Highlights
-
•
The rapid diagnosis of COVID-19 patients is essential to reduce the disease spread.
-
•
The detection limits between rapid antigen detection (RAD) test, viral culture and RT-PCR varied hugely.
-
•
The RAD test was 103 fold less sensitive than viral culture while RAD was 105 fold less sensitive than RT-PCR.
-
•
The RAD test detected between 11.1 % and 45.7 % of RT-PCR-positive samples from COVID-19 patients.
-
•
The RAD test serves only as adjunct to RT-PCR test because of potential for false-negative results.
Keywords: 2019 novel coronavirus, SARS-CoV-2, COVID-19, Rapid antigen detection, RT-PCR, Viral culture
Abstract
Background
The rapid diagnosis of Coronavirus Disease 2019 (COVID-19) patients is essential to reduce the disease spread. Rapid antigen detection (RAD) tests are available, however, there is scanty data on the performance of RAD tests.
Objective
To evaluate the performance of the commercially available BIOCREDIT COVID-19 Ag test and compare it with RT-PCR for detecting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) virus. Analytical sensitivity for the detection of SARS-CoV-2 virus was determined for the RAD test using viral culture and RT-PCR as reference methods. The RAD test was further evaluated using respiratory samples collected from confirmed COVID-19 patients. The results were compared with RT-PCR test.
Results
The detection limits between RAD test, viral culture and RT-PCR varied hugely. RAD was 103 fold less sensitive than viral culture while RAD was 105 fold less sensitive than RT-PCR. The RAD test detected between 11.1 % and 45.7 % of RT-PCR-positive samples from COVID-19 patients.
Conclusions
This study demonstrated that the RAD test serves only as adjunct to RT-PCR test because of potential for false-negative results.
1. Introduction
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a new type of coronavirus belonging to the genus β. On 11 Mar 2020, World Health Organization (WHO) declared Coronavirus Disease 2019 (COVID-19) as a pandemic [1]. Viral culture and RT-PCR are the gold standards in the diagnosis of SARS-CoV-2 infection [2]. However, it takes hours to detect the nucleic acid and days to isolate the virus. In addition, specialized instrument and expertise are required. For rapid diagnosis of SARS-CoV-2 infection, rapid antigen detection (RAD) tests for qualitative determination of SARS-CoV-2 antigen are available. RAD tests detect viral antigen by the immobilized coated SARS-CoV-2 antibody on the device. The test results of RAD can be interpreted without specialized instrument and available within 30 min. Hence, RAD tests can relieve the workload in diagnostic hospitals and laboratories and improve the turn-around time. However, according to WHO, the role of RAD tests for antigen detection for SARS−COV-2 needs to be evaluated and is not recommended for clinical diagnosis [3].
The purpose of this evaluation is to assess the diagnostic use of the commercially available BIOCREDIT COVID-19 Ag test. The aim of the first part of the study was to assess the limit of detection (LOD) between RAD test, viral culture and RT-PCR and the second part was to evaluate performance of RAD test in detecting SARS-CoV-2 virus in different types of respiratory samples.
2. Methods
2.1. Respiratory isolates
The SARS-CoV-2 culture isolate (strain hCoV-19/Hong Kong/VM20001097/2020, the first COVID-19 case detected in Hong Kong) was used to perform a serial tenfold dilution to determine LOD between different assays.
To evaluate the cross-reactivity of the RAD test, 13 non-SARS-CoV-2 respiratory virus isolates were tested. They were influenza A(H1pdm09), influenza A(H3), influenza B, adenovirus, coronavirus type OC43, coronavirus type 229E, parainfluenza virus type 1, parainfluenza virus type 2, parainfluenza virus type 3, parainfluenza virus type 4, respiratory syncytial virus, rhinovirus and enterovirus.
2.2. Respiratory samples
From February 1, 2020 to April 21, 2020, respiratory samples from individuals confirmed with SARS-CoV-2 infection by RT-PCR targeting the SARS-CoV-2 virus–specific RdRp gene were retrieved for this evaluation. Samples were placed in viral transport media (VTM) or Phosphate-Buffered Saline (PBS) for RNA extraction. The remaining part of the suspension was stored at −70 °C until use in this study. Virus concentrations in samples were estimated from cycle threshold (Ct) value.
The Public Health Laboratory Services Branch (PHLSB) in Hong Kong has been designated as WHO COVID-19 reference laboratory since April 2020 and all confirmed cases in Hong Kong were either diagnosed or confirmed by PHLSB [4]. Total number of 368 confirmed COVID-19 samples collected with sufficient quantity were available for this study in descending order: throat saliva (N = 122), nasopharyngeal swab and throat swab (NPS & TS, N = 103), nasopharyngeal aspirate and throat swab (NPA & TS, N = 81), sputum (N = 62).
