Abstract
Simple Summary
According to the tail types, sheep can be briefly classified into three groups (fat-tailed, fat-rumped, and thin-tailed sheep). In this study, we used these three typical breeds from Chinese indigenous sheep breeds to perform a genome scan for selective sweeps using Ovine Infinium HD SNP BeadChip genotype data. Results showed that 25 genomic regions exhibited selection signals and harbored 73 positional candidate genes. These genes were documented not only to be associated with tail fat formation, but also be related to reproduction, body conformation, and appearance. Our findings contributed to understanding genetic basis of fat deposition in sheep tail and provide a reference for developing new sheep breeds with an ideal tail type.
Abstract
It is a unique feature that fat can be deposited in sheep tails and rumps. To elucidate the genetic mechanism underlying this trait, we collected 120 individuals from three Chinese indigenous sheep breeds with extreme tail types, namely large fat-tailed sheep (n = 40), Altay sheep (n = 40), and Tibetan sheep (n = 40), and genotyped them using the Ovine Infinium HD SNP BeadChip. Then genomic scan for selection signatures was performed using the hapFLK. In total, we identified 25 genomic regions exhibiting evidence of having been under selection. Bioinformatic analysis of the genomic regions showed that selection signatures related to multiple candidate genes had a demonstrated role in phenotypic variation. Nine genes have documented association with sheep tail types, including WDR92, TBX12, WARS2, BMP2, VEGFA, PDGFD, HOXA10, ALX4, and ETAA1. Moreover, a number of genes were of particular interest, including RXFP2 associated with the presence/absence and morphology of horns; MITF involved in coat color; LIN52 and SYNDIG1L related to the number of teats; MSRB3 gene associated with ear sizes; LTBP2 considered as a positional candidate genes for number of ribs; JAZF1 regulating lipid metabolism; PGRMC2, SPAG17, TSHR, GTF2A1, and LARP1B implicated with reproductive traits. Our findings provide insights into fat tail formation and a reference for carrying out molecular breeding and conservation in sheep.
Keywords: tail type, selection signature, sheep, fat deposition
1. Introduction
As one of the first domesticated species, sheep was probably domesticated approximately 11,000 years ago in the fertile crescent. It is documented that wild ancestors of sheep had thin tails. Then, the fat-tailed or fat-rumped sheep was bred via artificial selection [1]. Therefore, artificial and natural selection has led to an increased prevalence of fat in sheep tails across generations. Sheep can be classified into three groups according to their tail types: fat-tailed, fat-rumped, and thin-tailed [2]. Fat stored in tails or rumps acts as an energy reserve to support migration and survival during cold winters so as to adapt to hostile environments [3]. In the world, more than 25% of sheep breeds are fat-rumped or fat-tailed [4].
Domestication and artificial selection pressures have generated a series of modern sheep breeds with different phenotypic characteristics. They can adapt to a variety of environments and produce special products of meat, milk, and fine wool [2]. Selection can increase the beneficial allele frequency over time and fix it within a population, which can carve some signals in the genome. Detecting these selection signals in genomic regions is of great importance in animal genetics. These genomic regions often harbor QTLs or genes affecting economically important traits. Therefore, selection signature detection is considered as one of the strategies for identifying candidate genes. Recently, numerous studies have used genotypic data to identify selection signatures for exploring the potential genetic mechanism of phenotype polymorphisms and adaption in sheep [5,6,7,8,9].
To elucidate the genetic mechanism of sheep tail types, a handful of studies have detected selection signatures. The first genome-wide scan of selective sweeps for thin- and fat-tailed sheep was conducted by Moradi et al. [1]. They revealed QTLs on chromosomes 5, 7, and X harboring candidate genes (namely, PPP2CA, SKP1, and TCF7). Wei et al. [10] clustered 10 Chinese indigenous sheep breeds into two subpopulations (fat-tailed and thin-tailed sheep) and identified three strong selective genomic windows containing two functional genes (PPP1CC and PDGFD) associated with tail types. Yuan et al. [11] further employed population differentiation methods to analyze these genotypic data and found candidate genomic regions spanning 6.24 Mb with several candidate genes therein (i.e., HOXA11, BMP2, PPP1CC, SP3, SP9, WDR92, PROKR1, and ETAA1) impacting on fat tail development. Moioli et al. [12] also utilized two fat-tailed breeds vs. 13 thin-tailed breeds to perform a genome-wide scan for selection signatures and detected BMP2 and VRTN genes as the most plausible genes associated with the fat-tailed phenotype in sheep. Ahbara et al. [13] divided Ethiopian indigenous sheep breeds into different groups for comparison and identified three genes (ALX4, HOXB13, BMP4) related to tail formation. All studies mentioned above divided many sheep breeds into two groups (fat tail and thin tail) to conduct selection signature detection. Therefore, it is hard to ignore the role of different genetic backgrounds.
