigma) and penicillin/streptomycin (#P0781 from Sigma, 100 units penicillin and 100 μg streptomycin/ml). For the generation of AKAP8L KO, HEK293A cells stably expressing FLAG-tagged full-length AKAP8L or amino acid region 247–646, a lentiviral vector (Addgene, #52962) encoding FLAG-AKAP8L full-length or FLAG-AKAP8L amino acid region 247–646 was transfected in HEK293A cells with packaging plasmids (Addgene, #12259 and #12260), and the produced virus was collected from the medium 48 h after transfection. AKAP8L KO HEK293A cells were then infected with the lentivirus, followed by selection of infected cells using blasticidin (5 μg/ml; Life Technologies REF A11139-03).
Antibodies
The following antibodies were purchased from Cell Signaling and used at the indicated dilution for Western blot analysis: S6K (#9202, 1:1000), phospho-S6K (#9234, 1:1000), 4EBP1 (#9452, 1:1500), Raptor (#2280, 1:1000), mLST8 (#3274, 1:1000), DEPTOR (#11816, 1:1000), PRAS40 (#2691, 1:1000), phospho-CREB (#9198S, 1:1000), CREB (#9197S, 1:1000), actin (#3700, 1:1000), FLAG (#2044, 1:1000), Myc (#2276, 1:2000), phospho-PKA substrate RRX(S*/T*) (#9624, 1:1000), phospho-Akt substrate RXX(S*/T*) (#9614, 1:1000), lamin A/C (#2032, 1:1000), and tubulin (#2144, 1:1000). FLAG (#F1804, 1:5000) was obtained from Sigma. HA (#sc-7392, 1:500) was from Santa Cruz Biotechnology, Inc. mCherry (#GTX128508, 1:1000) and AKAP8L (#GTX115831, 1:1000) were from GeneTex. LARP1 (#A302-087A, 1:1000) and AKAP8 (#A301-061A, 1:1000) were from Bethyl Laboratories. Horseradish peroxidase–linked secondary antibodies (#NXA931V anti-mouse (1:8000) or #NA934V anti-rabbit (1:4000)) were from GE Healthcare.
Plasmids
AKAP8L cDNA was cloned into a modified pcDNA-based vector (pCCF) with an N-terminal FLAG tag. FLAG-tagged AKAP8L truncations were based on domains described previously (49). HA-RHA was obtained from Sino Biological (#HG17921-NY).
Data availability
All data are contained within the article. Detailed procedures are available in the supporting information.
Supplementary Material
Acknowledgments
We thank members of the Buszczak and Mendell laboratories for help with polysome profiling. We are grateful to all members of the Jewell laboratory for insightful discussions. We thank Noemi Eckhoff and Greg Urquhart for technical help.
This work was supported by Cancer Prevention Research Institute of Texas (CPRIT) Scholar Recruitment of First-Time, Tenure-Track Faculty Member Grant RR150032; Cancer Prevention Research Institute of Texas (CPRIT) High-Impact/High-Risk Research Award RP160713; Welch Foundation Grant I-1927-20170325; the 2017 UT Southwestern President's Research Council Distinguished Researcher Award; American Cancer Society Institutional Research Grant ACS-IRG-17-174-13; and National Institutes of Health Grants R01GM129097-01 (to J. L. J.) and T32GM008203 (to C. H. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S9.
Author contributions—C. H. M. and J. L. J. investigation; C. H. M. and J. L. J. writing-original draft; C. H. M. and J. L. J. writing-review and editing; D. M. data curation; C. H. M. and J. L. J. conceptualization; J. L. J. supervision; J. L. J. visualization.
- mTOR
- mechanistic target of rapamycin, mammalian target of rapamycin
- S6K
- ribosomal S6 kinase
- 4EBP
- eIF4E-binding protein
- LARP1
- La-related protein 1
- ULK1
- Unc-51 like autophagy activating kinase 1
- GPCR
- G protein–coupled receptor
- PKA
- protein kinase A
- CREB
- cAMP-response element–binding protein
- AKAP
- A-kinase anchoring protein
- NES
- nuclear export sequence
- WCL
- whole-cell lysate
- RHA
- RNA helicase A
- KO
- knockout
- SKAR
- S6K1 Aly/REF-like substrate
- HEK
- human embryonic kidney.
