Abstract
Age is one of the key risk factors to develop malignant diseases leading to a high incidence of hepatic tumors in the elderly population. The only curative treatment for hepatic tumors is surgical removal, which initiates liver regeneration. However, liver regeneration is impaired with aging, leading to an increased surgical risk for the elderly patient. Due to the increased risk, those patients are potentially excluded from curative surgery. Aging impairs autophagy via lipofuscin accumulation and inhibition of autophagosome formation. Autophagy is a recycling mechanism for eukaryotic cells to maintain homeostasis. Its principal function is to degrade endogenous bio-macromolecules for recycling cellular substances. A number of recent studies have shown that the reduced regenerative capacity of the aged remnant liver can be restored by promoting autophagy. Autophagy can be activated via multiple mTOR-dependent and mTOR-independent pathways. However, inducing autophagy through the mTOR-dependent pathway alone severely impairs liver regeneration. In contrast, recent observations suggest that inducing autophagy via mTOR-independent pathways might be promising in promoting liver regeneration. Conclusion: Activation of autophagy via an mTOR-independent autophagy inducer is a potential therapy for promoting liver regeneration, especially in the elderly patients at risk.
Keywords: mTOR, AMPK, ULK1, TFEB, hepatectomy, hepatocyte, proliferation
1. Introduction
1.1. Aging Increases the Incidence of Malignancies
With the advancement of the health system, the lifespan of the population has increased significantly compared with the 1950s [1]. The incidence of cancer, an age-associated malignant transformation of cells, increased in parallel. Despite all developments, cancer-related mortality remains high. According to 2018 Global cancer statistics, the mortality of hepatic carcinoma was ranked second among various tumors in males [2].
1.2. Aging Increases the Risk of Liver Resection Due to Impaired Liver Regeneration
Aging in the human liver is causing morphological and physiological alterations, such as a decrease in liver volume and liver blood flow. Nonetheless, the aging liver can still maintain relatively normal metabolic functions under physiological conditions [3,4]. However, the regenerative capacity of the aging liver is significantly reduced compared with the young liver [5,6]. This age-related change has brought a dilemma to the clinical treatment of liver tumors in this subgroup of patients.
Liver resection and especially, extended resection is only performed in patients without obvious liver dysfunction, without severe dysfunction of other organs and without obvious impairment of liver regeneration. However, the mitotic capacity of the hepatocytes gradually declines with aging, leading to an impairment of liver regeneration in the elderly [6,7]. This impairment represents an additional risk for elderly patients, thereby possibly excluding these patients from curative surgery.
Patients with non-alcoholic fatty liver disease (NAFLD) or alcoholic cirrhosis have obviously impaired hepatic regenerative capacity [8,9]. Aging is often associated with similar morphological changes such as increased liver steatosis and fibrosis [10], which further limits the functional capacity of the aged remnant liver.
1.3. Aging Impairs Autophagy
Autophagy is an essential recycling mechanism of cellular components [11], which plays a crucial role in maintaining liver metabolism and also in promoting liver regeneration. Autophagy provides glucose, amino acids and free fatty acids to hepatocytes for maintaining their basal metabolic function [12]. In liver-specific Atg5 knockout (KO) mice, liver regeneration was substantially declined with an impaired postoperative mitotic capacity after 70% partial hepatectomy (PH) compared with control animals. In the regenerating liver of the KO-mice, the ability to reorganize damaged proteins and organelles during autophagy was diminished [13].
Recently, several studies indicated that autophagy is substantially lower in the aged liver compared to the young liver [5,14,15,16]. Aging leads to a decrease in the number and function of autophagosomes and causes lipofuscin accumulation. Lipofuscin accumulation reduces the efficacy of autophagy enzymes, resulting in a significant decrease in autophagy activity [17,18,19,20,21,22].
1.4. Impaired Regeneration of the Aged Liver is Related to Impaired Autophagy
Liver regeneration requires abundant energy and cellular substances for DNA replication and cell division [23]. Autophagy can effectively provide the needed substances during the regenerative period and remove dysfunctional organelles or aggregated proteins [11,13]. Both contribute to the coordinated proliferation of hepatocytes during the regenerative process.
Recent experiments have shown that there is a close link between autophagy and liver regeneration, but its role is discussed controversially [5,13,24,25,26,27,28,29]. Most studies have shown that a moderate induction of autophagy can promote liver regeneration, but some studies have reached the opposite conclusion. However, little is known about the impact of old age on the inter-related processes of autophagy and regeneration,
In this review, we want to
(1) clarify the relationship of intermingled molecular pathways of liver regeneration and autophagy in the aged liver and
(2) identify potential pharmacological strategies to induce autophagy and thereby restore the age-related impaired liver regeneration.
2. Liver Regeneration
2.1. The Powerful Regenerative Capacity of the Liver Is the Pathophysiological Basis for Successful Partial Hepatectomy
The liver consists of parenchymal cells (hepatocytes) and non-parenchymal cells (Kupffer cells, endothelial cells, epithelial cells, stellate cells and lymphocytes). Under normal physiological conditions, most of the hepatocytes are quiescent. The liver has the unique ability to switch from a quiescent to a proliferative state in response to a loss of liver cells due to surgery or chemical injury.
For example, hepatocytes enter into the cell cycle and start mitosis after partial hepatectomy (PH) (of various extents), portal vein ligation (PVL), acute toxic insult, viral infection and other types of stimuli.
Partial hepatectomy (PH) in rats or mice is a widely used model for studying liver regeneration. After 2/3PH, the residual hepatic tissue almost completely restores the original mass and function in about one week, demonstrating the amazing regenerative ability and compensatory functional capacity [30,31,32].
2.2. Under Most Circumstances Regeneration of the Liver Is Achieved by the Division of the Remaining Mature Hepatocytes
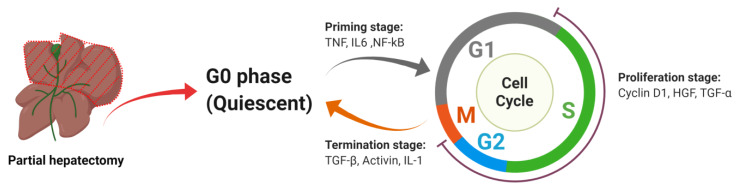
After loss of a substantial amount of liver mass, the remnant mature hepatocytes start to divide rapidly to regenerate the organ to approximately full size. Liver regeneration via proliferation of hepatocytes is a highly complex process, consisting of three stages: the priming stage, the proliferation stage and the termination stage. Each of the stages is controlled by specific transcription factors and cytokines resulting in a highly regulated process (Figure 1), which leads to restoration of the liver mass within days.
Figure 1.
Overview of liver regeneration stages. After partial hepatectomy, every stage is governed by specific transcription factors and cytokines. Under their tight regulation, hepatocytes undergo the process from initiation to termination of proliferation.
2.2.1. Priming Stage
Quiescent hepatocytes switch from G0 to G1 of the cell cycle within 4 h after partial hepatectomy both in rats and mice [33]. The cytokines TNF, IL6 and transcription factor NF-kB are three key regulators in this stage.
TNF
Tumor Necrosis Factor (TNF) is a multi-function cytokine. It is mainly produced by macrophages. The main effect of TNF is to regulate immune cells. It is involved in a variety of cellular processes, such as inflammatory responses, apoptosis, cell differentiation and proliferation.
TNF is secreted in the liver within 1 h in response to partial hepatectomy [34]. It binds to Tumor Necrosis Factor Receptor 1 (TNFR-1) in non-parenchymal cells in the liver and activates Nuclear Factor Kappa B (NF-kB). The activated NF-kB shifts to the nucleus to induce IL-6 expression, which in turn, activates Signal Transducer and Activator of Transcription 3 (STAT-3), Extracellular Signal-Regulated protein Kinases 1 and 2 (ERK1/2) pathways. Activated STAT3 enters the nucleus to initiate transcription of hepatocyte growth response genes. ERK1/2 can promote hepatocyte proliferation and DNA replication [35,36,37,38,39].
Talarmin [40] observed in a rat experiment that ERK activation occurred in the G1 phase in hepatocytes of liver-resected rats undergoing regeneration, but ERK activation was not found in livers from sham-operated rats. Yamada [41] established a TNFR-1 knockout model and observed severe impairment of liver regeneration in the mice, which confirms the importance of TNF signaling in liver regeneration.
IL6
Interleukin-6 (IL-6) is a pleiotropic cytokine that is also involved in several pathophysiological activities, such as immune responses, apoptosis and cell proliferation. It is considered as being an effective hepatocyte mitogen [42].
IL-6 first binds to the IL-6 receptor (IL-6R) to form a complex. Due to a change of configuration, it can bind to Glycoprotein 130 (gp130) to form a high-affinity complex with signal transduction properties [43,44,45,46]. After hepatectomy, Lipopolysaccharide (LPS) and other gut-derived factors activate Kupffer cells, resulting in a TNF-α-dependent secretion of IL-6 [36]. Subsequently, the elevation of IL-6 activates the transcription factors STAT3 and CCAAT/enhancer-binding protein beta (C/EBPβ)/nuclear factor-interleukin 6, leading to increased transcription of their target genes.
Lack of IL6 expression leads to an impairment of liver regeneration as observed in IL-6−/− mice. Liver morphology was characterized by ballooning degeneration and necrosis of hepatocytes. Furthermore, decreased DNA synthesis in hepatocytes and lack of STAT3 expression was observed in this study. Interestingly, this defect was limited to hepatocytes because the DNA synthesis seemed normal in IL-6−/− non-parenchymal cells [47]. Thus, the transcription factors and cytokines mentioned above might contribute to the transition of hepatocytes from G0 to G1 phase after liver resection [48,49,50,51,52,53,54].
NF-kB
Nuclear factor kappa B (NF-kB) is a transcription factor that is involved in the inflammatory response, apoptosis, cell survival and proliferation. It is a heterodimer, which consists of two components—RelA and p50. However, the heterodimer is inactive when IκB binds to the RelA subunit. Activation of NF-kB requires the elimination of IκB from the heterodimer. The IκB kinase (IKK) can phosphorylate IκB and trigger its degradation. Once it is degraded, the activated NF-kB heterodimer migrates to the nucleus, where it binds with the target gene to promote gene transcription [33,55,56].
NF-kB is activated within 30 min after partial hepatectomy [55,56,57]. Iimuro [58] observed that blocking NF-kB by a NF-kB inhibitor (Ad5IkB) during liver regeneration caused a large number of hepatocytes to undergo apoptosis, indicating that NF-kB plays a positive role in preventing apoptosis during the course of liver regeneration.
2.2.2. Proliferation Stage
Hepatocytes cross the restriction point of the G1 phase stimulated by mitogens. Then, they enter the synthesis and mitosis phase. HGF, TGF-α and Cyclin D1 play an important role in this stage.
Cyclin D1
Cyclin D1 is a key mediator for hepatocyte proliferation and is also one of the major markers of the hepatocyte entry into the cell cycle. Cyclin D1 expression is increased after 70% PH in animals. The Cyclin D1/Cyclin-dependent kinase 4 (CDK4) complex facilitates cells crossing the G1 restriction point and entering the G1/S transition phase in the cell cycle by phosphorylating Retinoblastoma (Rb) and E2F [33,59]. In Jeffrey’s observation [60], he found the expression of Cyclin D1 promoted hepatocytes to cross the G1 restriction point under the stimulation of epidermal growth factor (EGF). Furthermore, overexpression of Cyclin D1 in primary hepatocytes elevated the level of DNA replication even in the absence of growth factors.
HGF
Hepatocyte growth factor (HGF) is a potent growth factor mainly derived from activated hepatic stellate cells (HSC). HGF is a hepatocyte-specific mitogen that promotes hepatocyte proliferation and DNA synthesis [61,62,63]. Lindroos [64] found that the plasma level of HGF increased sharply after partial hepatectomy in rats. HGF was about 17-fold higher in liver-resected rats compared to normal animals, and HGF elevation did persist for more than 24 h.
