Abstract
Noroviruses are the main causative agents of acute viral gastroenteritis, but the host factors that restrict their replication remain poorly identified. Guanylate-binding proteins (GBPs) are interferon (IFN)-inducible GTPases that exert broad antiviral activity and are important mediators of host defenses against viral infections. Here, we show that both IFN-γ stimulation and murine norovirus (MNV) infection induce GBP2 expression in murine macrophages. Results from loss- and gain-of-function assays indicated that GBP2 is important for IFN-γ–dependent anti-MNV activity in murine macrophages. Ectopic expression of MNV receptor (CD300lf) in human HEK293T epithelial cells conferred susceptibility to MNV infection. Importantly, GBP2 potently inhibited MNV in these human epithelial cells. Results from mechanistic dissection experiments revealed that the N-terminal G domain of GBP2 mediates these anti-MNV effects. R48A and K51A substitutions in GBP2, associated with loss of GBP2 GTPase activity, attenuated the anti-MNV effects of GBP2. Finally, we found that nonstructural protein 7 (NS7) of MNV co-localizes with GBP2 and antagonizes the anti-MNV activity of GBP2. These findings reveal that GBP2 is an important mediator of host defenses against murine norovirus.
Keywords: viral replication, host defense, host-pathogen interaction, innate immunity, GTPase, antiviral restriction factor, guanylate-binding protein 2 (GBP2), interferon-stimulated gene (ISG), murine norovirus (MNV), non-structural protein 7 (NS7)
Introduction
Human norovirus (HuNV) infection is the major cause of epidemic nonbacterial gastroenteritis worldwide (1, 2). Currently, there is no vaccination or specific antiviral treatment available. Clinical management is limited to supportive care and oral rehydration, and HuNV imposes a heavy global health burden (3). Thus, defining improved anti-HuNV therapy represents an urgent clinical need. Research into HuNV infection, however, has been hampered by the lack of robust experimental models. Murine norovirus (MNV), capable of replicating in both cell culture and small-animal models, shares similar traits with HuNV in structural and genetic features and has thus been widely used as a surrogate model (4, 5). The recent discovery of the MNV receptor (CD300lf) has now enabled MNV infection in human cells by ectopically expressing this receptor. This has resulted in improved understanding of the mechanisms underlying viral replication and the identification of cellular factors as potential antiviral targets (6, 7).
Noroviruses are nonenveloped, positive single-stranded RNA viruses belonging to the Caliciviridae family. The genome is about 7.5 kb in length and encodes three or four ORFs (3, 8). The 5′-proximal ORF1 encodes a polyprotein that is post-translationally cleaved into six nonstructural proteins (NS1/2 to NS7). ORF2 and ORF3, encoding the major and minor structural proteins, are referred as VP1 and VP2, respectively, which are translated from a subgenomic RNA. VP2 has been reported to possess important functions in viral replication and virion stability but may also corrupt host immune response (9). Specific for MNV, ORF4 overlaps with ORF2 and produces an additional protein called virulence factor (VF1). VF1 has been reported to antagonize innate immune response to MNV infection (8, 9). MNV NS1/2 protein is associated with cell tropism and mediates resistance to interferon-λ (IFN-λ)-mediated clearance used for treating persistent viral infection (10). NS7 is the viral RNA-dependent RNA polymerase that can also modulate innate immune response (11, 12). However, the exact interactions of these viral proteins with host innate antiviral immunity remain poorly understood.
IFN-mediated innate immune responses provide the first line of host defense against viral infections. Specific viral components sensed by pathogen recognition receptors including Toll-like receptors and RIG-I-like receptors lead to IFN production (13, 14). The released IFNs bind to their cognate receptors to activate the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway, resulting in the transcription of hundreds of interferon-stimulated genes (ISGs). A subset of ISGs are considered as the ultimate antiviral effectors limiting viral replication and spread (15). Currently, only a few ISGs have been identified to inhibit MNV infection, including interferon regulatory factor 1 (IRF1) and interferon-stimulated gene 15 (ISG15) (16, 17). Thus, it is largely unknown which factors are important for effective cell-autonomous defense against MNV infection.
Interesting candidate molecules to act in the defense against MNV infection are the guanylate-binding proteins (GBPs). They are members of the superfamily of IFN-inducible GTPases with many typical characteristics of ISGs. These proteins are composed of three distinct domains, including the N-terminal globular GTPase domain containing all motifs responsible for nucleotide binding and hydrolysis (G domain), the following helical part presenting as the middle domain (M domain), and the C-terminal GTPase effector domain (E domain) (18, 19). To date, seven human GBPs and 11 murine GBPs as well as two mouse pseudogenes encoding GBPs have been identified (20, 21). Apart from host resistance against bacterial and protozoan pathogens (18, 22, 23), GBPs (e.g. GBP1, GBP2, and GBP5) have been reported to exert broad antiviral activity against HIV, Zika virus, hepatitis C virus (HCV), classical swine fever virus (CSFV), and influenza virus (24–27). They are capable of regulating inflammasome activation (28, 29), and this plays a role in controlling replication of rotavirus and influenza virus (30, 31). A recent study has indicated that IFN-inducible GTPases (including GBP2) are important for IFN-γ–mediated inhibition of MNV replication and help to block the formation of viral replication complexes (32). In this study, we investigated in detail the regulation and function of GBP2 in MNV infection, revealing an important role in orchestrating host response to MNV infection.
Results
IFN-γ stimulation up-regulates GBP2 expression in mouse macrophages
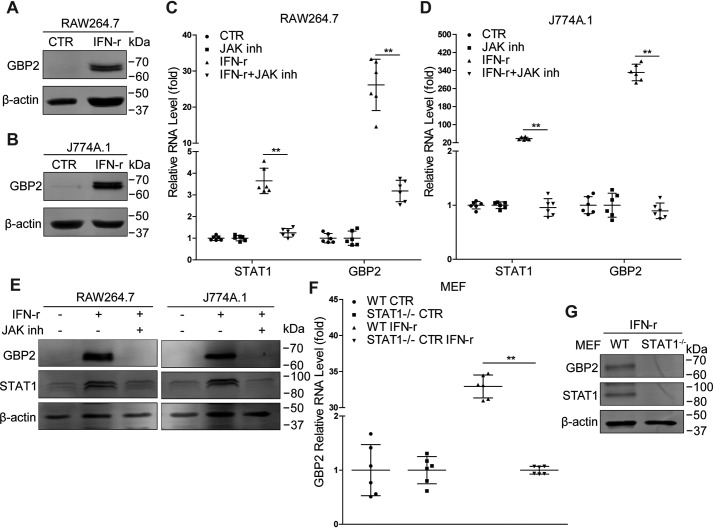
IFN-γ signaling can potently inhibit MNV replication and induce expression of GBPs. In this study, we observed up-regulation of GBP2 following IFN-γ treatment in RAW264.7 and J774A.1 cells (Fig. 1, A and B), two murine macrophage cell lines susceptible to MNV infection. This effect appears to be mediated by canonical JAK/STAT-mediated IFN signaling as the pharmacological compound JAK inhibitor 1 blocked IFN-γ–induced GBP2 expression at both mRNA (Fig. 1, C and D) and protein levels (Fig. 1E) in RAW264.7 and J774A.1 cells. Furthermore, IFN-γ failed to stimulate GBP2 expression in STAT1-deficient mouse embryonic fibroblasts (MEFs) (Fig. 1, F and G). We thus concluded that up-regulation of GBP2 is part of the canonical IFN-γ response.
Figure 1.