References
- 1. Ma X. M., and Blenis J. (2009) Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- 2. Menon S., and Manning B. D. (2008) Common corruption of the mTOR signaling network in human tumors. Oncogene 27, S43–S51 10.1038/onc.2009.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laplante M., and Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saxton R. A., and Sabatini D. M. (2017) mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- 5. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., and Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., and Sabatini D. M. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jewell J. L., Russell R. C., and Guan K. L. (2013) Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139 10.1038/nrm3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jewell J. L., and Guan K. L. (2013) Nutrient signaling to mTOR and cell growth. Trends Biochem. Sci. 38, 233–242 10.1016/j.tibs.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nandagopal N., and Roux P. P. (2015) Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation 3, e983402 10.4161/21690731.2014.983402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foster K. G., and Fingar D. C. (2010) Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J. Biol. Chem. 285, 14071–14077 10.1074/jbc.R109.094003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roux P. P., and Topisirovic I. (2012) Regulation of mRNA translation by signaling pathways. Cold Spring Harb. Perspect. Biol. 4, a012252 10.1101/cshperspect.a012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hay N., and Sonenberg N. (2004) Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 10.1101/gad.1212704 [DOI] [PubMed] [Google Scholar]
- 13. Dowling R. J., Topisirovic I., Alain T., Bidinosti M., Fonseca B. D., Petroulakis E., Wang X., Larsson O., Selvaraj A., Liu Y., Kozma S. C., Thomas G., and Sonenberg N. (2010) mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328, 1172–1176 10.1126/science.1187532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin T. A., Kong X., Haystead T. A., Pause A., Belsham G., Sonenberg N., and Lawrence J. C. Jr. (1994) PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266, 653–656 10.1126/science.7939721 [DOI] [PubMed] [Google Scholar]
- 15. Pause A., Belsham G. J., Gingras A. C., Donzé O., Lin T. A., Lawrence J. C. Jr., and Sonenberg N. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371, 762–767 10.1038/371762a0 [DOI] [PubMed] [Google Scholar]
- 16. Hsieh A. C., Liu Y., Edlind M. P., Ingolia N. T., Janes M. R., Sher A., Shi E. Y., Stumpf C. R., Christensen C., Bonham M. J., Wang S., Ren P., Martin M., Jessen K., Feldman M. E., et al. (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., and Sabatini D. M. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jefferies H. B., Reinhard C., Kozma S. C., and Thomas G. (1994) Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. U.S.A. 91, 4441–4445 10.1073/pnas.91.10.4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hong S., Freeberg M. A., Han T., Kamath A., Yao Y., Fukuda T., Suzuki T., Kim J. K., and Inoki K. (2017) LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. Elife 6, e25237 10.7554/eLife.25237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tcherkezian J., Cargnello M., Romeo Y., Huttlin E. L., Lavoie G., Gygi S. P., and Roux P. P. (2014) Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 28, 357–371 10.1101/gad.231407.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fonseca B. D., Zakaria C., Jia J. J., Graber T. E., Svitkin Y., Tahmasebi S., Healy D., Hoang H. D., Jensen J. M., Diao I. T., Lussier A., Dajadian C., Padmanabhan N., Wang W., Matta-Camacho E., et al. (2015) La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J. Biol. Chem. 290, 15996–16020 10.1074/jbc.M114.621730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Philippe L., Vasseur J. J., Debart F., and Thoreen C. C. (2018) La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 46, 1457–1469 10.1093/nar/gkx1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Egan D., Kim J., Shaw R. J., and Guan K. L. (2011) The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy 7, 643–644 10.4161/auto.7.6.15123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., and Gloriam D. E. (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug. Discov. 