TGF-α
Transforming growth factor alpha (TGF-α) is a member of the EGF ligand family and acts also as a mitogen for hepatocytes. It promotes hepatocyte proliferation by binding to EGF receptor (EGFR). TGF-α can significantly promote DNA synthesis in hepatocytes [65,66,67,68]. Mead [68] indicated that the TGF-α mRNA increased by about 9-fold in regenerating rat livers compared to normal livers during the peak of DNA synthesis. Treating primary hepatocytes with TGF-α resulted in a 13-fold increase of DNA synthesis.
2.2.3. Termination Stage
Once the volume and weight of the liver return to the preoperative level, the proliferation ceases. Hepatocytes terminate proliferation under the control of TGF-β1, Activin and IL-1.
TGF-β
Transforming Growth Factor beta (TGF-β) is mainly secreted by hepatic stellate cells in the liver. It can inhibit the synthesis of hepatocyte DNA. It can also induce apoptosis and contribute to size regulation during regeneration [69,70,71].
TGF-β family messenger RNA and protein levels are upregulated in quiescent hepatocytes where most cells are in the G0 phase. TGF-β is inhibited in the early stage of regeneration by up-regulation of TGF-β inhibitory proteins Ski-related novel protein N (SnoN) and Sloan-Kettering Institute (Ski). Down-regulation of the TGF-β receptors prompt hepatocytes to transition from G1 to S phase. However, it is restored during termination stage of liver regeneration, driving the hepatocytes into a quiescent state [36,72,73].
Activin
Activin is a member of the TGF-β family. As well as most other members of the family, it is a dimeric protein. It is involved in regulating cell differentiation and homeostasis [74,75].
Activin is a DNA synthesis inhibitor of hepatocytes [70,76]. Takamura [77] observed that Activin was reduced in the early stage of liver regeneration after partial hepatectomy, but it increased significantly at 120 h. Multiple studies showed it can induce hepatocyte growth arrest and apoptosis [76,78,79,80].
IL-1α/β
The IL-1 family contains 11 cytokines; the most studied cytokines are IL-1α and IL-1β. IL-1α/β are pro-inflammatory cytokines and they play an important role in the inflammatory response, cell proliferation and differentiation [81]. Numerous studies showed that IL-1α/β are negative regulators during liver regeneration.
IL-1α/β are secreted by Kupffer cells and other non-parenchymal cells in the regenerating liver [82]. Both cytokines inhibit hepatocyte proliferation induced by liver resection. Boulton [83] found that the expression of IL-1α mRNA was down-regulated in the early stage of liver regeneration at about 10 h postoperatively. However, IL-1α mRNA was up-regulated at 24 and 48 h when the proliferation intensity decreased in rat livers after liver resection. The IL-1β expression pattern was similar to IL-1α. Similarly, Nakamura [84] observed in-vitro that IL-1β significantly impaired DNA synthesis in primary rat hepatocytes stimulated by insulin and EGF. The above results show that IL-1α/β is involved in the terminating excessive proliferation of hepatocytes.
2.3. The Effects of Aging on Liver Morphology, Metabolism and Regeneration
Aging causes morphological and physiological changes in the liver, such as a decrease in hepatic volume and perfusion [4]. It leads to an increase in the size of hepatocytes, a decrease in the number of mitochondria and an increase in the number of binucleated cells [10,85]. However, the aging liver can maintain its metabolic function relatively well. Structural analysis of the liver showed that mitochondrial integrity did not undergo an obvious age-related change. No significant change in liver enzyme activity was observed [3,86].
In contrast, aging significantly impedes liver regeneration [10]. After partial hepatectomy, 90%–100% of the hepatocytes in the young liver enter the S phase, while only about 30% of the hepatocytes in the aged liver enter the S phase [87].
2.4. Liver Progenitor Cells may Enhance Liver Regeneration under Certain Circumstances
In addition to the division of mature hepatocytes, the first line of defense for liver regeneration, liver progenitor cells (LPCs) are also shown to participate in the course of animal liver regeneration [88,89]. This is called the second line of defense for liver regeneration (Table 1).
Table 1.
Differentiation between first and second line of regeneration (based on an analysis of specific reviews). The regenerative capacity of the liver is built on two lines of defense. The 1st line of defense: division of remaining mature hepatocytes; 2nd line of defense: mainly through division and differentiation of liver stem/progenitor cells.
| Modes of Liver Regeneration | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Year | Author | Conditions for 1st Line Regeneration | Conditions and Mechanism for 2nd Line Regeneration | Main Cell Type for 2nd Line of Regeneration (as Indicated by Author) | |||||
| Mild Liver Injury | CCL4 ΔΔ | PH | Severe Liver Injury | Impaired Proliferative Capacity of Mature HCs 🗶 | Chronic Liver Injury | Delayed Response to Hepatic Injury | |||
| 1996 | Snorri [91] | Y Δ | Y | OCs * | |||||
| 2001 | Nelson [95] | Y | Y | HSCs **/LPCs | |||||
| 2008 | Viebahn [96] | Y | Y | LPCs | |||||
| 2013 | Jan [97] | Not given | Y | LPCs | |||||
| 2013 | Ioannis [98] | Y | Y | Y | Y | Y | PCs *** | ||
| 2014 | THAN [92] | Y | Y | Y | Y | Y | LPCs | ||
| 2014 | Itoh [90] | Y | Y | Y | Y | LPCs | |||
| 2015 | Jan [99] | Y | Y | Y | LPCs | ||||
| 2016 | Minoru [100] | Y | Y | Y | LPCs | ||||
| 2017 | Veronika [101] | Not given | Y | LPCs | |||||
Δ The Y mark (Yes) indicates that the item is selected; ΔΔ CCL4: Carbon Tetrachloride. * OCs (Oval cells); ** HSCs (Hepatic stem cells); *** PCs (Progenitor cells); 🗶 HCs: Hepatocytes.
LPCs, also described by some authors as oval cells, are bipotent progenitor cells residing in the canal of Hering at the transition between bile canaliculi and bile ductuli. They can differentiate into hepatocytes and cholangiocytes [29,90].
This is described in two conditions: (1) when the liver is severely damaged, e.g., by the administration of 2-acetylaminofluorene administration before PH; (2) when the liver is subjected to chronic injury, e.g., by administration of a Choline-Deficient, Ethionine-supplemented (CDE) diet. In both cases, the remaining hepatocytes cannot meet the needs of liver regeneration. As a consequence, LPCs are activated and contribute to liver regeneration by renewing and differentiating into hepatocytes and cholangiocytes [91,92].
However, the contribution of LPCs for liver regeneration was questioned by some authors [93,94]. For example, using the mouse model of chronic liver injury induced by CDE diet, Schaub [93] could not trace hepatocytes which were clearly derived from liver stem cells (LSCs). One possibility they considered was that their model of liver injury was insufficient for LSCs activation.
2.5. The Effects of Aging on Liver Progenitor Cells
The ability of LPCs to respond to hepatic injury declines with age. Cheng observed that [89] LPCs in young mice can be activated and proliferate properly after CDE diet-induced hepatic injury. However, LPCs in old mice failed to respond to the injury, resulting in impaired liver regeneration. Further research showed that hepatic stellate cells in old mice secreted more Chemokine (C-X-C motif) ligand 7 (CXCL7) compared with young mice, which induced neutrophil infiltration in the liver. Neutrophil infiltration produced excessive reactive oxygen species (ROS), thus impairing the activation and proliferation of LPCs to limit the regeneration of liver.
Autophagy plays a vital role in maintaining the function of LPCs as indicated by three different observations. First, Cheng [29] found LPCs had higher autophagy activity compared with differentiated hepatocytes. After inhibiting autophagy by knocking down the Atg5 or Beclin1 genes, self-renewal, proliferation and hepatic-differentiation capacity of LPCs were significantly impaired. Second, inhibition of autophagy makes LPCs more vulnerable to senescence induced by Etoposide stimulation. In contrast, overexpression of Beclin 1 in Beclin1 knockdown (KD)-LPCs restored their hepatic-differentiation capacity and increased its resistance to Etoposide-induced senescence. Third, Ma [102] also observed a similar result that inhibiting the expression of Atg5 seriously hampered hepatic differentiation of LPCs. These results reflect the important role of autophagy in hepatic differentiation of LPCs.
3. Autophagy
After partial hepatectomy, the loss of liver mass triggers autophagy to provide the necessary energy, thereby creating an optimal environment for regeneration. However, hepatic autophagy levels gradually decline with age [5,14,15,16]. Induction of autophagy may be an effective way to promote the recovery of liver mass and physiological function.
3.1. Autophagy Is an Essential Mechanism for Eukaryotes to Recycle Intracellular Components
Three forms of autophagy are currently known: macroautophagy, microautophagy and chaperone-mediated autophagy. Here, we mainly discuss macroautophagy, which is closely related to liver regeneration.
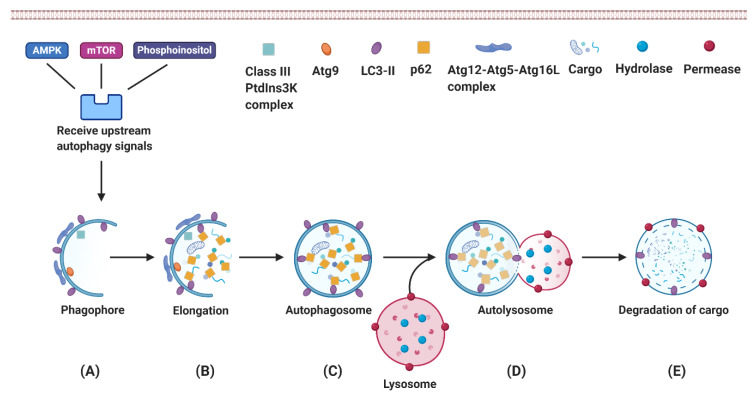
Macroautophagy is the most common type of autophagy (Figure 2). Activation of the autophagy signaling cascade leads to the formation of phagophore as the very first step. The phagophores derive from the omegasome, a cell compartment rich in phosphatidylinositol-3-phosphate (PI3P) on the endoplasmic reticulum. As a next step, phagophores continue to extend and capture autophagy cargo in the cytoplasm. They eventually form the bi-membrane layered autophagosome. Autophagosomes fuse with lysosomes to deliver the autophagy cargo into the lysosome. The cargo is digested by lysosomal hydrolases into amino acids, fatty acids and other basal components, which are then released by membrane permease for further reuse [103,104,105].
Figure 2.
The dynamic process of autophagy. Macroautophagy mainly includes the following steps: (A) Phagophore formation. Activation of the Class III PtdIns3K complex produces PI3P, which facilitates the nucleation of phagophores and regulates the degradation of autophagy cargo [106]. (B) Phagophore elongation and capture of degradation targets. The function of the transmembrane protein Atg9 is to deliver the membrane from donor organelles to the expanding phagophore [107]. The LC3-II and Atg12–Atg5–Atg16L complex promotes phagophore elongation [106]. (C) Autophagosome formation. p62 interacts with the autophagy cargo and delivers it to the autophagosome. (D) Fusion of the autophagosome with lysosome; (E) Degradation of the cargo.
Autophagy is divided into basal autophagy and induced autophagy. Basal low-level autophagy occurs constantly in most cells since it is vital for intracellular homeostasis and cell self-renewal. In contrast, induced high-level of autophagy is mainly observed in cells under stress, since it represents a protective response to compensate injury [105,108,109]. Under stress conditions, moderate-level autophagy contributes to cell survival but excessive autophagy may cause cell death [110,111].
3.2. Autophagy Plays an Essential Role in Hepatic Physiological Processes
Autophagy is one of the pro-survival processes allowing hepatocytes to resist cellular stressors such as anoxia and nutrient starvation [112]. Autophagy protects hepatocytes from damage and cell death by clearing damaged organelles and misfolded proteins generated in hepatic disease [113].
The liver is the largest metabolic organ in the human body, and autophagy plays a vital role in metabolic activities. Autophagy is involved in glucose, lipid and protein metabolism of the liver [114]. Liver autophagy converts amino acids into glucose through gluconeogenesis, a fundamental process for maintaining blood glucose level [115].