IFN-γ stimulation triggers GBP2 expression in mouse macrophages. A and B, Western blot analysis of GBP2 expression in RAW264.7 (A) and J774A.1 (B) cells that were treated or untreated with IFN-γ (100 units/ml) for 24 h. RAW264.7 and J774A.1 cells were treated or untreated with IFN-γ (100 units/ml) or JAK inhibitor 1 (10 μm) for 4 or 6 h, respectively. mRNA (C, n = 6; D, n = 6) and protein (E) levels of STAT1 and GBP2 were analyzed by qRT-PCR and Western blotting, respectively. F, WT and STAT1−/− MEFs were untreated or treated with IFN-γ (100 units/ml) for 6 h. The mRNA level of GBP2 was analyzed by qRT-PCR (n = 6). G, expression of STAT1 and GBP2 in WT and STAT1−/− MEFs treated with IFN-γ (100 units/ml) for 6 h were analyzed by Western blotting. Data in C and D were normalized to the untreated and JAK inhibitor–treated cells (both set as 1). Data in F were normalized to untreated WT and STAT1−/− MEFs, respectively (both set as 1). **, p < 0.01. β-Actin was used as a loading control. Error bars, S.D.
GBP2 enhances IFN-γ–mediated inhibition of MNV-1 replication in mouse macrophages
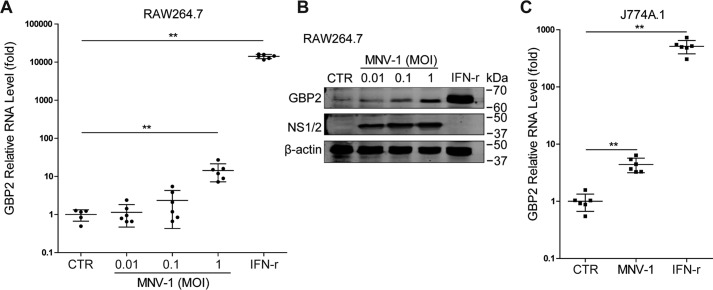
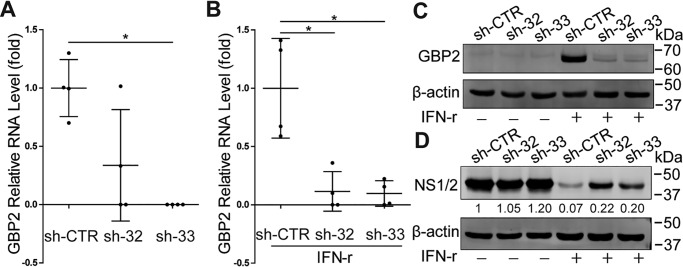
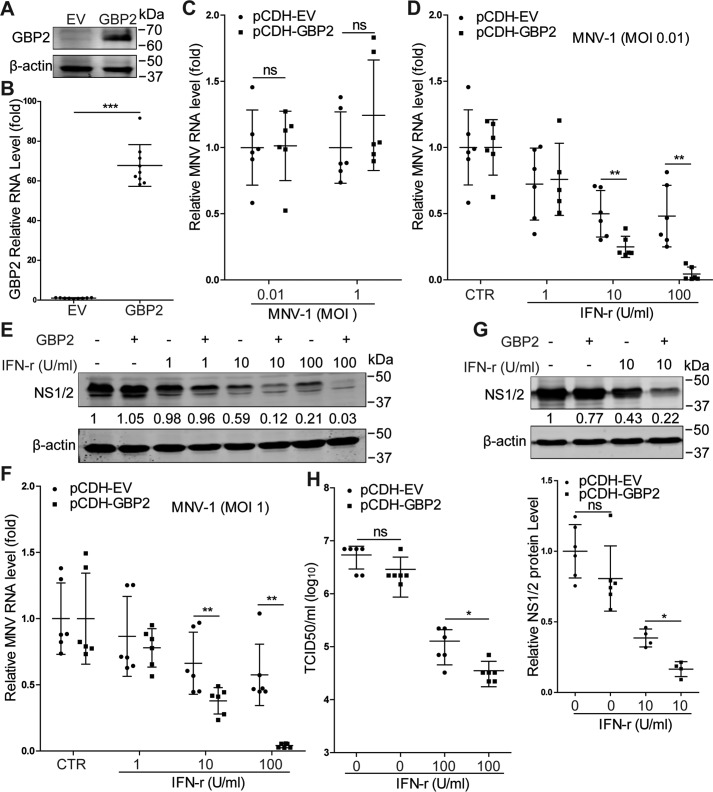
We next examined whether MNV infection could induce GBP2 expression. Interestingly, inoculation of both in RAW264.7 cells (Fig. 2, A and B) and J774A.1 cells (Fig. 2C) with MNV-1 up-regulated GBP2. To investigate the functional implication, we silenced GBP2 expression in RAW264.7 cells by lentiviral shRNAs. The efficacy of this approach was confirmed both with and without IFN-γ treatment (Fig. 3, A–C). Although GBP2 silence per se has no major effect on MNV-1 replication, this strategy decreased anti-MNV activity of IFN-γ (Fig. 3D). This is in accordance with a study demonstrating that MNV replicates to a high level in GBP2-deficient murine bone marrow–derived macrophages (BMDMs) in the presence of IFN-γ (33). Thus, GBP2 is necessary for the anti-MNV activity of IFN-γ. Furthermore, we succeeded with stable GBP2 overexpression employing a lentiviral vector (Fig. 4, A and B). This did not affect MNV-1 RNA level (Fig. 4C), but it augmented IFN-γ–mediated inhibition of MNV-1 RNA transcription and NS1/2 protein expression (Fig. 4, D–G) as well as the viral titers (Fig. 4H). Thus, GBP2 is an important mediator of IFN-γ–triggered inhibition of MNV-1 replication in murine macrophages.
Figure 2.
MNV-1 infection up-regulates GBP2 expression. RAW264.7 cells were infected with MNV-1 at the indicated MOIs or treated with IFN-γ (100 units/ml) for 24 h. A, the GBP2 mRNA level was analyzed by qRT-PCR (n = 5–6). B, expression of GBP2 and MNV NS1/2 were analyzed by Western blotting. C, J774A.1 cells were infected with MNV-1 at a MOI of 1 or treated with IFN-γ (100 units/ml) for 24 h. The mRNA level of GBP2 was analyzed by qRT-PCR (n = 6). Data were normalized to the untreated control (CTR, set as 1). **, p < 0.01. β-Actin was used as a loading control. Error bars, S.D.
Figure 3.
Knockdown of GBP2 attenuates IFN-γ–mediated inhibition of MNV-1 NS1/2 protein expression in mouse macrophages. A, qRT-PCR analysis of GBP2 knockdown by lentiviral shRNA vectors in RAW264.7 cells (n = 4). B, qRT-PCR analysis of GBP2 knockdown by lentiviral shRNA vectors in RAW264.7 cells that were stimulated with IFN-γ (100 units/ml) for 6 h (n = 4). C, Western blot analysis of GBP2 knockdown by lentiviral shRNA vectors in RAW264.7 cells without or with IFN-γ (100 units/ml) treatment. D, GBP2-knockdown RAW264.7 cells were infected with MNV-1 at a MOI of 1 for 1 h and then untreated or treated with IFN-γ (100 units/ml) for 24 h. The viral NS1/2 protein level was analyzed by Western blotting. Data in (A and B) were normalized to the control (CTR, set as 1). *, p < 0.05. β-Actin was used as a loading control. Error bars, S.D.
Figure 4.
GBP2 overexpression enhances IFN-γ–mediated inhibition of MNV-1 replication in mouse macrophages. A, HEK293T cells were transfected with pCDH-GBP2 (1 μg) or the empty vectors (EV) (1 μg) for 24 h. Western blot analysis of GBP2 expression by using rabbit anti-GBP2 antibody (1:1000). B, qRT-PCR analysis of GBP2 overexpression by lentiviral vectors in RAW264.7 cells (n = 9). C, qRT-PCR analysis of MNV RNA level (n = 6) in GBP2 stable expression cells that were infected with MNV-1 with the indicated MOIs. qRT-PCR (D) analysis of MNV RNA level (n = 5–6) and Western blotting (E) analysis of MNV NS1/2 expression in GBP2 stable expression cells that were infected with MNV-1 (MOI 0.01) for 1 h and then treated with the indicated concentrations of IFN-γ for 24 h. Shown is qRT-PCR (F) analysis of MNV RNA level (n = 6) and Western blotting (G) analysis (n = 4–6) of MNV NS1/2 expression in GBP2 stable expression cells that were infected with MNV-1 (MOI 1) for 1 h and then treated with the indicated concentrations of IFN-γ for 24 h. H, TCID50 assay analysis of viral titers in the supernatants from GBP2 stable expression cells that were infected with MNV-1 (MOI 1) for 1 h and then untreated or treated with IFN-γ for 24 h (n = 6). Data in B, C, G (bottom), and H were normalized to the untreated control (set as 1). Data in D and F were normalized to untreated empty virus and GBP2 stable expression cells, respectively (both set as 1). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. β-Actin was used as a loading control. Error bars, S.D.