16, 829–842 10.1038/nrd.2017.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malbon C. C., Tao J., and Wang H. Y. (2004) AKAPs (A-kinase anchoring proteins) and molecules that compose their G-protein-coupled receptor signalling complexes. Biochem. J. 379, 1–9 10.1042/bj20031648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanlon C. D., and Andrew D. J. (2015) Outside-in signaling—a brief review of GPCR signaling with a focus on the Drosophila GPCR family. J. Cell Sci. 128, 3533–3542 10.1242/jcs.175158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pavlos N. J., and Friedman P. A. (2017) GPCR signaling and trafficking: the long and short of it. Trends Endocrinol. Metab. 28, 213–226 10.1016/j.tem.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Francis S. H., and Corbin J. D. (1994) Structure and function of cyclic nucleotide-dependent protein kinases. Annu. Rev. Physiol. 56, 237–272 10.1146/annurev.ph.56.030194.001321 [DOI] [PubMed] [Google Scholar]
- 29. Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., and Sowadski J. M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 10.1126/science.1862342 [DOI] [PubMed] [Google Scholar]
- 30. Rubin C. S., Erlichman J., and Rosen O. M. (1972) Cyclic adenosine 3′,5′-monophosphate-dependent protein kinase of human erythrocyte membranes. J. Biol. Chem. 247, 6135–6139 [PubMed] [Google Scholar]
- 31. Sarkar D., Erlichman J., and Rubin C. S. (1984) Identification of a calmodulin-binding protein that co-purifies with the regulatory subunit of brain protein kinase II. J. Biol. Chem. 259, 9840–9846 [PubMed] [Google Scholar]
- 32. Nigg E. A., Schäfer G., Hilz H., and Eppenberger H. M. (1985) Cyclic-AMP-dependent protein kinase type II is associated with the Golgi complex and with centrosomes. Cell 41, 1039–1051 10.1016/S0092-8674(85)80084-2 [DOI] [PubMed] [Google Scholar]
- 33. Cadd G., and McKnight G. S. (1989) Distinct patterns of cAMP-dependent protein kinase gene expression in mouse brain. Neuron 3, 71–79 10.1016/0896-6273(89)90116-5 [DOI] [PubMed] [Google Scholar]
- 34. Salvatori S., Damiani E., Barhanin J., Furlan S., Salviati G., and Margreth A. (1990) Co-localization of the dihydropyridine receptor and the cyclic AMP-binding subunit of an intrinsic protein kinase to the junctional membrane of the transverse tubules of skeletal muscle. Biochem. J. 267, 679–687 10.1042/bj2670679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joachim S., and Schwoch G. (1990) Localization of cAMP-dependent protein kinase subunits along the secretory pathway in pancreatic and parotid acinar cells and accumulation of the catalytic subunit in parotid secretory granules following beta-adrenergic stimulation. Eur. J. Cell Biol. 51, 76–84 [PubMed] [Google Scholar]
- 36. Scott J. D. (1991) Cyclic nucleotide-dependent protein kinases. Pharmacol Ther. 50, 123–145 10.1016/0163-7258(91)90075-W [DOI] [PubMed] [Google Scholar]
- 37. Døskeland S. O., Maronde E., and Gjertsen B. T. (1993) The genetic subtypes of cAMP-dependent protein kinase—functionally different or redundant? Biochim. Biophys. Acta 1178, 249–258 10.1016/0167-4889(93)90201-Y [DOI] [PubMed] [Google Scholar]
- 38. Jahnsen T., Hedin L., Lohmann S. M., Walter U., and Richards J. S. (1986) The neural type II regulatory subunit of cAMP-dependent protein kinase is present and regulated by hormones in the rat ovary. J. Biol. Chem. 261, 6637–6639 [PubMed] [Google Scholar]
- 39. Delghandi M. P., Johannessen M., and Moens U. (2005) The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell. Signal. 17, 1343–1351 10.1016/j.cellsig.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 40. Wong W., and Scott J. D. (2004) AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5, 959–970 10.1038/nrm1527 [DOI] [PubMed] [Google Scholar]