The process whereby autophagy degrades lipid droplets is termed Lipophagy. The liver is the body’s second-largest “storeroom” of lipids. Lipophagy can degrade lipid droplets into free fatty acids (FFAs), and these FFAs are oxidized in mitochondria to produce Adenosine Triphosphate (ATP) [114,116]. In case of metabolic needs due to a lack of nutrients such as starvation or due to an increased demand as in regeneration, the autophagic cargo is degraded, resulting in an enhanced ATP production as needed for the given processes.
3.3. Autophagy Provides Energy as Needed for Liver Regeneration
It is well known that liver regeneration requires abundant ATP to support the energy-consuming process of cell division and growth [23]. Hepatocytes are rich in mitochondria to meet the energy needs such as their multitudinous metabolic activities and liver regeneration. However, liver resection can cause massive mitochondrial damage and reduce ATP production in hepatocytes, leading to a significantly reduced availability of ATP. Toshima [13] observed a substantial decline of ATP reserves as early as 6 h after mouse liver resection.
In the initial stage of liver regeneration, autophagy, especially mitochondrial selective autophagy (Mitophagy), is essential for maintaining healthy mitochondria that can produce ATP [105,117]. Mitochondria are highly dynamic organelles that continuously fuse, divide (termed mitochondrial dynamics) and form a network structure within the cells [118]. Mitochondrial dynamics manage mitochondrial morphology, distribution and quality [119,120,121]. During this process, mitophagy selectively degrades damaged or dysfunctional mitochondria to promote mitochondrial regeneration and maintain ATP synthesis [122,123,124]. This process provides the energy needed for liver regeneration.
3.4. Age-Related Alterations Impair Autophagy Activity
3.4.1. Age-Related Decline of AMPK Activation Impairs Autophagosome Formation
AMP-activated protein kinase (AMPK) is the main cellular energy sensor. It is activated when cells are in a low energy state indicated by an increased AMP/ATP ratio [125]. Activation of AMPK can effectively induce the onset of autophagy. However, aging causes the decrease of the activation capacity of AMPK, which impairs autophagosome formation, disrupts the maintenance of homeostasis in cells and further accelerates the aging process [20,21,22].
3.4.2. Age-Related Lipofuscin Accumulation in Lysosomes Reduces the Efficiency of the Degradation Process
Lipofuscin is an intracellular cross-linked polymer, consisting of protein residues. The formation of lipofuscin is mainly due to the iron-catalyzed oxidative damage of macromolecules. It is generally accumulating in lysosomes of most cells (including liver cells) with age [126,127].
The lysosome is one of the most important subcellular components in the process of autophagy. Lysosomes maintain an acidic microenvironment to ensure optimal operating conditions for lysosomal enzymes. However, some of the autophagic cargo remains hard to digest. For instance, some damaged and denatured proteins after being exposed to oxidative stress are not degraded, which leads to the formation of lipofuscin.
When lipofuscin-loaded lysosomes accumulate in senescent cells, most lysosomal enzymes are directed from the Golgi-apparatus to the lipofuscin-loaded lysosomes. However, the lysosomal enzymes cannot degrade lipofuscin. In consequence, delivery of enzymes to the lipofuscin-loaded lysosomes is not effective in terms of recycling the damaged proteins. This imbalanced distribution decreases the availability of autophagic enzymes in healthy non-lipofuscin loaded lysosomes [17,18,19], altogether resulting in a further impairment of the autophagy process.
Moreover, lipofuscin accumulation leads to a reduced turnover of damaged mitochondria, which results in an increase in reactive oxygen species. In turn, the oxidative stress further impedes autophagy via the additional impairment of lysosomal function [128,129,130,131,132].
In conclusion, the accumulation of lipofuscin leads to a decline in the lysosomal capability to break down the intracellular macromolecules, resulting in substantial impairment of autophagy.
3.5. Autophagy Can Be Modulated by Interfering with Key Signal Transduction Pathways
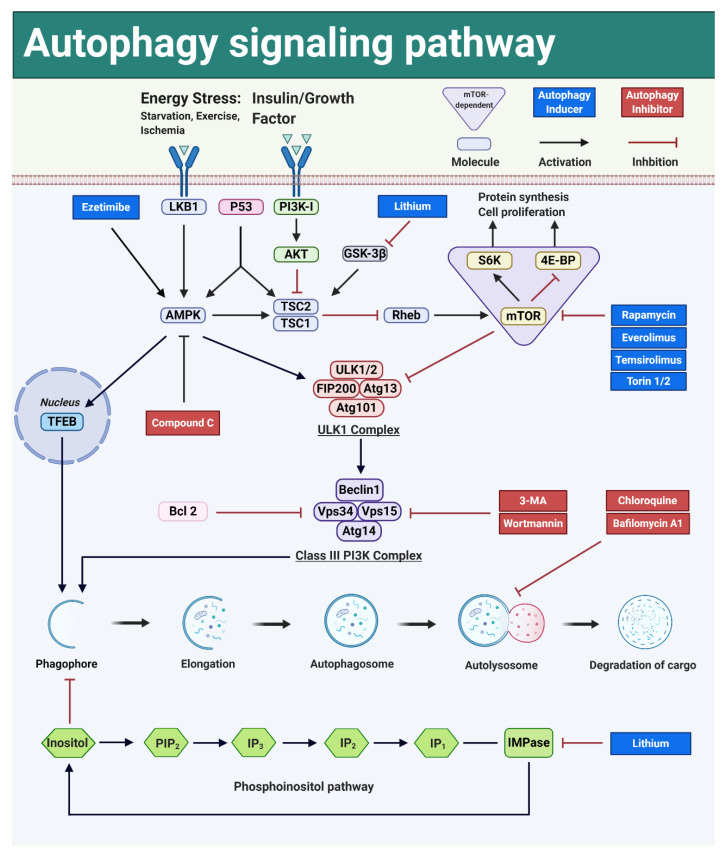
Autophagy is regulated via mammalian target of rapamycin (mTOR)-dependent and mTOR-independent signal transduction pathways (Figure 3). mTOR is a kinase centrally involved in the molecular control mechanism governing cell proliferation, immune responses and metabolic processes, but is also of utmost importance in autophagy [133].
Figure 3.
Signaling pathways and modulators involved in the regulation of macroautophagy. mTOR is a key regulator for both autophagy and cell proliferation. Autophagy can be activated in the mTOR-dependent and the mTOR-independent signaling pathways. Dark blue square: autophagy inducer; Dark red square: autophagy inhibitor.
The autophagy pathways are named based on the key molecules involved. Currently, three mTOR-dependent pathways are considered to be important: the PI3K-AKT-mTOR pathway, the LKB1-AMPK-mTOR pathway and the P53-AMPK-mTOR pathway. Furthermore, also three, but less well-explored mTOR-independent pathways are of importance: the AMPK-ULK1 pathway, the AMPK-TFEB pathway and the Phosphoinositol pathway.
With the deepening of autophagy research, an increasing number of autophagy inducers are available (Table 2). According to their mechanism of action on autophagy, they are divided into mTOR-dependent and mTOR-independent autophagy inducers [134,135].
Table 2.
| Commonly Used Autophagy Inducers | ||
|---|---|---|
| Autophagy Inducers | Mode of ACTION | mTOR-Dependent |
| Rapamycin | mTOR inhibitor | Yes |
| Everolimus | mTOR inhibitor | Yes |
| Temsirolimus | mTOR inhibitor | Yes |
| Torins | mTOR inhibitor | Yes |
| Perifosine | AKT inhibitor | Yes |
| Ezetimibe | AMPK activator; MAPK/ERK inhibitor | No |
| Carbamazepine | Ins and IP3 * inhibitor | No |
| Sodium valproate | Ins and IP3 * inhibitor | No |
| Xestospongin B | IP3 receptor inhibitor | No |
| Xestospongin C | IP3 receptor inhibitor | No |
| Lithium chloride | IMPase ** inhibitor | No |
| Trehalose | Glucose transporter inhibitor; AMPK activator | No |
| Amiodarone | Calcium channel blocker | No |
* Ins: Inositol; IP3: Inositol 1,4,5-trisphosphate. ** IMPase: Inositol monophosphatase.
3.5.1. Autophagy Can Be Activated by Interfering with the mTOR-Dependent Signal Transduction Pathways
In the following paragraphs, we are first introducing the pathways, followed by explaining the mechanism of action of key drugs and are then discussing their potential to modulate liver regeneration.
PI3K-AKT-mTOR
The best described pathway is the PI3K-AKT-mTOR pathway. The Phosphatidylinositol 3-Kinase (PI3K) protein family is involved in the regulation of autophagy, but also in various other cellular functions such as cell proliferation, differentiation and apoptosis.
Activation of PI3K via phosphorylation leads to the generation of a second messenger called phosphatidylinositol-3,4,5-triphosphate (PIP3), which binds to signaling kinases AKT and PDK1 (phosphoinositide-dependent kinase-1). AKT is phosphorylated by PDK1. Activated AKT can directly phosphorylate mTOR and thereby activate mTOR to inhibit autophagy. Moreover, AKT enriches the GTP-binding protein-Ras homolog enriched in brain (Rheb) by phosphorylating Tuberous sclerosis complex 1/2 (TSC1/2), then up-regulating mTOR to inhibit autophagy [141,142,143].
mTOR is capable of interacting with multiple binding proteins to form two complexes with different structures and functions—named mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2).
mTORC1 regulates several pathways that together determine the size of the cell. In mammals, it is primarily responsible for the regulation of cell proliferation and autophagy, but also for protein synthesis and ribosome biosynthesis.
mTORC1 is a negative regulator of autophagy. When nutrients such as amino acids are abundant, the activity of mTORC1 is elevated to down-regulate autophagy. In the absence of nutrients (especially glucose), AMPK is up-regulated, which causes mTOR to be down-regulated, thereby inducing autophagy. mTOR acts on two important downstream proteins: S6K1 (p70S6 kinase 1) and eIF4E binding protein (4E-BP). Their main role is to regulate cell growth, proliferation and protein synthesis [141,144,145].
mTORC2 is relatively insensitive to Rapamycin, it mainly regulates the actin cytoskeleton to organize the cell shape and modulates cell metabolism [146].
LKB1-AMPK-mTOR
The LKB1-AMPK-mTOR pathway is involved in the regulation of autophagy, metabolism and cell growth. The liver kinase B1 (LKB1) is a tumor suppressor that acts on the upstream kinase of AMPK. LKB1 can phosphorylate and activate AMPK, which is a key signaling molecule in autophagy as well as in glucose and lipid metabolism [147,148].
AMPK is considered to be a major energy-sensing kinase that activates a variety of catabolic processes. mTORC1 senses the energy state of cells through AMPK, which is activated in response to low intracellular energy levels. Activated AMPK enhances autophagy to promote the production of ATP [146].
Previously, AMPK was thought to regulate mammalian autophagy through the mTOR pathway. AMPK phosphorylation of TSC2 indirectly leads to inhibition of mTOR by inactivating Rheb. Inhibition of mTOR restores the activity of UNC-51-like kinase 1 (ULK-1)—a key autophagy promoter—thereby activating autophagy [149,150,151,152,153,154].
p53-AMPK-mTOR
The p53-AMPK-mTOR pathway is mainly involved in the regulation of autophagy and cell growth. p53—a human tumor suppressor protein—activates AMPK, leading to the inhibition of mTOR [155].
Evidence is accumulating that p53 may regulate autophagy in two-ways according to the subcellular localization. In the nucleus, active p53 tetramers bind to a promoter region of multiple genes encoding for autophagy regulators such as AMPK, TSC2 and death-associated protein kinase 1 (DAPK-1). In contrast, cytoplasmic p53 inhibits autophagy, but its related mechanism remains unclear [156].
Furthermore, p53 can indirectly affect cell growth and proliferation by activating cell cycle regulators. For example, p53 activates its downstream factor p21, which is a Cyclin-dependent kinase (CDK) inhibitor that causes cell cycle arrest [156].
mTOR-Dependent Autophagy Induction and Liver Regeneration
The mTOR-dependent signaling pathways are not only crucial for autophagy, but are also of utmost importance for cell proliferation.