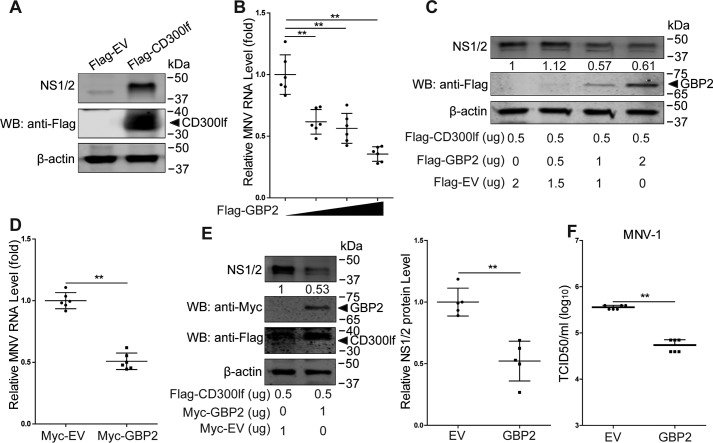
GBP2 overexpression restricts MNV-1 replication in human epithelial cells
Discovery of the MNV receptor (CD300lf) has enabled MNV infection in human cells by ectopic expression of this receptor (6). We found that transfection with FLAG-tagged CD300lf allowed MNV-1 replication in human HEK293T cells (Fig. 5A). Studies have reported that the anti-MNV ability of IFN-γ is impaired in human HAP1 cells with complete loss of GBPs (33). To further examine whether GBP2 exerts anti-norovirus activity in human cells, HEK293T cells were co-transfected with FLAG-tagged CD300lf and different concentrations of GBP2 vectors for 24 h and subsequently infected with MNV-1 for another 24 h. Mirroring the results in murine macrophages, GBP2 overexpression alone can inhibit MNV-1, shown at both viral RNA (Fig. 5B) and protein levels (Fig. 5C). Furthermore, transfection with Myc-tagged GBP2 into CD300lf-expressing HEK293T cells confirmed the inhibitory effects on MNV-1 (Fig. 5, D–F). Thus, a role for GBP2 in constraining MNV replication is not restricted to murine macrophages but also extends to human epithelial cells.
Figure 5.
GBP2 restricts MNV-1 replication in human epithelial cells. A, HEK2923T cells were transfected with the pFLAG-CD300lf (0.5 μg) or the empty vectors (EV) (0.5 μg) for 24 h and then infected with MNV-1 at an MOI of 1 for 24 h. Shown is Western blot (WB) analysis of MNV-1 infection by detecting the viral NS1/2 protein. B and C, qRT-PCR analysis of MNV RNA (B) (n = 6) and Western blot analysis of MNV NS1/2 and GBP2 proteins (C) in HEK293T cells that were transfected with pFLAG-CD300lf (0.5 μg) and pFLAG-GBP2 or empty vectors with the indicated concentrations and then infected with MNV-1 at an MOI of 1 for 24 h. HEK293T cells were transfected with pFLAG-CD300lf (0.5 μg) and pMyc-GBP2 (1 μg) or empty vectors (1 μg) for 24 h and then infected with MNV-1 at an MOI of 1 for 24 h. The viral RNA and NS1/2 protein levels were analyzed by qRT-PCR (D) (n = 6) and Western blotting (E) (n = 5), respectively. F, HEK293T cells were transfected with pFLAG-CD300lf (0.5 μg) and pMyc-GBP2 (1 μg) or empty vectors (1 μg) and then infected with MNV-1 at an MOI of 1 for 24 h. Viral titers in the supernatants were analyzed by a TCID50 assay (n = 6). Data in B, D, E (right) and F were normalized to the empty virus control (set as 1). **, p < 0.01. β-Actin was used as a loading control. Error bars, S.D.
Of note, several ISGs with broad antiviral activity, including IRF1, RIG-I, and MDA5, have been shown to activate the transcription of many other ISGs (15, 33–35). To address whether GBP2 also exerts its action in a similar manner, we measured the transcriptional level of several selected ISGs in RAW264.7 cells in the absence or presence of stable GBP2 overexpression. Overexpression of GBP2 did not affect the expression of these tested ISGs (Fig. S1A). This was further confirmed in HEK293T cells (Fig. S1B). These results suggest that the antiviral activity of GBP2 is independent of ISG induction.
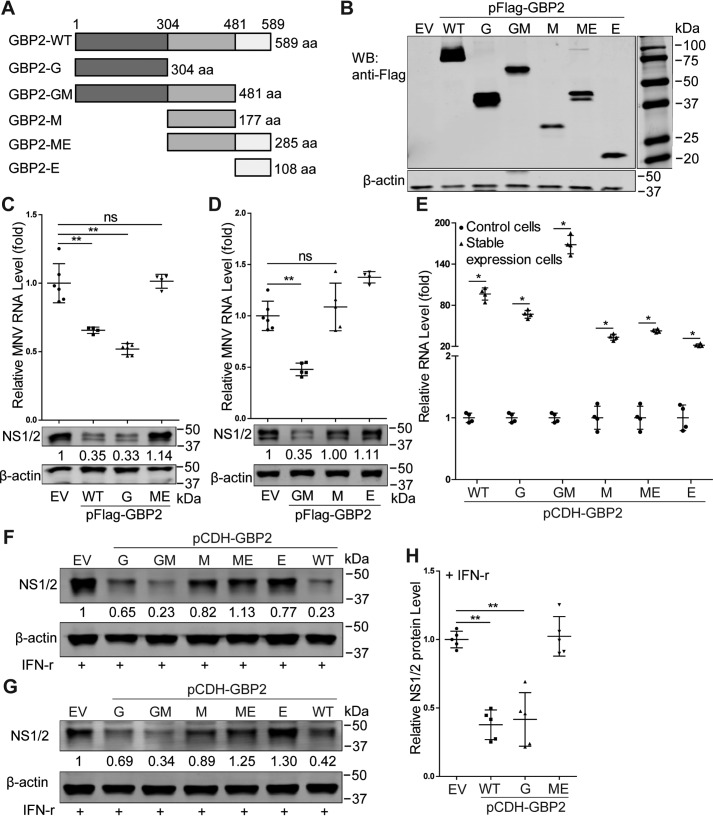
The N terminus of GBP2 is essential for the anti-MNV activity
Structural analysis of human GBP1 has revealed three distinct domains, including the N-terminal globular GTPase domain (G domain), the following two-helix part presenting as the middle domain (M domain), and the C-terminal GTPase effector domain (E domain) (19). Based on the human GBP1 structure, we predicted and modeled the mouse GBP2 protein structure (Fig. S2) by using the online software SWISS-MODEL (RRID:SCR_018123) and mapped GBP2 into three corresponding domains (Fig. 6A). To investigate which domain is responsible for the antiviral actions, we constructed FLAG-tagged truncated mutants of GBP2 and verified their expression in HEK293T cells (Fig. 6B). Their anti-MNV activities were examined in HEK293T cells transfected with FLAG-tagged CD300lf. We found that the G and GM domains of GBP2 inhibited viral replication shown at both viral RNA and protein levels (Fig. 6, C and D). In contrast, the M, ME, and E domains did not exert anti-MNV activity (Fig. 6, C and D). To further confirm, we established stable expression of GBP2 truncated mutants in RAW264.7 cells (Fig. 6E). These cells were infected with MNV-1 and then treated with IFN-γ for 24 h. We found that the G and GM domains of GBP2 enhanced IFN-γ–mediated anti-MNV activity, whereas the remaining domains appeared not to influence viral replication (Fig. 6, F–H). Taken together, the N terminus of GBP2 is indispensable for restricting MNV-1 replication in human cells and augmenting IFN-γ–mediated anti-MNV ability in murine macrophages.