In Chinese indigenous sheep breeds, three typical sheep breeds can be found. Large fat-tailed Han sheep have the largest tails (Figure 1a), Altay sheep have two large bulks of fat on the buttocks (Figure 1b), and Tibetan sheep are thin-tailed (Figure 1c). In this study, to elucidate the mechanism of tail fat deposition in sheep, we used hapFLK to detect selection signatures in these three Chinese indigenous sheep breeds with extreme tail types based on the Ovine Infinium HD SNP BeadChip (600K) genotype data. Furthermore, we annotate the genomic regions with selection signatures to explore the potential biological functions of the candidate genes under selection using bioinformatic analyses.
Figure 1.
Chinese indigenous sheep breeds with three different tail types: (a) Large-Tailed Han sheep, (b) Altay sheep, and (c) Tibetan sheep.
2. Materials and Methods
2.1. Ethnic Statement
All animal handling procedures were performed in strict accordance with the guidelines proposed by the China Animal Welfare and the Ministry of Agriculture of the People’s Republic of China. All animal experiments were approved by the Chinese Academy of Agricultural Sciences (CAAS, Permit Number: 2014-0035).
2.2. Genotype Data
In this study, a total of 120 individuals from three Chinese indigenous sheep breeds (40 Large-Tailed Han sheep, 40 Altay sheep, and 40 Tibetan sheep) were obtained. The information on all the individuals has been reported in our previous studies [14,15]. For the article to be self-contained, these three sheep populations are described here in detail. Large-Tailed Han sheep were collected from Liaocheng city in Shangdong Province, Altay sheep from Altay city in Xinjiang Uighur autonomous region, and Tibetan Sheep from Tianzhu county in Gansu Province. Their phenotypic characteristics and environmental variables are summarized in Table 1.
Table 1.
Description of phenotypic characteristics and environmental variables of three sheep breeds sampled in this study.
| Large-Tailed Han Sheep | Altay Sheep | Tibetan Sheep | |
|---|---|---|---|
| Tail type | Long fat-tail | Fat-rumped | Long short-tail |
| No. individual | 40 | 40 | 40 |
| Body weight (average kg) | 70.4 kg in male and 60.2 kg in female | 98.3 kg in male and 77.1 kg in female | 51 kg in male and 43.6 kg in female |
| Coat color | White | Brown | White body, heads and feet in miscellaneous colors |
| Horn | Most males have spiral horns and most females have ginger-shaped horns | Males have large horns, most females have horns | Males have large horns, females have horns |
| Region | Liaocheng county, Shan Dong Province | Fuhai county, Xinjiang Uighur autonomous region | Tianzhu county, Gansu Province |
| Geographical coordinates | N35°47′–37°02’ and E115°16´–116°32´ | N40°00´–48°10´ and E87 °00´–89°04´ | N36°31´–37°55´and E102 °07´–103°46´ |
| Climate | Warm temperate continental monsoon climate | Continental arid climate | Continental plateau monsoon climate |
| Altitude (m) | 22–49 | 386–3332 | 2040–4874 |
| Temperature(°C) | −22.3–41.8 | −42.7–40 | −8–4 |
| Use | Meat-fat dual type | Meat-fat dual and coarse wool type | Coarse wool type |
Note: The data were derived from animal genetic resources in China for sheep and goats [2].
The genomic DNA of samples were genotyped with the Ovine Infinium HD SNP BeadChip (Illumina Inc., San Diego, CA, USA), which contains 606,006 SNPs spanning the whole ovine genome. The detailed criteria to control the quality of the whole genotype data are also followed as according to our previous study [14]. After filtering, 500,593 SNPs were available with a mean distance of 4.89 kb between adjacent SNPs, and 115 animals including 40 Altay sheep, 35 Large-Tailed Han sheep and 40 Tibetan sheep were retained for further analysis. The sporadic missing genotypes were imputed, and genotypes were phased using BEAGLE software [16].
2.3. Population Structure Analysis
To investigate genetic relationships between individuals and population, principal component analysis (PCA) was performed using the commands of make-grm and pca in GCTA software [17].
2.4. Selection Signature Detection Using hapFLK
In this study, hapFLK was performed to detect the selection signatures in three typical sheep population with obviously different tail types. The hapFLK proposed by Fariello et al. [18] is a haplotype-based approach applied to unphased genotypic data. This method was based on differences haplotype frequencies between populations using fastPHASE 1.4 to estimate the haplotype information. In this study, no outgroups were defined, 10 clusters (−K 10) were used for the fastPHASE model and the hapFLK statistic was computed for 20 EM runs to fit the LD model (–nfit = 20). The hapFLK 1.4.0 program version can be available at forgedga.jouy.inra.fr/projects/hapflk/files.
Since hapFLK values for each SNP on the entire genome approximately follow the normal distribution, hapFLK values were further standardized to calculate the p-values. The formula is as follows:
| (1) |
where Mean(hapFLK) and SD(hapFLK) are the mean and standard error of all hapFLK values in the whole genome, respectively. The adjusted hapFLK follows a standardized normal distribution. Moreover, 0.01 significance level and Bonferroni correction were taken into account to calculate the genome-wide significance level. After adjustment, the threshold value was equal to −log10 (p-value) = 7.77 (0.01/500,593 independent tests).
2.5. Gene Annotation and Enrichment Analysis
Genes located on the selection regions were obtained from the Ensembl Genes 64 Database using BioMart software based on the Ovis aries (Oar_v3.1) gene sequence assembly. To learn the biological functions of annotated genes, we carried out a comprehensive literature search including information from other species.
