Abstract
Chikungunya fever is a re-emerging zoonotic disease caused by chikungunya virus (CHIKV), a member of the Alphavirus genus in the Togaviridae family. Only a few studies have reported on the host factors required for intracellular CHIKV trafficking. Here, we conducted an imaging-based siRNA screen to identify human host factors for intracellular trafficking that are involved in CHIKV infection, examined their interactions with CHIKV proteins, and investigated the contributions of these proteins to CHIKV infection. The results of the siRNA screen revealed that host endosomal sorting complexes required for transport (ESCRT) proteins are recruited during CHIKV infection. Co-immunoprecipitation analyses revealed that both structural and nonstructural CHIKV proteins interact with hepatocyte growth factor–regulated tyrosine kinase substrate (HGS), a component of the ESCRT-0 complex. We also observed that HGS co-localizes with the E2 protein of CHIKV and with dsRNA, a marker of the replicated CHIKV genome. Results from gene knockdown analyses indicated that, along with other ESCRT factors, HGS facilitates both genome replication and post-translational steps during CHIKV infection. Moreover, we show that ESCRT factors are also required for infections with other alphaviruses. We conclude that during CHIKV infection, several ESCRT factors are recruited via HGS and are involved in viral genome replication and post-translational processing of viral proteins.
Keywords: Chikungunya virus, endosomal sorting complexes required for transport (ESCRT), alphavirus, trafficking, viral replication, hepatocyte growth factor-regulated tyrosine kinase substrate (HGS), viral release, host factor, siRNA screen, particle release, host-pathogen interaction
Introduction
Chikungunya fever (CHIKF) is a re-emerging zoonotic disease caused by Chikungunya virus (CHIKV), a member of the genus Alphavirus, family Togaviridae (1). The major symptoms of CHIKF are an acute febrile illness accompanied by arthralgia and rash (2–4). Arthralgias may persist, and relapse, for weeks to months and even years, consequently reducing the quality of life of CHIKF patients (5, 6). During the major outbreaks of CHIKF in the Indian Ocean islands in 2005, following the re-emergence of CHIKF in Kenya (7), CHIKF became disseminated worldwide via thousands of infected travelers (8). To date, CHIKF cases have been identified in over 60 countries, including outbreaks in Europe and the Americas (9–11). Effective control measures for CHIKF are desired; however, no commercial vaccines or effective antiviral treatments currently exist. To develop new treatment strategies for CHIKV infection, a better understanding of the molecular mechanisms underlying CHIKV infection is required.
The genome of CHIKV is a positive sense, single-stranded RNA composed of two open reading frames, encoding both nonstructural and structural polyproteins. The nonstructural proteins (nsPs), consisting of nsP1–4, are required for transcription and replication of viral RNA (12, 13). The structural proteins (sPs), comprising the capsid (C) and envelope glycoproteins (E2 and E1), are the main constituents of virus particles and facilitate receptor binding and cellular entry (12, 14). The CHIKV replication cycle begins with cellular attachment and entry via endocytosis (15). The post-entry steps of CHIKV have not been fully characterized, but genome replication may take place both at the plasma membrane and type I cytopathic vacuoles (CPV-I), based on data obtained with other alphaviruses (e.g. Semliki Forest virus (SFV)) (16, 17). Colocalization of SFV nsPs and newly synthesized viral RNAs is initially observed in spherule structures near the plasma membrane (16). Then SFV replication complexes are thought to be transported to CPV-I originating from late endosomes and lysosomes (16, 17). At the late stages of the viral infection, the newly synthesized E1/E2 glycoproteins are transported to the endoplasmic reticulum, trans-Golgi network (TGN), TGN-derived CPV-II, and finally the plasma membrane (18–20). Following the formation of nucleocapsid by binding of the alphaviral C protein and RNA (21, 22), virus particles are assembled and released from the plasma membrane (20). Both CPV-I and -II have been observed in CHIKV-infected cells, and CHIKV has been shown to bud from the plasma membrane (12, 23). Although alphaviruses seem to be transported by a dynamic and unique subcellular trafficking machinery, little is known about the host factors involved in the intracellular CHIKV-trafficking processes (24, 25).
A variety of enveloped viruses can exploit the cellular endosomal sorting complexes required for transport (ESCRT) in their infection steps (26). In the ESCRT pathway, which is highly conserved across Eukarya, ubiquitinated proteins are sorted into the multivesicular bodies by deforming the membrane inward. A series of four distinct complexes (ESCRT-0, -I, -II, and -III) and VPS4 proteins are sequentially recruited for membrane deformation (27). Because HIV-1 was initially reported to engage the ESCRT pathway to acquire its envelope and pinch off viral particles during the budding step, other enveloped viruses have been shown to employ this pathway for their budding steps (26). Whereas VPS4 is required for the budding of a variety of viruses, SFV was shown to bud from cells independent of both VPS4 and ubiquitin, suggesting a possible ESCRT-independent budding process (28). However, the putative roles of ESCRT factors in infection of other alphaviruses have not been examined.
The molecular mechanisms of viral trafficking by ESCRT factors remain to be elucidated. Herpes simplex virus, hepatitis C virus, dengue virus (DENV), and Japanese encephalitis virus utilize ESCRT factors, whereas their late assembly domains (L domains) known to serve as a viral motif to recruit ESCRT factors are still undiscovered (29–36). VPS4-independent ESCRT pathways have been described in DENV and Japanese encephalitis virus infection (33). Among viruses, even within the same families, interactions between viruses and ESCRT factors appear different (26). These studies suggest that the role of ESCRT factors in viral infection differs from virus to virus.
Genome-wide screens using CHIKV-infected cells have identified a variety of host factors required for CHIKV infection; however, host trafficking factors have not been fully characterized (37, 38). In this study, we have identified components of the ESCRT machinery required for CHIKV infection through siRNA screens. Subsequently, we have characterized the interaction of the identified ESCRT factors with CHIKV and a role for ESCRT factors in CHIKV infection. Our findings clearly demonstrated the importance of the host ESCRT trafficking machinery in the CHIKV replication cycle.
Results
Imaging-based siRNA screens demonstrate that ESCRT factors are required for CHIKV infection
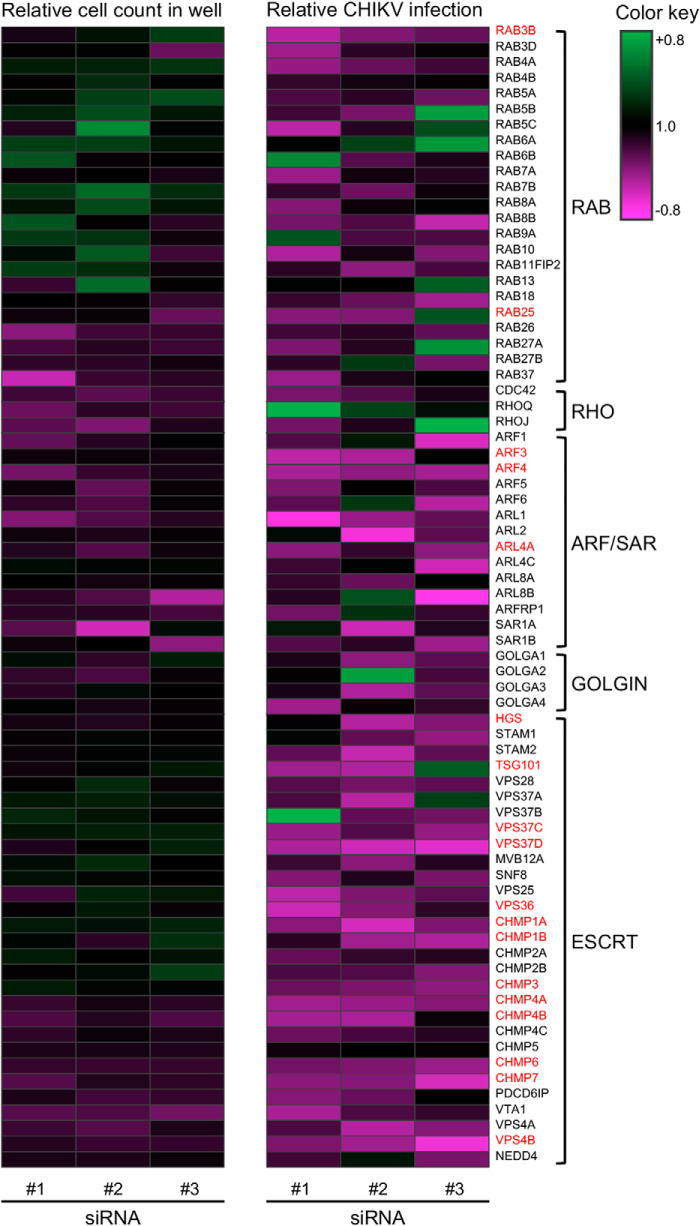
To identify the trafficking genes required for CHIKV infection, we performed imaging-based siRNA screens. siRNAs against 73 trafficking genes belonging to the RAB, RHO, ARF/SAR, GOLGIN, and ESCRT families were selected for the screen. HEK293T cells were reverse-transfected with siRNA, cultured for 48 h, and then infected with CHIKV. The CHIKV infection rate was determined by dividing the number of CHIKV-positive cells by total cells. Three different siRNAs targeting the same gene product were examined independently. The heatmap was obtained by the relative cell count and relative infection rate in the presence of each siRNA (Fig. 1 and Table S1).
Figure 1.
Heatmap for the quantitative imaging-based siRNA screen of CHIKV infection. HEK293T cells were individually reverse-transfected with three different siRNAs targeting 73 host trafficking factors and inoculated with CHIKV at an MOI of 0.1. Infection rates were determined by dividing the number of CHIKV-positive cells by total numbers of cells following IFA performed at 24 hpi. The ratio is normalized against control siRNA-reverse-transfected cells in the same plates and demonstrated according to the color key. The genes validated as hits are highlighted in red.
For the siRNA screen, siRNA treatment was considered as a positive effect if the relative CHIKV infection rate of each siRNA treatment was <60% of control siRNA treatments and the relative cell count numbers of each siRNA treatment were >80% those of control siRNA treatments. Then we selected the gene as a hit if at least two of the three siRNAs targeting the same gene product had positive effects, to exclude potential off-target effects of each siRNA. As a result of the siRNA screen, we found 18 hits, including RAB (2 of 23), ARF/SAR (3 of 13), and ESCRT (13 of 29) genes, highlighted in red (Fig. 1). ARF3 and ARF4 in the ARF/SAR family were validated as hits, consistent with a previous report demonstrating the requirement of ARFs in CHIKV replication (39). Strikingly, 13 of 29 of the investigated ESCRT genes were related to the CHIKV infection, and those genes belonged to multiple ESCRT complexes, including ESCRT-0, -I, -II, -III, and VPS4. Moreover, siRNAs against some ESCRT genes, such as VPS37D and VPS4B, had great effects on CHIKV infection without the reduction of cell numbers. These results suggested that a series of ESCRT factors are involved in CHIKV infection. Because budding of SFV was considered independent of the ESCRT pathway, interactions between ESCRT factors and CHIKV infection have not been examined previously (28). Therefore, we focused our investigations on the ESCRT factors and examined their specific roles in CHIKV infection.