Figure 6.
The N terminus of GBP2 is essential for its anti-MNV activity. A, schematic representation of the protein domains of GBP2. B, expression of full-length or truncated forms of GBP2. The indicated expression plasmids were transfected into HEK293T cells for 24 h, and the cell lysates were analyzed by Western blotting (WB) using antibodies against FLAG tag and β-actin. C and D, qRT-PCR (n = 4–6) and Western blot analysis of MNV RNA level and NS1/2 protein in HEK293T cells that were transfected with pFLAG-CD300lf (0.5 μg) and the indicated plasmids (1 μg) for 24 h and then infected with MNV-1 (MOI 1) for another 24 h. E, identification of stable expression of GBP2 truncated mutants by lentiviral vectors in RAW264.7 cells by qRT-PCR assay (n = 4). Shown is Western blot analysis of MNV NS1/2 protein level in RAW264.7 cells with stable expression of GBP2 truncated mutants by lentiviral vectors that were infected with MNV-1 at an MOI of 0.01 (F) or at an MOI of 1 (G) for 1 h and then treated with IFN-γ (10 units/ml) for 24 h. H, viral NS1/2 protein expression by Western blotting was statistically analyzed (n = 5). Data in C, D, E, and H were normalized to the empty virus (EV) control (set as 1). *, p < 0.05; **, p < 0.01; ns, not significant. β-Actin was used as a loading control. aa, amino acids. Error bars, S.D.
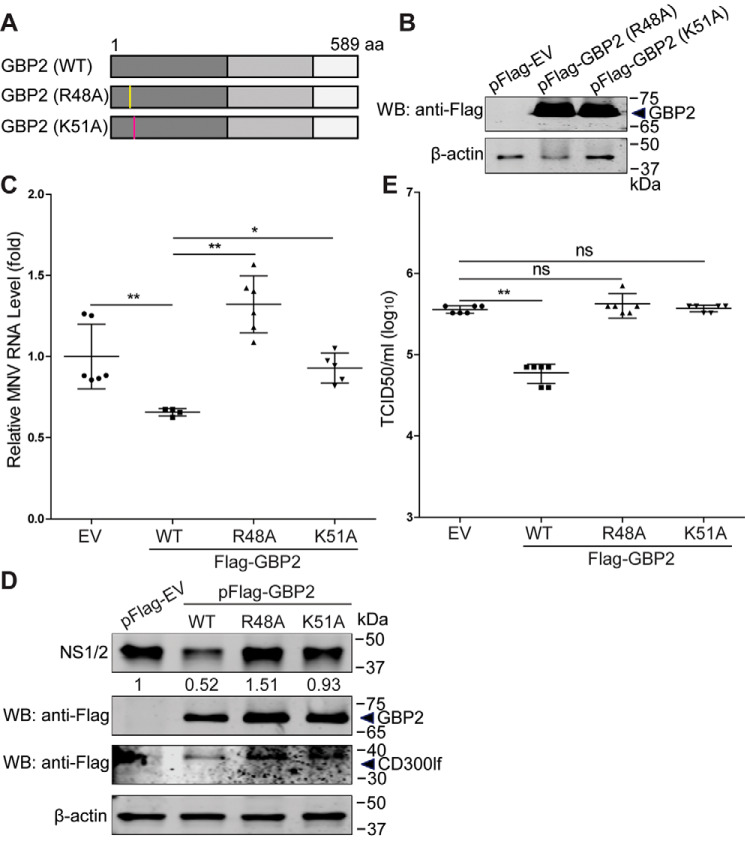
The Arg-48 and Lys-51 residues are critical for GBP2-mediated anti-MNV activity
It has been reported that the GTPase activity of GBPs is important for the antiviral activity (27, 36). The Arg-48 and Lys-51 residues are important for its GTPase activity of both hGBP1 (36, 37) and mGBP2 (38). Thus, we constructed two GBP2 mutants, including GBP2 (R48A) and GBP2 (K51A) (Fig. 7A), and confirmed their expression in HEK293T cells (Fig. 7B). To determine the antiviral effects of these two GBP2 mutants, we transfected the constructed mutants and viral receptor vectors into HEK293T cells and then infected with MNV-1 for 24 h. Compared with the WT GBP2, these two mutants failed to restrict MNV-1 at both viral RNA (Fig. 7C) and NS1/2 protein levels (Fig. 7D). Measuring viral titers in the supernatants of infected cells further confirmed these results (Fig. 7E). Thus, the Arg-48 and Lys-51 residues of GBP2 are essential for its anti-MNV activity.
Figure 7.
The Arg-48 and Lys-51 residues of GBP2 are critical for its anti-MNV activity. A, schematic representation of the GBP2 mutations. B, expression of GBP2 mutations. The indicated expression plasmids were transfected into HEK293T cells for 24 h, and the cell lysates were analyzed by Western blotting (WB) using antibodies against FLAG tag and β-actin. HEK293T cells were transfected with pFLAG-CD300lf (0.5 μg) and pFLAG-GBP2 (WT), pFLAG-GBP2 (R48A), pFLAG-GBP2 (K51A), or empty vectors for 24 h and then infected with MNV-1 at an MOI of 1 for 24 h. The MNV RNA (C) (n = 4–6) and NS1/2 protein (D) levels were analyzed by qRT-PCR and Western blotting, respectively. E, the viral titers in the supernatants were examined by a TCID50 assay (n = 6). Data in C and E were normalized to the empty virus (EV) control (set as 1). *, p < 0.05; **, p < 0.01; ns, not significant. β-Actin was used as a loading control. Error bars, S.D.
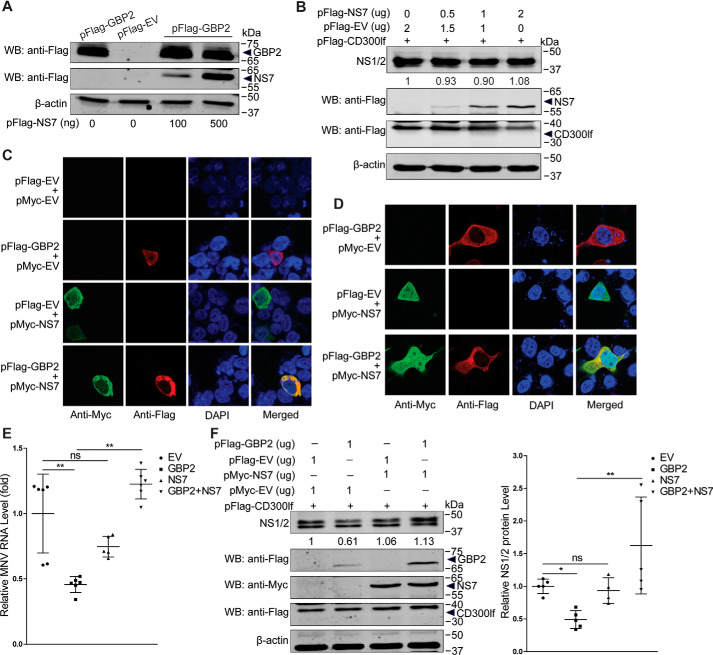
MNV NS7 antagonizes the antiviral activity of GBP2
Studies have reported that the viral replicase has a negative regulation on porcine GBP1-mediated inhibition of CSFV (27) and human GBP1-mediated restriction of HCV (36). Thus, we next determined whether the MNV replicase NS7 has a role on GBP2-mediated antiviral activity. We co-transfected pFLAG-GBP2 and pFLAG-NS7 into HEK293T cells. We found that GBP2 expression was not affected by NS7 expression (Fig. 8A), and NS7 did not affect viral NS1/2 protein expression (Fig. 8B). Moreover, confocal microscopy indicated that besides localization in the nucleus, NS7 could co-localize with GBP2 in the cytoplasm of the cells (Fig. 8, C and D). To further investigate NS7 on GBP2-mediated antiviral activity, we co-transfected pFLAG-CD300lf, pFLAG-GBP2, and pMyc-NS7 into HEK293T cells and then infected with MNV-1 for 24 h. We found that NS7 attenuated GBP2-mediated anti-MNV effects shown at both viral RNA and NS1/2 protein levels (Fig. 8, E and F). These results indicated a potential antagonistic effects of NS7 on GBP2-mediated anti-MNV activity.