Rapamycin, a macrolide immunosuppressant, is a typical mTOR inhibitor. It has been used to combat fungal infections and to suppress immune functions in organ transplant recipients [157]. Nowadays, it has attracted much attention as an mTOR-dependent autophagy inducer. It can induce autophagy in yeast even under nutrient-rich conditions [158]. Rapamycin activates autophagy by inhibiting the activity of mTOR. It inhibits mTORC1 via forming a compound with FK506 binding protein 12. The complex acts on downstream proteins to inhibit protein synthesis and leads to cell cycle arrest by preventing the transition from G1 to S phase [146,159]. This anti-proliferative effect has been confirmed in several studies of liver regeneration after partial hepatectomy [160,161]. Similar effects were observed when using other mTOR-dependent inducers such as Temsirolimus [27].
In conclusion, mTOR inhibitors can induce autophagy via the mTOR-dependent pathway. However, this may severely impede cell proliferation. Therefore, mTOR-dependent autophagy inducers are not suitable for promoting liver regeneration.
3.5.2. mTOR-Independent Autophagy Pathways Are Not Interrelated with Cell Proliferation Pathways
Inducing autophagy without inhibiting cell proliferation as needed for liver regeneration is the better alternative. Here, modulating the mTOR-independent pathways may have the potential for promoting liver regeneration. As in the previous chapter, we are first introducing the pathways, then present potential mTOR-independent autophagy inducers and discuss their potential to augment regeneration.
AMPK-ULK1
Recent studies have shown that there is another pathway downstream of AMPK influencing autophagy, the AMPK-ULK1 pathway. AMPK directly phosphorylates ULK1, which in turn, initiates the formation of the autophagosomes [162,163].
Several observations support this mechanism:
First, Guha [164] elucidated this pathway when studying the impact of IPMK on autophagy. Inositol Polyphosphate Multikinase (IPMK) is a potent enzyme that catalyzes the production of inositol polyphosphate and phosphatidylinositol. IPMK deletion in mouse embryonic fibroblasts (MEFs) abrogated the AMPK-related ULK phosphorylation, thereby decreasing the autophagy level. The deletion of IMPK down-regulated autophagy and suppressed hepatocyte proliferation by approximately 50% in a mouse liver regeneration experiment using the Carbon Tetrachloride (CCL4) model.
Second, Liu further clarified this pathway when investigating the impact of young plasma on age-impaired autophagy. He [5] first observed in aged rats that treatment with young plasma promoted impaired liver regeneration through the restoration of age-impaired autophagy. In a subsequent study [14], he revealed in a rat model of liver ischemia-reperfusion injury (IRI) that young plasma restored age-impaired autophagy at least partially through the AMPK-ULK1 signaling pathway. In old rats (22 months) treated with young plasma, AMPK phosphorylation was increased, leading to ULK1 activation. Furthermore, in the corresponding cell culture experiment, inhibition of AMPK activity with an AMPK inhibitor (Compound C) prevented the activation of ULK1 and thereby prevented the initiation of autophagy induced by young serum. In other words, treatment with the AMPK inhibitor abolished the protective effect of young serum on hypoxia/re-oxygenation injury in aged hepatocytes, supporting the key role of autophagy for this effect [14]. Taken together, his observations suggest that induction of autophagy via the AMPK-ULK1 pathway may also have the potential for enhancing liver regeneration.
AMPK-TFEB
The transcription factor EB (TFEB) is one of the critical components in lysosomal biogenesis and autophagy. Furthermore, TFEB regulates the expression of genes involved in different phases of the autophagy process [165,166].
Kim [167] observed that TFEB-induced autophagy was dependent on the AMPK pathway. After inhibiting AMPK activity via an AMPK-specific small interfering RNA (siRNA), autophagy was no longer activated through the TFEB pathway.
The activity of the TFEB is mainly affected by its subcellular localization. In the cytoplasm, phosphorylated TFEB suppresses the transcriptional activation of its target genes (e.g., BECN1, UVRAG and RAB7). Once TFEB is transferred to the nucleus, as in starvation or in lysosomal storage disease (e.g., Gaucher disease), it is dephosphorylated and binds to the target genes [165,166], which promotes autophagosome formation and its fusion with lysosomes.
Phosphoinositol Pathway
Autophagy can also be activated via decreasing Inositol or Inositol 1,4,5-trisphosphate (IP3) levels [168]. Inositol 1,4,5-trisphosphate (IP3) negatively regulates autophagy, but the specific mechanism is still unclear.
Two potential mechanisms, both proposed by the group of Kania, are currently discussed. On the one hand, Kania [169] proposed that the endoplasmic reticulum (ER), the organelle storing most of the intracellular Ca2+, can release Ca2+ after IP3 binds to membraneous IP3 receptor on its surface. This step is considered to be the prerequisite for maintaining the mitochondrial energy state. Providing Ca2+ to mitochondria promotes the production of Nicotinamide adenine dinucleotide (NADH) and ATP. In contrast, inhibition of IP3 or IP3 receptor through pharmacological modulation or using gene knockout resulted in a reduced ATP production. Decreased energy levels activate AMPK and induce autophagy via an mTOR-independent mechanism to maintain energy homeostasis [170].
On the other hand, Kania also proposed, that autophagy can be induced by inhibition of IP3 receptor. Beclin 1 promotes autophagy by forming a class III PI3K complex with Atg14, Vps34, Vps15 [138,169], this complex is essential for autophagosome formation. IP3 receptor can bind to Beclin 1 since IP3 receptor may become a target of Beclin 1 recruitment, which diminishes the probability of Beclin 1 for promoting autophagy [171].
mTOR-Independent Autophagy Inducers and Liver Regeneration
Currently a number of mTOR-independent autophagy inducers are available. Based on this pathway analysis, mTOR-independent autophagy inducers should not interfere with liver regeneration. For two of those drugs, recent studies confirmed that inducing autophagy via the mTOR-independent pathways indeed did promote liver regeneration in liver-resection models.
Carbamazepine is a common anticonvulsant, which is clinically often used to treat epilepsy and neuropathic pain. Carbamazepine can induce autophagy through depletion of cellular inositol and AMPK activation. The depletion of cytosolic inositol leads to a reduction in basal IP3, which reduces ATP production by impeding mitochondrial Ca2+ entry. Then, the decreased ATP concentration can activate the AMPK-ULK1 pathway to induce autophagy [172,173].
Kawaguchi [27] recently presented the impact of Carbamazepine-treatment on liver regeneration after partial hepatectomy in mice. Treatment with Carbamazepine significantly promoted hepatocyte proliferation by activating mTOR and its downstream factors S6K. Compared with the animals in the control group, the Liver to Body Weight Ratio (LBWR), Proliferating Cell Nuclear Antigen (PCNA), Ki-67 and 5-Bromo-2′-Deoxyuridine (BrdU) index of Carbamazepine-treatment animals increased significantly on the post-operation day 2 (POD2).
Amiodarone is an antiarrhythmic drug. It is commonly used to treat tachycardia and ventricular fibrillation. It is currently gaining attention as an autophagy inducer. Amiodarone-treatment blocks extracellular Ca2+ afflux via inhibiting L-type Ca2+ channels on the plasma membrane, thereby reducing the intracellular Ca2+ level [140]. Decreasing the intracellular Ca2+ level can activate autophagy [174,175].
Lin [26] observed in a mouse model of PH that Amiodarone can substantially increase autophagy flux via the mTOR-independent pathway and enhance liver regeneration. The authors observed significantly higher LC3-II levels in animals of the amiodarone-treated group compared to the control group, but significantly lower p62 levels. The LBWR and Ki-67 index increased substantially in these mice, indicating an enhanced hepatic proliferative response. In contrast, inhibition of autophagy by Atg7 knockdown or Chloroquine pretreatment caused a significant impairment in liver regeneration.
However, for a number of other mTOR-independent autophagy inducers, the impact on liver regeneration was not yet demonstrated. Based on their mode of action, peri-operative treatment with these drugs should also promote liver regeneration.
Ezetimibe is a Niemann-Pick C1-Like 1 (NPC1L1) inhibitor, which is currently used for the treatment of hypercholesterolemia. It has been proven to improve hepatic steatosis and protect hepatocytes from excessive apoptosis by activating autophagy [167,176,177].
Kim [167] observed that Ezetimibe improved lipid metabolism and reduced hepatocyte apoptosis by inducing autophagy. Ezetimibe promoted autophagy via AMPK activation and TFEB nuclear translocation. TSC2 is a negative modulator of mTOR. Knocking out TSC2 can activate mTOR. It is noteworthy that Ezetimibe still effectively induced autophagy in the TSC2−/− MEFs. Treatment with Ezetimibe upregulated p-AMPK in the cells. Further experiments revealed that under the mTOR-activated condition, Ezetimibe-treatment promoted TFEB nuclear translocation via inhibition of Mitogen-Activated Protein Kinase (MAPK)/Extracellular Signal-Regulated Kinase (ERK) signaling in primary hepatocytes.
Moreover, Yu [176] observed in a rat model of middle cerebral artery occlusion that Ezetimibe promoted the phosphorylation of AMPK, which induced autophagy through directly activating ULK1. Consequently, enhanced autophagy alleviated neuronal apoptosis. Pretreatment of the rats with an AMPK inhibitor (Dorsomorphin) inhibited autophagy and abolished the favorable effects of Ezetimibe. Given that Ezetimibe can induce autophagy and effectively promote hepatocyte survival, it deserves further study regarding its value in liver regeneration.
Lithium is widely used in the treatment of mental disorders such as manic depression. Lithium can activate autophagy without reducing the activity of mTOR and its downstream factors p-S6K1 and p-4E-BP1. It activates autophagy by the inhibition of inositol monophosphatase, causing the depletion of inositol and reduces IP3 levels [168].
Moreover, Lithium inhibits Glycogen Synthase Kinase-3 Beta (GSK-3β) signaling, which activates mTOR by inhibiting TSC2. Nonetheless, Lithium can effectively induce autophagy via the Phosphoinositol pathway in mTOR-activated condition, since these pathways are independent from each other [178]. In conclusion, it is also of high interest to explore the potential of Lithium for promoting liver regeneration.
4. Interactions between Autophagy, Liver Regeneration and Aging
In summary, aging leads to the accumulation of lipofuscin in the lysosome, which impairs the efficiency of autophagic enzymes [17,18,19]. Moreover, aging causes a significant decrease in the number of autophagosomes, which may be related to the decline of activation capacity of AMPK. It further reduces autophagy activity [20,21,22].
As said before, liver resection not only triggers liver regeneration, but also induces autophagy of hepatocytes. Autophagy plays a crucial role in liver regeneration (Table 3) [5,13,26,28,29,164]. Liver regeneration requires abundant energy [23], which is among others generated by recycling intracellular macromolecules derived from organelles damaged during hepatectomy.
Table 3.