3. Results
3.1. Population Genetic Structure
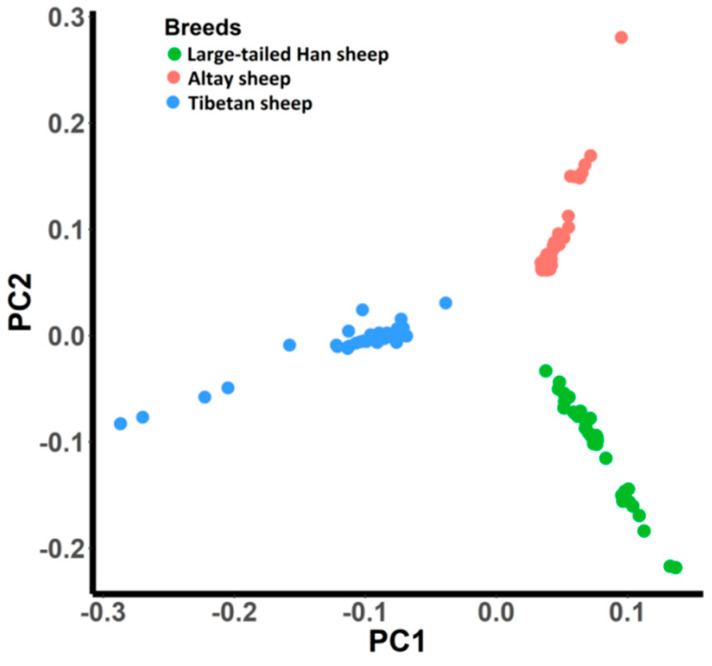
PCA analysis uses all individuals (n = 115) and 500, 593 autosomal markers to assess whether all animals were divided into three groups adhere to different tail types. The first two principal components (PC1 and PC2) accounted for 3.3% and 2.7% of the variation, respectively, and divided all animals appropriate into three groups, as shown in Figure 2.
Figure 2.
Principal component analysis (PCA) for population structure of three Chinese indigenous sheep breeds with extreme tail types. The first (PC1) and second (PC2) components were plotted.
3.2. Identifying the Genomic Regions with Selection Signals Using hapFLK
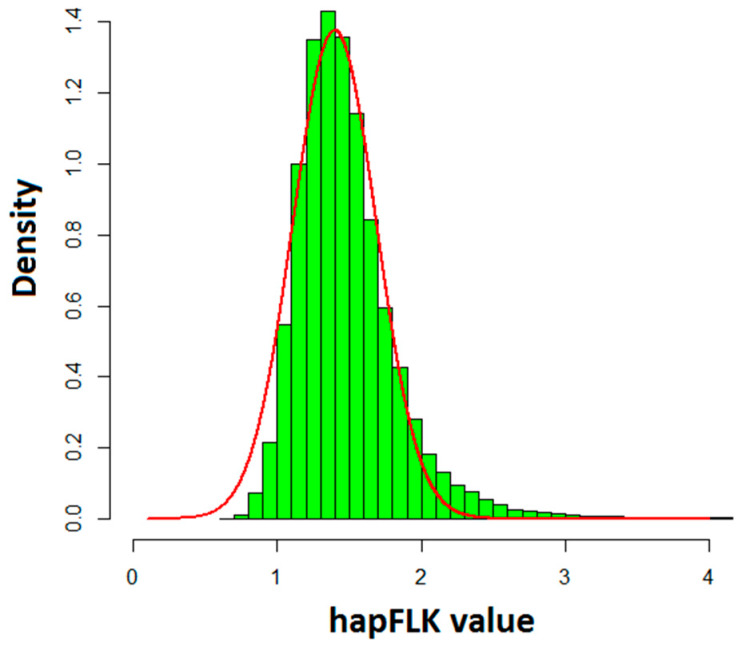
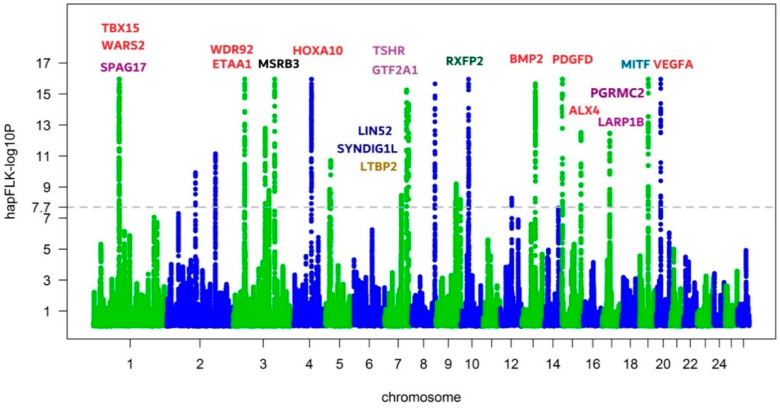
The hapFLK statistic was implemented to find genomic regions under selection. As shown in Figure 3, the distribution of hapFLK values approximately follows the normal distribution. p-values for hapFLK calculated using an empirical method is appropriate because of ascertainment bias in the SNP panel [18]. Therefore, the p-value of each SNP was obtained from a normal distribution. Figure 4 shown the −logp-values in genome indicated that several regions suffered strong selection. To construct one genomic region with selection signals, the adjacent significant SNPs were grouped together.
