Abstract
The 3D architecture of the genome governs its maintenance, expression and transmission. The cohesin complex organises the genome by topologically linking distant loci and is highly enriched in specialized chromosomal domains surrounding centromeres, called pericentromeres1–6. Here we report the 3D structure of budding yeast pericentromeres and establish the relationship between genome organisation and function. We find that convergent genes mark pericentromere borders and, together with core centromeres, define their structure and function by positioning cohesin. Centromeres load cohesin and convergent genes at pericentromere borders trap it. Each side of the pericentromere is organised into a looped conformation, with border convergent genes at the base. Microtubule attachment extends a single pericentromere loop, size-limited by convergent genes at its borders. Re-orienting genes at borders into a tandem configuration repositions cohesin, enlarges the pericentromere and impairs chromosome biorientation in mitosis. Thus, the linear arrangement of transcriptional units together with targeted cohesin loading shapes pericentromeres into a structure competent for chromosome segregation. Our results reveal the architecture of the chromosomal region within which kinetochores are embedded and the re-structuring caused by microtubule attachment. Furthermore, we establish a direct, causal relationship between 3D genome organization of a specific chromosomal domain and cellular function.
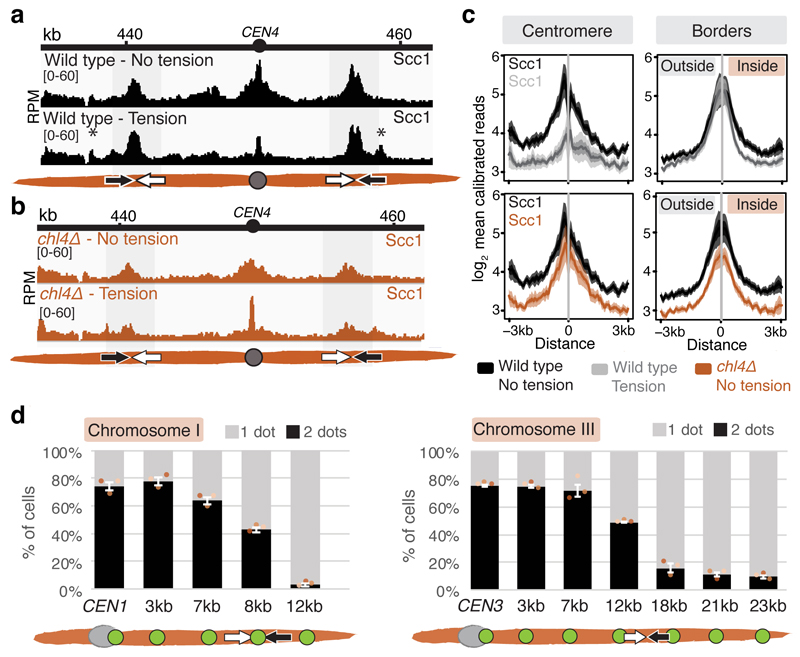
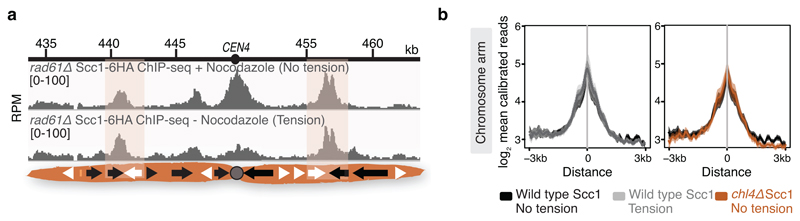
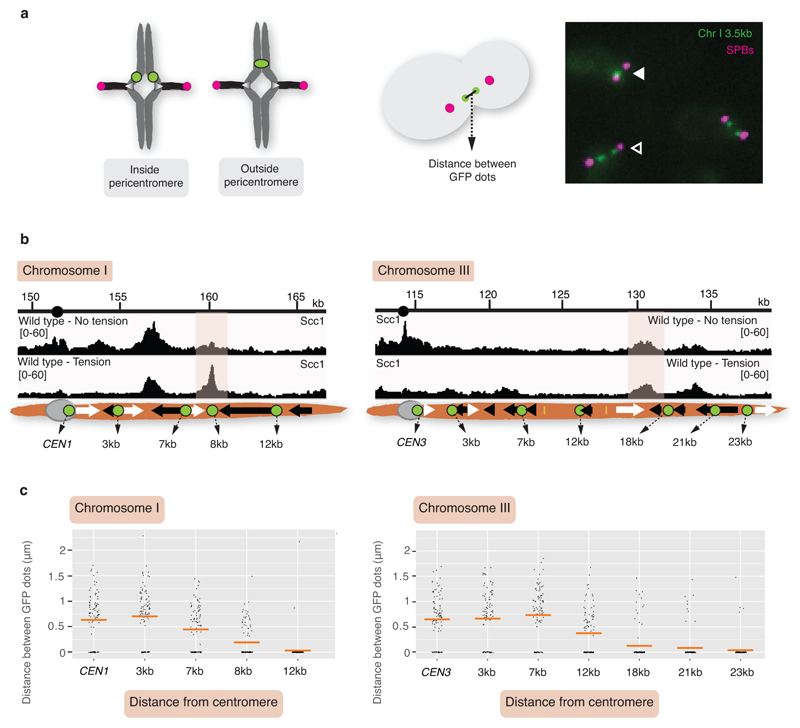
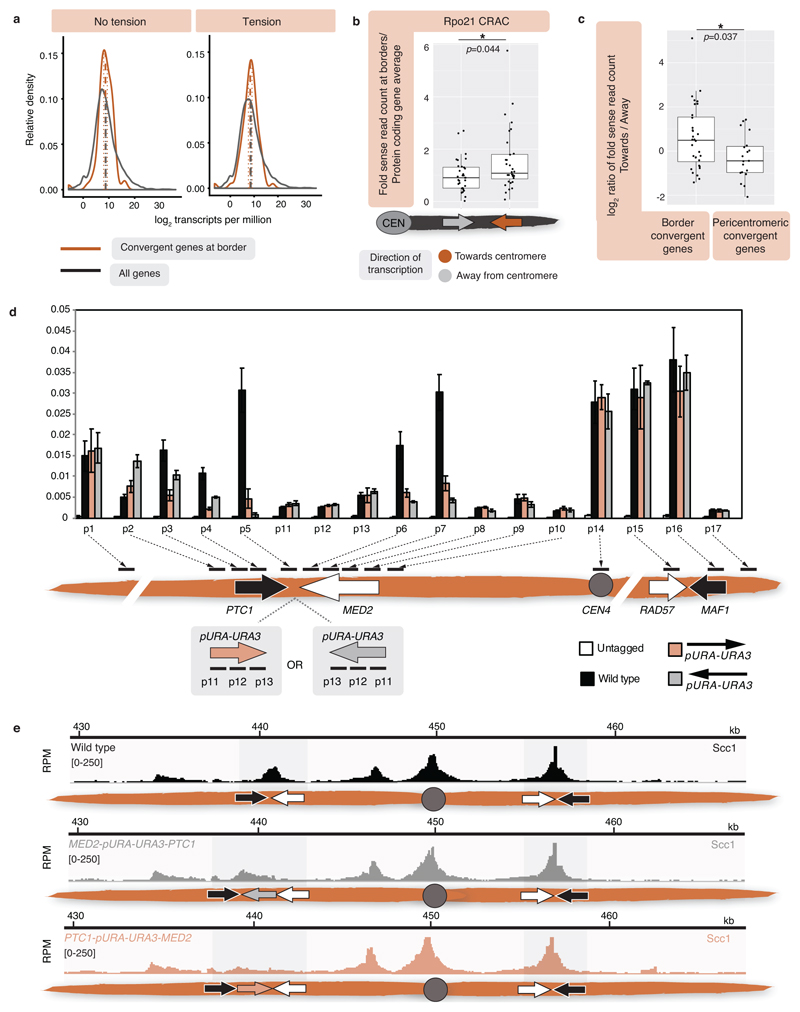
To map pericentromere domains, we arrested cells in metaphase either in the presence or absence of microtubules and analysed cohesin (Scc1) localization by calibrated ChIP-Seq. While cohesin peaks on chromosome arms were comparable, signal was reduced over ~15kb surrounding centromeres in the presence of microtubule-dependent tension, as reported6–8 (Fig. 1a). Wpl1/Rad61 promotes cohesin turnover prior to metaphase9, but is dispensable for the tension-dependent reduction in pericentromeric cohesin (Extended Data Fig. 1a), suggesting passive removal. Interestingly, prominent peaks flanking centromeres persisted in the presence of tension, and additional peaks appeared further away from some centromeres (Fig. 1a, asterisks). Pericentromeric cohesin enrichment occurs through specific targeting of cohesin loading to the centromere by a direct interaction between the Ctf19 inner kinetochore subcomplex and the Scc2/Scc4 cohesin loader10,11. Current models posit that cohesin accumulates at positions distinct from its loading sites12. Indeed, abolishing kinetochore-driven cohesin loading (by deletion of CHL45, encoding a Ctf19 complex component), diminished the cohesin peaks flanking centromeres (Fig. 1b), suggesting that some centromere-loaded cohesin collects at these regions. We henceforth denote these centromere-flanking regions that retain high levels of cohesin under tension and mark the limits of the pericentromere as “borders”. Aligning pericentromere borders from all 16 chromosomes, using the centre of the first cohesin peak that persists under tension, confirmed that while cohesin at centromeres is diminished under tension, cohesin at borders or chromosome arm peaks is not, and that Chl4 promotes cohesin association with centromeres and borders, but not chromosome arms (Fig. 1c, Extended Data Fig. 1b).