Figure 8.
MNV NS7 antagonizes GBP2-mediated anti-MNV activity. A, HEK293T cells were co-transfected with pFLAG-GBP2 (1 μg) and pFLAG-EV (1 μg) or pFLAG-NS7 with the indicated concentrations for 24 h. The cell lysates were collected for analysis by Western blotting (WB). B, HEK293T cells were co-transfected with pFLAG-CD300lf (0.5 μg) and pFLAG-EV or pFLAG-NS7 with the indicated concentrations for 24 h and then infected with MNV-1 (MOI 1) for 24 h. The cell lysates were collected for analysis by Western blotting using antibodies against the NS1/2, FLAG tag, and β-actin. Co-localization of GBP2 with MNV NS7 was examined. Expression plasmids pFLAG-GBP2 (1 μg) and pMyc-NS7 (1 μg) or the vectors (1 μg) were co-transfected into HEK293T (C) and COS-1 (D) cells and subjected to a confocal assay, respectively. HEK293T cells were cotransfected with pFLAG-CD300lf (0.5 μg), pFLAG-GBP2, pMyc-NS7, or the empty vectors with the indicated concentrations for 24 h and then infected with MNV-1 (MOI 1) for 24 h. The total RNA and cell lysates were collected and analyzed by qRT-PCR (E) (n = 5–6) and Western blotting (F) (n = 4–5). Data in E and F (right) were normalized to the empty virus (EV) control (set as 1). *, p < 0.05; **, p < 0.01; ns, not significant. β-Actin was used as a loading control. Error bars, S.D.
Discussion
GBPs as a group of IFN-induced proteins are essential for innate immune response against intracellular bacterial, viral, and protozoan pathogens (18, 22, 25). With respect to viruses, it has been shown that human GBP1 can restrict replication of vesicular stomatitis virus and encephalomyocarditis virus (39), whereas several GBPs have been linked to host defense against HIV, CSFV, and influenza virus (25, 27, 40). A recent study suggested potential interactions between this family of GTPases and norovirus replication (32). Here, we further show that GBP2 effectively responds to and defends murine norovirus infection. Our findings fit well with the increasing momentum and support for the notion that IFN-inducible GTPases are crucial for cell-autonomous host defense against viral infection in general and norovirus in particular (7). We first demonstrated that GBP2 mediates IFN-γ–triggered anti-MNV activity in murine macrophages, whereas GBP2 alone is not sufficient to inhibit MNV. By exploiting ectopic expression of the viral receptors (6), we conferred susceptibility of human HEK293T cells to MNV infection. We further demonstrated that GBP2 alone is sufficient to potently inhibit MNV in human epithelial cells, without requiring the presence of IFN-γ. We speculate that the disparity in requiring IFN-γ may be attributed to the differences in species, cell types, and the expression patterns of GBP2.
Although ISGs are known as antiviral effectors, their modes of actions are diverse, including direct and indirect antiviral actions. Some ISGs have been linked to immunity against norovirus infection. IRF1 has been reported to contribute to IFN-γ–mediated inhibition of MNV replication in macrophages (16). This may be an indirect effect, as shown in the setting of hepatitis E virus (HEV) infection. IRF1 activates STAT1 to induce the expression of a wide range of ISGs that eventually inhibit HEV replication (33). ISG15 inhibits an early step of the MNV life cycle upstream of viral genome transcription (17). GBP1 has been reported to restrict DENV replication by modulating NF-κB activity, leading to the production of antiviral and pro-inflammatory cytokines (41). As seen in this study, the inhibition of MNV replication by GBP2 appears to be independent of ISG induction. Interestingly, recent studies have shown an association between GBPs and inflammasome activation. The inflammasome machinery is essential for host defense against viral pathogens (28, 29). MNV infection triggers NLRP3 inflammasome activation in primary BMDMs with STAT1 deficiency or in TLR2-primed BMDMs (42). MNV infection persists much longer in NLRP6-deficient compared with WT mice (43). It is thus tempting to suggest that GBP2 functions through inflammasome activation, and this scenario should be investigated in future experimentation.
Structurally, hGBP1 can be mapped into three domains with distinct functionality (19). The N-terminal domain of GBP1 is responsible for the antiviral activity against influenza A virus, HCV, and CSFV infections (27, 36, 37). The C-terminal domain of mGBP2 dictates the recruitment to the Toxoplasma gondii parasitophorous vacuole and contributes to control of its replication (18). Based on the hGBP1 structure (Protein Data Bank entry 1F5N) (19), we modeled the different domains of mouse GBP2, including the G, M, and E domains. By constructing the truncated mutants of GBP2, we found that the N-terminal G-domain is important for the anti-MNV activity in human epithelial cells and for augmenting IFN-γ–mediated anti-MNV response in murine macrophages.
The Arg-48 and Lys-51 residues within the N-terminal domain are essential for the GTPase activity and antiviral function of GBP1 (27, 36). Arg-48 in the P-loop is highly conserved across different GBPs and functions as a GTPase-activating “arginine finger” involved in the multimerization process. The R48A mutant has much weaker GTPase activity (38). Lys-51 (K51A) mutation in mouse GBP2 leads to a nearly complete loss of function, including hydrolysis, dimerization, and nucleotide binding (38). In this study, we found that the R48A and K51A mutants attenuate the anti-MNV effects of GBP2, suggesting the potential requirement of GTPase activity. Viruses have developed sophisticated strategies to evade host defense (44). MNV NS1/2 interacts with host protein VAPA to enhance viral replication (45), and NS3 interacts with microtubule-associated protein GEF-H1, which plays a role in immune detection of viral replication (46). MNV NS7 is the viral replicase and catalyzes replication of the viral genome. NS7 presents a diffused pattern both in cell cytoplasm and nucleus (47, 48). In this study, we revealed that NS7 co-localizes with GBP2 in the cytoplasm by transient expression and antagonizes GBP2-mediated anti-MNV activity. Viral replicases including NS5B of HCV and NS5A of CSFV, and NS1 of influenza A virus have been reported to interact with GBP1 (36, 37) and attenuate GBP1-mediated antiviral activity (27, 36). Thus, the potential interaction of NS7 with GBP2 and the possible inhibitory effect on GTPase activity of GBP2 will be interesting subjects for further study.
In summary, MNV-1 infection activates the expression of GBP2, an IFN-inducible GTPase. GBP2 orchestrates innate immune defense against MNV independent of its N terminus. However, MNV NS7 can co-localize with GBP2 in the cytoplasm and antagonize GBP2-mediated anti-MNV activity. These findings shed new light on norovirus-host interactions and shall be helpful for better understanding the pathogenesis and developing new antiviral strategies.
Experimental procedures
Reagents
Mouse IFN-γ (ab9922, Abcam) was dissolved in PBS. Stocks of JAK inhibitor 1 (SC-204021, Santa Cruz Biotechnology, Inc.) were dissolved in DMSO (Sigma) with a final concentration of 5 mg/ml. Puromycin (P8833, Sigma) was dissolved in PBS with a final concentration of 10 mg/ml. The Q5® site-directed mutagenesis kit (New England Biolabs) was used. GBP2 antibody (11854-1-AP) was purchased from Proteintech. STAT1 (catalog no. 9172) antibody was purchased from Cell Signaling Technology. Rabbit polyclonal antisera to MNV NS1/2 was kindly provided by Prof. Vernon K. Ward (School of Biomedical Sciences, University of Otago, New Zealand) (49). β-Actin antibody (catalog no. sc-47778) was purchased from Santa Cruz Biotechnology. Secondary antibodies, including IRDye® 800CW-conjugated goat anti-rabbit and goat anti-mouse IgGs (LI-COR Biosciences, Lincoln, NE, USA) and anti-rabbit IgG(H+L), F(ab′)2 fragment (Alexa Fluor® 488 conjugate) and anti-mouse IgG(H+L), F(ab′)2 fragment (Alexa Fluor 594 conjugate) were used, as appropriate.