Controversy about role of autophagy in influencing regeneration.
| (a). Scientific Evidence that Activating Autophagy Is Promoting Liver Regeneration | |||||||
|---|---|---|---|---|---|---|---|
| Author Year |
Research Model | Pathway | Autophagy Modulation | Parameters Indicating | |||
| Enhanced Autophagy | Enhanced Regeneration | Reduced Autophagy | Reduced Regeneration | ||||
| Takeo [13] 2014 |
Mice | Not investigated | Atg5-KO | LC3-II: --- p62: +++ |
BrdU: --- | ||
| Lin [26] 2015 |
C57BL/6 mice, Male | mTOR- independent |
Amiodarone | LC3-II: +++ p62: --- |
LBWR *: +++ Ki-67: +++ |
||
| Atg7-KD | LC3-II: --- | LBWR *: --- Ki-67: --- |
|||||
| Cheng [29] 2015 |
Liver progenitor cells | Not investigated | Overexpression of Beclin1 | LC3-II: +++ | PAS ×: +++ | ||
|
Beclin1-KD; Atg5-KD |
LC3-II: --- p62: +++ |
CCK-8: --- PAS: --- |
|||||
| Liu [5] 2018 |
SD rats, Male, 3m, 22m; Primary rat hepatocytes |
Not investigated | Young plasma | LC3-II: +++ p62: --- |
Ki-67: +++ | ||
| 3-Methyladenine; Wortmannin |
LC3-II: --- | Ki-67: --- | |||||
| Jia [28] 2019 |
SD rats, Male, 5w | Not investigated | 70% Portal Vein Ligation | LC3-II: +++ | Cyclin D1: +++ | ||
| Guha [164] 2019 |
Mice; MEFs; HEK293T cells. |
IPMK-AMPK-ULK1; IPMK-AMPK-SIRT1 |
IPMK-KO | LC3-II: --- | Ki-67: --- Edu: --- |
||
| (b). Scientific Evidence that Inducing Autophagy Is Inhibiting Liver Regeneration | |||||||
|
Author
Year |
Research Model | Pathway | Autophagy Modulation | Parameters Indicating | |||
| Enhanced Autophagy | Reduced Regeneration | Reduced Autophagy | Enhanced Regeneration | ||||
| Jiang [161] 2001 |
Harlan SD rats, Male | mTOR- dependent |
Rapamycin | Not investigated | LWRR **: --- | ||
| Palme [159] 2008 |
Lewis rats, Male | mTOR- dependent |
Rapamycin | Not investigated | Ki-67: --- | ||
| Fouraschen [160] 2013 |
C57BL/6 mice Male, 12–15w |
mTOR- dependent |
Rapamycin and Steroid dexamethasone |
LC3-II: +++ | BrdU: --- PCNA: --- LWRR **: --- |
||
| Kawaguchi [27] 2013 | C57BL/6J mice, Male, 6–8w | mTOR- dependent |
Temsirolimus | Not investigated | LBWR *: --- PCNA: --- |
||
| Shi [25] 2018 |
Balb/c mice, 6–8w | mTOR- dependent |
Rapamycin | LC3-II: +++ | LBWR *: --- PCNA: --- |
||
| 3-Methyladenine; | LC3-II: --- | LBWR *: +++ PCNA: +++ |
|||||
| ASPP2-haploinsufficient | LC3-II: --- p62: +++ |
LBWR *: +++ PCNA: +++ |
|||||
+++: Increase; ---: Decrease. * LBWR: Liver to body weight ratio. ** LWRR: Liver weight recovery rate. × used for estimating the capacity of LPCs differentiated into hepatocytes.
Autophagy activity in aged liver is significantly reduced compared to young liver [5,14,15]. Therefore, improving autophagy through pharmacological intervention seems to be an effective treatment to promote regeneration in senescent livers. The mTOR pathway is the most common autophagy-related pathway. However, the mTOR pathway is not only the key regulatory pathway for autophagy, but also the pathway that modulates cell proliferation. Inhibition of mTOR activity can induce autophagy, but inhibits cell proliferation at the same time. In the case of liver resection, inhibition of cell proliferation is detrimental, since it causes impairment of liver regeneration, as described in detail above.
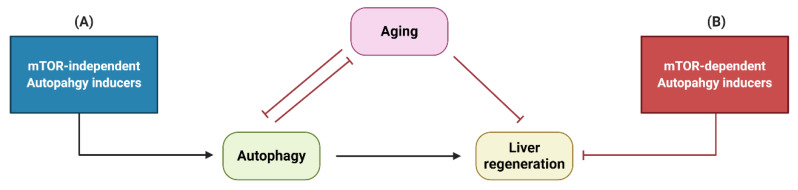
Therefore, modulation of autophagy via the mTOR-independent pathway is a better strategy (Figure 4). Strikingly different drugs such as Carbamazepine, Amiodarone, Ezetimibe and Lithium induce autophagy via these pathways. They are of documented or putative benefit for enhancing liver regeneration and should be explored in more depth. This is of special importance for the elderly population, where liver regeneration is already impaired, in part due to the age-dependent decrease of autophagic activity.
Figure 4.
The relationship between autophagy, liver regeneration and aging. (A) mTOR-independent autophagy inducers can promote liver regeneration, while (B) mTOR-dependent autophagy inducers inhibit liver regeneration.
5. Conclusions
At present, many aging patients with malignant liver disease cannot be treated effectively because of the aging-related impairment of liver regeneration. Aging-related changes also lead to decreased autophagy activity, which is an important cause for insufficient liver regeneration. Age-specific strategies to promote liver regeneration for these patients at risk are needed.
Evidence is accumulating that the modulation of autophagy via pharmacological intervention is an effective approach to promote liver regeneration. This is of utmost benefit for aging patients with impaired autophagy. However, choosing the appropriate autophagy pathway to activate autophagy is crucial. Inducing autophagy by the mTOR-dependent pathway alone is detrimental to liver regeneration. In contrast, activation of autophagy via the mTOR-independent pathway does not affect cell proliferation. Therefore, mTOR-independent autophagy inducers such as Carbamazepine, Amiodarone, Ezetimibe and Lithium should be further explored to promote liver regeneration in the elderly patients.
Abbreviations
| AMPK | AMP-activated protein kinase |
| AMP | Adenosine monophosphate |
| ASPP2 | Apoptosis-stimulating protein two of p53 |
| ATP | Adenosine triphosphate |
| Atg5 | Autophagy related 5 protein |
| Atg7 | Autophagy related 7 protein |
| Atg13 | Autophagy related 13 protein |
| Bcl2 | B-cell CLL/lymphoma 2 |
| BrdU | 5-Bromo-2′-Deoxyuridine |
| CDE | Choline-Deficient, Ethionine-supplemented |
| CDK | Cyclin-dependent kinase |
| CXCL7 | Chemokine (C-X-C motif) ligand 7 |
| C/EBP | CCAAT enhancer binding protein |
| DAPK-1 | Death-associated protein kinase 1 |
| DNA | Deoxyribonucleic Acid |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| ER | Endoplasmic reticulum |
| ERK1/2 | Extracellular signal-regulated kinase 1/2 |
| FFAs | Free fatty acids |
| GSK-3β | Glycogen synthase kinase 3 beta |
| HCs | Hepatocytes |
| HEK | Human embryonic kidney |
| HGF | Hepatocyte growth factor |
| HSC | Hepatic stellate cells |
| HSCs | Hepatic stem cells |
| IL-1 | Interleukin 1 |
| IL-6 | Interleukin 6 |
| IL-6R | Interleukin 6 receptor |
| IMPase | Inositol monophosphatase |
| Ins | Inositol |
| IPMK | Inositol Polyphosphate Multikinase |
| IRI | Ischemia-reperfusion injury |
| KD | Knockdown |
| KO | Knockout |
| LBWR | Liver to body weight ratio |
| LC3 | Microtubule-associated protein 1A/1B-light chain 3 |
| LKB1 | Liver kinase B1 |
| LPCs | Liver progenitor cells |
| LPS | Lipopolysaccharide |
| LSCs | Liver stem cells |
| LWRR | Liver weight recovery rate |
| MAPKs | Mitogen-activated protein kinases |
| MEFs | Mouse Embryonic Fibroblasts |
| mTOR | mammalian Target of Rapamycin |
| mTORC1 | mammalian Target of Rapamycin complex 1 |
| mTORC2 | mammalian Target of Rapamycin complex 2 |
| NADH | Nicotinamide adenine dinucleotide |
| NAFLD | Non-alcoholic fatty liver disease |
| NF-kB | Nuclear factor kappa B |
| NPC1L1 | Niemann-Pick C1-Like 1 |
| OCs | Oval cells |
| PCs | Progenitor cells |
| PCNA | Proliferating cell nuclear antigen |
| PDK1 | Phosphoinositide dependent kinase 1 |
| PH | Partial hepatectomy |
| PIP3 | Phosphatidylinositol-3,4,5-triphosphate |
| PI3K | Phosphatidylinositol 3-kinase |
| PVL | Portal vein ligation |
| Rb | Retinoblastoma |
| Rheb | Ras homolog enriched in brain |
| ROS | Reactive oxygen species |
| SD | Sprague Dawley |
| S6K1 | p70S6 kinase 1 |
| SIRT1 | Silent information regulator 1 |
| Ski | Sloan-Kettering Institute |
| SnoN | Ski-related novel protein N |
| STAT-3 | Signal transducer and activator of transcription 3 |
| TFEB | Transcription Factor EB |
| TGF-α | Transforming growth factor alpha |
| TGF-β | Transforming growth factor beta |
| TNF-α | Tumor necrosis factor alpha |
| TNFR-1 | Tumor necrosis factor receptor 1 |
| TSC1/2 | Tuberous sclerosis complex 1/2 |
| ULK1 | Unc-51 like autophagy activating kinase 1 |
| 3-MA | 3-methyladenine |
| 4E-BP | eIF4E binding protein |
Author Contributions
Writing—original draft preparation, F.X.; conceptualization, F.X. and U.D.; writing—review and editing, C.H., H.-M.T., O.D. and U.D.; project administration, U.D.; funding acquisition, U.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the project “Jena School for Ageing Medicine (JSAM)” of Else Kröner-Fresenius-Stiftung (EKFS).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Roser M. Life Expectancy. [(accessed on 27 February 2019)];2019 Available online: https://ourworldindata.org/life-expect.