Fig. 1. Convergent genes mark pericentromere borders.
Representative cohesin (Scc1) enrichment in wild type (n=3) (a) and chl4Δ (n=2) (b), arrested in metaphase in no tension or tension condition. Pericentromere borders are shaded in grey, black and white arrows indicate convergent genes at borders, asterisk indicates additional cohesin peak under tension. c, Mean calibrated ChIP-Seq reads (solid line), standard error (dark shading) and 95% confidence interval (light shading) at all 32 borders and 16 centromeres. d, Separation of tetO/TetR-GFP markers at indicated distances from CEN1 (left panel) or CEN3 (right panel) in metaphase. For each biological replicate (data points, n=3), 200 cells were scored. Bars show mean ±s.e.m.
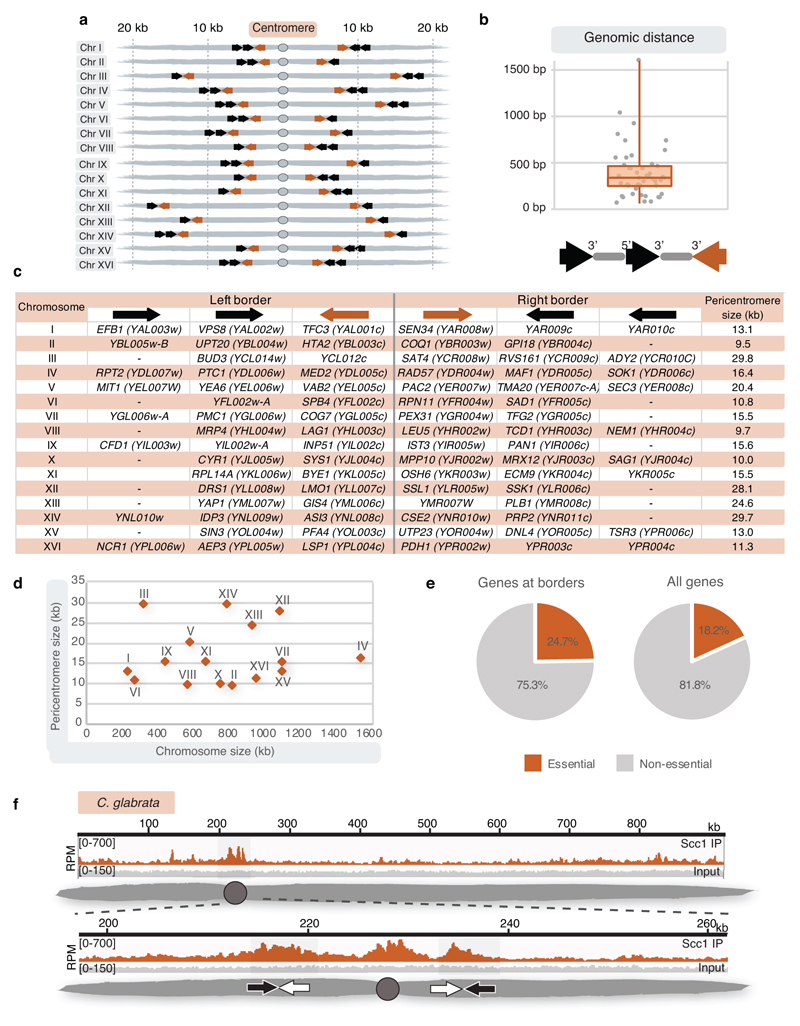
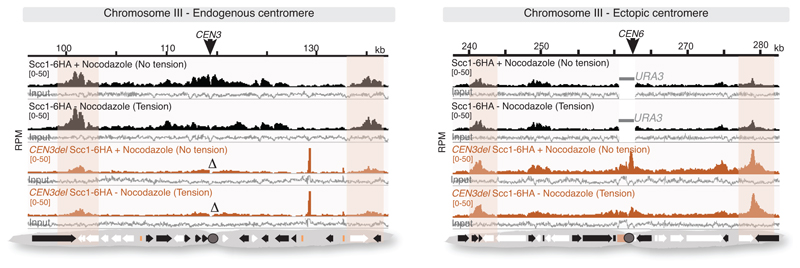
Closer inspection of all pericentromere borders revealed the presence of convergent gene pairs, known sites of cohesin accumulation12, typically symmetrically arranged around the centromere and often associated with an additional, centromere-oriented gene on the distal side (Extended Data Fig. 2a, b). Pericentromere size, as measured by distance between borders, ranges from 9.7 kb (chromosome II) to 29.8 kb (chromosome III) with a mean of ~17 kb, and shows no correlation with chromosome size (Extended Data Fig. 2c, d). Border convergent gene pairs are more frequently essential than S. cerevisiae genes overall and also found at Candida glabrata pericentromeres13 (Extended Data Fig. 2e, f), suggesting a conserved functional arrangement. To determine whether any convergent gene pair has potential for border function, we analysed a strain with an endogenous centromere (CEN3) removed and an ectopic centromere (CEN6) inserted on the arm of chromosome III3. This showed loss of cohesin enrichment at the endogenous pericentromere and borders, and tension-sensitive accumulation of cohesin surrounding ectopic CEN6 on the arm of chromosome III (Extended Data Fig. 3). Interestingly, convergent gene pairs flanking the ectopic centromere showed increased cohesin that persisted under tension, similar to endogenous pericentromere borders (Extended Data Fig. 3).
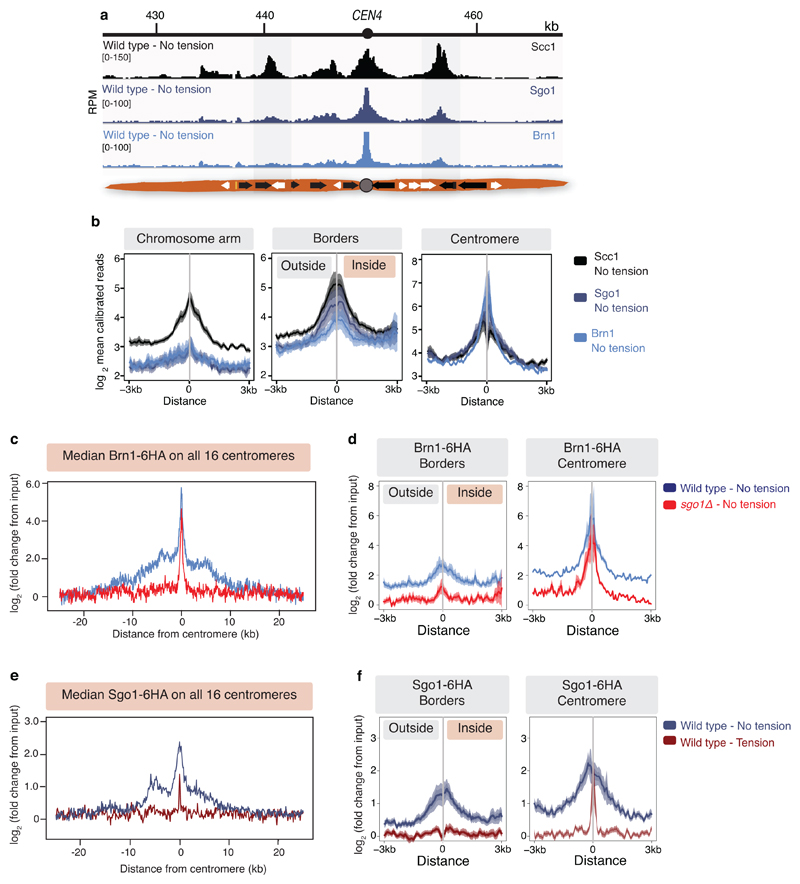
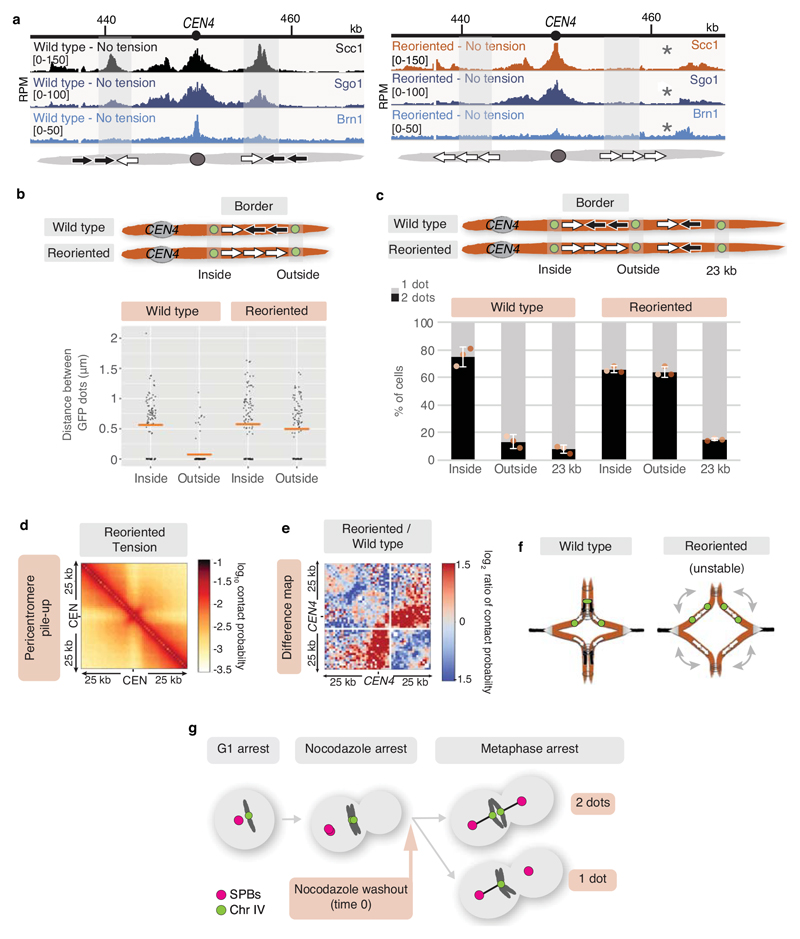
The pericentromeric adaptor protein, shugoshin (Sgo1), promotes sister kinetochore biorientation and proper chromosome segregation, in part by recruiting the chromosome-organising protein condensin to pericentromeres14,15 and dissociates in a tension-dependent manner upon biorientation8. Pericentromere borders show enrichment for both Sgo1 and condensin (Brn1) (Extended Data Fig. 4a, b), and condensin at borders, but not core centromeres, is dependent on Sgo1 (Extended Data Fig. 4c, d). Moreover, tension-sensitive Sgo1 resides predominantly at borders (Extended Data Fig. 4e, f), implying that pericentromere borders may elicit the signal that indicates tension-generating biorientation has been achieved.