The ESCRT factor HGS interacts with CHIKV sPs and nsPs
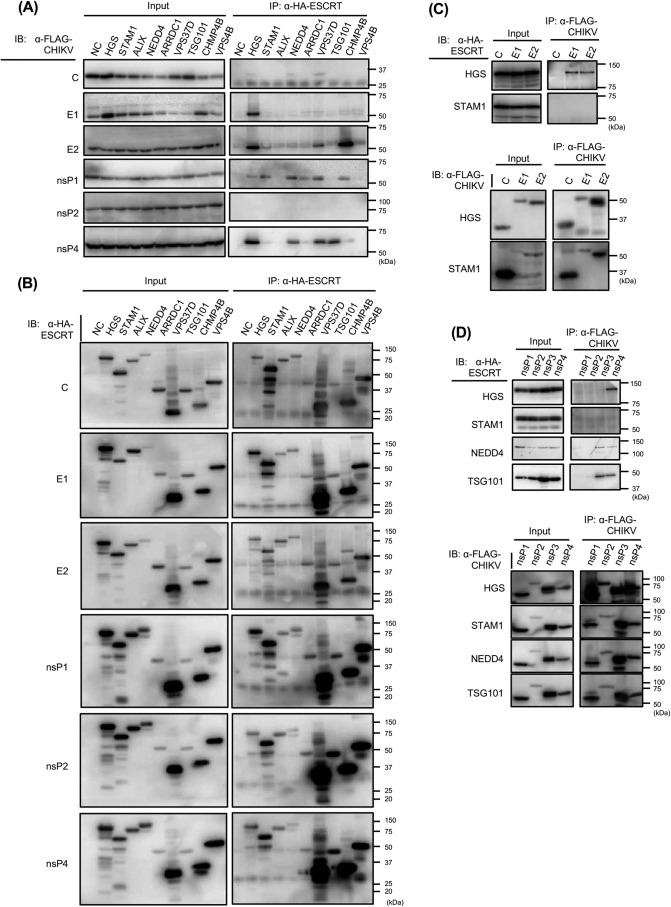
To identify the ESCRT partners physically interacting with CHIKV proteins, we exogenously introduced HA-tagged host factors and FLAG-tagged CHIKV proteins and thereafter performed anti-HA co-immunoprecipitation (co-IP). Following the co-IP experiments with anti-HA antibody, precipitated CHIKV proteins combined with HA-tagged ESCRT proteins were detected with anti-FLAG antibody. We selected eight ESCRT-related factors in these studies. HGS, also known as Hrs, and STAM1 belong to the ESCRT-0 complex and act as a sorting machinery that recognizes ubiquitinated receptors (27). TSG101 (ESCRT-I) and ESCRT-associated proteins (ALIX and NEDD4 (E3 ubiquitin protein ligase)) are reported to bind to viral L domains (26). VPS37D (ESCRT-I), CHMP4B (ESCRT-III), and VPS4B (VPS4 proteins) were considered as potential interactors with CHIKV proteins, because siRNA treatments against them markedly reduced CHIKV infection rates with little reduction of the relative cell count in the siRNA screen (Fig. 1). In addition, we also examined the arrestin domain–containing protein ARRDC1, which can recruit ESCRT factors at the plasma membrane in an L domain–dependent viral budding process (40). A total of seven CHIKV proteins were examined (C, E1, E2, and nsP1–4).
Immunoblot analyses of IP fractions showed that HGS strongly interacted with E1, E2, and nsP4 (Fig. 2, A and B). The co-IP results of nsP3 fractions were excluded, because exogenously expressed FLAG-nsP3 was nonspecifically precipitated with anti-HA antibody (data not shown). The obtained results were confirmed by reciprocal immunoprecipitation with FLAG-tagged CHIKV proteins and detection of HA-tagged ESCRT proteins with anti-HA antibody. Co-IP results showed that HGS interacted with E1, E2, and nsP4 (Fig. 2, C and D). In addition, specific bands for NEDD4 were detected in the nsP1, nsP3, and nsP4 fractions, and TSG101 bands were detected in the nsP3 and nsP4 fractions (Fig. 2D). We excluded the results of VPS37D and CHMP4B fractions, because exogenously expressed HA-VPS37D and CHMP4B were nonspecifically precipitated with anti-FLAG antibody (data not shown). These results showed that HGS interacted with both sPs and nsPs of CHIKV, suggesting that HGS may have a critical role in multiple steps of CHIKV infection.
Figure 2.
Interactions between CHIKV and ESCRT proteins analyzed by co-IP. HA-tagged host factors and FLAG-tagged CHIKV proteins were exogenously introduced into HEK293T cells. A and B, anti-HA mAb was utilized for IP. Only FLAG-tagged CHIKV protein-expressing plasmid was transfected into the cells as a negative control (NC). A, individual FLAG-tagged CHIKV protein in each fraction was detected by immunoblotting (IB) using the cell lysates. B, individual HA-tagged host factor in each fraction was detected by immunoblotting using the cell lysates. C and D, reciprocally with A and B, anti-FLAG mAb was utilized for IP, and anti-HA mAb was utilized for detection of HA-tagged ESCRT proteins. CHIKV structural proteins (C) and CHIKV nonstructural proteins (D) were examined by IP.
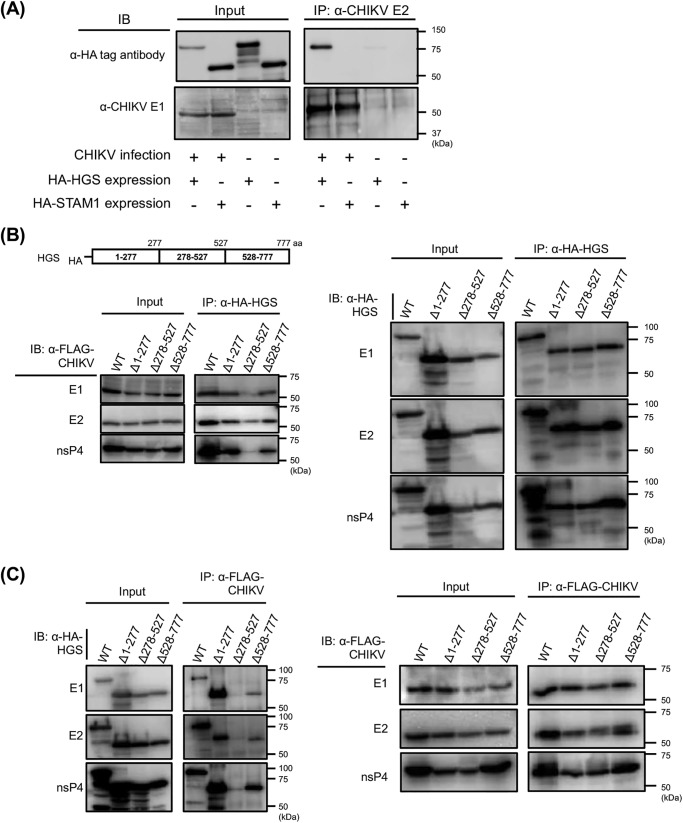
Then we examined the interaction of HGS and CHIKV E2 in CHIKV-infected cells. HEK293T cells were transfected with either HA-HGS– or HA-STAM1–expressing plasmid and thereafter inoculated with CHIKV. HA-tagged HGS but not STAM1 was recovered from the immunoprecipitates of CHIKV-infected cells with anti-CHIKV E2 antibody (Fig. 3A). These results suggest that CHIKV E2 interacts with HGS in CHIKV-infected cells. To further characterize the interactions of HGS with CHIKV proteins, we searched the HGS region responsible for binding to CHIKV proteins using truncated HGS mutants lacking amino acid residues 1–277, 278–527, or 528–777 (Δ1–277, Δ278–527, or Δ528–777, respectively). HA-tagged truncated HGS mutants were immunoprecipitated, followed by detection of CHIKV proteins. The IP fractions indicated that HGS Δ278–527 had markedly reduced binding ability to CHIKV E1, E2, and nsP4 compared with full-length HGS protein (WT), Δ1–277, and Δ528–777 (Fig. 3B). The reciprocal experiments confirmed that HGS Δ278–527 also failed to bind to CHIKV E1, E2, and nsP4 (Fig. 3C). These results suggested that the amino acid residues 278–527 of HGS play an important role in binding to CHIKV sPs and nsPs.
Figure 3.
Interactions between CHIKV and HGS analyzed by co-IP. A, either HA-tagged HGS or HA-tagged STAM1 was exogenously introduced into HEK293T cells, following CHIKV infection. Interactions between CHIKV and either HGS or STAM1 were analyzed by co-IP. Anti-CHIKV E2 mAb (5.5G9) was utilized for IP, and either anti-HA tag or anti-CHIKV-E1 antibody (CK47) was used to detect each ESCRT proteins or CHIKV E1 protein. B, genome organization of HGS is shown in the schematic at the top. HGS truncated mutants were lacking either amino acid residues 1–277, 278–527, or 528–777 from WT HA-tagged HGS, designated as Δ1–277, Δ278–527, or Δ528–777, respectively. Interactions between CHIKV and truncated HGS mutants were analyzed by co-IP. HA-tagged truncated HGS mutants or full-length HGS, and FLAG-tagged CHIKV proteins were exogenously introduced into HEK293T cells. Following anti-HA IP, individual FLAG-tagged CHIKV proteins (left) or individual HA-tagged host factors (right) in each fraction were detected by immunoblotting (IB) using the cell lysates. C, reciprocally with Fig. 3B, anti-FLAG mAb was utilized for IP, and anti-HA mAb was utilized for detection of HA-tagged HGS mutants.
CHIKV E2 and dsRNA colocalize with HGS in CHIKV-infected cells
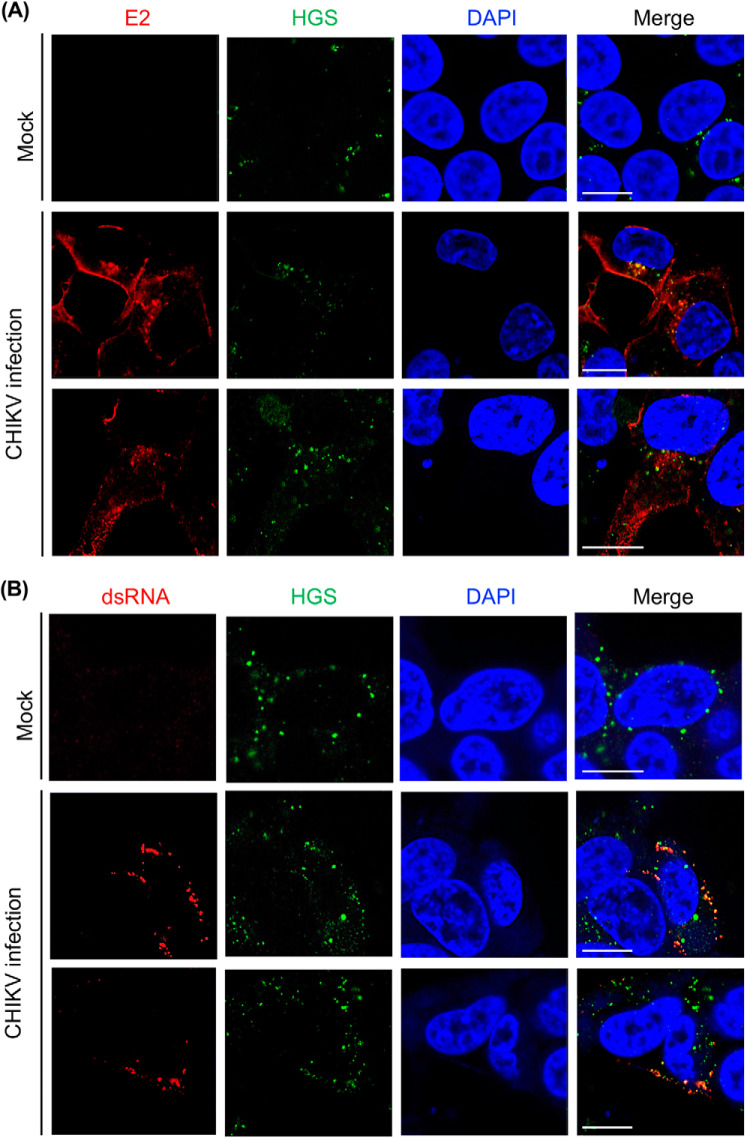
Next, we examined the interactions of HGS and CHIKV in CHIKV-infected cells by confocal microscopy. As commercially available antibodies failed to visualize endogenous HGS in Huh-7 cells, probably due to the low level of endogenous HGS protein that has been reported previously (41), we generated Huh-7 cells stably expressing HGS (HGS-Huh-7 cells). We then confirmed that the distribution of HGS was similar to the distribution of endogenous HGS in HeLa cells, as shown in the previous report (42); the dotlike signals of HGS were detected in the cytoplasm of mock-infected HGS-Huh-7 cells (Fig. 4A). In addition, the morphology and growth activity of HGS-Huh-7 cells was similar to the parent HGS cells.