Cells and viruses
RAW264.7, J774A.1, COS-1, and human embryonic kidney (HEK293T) cells were cultured in Dulbecco's modified Eagle's medium (Lonza Verviers, Verviers, Belgium) supplemented with 10% (v/v) heat-inactivated fetal calf serum (Hyclone, Logan, UT, USA), 100 μg/ml streptomycin, and 100 IU/ml penicillin. MNV-1 (murine norovirus strain MNV-1.CW1) (4) was produced by consecutively inoculating the virus (kindly provided by Prof. Herbert Virgin (Washington University School of Medicine) into RAW264.7 cells. The MNV-1 cultures were purified, aliquoted, and stored at −80 °C for all subsequent experiments. The MNV-1 stock was quantified three independent times by the 50% tissue culture infective dose (TCID50).
TCID50
MNV-1 was quantified by a TCID50 assay. Briefly, 10-fold dilutions of MNV-1 were inoculated into RAW264.7 cells grown in a 96-well tissue culture plate at 1,000 cells/well. The plate was incubated at 37 °C for another 5 days, followed by observing the cytopathic effect of each well under a light scope. TCID50 was calculated by using the Reed–Muench method.
Plasmid construction and cell transfection
The full-length mouse GBP2 gene was amplified from IFN-γ–stimulated RAW264.7 cells and cloned into pcDNA3.1/FLAG-HA (Addgene), pcDNA3.1/Myc-His (provided by Dr. Shuaiyang Zhao, Chinese Academy of Agricultural Sciences), and the lentiviral vector pCDH-CMV-MCS-EF1-GFP-T2A-Puro (Sanbio BV) to generate pFLAG-GBP2, pMyc-GBP2, and pCDH-GBP2, respectively. The truncated mutants of GBP2 were further amplified and cloned into the FLAG-tagged and lentiviral vectors, respectively. The MNV NS7 gene was amplified from cDNA that was extracted from MNV-1–infected RAW264.7 cells and cloned into FLAG- and Myc-tagged empty vectors, respectively. The FLAG-CD300lf vector was kindly provided by Prof. Herbert Virgin (6). All primer sequences used for plasmid construction are listed in Table S1.
HEK293T cells were transfected with various plasmids at the indicated concentrations using FuGENE HD transfection reagent (catalogue no. E2311; Promega) according to the manufacturer's instructions. Where necessary, the appropriate empty vector was used to maintain a constant amount of plasmid DNA per transfection. At 6 h post-transfection, fresh Dulbecco's modified Eagle's medium containing 10% FCS replaced the transfection mixture, and the cells were incubated at 37 °C.
Silencing or overexpressing mouse GBP2 by lentiviral vectors
Lentiviral pLKO.1 knockdown vectors (Sigma–Aldrich) targeting mouse GBP2 were obtained from the Erasmus Biomics Center. The lentiviral pseudoparticles were produced in HEK293T cells as described previously (50). After a pilot study, the shRNA vectors exerting optimal gene knockdown were selected. These shRNA sequences are listed in Table S2. Stable gene knockdown cells were generated after lentiviral vector transduction and puromycin (5 μg/ml; Sigma) selection. For stable expression, GBP2 WT and truncated overexpression lentiviral vectors were used to generate the GBP2 stable expression RAW264.7 cell lines. Meanwhile, control shRNA and the lentiviral empty vectors were also used as control, respectively.
qRT-PCR
Total RNA was isolated with a Macherey NucleoSpin RNA II Kit (Bioke, Leiden, The Netherlands) and quantified with a Nanodrop ND-1000 (Wilmington, DE). cDNA was synthesized from 500 ng of RNA using a cDNA synthesis kit (TaKaRa Bio, Inc., Shiga, Japan). The cDNA of all targeted gene transcripts were quantified by SYBR Green–based (Applied Biosystems) real-time PCR on the StepOnePlusTM system (Thermo Fisher Scientific) according to the manufacturer's instructions. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and murine GAPDH genes were used as reference genes to normalize gene expression. The relative expression of targeted gene was calculated as 2−ΔΔCT, where ΔΔCT = ΔCTsample − ΔCTcontrol (ΔCT = CT(targeted gene) − CT(GAPDH)). All primer sequences are listed in Table S3.
Western blotting
Cultured cells were lysed in Laemmli sample buffer containing 0.1 m DTT and heated 5 min at 95 °C and then loaded onto a 10% SDS-polyacrylamide gel. Proteins were electrophoretically transferred onto a polyvinylidene difluoride membrane (pore size, 0.45 μm; Invitrogen) for 2 h with an electric current of 250 mA. Subsequently, the membrane was blocked with a mixture of 2.5 ml of blocking buffer (Odyssey) and 2.5 ml of PBS containing 0.05% Tween 20 for 1 h, followed by overnight incubation with primary antibodies (1:1000) at 4 °C. The membrane was washed three times and then incubated with IRDye-conjugated secondary antibody (1:5000) for 1 h. After washing three times, protein bands were detected with the Odyssey 3.0 IR Imaging System (LI-COR Biosciences).
Confocal fluorescence microscopy
HEK293T and COS-1 cells (3 × 104 cells/well) were cotransfected with pFLAG-GBP2 and pMyc-NS7 (1 μg/each) into a μ-slide 8-well chamber (catalog no. 80826, ibidi GmbH) at 37 °C for 24 h. The cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.2% Triton X-100, blocked with 5% skim milk for 1 h, reacted with the appropriate antibody, and stained with 4′,6-diamidino-2-phenylindole. Antibodies used in this study were mouse anti-FLAG mAb (F1804, Sigma–Aldrich), rabbit anti-Myc polyclonal antibody (Cell Signaling), and anti-rabbit IgG(H+L), F(ab′)2 fragment (Alexa Fluor® 488 conjugate) or anti-mouse IgG(H+L), F(ab′)2 fragment (Alexa Fluor 594 conjugate) secondary antibodies. Imaging was performed on a Leica SP5 confocal microscopy using a ×63 oil objective.
Statistical analysis
Data are presented as the mean ± S.D. Comparisons between groups were performed with the Mann–Whitney test using GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA). Differences were considered significant at a p value of <0.05.
Data availability
All of the data are contained within the article and supporting materials.
Supplementary Material
Acknowledgments
We gratefully acknowledge Prof. Herbert W. Virgin (Washington University, St Louis, MO, USA) for providing the MNV-1 and FLAG-tagged CD300lf plasmid; Prof. Vernon K. Ward (School of Biomedical Sciences, University of Otago, New Zealand) for providing rabbit polyclonal antisera to MNV NS1–2; Dr. Shuaiyang Zhao (Chinese Academy of Agricultural Sciences) for providing the pcDNA3.1/Myc-His vector; Prof. Andrea Kröger (Helmholtz Centre for Infection Research) for providing the WT and STAT1−/− MEFs; and Dr. Ron Smits (Erasmus MC-University Medical Center, Rotterdam, The Netherlands) for providing the COS-1 cells.
This article contains supporting information.
Author contributions—P. Y. and Q. P. conceptualization; P. Y., Yang L., Yunlong L., and Z. M. formal analysis; P. Y. investigation; P. Y., Yang L., Yunlong L., and Z. M. methodology; P. Y. writing-original draft; P. Y., M. P. P., and Q. P. writing-review and editing; M. P. P. and Q. P. supervision; Q. P. funding acquisition; Q. P. validation; Q. P. project administration.