Paradoxically, despite the high levels of cohesin, the attachment of sister kinetochores to opposite poles at metaphase causes the separation of sister centromeres, but not chromosomal arms16–18. If borders define the limits of pericentromeres by trapping cohesin to resist the separation of sister chromatids at metaphase, then fluorescent tetO/TetR-GFP markers within the pericentromere are expected to split into two foci at metaphase, while markers outside the border should appear as a single focus (Extended Data Fig. 5a). We selected chromosome I, with its clearly delineated border cohesin peaks indicating a small (13.1kb) pericentromere, and chromosome III, with less defined tension-insensitive cohesin peaks, inferring a large pericentromere (~29.8 kb), for further analysis (Extended Data Fig. 5b). This difference in pericentromere size predicts differential behaviour of GFP foci at equivalent distances from their centromeres. Indeed, while a GFP marker 12kb from CEN1 was almost always observed as a single focus at metaphase, a marker 12kb from CEN3 frequently split into two foci and, for CEN3, single foci became predominant only when markers were positioned 23kb from the centromere (Fig. 1d, Extended Data Fig. 5c). However, a marker 18 kb from CEN3, outside the annotated border, splits in ~10% of cells (Fig. 1d). Similarly, on chromosome I, the second peak of cohesin that persists in the presence of tension appears predominant in border function because a marker at 7kb separates in ~58% of cells, while a marker at 8kb, within a second, distal cohesin peak, separates in only ~30 % of cells (Fig. 1d, Extended Data Fig. 5c, d). Therefore, while preferred pericentromere borders exist, they are not fail-safe and alternative sites of cohesin accumulation lead to cell-to-cell variability in the extent of sister chromatid separation at metaphase.
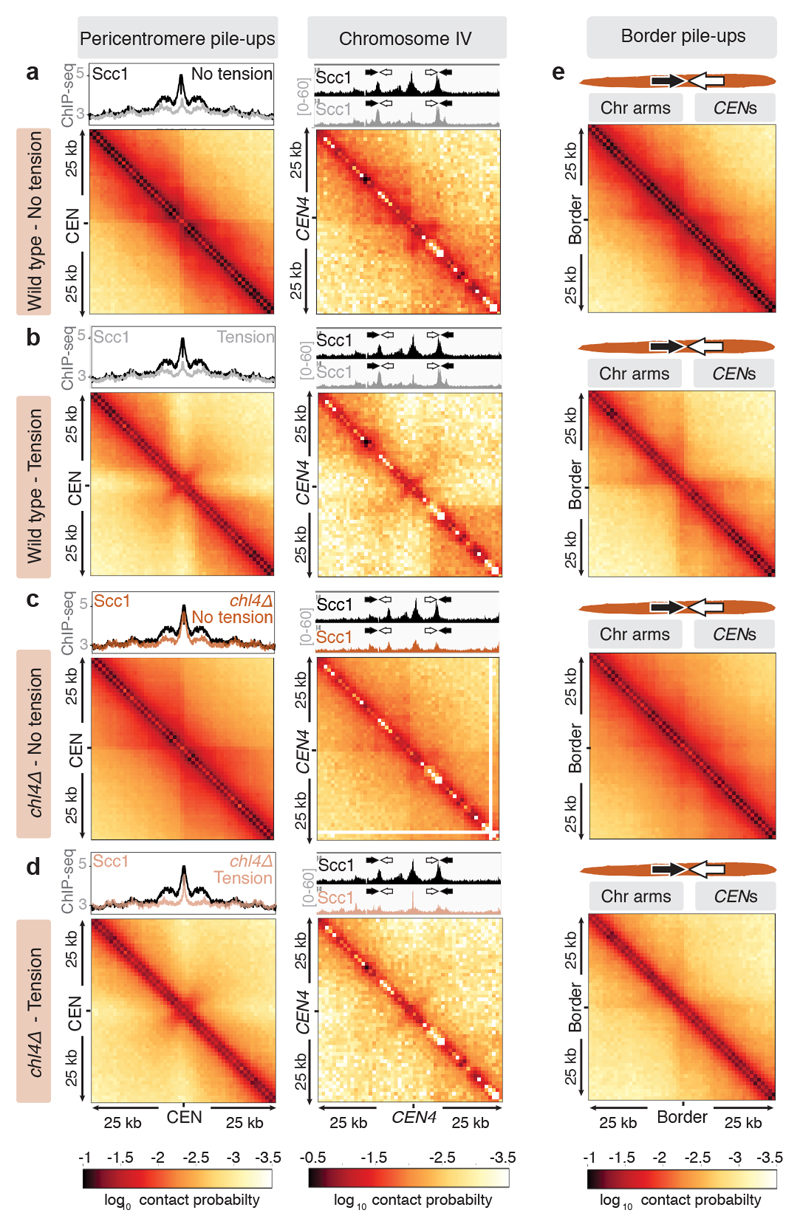
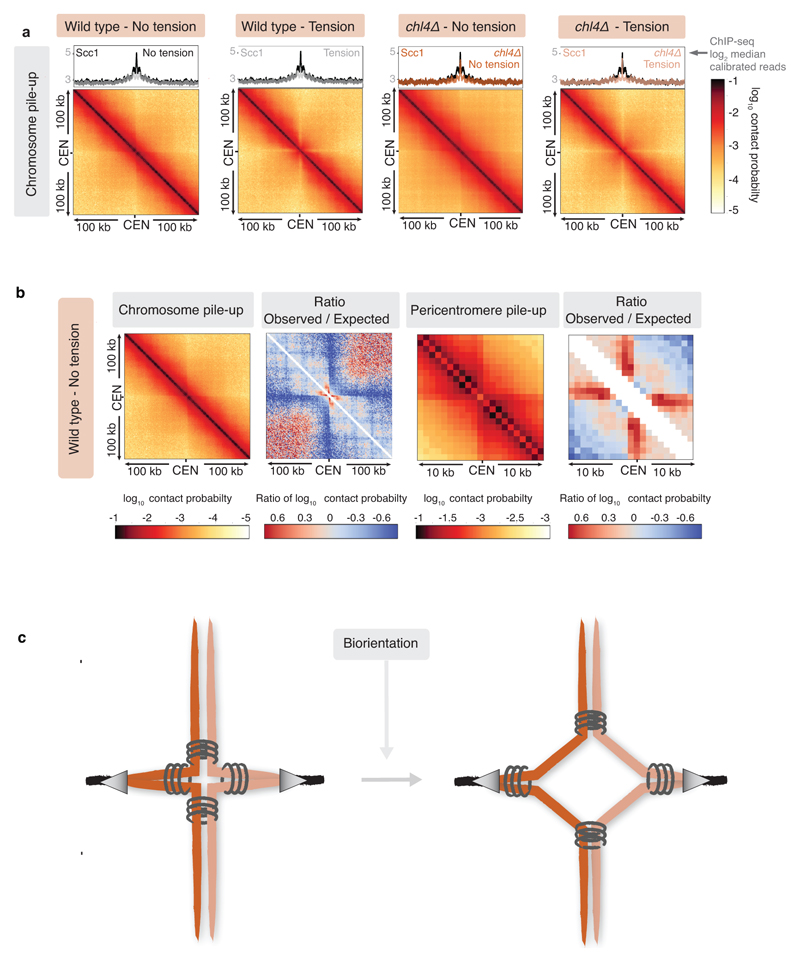
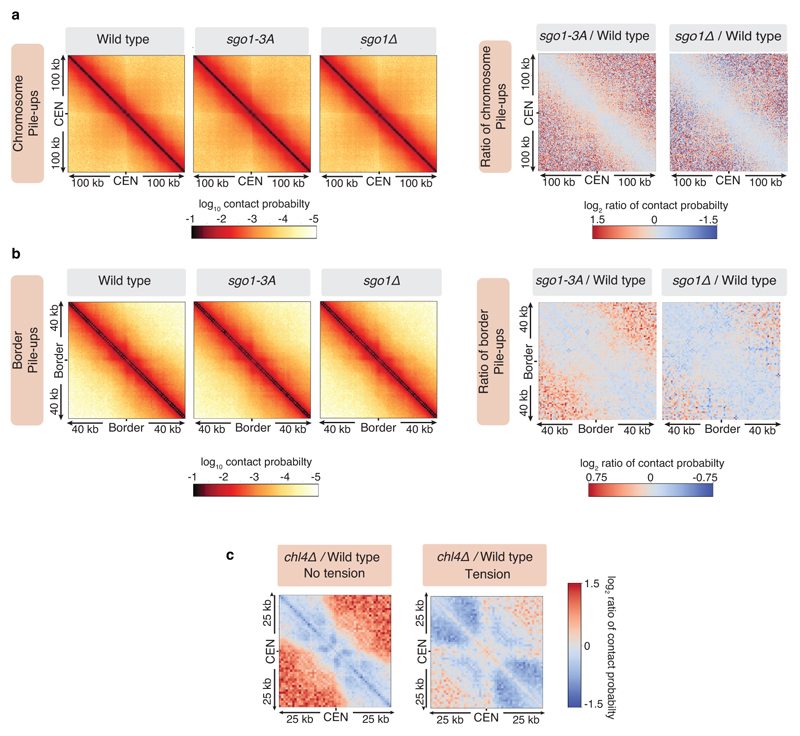
Our data suggest that cohesin accumulation at borders defines the domain of chromosomal separation under tension, which we hypothesise defines the structure of the pericentromere. A previous 3C study observed contacts between the left and right flanks of pericentromere III and it was suggested to form an intra-molecular loop, extending between 11.5kb and 25 kb19. Although this predicted pericentromere size is consistent with our mapping and functional analysis (Fig. 1a, d, Extended Data Fig. 5), the role of borders remains unclear. To determine pericentromere structure globally and the effect of spindle tension, we performed high resolution Hi-C analysis on metaphase-arrested cells both in the absence (no tension) or presence (tension) of microtubules to capture unbiased genome-wide interactions. In the absence of tension, consistent with cis-looping in mitosis20,21, centromere-centered pile-up contact maps of all chromosomes showed a high frequency of cis contacts along chromosome arm regions with core centromeres acting as strong insulators (Fig. 2a, left panel, Extended Data Fig. 6a). The lower than expected frequency of contacts on the diagonal, between the left and right side of the centromere argues against the presence of the previously proposed single intramolecular loop across both sides of the pericentromere19 (Extended Data Fig. 6b, c). Instead, examination of individual pericentromeres or pile-ups revealed that each side of the core centromere made frequent contacts with the pericentromere on the same side, extending as far as the border, 5-10kb away (Fig. 2a, right panel, Extended Data Fig. 7). Both cohesin and condensin can extrude DNA loops in vitro22–24 and the characteristic Hi-C stripe protruding from the core-centromere is suggestive of extrusion of a chromatin loop by a centromere-anchored factor25,26 (Fig. 2a, right panel, Extended Data Fig. 7). There is also evidence of longer (20-30kb) cis looping emanating from directly adjacent to the core centromere into either chromosome arm (Extended Data Fig. 6a). This is consistent with the notion that the usage of convergent gene pairs as boundaries is somewhat stochastic (Fig. 1d). Interestingly, the strongest Hi-C signal occurs where there is the greatest cohesin density at pericentromere borders (Fig 2a, Scc1 traces). In contrast, pericentromeric condensin does not appear to be critical for pericentromere structure in the absence of tension. Hi-C maps of sgo1Δ which reduces pericentromeric condensin or sgo1-3A, which although failing to bind PP2A, recruits condensin normally14,15, showed pericentromeric structures that were virtually indistinguishable from wild type (Extended Data Fig. 8a, b).