Figure 4.
HGS colocalizes with CHIKV E2 and dsRNA. A, CHIKV (MOI = 10) was inoculated into HGS-Huh-7 cells. At 12 hpi, CHIKV- or mock-infected HGS-Huh-7 cells were fixed and costained for anti-CHIKV E2 (red), anti-HGS (green), and DAPI (blue). B, CHIKV (MOI = 1.0) was inoculated into HGS-Huh-7 cells. At 24 hpi, CHIKV- or mock-infected HGS-Huh-7 cells were fixed and costained for anti-dsRNA (red), anti-HGS (green), and DAPI (blue). The cells were observed by confocal microscopy (Zeiss 780 LSM confocal microscope). Scale bars, 10 μm.
Using HGS-Huh-7 cells, we first examined the localization of HGS and CHIKV E2 at 12 h post-infection (hpi) of CHIKV. As shown in the middle and bottom panels of Fig. 4A, colocalization of HGS and CHIKV E2 was observed in the cytoplasm of CHIKV-infected HGS-Huh-7 cells. Next, we examined colocalization of the HGS and CHIKV replication site. To achieve this, we applied anti-dsRNA antibody, as dsRNA is exclusively expressed in the CHIKV replication site in CHIKV-infected cells (43). Colocalization of HGS and dsRNA in the cytoplasm of CHIKV-infected cells was confirmed (middle and bottom panels, Fig. 4B). Taken together, the ESCRT protein, HGS, colocalized with CHIKV structural protein and replication sites in CHIKV-infected cells. Thus, we next examined CHIKV infection efficiency in HGS-knockdown (HGS-KD) cells.
KD of ESCRT factor, HGS, reduces CHIKV titers
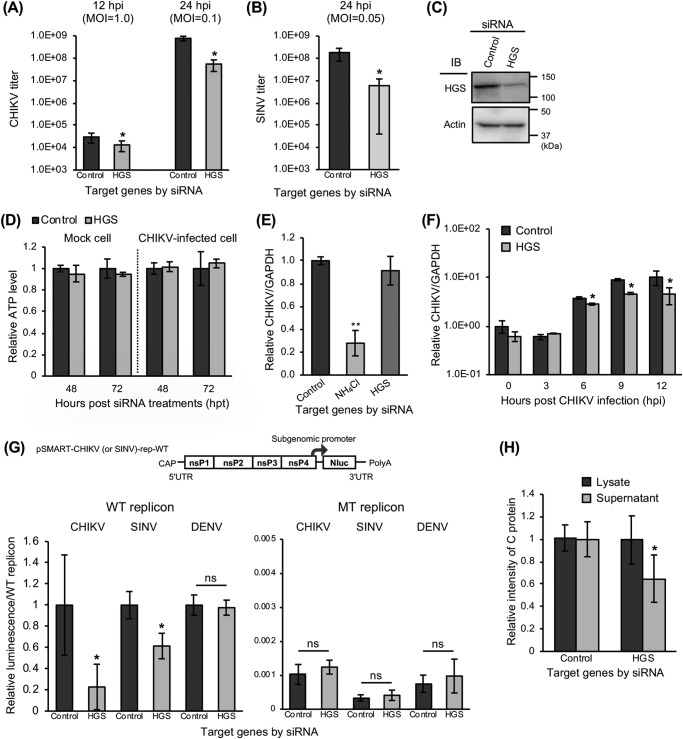
To validate the involvement of HGS in CHIKV infection, virus titers were measured in HGS-KD HEK293T cell cultures. From the set of three different siRNAs targeting HGS, a single siRNA (siRNA#3 in Table S1) was selected. We then performed plaque assays to determine CHIKV titers with cell supernatants collected at 12 and 24 hpi. The KD of HGS significantly reduced CHIKV titers at 12 hpi and showed a >10-fold reduction of CHIKV titers at 24 hpi (Fig. 5A). Furthermore, to examine the requirement of HGS in other alphavirus infection, titers of Sindbis virus (SINV) were also measured at 24 hpi in HGS-KD cells by plaque assay. The KD of HGS also inhibited infection of SINV >10-fold (Fig. 5B). The KD of HGS was confirmed by immunoblotting with siRNA-treated cell lysates (Fig. 5C). We confirmed that the KD of HGS did not affect cell viabilities during gene KD analyses, by measuring cellular ATP levels of HGS-KD mock– and CHIKV–infected cells 48 and 72 h after siRNA treatments (Fig. 5D). In these experiments, we clearly demonstrated that the KD of HGS significantly inhibited infection of both CHIKV and SINV.
Figure 5.
Significant inhibition of genome replication and post-translational steps of CHIKV infection by KD of HGS. A and B, following the reverse transfection of siRNAs against HGS, HEK293T cells were inoculated with CHIKV (MOI = 1.0 or 0.1) (A) or SINV (MOI = 0.05) (B). Virus titers were determined by plaque assay using cell supernatants collected at the indicated time points. C, protein expression levels of HGS and β-actin (control) in siRNA-treated cells were determined by immunoblotting (IB). D, relative viability of the siRNA-treated cell was evaluated by measuring cellular ATP levels 48 and 72 h after siRNA treatments and normalizing with control siRNA-treated cells. The results of mock cells are shown on the left, and CHIKV-infected cells are shown on the right. E, intracellular viral RNA levels were examined at 3 hpi of CHIKV infection (MOI = 0.1) into siRNA-treated HEK293T cells. NH4Cl-treated cells were used as a control. F, time course of CHIKV genome expression in siRNA-treated HEK293T cells inoculated with CHIKV (MOI = 0.1) for 12 h after CHIKV infection. Intracellular viral RNA levels were measured at the indicated time points. E and F, expression of viral RNA and GAPDH mRNA was determined by qRT-PCR using total RNAs extracted from trypsinized CHIKV-infected cells. G, genome organization of WT CHIKV (or SINV) replicon encoding NanoLuc (Nluc) is shown in the schematic at the top. HEK293T cells were transfected with siRNAs, incubated for 48 h, and transfected with replicon-expressing plasmids (WT CHIKV, SINV, or DENV replicon– or RNA-dependent RNA polymerase-inactivated mutant (MT) CHIKV, SINV, or DENV replicon–expressing plasmids). The luciferase activities, which represent replication efficiency, were measured at 24 hpt of replicon-expressing plasmids and normalized to each WT replicon. H, HEK293T cells were transfected with siRNAs and incubated for 48 h. Then, the cells were transfected with CHIKV-VLP-expressing plasmids. At 48 hpt of CHIKV-VLP–expressing plasmids, the culture supernatants and cells were collected and lysed with TNE lysis buffers. The amount of C protein, representing the amount of CHIKV-VLP in both lysate and supernatant samples, was then analyzed by the intensity of specific bands observed by immunoblotting. All relative data were normalized against control cells and represented as means ± S.D. (error bars), collected in triplicate and independently at least two times (D, E, F, and H). Statistical significance was assessed using Student's t test (A, B, D, F, G, and H) or one-way ANOVA with Dunnett's test (E) and is indicated by asterisks (*, p < 0.05; **, p < 0.001; ns, not significant, compared between control cells).
HGS is essential for CHIKV genome replication and post-translational steps
We further investigated which phase(s) of CHIKV replication cycle depends on HGS, using HGS-KD cells. First, we measured intracellular CHIKV RNA levels at 3 hpi by reverse transcription quantitative PCR (qRT-PCR), to focus on the early phase of the CHIKV replication cycle (0–3 hpi). NH4Cl-treated cells, which inhibit the endosomal acidification, were employed for the assay control. We confirmed that expression levels of intracellular CHIKV RNA after trypsin treatments were significantly decreased under the NH4Cl treatment at 3, 6, and 12 hpi, indicating that NH4Cl treatment inhibited amplification of CHIKV RNA (Fig. S1). Unlike NH4Cl treatment, the KD of HGS had no significant effect on intracellular viral RNA level at 3 hpi (Fig. 5E).
Second, we examined the effect of HGS on the viral genome replication steps. Intracellular CHIKV genomic and subgenomic RNA levels in HGS-KD cells were measured at various time points from 0 to 12 hpi by qRT-PCR with primer sets targeting the CHIKV E1 region. An increase in intracellular viral RNA levels was found from 6 hpi (Fig. 5F). Whereas there was no difference in intracellular viral RNA levels between HGS-KD cells and control siRNA- treated cells until 6 hpi, the KD of HGS significantly reduced viral RNA levels at 9 and 12 hpi (Fig. 5F). These results demonstrate that HGS plays a role in the replication of CHIKV RNA. In addition, we quantified intracellular viral genomic RNA levels selectively using primer sets targeting the CHIKV nsP4 region (Fig. S2). The KD of HGS significantly affected replication of CHIKV RNA at 6 and 12 hpi, indicating that HGS may be involved in replication of CHIKV genomic RNA.
We also examined CHIKV replication efficiency using the CHIKV subgenomic replicon system, encoding the luciferase gene (44). The relative luciferase intensity of a WT CHIKV subgenomic replicon was significantly inhibited by KD of HGS at 24 h post-transfection (hpt) of the CHIKV replicon RNA (Fig. 5G). Meanwhile, an RNA-dependent RNA polymerase (RdRp)-inactivated mutant type (MT) CHIKV subgenomic replicon prepared by the introduction of a GAA substitution at nsP4 was not affected by HGS-KD (Fig. 5G). These results indicate that HGS is involved in replication of CHIKV subgenomic RNA. We further examined the role of HGS in the replication of SINV using a newly established SINV subgenomic replicon system, which consists of a WT and a RdRp-inactivated MT SINV replicon, encoding the luciferase gene. We first confirmed that luminescence of WT SINV subgenomic replicon was significantly higher than the RdRp-inactivated MT SINV replicon from 12 to 96 hpt (Fig. S3). At the same time, the replication efficiency of the flavivirus DENV was also investigated using a previously established DENV subgenomic replicon system, which also consists of WT and RdRp-inactivated MT replicons, encoding the luciferase gene (45). As shown in Fig. 5G, the KD of HGS reduced relative luminescence from WT SINV subgenomic replicon, whereas the KD of HGS did not affect those of the MT SINV replicon or DENV subgenomic replicons compared with control cells. Taken together, HGS specifically regulated replication activities of CHIKV genomic and subgenomic RNA and SINV subgenomic RNA.