- 41. Kapiloff M. S., Rigatti M., and Dodge-Kafka K. L. (2014) Architectural and functional roles of A kinase-anchoring proteins in cAMP microdomains. J. Gen. Physiol. 143, 9–15 10.1085/jgp.201311020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Esseltine J. L., and Scott J. D. (2013) AKAP signaling complexes: pointing towards the next generation of therapeutic targets? Trends Pharmacol. Sci. 34, 648–655 10.1016/j.tips.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pérez López I., Cariolato L., Maric D., Gillet L., Abriel H., and Diviani D. (2013) A-kinase anchoring protein Lbc coordinates a p38 activating signaling complex controlling compensatory cardiac hypertrophy. Mol. Cell. Biol. 33, 2903–2917 10.1128/MCB.00031-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rinaldi L., Sepe M., Delle Donne R., Conte K., Arcella A., Borzacchiello D., Amente S., De Vita F., Porpora M., Garbi C., Oliva M. A., Procaccini C., Faicchia D., Matarese G., Zito Marino F., et al. (2017) Mitochondrial AKAP1 supports mTOR pathway and tumor growth. Cell Death Dis. 8, e2842 10.1038/cddis.2017.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jewell J. L., Fu V., Hong A. W., Yu F. X., Meng D., Melick C. H., Wang H., Lam W. M., Yuan H. X., Taylor S. S., and Guan K. L. (2019) GPCR signaling inhibits mTORC1 via PKA phosphorylation of Raptor. Elife 8, e43038 10.7554/eLife.43038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orstavik S., Eide T., Collas P., Han I. O., Tasken K., Kieff E., Jahnsen T., and Skålhegg B. S. (2000) Identification, cloning and characterization of a novel nuclear protein, HA95, homologous to A-kinase anchoring protein 95. Biol. Cell 92, 27–37 10.1016/S0248-4900(00)88761-4 [DOI] [PubMed] [Google Scholar]
- 47. Ryan K. J., and Wente S. R. (2000) The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12, 361–371 10.1016/S0955-0674(00)00101-0 [DOI] [PubMed] [Google Scholar]
- 48. Koyama S., Yu H., Dalgarno D. C., Shin T. B., Zydowsky L. D., and Schreiber S. L. (1993) Structure of the PI3K SH3 domain and analysis of the SH3 family. Cell 72, 945–952 10.1016/0092-8674(93)90582-B [DOI] [PubMed] [Google Scholar]
- 49. Yang J. P., Tang H., Reddy T. R., and Wong-Staal F. (2001) Mapping the functional domains of HAP95, a protein that binds RNA helicase A and activates the constitutive transport element of type D retroviruses. J. Biol. Chem. 276, 30694–30700 10.1074/jbc.M102809200 [DOI] [PubMed] [Google Scholar]
- 50. Martins S. B., Eide T., Steen R. L., Jahnsen T., Skålhegg B. S., and Collas P. (2000) HA95 is a protein of the chromatin and nuclear matrix regulating nuclear envelope dynamics. J. Cell Sci. 113, 3703–3713 [DOI] [PubMed] [Google Scholar]
- 51. Martins S., Eikvar S., Furukawa K., and Collas P. (2003) HA95 and LAP2 β mediate a novel chromatin-nuclear envelope interaction implicated in initiation of DNA replication. J. Cell Biol. 160, 177–188 10.1083/jcb.200210026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Westberg C., Yang J. P., Tang H., Reddy T. R., and Wong-Staal F. (2000) A novel shuttle protein binds to RNA helicase A and activates the retroviral constitutive transport element. J. Biol. Chem. 275, 21396–21401 10.1074/jbc.M909887199 [DOI] [PubMed] [Google Scholar]
- 53. Coghlan V. M., Langeberg L. K., Fernandez A., Lamb N. J., and Scott J. D. (1994) Cloning and characterization of AKAP 95, a nuclear protein that associates with the regulatory subunit of type II cAMP-dependent protein kinase. J. Biol. Chem. 269, 7658–7665 [PubMed] [Google Scholar]
- 54. Eide T., Coghlan V., Orstavik S., Holsve C., Solberg R., Skâlhegg B. S., Lamb N. J., Langeberg L., Fernandez A., Scott J. D., Jahnsen T., and Taskén K. (1998) Molecular cloning, chromosomal localization, and cell cycle-dependent subcellular distribution of the A-kinase anchoring protein, AKAP95. Exp. Cell Res. 238, 305–316 10.1006/excr.1997.3855 [DOI] [PubMed] [Google Scholar]
- 55. Zhu X., Zelmer A., and Wellmann S. (2017) Visualization of protein-protein interaction in nuclear and cytoplasmic fractions by co-immunoprecipitation and in situ proximity ligation assay. J. Vis. Exp. 10.3791/55218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baroni M. D., Martegani E., Monti P., and Alberghina L. (1989) Cell size modulation by CDC25 and RAS2 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 2715–2723 10.1128/MCB.9.6.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tokiwa G., Tyers M., Volpe T., and Futcher B. (1994) Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature 371, 342–345 10.1038/371342a0 [DOI] [PubMed] [Google Scholar]