Fig. 2. The pericentromere is a multi-looped structure in mitosis which extends to an open V shape under tension.
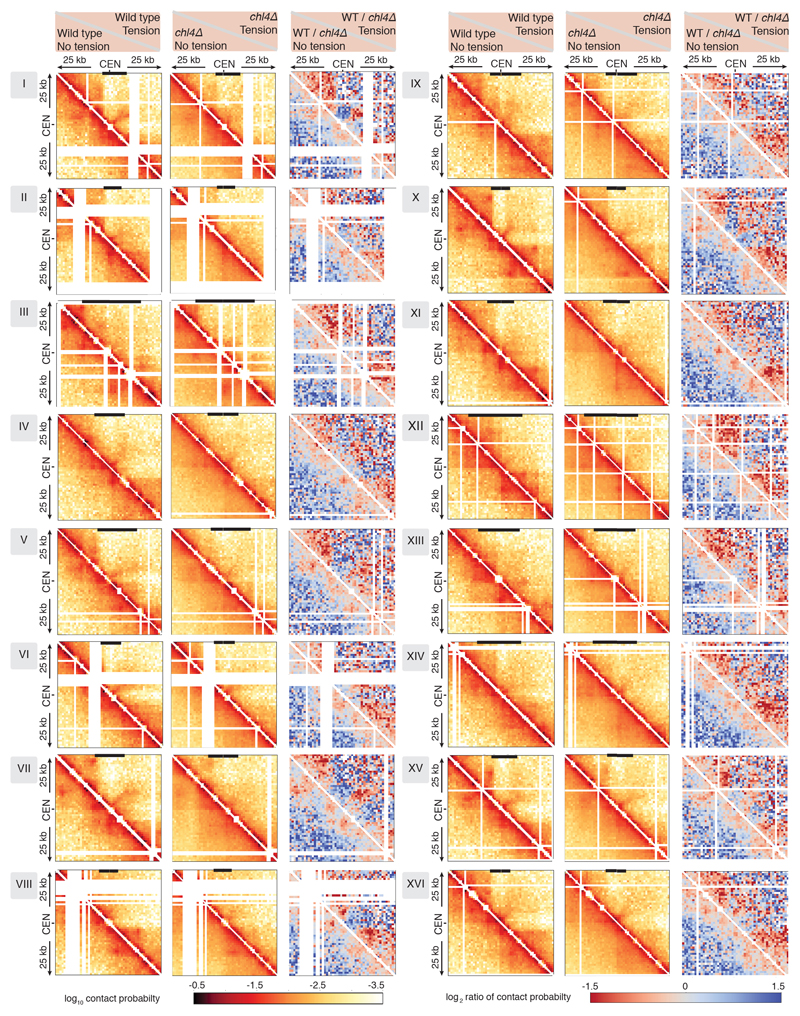
Hi-C of wild type and chl4Δ arrested in metaphase. a, b, c, d, Pile-ups (bin size 1kb) of cis contacts 25 kb surrounding all 16 centromeres (left panels), and contact maps for pericentromere IV (right panels) are shown for wild type and chl4Δ in the absence (a, c) or presence (b, d) of tension. Median calibrated Scc1 ChIP-Seq signal around all 16 centromeres, or for chromosome IV is shown above. Arrows denote border convergent genes. e, Pile-ups (1kb bins) of cis contacts (25 kb) surrounding the 32 pericentromere borders are shown.
The presence of spindle tension changed the conformation of pericentromeres radically, while chromosome arm conformation was unchanged (Fig. 2b, Extended Data Fig. 6a). Under tension, the centromeres were no longer the point of chromosome arm insulation and instead border regions formed chromosomal arm loop boundaries ~ 5-10kb from the core centromeres (Fig. 2b). Inside the borders, the frequency of contacts within, and reaching out of, pericentromeres, was substantially reduced with a new conformation definable. Contacts across individual centromeres describe an open loop or V-shaped structure with the core centromere at the apex and the borders at the tips (Fig. 2b, right panel; Extended Data Fig. 7). Therefore, borders mark the boundary between the pericentromere open loop and the cis-loop chromosome arm conformation.
To determine whether cohesin confers boundary function at borders we analysed chl4Δ cells (Fig. 2c, d), in which cohesin enrichment at pericentromere borders is reduced (Fig. 1b, c). Hi-C maps of chl4Δ in the presence of tension revealed a reduction in both boundary function at borders and the strength of centromere-proximal loops, as evidenced by less distinct lines and spots on individual chromosomes (Fig. 2d; Extended Data Fig. 6a; Extended Data Fig. 7, particularly evident on chromosomes VII, X, XV), consistent with the increased inter-sister centromere distance at metaphase in chl4Δ5. In the absence of tension in chl4Δ, centromere proximal loop structures were less defined, indicating that kinetochore-driven cohesin loading promotes cis-looping from centromeres in addition to establishing boundary function at borders (Fig. 2c; Extended Data Fig. 6a; Extended Data Fig. 7). The maintenance of residual loop structures in chl4Δ lacking tension is consistent with the presence of residual, non-cohesive, cohesin at centromeres of these cells4,5,11. Centering the pile-ups on the borders themselves revealed strong isolation of domains proximal and distal to the centromere, which sharpens under tension but is less distinct in chl4Δ (Fig. 2e), confirming their boundary function and dependence on CHL4. Vertical and horizontal lines 25,26 are also suggestive of loop extrusion emanating from borders.
Which property of borders enables the structural organization of the pericentromere? Since cohesin localization is altered by transcription11,27,28, we hypothesized that convergent transcription of border gene pairs leads to cohesin retention which results in robust inter-sister chromatid linkages that isolate domains and resist spindle forces. Indeed, convergent genes at borders show a narrower RNA-Seq density distribution compared to all genes, suggesting moderate expression on average (Extended Data Fig. 9a). Analysis of transcriptome-wide RNA polymerase II binding site data29 revealed that active transcription at convergent gene pairs is typically higher towards, rather than away from, the centromere (Extended Data Fig. 9b). Conversely, convergent genes within the pericentromere show higher expression away from, rather than towards the centromere (Extended Data Fig. 9c). Consistent with transcription-dependent cohesin positioning11,27,28, insertion of a URA3 cassette between convergent genes at the left border on chromosome IV led to re-distribution of cohesin in the direction of transcription (Extended Data Fig. 9d, e).