Funding and additional information—This research is supported by China Scholarship Council Ph.D. Fellowships 201708620177 (to P. Y.), 201703250073 (Yang L.), 201708530243 (Yunlong L.), and 201708530234 (Z. M.) and Netherlands Organization for Scientific Research VIDI Grant 91719300 (to Q. P.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- HuNV
- human norovirus
- MNV
- murine norovirus
- IFN
- interferon
- JAK
- Janus kinase
- STAT
- signal transducers and activators of transcription
- ISG
- interferon-stimulated gene
- GBP
- guanylate-binding proteins
- HCV
- hepatitis C virus
- HEV
- hepatitis E virus
- CSFV
- classical swine fever virus
- MEF
- mouse embryo fibroblast
- BMDM
- bone marrow–derived macrophage
- TCID50
- 50% tissue culture infective dose
- qRT-PCR
- quantitative RT-PCR
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MOI
- multiplicity of infection
- shRNA
- short hairpin RNA.
References
- 1. Glass R. I., Parashar U. D., and Estes M. K. (2009) Norovirus gastroenteritis. N. Eng. J. Med. 361, 1776–1785 10.1056/NEJMra0804575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bok K., and Green K. Y. (2012) Norovirus gastroenteritis in immunocompromised patients. N. Eng. J. Med. 367, 2126–2132 10.1056/NEJMra1207742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karst S. M., Wobus C. E., Goodfellow I. G., Green K. Y., and Virgin H. W. (2014) Advances in norovirus biology. Cell Host Microbe 15, 668–680 10.1016/j.chom.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wobus C. E., Karst S. M., Thackray L. B., Chang K.-O., Sosnovtsev S. V., Belliot G., Krug A., Mackenzie J. M., Green K. Y., and Virgin H. W. (2004) Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2, e432 10.1371/journal.pbio.0020432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wobus C. E., Thackray L. B., and Virgin H. W. 4th (2006) Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80, 5104–5112 10.1128/JVI.02346-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Orchard R. C., Wilen C. B., Doench J. G., Baldridge M. T., McCune B. T., Lee Y.-C. J., Lee S., Pruett-Miller S. M., Nelson C. A., Fremont D. H., and Virgin H. W. (2016) Discovery of a proteinaceous cellular receptor for a norovirus. Science 353, 933–936 10.1126/science.aaf1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orchard R. C., Sullender M. E., Dunlap B. F., Balce D. R., Doench J. G., and Virgin H. W. (2019) Identification of antinorovirus genes in human cells using genome-wide CRISPR activation screening. J. Virol. 93, e01324–18 10.1128/JVI.01324-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McFadden N., Bailey D., Carrara G., Benson A., Chaudhry Y., Shortland A., Heeney J., Yarovinsky F., Simmonds P., and Macdonald A., and Goodfellow I. (2011) Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 7, e1002413 10.1371/journal.ppat.1002413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu S., Regev D., Watanabe M., Hickman D., Moussatche N., Jesus D. M., Kahan S. M., Napthine S., Brierley I., and Hunter R. N. 3rd, Devabhaktuni D., Jones M. K., and Karst S. M. (2013) Identification of immune and viral correlates of norovirus protective immunity through comparative study of intra-cluster norovirus strains. PLoS Pathog. 9, e1003592 10.1371/journal.ppat.1003592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee S., Liu H., Wilen C. B., Sychev Z. E., Desai C., Hykes B. L. Jr, Orchard R. C., McCune B. T., Kim K. W., Nice T. J., Handley S. A., Baldridge M. T., Amarasinghe G. K., and Virgin H. W. (2019) A secreted viral nonstructural protein determines intestinal norovirus pathogenesis. Cell Host Microbe 25, 845–857.e5 10.1016/j.chom.2019.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Högbom M., Jäger K., Robel I., Unge T., and Rohayem J. (2009) The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J. Gen. Virol. 90, 281–291 10.1099/vir.0.005629-0 [DOI] [PubMed] [Google Scholar]
- 12. Subba-Reddy C. V., Goodfellow I., and Kao C. C. (2011) VPg-primed RNA synthesis of norovirus RNA-dependent RNA polymerases by using a novel cell-based assay. J. Virol. 85, 13027–13037 10.1128/JVI.06191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu J., and Chen Z. J. (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 10.1146/annurev-immunol-032713-120156 [DOI] [PubMed] [Google Scholar]
- 14. Wang W., Xu L., Su J., Peppelenbosch M. P., and Pan Q. (2017) Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol. 25, 573–584 10.1016/j.tim.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schoggins J. W., Wilson S. J., Panis M., Murphy M. Y., Jones C. T., Bieniasz P., and Rice C. M. (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maloney N. S., Thackray L. B., Goel G., Hwang S., Duan E., Vachharajani P., Xavier R., and Virgin H. W. (2012) Essential cell autonomous role for interferon regulatory factor 1 in interferon-γ-mediated inhibition of norovirus replication in macrophages. J. Virol. 86, 12655–12664 10.1128/JVI.01564-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez M. R., Monte K., Thackray L. B., and Lenschow D. J. (2014) ISG15 functions as an interferon-mediated antiviral effector early in the murine norovirus life cycle. J. Virol. 88, 9277–9286 10.1128/JVI.01422-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Degrandi D., Kravets E., Konermann C., Beuter-Gunia C., Klümpers V., Lahme S., Wischmann E., Mausberg A. K., Beer-Hammer S., and Pfeffer K. (2013) Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc. Natl. Acad. Sci. U.S.A. 110, 294–299 10.1073/pnas.1205635110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prakash B., Praefcke G. J. K., Renault L., Wittinghofer A., and Herrmann C. (2000) Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403, 567–571 10.1038/35000617 [DOI] [PubMed] [Google Scholar]
- 20. Kresse A., Konermann C., Degrandi D., Beuter-Gunia C., Wuerthner J., Pfeffer K., and Beer S. (2008) Analyses of murine GBP homology clusters based on in silico, in vitro and in vivo studies. BMC Genomics 9, 158 10.1186/1471-2164-9-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Man S. M., Place D. E., Kuriakose T., and Kanneganti T. D. (2017) Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation. J. Leukocyte Biol. 101, 143–150 10.1189/jlb.4MR0516-223R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wandel M. P., Pathe C., Werner E. I., Ellison C. J., Boyle K. B., von der Malsburg A., Rohde J., and Randow F. (2017) GBPs inhibit motility of Shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9.8. Cell Host Microbe 22, 507–518.e5 10.1016/j.chom.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li P., Jiang W., Yu Q., Liu W., Zhou P., Li J., Xu J., Xu B., Wang F., and Shao F. (2017) Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551, 378–383 10.1038/nature24467 [DOI] [PubMed] [Google Scholar]
- 24. Braun E., Hotter D., Koepke L., Zech F., Gross R., Sparrer K. M. J., Müller J. A., Pfaller C. K., Heusinger E., and Wombacher R., Sutter K., Dittmer U., Winkler M., Simmons G., Jakobsen M. R., et al. (2019) Guanylate-binding proteins 2 and 5 exert broad antiviral activity by inhibiting furin-mediated processing of viral envelope proteins. Cell Rep. 27, 2092–2104.e10 10.1016/j.celrep.2019.04.063 [DOI] [PubMed] [Google Scholar]
- 25. Krapp C., Hotter D., Gawanbacht A., McLaren P. J., Kluge S. F., Stürzel C. M., Mack K., Reith E., Engelhart S., Ciuffi A., Hornung V., Sauter D., Telenti A., and Kirchhoff F. (2016) Guanylate binding protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe 19, 504–514 10.1016/j.chom.2016.02.019 [DOI] [PubMed] [Google Scholar]