Finally, we investigated the post-translational steps of CHIKV infection using the CHIKV-VLP system. To examine whether the detection of sPs in the supernatants of CHIKV sP-expressing cells reflect the release of particles, the supernatants from CHIKV sP-expressing cells were fractionated through sucrose gradients (0–50%) into 24 fractions (Fig. S4, A and B). Subsequently, each fraction was applied to immunoblotting using an anti-CHIKV 181/25 polyclonal antibody (pAb) for CHIKV sPs and mAb 5.5G9 for the C protein. Both CHIKV E1 and E2 at 50 kDa and C at 30 kDa were clearly detected in fractions 19–23, demonstrating that CHIKV-VLP existed in the supernatants from the CHIKV sP-expressing cells (Fig. S4B). We confirmed that the detection of CHIKV C protein in the supernatants reflects the extracellular release of CHIKV-VLP. Using this CHIKV-VLP system, we examined release efficiency of CHIKV-VLP from HGS-KD cells. The relative intensity of C protein expression in these cells was similar to that in the cellular lysates from control siRNA-treated and HGS-KD cells; however, the levels of C protein detected in supernatants from HGS-KD cells were significantly decreased (Fig. 5H). These results indicate that HGS contributes to efficient release of CHIKV-VLP from cells.
Taken together, HGS, a member of the ESCRT proteins, plays an important role in CHIKV and SINV infection. Next, we examined the effects of other ESCRT factors in CHIKV infection.
Selected ESCRT factors are essential for the replication and post-translational steps in CHIKV infection
We further examined the role and requirement of other ESCRT factors in CHIKV infection. We selected four ESCRT factors (VPS37D, TSG101, CHMP4B, and VPS4B) that were considered to be involved in CHIKV infection, as siRNA treatment against them inhibited CHIKV infection in the imaging-based siRNA screen (Fig. 1).
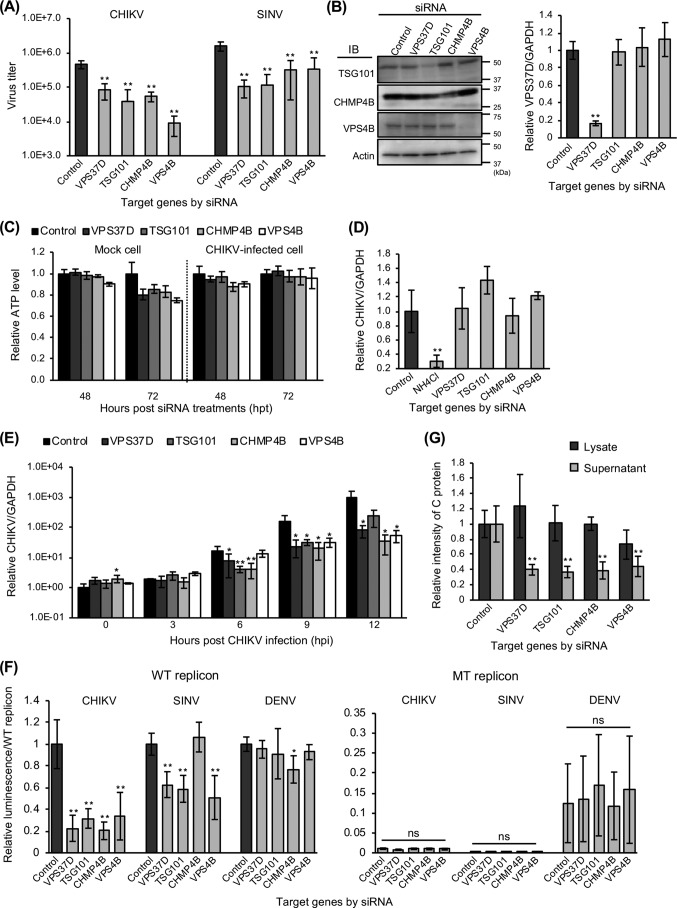
First, we confirmed that the four selected ESCRT factors were involved in CHIKV as well as SINV infection by titration of ESCRT factor-KD cells. CHIKV and SINV titers in the supernatants from virus-infected cells were significantly reduced by KD of the ESCRT factors (Fig. 6A). We validated the KD of TSG101, CHMP4B, and VPS4B by immunoblotting with siRNA-treated cell lysates and the KD of VPS37D by qRT-PCR with RNA extracted from siRNA-treated cells (Fig. 6B). These results suggested that four selected ESCRT factors are required for infection of both CHIKV and SINV. Whereas the KD of four factors did not induce cytotoxity in CHIKV-infected cells at 48 and 72 h post-siRNA treatments, it reduced cellular ATP levels >20% at 72 h after siRNA treatment in mock cells (Fig. 6C).
Figure 6.
KD of ESCRT factors inhibits genome replication and release steps of CHIKV infection. A, following the reverse transfection of siRNAs against four ESCRT factors, HEK293T cells were infected with CHIKV (MOI = 0. 1) for 48 h or SINV (MOI = 0.05) for 24 h. CHIKV titers were then determined by plaque assay using the cell supernatants. B, protein expression levels of TSG101, CHMP4B, VPS4B, and β-actin (control) in siRNA-treated cells were determined by immunoblotting (IB). The relative expression levels of VPS37D to GAPDH mRNA were determined by qRT-PCR using the total RNAs extracted from siRNA-treated cells. C, relative viability of siRNA-treated cells was evaluated by measuring cellular ATP levels from 48 and 72 h after siRNA treatments and normalizing with control siRNA-treated cells. D, intracellular viral RNA levels were examined at 2 hpi of CHIKV (MOI = 1.0) into siRNA-treated HEK293T cells. NH4Cl-treated cells were used as control. E, time course of CHIKV genome expression in siRNA-treated HEK293T cells inoculated with CHIKV (MOI = 0.1) for 12 h. Intracellular viral RNA levels were measured at the indicated time points. D and E, expression of viral RNA and GAPDH mRNA was determined by qRT-PCR using the total RNAs extracted from trypsinized CHIKV-infected cells. F, HEK293T cells were transfected with siRNAs and CHIKV, SINV, or DENV replicon-expressing plasmids. The luciferase activities were measured at 24 hpt of replicon-expressing plasmids and normalized to each WT replicon. G, HEK293T cells were transfected with siRNAs and CHIKV-VLP–expressing plasmids. At 48 hpt of CHIKV-VLP–expressing plasmids, the culture supernatants and cells were collected and lysed with TNE lysis buffer. The amount of C proteins in both lysate and supernatant samples was then analyzed by immunoblotting. All relative data were normalized against control siRNA-reverse-transfected cells and are represented as means ± S.D. (error bars), collected in triplicate and independently at least two times (B, D, E, and G). Statistical significance was assessed using one-way ANOVA with Dunnett's test and is indicated by asterisks (*, p < 0.05; **, p < 0.001; ns, not significant, compared between control siRNA-treated cells).
We next examined the effect of ESCRT factors in the early phases of CHIKV replication. The KD of each ESCRT factor did not reduce intracellular viral RNA level at 2 hpi, whereas NH4Cl treatment significantly reduced the level of intracellular CHIKV RNA (Fig. 6D). We also performed a time course assay in the presence of siRNAs targeting these four ESCRT factors. At 0 and 3 hpi, these four ESCRT factor-KD cells did not decrease intracellular viral RNA levels (Fig. 6E), similar to HGS. Increases in intracellular viral RNA levels were observed in control siRNA-treated cells from 3 to 12 hpi, suggesting that CHIKV genome replication started prior to this time point, and inhibition of CHIKV increases in RNA levels by the KD of these four ESCRT factors were observed at 6, 9, and/or 12 hpi, demonstrating that these four ESCRT factors play a role in CHIKV replication (Fig. 6E). Although siRNA treatment against four ESCRT factors had a cytotoxic effect, it did not affect the early phases of CHIKV infection. Taken together, these four ESCRT factors may play a specific role in post-entry steps (Fig. 6E). We also investigated intracellular CHIKV RNA levels using primer sets targeting the CHIKV nsP4 region, to focus on replication steps of genomic RNA (Fig. S2). The KD of four ESCRT factors significantly inhibited replication of CHIKV genomic RNA at 6 and/or 12 hpi.
We therefore examined replication activities of CHIKV as well as SINV and DENV in ESCRT factor-KD cells using CHIKV, SINV and DENV subgenomic replicon. The results of CHIKV subgenomic replicon assays demonstrated that the KD of these four ESCRT factors significantly reduced the relative luminescence of the WT CHIKV subgenomic replicon, whereas there was no effect on RdRp-inactivated MT CHIKV subgenomic replicon (Fig. 6F). The KD of VPS37D, TSG101, and VPS4B also decreased the relative luminescence of WT SINV subgenomic replicon but did not decrease WT DENV subgenomic replicon activities (Fig. 6F). These results demonstrated that VPS37D, TSG101, and VPS4B regulate genome replication steps of CHIKV and SINV specifically. In addition, CHMP4B may be involved in replication of CHIKV.
Finally, we examined the post-translational steps of CHIKV infection in ESCRT factor-KD cells using the CHIKV-VLP system. The KD of the examined ESCRT factors apparently reduced the relative intensity of C protein in supernatants but not in lysates (Fig. 6G). Taken together, ESCRT factors, including VPS37D, TSG101, CHMP4B, and VPS4B have a pivotal role in the replication and the post-translational steps of CHIKV as well as HGS (ESCRT-0). In addition, HGS, VPS37D, TSG101, and VPS4 are involved in replication of SINV, but not of DENV.
ESCRT factors are required for CHIKV infection as well as SFV
We also examined whether CHIKV infection depends on VPS4B by using dominant-negative VPS4B-expressing cells at the same time with SFV and vesicular stomatitis virus (VSV).
The exogenous expression of WT VPS4B (VPS4B WT) or ATP hydrolysis–deficient VPS4B E235Q mutant (VPS4BEQ) was induced by a doxycycline-inducible gene expression system (see “Experimental procedures” for details). After doxycycline induction for 24 h, VPS4B-expressing HEK293T cells were infected with viruses, and then the supernatants were collected from the virus-infected cells at the indicated time points (Fig. S5). The virus titers were determined by plaque assay.
The induction of VPS4BEQ expression decreased CHIKV titers at 12 and 24 hpi, whereas induction of VPS4B WT expression did not decrease titers (Fig. S5). The expression of VPS4BEQ also reduced titers of SFV and VSV at 7 and 12 hpi, whereas expression of VPS4B WT did not reduce them. These results clearly demonstrate that multiple ESCRT factors regulate propagation of some alphaviruses, including CHIKV.