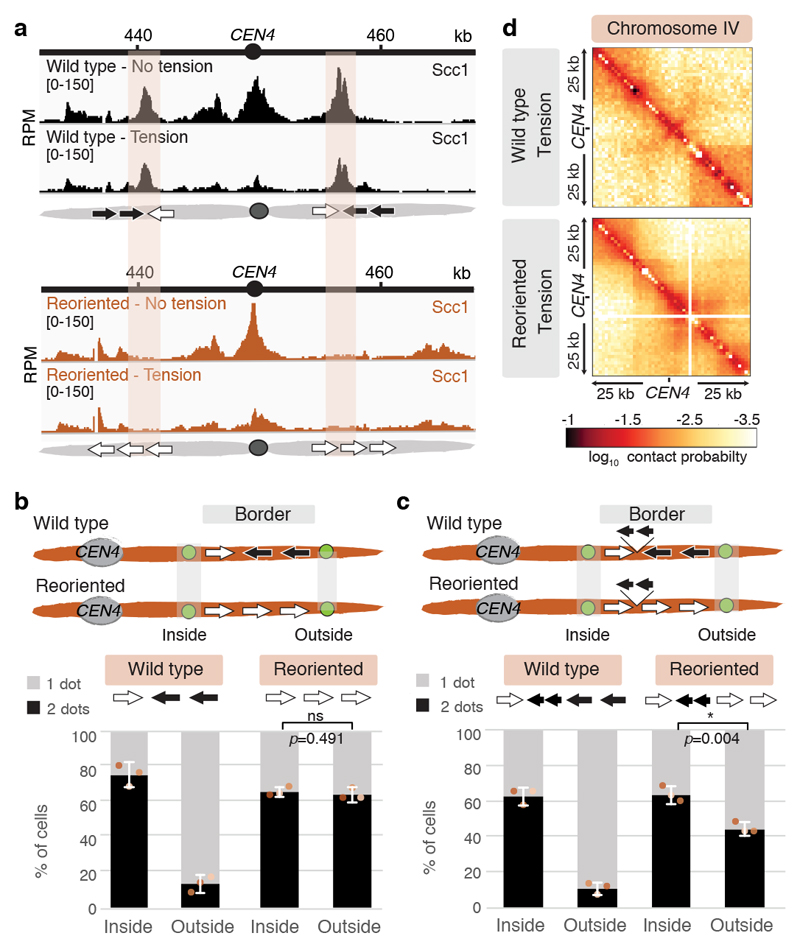
If convergent genes at borders define pericentromere boundaries, re-orienting gene pairs into a tandem arrangement might affect pericentromere behaviour. We engineered such a “reoriented” strain where tandem border gene pairs on both sides of CEN4 transcribe away from the centromere. The reoriented chromosome IV lost cohesin peaks at pericentromere borders, while additional cohesin peaks emerged further away from the centromere, potentially forming new borders (Fig. 3a). Both Sgo1 and condensin (Brn1) associate with the “new” borders only on reoriented chromosome IV (Extended Data Fig. 10a).
Fig. 3. Gene orientation determines pericentromere size.
Border convergent genes on chromosome IV were reversed to tandem orientation (“reoriented”). a, Representative cohesin enrichment in wild type and reoriented strains (n=2). Shading indicates wild type pericentromere border position. Black and white arrows in schematics indicate genes transcribed towards and away from the centromere, respectively. b, Separation of tetO/TetR-GFP markers at the indicated positions in metaphase. c, Separation of tetO/TetR-GFP markers after insertion of two short model genes downstream of the first border genes on both sides of pericentromere IV, in wild type and reoriented strains. In b and c, for each biological replicate (data points, n=3), 200 cells were scored. Bars show mean ±s.e.m.; unpaired two-tailed t-test, * p<0.05, ns p>0.05. d, Hi-C maps of pericentromere IV (n=1).
Since orienting the original border genes in tandem causes other, centromere-distal regions, to act as borders, pericentromere size is expected to increase resulting in an expansion of the region of sister chromatid separation at metaphase. Consistently, a tetO-GFP marker outside the original border, which infrequently separated in wild type metaphase cells, was separated in the reoriented strain to a similar extent to a marker inside the original border (Fig. 3b, Extended Data Fig. 10b). However, a marker outside the new border, ~23kb away from the centromere was infrequently separated in both wild type and reoriented strains (Extended Data Fig. 10c). Insertion of a pair of tandemly arranged model genes oriented towards the centromere to restore the convergent gene arrangement at the borders partially rescued separation of the “outside” marker (Fig. 3c). Therefore, convergent genes set the boundaries at pericentromere borders. Hi-C of the reoriented strain in metaphase in the presence of tension revealed a striking change in structure, specifically of pericentromere IV (Fig. 3d, Extended Data Fig. 10d, e). Boundaries at the original border positions were lost, while centromere-distal regions displayed increased contact frequency, consistent with an expansion of the pericentromeric domain. The typical V-shape of endogenous pericentromeres was less apparent: frequent asymmetric contacts were observed within the pericentromere and the centromere lost its strong insulating effect (Fig. 3d; Extended Data Figure 10e). Therefore, border gene reorientation results in a more open, structurally disorganised, pericentromeric structure (Extended Data, Fig. 10f).
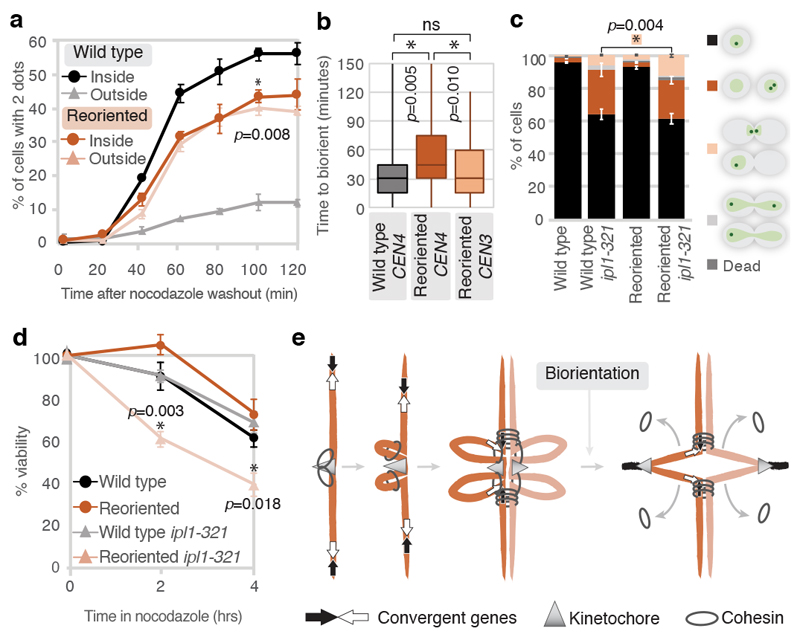
To determine the functional importance of pericentromere boundaries we assayed sister chromatid biorientation upon metaphase spindle re-formation after washing out microtubule-depolymerising drugs (Extended Data Fig. 10g). Compared to wild type chromosome IV, reoriented chromosome IV showed a delay in, and reduced frequency of, sister kinetochore biorientation (Fig. 4a), consistent with collapse of the V-shape structure indicated by Hi-C. Live cell imaging confirmed that the biorientation delay was specific to the reoriented chromosome IV, since chromosome III (CEN3-GFP) in this strain bioriented with similar efficiency to CEN4-GFP in wild type cells (Fig. 4b). Inefficient sister kinetochore biorientation of the reoriented chromosome might lead to greater reliance on the Aurora B (Ipl1)-dependent error correction process30,31. In the temperature-sensitive ipl1-321 background under semi-permissive conditions, after a single cell cycle, the reoriented chromosome IV strain showed a modest decrease in G1 cells that had inherited a single CEN4-GFP focus and an accumulation of cells in mitosis (Fig. 4c). Strikingly, reoriented chromosome IV ipl1-321 cells, grown at the permissive temperature, showed a pronounced loss of viability after microtubule depolymerization (Fig. 4d). Therefore, convergent genes at borders structure pericentromeres to enable efficient sister kinetochore biorientation and proficient error correction which is critical for cellular fitness.
Fig. 4. Gene orientation affects biorientation efficiency and cell viability.
a, Sister kinetochore biorientation following spindle reassembly. Percentage of cells with separated GFP foci (n=200 at each timepoint), mean of 3 biological replicates ±s.e.m. is shown; unpaired two-tailed t-test, * p<0.05. b, Biorientation time (from SPB separation until centromere-linked GFP dot splitting) in live cells. Centre line, median; box limits, second and third quartile; whiskers, first and fourth quartiles for 120 cells split equally across two biological replicates; two-sided Mann-Whitney test, *, p<0.05. c, Categorization of 500 cells based on morphology and GFP foci number following partial ipl1-321 inactivation. Data show mean of 3 biological replicates ±s.e.m.; unpaired two-tailed t-test, * p<0.05. d, Cell viability following nocodazole treatment after plating 1,000 cells at each timepoint. Data are mean of 3 biological replicates ±s.e.m.; unpaired two-tailed t-test, *, p<0.05 (p-values refer to wild type ipl1-321 vs. reoriented ipl1-321). e, Model of pericentromere structure. Cohesin loaded at kinetochores extrudes a chromatin loop on either side of the centromere until halted by convergent genes at pericentromere borders. When biorientation extends pericentromeric chromatin outwards, intra-sister chromatid cohesin at the base of loops is passively removed from chromosomes, while inter-sister chromatid cohesin is trapped at borders, converting centromere-flanking cis-loops to a V-shaped structure.