Figure 3.
Distribution of hapFLK values over the sheep autosome. The histogram in green is the distribution of hapFLK values. The red line is the curve of normal distribution.
Figure 4.
Manhattan plot of hapFLK values on the sheep autosome. The gray horizontal line denotes 0.01 genome-wide threshold (−log10(0.01/500,593) = 7.77).
3.3. Candidate Genes Annotated in Selection Regions
In total, we identified 25 genomic regions with selective signals, as shown in Table 2. The total length of genomic regions is 9.7 Mb. The largest length of genomic region is located on OAR10, while the genomic region on OAR1 contains the largest number of SNPs.
Table 2.
Genes located in genomic regions showing evidence of selection.
| Chr | Start | End | No. SNP | Size (kb) | Candidate Genes |
|---|---|---|---|---|---|
| 1 | 94756172 | 96112605 | 224 | 1356.433 | SPAG17, RF00026, TBX15, WARS2, RF00100, HAO2, HSD3B1 |
| 2 | 105901381 | 105961277 | 11 | 59.896 | - |
| 2 | 183012215 | 183196527 | 34 | 184.312 | - |
| 3 | 39961457 | 40797601 | 172 | 836.144 | PNO1, WDR92, C1D, ETAA1 |
| 3 | 118322493 | 118505234 | 38 | 182.741 | METTL25, RF00001 |
| 3 | 132458013 | 132563310 | 26 | 105.297 | - |
| 3 | 154055122 | 154324519 | 58 | 269.397 | MSRB3 |
| 4 | 68223979 | 69165823 | 183 | 941.844 | JAZF1, TAX1BP1, EVX1, RF00275, RF01975, RF01976, RF01978, RF01979, RF02040, RF02041, RF02042, RF02043, RF02137, RF02138, RF02139, RF02140, RF02141, RF02142, RF02143, HOXA1, HOXA2, HOXA3, HOXA6, HOXA10, SKAP2 |
| 5 | 16293532 | 16380709 | 19 | 87.177 | ASFB2 |
| 5 | 20143416 | 20250955 | 12 | 107.539 | - |
| 7 | 63493868 | 63648665 | 16 | 154.797 | - |
| 7 | 82373886 | 82795821 | 89 | 421.935 | LIN52, VSX2, ABCD4, SYNDIG1L, NPC2, RF00004, LTBP2 |
| 7 | 89260877 | 89527917 | 51 | 267.04 | TSHR, GTF2A1, RF00600 |
| 8 | 87547094 | 88056486 | 108 | 509.392 | TBXT, MPC1, RPS6KA2 |
| 9 | 75623521 | 75637947 | 2 | 14.426 | - |
| 9 | 77403165 | 77840673 | 47 | 437.508 | VPS13B |
| 9 | 93544798 | 93558759 | 3 | 13.961 | SLC10A5, ENPP2 |
| 10 | 29320784 | 30719200 | 88 | 1398.416 | RXFP2, B3GLCT, HSPH1, TEX26, MEDAG, ALOX5AP, USPL1 |
| 12 | 42361453 | 42380380 | 8 | 18.927 | H6PD |
| 13 | 48400700 | 49177821 | 81 | 777.121 | BMP2 |
| 15 | 3434222 | 3945775 | 68 | 511.553 | PDGFD |
| 15 | 72489270 | 72637084 | 28 | 147.814 | EXT2, RF00026, ALX4 |
| 17 | 29091651 | 29420825 | 44 | 329.174 | PGRMC2, LARP1B |
| 19 | 31539577 | 31850022 | 64 | 310.445 | MITF |
| 20 | 17281001 | 17555177 | 44 | 274.176 | VEGFA |
These genomic regions harbored 73 positional candidate genes. Out of these, six genes including WDR92, BMP2, PDGFD, HOXA10, ALX4, and ETAA1 have been reported as implicated with tail formation in sheep [11,13,19,20]. Moreover, three genes (VEGFA, TBX12, and WARS2) have been associated with fat distribution in waist and hip in human [21], which indicate these genes may play important role in fat deposition in sheep tails.
Several genes were also identified to be associated with other economically important traits. RXFP2 has been identified as being involved in horn morphology and was considered as the major effect gene in sheep [22,23]. PGRMC2 and SPAG17 have documented associated with fertility [24,25]. TSHR and GTF2A1 genes are associated with photoperiod control of reproduction [26,27]. LIN52 and SYNDIG1L were showed related to the number of teats in pig [28,29]. LARP1B is the primary candidate gene associated with milk fatty acids in dairy cattle [30]. LTBP2 is a positional candidate gene for thoracic vertebrae in pigs [31]. MSRB3 gene has shown to be the causal gene of ear size [32]. JAZF1 can regulate lipid metabolism [33].