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Anantharaju A., Feller A., Chedid A. Aging Liver. Gerontology. 2002;48:343–353. doi: 10.1159/000065506. [DOI] [PubMed] [Google Scholar]
- 4.Wynne H.A., Cope L.H., Mutch E., Rawlins M.D., Woodhouse K.W., James O.F. The effect of age upon liver volume and apparent liver blood flow in healthy man. Hepatology. 1989;9:297–301. doi: 10.1002/hep.1840090222. [DOI] [PubMed] [Google Scholar]
- 5.Liu A., Guo E., Yang J., Yang Y., Liu S., Jiang X., Hu Q., Dirsch O., Dahmen U., Zhang C., et al. Young plasma reverses age-dependent alterations in hepatic function through the restoration of autophagy. Aging Cell. 2018;17:e12708. doi: 10.1111/acel.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pibiri M. Liver regeneration in aged mice: New insights. Aging. 2018;10:1801–1824. doi: 10.18632/aging.101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu C.Z., Ikemoto T., Utsunomiya T., Yamada S., Morine Y., Imura S., Arakawa Y., Takasu C., Ishikawa D., Shimada M. Senescence-related genes possibly responsible for poor liver regeneration after hepatectomy in elderly patients. J. Gastroenterol. Hepatol. 2014;29:1102–1108. doi: 10.1111/jgh.12468. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J.W., Xu H.Y., Li Y., Gong L.L., Zheng G., Wang X.F., Luan W.J., Li S.L., Ma F.X., Ni L.H., et al. NAFLD Induction Delays Postoperative Liver Regeneration of ALPPS in Rats. Dig. Dis. Sci. 2019;64:456–468. doi: 10.1007/s10620-018-5346-3. [DOI] [PubMed] [Google Scholar]
- 9.Horiguchi N., Ishac E.J.N., Gao B. Liver regeneration is suppressed in alcoholic cirrhosis: Correlation with decreased STAT3 activation. Alcohol. 2007;41:271–280. doi: 10.1016/j.alcohol.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheedfar F., Di Biase S., Koonen D., Vinciguerra M. Liver diseases and aging: Friends or foes? Aging Cell. 2013;12:950–954. doi: 10.1111/acel.12128. [DOI] [PubMed] [Google Scholar]
- 11.Chun Y., Kim J. Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life. Cells. 2018;7:278. doi: 10.3390/cells7120278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiskirchen R., Tacke F. Relevance of Autophagy in Parenchymal and Non-Parenchymal Liver Cells for Health and Disease. Cells. 2019;8:16. doi: 10.3390/cells8010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toshima T., Shirabe K., Fukuhara T., Ikegami T., Yoshizumi T., Soejima Y., Ikeda T., Okano S., Maehara Y. Suppression of Autophagy During Liver Regeneration Impairs Energy Charge and Hepatocyte Senescence in Mice. Hepatology. 2014;60:290–300. doi: 10.1002/hep.27140. [DOI] [PubMed] [Google Scholar]
- 14.Liu A., Yang J., Hu Q., Dirsch O., Dahmen U., Zhang C., Gewirtz D.A., Fang H., Sun J. Young plasma attenuates age-dependent liver ischemia reperfusion injury. FASEB J. 2019;33:3063–3073. doi: 10.1096/fj.201801234R. [DOI] [PubMed] [Google Scholar]
- 15.Escobar K.A., Cole N.H., Mermier C.M., VanDusseldorp T.A. Autophagy and aging: Maintaining the proteome through exercise and caloric restriction. Aging Cell. 2019;18:e12876. doi: 10.1111/acel.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bi J., Yang L., Wang T., Zhang J., Li T., Ren Y., Wu R. Irisin Improves Autophagy of Aged Hepatocytes via Increasing Telomerase Activity in Liver Injury. Oxidative Med. Cell. Longev. 2020;2020:6946037. doi: 10.1155/2020/6946037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terman A., Brunk U.T. Autophagy in cardiac myocyte homeostasis, aging, and pathology. Cardiovasc. Res. 2005;68:355–365. doi: 10.1016/j.cardiores.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Cuervo A.M., Bergamini E., Brunk U.T., Dröge W., Ffrench M., Terman A. Autophagy and Aging: The Importance of Maintaining “Clean” Cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 19.Brunk U.T., Terman A. The mitochondrial-lysosomal axis theory of aging—Accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur. J. Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 20.Reznick R.M., Zong H., Li J., Morino K., Moore I.K., Yu H.J., Liu Z.-X., Dong J., Mustard K.J., Hawley S.A., et al. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab. 2007;5:151–156. doi: 10.1016/j.cmet.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salminen A., Kaarniranta K., Kauppinen A. Age-related changes in AMPK activation: Role for AMPK phosphatases and inhibitory phosphorylation by upstream signaling pathways. Ageing Res. Rev. 2016;28:15–26. doi: 10.1016/j.arr.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Salminen A., Kaarniranta K. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res. Rev. 2012;11:230–241. doi: 10.1016/j.arr.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Alexandrino H., Rolo A., Tralhão J.G., Castro e Sousa F., Palmeira C. Mitochondria in liver regeneration: energy metabolism and posthepatectomy liver dysfunction. In: Oliveira P.J., editor. Mitochondrial Biology and Experimental Therapeutics. Springer International Publishing; Cham, Switzerland: 2018. pp. 127–152. [Google Scholar]
- 24.Enkhbold C., Morine Y., Utsunomiya T., Imura S., Ikemoto T., Arakawa Y., Saito Y., Yamada S., Ishikawa D., Shimada M. Dysfunction of liver regeneration in aged liver after partial hepatectomy. J. Gastroenterol. Hepatol. 2015;30:1217–1224. doi: 10.1111/jgh.12930. [DOI] [PubMed] [Google Scholar]
- 25.Shi H., Zhang Y., Ji J., Xu P., Shi H., Yue X., Ren F., Chen Y., Duan Z., Chen D. Deficiency of apoptosis-stimulating protein two of p53 promotes liver regeneration in mice by activating mammalian target of rapamycin. Sci. Rep. 2018;8:17927. doi: 10.1038/s41598-018-36208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C.W., Chen Y.S., Lin C.C., Chen Y.J., Lo G.H., Lee P.H., Kuo P.L., Dai C.Y., Huang J.F., Chung W.L., et al. Amiodarone as an autophagy promoter reduces liver injury and enhances liver regeneration and survival in mice after partial hepatectomy. Sci. Rep. 2015;5:15807. doi: 10.1038/srep15807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi T., Kodama T., Hikita H., Tanaka S., Shigekawa M., Nawa T., Shimizu S., Li W., Miyagi T., Hiramatsu N., et al. Carbamazepine promotes liver regeneration and survival in mice. J. Hepatol. 2013;59:1239–1245. doi: 10.1016/j.jhep.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Jia C.J., Sun H., Dai C.L. Autophagy Contributes to Liver Regeneration After Portal Vein Ligation in Rats. Med. Sci. Monit. 2019;25:5674–5682. doi: 10.12659/MSM.915404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Y.J., Wang B., Zhou H., Dang S.P., Jin M., Shi Y.F., Hao L., Yang Z.X., Zhang Y.Y. Autophagy is Required for the Maintenance of Liver Progenitor Cell Functionality. Cell Physiol. Biochem. 2015;36:1163–1174. doi: 10.1159/000430287. [DOI] [PubMed] [Google Scholar]
- 30.Gebhardt R., Baldysiak-Figiel A., Krugel V., Ueberham E., Gaunitz F. Hepatocellular expression of glutamine synthetase: An indicator of morphogen actions as master regulators of zonation in adult liver. Prog. Histochem. Cyto. 2007;41:201–266. doi: 10.1016/j.proghi.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Yang L.L., Luo Y. Liver regeneration after partial hepatectomy. Shijie Huaren Xiaohua Zazhi. 2016;24:67–74. doi: 10.11569/wcjd.v24.i1.67. [DOI] [Google Scholar]
- 32.Michalopoulos G.K., DeFrances M.C. Liver Regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 33.Fausto N. Liver regeneration. J. Hepatol. 2000;32:19–31. doi: 10.1016/S0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 34.Loffreda S., Rai R., Yang S.Q., Lin H.Z., Diehl A.M. Bile ducts and portal and central veins are major producers of tumor necrosis factor alpha in regenerating rat liver. Gastroenterology. 1997;112:2089–2098. doi: 10.1053/gast.1997.v112.pm9178702. [DOI] [PubMed] [Google Scholar]
- 35.Li W., Liang X.P., Kellendonk C., Poli V., Taub R. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J. Biol. Chem. 2002;277:28411–28417. doi: 10.1074/jbc.M202807200. [DOI] [PubMed] [Google Scholar]
- 36.Taub R. Liver regeneration: From myth to mechanism. Nat. Rev. Mol. Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 37.Souissi I., Najjar I., Ah-Koon L., Schischmanoff P.O., Lesage D., Le Coquil S., Roger C., Dusanter-Fourt I., Varin-Blank N., Cao A., et al. A STAT3-decoy oligonucleotide induces cell death in a human colorectal carcinoma cell line by blocking nuclear transfer of STAT3 and STAT3-bound NF-κB. BMC Cell Biol. 2011;12:14. doi: 10.1186/1471-2121-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L., McBride K.M., Reich N.C. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc. Natl. Acad. Sci. USA. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guégan J.-P., Frémin C., Baffet G. The MAPK MEK1/2-ERK1/2 Pathway and Its Implication in Hepatocyte Cell Cycle Control. Int. J. Hepatol. 2012;2012:328372. doi: 10.1155/2012/328372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talarmin H., Rescan C., Cariou S., Glaise D., Zanninelli G., Bilodeau M., Loyer P., Guguen-Guillouzo C., Baffet G. The mitogen-activated protein kinase kinase/extracellular signal-regulated kinase cascade activation is a key signalling pathway involved in the regulation of G(1) phase progression in proliferating hepatocytes. Mol. Cell Biol. 1999;19:6003–6011. doi: 10.1128/MCB.19.9.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada Y., Kirillova I., Peschon J.J., Fausto N. Initiation of liver growth by tumor necrosis factor: Deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc. Natl. Acad. Sci. USA. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt-Arras D., Rose-John S. IL-6 pathway in the liver: From physiopathology to therapy. J. Hepatol. 2016;64:1403–1415. doi: 10.1016/j.jhep.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Yamasaki K., Taga T., Hirata Y., Yawata H., Kawanishi Y., Seed B., Taniguchi T., Hirano T., Kishimoto T. Cloning and Expression of the Human Interleukin-6 (Bsf-2/Ifn-Beta-2) Receptor. Science. 1988;241:825–828. doi: 10.1126/science.3136546. [DOI] [PubMed] [Google Scholar]
- 44.Taga T., Hibi M., Hirata Y., Yamasaki K., Yasukawa K., Matsuda T., Hirano T., Kishimoto T. Interleukin-6 Triggers the Association of Its Receptor with a Possible Signal Transducer, Gp130. Cell. 1989;58:573–581. doi: 10.1016/0092-8674(89)90438-8. [DOI] [PubMed] [Google Scholar]
- 45.Hibi M., Murakami M., Saito M., Hirano T., Taga T., Kishimoto T. Molecular-Cloning and Expression of an Il-6 Signal Transducer, Gp130. Cell. 1990;63:1149–1157. doi: 10.1016/0092-8674(90)90411-7. [DOI] [PubMed] [Google Scholar]
- 46.Taga T., Kishimoto T. gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- 47.Cressman D.E., Greenbaum L.E., DeAngelis R.A., Ciliberto G., Furth E.E., Poli V., Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 48.Streetz K.L., Luedde T., Manns M.P., Trautwein C. Interleukin 6 and liver regeneration. Gut. 2000;47:309–312. doi: 10.1136/gut.47.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., Kishimoto T. A Nuclear Factor for Il-6 Expression (Nf-Il6) Is a Member of a C/Ebp Family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong Z., Wen Z., Darnell J. Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 51.Trautwein C., Caelles C., Vandergeer P., Hunter T., Karin M., Chojkier M. Transactivation by Nf-Il6 Lap Is Enhanced by Phosphorylation of Its Activation Domain. Nature. 1993;364:544–547. doi: 10.1038/364544a0. [DOI] [PubMed] [Google Scholar]
- 52.Cressman D.E., Diamond R.H., Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology. 1995;21:1443–1449. doi: 10.1002/hep.1840210531. [DOI] [PubMed] [Google Scholar]
- 53.Trautwein C., Rakemann T., Niehof M., RoseJohn S., Manns M.P. Acute-phase response factor, increased binding, and target gene transcription during liver regeneration. Gastroenterology. 1996;110:1854–1862. doi: 10.1053/gast.1996.v110.pm8964411. [DOI] [PubMed] [Google Scholar]
- 54.Niehof M., Manns M.P., Trautwein C. CREB controls LAP/C/EBP beta transcription. Mol. Cell Biol. 1997;17:3600–3613. doi: 10.1128/MCB.17.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tewari M., Dobrzanski P., Mohn K.L., Cressman D.E., Hsu J.C., Bravo R., Taub R. Rapid Induction in Regenerating Liver of Rl/If-1 (an I-Kappa-B That Inhibits Nf-Kappa-B, Relb-P50, and C-Rel-P50) and Phf, a Novel Kappa-B Site-Binding Complex. Mol. Cell Biol. 1992;12:2898–2908. doi: 10.1128/MCB.12.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fitzgerald M.J., Webber E.M., Donovan J.R., Fausto N. Rapid DNA-Binding by Nuclear Factor Kappa-B in Hepatocytes at the Start of Liver-Regeneration. Cell Growth Differ. 1995;6:417–427. [PubMed] [Google Scholar]