Discussion
A variety of enveloped viruses exploit the ESCRT machinery to promote budding of their infectious particles from cell membranes (26); however, similar roles have not been reported for alphaviruses, including CHIKV. In this study, our specific siRNA screen targeting trafficking genes demonstrated that ESCRT factors are indeed required for efficient CHIKV infection. By co-IP analyses, we found that HGS in ESCRT-0 forms a complex with CHIKV E1, E2, or nsP4. Confocal analyses showed that HGS colocalized with E2 or dsRNA in CHIKV-infected cells. Functional analyses indicated that the KD of ESCRT factors inhibited replication and post-translational steps of CHIKV infection, although ESCRT machinery was reported to be utilized mainly for budding of viruses. Here, we propose for the first time that CHIKV requires ESCRT factors during the intracellular replication cycle.
Because there are no known L domains in CHIKV proteins, the nsPs and sPs of CHIKV would appear to interact with HGS in an L domain–independent manner. Prior studies suggest that HGS recognizes ubiquitinated cargo by its ubiquitin-interacting motif (UIM) for sorting into the multivesicular bodies (46). For instance, polyubiquitination of hepatitis C virus NS2 has been reported to mediate binding to the HGS UIM and access into the ESCRT network without viral L domains (29, 30). Our co-IP experiments determined that amino acid residues 278–527 of HGS were critical for association with the CHIKV E1, E2, and nsP4 (Fig. 3, B and C). The putative HGS UIM domain is between residues 258 and 277 (46) and immediately adjacent to residues 278–527; therefore, it is possible that deletion of residues 278–527 affected the UIM domain of HGS, and the interactions of HGS with CHIKV may depend upon ubiquitination. Further investigation using HGS mutants lacking several amino acids within 258–277 is needed to determine HGS-binding sites to CHIKV proteins. In addition to HGS, CHIKV nsPs were shown to interact with TSG101 and NEDD4 (Fig. 2D), indicating that CHIKV nsPs may interact with ESCRT factors similar to the viral proteins encoded by retroviruses and filoviruses, which have been reported to have more than one binding domain for ESCRT factors (47–49). To date, most ESCRT studies have focused on ubiquitin and certain other factors, such as L domain–binding proteins and VPS4, at the budding steps of virus particles; however, we have demonstrated unique interaction mechanisms of ESCRT factors independent of viral L domains in CHIKV infection, indicating the importance of examining detailed mechanisms in individual viruses.
Initial siRNA screens demonstrated that multiple ESCRT factors were related to CHIKV infection (Fig. 1); moreover, at least five ESCRT factors in different ESCRT complexes would be demonstrated to regulate the same CHIKV propagation steps (Figs. 5 and 6). Hence, it is possible that a series of ESCRT components may be consequently recruited after recognition of CHIKV by HGS, and they play a role in the invagination of the host membranes and biogenesis of vacuoles during the viral replication cycle. Interaction of HGS with structural proteins and inhibition of CHIKV-VLP release by KD of HGS indicate that ESCRT factors may mediate assembly and budding of CHIKV similar to the budding of HIV. Interestingly, our evidence also implicates the ESCRT machinery in regulation of the CHIKV replication. This finding is not entirely surprising because brome mosaic virus, belonging to the alphavirus-like superfamily, as well as tomato bushy stunt virus in the family Tombusviridae, have been reported to require particular ESCRT factors for formation of their RNA replication compartments (50–52). Therefore, we speculate that the KD of ESCRT factors inhibited proper formation of plasma membrane–derived replication complex spherules and/or CPV-I, in which genome replication of alphaviruses occurs, resulting in a decrease of the replication efficiency of CHIKV. Colocalization of dsRNA and HGS in the cytoplasm in the CHIKV-infected cells provides evidence in support of this hypothesis (Fig. 4B). Because the biogenesis of the unique spherules in CHIKV replication remains poorly characterized, high-resolution EM-based approaches will be needed to resolve this. It has been reported that ESCRT factors regulate various virus propagation steps relating to membrane compartments: STAM1 in ESCRT-0 interacts with TIM-1, an authentic DENV entry receptor (53), and ESCRT-III factors are recruited for the nuclear export of herpes simplex virus-1 (54). These findings indicate that comprehensive study of intracellular trafficking of viruses will be essential for understanding the precise roles of ESCRT factors in virus infection.
In consideration of the profound impact of CHIKV infection on public health, a deeper understanding of the intracellular CHIKV replication cycle is demanded. Currently, there is little information describing CHIKV replication, assembly, and budding steps; therefore, knowledge of the CHIKV replication cycle has largely been deduced from data obtained with other alphaviruses (20). However, the mechanism(s) of viral replication in association with host factors seems to be different even in the same genus. Follow-up studies to clarify the detailed mechanisms of inhibition of alphavirus infection are necessary. In the present study, we propose that ESCRT components are the key factors regulating at least two different steps of CHIKV propagation. Our findings will open molecular studies of CHIKV to broaden our knowledge regarding the pathogenesis and proliferation mechanisms of CHIKV, and we consider that further analysis of CHIKV-ESCRT interactions will provide a basis for the rational design of effective antiviral strategies.
Experimental procedures
Cells and viruses
HEK293T cells were maintained in high-glucose Dulbecco's modified Eagle's media (DMEM) (Sigma), containing 10% fetal bovine serum (FBS) (Sigma), 2 mm l-glutamine (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin (Sigma). For VPS4B-expressing HEK293T cells, Tet system–approved FBS (Clontech) was used. Vero cells, Huh-7 cells, and HGS-Huh-7 cells were grown in DMEM (Nissui) supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. All cells were cultured at 37 °C under 5% CO2.
CHIKV-SL10571 (AB455494) was kindly provided by Dr. Tomohiko Takasaki (National Institute of Infectious Diseases) (55). SFV (original strain) was kindly provided by Dr. Masahiro Hayashi (National Institute of Infectious Diseases). All experiments involving CHIKV and SFV were performed in biosafety level-3 laboratories, following standard biosafety protocols approved by the Research Center for Zoonosis Control, Hokkaido University. SINV strain AR339 was obtained from the American Type Culture Collection, and VSV (Indiana strain) from our repository was used. All viruses were initially amplified in Vero cells, and supernatants from the virus-infected cells were harvested and stored at −80 °C until use.
Antibodies
To detect CHIKV antigen, mouse anti-CHIKV mAbs 5.5G9 and 5.2B2 recognizing CHIKV C and E2 protein in each were employed as described previously (56, 57). The mAbs were able to detect CHIKV antigens by immunoblotting, immunofluorescence assay (IFA), and ELISA. Mouse anti-CHIKV mAb CK47, which recognizes domain III within CHIKV E1, was kindly provided by Dr. Tatsuo Shioda (Department of Viral Infections, Research Institute for Microbial Diseases, Osaka University) (58). Other antibodies were purchased as follows: rabbit anti-CHIKV 181/25 pAb (04-0008, IBT Bioservices); mouse anti-dsRNA mAb J2 (10010200, English & Scientific Consulting); rabbit anti-CHMP4B and anti-VPS4B pAbs (ab105767 and ab224736, Abcam); rabbit anti-HGS pAb (PA5-27491, Thermo Fisher Scientific); rabbit anti-TSG101 pAb (14497-1-AP, Proteintech); mouse anti-FLAG M2 mAb, mouse anti-HA mAb HA-7 and mouse horseradish peroxidase (HRP)-conjugated anti-FLAG M2 mAb (F3165, H9658, and A8592, Sigma); and mouse HRP-conjugated anti-HA tag mAb and rabbit HRP-conjugated anti-β-actin pAb (M180-7 and PM053-7, MBL).
Plasmids
FLAG-tagged CHIKV protein-expressing plasmids were generated by PCR-amplified CHIKV genome from CHIKV SL10571 into a pCXSN expression vector, which was created by removing the Myc tag from pCMV-Myc (Clontech) and adding XhoI, SalI, and NotI recognition sites (59). HA-tagged ESCRT protein–expressing plasmids were also generated by PCR-amplified ESCRT genome from total RNAs of HEK293T cells into a pCXSN expression vector. The HA-tagged HGS truncation mutant-expressing plasmids were derived from the WT HGS backbone by deleting either amino acids 1–277, 278–527, or 528–777 by PCR amplification.
CHIKV-VLP–expressing plasmid (pCXSN-CHIKV-VLP) was constructed as described previously (60). The only CHIKV sP genes (C-E3-E2-6K-E1) from the SL10571 strain were inserted into a pCXSN expression vector. CHIKV-VLPs were released in culture supernatants of pCXSN-CHIKV-VLP plasmid-transfected cells and were unable to produce progeny particles because of a lack of nsP genes. All plasmids driven by pCXSN expression vector were transfected to the HEK293T or Huh-7 cells using Lipofectamine 3000 (Thermo Fisher Scientific).
CHIKV subgenomic replicon–expressing plasmid (pSMART-CHIKV-rep-WT) and RdRp-inactivated subgenomic replicon–expressing plasmid (pSMART-CHIKV-rep-MT), which has no RdRp activity, were used as negative control as previously described (44). SINV subgenomic replicon–expressing plasmid (pSMART-SINV-rep-WT) and RdRp-inactivated subgenomic replicon–expressing plasmid (pSMART-SINV-rep-MT) were also constructed in a similar fashion to the CHIKV subgenomic replicon–expressing plasmids. Briefly, the full-length genome sequence of SINV AR339 strain was inserted into the pSMART-LCKan vector (Lucigen), and thereafter, the sequences encoding the structural polyprotein were replaced by a NanoLuc luciferase sequence derived from pNL2.1 vector (Promega). The pSMART-SINV-rep-MT had introduced amino acid mutations from GDD to GAA in motif C of pSMART-SINV-rep-WT, which is the catalytic core of the RdRp (the GDD motif); therefore, luciferase activity represented genome replication efficiency.
DENV subgenomic reporter replicon and an RdRp-inactivated DENV replicon, which has no detective RdRp activity, were used as described previously (45). The DENV replicon plasmid was transfected into HEK293T cells using Lipofectamine 3000. The sequences of all the above plasmids were confirmed by Sanger sequencing (ABI 3100).
Establishment of an Huh-7 cell line overexpressing HGS
The HGS-Huh-7 cell line was obtained by a retrovirus vector pseudotyped with the vesicular stomatitis virus G glycoprotein (VSV-G), using Platinum-E packaging cells (61). Briefly, HGS-expressing plasmid (pMX-HGS) was constructed by inserting the HGS gene into pMX-IRES-bd vector. The Platinum-E packaging cell was then transfected with pMX-HGS and pCMV-VSV-G by FuGENE HD transfection reagent (Promega). The supernatants containing retrovirus vector were collected after incubation of the cells at 37 °C for 96 h. The HGS-Huh-7 cell was established by infecting Huh-7 cells with the collected retroviral vector and cultured in the presence of 10 μg/ml blasticidin S HCl (Invitrogen). The HGS-Huh-7 cells were maintained by the same methods as used for Huh-7 cells.
RNAi
Silencer Select siRNAs, targeting 73 trafficking genes, and control siRNA were purchased from Thermo Fisher Scientific. For an imaging-based siRNA screen, a set of three different siRNAs targeting the same genes was prepared to rule out potential off-target effects of individual siRNAs. HEK293T cells were used for the RNAi assays because of their permissiveness to CHIKV infection and to high-efficiency of siRNA transfection. In all RNAi assays, siRNAs (at 10 nm final concentration) were reverse-transfected into the HEK293T cells using Lipofectamine RNAiMAX (Thermo Fisher Scientific) 48 h before virus infection or plasmid transfection.