4. Discussion
Among multiple populations, common selection signatures can be detected using the fixation index (FST). However, hidden population substructures exist in multiple breeds, which cannot be taken into account by FST, due to its simplicity, and can lead to false positives. Moreover, these substructures are common in domestic animal populations. To address this limitation, a haplotype-based approach (hapFLK) was proposed that accommodates population substructures [18]. The hapFLK was implemented to detect selection signatures through haplotype differentiation among hierarchically structured populations. Hence, hapFLK can integrate the information from differences in haplotype allele frequencies among populations, the local haplotype data, and population substructures to identify genomic regions showing under selection. Furthermore, this approach can be applied unphased genotypic data and does not need information from the ancestral allele at each locus. Thus, hapFLK can be identified recent positive selection that results in a beneficial mutation residing on a haplotype with high frequency that is longer than average. In this study, we also employed the stringent significant threshold value (−log10(0.01/500935) = 7.7) to further control the positive signal. Therefore, the genomic regions undergoing selection would be more reliable.
In the present study, we selected Chinese indigenous sheep breeds with obviously different tail types. Large-Tailed Han sheep have the largest and fattiest tails in Chinese indigenous sheep breeds. Their tails sag below the hock and drag on the ground with circular shapes at the end of tails (Figure 1a). Altay sheep can deposit fat on the rump, which forms substantial buttocks (Figure 1b). This is another type of fat tail (fat-rumped). Meanwhile, Tibetan sheep have thin tails with a shape similar to that of a stick (Figure 1c). In addition, these three sheep breeds also live in a diverse range of ecological environments. Large-Tailed Han sheep are primarily distributed in the north Chinese plain, which has four distinctive seasons. Altay sheep live in the Gobi Desert, where the temperature ranges from −42.7 °C to +40.0 °C, and the ground is snow-covered for 200–250 days per year. Altay sheep can use the fat on their rumps to adapt to the harsh environment. Thin-tailed Tibetan sheep live in the Tibetan plateau at an altitude of 3000–5000 m. Therefore, in present study, we identified the candidate genes related not only to tail type formation but also to other economically important traits.
In the present study, we identified nine important candidate genes of tail formation in sheep (WDR92, TBX12, WARS2, BMP2, VEGFA, PDGFD, HOXA10, ALX4, and ETAA1). WDR92 is a WD40 repeat protein that has several biological functions. Remarkably, as a WD40 repeat protein, WIPI1 has a seven-bladed propeller structure and encompasses a conserved motif to interact with phospholipids [34]. WIPI1 is also differentially expressed between adipocyte tissue from Kazak sheep’s and Tibetan sheep’s fat tails [35]. ETAA1 plays an important role in fat distribution in fat-tailed sheep [11] and humans [36]. WDR92 and ETAA1 are also shown to be differentially expressed in the adipocyte tissue of sheep populations according to their different tail types [19]. The BMP2 gene was shown to be differentially expressed in tail adipose tissue between fat-tailed and thin-tailed sheep using RNA-seq [20]. Members of the PDGF family promote proliferation and inhibit the differentiation of preadipocytes [19,20]. Thus, PDGFD played a pivotal role in sheep adipose tissue and is hypothesized to be one of candidate genes leading to fat tail formation. In the current study, ALX4 was identified in the candidate region at chr15:72.5–72.6 Mb. Mutations are involved in the development of limbs and skeleton [6], and its protein has been shown to bind proteins from the HOXA clusters [37]. As one of HOXA cluster genes, HOXA10 has showed to be differentially expressed between subcutaneous-, visceral- and tail-fat tissues by RNA-seq [38]. Moreover, other three genes (VEGFA, TBX12, and WARS2) have been associated with fat distribution in waist and hip in human [21], which indicate these three genes play important role in fat deposition in sheep tails. These findings indicate that these nine genes may be important candidate genes related to tail-type formation. Their functional verification needs further investigation and validation using full genome sequences and expression studies in the future.
To dissect the genetic basis of fat deposition in sheep, genome-wide association studies have also been performed directly using the fat-tail phenotypes and genotype data [39,40]. However, no common genes were identified by two strategies. This may be due to two main factors. One is the different principles of identifying candidate genes. Another is the use of different populations. However, the results by these two strategies suggest that the tail phenotype in sheep is most probably a complex trait.
In this study, the regions under selection contain other genes shown to be involved in agriculturally important traits. One of the most striking selective sweeps occurred at the locus for RXFP2, which has been shown to be associated with horn morphology and is considered as a major effect gene in sheep [22,23,41]. In the Large-Tailed Han sheep population, most males have spiral horns and most females have horns that are ginger-shaped or polled [2]. This gene was also identified by other studies on selection signature detection [6,10,42,43,44]. Furthermore, we found an interesting peak on OAR3:154.1~154.3 Mb harboring the MSRB3 gene which is linked to ear size in sheep [10]. In the Altay population, few individuals have the small ear, and ear size morphologies exist in these three sheep breeds [2]. MSRB3 was also identified as a candidate gene for ear morphology in dogs [45] and pigs [32]. Therefore, the MSRB3 gene was hypothesized to be candidate gene for ear size in sheep. In line with a remarkable difference in coat color among these three sheep populations, we detected one potential candidate gene, MITF, known to be associated with pigmentation in sheep [5,46]. LTBP2 was considered as a positional candidate gene for thoracic vertebrae in pigs [31]. JAZF1 can regulate lipid metabolism [33] and is shown to be associated with height in human [47]. Moreover, this gene was also identified by selection signature in cattle [48]. The region on OAR7: 89.2~89.5 Mb harbored two important genes (TSHR and GTF2A1). The TSHR gene has a pivotal role in metabolic regulation and photoperiod control of reproduction in chicken [49] and also affects the seasonal reproduction of sheep and goats [50]. The GTF2A1 gene is supposedly associated with the function of ovary and uterus and, hence, influenced egg production [27]. It is possible that selection for these variants could be associated with photoperiod control of reproduction in Chinese short fat-tailed sheep. PGRMC2 and SPAG17 have been documented as being associated with fertility [24,25]. LIN52 and SYNDIG1L were shown to be associated with the number of teats in pig [28,29]. LARP1B is the primary candidate gene associated with milk fatty acids in dairy cattle [30]. These genes could be relevant for reproduction in these sheep.