- 58. Gutzkow K. B., Låhne H. U., Naderi S., Torgersen K. M., Skålhegg B., Koketsu M., Uehara Y., and Blomhoff H. K. (2003) Cyclic AMP inhibits translation of cyclin D3 in T lymphocytes at the level of elongation by inducing eEF2-phosphorylation. Cell. Signal. 15, 871–881 10.1016/S0898-6568(03)00038-X [DOI] [PubMed] [Google Scholar]
- 59. Han I., Xue Y., Harada S., Orstavik S., Skalhegg B., and Kieff E. (2002) Protein kinase A associates with HA95 and affects transcriptional coactivation by Epstein-Barr virus nuclear proteins. Mol. Cell. Biol. 22, 2136–2146 10.1128/MCB.22.7.2136-2146.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hartman T. R., Qian S., Bolinger C., Fernandez S., Schoenberg D. R., and Boris-Lawrie K. (2006) RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 13, 509–516 10.1038/nsmb1092 [DOI] [PubMed] [Google Scholar]
- 61. Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W. C., and Sonenberg N. (2011) mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12, 235–245 10.1038/nrm3083 [DOI] [PubMed] [Google Scholar]
- 62. Avni D., Biberman Y., and Meyuhas O. (1997) The 5′ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 25, 995–1001 10.1093/nar/25.5.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fingar D. C., Salama S., Tsou C., Harlow E., and Blenis J. (2002) Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16, 1472–1487 10.1101/gad.995802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chung J., Kuo C. J., Crabtree G. R., and Blenis J. (1992) Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69, 1227–1236 10.1016/0092-8674(92)90643-Q [DOI] [PubMed] [Google Scholar]
- 65. Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., and Hall M. N. (1996) TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7, 25–42 10.1091/mbc.7.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Beretta L., Gingras A. C., Svitkin Y. V., Hall M. N., and Sonenberg N. (1996) Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 15, 658–664 10.1002/j.1460-2075.1996.tb00398.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jefferies H. B., Fumagalli S., Dennis P. B., Reinhard C., Pearson R. B., and Thomas G. (1997) Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16, 3693–3704 10.1093/emboj/16.12.3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Richardson C. J., Bröenstrup M., Fingar D. C., Jülich K., Ballif B. A., Gygi S., and Blenis J. (2004) SKAR is a specific target of S6 kinase 1 in cell growth control. Curr. Biol. 14, 1540–1549 10.1016/j.cub.2004.08.061 [DOI] [PubMed] [Google Scholar]
- 69. Ma X. M., Yoon S. O., Richardson C. J., Jülich K., and Blenis J. (2008) SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133, 303–313 10.1016/j.cell.2008.02.031 [DOI] [PubMed] [Google Scholar]
- 70. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., and Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 10.1126/science.1199498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yu Y., Yoon S. O., Poulogiannis G., Yang Q., Ma X. M., Villén J., Kubica N., Hoffman G. R., Cantley L. C., Gygi S. P., and Blenis J. (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 10.1126/science.1199484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang Y., Zhang Y., and Yu Y. (2017) Global phosphoproteomic analysis of insulin/Akt/mTORC1/S6K signaling in rat hepatocytes. J. Proteome Res. 16, 2825–2835 10.1021/acs.jproteome.7b00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mavrakis M., Lippincott-Schwartz J., Stratakis C. A., and Bossis I. (2006) Depletion of type IA regulatory subunit (RIalpha) of protein kinase A (PKA) in mammalian cells and tissues activates mTOR and causes autophagic deficiency. Hum. Mol. Genet. 15, 2962–2971 10.1093/hmg/ddl239 [DOI] [PubMed] [Google Scholar]
- 74. Yu F. X., Zhang Y., Park H. W., Jewell J. L., Chen Q., Deng Y., Pan D., Taylor S. S., Lai Z. C., and Guan K. L. (2013) Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 27, 1223–1232 10.1101/gad.219402.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article. Detailed procedures are available in the supporting information.