2.3. RAD kits for SARS−CoV-2 detection
We evaluated the only commercially available RAD kit in Hong Kong at the time of starting the evaluation, BIOCREDIT COVID-19 Ag for the diagnosis of SARS-CoV-2 infection.
2.4. BIOCREDIT COVID-19 Ag
The intended use for the BIOCREDIT COVID-19 Ag kit is for nasopharyngeal swab sample. As NPA & TS, NPS & TS, sputum and throat saliva had either been eluted in VTM or suspended in PBS, the test was carried out with modified sample processing methods.
2.5. Sample processing by BIOCREDIT COVID-19 Ag
The recommended sample volume by the BIOCREDIT COVID-19 Ag kit was 90–150 μL. To unify the sample volume, 100 μL sample volume was used. We evaluated two sample processing methods based on the nature of the samples.
2.5.1. For less viscous samples
These samples do not need preparation, 100 μL sample was added directly into a sample well of the device.
2.5.2. For viscous samples
The swab provided by the kit was used to collect the samples and the swab was immersed in the provided assay diluent tube. The subsequent procedures were carried out according to the manufacturer’s instructions.
In an effort to compare the performance of these two sample processing methods (i.e. methods for handling less viscous and viscous samples), LOD was determined using a serial tenfold dilution of virus. The results were then compared with viral culture and RT-PCR.
2.6. Viral culture for SARS-CoV-2 virus
Viral culture was conducted by inoculating samples onto Vero E6 cells. When virus-induced cytopathic effect was examined, identification of SARS-CoV-2 virus in culture fluid was confirmed by the RT-PCR.
2.7. RT-PCR for SARS-CoV-2 virus
The in-house developed RT-PCR was used to detect the presence of SARS-CoV-2 virus nucleic acid in all samples. It was conducted using NxtScript Enzyme and Master Mix (Roche Diagnostics GmbH, Germany). Each 10 μL reaction mixture contained 5 μL RNA samples, 2 μL Reaction Mix (5X), 0.06 μL Adpta Taq DNA polymerase (50U/μL), 0.05 μL NxtScript RT Enzyme (85U/μL), 0.9 μL volume of working primer/probe mix and nuclease-free water to obtain a final volume of 10 μL. The working primer/probe mix was prepared by mixing equal volume of forward primer, NCOV-F4: 5′-GTTGGACTGAGACTGACCTTAC-3′ (10 μM); reverse primer, NCOV-R4: 5′−CCCTAGGATTCTTGATGGATCTG-3′ (10 μM); and probe, NCOV-P4: 5′-FAM-ACAGGGTGATGATTATGTGTACCTTCCT-BHQ1−3’ (10 μM). The reverse transcription, amplification was performed in the LC480 System (Roche Diagnostics GmbH, Germany) according to the following program: 1 cycle of 50 °C for 10 min, 1 cycle of 95 °C for 30 s, 40 cycles of 95 °C for 10 s and 56 °C for 30 s; and holding at 4 °C.
3. Results
The LOD of the RAD test was 1000 fold less sensitive than viral culture when 100 μL sample was added directly into a sample well of the device (RAD: 10−2; viral culture: 10−5). RT-PCR is the most sensitive assay for detecting SARS-CoV-2 virus (10−7).
For the RAD test, there were marked differences in sensitivity when using the provided swab to transfer the sample to the assay diluent tube and then adding 100 μL of the suspension into a sample well. The fold difference between two RAD sample processing methods was at least 100 fold (Table 1 ). It seemed to be related to the volume of sample used and the dilution effect in the assay diluent tube.
Table 1.
Comparison of RT-PCR, viral culture and rapid antigen detection (RAD) test for the limit of detection of SARS-CoV-2 virus.
| Dilutionb | Test resultsa |
|||
|---|---|---|---|---|
| RT-PCRc | Viral culture | RAD test, sample processing methodd for: |
||
| less viscous samples | viscous samples | |||
| 10−1 | ND | ND | POS | NEG |
| 10−2 | ND | POS | POS | NEG |
| 10−3 | ND | POS | NEG | NEG |
| 10−4 | 25.17 | POS | NEG | NEG |
| 10−5 | 28.47 | POS | NEG | NEG |
| 10−6 | 31.08 | NEG | NEG | ND |
| 10−7 | 36.41 | NEG | ND | ND |
| 10−8 | NEG | NEG | ND | ND |
ND, not done; POS, positive; NEG, negative.
Serial tenfold dilution of the SARS-CoV-2 culture isolate, hCoV-19/Hong Kong/VM20001097/2020 (case 1 of the Hong Kong patient).
Ct values showing mean of two runs of the SARS-CoV-2 virus-specific RT-PCR.