5. Conclusion
We used Ovine Infinium HD SNP BeadChip genotype data to carry out a genome scan for selective sweeps in Chinese indigenous sheep breeds with extreme tail types. In total, 25 genomic regions were identified under selection which harbored 73 candidate genes. These genes are not only associated with tail fat formation but also with reproduction, body conformation, and appearance. Our findings contribute to the identification of candidate genes underlying important traits in different types of sheep breeds and provide a reference for developing new sheep breeds with an ideal tail type.
Acknowledgments
The authors would like to thank Youji Ma, Guoping Zhang and Dong Xu for providing the sheep samples.
Author Contributions
Conception and design of study, F.Z., L.D. and L.W.; analysis and interpretation of data, F.Z., T.D., L.S., W.W. and Q.Z.; drafting the manuscript, F.Z. All authors read and approved the final manuscript.
Funding
This research was funded by the Natural Science Foundations of China, grant number No. 31572357 and the National Science and Technology Pillar Program, grant number No. 2015BAD03B0503.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Moradi M.H., Nejati-Javaremi A., Moradi-Shahrbabak M., Dodds K.G., McEwan J.C. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet. 2012;13:10. doi: 10.1186/1471-2156-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CNCAGR (China National Commision of Animal Genetic Resource) Animal Genetic Resources in China-Sheep and Goats. China Agriculture Press; Beijing, China: 2012. (In Chinese) [Google Scholar]
- 3.Ermias E., Yami A., Rege J. Fat deposition in tropical sheep as adaptive attribute to periodic feed fluctuation. J. Anim. Breed. Genet. 2002;119:235–246. doi: 10.1046/j.1439-0388.2002.00344.x. [DOI] [Google Scholar]
- 4.Safdarian M., Zamiri M.J., Hashemi M., Noorolahi H. Relationships of fat-tail dimensions with fat-tail weight and carcass characteristics at different slaughter weights of Torki-Ghashghaii sheep. Meat Sci. 2008;80:686–689. doi: 10.1016/j.meatsci.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Kijas J.W., Lenstra J.A., Hayes B., Boitard S., Porto Neto L.R., San Cristobal M., Servin B., McCulloch R., Whan V., Gietzen K., et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012;10:e1001258. doi: 10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fariello M.I., Servin B., Tosser-Klopp G., Rupp R., Moreno C., International Sheep Genomics C., San Cristobal M., Boitard S. Selection signatures in worldwide sheep populations. PLoS ONE. 2014;9:e103813. doi: 10.1371/journal.pone.0103813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz-Larranaga O., Langa J., Rendo F., Manzano C., Iriondo M., Estonba A. Genomic selection signatures in sheep from the Western Pyrenees. Genet. Sel. Evol. 2018;50:9. doi: 10.1186/s12711-018-0378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Li W.R., Lv F.H., He S.G., Tian S.L., Peng W.F., Sun Y.W., Zhao Y.X., Tu X.L., Zhang M., et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016;33:2576–2592. doi: 10.1093/molbev/msw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao F., Wei C., Zhang L., Liu J., Wang G., Zeng T., Du L. A genome scan of recent positive selection signatures in three sheep populations. J. Integr. Agric. 2016;15:162–174. doi: 10.1016/S2095-3119(15)61080-2. [DOI] [Google Scholar]
- 10.Wei C., Wang H., Liu G., Wu M., Cao J., Liu Z., Liu R., Zhao F., Zhang L., Lu J., et al. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genom. 2015;16:194. doi: 10.1186/s12864-015-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan Z., Liu E., Liu Z., Kijas J.W., Zhu C., Hu S., Ma X., Zhang L., Du L., Wang H., et al. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet. 2017;48:55–66. doi: 10.1111/age.12477. [DOI] [PubMed] [Google Scholar]
- 12.Moioli B., Pilla F., Ciani E. Signatures of selection identify loci associated with fat tail in sheep. J. Anim. Sci. 2015;93:4660–4669. doi: 10.2527/jas.2015-9389. [DOI] [PubMed] [Google Scholar]