IFA
CHIKV-infected cells were fixed with 4% paraformaldehyde (Nacalai Tesque). After fixation, the cells were permeabilized with PBS (Nissui) containing 0.1% Triton X-100 (Nacalai Tesque) for 10 min, blocked with 1% BSA fraction V (Sigma) in PBS, and then reacted with the indicated primary antibodies in PBS overnight at 4 °C. After washing with PBS three times, cells were incubated with a 1:1,000 dilution of goat anti-mouse IgG Alexa Fluor 488– or 594–conjugated or anti-rabbit IgG Alexa Fluor 488–conjugated secondary antibodies (Thermo Fisher Scientific) in PBS for 1 h at room temperature. The cells were then incubated with DAPI (Thermo Fisher Scientific) (1:2,000 dilution) for 10 min. Immunopositive signals were confirmed under a fluorescent microscope (Olympus) with appropriate barrier and excitation filters.
Imaging-based siRNA screen
siRNAs were reverse-transfected into HEK293T cells in 96-well plates. Control siRNA reverse-transfected cells were prepared in all screening plates. At 24 hpi of CHIKV (multiplicity of infection (MOI) = 0.1), IFA was performed with mAb 5.5G9 recognizing C protein and DAPI. Quantitative imaging data for the screen was obtained by an IN Cell Analyzer 2000 (GE Healthcare), and the obtained images were analyzed using the IN Cell developer Toolbox software (GE Healthcare). Relative infection rates were then calculated by dividing the number of CHIKV-positive cells recognized with mAb 5.5G9 by the total number of cell nuclei stained with DAPI.
Co-IP
HEK293T cells were co-transfected with an HA-tagged ESCRT protein-expressing plasmid and a FLAG-tagged CHIKV protein-expressing plasmid. As negative control, only CHIKV protein-expressing plasmid was transfected into HEK293T cells. In the co-IP experiments of CHIKV-infected cells, HEK293T cells were transfected with HA-tagged ESCRT-expressing plasmids and then infected with CHIKV (MOI = 1.0). The cells were lysed in either high-NP-40 cell lysis buffer (1% NP-40, 250 mm NaCl, and 50 mm Tris-HCl (pH 7.4)) or low-NP-40 cell lysis buffer (0.125% NP-40, 250 mm NaCl, and 50 mm Tris-HCl (pH 7.4)) for CHIKV sP- or nsP-expressing cells, respectively. Both lysis buffers contained complete protease inhibitor mixture (Roche Diagnostics). The lysates were incubated at 4 °C overnight with Dynabeads Protein G (Thermo Fisher Scientific) and the indicated antibodies. Cell lysate immunoprecipitates were analyzed by SDS-PAGE and immunoblotting using HRP-conjugated mAbs.
Immunoblotting
Samples were lysed with an indicated lysis buffer, mixed with the equal amount of SDS-PAGE sample buffer (0.5 m Tris-HCl (pH 6.8), 10% SDS, and 11% glycerol) and fractionated by SDS-PAGE. Then separated proteins were transferred to a polyvinylidene difluoride filter (Merck Millipore) using a semidry system (Bio-Rad).
The membrane was blocked with 5% skimmed milk (Megmilk Snow Brand) in TBS (50 mm Tris-HCl (pH 7.5) and 150 mm NaCl) containing 0.1% Tween 20 (TBST) for 1 h at room temperature. The membrane was then washed with TBST and subsequently incubated for 1 h at room temperature with the indicated primary antibody in TBST containing 5% skimmed milk. The membrane was then washed three times with TBST and treated with TBST containing goat anti-rabbit or mouse IgG HRP-conjugated secondary antibody (Cell Signaling Technology, Tokyo, Japan) (1:1,000 dilution) for 1 h at room temperature. After washing three times with TBST, the proteins were detected using Immobilon Western HRP-Substrate (Merck Millipore), according to the manufacturer's instructions. The chemiluminescence signals were visualized with the VersaDoc 5000MP imager (Bio-Rad), and obtained images were analyzed using Quantity One software (Bio-Rad).
Confocal microscopy
To detect localization of HGS, HGS-Huh-7 cells were cultured on chambered coverglasses. IFA was performed with CHIKV-infected cells using the indicated antibodies. Confocal images of CHIKV-infected cells were acquired by a Zeiss 780 LSM confocal microscope (Carl Zeiss), and colocalization was analyzed using ZEN 2011 software (Carl Zeiss).
Plaque assay
CHIKV (MOI = 1.0 or 0.1) or SINV (MOI = 0.05) was infected to siRNA reverse-transfected HEK293T cells in 24-well plates. Culture supernatants from virus-infected cells were inoculated onto Vero cells in 24-well plates after 10-fold serial dilution with DMEM containing 2% FBS. Then the cells were incubated for 1 h to allow virus attachment. After removal of virus-containing supernatants, cells were added with overlay media (Eagle's minimum essential media (Nissui) containing 5% FBS and 1.25% methyl cellulose) and incubated for 3 days. To count plaque numbers, cells were fixed by buffered formalin and stained by 1% crystal violet solution in 70% ethanol. The titers (pfu/ml) were determined by taking the plaque numbers for a dilution and the inverse of the total dilution factor. The relative CHIKV titers were normalized against control siRNA-reverse-transfected cells.
Validation of RNAi
The expression levels of targeted genes of siRNA-treated cells were examined by immunoblotting. At 48 h after reverse transfection of siRNAs, HEK293T cells in 24-well plates were collected. Collected cells were then lysed with TNE lysis buffer (1% NP-40, 150 mm NaCl, 5 mm EDTA, 20 mm Tris-HCl (pH 7.5), and 10% glycerol) supplemented with complete Protease inhibitor mixture (Roche Diagnostics). Then cell lysate immunoprecipitates were subjected to SDS-PAGE followed by immunoblotting. The expression levels of VPS37D were analyzed by qRT-PCR with the collected cell samples. Cell viabilities of the gene knockdown cells were assessed by the CellTiter-Glo assay (Promega), according to the manufacturer's instructions. In brief, at 48 and 72 h after reverse transfection of siRNAs (at 10 nm final concentration) into HEK293T cells in 96-well plates, CellTiter-Glo reagent (100 μl) was added to the cells. After a 10-min incubation at room temperature, luminescence indicating the amount of ATP present was recorded by using a GloMax 96 microplate luminometer (Promega). The ATP levels were normalized against control siRNA-reverse-transfected cells.
RNA extraction and qRT-PCR
For evaluation of the intracellular CHIKV RNA levels, CHIKV-infected cells were collected and treated with 0.25% trypsin EDTA (Wako) at 37 °C for 5 min to detach extra CHIKV on the cell surface. After trypsinized cells were collected to microcentrifuge tubes, the cells were washed three times by pelleting. For pelleting, we repeated centrifugation 1,500 × g for 2 min, carefully removing the PBS and resuspending pellets in 400 μl of PBS. Following the final PBS wash and removal of PBS, the cell pellets were subjected to RNA extraction using TRIzol (Thermo Fisher Scientific) according to the manufacturer's protocol. CHIKV RNA levels in CHIKV-infected cells were then determined by qRT-PCR for CHIKV with extracted RNA, primers, and probes using TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific), following the manufacturer's protocol. To amplify CHIKV E1 region, a primer set (CHIKV UBIV F and CHIKV UBIV R) was used as described previously (62). To amplify the CHIKV nsP4 region, a primer set (CHIKV 6856 and CHIKV 6981) was used as described previously (63).
Expression levels of VPS37D in siRNA-treated cells were also determined by qRT-PCR. Total RNAs were extracted from the siRNA-treated HEK293T cells using TRIzol, according to the manufacturer's protocol. qRT-PCR was performed using the One Step TB Green PrimeScript PLUS RT-PCR Kit (Takara) with extracted RNA and the following primer set: vps37d-386F (5′-TTCGTGAGGTGGCCGAGAAC-3′) and vps37d-524R (5′-TCCATCTGCTCCTCTTGCCT-3′).
The relative expression levels of target mRNA were normalized against control siRNA-reverse-transfected cells with GAPDH employed as an internal standard. The mRNA levels of the GAPDH gene were analyzed using human GAPD (GAPDH) endogenous control (FAM dye/MGB probe, non-primer-limited) (Thermo Fisher Scientific).
Evaluation of the early phase of CHIKV infection
CHIKV was allowed to bind to HEK293T cells in 24-well plates at 4 °C for 1 h. Subsequently, cells were shifted to 37 °C for 3 h to allow endocytosis in new media. As a control, we prepared cells incubated in the new media containing NH4Cl (at 30 mm final concentration). After removing the culture supernatants and washing with PBS, cell samples were collected and subjected to RNA extraction followed by trypsin treatment. Then qRT-PCR for CHIKV was performed with the extracted RNAs to determine intracellular CHIKV RNA levels.
Time course assay
CHIKV was inoculated into HEK293T cells in 24-well plates. After incubation at 4 °C for 1 h to allow CHIKV to attach to the cells, the cells were incubated at 37 °C in new media. The cell samples were collected at 0, 3, 6, 9, and 12 hpi. Trypsin treatment followed by RNA extraction was performed with the collected samples. Amplification of CHIKV RNA was determined by qRT-PCR for CHIKV with extracted RNA.
Reverse transcription for replicon RNA and replicon assay
Either CHIKV replicon plasmids (pSMART-CHIKV-rep-WT and pSMART-CHIKV-rep-MT) or SINV replicon plasmids (pSMART-SINV-rep-WT and pSMART-SINV-rep-MT) were linearized with either NotI or XbaI, respectively. Then linearized plasmids were in vitro transcribed with the mMESSAGE mMACHINE T7 transcription kit (Thermo Fisher Scientific) according to the manufacturer's protocol. HEK293T cells in 96-well plates were reverse-transfected with siRNA, incubated for 48 h, and transfected with reverse-transcribed RNAs (100 ng) from replicon plasmids by the Lipofectamine Messenger MAX (Thermo Fisher Scientific). At the same time point, DENV replicon plasmids (20 ng) were also transfected into HEK293T cells. At the indicated time points, NanoLuc activities were measured using the Nano-Glo luciferase assay kit (Promega) and the GloMax 96 microplate luminometer with cell lysates, following the manufacturer's instructions.
CHIKV-VLP release assay
HEK293T cells were prepared in 24-well plates. The cells were then transfected with the CHIKV-VLP–expressing plasmid (pCXSN-CHIKV-VLP) (200 ng). At 48 hpt, the culture supernatants were collected and mixed with SDS-PAGE sample buffers. The plasmid-transfected cells were lysed with TNE lysis buffer and mixed with SDS-PAGE sample buffer. Both lysate and supernatant samples were then analyzed by immunoblotting using mAb 5.5G9. The amount of CHIKV-VLP was estimated as the intensity of the specific C protein band. Expression of sPs in the cells was confirmed by the relative intensity of C protein in lysates, and the release efficiency of CHIKV-VLP was evaluated in supernatants.