Less viscous samples: 100 μL sample was added directly into a sample well of the device; viscous samples: the swab provided was used to transfer the sample to the assay diluent tube, then 100 μL of the suspension was added into a sample well of the device.
As shown in Table 1, the LOD for the RAD test was 10−2, the corresponding Ct value of the samples that can be detected by RT-PCR were estimated to be 18.57. This Ct value was estimated from the 10−4 dilution of virus (i.e. 10−4 Ct = 25.17, 10−2: 25.17−6.6 = 18.57). Respiratory samples of various Ct values up to ∼30 were selected (which is close to the limit of detection for viral culture).
A total of 35 samples each for NPA & TS, NPS & TS were selected to evaluate the RAD test. Since sputum and throat saliva are rarely used for the RAD tests, an additional 10 more samples each for sputum and throat saliva were selected.
We retrospectively tested 160 RT-PCR-positive respiratory samples from 152 different patients for the RAD test. All 70 NPA & TS and NPS & TS samples were handled with ‘less viscous samples’ processing method. For the 90 sputum and throat saliva samples, 83 samples were handled with ‘less viscous samples’ processing method, seven samples (sputum = 3, throat saliva = 4) were handled with ‘viscous samples’ processing method because of the viscous nature of these respiratory samples.
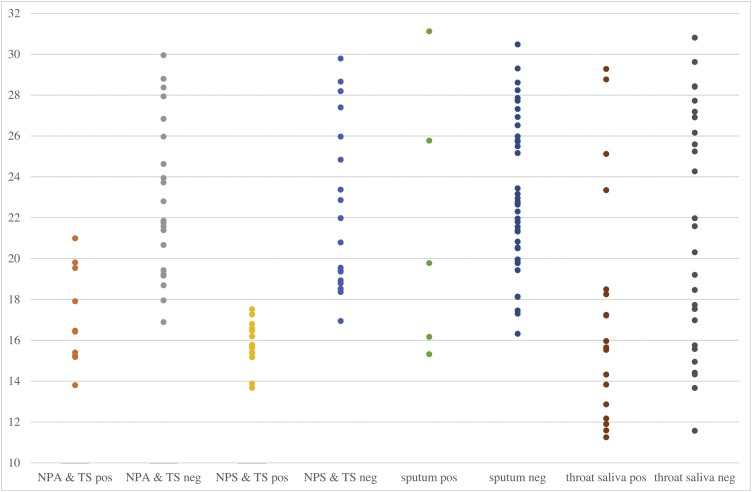
Respiratory samples were divided into two groups based on the Ct values. Samples with Ct values <18.57 were classified as ‘high viral load’ while samples >18.57 were classified as ‘normal viral load’. Of the 160 samples tested, 64 and 96 samples were classified as ‘high viral load’ and ‘normal viral load’ samples respectively. The corresponding mean Ct value, Ct value range and sensitivity for NPA & TS, NPS & TS, sputum and throat saliva samples among these two group of samples were shown in Table 2 . Review of the Ct values showed that samples missed by the RAD test had relatively high Ct values (Fig. 1 ). The RAD test detected between 28.6 % and 81.8 % of RT-PCR-positive high viral load samples from COVID-19 patients. However, it only detected between 0–21.1 % for normal viral load samples.
Table 2.
Performance characteristics of the rapid antigen detection test for the presence of SARS-CoV-2 virus in 160 respiratory samplesa.
| Sample type |
All samples (N = 160) |
High viral load samples (N = 64) |
Normal viral load samples (N = 96) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ct value |
No. of samples |
Ct value |
No. of samples |
Ct value |
No. of samples |
||||||||||
| mean | range | tested | positive | sensitivity | mean | range | tested | positive | sensitivity | mean | range | tested | positive | sensitivity | |
| NPA & TS | 20.72 | 13.80−29.95 | 35 | 12 | 34.3% | 16.08 | 13.80−17.95 | 11 | 9 | 81.8 % | 22.84 | 18.69−29.95 | 24 | 3 | 12.5% |
| NPS & TS | 19.35 | 13.67−29.79 | 35 | 16 | 45.7 % | 16.36 | 13.67−18.52 | 20 | 16 | 80.0% | 23.33 | 18.79−29.79 | 15 | 0 | 0.0% |
| sputum | 22.91 | 15.32−31.12 | 45 | 5 | 11.1 % | 16.98 | 15.32−18.14 | 7 | 2 | 28.6 % | 24.00 | 19.43−31.12 | 38 | 3 | 7.9% |
| throat saliva | 19.65 | 11.25−30.81 | 45 | 18 | 40.0% | 15.16 | 11.25−18.49 | 26 | 14 | 53.8% | 25.79 | 19.20−30.81 | 19 | 4 | 21.1 % |
High viral load samples means samples with Ct values <18.57 of SARS-CoV-2 virus–specific RT-PCR while normal viral load samples mean samples with Ct values >18.57.