- 13.Ahbara A., Bahbahani H., Almathen F., Al Abri M., Agoub M.O., Abeba A., Kebede A., Musa H.H., Mastrangelo S., Pilla F., et al. Genome-Wide Variation, Candidate Regions and Genes Associated with Fat Deposition and Tail Morphology in Ethiopian Indigenous Sheep. Front. Genet. 2018;9:699. doi: 10.3389/fgene.2018.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu C., Fan H., Yuan Z., Hu S., Ma X., Xuan J., Wang H., Zhang L., Wei C., Zhang Q., et al. Genome-wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays. Sci. Rep. 2016;6:27822. doi: 10.1038/srep27822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu C., Li M., Qin S., Zhao F., Fang S. Detection of copy number variation and selection signatures on the X chromosome of Chinese indigenous sheep with different tail types. Asian-Australas J. Anim. Sci. 2020 doi: 10.5713/ajas.18.0661. Online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fariello M.I., Boitard S., Naya H., SanCristobal M., Servin B. Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics. 2013;193:929–941. doi: 10.1534/genetics.112.147231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma L., Li Z., Cai Y., Xu H., Yang R., Lan X. Genetic variants in fat- and short-tailed sheep from high-throughput RNA-sequencing data. Anim. Genet. 2018;49:483–487. doi: 10.1111/age.12699. [DOI] [PubMed] [Google Scholar]
- 20.Pan Z., Li S., Liu Q., Wang Z., Zhou Z., Di R., An X., Miao B., Wang X., Hu W., et al. Rapid evolution of a retro-transposable hotspot of ovine genome underlies the alteration of BMP2 expression and development of fat tails. BMC Genom. 2019;20:261. doi: 10.1186/s12864-019-5620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K., Magi R., et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston S.E., McEwan J.C., Pickering N.K., Kijas J.W., Beraldi D., Pilkington J.G., Pemberton J.M., Slate J. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol. Ecol. 2011;20:2555–2566. doi: 10.1111/j.1365-294X.2011.05076.x. [DOI] [PubMed] [Google Scholar]
- 23.Wiedemar N., Drogemuller C. A 1.8-kb insertion in the 3′-UTR of RXFP2 is associated with polledness in sheep. Anim. Genet. 2015;46:457–461. doi: 10.1111/age.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M., Su G., Fu J., Wang A., Liu J.F., Lund M.S., Guldbrandtsen B. Introgression of Chinese haplotypes contributed to the improvement of Danish Duroc pigs. Evol. Appl. 2019;12:292–300. doi: 10.1111/eva.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., Sha Y.W., Mei L.B., Ji Z.Y., Qiu P.P., Ji H., Li P., Wang T., Li L. A familial study of twins with severe asthenozoospermia identified a homozygous SPAG17 mutation by whole-exome sequencing. Clin. Genet. 2018;93:345–349. doi: 10.1111/cge.13059. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z., Ji Z., Wang G., Chao T., Hou L., Wang J. Genome-wide analysis reveals signatures of selection for important traits in domestic sheep from different ecoregions. BMC Genom. 2016;17:863. doi: 10.1186/s12864-016-3212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J., Sun C., Dou T., Yi G., Qu L., Qu L., Wang K., Yang N. Identification of Promising Mutants Associated with Egg Production Traits Revealed by Genome-Wide Association Study. PLoS ONE. 2015;10:e0140615. doi: 10.1371/journal.pone.0140615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan C., Wu Z., Ren J., Huang Z., Liu D., He X., Prakapenka D., Zhang R., Li N., Da Y., et al. Genome-wide association study and accuracy of genomic prediction for teat number in Duroc pigs using genotyping-by-sequencing. Genet. Sel. Evol. 2017;49:35. doi: 10.1186/s12711-017-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verardo L.L., Silva F.F., Lopes M.S., Madsen O., Bastiaansen J.W., Knol E.F., Kelly M., Varona L., Lopes P.S., Guimaraes S.E. Revealing new candidate genes for reproductive traits in pigs: Combining Bayesian GWAS and functional pathways. Genet. Sel. Evol. 2016;48:9. doi: 10.1186/s12711-016-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duchemin S.I., Bovenhuis H., Megens H.J., Van Arendonk J.A.M., Visker M. Fine-mapping of BTA17 using imputed sequences for associations with de novo synthesized fatty acids in bovine milk. J. Dairy Sci. 2017;100:9125–9135. doi: 10.3168/jds.2017-12965. [DOI] [PubMed] [Google Scholar]
- 31.Park H.B., Han S.H., Lee J.B., Cho I.C. Rapid Communication: High-resolution quantitative trait loci analysis identifies LTBP2 encoding latent transforming growth factor beta binding protein 2 associated with thoracic vertebrae number in a large F2 intercross between Landrace and Korean native pigs. J. Anim. Sci. 2017;95:1957–1962. doi: 10.2527/jas.2017.1390. [DOI] [PubMed] [Google Scholar]
- 32.Chen C., Liu C., Xiong X., Fang S., Yang H., Zhang Z., Ren J., Guo Y., Huang L. Copy number variation in the MSRB3 gene enlarges porcine ear size through a mechanism involving miR-584-5p. Genet. Sel. Evol. 2018;50:72. doi: 10.1186/s12711-018-0442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ming G.F., Xiao D., Gong W.J., Liu H.X., Liu J., Zhou H.H., Liu Z.Q. JAZF1 can regulate the expression of lipid metabolic genes and inhibit lipid accumulation in adipocytes. Biochem. Biophys. Res. Commun. 2014;445:673–680. doi: 10.1016/j.bbrc.2014.02.088. [DOI] [PubMed] [Google Scholar]