We find that targeted cohesin loading at centromeres, and trapping between convergent genes at borders, fold the pericentromere into a multi-looped structure (Fig. 4e). This conformation is likely the product of loop extrusion anchored on each side of the centromere, with borders acting to restrict loop size. Consequently, each centromere is isolated from its two flanking pericentromeric regions, providing structural integrity to support the establishment of sister kinetochore biorientation. The resultant pulling forces extend pericentromeric chromatin outwards until cohesin stalling by convergent transcription at borders prevents further unzipping of the sister chromatids. In the absence of either convergent genes (reoriented strain) or efficient cohesin loading at centromeres (chl4Δ), borders are unable to provide the robust cohesion to resist pulling forces at metaphase and further unzipping occurs.
The suggestion that cohesin makes intra-sister chromatid linkages between two sides of the pericentromere19 (Extended Data Fig. 6c) is difficult to reconcile with the strong isolation of these regions in the absence of tension (Fig. 2a), or the observation that cohesin is passively removed within the pericentromere when tension is applied (Fig. 1a). Instead, we favour the idea that while some pericentromeric cohesin entraps sister chromatids to provide cohesion, other cohesin molecules make intra-sister chromatid interactions on either side of the centromere to extrude chromatid loops. While spindle forces will pull chromatin through the inter-sister-chromatid-entrapping cohesin until they are halted by the transcriptional machinery at borders, intra-sister chromatid loop-extruding cohesin will be evicted from the chromosomes, consistent with passive removal (Fig. 4e).
We show that targeted cohesin loading collaborates with the linear organisation of genes to fold a chromosomal domain into a structure competent for chromosome segregation. Non-coding transcription and enrichment of cohesin are features of centromeric regions in many organisms, suggesting general principles may underlie their structure32. Potentially, the linear order of transcriptional units throughout a genome has evolved in such a way to broadly influence its function by locally controlling its architecture.
Methods
Yeast strains and plasmids
All yeast strains were derivatives of w303 and are listed in Supplemental Table 1. Plasmids generated in this study are listed in Supplemental Table 2. For calibrated ChIP-Seq we used Schizosaccharomyces pombe strain spAM635 (h- rad21-6HA::KanMX6) was used. The yeast strain carrying chromosome III with an ectopic centromere was described previously3. To visualize chromosomal loci, tetOs were integrated at defined sites on chromosome I, III and IV after cloning of the appropriate region into pRS306(tetOx224) (Supplemental Table 2). URA3 was inserted between convergent gene pairs by a PCR-directed approach. To reorient potential border genes on chromosome IV, the gene cassette including its promoter were cloned into a plasmid (Supplemental Table 2), upstream of KanMX, flanked by LoxP sites. Plasmids were used as template for PCR, which was used for transformation, to insert the gene and its promoter in the opposite orientation, together with LoxP-KanMX6-LoxP. Insertion in the desired orientation was confirmed by PCR. The marker was then excised by Cre-mediated recombination. Plasmids for rescue constructs were assembled using 5-fragment Gibson assembly, and the resulting pURA3::ABIx2-V5::TRP1 and pURA3::PYLx2-FLAG::HISMX6 were inserted at chromosome IV pericentromere borders by a PCR-directed approach.
Growth conditions
Cells carrying pMET-CDC20 were arrested in metaphase in the presence and absence of tension as described by8. Briefly, cultures were arrested in G1 in synthetic medium lacking methionine (SC/-Met/D) with alpha factor (5 μg/ml) for 1.5 h, before re-adding alpha factor to 2.5 μg/ml and shaking for a further 1.5 h. Cells were washed with rich medium lacking glucose (YEP) and released into rich medium containing 8 μM methionine (YPDA/Met). Methionine was re-added at 4 μM every hour. To achieve a metaphase arrest in the absence of microtubules (no tension), cells were released from G1 into YPDA/Met medium containing 15 μg/ml nocodazole and 30 μg/ml benomyl. Nocadazole was re-added at 7.5 μg/ml every hour. For both tension and no tension (nocodazole) conditions, cells were harvested 2 h after release from G1. For biorientation assays, cells were arrested in the absence of tension as above, after 2 h nocodazole was washed out by filtering and washed with rich medium lacking glucose, before cultures were released into YPDA +Met to allow spindles to reform while maintaining the metaphase arrest. Samples were taken at 20 min intervals and scored blind. To arrest cells lacking the pMET-CDC20 construct in metaphase in the absence of spindle tension, cycling cells (OD600=0.2) were treated with 15 μg/ml nocodazole and 30 μg/ml benomyl; every hour, 7.5 μg/ml nocodazole was added and cells were harvested after a total of 3 h. For partial ipl1-321 inactivation, cultures were arrested in G1 at room temperature with alpha factor (5 μg/ml) for 1.5 h, before re-adding alpha factor to 2.5 μg/ml and shaking for a further 1.5 h. Cells were washed with rich medium lacking glucose (YEP) and released into rich medium (YPDA) pre-warmed to 32°C. Alpha-factor was re-added to block cells in the next G1 when small buds started to appear. For viability assays, cultures in exponential phase were diluted to OD600=0.2 and for each condition approximately 1000 cells were plated over 6 YPDA plates before (0 h) and 2 or 4 hours after the addition of 15 μg/ml nocodazole. Nocodazole was re-added at 7.5 μg/ml every 90 minutes. Cells on plates were grown at 25°C for 2 days, then the number of colonies were scored. Viability was calculated as a percentage of colony count at 0 h.
Chromatin immunoprecipitation, ChIP-Seq and data analysis
ChIP-qPCR and ChIP-Seq was carried out as described previously14 except that cells were fixed for 30 minutes, and that DNA from purified chromatin was recovered using a PCR purification kit (Promega). Sequencing libraries were generated using standard methods and samples were sequenced on a MiniSeq instrument (Illumina) with the exception of data shown in Extended Data Fig. 3, 4 where libraries were prepared and sequenced by the EMBL Genomics Core Facility. ChIP-Seq data used to generate Extended Data Fig. 4c, d was published previously14. Scripts, data files, and workflows used to analyse the data and prepare the ChIP-Seq figures can be found on the github repository at https://github.com/AlastairKerr. For the strains where the centromere was repositioned, or URA3 cassette was inserted at borders, or where gene orientation at pericentromere borders is reversed the corresponding reference genome sequence was assembled in silico and the appropriate reference was used to map sequencing reads for each strain. Plots showing averages of all centromeres were generated using Seqplots33. Read counts were normalized to reads per million mapped (RPM) and the ratio of ChIP reads to input was calculated. The mean or the median value was determined for all 16 chromosomes per 50bp window and its log2 value is graphed. Mean values are shown for the +/- 3kb plots; the +/- 25kb plots use median values. Reference genome coordinates used to centre Seqplots are give in Supplemental Table 3. To allow quantitative comparison between different conditions all ChIP-Seq, with the exception of the data shown in Extended Data Fig. 3 and Extended Data Fig. 4c-f, was calibrated with an internal reference by modifying the procedure described by13. Rather than Candida glabrata, S. pombe carrying Rad21-6HA was used as the calibration genome. Briefly, for each IP, 100 ml of S. pombe cells were grown in YES to OD595=0.25-0.3 and fixed by addition of 1/10 volume of 11% formaldehyde in diluent (0.143 M NaCl, 1.43 mM EDTA, 71.43 mM HEPES-KOH) with gentle agitation for 30 minutes. Cell pellets were washed twice with 10 ml cold TBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) and once with 10 ml cold FA lysis buffer (100 mM Hepes-KOH, pH 7.5, 300 mM NaCl, 2 mM EDTA, 2% Triton X-100, 0.2% Na Deoxycholate)/0.1% SDS, frozen in liquid nitrogen and stored at -80°C. S. pombe cell pellets were resuspended in 400 μL of cold 1x FA lysis buffer/0.5 % SDS containing 1x complete protease inhibitor cocktail (Roche) and 1 mM PMSF and mixed with thawed S. cerevisiae pellet (approximately 100 ml cells OD600=0.4). ChIP and sequencing was performed as described above. Calculation of Occupancy Ratio (OR) and data analysis was performed as described in 13. The number of reads at each position were normalized to the total number of reads for each sample (RPM: Reads Per Million), multiplied by the occupancy ratio (OR) and shown in the Integrated Genome Viewer from the Broad Institute. For ChIP-Seq, data from a representative experiment is show with n= referring to the number of biological replicates performed overall. Data was comparable in all cases. Cohesin ChIP-Seq in wild type and reoriented strains shown in Fig. 3a has been performed in 3 biological replicates, using different epitopes for immunoprecipitation and different strain genotypes. Primers used for qPCR analysis are given in Supplemental Table 4.