Density gradient sedimentation analysis
Release of CHIKV-VLPs in the supernatant of the pCXSN-CHIKV-VLP–transfected cells was confirmed by density gradient sedimentation analysis. HEK293T cells in a 100-mm TC-treated culture dish were transfected with 20 μg of pCXSN-CHIKV-VLP plasmid and then the cells were harvested at 37 °C for 96 h. Then the culture supernatant was collected and filtered through a 0.45-μm pore-sized filter. Thereafter, the sample was centrifuged at 1,500 × g for 3 min to remove cells and debris, followed by concentration using ultracentrifugation at 30,000 rpm for 2 h in a SW32 Ti rotor (Beckman Coulter). The CHIKV-VLP pellet was resuspended with 1 ml of PBS and placed at 4 °C for 30 min. For sedimentation, 10, 20, 30, 40, and 50% sucrose in PBS was prepared. Sucrose-buffer solution (50%, 2.2 ml) was added to the bottom of the centrifugation tube. Then the same volume of 40% sucrose-buffer solution was layered. This procedure was repeated with the same volumes of the 30, 20, and 10% sucrose-buffer solutions. After placing the tubes at 4 °C for 30 min at 4 °C, the resuspended CHIKV-VLP sample was layered carefully on the top of the gradient. This gradient was centrifuged at 38,000 rpm in a SW41 rotor (Beckman Coulter) at 4 °C for 14 h and then fractionated from the top into 24 fractions (500 μl each). All fractions (20 μl) were analyzed by SDS-PAGE and immunoblotting using anti-CHIKV pAb or mAb 5.5G9.
Generation and infection of tetracycline-inducible VPS4B-expressing cells
The ATP hydrolysis–deficient VPS4B E235Q mutant (VPS4BEQ) was derived from the VPS4B WT backbone by PCR mutagenesis. Tetracycline-inducible stable cell lines expressing WT or mutant HA-tagged VPS4B with Tet-On 3G transactivator protein were generated using the Retro-X Tet-On 3G inducible expression system (Clontech). Briefly, HA-tagged VPS4B-expressing plasmids (pRetroX-TRE3G-VPS4B WT and pRetroX-TRE3G-VPS4BEQ) were constructed by inserting VPS4B genes into RetroX-TRE3G vector. The above plasmids and pCMV-VSV-G were transfected into GP2-293 packaging cells by FuGENE HD transfection reagent (Promega). TRE3G-VPS4B retroviruses were collected from the culture media 48 h after transfection. HEK293T cells were then co-infected with TRE3G-VPS4B retrovirus and Tet-On 3G retrovirus, expressing Tet-On 3G transactivator protein. Tetracycline-inducible VPS4B-stable expressing HEK293T cell lines were selected under G418 and puromycin treatments.
Tetracycline-inducible VPS4B-expressing HEK293T cells in 24-well plates were cultured with or without doxycycline hydrochloride (Sigma) for 24 h and then infected with CHIKV, SFV, or VSV (MOI = 0.1). After incubation at 4 °C for 1 h, the cells were incubated at 37 °C in new media. The culture supernatants were collected from the virus-infected cells at the indicated time points. The viral titers in the supernatant samples were determined by a plaque assay.
Statistical analysis and normalization
All assays were performed in triplicate and independently at least two times. The data were expressed as means ± S.D. Statistical significance was determined by the two-tailed Student's t test or one-way ANOVA with Dunnett's test. p values <0.05 or <0.001 were considered significant and are indicated by a single asterisk or double asterisks, respectively. The relative data were normalized against control siRNA-reverse-transfected cells.
Data availability
All of the data are contained within the article.
Supplementary Material
Acknowledgments
We greatly appreciate Dr. Tomohiko Takasaki (National Institute of Infectious Diseases, Tokyo, Japan), Masahiro Hayashi (National Institute of Infectious Diseases, Tokyo, Japan), and Tatsuo Shioda (Department of Viral Infections, Research Institute for Microbial Diseases, Osaka University) for providing viruses or antibody employed in these studies. We also thank Mikiko Ishibashi (Department of Molecular Virology, Research Institute for Microbial Diseases, Osaka University) for assistance.
This article contains supporting information.
Author contributions—S. T. conceptualization; S. T. data curation; S. T. formal analysis; S. T. validation; S. T. investigation; S. T. visualization; S. T. methodology; S. T. writing-original draft; S. T. project administration; Y. O., M. S., and H. S. supervision; K. T., Y. W., J. H.-P., R. A. H., A. T., T. F., and Y. M. resources; M. C., J. H.-P., R. A. H., and W. W. H. writing-review and editing.
Funding and additional information—This study was supported by grants for Scientific Research on Innovative Areas and International Group from the MEXT/JSPS KAKENHI (JP16H06431, JP16H06429, JP16K21723), and the Japan Initiative for Global Research Network on Infectious Diseases, from the Japan Agency for Medical Research and Development (JP19fm0108008).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- CHIKF
- chikungunya fever
- CHIKV
- chikungunya virus
- nsP
- nonstructural protein
- sP
- structural protein
- CPV-I
- type I cytopathic vacuole
- SFV
- Semliki Forest virus
- TGN
- trans-Golgi network
- ESCRT
- endosomal sorting complexes required for transport
- DENV
- dengue virus
- HA
- hemagglutinin
- IP
- immunoprecipitation
- hpi
- hours post-infection
- KD
- knockdown
- SINV
- Sindbis virus
- qRT-PCR
- quantitative RT-PCR
- hpt
- hours post-transfection
- RdRp
- RNA-dependent RNA polymerase
- pAb
- polyclonal antibody
- VSV
- vesicular stomatitis virus
- VSV-G
- vesicular stomatitis virus G glycoprotein
- UIM
- ubiquitin-interacting motif
- DMEM
- Dulbecco's modified Eagle's media
- FBS
- fetal bovine serum
- IFA
- immunofluorescence assay
- HRP
- horseradish peroxidase
- DAPI
- 4′,6-diamidino-2-phenylindole
- MOI
- multiplicity of infection
- NP-40
- Nonidet P-40
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
- analysis of variance
- HGS
- hepatocyte growth factor–regulated tyrosine kinase substrate.
References
- 1. Chen R., Mukhopadhyay S., Merits A., Bolling B., Nasar F., Coffey L. L., Powers A., Weaver S. C., and Ictv Report Consortium. (2018) ICTV Virus Taxonomy Profile: Togaviridae. J. Gen. Virol. 99, 761–762 10.1099/jgv.0.001072 [DOI] [PubMed] [Google Scholar]
- 2. Pialoux G., Gaüzère B. A., Jauréguiberry S., and Strobel M. (2007) Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 7, 319–327 10.1016/S1473-3099(07)70107-X [DOI] [PubMed] [Google Scholar]
- 3. Couderc T., and Lecuit M. (2015) Chikungunya virus pathogenesis: from bedside to bench. Antiviral Res. 121, 120–131 10.1016/j.antiviral.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 4. Borgherini G., Poubeau P., Staikowsky F., Lory M., Le Moullec N., Becquart J. P., Wengling C., Michault A., and Paganin F. (2007) Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin. Infect. Dis. 44, 1401–1407 10.1086/517537 [DOI] [PubMed] [Google Scholar]
- 5. Borgherini G., Poubeau P., Jossaume A., Gouix A., Cotte L., Michault A., Arvin-Berod C., and Paganin F. (2008) Persistent arthralgia associated with chikungunya virus: a study of 88 adult patients on reunion island. Clin. Infect. Dis. 47, 469–475 10.1086/590003 [DOI] [PubMed] [Google Scholar]
- 6. Schilte C., Staikowsky F., Staikovsky F., Couderc T., Madec Y., Carpentier F., Kassab S., Albert M. L., Lecuit M., and Michault A. (2013) Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl. Trop. Dis. 7, e2137 10.1371/journal.pntd.0002137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chretien J. P., Anyamba A., Bedno S. A., Breiman R. F., Sang R., Sergon K., Powers A. M., Onyango C. O., Small J., Tucker C. J., and Linthicum K. J. (2007) Drought-associated chikungunya emergence along coastal East Africa. Am. J. Trop. Med. Hyg. 76, 405–407 10.4269/ajtmh.2007.76.405 [DOI] [PubMed] [Google Scholar]
- 8. Weaver S. C., and Forrester N. L. (2015) Chikungunya: evolutionary history and recent epidemic spread. Antiviral Res. 120, 32–39 10.1016/j.antiviral.2015.04.016 [DOI] [PubMed] [Google Scholar]
- 9. Rezza G., Nicoletti L., Angelini R., Romi R., Finarelli A. C., Panning M., Cordioli P., Fortuna C., Boros S., Magurano F., Silvi G., Angelini P., Dottori M., Ciufolini M. G., Majori G. C., et al. (2007) Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370, 1840–1846 10.1016/S0140-6736(07)61779-6 [DOI] [PubMed] [Google Scholar]
- 10. Grandadam M., Caro V., Plumet S., Thiberge J. M., Souarès Y., Failloux A. B., Tolou H. J., Budelot M., Cosserat D., Leparc-Goffart I., and Desprès P. (2011) Chikungunya virus, southeastern France. Emerg. Infect. Dis. 17, 910–913 10.3201/eid1705.101873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leparc-Goffart I., Nougairede A., Cassadou S., Prat C., and de Lamballerie X. (2014) Chikungunya in the Americas. Lancet 383, 514 10.1016/S0140-6736(14)60185-9 [DOI] [PubMed] [Google Scholar]
- 12. Solignat M., Gay B., Higgs S., Briant L., and Devaux C. (2009) Replication cycle of chikungunya: a re-emerging arbovirus. Virology 393, 183–197 10.1016/j.virol.2009.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rupp J. C., Sokoloski K. J., Gebhart N. N., and Hardy R. W. (2015) Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 96, 2483–2500 10.1099/jgv.0.000249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voss J. E., Vaney M. C., Duquerroy S., Vonrhein C., Girard-Blanc C., Crublet E., Thompson A., Bricogne G., and Rey F. A. (2010) Glycoprotein organization of chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712 10.1038/nature09555 [DOI] [PubMed] [Google Scholar]