Fig. 1.
The results of the 160 respiratory samples tested for rapid antigen detection test and the corresponding Ct values of SARS-CoV-2 virus–specific RT-PCR.
In the cross-reactivity test using virus isolates, all were tested negative by the RAD test.
4. Discussion
In this study, we determined the performance characteristics of the RAD test, the BIOCREDIT COVID-19 Ag, for detecting SARS-CoV-2 virus in respiratory samples and compared the results with RT-PCR as the gold standard.
Although our data indicated that RAD test was capable of detecting SARS-CoV-2 virus in NPA & TS, NPS & TS, sputum and throat saliva with different sensitivities, this method was less sensitive than RT-PCR. Consequently, the negative results from this RAD method cannot exclude SARS-CoV-2 virus infection confidently and thus results should be verified by further RT-PCR testing. Additionally, the low prevalence of high viral load samples further limits the use of RAD test in clinical setting. At the time of writing this report (May 2020), among the SARS-CoV-2 positive samples received in PHLSB, only 16.6 % (218/1311) were high viral load (Ct<18.57 = 218: NPA & TS = 80, NPS & TS = 68, sputum = 13, throat saliva = 57; Ct>18.57 = 1093: NPA & TS = 268, NPS & TS = 318, sputum = 101, throat saliva = 406).
The limitations of this study include the fact that we employed a modified sample processing method to perform the RAD test. We added some samples directly into a sample well of the device instead of using the swab provided by the kit to collect the samples. However, we were capable of determining the viral load dependent effect on its sensitivity.
Other limitations of our study include the fact that the samples were refrigerated after completion of RT-PCR and only tested for this study after storage a period of time which may have led to antigen degradation. However, because our findings of viral load dependent effect for NPA & TS and NPS & TS samples were consistent with those of the test results using serial tenfold dilution of the SARS-CoV-2 culture isolate, the antigen degradation is unlikely. The maximum Ct values for NPA & TS, NPS & TS samples that were detectable by the RAD test were 20.99 and 17.52, respectively. The LOD for RAD test using serial tenfold dilution of the SARS-CoV-2 culture isolate was estimated to be 18.57.
In summary, it was shown that testing of patients suspected of SARS-CoV-2 infection with antigen-based assay may produce more false negative results in clinical practice. A recent study evaluating another RAD test showed that an overall sensitivity of 30.2 % was found for the 106 SARS-CoV-2 RT-PCR positive samples [5]. Application of such assays alone in clinical settings is not recommended in favor of continued molecular diagnostics. The balancing between cost, turnaround time, ease of performance and sensitivity in adopting antigen-based assay should be considered [6].
CRediT authorship contribution statement
Gannon CK Mak: Conceptualization, Methodology, Validation, Investigation, Writing - original draft, Writing - review & editing. Peter KC Cheng: Methodology, Validation, Investigation, Writing - review & editing. Stephen SY Lau: Validation, Investigation. Kitty KY Wong: Validation, Investigation. CS Lau: Resources, Supervision. Edman TK Lam: Supervision. Rickjason CW Chan: Supervision, Writing - original draft, Writing - review & editing. Dominic NC Tsang: Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
None.
Acknowledgement
We thank all staff of the Microbiology Division, Public Health Laboratory Services Branch, and Communicable Disease Branch, Centre for Health Protection, for technical assistance and epidemiological information during the current SARS-CoV-2 pandemic.
References
- 1.WHO . 2020. Coronavirus Disease (COVID-2019) Situation Report – 51.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_10 11 March.Available from: [Google Scholar]
- 2.WHO . 2020. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases: Interim Guidance, 2 March.https://apps.who.int/iris/handle/10665/331329 Available from: [Google Scholar]
- 3.WHO . 2020. Laboratory Testing Strategy Recommendations for COVID-19: Interim Guidance.https://apps.who.int/iris/handle/10665/331509 21 March. Available from: [Google Scholar]
- 4.WHO . 2020. WHO Reference Laboratories Providing Confirmatory Testing for COVID-19.https://www.who.int/who-documents-detail/who-reference-laboratories-providing-confirmatory-testing-for-covid-19 (last updated: 29 April 2020). Available from: [Google Scholar]
- 5.Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;(May):21. doi: 10.1016/j.jcv.2020.104455. 2020 published online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chauhan N., Narang J., Pundir S., Singh S., Pundir C.S. Laboratory diagnosis of swine flu: a review. Artif. Cells Nanomed. Biotechnol. 2013;41(June 3):189–195. doi: 10.3109/10731199.2012.716063. [DOI] [PubMed] [Google Scholar]