- 57.Black D., Lyman S., Heider T.R., Behrns K.E. Molecular and cellular features of hepatic regeneration. J. Surg. Res. 2004;117:306–315. doi: 10.1016/j.jss.2003.10.026. [DOI] [PubMed] [Google Scholar]
- 58.Iimuro Y., Nishiura T., Hellerbrand C., Behrns K.E., Schoonhoven R., Grisham J.W., Brenner D.A. NFkappaB prevents apoptosis and liver dysfunction during liver regeneration. J. Clin. Invest. 1998;101:802–811. doi: 10.1172/JCI483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loyer P., Cariou S., Glaise D., Bilodeau M., Baffet G., GuguenGuillouzo C. Growth factor dependence of progression through G(1) and S phases of adult rat hepatocytes in vitro—Evidence of a mitogen restriction point in mid-late G(1) J. Biol. Chem. 1996;271:11484–11492. doi: 10.1074/jbc.271.19.11484. [DOI] [PubMed] [Google Scholar]
- 60.Albrecht J.H., Hansen L.K. Cyclin D1 promotes mitogen-independent cell cycle progression in hepatocytes. Cell Growth Differ. 1999;10:397–404. [PubMed] [Google Scholar]
- 61.Huh C.G., Factor V.M., Sanchez A., Uchida K., Conner E.A., Thorgeirsson S.S. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc. Natl. Acad. Sci. USA. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura T., Mizuno S. The discovery of Hepatocyte Growth Factor (HGF) and its significance for cell biology, life sciences and clinical medicine. Proc. Jpn. Acad. B-Phys. 2010;86:588–610. doi: 10.2183/pjab.86.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michalopoulos G. HGF and liver regeneration. Gastroenterol. Jpn. 1993;28(Suppl. 4):36–39. doi: 10.1007/BF02782887. [DOI] [PubMed] [Google Scholar]
- 64.Lindroos P.M., Zarnegar R., Michalopoulos G.K. Hepatocyte growth factor (hepatopoietin A) rapidly increases in plasma before DNA synthesis and liver regeneration stimulated by partial hepatectomy and carbon tetrachloride administration. Hepatology. 1991;13:743–750. doi: 10.1002/hep.1840130422. [DOI] [PubMed] [Google Scholar]
- 65.Zambreg I., Assouline B., Housset C., Schiffer E. Overexpression of TGF-α and EGFR, a key event in liver carcinogenesis, is induced by hypoxia specifically in hepatocytes. Gastroenterol. Hepatol. 2019;4:1–4. doi: 10.15761/GHE.1000183. [DOI] [Google Scholar]
- 66.Michalopoulos G.K., Khan Z. Liver regeneration, growth factors, and amphiregulin. Gastroenterology. 2005;128:503–506. doi: 10.1053/j.gastro.2004.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomiya T., Ogata I., Fujiwara K. Transforming growth factor alpha levels in liver and blood correlate better than hepatocyte growth factor with hepatocyte proliferation during liver regeneration. Am. J. Pathol. 1998;153:955–961. doi: 10.1016/S0002-9440(10)65637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mead J.E., Fausto N. Transforming growth factor alpha may be a physiological regulator of liver regeneration by means of an autocrine mechanism. Proc. Natl. Acad. Sci. USA. 1989;86:1558–1562. doi: 10.1073/pnas.86.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Breitkopf K., Godoy P., Ciuclan L., Singer M.V., Dooley S. TGF-beta/Smad signaling in the injured liver. Zeitschrift für Gastroenterol. 2006;44:57–66. doi: 10.1055/s-2005-858989. [DOI] [PubMed] [Google Scholar]
- 70.Tao Y.C., Wang M.L., Chen E.Q., Tang H. Liver Regeneration: Analysis of the Main Relevant Signaling Molecules. Mediat Inflamm. 2017;2017:4256352. doi: 10.1155/2017/4256352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schon H.-T., Weiskirchen R. Immunomodulatory effects of transforming growth factor-β in the liver. Hepatobiliary Surg. Nutr. 2014;3:386–406. doi: 10.3978/j.issn.2304-3881.2014.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Braun L., Mead J.E., Panzica M., Mikumo R., Bell G.I., Fausto N. Transforming growth factor beta mRNA increases during liver regeneration: A possible paracrine mechanism of growth regulation. Proc. Natl. Acad. Sci. USA. 1988;85:1539–1543. doi: 10.1073/pnas.85.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macias-Silva M., Li W., Leu J.I., Crissey M.A.S., Taub R. Up-regulated transcriptional repressors SnoN and Ski bind Smad proteins to antagonize transforming growth factor-beta signals during liver regeneration. J. Biol. Chem. 2002;277:28483–28490. doi: 10.1074/jbc.M202403200. [DOI] [PubMed] [Google Scholar]
- 74.Werner S., Alzheimer C. Roles of activin in tissue repair, fibrosis, and inflammatory disease. Cytokine Growth Factor Rev. 2006;17:157–171. doi: 10.1016/j.cytogfr.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 75.Wang X., Fischer G., Hyvönen M. Structure and activation of pro-activin A. Nat. Commun. 2016;7:12052. doi: 10.1038/ncomms12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zimmermann A. Regulation of liver regeneration. Nephrol. Dial. Transplant. 2004;19(Suppl. 4):iv6–iv10. doi: 10.1093/ndt/gfh1034. [DOI] [PubMed] [Google Scholar]
- 77.Takamura K., Tsuchida K., Miyake H., Tashiro S., Sugino H. Activin and activin receptor expression changes in liver regeneration in rat. J. Surg. Res. 2005;126:3–11. doi: 10.1016/j.jss.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 78.Chen L., Zhang W., Liang H.-F., Zhou Q.-F., Ding Z.-Y., Yang H.-Q., Liu W.-B., Wu Y.-H., Man Q., Zhang B.-X., et al. Activin A induces growth arrest through a SMAD-dependent pathway in hepatic progenitor cells. Cell Commun. Signal. 2014;12:18. doi: 10.1186/1478-811X-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hully J.R., Chang L., Schwall R.H., Widmer H.R., Terrell T.G., Gillett N.A. Induction of Apoptosis in the Murine Liver with Recombinant Human Activin-A. Hepatology. 1994;20:854–862. doi: 10.1002/hep.1840200413. [DOI] [PubMed] [Google Scholar]
- 80.Schwall R.H., Robbins K., Jardieu P., Chang L., La C., Terrel T.G. Activin induces cell death in hepatocytes in vivo and in vitro. Hepatology. 1993;18:347–356. doi: 10.1016/0270-9139(93)90018-I. [DOI] [PubMed] [Google Scholar]
- 81.Wang X., Fu S., Wang Y., Yu P., Hu J., Gu W., Xu X.-M., Lu P. Interleukin-1β mediates proliferation and differentiation of multipotent neural precursor cells through the activation of SAPK/JNK pathway. Mol. Cell. Neurosci. 2007;36:343–354. doi: 10.1016/j.mcn.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 82.Goss J.A., Mangino M.J., Flye M.W. Kupffer cell autoregulation of IL-1 production by PGE2 during hepatic regeneration. J. Surg. Res. 1992;52:422–428. doi: 10.1016/0022-4804(92)90306-K. [DOI] [PubMed] [Google Scholar]
- 83.Boulton R., Woodman A., Calnan D., Selden C., Tam F., Hodgson H. Nonparenchymal cells from regenerating rat liver generate interleukin- 1alpha and -1beta: A mechanism of negative regulation of hepatocyte proliferation. Hepatology. 1997;26:49–58. doi: 10.1053/jhep.1997.v26.pm0009214451. [DOI] [PubMed] [Google Scholar]
- 84.Nakamura T., Arakaki R., Ichihara A. Interleukin-1-Beta Is a Potent Growth Inhibitor of Adult-Rat Hepatocytes in Primary Culture. Exp. Cell Res. 1988;179:488–497. doi: 10.1016/0014-4827(88)90286-8. [DOI] [PubMed] [Google Scholar]
- 85.Gan L., Chitturi S., Farrell G.C. Mechanisms and implications of age-related changes in the liver: Nonalcoholic Fatty liver disease in the elderly. Curr. Gerontol. Geriatr. Res. 2011;2011:831536. doi: 10.1155/2011/831536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmucker D.L. Aging and the liver: An update. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 1998;53:B315–B321. doi: 10.1093/gerona/53A.5.B315. [DOI] [PubMed] [Google Scholar]
- 87.Schmucker D.L., Sanchez H. Liver regeneration and aging: A current perspective. Curr. Gerontol. Geriatr. Res. 2011;2011:526379. doi: 10.1155/2011/526379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Evarts R.P., Nagy P., Marsden E., Thorgeirsson S.S. A precursor—Product relationship exists between oval cells and hepatocytes in rat liver. Carcinogenesis. 1987;8:1737–1740. doi: 10.1093/carcin/8.11.1737. [DOI] [PubMed] [Google Scholar]
- 89.Cheng Y., Wang X., Wang B., Zhou H., Dang S., Shi Y., Hao L., Luo Q., Jin M., Zhou Q., et al. Aging-associated oxidative stress inhibits liver progenitor cell activation in mice. Aging. 2017;9:1359–1374. doi: 10.18632/aging.101232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Itoh T., Miyajima A. Liver regeneration by stem/progenitor cells. Hepatology. 2014;59:1617–1626. doi: 10.1002/hep.26753. [DOI] [PubMed] [Google Scholar]
- 91.Thorgeirsson S.S. Hepatic stem cells in liver regeneration. FASEB J. 1996;10:1249–1256. doi: 10.1096/fasebj.10.11.8836038. [DOI] [PubMed] [Google Scholar]
- 92.Than N.N., Newsome P.N. Stem cells for liver regeneration. QJM Mon. J. Assoc. Physicians. 2014;107:417–421. doi: 10.1093/qjmed/hcu013. [DOI] [PubMed] [Google Scholar]
- 93.Schaub J.R., Malato Y., Gormond C., Willenbring H. Evidence against a Stem Cell Origin of New Hepatocytes in a Common Mouse Model of Chronic Liver Injury. Cell Rep. 2014;8:933–939. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gilgenkrantz H., Collin de l’Hortet A. Understanding Liver Regeneration: From Mechanisms to Regenerative Medicine. Am. J. Pathol. 2018;188:1316–1327. doi: 10.1016/j.ajpath.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 95.Fausto N. Liver regeneration: From laboratory to clinic. Liver Transplant. 2001;7:835–844. doi: 10.1053/jlts.2001.27865. [DOI] [PubMed] [Google Scholar]
- 96.Viebahn C.S., Yeoh G.C. What fires prometheus? The link between inflammation and regeneration following chronic liver injury. Int. J. Biochem. Cell Biol. 2008;40:855–873. doi: 10.1016/j.biocel.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 97.Best J., Dollé L., Manka P., Coombes J., van Grunsven L., Syn W.-K. Role of liver progenitors in acute liver injury. Front. Physiol. 2013;4:258. doi: 10.3389/fphys.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drosos I., Kolios G. Stem Cells in Liver Regeneration and Their Potential Clinical Applications. Stem Cell Rev. Rep. 2013;9:668–684. doi: 10.1007/s12015-013-9437-4. [DOI] [PubMed] [Google Scholar]
- 99.Best J., Manka P., Syn W.-K., Dollé L., van Grunsven L.A., Canbay A. Role of liver progenitors in liver regeneration. Hepatobiliary Surg. Nutr. 2015;4:48–58. doi: 10.3978/j.issn.2304-3881.2015.01.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tanaka M., Miyajima A. Liver regeneration and fibrosis after inflammation. Inflamm. Regen. 2016;36:19. doi: 10.1186/s41232-016-0025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lukacs-Kornek V., Lammert F. The progenitor cell dilemma: Cellular and functional heterogeneity in assistance or escalation of liver injury. J. Hepatol. 2017;66:619–630. doi: 10.1016/j.jhep.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 102.Ma Z., Li F., Chen L., Gu T., Zhang Q., Qu Y., Xu M., Cai X., Lu L. Autophagy promotes hepatic differentiation of hepatic progenitor cells by regulating the Wnt/β-catenin signaling pathway. J. Mol. Histol. 2019;50:75–90. doi: 10.1007/s10735-018-9808-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mizushima N., Komatsu M. Autophagy: Renovation of Cells and Tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 104.Ravikumar B., Sarkar S., Davies J.E., Futter M., Garcia-Arencibia M., Green-Thompson Z.W., Jimenez-Sanchez M., Korolchuk V.I., Lichtenberg M., Luo S.Q., et al. Regulation of Mammalian Autophagy in Physiology and Pathophysiology. Physiol. Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 105.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 106.Zhi-Hong Liu J.C.H. Podocytopathy. Contrib. Nephrol. Basel Karger. 2014;183:83–100. [Google Scholar]
- 107.Feng Y., Klionsky D.J. Autophagic membrane delivery through ATG9. Cell Res. 2017;27:161–162. doi: 10.1038/cr.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mizushima N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005;12:1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 109.Mizushima N., Levine B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su Z.Y., Yang Z.Z., Xu Y.Q., Chen Y.B., Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shen H.M., Codogno P. Autophagic cell death Loch Ness monster or endangered species? Autophagy. 2011;7:457–465. doi: 10.4161/auto.7.5.14226. [DOI] [PubMed] [Google Scholar]