- 15. van Duijl-Richter M. K., Hoornweg T. E., Rodenhuis-Zybert I. A., and Smit J. M. (2015) Early events in chikungunya virus infection—from virus cell binding to membrane fusion. Viruses 7, 3647–3674 10.3390/v7072792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spuul P., Balistreri G., Kääriäinen L., and Ahola T. (2010) Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 84, 7543–7557 10.1128/JVI.00477-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Froshauer S., Kartenbeck J., and Helenius A. (1988) Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107, 2075–2086 10.1083/jcb.107.6.2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Curtis I., and Simons K. (1988) Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc. Natl. Acad. Sci. U.S.A. 85, 8052–8056 10.1073/pnas.85.21.8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Griffiths G., Quinn P., and Warren G. (1983) Dissection of the Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus. J. Cell Biol. 96, 835–850 10.1083/jcb.96.3.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Griffin D. E. (2013) Alphaviruses. in Fields Virology, 6th Ed. (Knipe D. M., and Howley P. M., eds) pp. 651–686, Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 21. Geigenmüller-Gnirke U., Nitschko H., and Schlesinger S. (1993) Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J. Virol. 67, 1620–1626 10.1128/JVI.67.3.1620-1626.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perera R., Owen K. E., Tellinghuisen T. L., Gorbalenya A. E., and Kuhn R. J. (2001) Alphavirus nucleocapsid protein contains a putative coiled coil α-helix important for core assembly. J. Virol. 75, 1–10 10.1128/JVI.75.1.1-10.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen K. C., Kam Y. W., Lin R. T., Ng M. M., Ng L. F., and Chu J. J. (2013) Comparative analysis of the genome sequences and replication profiles of chikungunya virus isolates within the East, Central and South African (ECSA) lineage. Virol. J. 10, 169 10.1186/1743-422X-10-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernard E., Solignat M., Gay B., Chazal N., Higgs S., Devaux C., and Briant L. (2010) Endocytosis of chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PLoS ONE 5, e11479 10.1371/journal.pone.0011479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas S., Rai J., John L., Schaefer S., Pützer B. M., and Herchenröder O. (2013) Chikungunya virus capsid protein contains nuclear import and export signals. Virol. J. 10, 269 10.1186/1743-422X-10-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Votteler J., and Sundquist W. I. (2013) Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241 10.1016/j.chom.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Henne W. M., Buchkovich N. J., and Emr S. D. (2011) The ESCRT pathway. Dev. Cell 21, 77–91 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- 28. Taylor G. M., Hanson P. I., and Kielian M. (2007) Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J. Virol. 81, 13631–13639 10.1128/JVI.01688-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barouch-Bentov R., Neveu G., Xiao F., Beer M., Bekerman E., Schor S., Campbell J., Boonyaratanakornkit J., Lindenbach B., Lu A., Jacob Y., and Einav S. (2016) Hepatitis C virus proteins interact with the endosomal sorting complex required for transport (ESCRT) machinery via ubiquitination to facilitate viral envelopment. MBio 7, e01456–16 10.1128/mBio.01456-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar S., Barouch-Bentov R., Xiao F., Schor S., Pu S., Biquand E., Lu A., Lindenbach B. D., Jacob Y., Demeret C., and Einav S. (2019) MARCH8 ubiquitinates the hepatitis C virus nonstructural 2 protein and mediates viral envelopment. Cell Rep. 26, 1800–1814.e5 10.1016/j.celrep.2019.01.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pattanakitsakul S. N., Poungsawai J., Kanlaya R., Sinchaikul S., Chen S. T., and Thongboonkerd V. (2010) Association of Alix with late endosomal lysobisphosphatidic acid is important for dengue virus infection in human endothelial cells. J. Proteome Res. 9, 4640–4648 10.1021/pr100357f [DOI] [PubMed] [Google Scholar]
- 32. Chiou C. T., Hu C. A., Chen P. H., Liao C. L., Lin Y. L., and Wang J. J. (2003) Association of Japanese encephalitis virus NS3 protein with microtubules and tumour susceptibility gene 101 (TSG101) protein. J. Gen. Virol. 84, 2795–2805 10.1099/vir.0.19201-0 [DOI] [PubMed] [Google Scholar]
- 33. Tabata K., Arimoto M., Arakawa M., Nara A., Saito K., Omori H., Arai A., Ishikawa T., Konishi E., Suzuki R., Matsuura Y., and Morita E. (2016) Unique requirement for ESCRT factors in flavivirus particle formation on the endoplasmic reticulum. Cell Rep. 16, 2339–2347 10.1016/j.celrep.2016.07.068 [DOI] [PubMed] [Google Scholar]
- 34. Crump C. M., Yates C., and Minson T. (2007) Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81, 7380–7387 10.1128/JVI.00222-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Calistri A., Sette P., Salata C., Cancellotti E., Forghieri C., Comin A., Göttlinger H., Campadelli-Fiume G., Palù G., and Parolin C. (2007) Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 81, 11468–11478 10.1128/JVI.01364-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pawliczek T., and Crump C. M. (2009) Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83, 11254–11264 10.1128/JVI.00574-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang R., Kim A. S., Fox J. M., Nair S., Basore K., Klimstra W. B., Rimkunas R., Fong R. H., Lin H., Poddar S., Crowe J. E. Jr., Doranz B. J., Fremont D. H., and Diamond M. S. (2018) Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574 10.1038/s41586-018-0121-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karlas A., Berre S., Couderc T., Varjak M., Braun P., Meyer M., Gangneux N., Karo-Astover L., Weege F., Raftery M., Schönrich G., Klemm U., Wurzlbauer A., Bracher F., Merits A., Meyer T. F., and Lecuit M. (2016) A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat. Commun. 7, 11320 10.1038/ncomms11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang N., and Zhang L. (2017) Key components of COPI and COPII machineries are required for chikungunya virus replication. Biochem. Biophys. Res. Commun. 493, 1190–1196 10.1016/j.bbrc.2017.09.142 [DOI] [PubMed] [Google Scholar]
- 40. Rauch S., and Martin-Serrano J. (2011) Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J. Virol. 85, 3546–3556 10.1128/JVI.02045-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chou S. F., Tsai M. L., Huang J. Y., Chang Y. S., and Shih C. (2015) The dual role of an ESCRT-0 component HGS in HBV transcription and naked capsid secretion. PLoS Pathog. 11, e1005123 10.1371/journal.ppat.1005123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Komada M., Masaki R., Yamamoto A., and Kitamura N. (1997) Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J. Biol. Chem. 272, 20538–20544 10.1074/jbc.272.33.20538 [DOI] [PubMed] [Google Scholar]
- 43. Remenyi R., Gao Y., Hughes R. E., Curd A., Zothner C., Peckham M., Merits A., and Harris M. (2018) Persistent replication of a chikungunya virus replicon in human cells is associated with presence of stable cytoplasmic granules containing nonstructural protein 3. J. Virol. 92, e00477–18 10.1128/JVI.00477-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wada Y., Orba Y., Sasaki M., Kobayashi S., Carr M. J., Nobori H., Sato A., Hall W. W., and Sawa H. (2017) Discovery of a novel antiviral agent targeting the nonstructural protein 4 (nsP4) of chikungunya virus. Virology 505, 102–112 10.1016/j.virol.2017.02.014 [DOI] [PubMed] [Google Scholar]
- 45. Nobori H., Toba S., Yoshida R., Hall W. W., Orba Y., Sawa H., and Sato A. (2018) Identification of Compound-B, a novel anti-dengue virus agent targeting the non-structural protein 4A. Antiviral Res. 155, 60–66 10.1016/j.antiviral.2018.05.003 [DOI] [PubMed] [Google Scholar]
- 46. Hirano S., Kawasaki M., Ura H., Kato R., Raiborg C., Stenmark H., and Wakatsuki S. (2006) Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat. Struct. Mol. Biol. 13, 272–277 10.1038/nsmb1051 [DOI] [PubMed] [Google Scholar]
- 47. Liu Y., and Harty R. N. (2010) Viral and host proteins that modulate filovirus budding. Future Virol. 5, 481–491 10.2217/fvl.10.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gordon T. B., Hayward J. A., Marsh G. A., Baker M. L., and Tachedjian G. (2019) Host and viral proteins modulating Ebola and Marburg virus egress. Viruses 11, E25 10.3390/v11010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fujii K., Hurley J. H., and Freed E. O. (2007) Beyond Tsg101: the role of Alix in “ESCRTing” HIV-1. Nat. Rev. Microbiol. 5, 912–916 10.1038/nrmicro1790 [DOI] [PubMed] [Google Scholar]
- 50. Barajas D., Jiang Y., and Nagy P. D. (2009) A unique role for the host ESCRT proteins in replication of tomato bushy stunt virus. PLoS Pathog. 5, e1000705 10.1371/journal.ppat.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Diaz A., Zhang J., Ollwerther A., Wang X., and Ahlquist P. (2015) Correction: Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog. 11, e1004845 10.1371/journal.ppat.1004845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barajas D., Martín I. F., Pogany J., Risco C., and Nagy P. D. (2014) Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of tomato bushy stunt virus replicase. PLoS Pathog. 10, e1004087 10.1371/journal.ppat.1004087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dejarnac O., Hafirassou M. L., Chazal M., Versapuech M., Gaillard J., Perera-Lecoin M., Umana-Diaz C., Bonnet-Madin L., Carnec X., Tinevez J. Y., Delaugerre C., Schwartz O., Roingeard P., Jouvenet N., Berlioz-Torrent C., et al. (2018) TIM-1 ubiquitination mediates dengue virus entry. Cell Rep. 23, 1779–1793 10.1016/j.celrep.2018.04.013 [DOI] [PubMed] [Google Scholar]
- 54. Arii J., Watanabe M., Maeda F., Tokai-Nishizumi N., Chihara T., Miura M., Maruzuru Y., Koyanagi N., Kato A., and Kawaguchi Y. (2018) ESCRT-III mediates budding across the inner nuclear membrane and regulates its integrity. Nat. Commun. 9, 3379 10.1038/s41467-018-05889-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lim C. K., Nishibori T., Watanabe K., Ito M., Kotaki A., Tanaka K., Kurane I., and Takasaki T. (2009) Chikungunya virus isolated from a returnee to Japan from Sri Lanka: isolation of two sub-strains with different characteristics. Am. J. Trop. Med. Hyg. 81, 865–868 10.4269/ajtmh.2009.09-0009 [DOI] [PubMed] [Google Scholar]
- 56. Goh L. Y., Hobson-Peters J., Prow N. A., Gardner J., Bielefeldt-Ohmann H., Pyke A. T., Suhrbier A., and Hall R. A. (2013) Neutralizing monoclonal antibodies to the E2 protein of chikungunya virus protects against disease in a mouse model. Clin. Immunol. 149, 487–497 10.1016/j.clim.2013.10.004 [DOI] [PubMed] [Google Scholar]
- 57. Goh L. Y. H., Hobson-Peters J., Prow N. A., Gardner J., Bielefeldt-Ohmann H., Suhrbier A., and Hall R. A. (2015) Monoclonal antibodies specific for the capsid protein of chikungunya virus suitable for multiple applications. J. Gen. Virol. 96, 507–512 10.1099/jgv.0.000002 [DOI] [PubMed] [Google Scholar]
- 58. Masrinoul P., Puiprom O., Tanaka A., Kuwahara M., Chaichana P., Ikuta K., Ramasoota P., and Okabayashi T. (2014) Monoclonal antibody targeting chikungunya virus envelope 1 protein inhibits virus release. Virology 464, 111–117 10.1016/j.virol.2014.05.038 [DOI] [PubMed] [Google Scholar]
- 59. Sasaki M., Hasebe R., Makino Y., Suzuki T., Fukushi H., Okamoto M., Matsuda K., Taniyama H., Sawa H., and Kimura T. (2011) Equine major histocompatibility complex class I molecules act as entry receptors that bind to equine herpesvirus-1 glycoprotein D. Genes Cells 16, 343–357 10.1111/j.1365-2443.2011.01491.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Akahata W., Yang Z. Y., Andersen H., Sun S., Holdaway H. A., Kong W. P., Lewis M. G., Higgs S., Rossmann M. G., Rao S., and Nabel G. J. (2010) A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 16, 334–338 10.1038/nm.2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morita S., Kojima T., and Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- 62. Andriamandimby S. F., Heraud J. M., Randrianasolo L., Rafisandratantsoa J. T., Andriamamonjy S., and Richard V. (2013) Dried-blood spots: a cost-effective field method for the detection of Chikungunya virus circulation in remote areas. PLoS Negl. Trop. Dis. 7, e2339 10.1371/journal.pntd.0002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lanciotti R. S., Kosoy O. L., Laven J. J., Panella A. J., Velez J. O., Lambert A. J., and Campbell G. L. (2007) Chikungunya virus in US travelers returning from India, 2006. Emerg. Infect. Dis. 13, 764–767 10.3201/eid1305.070015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data are contained within the article.