Microscopy
Cells were fixed in formaldehyde for visualization of TetR-GFP and Spc42-tdTomato foci. Yeast were mounted onto a glass slide and imaged on a Zeiss Axio Imager Z1 equipped with a x100 α Plan Fluar/1.45 NA (oil) objective lens. Images were recorded using a Photometrics Evolve EMCCD camera (Photometrics, Tucson, USA) controlled using MicroManager 1.4 aquisition software (US National Institutes of Health). The fluorescent intensity and distance between the GFP foci were measured using a custom ImageJ plugin that can be found on the github repository https://github.com/dkelly604/CellClicker_. Live-cell imaging was performed on a Zeiss Axio Observer Z1 (Zeiss UK, Cambridge) equipped with a Hamamatsu Flash 4 sCMOS camera, Prior motorised stage and Zen 2.3 acquisition software. Cells were imaged at 25°C using CellASIC ONIX microfluidics plates, with images taken at 15min intervals.
RNA isolation and RNA-Seq
Cell pellets were lysed by bead-beating in RLT buffer (Qiagen) and RNA was isolated using the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions except that on-column DNA digestion was performed using the Qiagen DNase digestion kit. RNA concentration was determined by nanodrop. For cDNA synthesis for qRT-PCR, 12ng purified total DNA, diluted in HyClone dH2O and 10 mM Oligo(dT)15 primer (Roche) or 1.5 mM gene-specific reverse primer were incubated at 65°C for 10 min before placing on ice to denature RNA. Subsequently, 4 μl 5xTranscriptor RT reaction buffer (Roche), 0.5 μl RNase OUT (Fisher), 1 μM dNTPs and 0.5 μl Transcriptor reverse transcriptase plus Hyclone dH2O were added to 20 μl and incubated at 55 °C for 3 h before heat inactivation of Transcriptor Reverse Transcriptase at 85 °C for 5 min. RNA was depleted of rRNA and libraries prepared for sequencing by Genecore, EMBL. Sequencing was also performed by Genecore on an Illumina Next Seq 500 with a read l length of 75 and multiplexed with a pool size of 4.
Hi-C library preparation and data analysis
Hi-C protocol was modified from20,34,35. Cells were cultured, fixed and lysed as described in34. Briefly, 200 ml of cells at OD~0.6 carrying pMET-CDC20 were arrested in metaphase at 25 °C in the presence and absence of tension as described above, fixed with 3% formaldehyde for 20 minutes at 25 °C at 250 rpm, and the reaction was quenched for 5 minutes by the addition of 0.35 M glycine (final concentration). Cells were washed with cold water, resuspended in 5 ml 1x NEBuffer 2 and frozen in liquid nitrogen. Lysates were prepared by grinding the frozen pellet in a chilled mortar with a pestle for 15 minutes and 1/10th of the initial pellet weight (~0.5 g) was taken for further processing. Restriction enzyme digestion (DpnII), filling-in, ligation, crosslink reversal, DNA concentration and purification and biotin removal were carried out as described in35. DNA was then fragmented on a Bioruptor Plus sonication device (Diagenode) for a total of 2x 30 cycles 30 seconds on/off at High setting. Following DNA end repair and A-tailing using T4 DNA polymerase, T4 Polynucleotide Kinase and Klenow fragment DNA polymerase I (as in35), Hi-C libraries were fractionated using Ampure XP beads as previously described in 34. Biotin pull-down, adapter ligation (NextFlex, Bioo Scientific) and sequencing (EMBL Core Genomics Facility, Heidelberg, Germany) were carried out as in20. Hi-C read numbers are given in Supplemental Table 5.
For Hi-C data analysis, Fastq reads were aligned to sacCer3 reference genome using HiC-Pro v2.11.136 bowtie2 v2.3.4.1 (--very-sensitive -L 30 --score-min L,-0.6,-0.2 --end-to-end --reorder), removing singleton, multi hit and duplicated reads. Read pairs were assigned to restriction fragment (DpnII) and invalid pairs filtered out. Valid interaction pairs were converted into the .cool contact matrix format using the cooler library, and matrixes balanced using Iterative correction down to one kilobase resolution. Multi-resolution cool files were uploaded onto a local HiGlass37 server for visualisation, cooler show was also used to generate individual plots for each chromosome. White stripes on plots represent regions where data was lost stochastically during mapping due to stringency settings filtering out reads. In the case of the reoriented chromosome, the presence of 2-3 small LoxP “scars” are likely to impact mapping in these regions and may account for the observed data loss. To generate pileups at centromeres/pericentromeric borders, the cooltools library was used, cool matrixes were binned at one kilobase resolutions. Plots were created around the midpoint of centromeres with ten/twenty-five/one-hundred kilobase flanks on each side, or around the midpoint of borders with forty kilobase flanks, showing the log10 mean interaction frequency using a colour map similar to HiGlass ‘fall’. All centromere/pericentromere annotations were duplicated in both the forward/reverse strand orientations to create an image which is mirror symmetrical. The ratio pile ups between samples were created in a similar fashion plotting the log2 difference between samples in the ‘coolwarm’ colour map, i.e. A/B; red signifying increased contacts in A relative to B and blue decreased contacts in B relative to A. Scripts are available at (https://github.com/danrobertson87/Paldi_2019).
Extended Data
Extended Data Fig. 1. Tension-dependent cohesin removal at metaphase is restricted to the pericentromere and occurs independently of Wpl1/Rad61.
a, Scc1-6HA calibrated ChIP-Seq profiles for the pericentromeric region of chromosome IV are shown for rad61Δ cells arrested in metaphase, in the absence and presence of spindle tension (n=1). b, Mean calibrated ChIP reads (solid line), standard error (dark shading) and 95% confidence interval (light shading) at a pericentromere-proximal cohesin site on each chromosome arm (n=32 sites) for wild type and chl4Δ, either in the presence or absence of tension.
Extended Data Fig. 2. Overview of border gene organization and pericentromere size.
a, Schematic shows the positions of convergent gene pairs flanking centromeres. Grey ovals represent the centromere, convergent gene pairs at the borders are indicated by arrows. b, Genomic distance between the 3’ ends of convergent genes at borders, as well as 3’ and 5’ ends of the two genes transcribed towards centromeres, indicated by grey lines between arrows that denote genes (n=49). Centre line, median; box limits, second and third quartile; whiskers, first and fourth quartile. c, Table of convergent genes identified at pericentromere borders for each chromosome, along with the corresponding pericentromere size. Borders were defined as the innermost cohesin peak near the centromere that persisted in the presence of tension. d, Pericentromere size determined in c plotted against chromosome size. e, Percentage of genes essential for growth on rich glucose media among genes at borders and among all genes. f, Scc1 ChIP-Seq in asynchronous Candida glabrata cells, showing chromosome F from13.
Extended Data Fig. 3. An ectopic centromere establishes new borders at convergent genes on a chromosome arm.
Cohesin (Scc1) ChIP-Seq profiles for the region surrounding the endogenous centromere on chromosome III (left panel) and for a ~50 kb region of chromosome III surrounding the neo-centromeric arm site (right panel) are shown (n=1). Regions of tension-insensitive cohesin peaks at convergent sites flanking the endogenous and ectopic centromeres are highlighted.
Extended Data Fig. 4. Shugoshin and condensin localise to pericentromere borders.
a, Representative cohesin (Scc1), shugoshin (Sgo1) and condensin (Brn1) enrichment in metaphase-arrested cells in the presence of nocodazole in the pericentromeric region of chromosome IV (n=2, immunoprecipitation was performed using proteins tagged with different epitopes in the two biological replicates). b, Plots show median calibrated ChIP reads (solid line), standard error (dark shading) and 95% confidence interval (light shading) at all 32 borders and 16 centromeres. For comparison, similar plots for the next convergent gene site on each chromosome arm are shown. c-d, Condensin associates with pericentromere borders in a Sgo1-dependent manner in cells arrested in metaphase in the absence of tension. c, ChIP-Seq data used was previously published in16. Median condensin (Brn1) signal across a 50kb region surrounding all 16 centromeres. d, Mean Brn1 signal centred around all 32 borders (left panel) or 16 centromeres (right panel). Sgo1 is removed from the borders, but not core centromeres, in response to spindle tension (solid line – median, dark shading – standard error, light shading – 95% confidence interval). e, Median Sgo1 enrichment by ChIP-Seq plotted over a 50kb region surrounding all 16 centromeres in metaphase-arrested cells in the presence or absence of tension. f, Mean Sgo1 signal centred around all 32 borders (left panel) or 16 centromeres (right panel) (solid line – median, dark shading – standard error, light shading – 95% confidence interval).
Extended Data Fig. 5. Pericentromere borders resist sister chromatid separation under tension.
a, Assay to measure separation of loci on sister chromatids in metaphase arrested cells. Cells carry tetO/TetR-GFP foci integrated at various positions, Spc42-tdTomato foci to mark spindle pole bodies and are arrested in metaphase by Cdc20 depletion. Left schematic shows expected separation of GFP foci positioned inside and outside pericentromere loci. Green dots, tetO/TetR-GFP foci, Red dots, spindle pole bodies. Representative image is shown to the right (n=3 biological replicates). White and black arrows mark cells with a single GFP focus or split foci, respectively. b, Position of GFP foci and corresponding calibrated Scc1-6HA ChIP-Seq profiles (n=3) are shown for chromosome I and III. c, Cells carrying tetO/TetR-GFP foci integrated at various positions were arrested in metaphase and distance between GFP dots were measured in 100 cells. Horizontal lines indicate mean.