- 34.Proikas-Cezanne T., Waddell S., Gaugel A., Frickey T., Lupas A., Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Zhou G., Xu X., Geng R., Zhou J., Yang Y., Yang Z., Chen Y. Transcriptome profile analysis of adipose tissues from fat and short-tailed sheep. Gene. 2014;549:252–257. doi: 10.1016/j.gene.2014.07.072. [DOI] [PubMed] [Google Scholar]
- 36.Liu C.T., Monda K.L., Taylor K.C., Lange L., Demerath E.W., Palmas W., Wojczynski M.K., Ellis J.C., Vitolins M.Z., Liu S., et al. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genet. 2013;9:e1003681. doi: 10.1371/journal.pgen.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravasi T., Suzuki H., Cannistraci C.V., Katayama S., Bajic V.B., Tan K., Akalin A., Schmeier S., Kanamori-Katayama M., Bertin N., et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang D., Zhou G., Zhou S., Zeng J., Wang X., Jiang Y., Yang Y., Chen Y. Comparative transcriptome analysis reveals potentially novel roles of Homeobox genes in adipose deposition in fat-tailed sheep. Sci. Rep. 2017;7:14491. doi: 10.1038/s41598-017-14967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu S.S., Ren X., Yang G.L., Xie X.L., Zhao Y.X., Zhang M., Shen Z.Q., Ren Y.L., Gao L., Shen M., et al. Genome-wide association analysis identifies the genetic basis of fat deposition in the tails of sheep (Ovis aries) Anim. Genet. 2017;48:560–569. doi: 10.1111/age.12572. [DOI] [PubMed] [Google Scholar]
- 40.Zhang T., Gao H., Sahana G., Zan Y., Fan H., Liu J., Shi L., Wang H., Du L., Wang L., et al. Genome-wide association studies revealed candidate genes for tail fat deposition and body size in the Hulun Buir sheep. J. Anim. Breed. Genet. 2019;136:362–370. doi: 10.1111/jbg.12402. [DOI] [PubMed] [Google Scholar]
- 41.Johnston S.E., Gratten J., Berenos C., Pilkington J.G., Clutton-Brock T.H., Pemberton J.M., Slate J. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature. 2013;502:93–95. doi: 10.1038/nature12489. [DOI] [PubMed] [Google Scholar]
- 42.de Simoni Gouveia J.J., Paiva S.R., McManus C.M., Caetano A.R., Kijas J.W., Facó O., Azevedo H.C., de Araujo A.M., de Souza C.J.H., Yamagishi M.E.B. Genome-wide search for signatures of selection in three major Brazilian locally adapted sheep breeds. Livest. Sci. 2017;197:36–45. doi: 10.1016/j.livsci.2017.01.006. [DOI] [Google Scholar]
- 43.Kijas J.W. Haplotype-based analysis of selective sweeps in sheep. Genome. 2014;57:433–437. doi: 10.1139/gen-2014-0049. [DOI] [PubMed] [Google Scholar]
- 44.Pan Z., Li S., Liu Q., Wang Z., Zhou Z., Di R., Miao B., Hu W., Wang X., Hu X., et al. Whole-genome sequences of 89 Chinese sheep suggest role of RXFP2 in the development of unique horn phenotype as response to semi-feralization. Gigascience. 2018;7:giy019. doi: 10.1093/gigascience/giy019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyko A.R., Quignon P., Li L., Schoenebeck J.J., Degenhardt J.D., Lohmueller K.E., Zhao K., Brisbin A., Parker H.G., vonHoldt B.M., et al. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M.H., Tiirikka T., Kantanen J. A genome-wide scan study identifies a single nucleotide substitution in ASIP associated with white versus non-white coat-colour variation in sheep (Ovis aries) Heredity. 2014;112:122–131. doi: 10.1038/hdy.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansson A., Marroni F., Hayward C., Franklin C.S., Kirichenko A.V., Jonasson I., Hicks A.A., Vitart V., Isaacs A., Axenovich T., et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum. Mol. Genet. 2009;18:373–380. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao F., McParland S., Kearney F., Du L., Berry D.P. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genet. Sel. Evol. 2015;47:49. doi: 10.1186/s12711-015-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubin C.J., Zody M.C., Eriksson J., Meadows J.R., Sherwood E., Webster M.T., Jiang L., Ingman M., Sharpe T., Ka S., et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 50.Huang D.W., Wang J.X., Liu Q.Y., Chu M.X., Di R., He J.N., Cao G.L., Fang L., Feng T., Li N. Analysis on DNA sequence of TSHB gene and its association with reproductive seasonality in goats. Mol. Biol. Rep. 2013;40:1893–1904. doi: 10.1007/s11033-012-2245-0. [DOI] [PubMed] [Google Scholar]