- 112.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ke P.-Y. Diverse Functions of Autophagy in Liver Physiology and Liver Diseases. Int. J. Mol. Sci. 2019;20:300. doi: 10.3390/ijms20020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Czaja M.J., Ding W.X., Donohue T.M., Friedman S.L., Kim J.S., Komatsu M., Lemasters J.J., Lemoine A., Lin J.D., Ou J.H.J., et al. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131–1158. doi: 10.4161/auto.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ezaki J., Matsumoto N., Takeda-Ezaki M., Komatsu M., Takahashi K., Hiraoka Y., Taka H., Fujimura T., Takehana K., Yoshida M., et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martinez-Lopez N., Singh R. Autophagy and Lipid Droplets in the Liver. Annu. Rev. Nutr. 2015;35:215–237. doi: 10.1146/annurev-nutr-071813-105336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sato T., Yamashina S., Izumi K., Ueno T., Koike M., Ikejima K., Peters C., Watanabe S. Cathepsin L-deficiency enhances liver regeneration after partial hepatectomy. Life Sci. 2019;221:293–300. doi: 10.1016/j.lfs.2019.02.040. [DOI] [PubMed] [Google Scholar]
- 118.Hales K.G. Mitochondrial Fusion and Division. Nat. Educ. 2010;3:12. [Google Scholar]
- 119.Tilokani L., Nagashima S., Paupe V., Prudent J. Mitochondrial dynamics: Overview of molecular mechanisms. Essays Biochem. 2018;62:341–360. doi: 10.1042/EBC20170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eisner V., Picard M., Hajnóczky G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat Cell Biol. 2018;20:755–765. doi: 10.1038/s41556-018-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lackner L.L. Shaping the dynamic mitochondrial network. BMC Biol. 2014;12:35. doi: 10.1186/1741-7007-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pickles S., Vigié P., Youle R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 124.Ke P.Y. Mitophagy in the Pathogenesis of Liver Diseases. Cells. 2020;9:831. doi: 10.3390/cells9040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hardie D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 126.Schmucker D.L., Sachs H. Quantifying dense bodies and lipofuscin during aging: A morphologist’s perspective. Arch. Gerontol. Geriatr. 2002;34:249–261. doi: 10.1016/S0167-4943(01)00218-7. [DOI] [PubMed] [Google Scholar]
- 127.Terman A., Brunk U.T. Lipofuscin: Mechanisms of formation and increase with age. Acta Pathol. Microbiol. Et Immunol. Scand. 1998;106:265–276. doi: 10.1111/j.1699-0463.1998.tb01346.x. [DOI] [PubMed] [Google Scholar]
- 128.Terman A., Kurz T., Navratil M., Arriaga E.A., Brunk U.T. Mitochondrial Turnover and Aging of Long-Lived Postmitotic Cells: The Mitochondrial-Lysosomal Axis Theory of Aging. Antioxid. Redox. Sign. 2010;12:503–535. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Perse M., Injac R., Erman A. Oxidative Status and Lipofuscin Accumulation in Urothelial Cells of Bladder in Aging Mice. PLoS ONE. 2013;8:e59638. doi: 10.1371/journal.pone.0059638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Terman A., Gustafsson B., Brunk U.T. Autophagy, organelles and ageing. J. Pathol. 2007;211:134–143. doi: 10.1002/path.2094. [DOI] [PubMed] [Google Scholar]
- 131.Moreno-Garcia A., Kun A., Calero O., Medina M., Calero M. An Overview of the Role of Lipofuscin in Age-Related Neurodegeneration. Front. Neurosci. 2018;12:464. doi: 10.3389/fnins.2018.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yuan Y.J., Chen Y.N., Peng T.Q., Li L., Zhu W.Z., Liu F., Liu S.Y., An X.X., Luo R.X., Cheng J.Q., et al. Mitochondrial ROS-induced lysosomal dysfunction impairs autophagic flux and contributes to M1 macrophage polarization in a diabetic condition. Clin Sci. 2019;133:1759–1777. doi: 10.1042/CS20190672. [DOI] [PubMed] [Google Scholar]
- 133.Laplante M., Sabatini D.M. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim Y.C., Guan K.L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fleming A., Noda T., Yoshimori T., Rubinsztein D.C. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat. Chem. Biol. 2011;7:9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 136.Leidal A.M., Levine B., Debnath J. Autophagy and the cell biology of age-related disease. Nat. Cell Biol. 2018;20:1338–1348. doi: 10.1038/s41556-018-0235-8. [DOI] [PubMed] [Google Scholar]
- 137.Byun S., Lee E., Lee K.W. Therapeutic Implications of Autophagy Inducers in Immunological Disorders, Infection, and Cancer. Int. J. Mol. Sci. 2017;18:1959. doi: 10.3390/ijms18091959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vicencio J.M., Ortiz C., Criollo A., Jones A.W.E., Kepp O., Galluzzi L., Joza N., Vitale I., Morselli E., Tailler M., et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 139.Levine B., Packer M., Codogno P. Development of autophagy inducers in clinical medicine. J. Clin. Invest. 2015;125:14–24. doi: 10.1172/JCI73938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sarkar S., Ravikumar B., Floto R.A., Rubinsztein D.C. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 141.Li Y., Inoki K., Yeung R., Guan K.L. Regulation of TSC2 by 14-3-3 binding. J. Biol. Chem. 2002;277:44593–44596. doi: 10.1074/jbc.C200510200. [DOI] [PubMed] [Google Scholar]
- 142.Nellist M., Goedbloed M.A., de Winter C., Verhaaf B., Jankie A., Reuser A.J.J., van den Ouweland A.M.W., van der Sluijs P., Halley D.J.J. Identification and characterization of the interaction between tuberin and 14-3-3 zeta. J. Biol. Chem. 2002;277:39417–39424. doi: 10.1074/jbc.M204802200. [DOI] [PubMed] [Google Scholar]
- 143.Nixon R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 144.Corradetti M.N., Guan K.L. Upstream of the mammalian target of rapamycin: Do all roads pass through mTOR? Oncogene. 2006;25:6347–6360. doi: 10.1038/sj.onc.1209885. [DOI] [PubMed] [Google Scholar]
- 145.Saxton R.A., Sabatini D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;127:5–19. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 147.Egan D.F., Shackelford D.B., Mihaylova M.M., Gelino S., Kohnz R.A., Mair W., Vasquez D.S., Joshi A., Gwinn D.M., Taylor R., et al. Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shackelford D.B., Shaw R.J. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ganley I.G., du Lam H., Wang J., Ding X., Chen S., Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., Kim D.H. ULK-Atg13-FIP200 Complexes Mediate mTOR Signaling to the Autophagy Machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.e08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.e08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kamada Y., Yoshino K., Kondo C., Kawamata T., Oshiro N., Yonezawa K., Ohsumi Y. Tor Directly Controls the Atg1 Kinase Complex to Regulate Autophagy. Mol. Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rosso P., Fioramonti M., Fracassi A., Marangoni M., Taglietti V., Siteni S., Segatto M. AMPK in the central nervous system: Physiological roles and pathological implications. Res. Rep. Biol. 2016;7:1–13. [Google Scholar]
- 154.Inoki K., Zhu T.Q., Guan K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 155.Feng Z., Zhang H., Levine A.J., Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Maiuri M.C., Galluzzi L., Morselli E., Kepp O., Malik S.A., Kroemer G. Autophagy regulation by p53. Curr. Opin. Cell Biol. 2010;22:181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 157.Baroja-Mazo A., Revilla-Nuin B., Ramírez P., Pons J.A. Immunosuppressive potency of mechanistic target of rapamycin inhibitors in solid-organ transplantation. World J. Transpl. 2016;6:183–192. doi: 10.5500/wjt.v6.i1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 159.Palmes D., Zibert A., Budny T., Bahde R., Minin E., Kebschull L., Spiegel H.U. Impact of rapamycin on liver regeneration. Virchows Arch. Int. J. Pathol. 2008;452:545–557. doi: 10.1007/s00428-008-0604-y. [DOI] [PubMed] [Google Scholar]
- 160.Fouraschen S.M., de Ruiter P.E., Kwekkeboom J., de Bruin R.W., Kazemier G., Metselaar H.J., Tilanus H.W., van der Laan L.J., de Jonge J. mTOR signaling in liver regeneration: Rapamycin combined with growth factor treatment. World J. Transpl. 2013;3:36–47. doi: 10.5500/wjt.v3.i3.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jiang Y.P., Ballou L.M., Lin R.Z. Rapamycin-insensitive regulation of 4e-BP1 in regenerating rat liver. J. Biol. Chem. 2001;276:10943–10951. doi: 10.1074/jbc.M007758200. [DOI] [PubMed] [Google Scholar]
- 162.Roach P.J. AMPK -> ULK1 -> Autophagy. Mol. Cell Biol. 2011;31:3082–3084. doi: 10.1128/MCB.05565-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guha P., Tyagi R., Chowdhury S., Reilly L., Fu C., Xu R., Resnick A.C., Snyder S.H. IPMK Mediates Activation of ULK Signaling and Transcriptional Regulation of Autophagy Linked to Liver Inflammation and Regeneration. Cell Rep. 2019;26:2692–2703.e7. doi: 10.1016/j.celrep.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Settembre C., Di Malta C., Polito V.A., Garcia-Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB Links Autophagy to Lysosomal Biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Di Malta C., Cinque L., Settembre C. Transcriptional Regulation of Autophagy: Mechanisms and Diseases. Front Cell. Dev. Biol. 2019;7:114. doi: 10.3389/fcell.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim S.H., Kim G., Han D.H., Lee M., Kim I., Kim B., Kim K.H., Song Y.M., Yoo J.E., Wang H.J., et al. Ezetimibe ameliorates steatohepatitis via AMP activated protein kinase-TFEB-mediated activation of autophagy and NLRP3 inflammasome inhibition. Autophagy. 2017;13:1767–1781. doi: 10.1080/15548627.2017.1356977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sarkar S., Floto R.A., Berger Z., Imarisio S., Cordenier A., Pasco M., Cook L.J., Rubinsztein D.C. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kania E., Roest G., Vervliet T., Parys J.B., Bultynck G. IP3 Receptor-Mediated Calcium Signaling and Its Role in Autophagy in Cancer. Front. Oncol. 2017;7:140. doi: 10.3389/fonc.2017.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cárdenas C., Miller R.A., Smith I., Bui T., Molgó J., Müller M., Vais H., Cheung K.-H., Yang J., Parker I., et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rubinsztein D.C., Shpilka T., Elazar Z. Mechanisms of Autophagosome Biogenesis. Curr. Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 172.Schiebler M., Brown K., Hegyi K., Newton S.M., Renna M., Hepburn L., Klapholz C., Coulter S., Obregon-Henao A., Tamayo M.H., et al. Functional drug screening reveals anticonvulsants as enhancers of mTOR-independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO Mol. Med. 2015;7:127–139. doi: 10.15252/emmm.201404137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang J.-J., Zhou Q.-M., Chen S., Le W.-D. Repurposing carbamazepine for the treatment of amyotrophic lateral sclerosis in SOD1-G93A mouse model. CNS Neurosci. 2018;24:1163–1174. doi: 10.1111/cns.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bootman M., Chehab T., Bultynck G., Parys J., Rietdorf K. The regulation of autophagy by calcium signals: Do we have a consensus? Cell Calcium. 2017;70:32–46. doi: 10.1016/j.ceca.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 175.Chen X., Li M., Chen D.H., Gao W.T., Guan J.L., Komatsu M., Yin X.M. Autophagy Induced by Calcium Phosphate Precipitates Involves Endoplasmic Reticulum Membranes in Autophagosome Biogenesis. PLoS ONE. 2012;7:e52347. doi: 10.1371/journal.pone.0052347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yu J., Li X., Matei N., McBride D., Tang J., Yan M., Zhang J.H. Ezetimibe, a NPC1L1 inhibitor, attenuates neuronal apoptosis through AMPK dependent autophagy activation after MCAO in rats. Exp. Neurol. 2018;307:12–23. doi: 10.1016/j.expneurol.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 177.Chang E., Kim L., Park S.E., Rhee E.J., Lee W.Y., Oh K.W., Park C.Y. Ezetimibe improves hepatic steatosis in relation to autophagy in obese and diabetic rats. World J. Gastroenterol. 2015;21:7754–7763. doi: 10.3748/wjg.v21.i25.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sarkar S., Krishna G., Imarisio S., Saiki S., O’Kane C.J., Rubinsztein D.C. A rational mechanism for combination treatment of Huntington’s disease using lithium and rapamycin. Hum. Mol. Genet. 2007;17:170–178. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]