- 26. Itsui Y., Sakamoto N., Kurosaki M., Kanazawa N., Tanabe Y., Koyama T., Takeda Y., Nakagawa M., Kakinuma S., Sekine Y., Maekawa S., Enomoto N., and Watanabe M. (2006) Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J. Viral Hepat. 13, 690–700 10.1111/j.1365-2893.2006.00732.x [DOI] [PubMed] [Google Scholar]
- 27. Li L. F., Yu J., Li Y., Wang J., Li S., Zhang L., Xia S. L., Yang Q., Wang X., and Yu S., Luo Y., Sun Y., Zhu Y., Munir M., and Qiu H. J. (2016) Guanylate-binding protein 1, an interferon-induced GTPase, exerts an antiviral activity against classical swine fever virus depending on its GTPase activity. J. Virol. 90, 4412–4426 10.1128/JVI.02718-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shenoy A. R., Wellington D. A., Kumar P., Kassa H., Booth C. J., Cresswell P., and MacMicking J. D. (2012) GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science 336, 481–485 10.1126/science.1217141 [DOI] [PubMed] [Google Scholar]
- 29. Meunier E., Wallet P., Dreier R. F., Costanzo S., Anton L., Rühl S., Dussurgey S., Dick M. S., Kistner A., Rigard M., Degrandi D., Pfeffer K., Yamamoto M., Henry T., and Broz P. (2015) Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16, 476–484 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ichinohe T., Lee H. K., Ogura Y., Flavell R., and Iwasaki A. (2009) Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206, 79–87 10.1084/jem.20081667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu S., Ding S., Wang P., Wei Z., Pan W., Palm N. W., Yang Y., Yu H., Li H.-B., and Wang G., Li H. B., Wang G., Lei X., de Zoete M. R., Zhao J., Zheng Y., Chen H., et al. (2017) Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 546, 667–670 10.1038/nature22967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Biering S. B., Choi J., Halstrom R. A., Brown H. M., Beatty W. L., Lee S., McCune B. T., Dominici E., Williams L. E., Orchard R. C., Wilen C. B., Yamamoto M., Coers J., Taylor G. A., and Hwang S. (2017) Viral replication complexes are targeted by LC3-guided interferon-inducible GTPases. Cell Host Microbe 22, 74–85.e7 10.1016/j.chom.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu L., Zhou X., Wang W., Wang Y., Yin Y., Laan L. J. W. v. d., Sprengers D., Metselaar H. J., Peppelenbosch M. P., and Pan Q. (2016) IFN regulatory factor 1 restricts hepatitis E virus replication by activating STAT1 to induce antiviral IFN-stimulated genes. FASEB J. 30, 3352–3367 10.1096/fj.201600356R [DOI] [PubMed] [Google Scholar]
- 34. Dang W., Xu L., Yin Y., Chen S., Wang W., Hakim M. S., Chang K. O., Peppelenbosch M. P., and Pan Q. (2018) IRF-1, RIG-I and MDA5 display potent antiviral activities against norovirus coordinately induced by different types of interferons. Antiviral Res. 155, 48–59 10.1016/j.antiviral.2018.05.004 [DOI] [PubMed] [Google Scholar]
- 35. Xu L., Wang W., Li Y., Zhou X., Yin Y., Wang Y., de Man R. A., van der Laan L. J. W., Huang F., Kamar N., Peppelenbosch M. P., and Pan Q. (2017) RIG-I is a key antiviral interferon-stimulated gene against hepatitis E virus regardless of interferon production. Hepatology 65, 1823–1839 10.1002/hep.29105 [DOI] [PubMed] [Google Scholar]
- 36. Itsui Y., Sakamoto N., Kakinuma S., Nakagawa M., Sekine-Osajima Y., Tasaka-Fujita M., Nishimura-Sakurai Y., Suda G., Karakama Y., Mishima K., Yamamoto M., Watanabe T., Ueyama M., Funaoka Y., Azuma S., and Watanabe M. (2009) Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50, 1727–1737 10.1002/hep.23195 [DOI] [PubMed] [Google Scholar]
- 37. Zhu Z., Shi Z., Yan W., Wei J., Shao D., Deng X., Wang S., Li B., Tong G., and Ma Z. (2013) Nonstructural protein 1 of influenza A virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity. PLoS One 8, e55920 10.1371/journal.pone.0055920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kravets E., Degrandi D., Weidtkamp-Peters S., Ries B., Konermann C., Felekyan S., Dargazanli J. M., Praefcke G. J. K., Seidel C. A. M., Schmitt L., Praefcke G. J., Seidel C. A., Schmitt L., Smits S. H., and Pfeffer K. (2012) The GTPase activity of murine guanylate-binding protein 2 (mGBP2) controls the intracellular localization and recruitment to the parasitophorous vacuole of Toxoplasma gondii. J. Biol. Chem. 287, 27452–27466 10.1074/jbc.M112.379636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anderson S. L., Carton J. M., Lou J., Xing L., and Rubin B. Y. (1999) Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256, 8–14 10.1006/viro.1999.9614 [DOI] [PubMed] [Google Scholar]
- 40. Feng J., Cao Z., Wang L., Wan Y., Peng N., Wang Q., Chen X., Zhou Y., and Zhu Y. (2017) Inducible GBP5 mediates the antiviral response via interferon-related pathways during influenza a virus infection. J. Innate Immun. 9, 419–435 10.1159/000460294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pan W., Zuo X., Feng T., Shi X., and Dai J. (2012) Guanylate-binding protein 1 participates in cellular antiviral response to dengue virus. Virol. J. 9, 292 10.1186/1743-422X-9-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dubois H., Sorgeloos F., Sarvestani S. T., Martens L., Saeys Y., Mackenzie J. M., Lamkanfi M., van Loo G., Goodfellow I., and Wullaert A. (2019) Nlrp3 inflammasome activation and Gasdermin D-driven pyroptosis are immunopathogenic upon gastrointestinal norovirus infection. PLoS Pathog. 15, e1007709 10.1371/journal.ppat.1007709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang P., Zhu S., Yang L., Cui S., Pan W., Jackson R., Zheng Y., Rongvaux A., Sun Q., and Yang G., Gao S., Lin R., You F., Flavell R., and Fikrig E. (2015) Nlrp6 regulates intestinal antiviral innate immunity. Science 350, 826–830 10.1126/science.aab3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y., Qu C., Yu P., Ou X., Pan Q., and Wang W. (2019) The interplay between host innate immunity and hepatitis E virus. Viruses 11, E541 10.3390/v11060541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCune B. T., Tang W., Lu J., Eaglesham J. B., Thorne L., Mayer A. E., Condiff E., Nice T. J., Goodfellow I., Krezel A. M., and Virgin H. W. (2017) Noroviruses co-opt the function of host proteins VAPA and VAPB for replication via a phenylalanine-phenylalanine-acidic-tract-motif mimic in nonstructural viral protein NS1/2. MBio 8, e00668–17 10.1128/mBio.00668-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fritzlar S., White P. A., and Mackenzie J. M. (2019) The microtubule-associated innate immune sensor GEF-H1 does not influence mouse norovirus replication in murine macrophages. Viruses 11, E47 10.3390/v11010047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fernandez-Vega V., Sosnovtsev S. V., Belliot G., King A. D., Mitra T., Gorbalenya A., and Green K. Y. (2004) Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J. Virol. 78, 4827–4837 10.1128/JVI.78.9.4827-4837.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hyde J. L., and Mackenzie J. M. (2010) Subcellular localization of the MNV-1 ORF1 proteins and their potential roles in the formation of the MNV-1 replication complex. Virology 406, 138–148 10.1016/j.virol.2010.06.047 [DOI] [PubMed] [Google Scholar]
- 49. Davies C., Brown C. M., Westphal D., Ward J. M., and Ward V. K. (2015) Murine norovirus replication induces G0/G1 cell cycle arrest in asynchronously growing cells. J. Virol. 89, 6057–6066 10.1128/JVI.03673-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Y., Zhou X., Debing Y., Chen K., Van Der Laan L. J., Neyts J., Janssen H. L., Metselaar H. J., Peppelenbosch M. P., and Pan Q. (2014) Calcineurin inhibitors stimulate and mycophenolic acid inhibits replication of hepatitis E virus. Gastroenterology 146, 1775–1783 10.1053/j.gastro.2014.02.036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data are contained within the article and supporting materials.