Extended Data Fig. 6. C-looping and alternative model for pericentromeric chromosome conformation adapted from21.
a, Pile-ups (bin size 1kb) of cis contacts 100 kb surrounding all 16 centromeres for wild type and chl4Δ cells in the absence or presence of spindle tension. b, Pile-ups of cis contacts 100kb surrounding centromeres (left panel), ratio of expected/observed signal (second panel), pericentromere pile-up (third panel, 10kb surrounding centromeres) and its ratio of expected/observed signal (right panel) is shown for wild type cells in the absence of spindle tension (n=16). c, Model for centromere-flanking pericentromeric chromatin forming an intramolecular loop in which cohesin bridges the two sides of the pericentromere flanking the centromere, whereas chromosome arms are paired intermolecularly between sister chromatids, resulting in a cruciform chromosome conformation.
Extended Data Fig. 7. Changes in pericentromere structure on individual chromosomes in response to tension and in the absence of pericentromeric cohesin.
Hi-C contact maps (1kb bin) over a 50kb region surrounding all centromeres in wild type (left panels) cells without tension (bottom half of heatmap) and with tension (top half of heatmap), and in chl4Δ (middle panels) without tension (bottom half) and with tension (top half) (n=1). Right panels show log2 ratio between wild type and chl4Δ, without tension (bottom half) and with tension (top half). The extent of the pericentromere for each chromosome is marked by black bars on top of the contact maps.
Extended Data Fig. 8. Pericentromere structure depends on pericentromeric cohesin rather than condensin.
Hi-C analysis of sgo1-3A and sgo1Δ in metaphase-arrested cells in the absence of tension reveals similar patterns to wild type. Data for wild type was reproduced from Fig. 3a for comparison. a, Pile-ups (bin size 1kb) of cis contacts surrounding all 16 centromeres in absence of spindle tension for the indicated strains (left three panels) or log2 ratio maps between wild type and sgo1-3A or sgo1Δ (right two panels) detect little change. b, Pile-ups (bin size 1kb) and log2 ratio maps of cis contacts surrounding all 32 borders. c, Log2 ratio between 25 kb pile-ups centered on the centromere in wild type and chl4Δ cells, in the absence (left) and presence (right) of tension (n=16).
Extended Data Fig. 9. Transcription at pericentromere borders influences cohesin position.
a, Genes at pericentromere borders are moderately transcribed on average. Relative RNA density for convergent border gene pairs compared to all genes is shown for no tension and tension conditions (n=1). RNA-Seq for wild type cells arrested in metaphase in the presence or absence of tension. Dashed lines indicate mean, dotted line mark 95% confidence interval. b, Boxplot of transcription levels of genes at pericentromere borders based on RNA polymerase II (Rpo21) Cross-linking and analysis of cDNA (CRAC) from29. Rpo21 CRAC sense read counts of genes at borders were normalized to the protein coding gene average and genes at pericentromere borders were grouped by their relative orientation to centromeres. Data points correspond to the mean of three biological repeats. Centre line, median; box limits, second and third quartile; whiskers, first and fourth quartile (non-normal distribution, Shapiro-Wilk; *, p<0.05, two-sided Mann-Whitney test). c, Boxplot of relative transcription levels of genes transcribed towards and away from centromeres, at pericentromere borders and at non-border convergent genes inside pericentromeres as in b. d, e, Insertion of a URA3 cassette between a convergent gene pair shifts the localization of cohesin in the direction of transcription. URA3 was integrated in either orientation between the convergent gene pairs at the left pericentromere border on chromosome IV and cohesin (Scc1) ChIP-qPCR (n=3, bars show mean ±s.e.m.) using primers at the indicated positions (d) and ChIP-Seq (n=1) (e) was performed.
Extended Data Fig. 10. Gene reorientation at borders impacts pericentromeric protein localization, sister chromatid separation and chromosome conformation.
a, Cohesin (Scc1), shugoshin (Sgo1) and condensin (Brn1) enrichment in metaphase-arrested cells in the presence of nocodazole in the pericentromeric region of wild type and reoriented chromosome IV is shown (n=2, immunoprecipitation was performed using proteins tagged with different epitopes in the two replicates). Asterisks indicate new peaks in reoriented strain. b, Distance between GFP foci does not change following gene reorientation. 100 cells were measured, horizontal lines indicate mean. c, The region of separation upon gene reorientation does not extend beyond the next convergent gene pair. Strains with tetO arrays integrated at the indicated positions were arrested in metaphase and the percentage of cells with 2 GFP foci was determined. For each biological replicate (data points, n=3), 200 cells were scored, data are mean ±s.e.m. d, Pile-up of cis contacts across all 16 pericentromeres in reoriented strain, in the presence of spindle tension. e, Log2 ratio map of Hi-C signal in pericentromere IV in wild type and reoriented strains (n=1). f, Model for pericentromere expansion and disorganisation in the absence of convergent genes. g, Schematic of sister kinetochore biorientation following spindle re-polymerisation. Cells carrying the indicated chromosomal GFP labels, Spc42-tdTomato and pMET-CDC20 were released from G1 into a metaphase arrest by depletion of Cdc20 in the presence of nocodazole. Nocadazole was washed out while maintaining metaphase arrest and the percentage of cells with 2 GFP foci was scored over time.
Supplementary Material
Acknowledgments
We are grateful to Bianka Baying and Vladimir Benes (Genecore, EMBL) for NGS and library preparation and to Weronika Borek for help with statistical analysis. We thank Robin Allshire, Stefan Bresson and David Tollervey for helpful discussions, Paul Megee for yeast strains and Weronika Borek, Stephen Hinshaw, Vasso Makrantoni and Pierre Romé for comments on the manuscript. This study was funded by a Wellcome Senior Research Fellowship [107827] (AM, BA, FP), a Wellcome PhD studentship [109091] (FP), core funding for the Wellcome Centre for Cell Biology [203149] (AM, FP, BA, DR, AK, DK), an ERC Consolidator Award [311336] (MJN, SS) and a Wellcome Trust Investigator Award [200843] (MJN, SS).
Footnotes
Author contributions
AM, FP and BA conceived the study. FP performed Hi-C, ChIP-Seq, microscopy and viability experiments. BA performed ChIP-Seq and ChIP-qPCR experiments. FP, BA and AM generated reagents and analysed data. DR, AK and SS performed bioinformatics analyses. SS developed protocols; DK wrote custom code for image analysis; FP, AM, SS, JB and MN interpreted Hi-C data. AM supervised the study. AM and FP wrote the paper, with input from all authors.
Competing interests The authors declare no competing interests.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
Sequencing datasets are available at GEO, accession number GSE104135. Other data generated is included in this published article as source data.
Code availability
ImageJ plugin to measure the fluorescent intensity and distance between the GFP foci can be found on the github repository https://github.com/dkelly604/CellClicker_. Scripts for Hi-C data analysis are available at https://github.com/danrobertson87/Paldi_2019.
References
- 1.Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nature reviews. 2018;19:789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–3827. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber SA, et al. The kinetochore is an enhancer of pericentric cohesin binding. PLoS biology. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernius J, et al. Cohesin-dependent association of scc2/4 with the centromere initiates pericentromeric cohesion establishment. Curr Biol. 2013;23:599–606. doi: 10.1016/j.cub.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernius J, Marston AL. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 2009;5:e1000629. doi: 10.1371/journal.pgen.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ocampo-Hafalla MT, Katou Y, Shirahige K, Uhlmann F. Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma. 2007;116:531–544. doi: 10.1007/s00412-007-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nerusheva OO, Galander S, Fernius J, Kelly D, Marston AL. Tension-dependent removal of pericentromeric shugoshin is an indicator of sister chromosome biorientation. Genes Dev. 2014;28:1291–1309. doi: 10.1101/gad.240291.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F. Budding yeast wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol. 2013;23:64–69. doi: 10.1016/j.cub.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 10.Hinshaw SM, Makrantoni V, Kerr A, Marston AL, Harrison SC. Structural evidence for Scc4-dependent localization of cohesin loading. Elife. 2015;4:e06057. doi: 10.7554/eLife.06057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinshaw SM, Makrantoni V, Harrison SC, Marston AL. The Kinetochore Receptor for the Cohesin Loading Complex. Cell. 2017;171:72–84.e13. doi: 10.1016/j.cell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu B, et al. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic acids research. 2015;43:e132. doi: 10.1093/nar/gkv670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verzijlbergen KF, et al. Shugoshin biases chromosomes for biorientation through condensin recruitment to the pericentromere. Elife. 2014;3:e01374. doi: 10.7554/eLife.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peplowska K, Wallek AU, Storchová Z. Sgo1 regulates both condensin and ipl1/aurora B to promote chromosome biorientation. PLoS Genet. 2014;10:e1004411. doi: 10.1371/journal.pgen.1004411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 17.Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature cell biology. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 19.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schalbetter SA, et al. SMC complexes differentially compact mitotic chromosomes according to genomic context. Nature cell biology. 2017;19:1071–1080. doi: 10.1038/ncb3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazar-Stefanita L, et al. Cohesins and condensins orchestrate the 4D dynamics of yeast chromosomes during the cell cycle. EMBO J. 2017;36:2684–2697. doi: 10.15252/embj.201797342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganji M, et al. Real-time imaging of DNA loop extrusion by condensin. Science. 2018;360:102–105. doi: 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson IF, et al. DNA loop extrusion by human cohesin. Science. 2019;366:1338–1345. doi: 10.1126/science.aaz3418. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. Human cohesin compacts DNA by loop extrusion. Science. 2019;366:1345–1349. doi: 10.1126/science.aaz4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vian L, et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell. 2018;173:1165–1178.e20. doi: 10.1016/j.cell.2018.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bausch C, et al. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Molecular and cellular biology. 2007;27:8522–8532. doi: 10.1128/MCB.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocampo-Hafalla M, Muñoz S, Samora CP, Uhlmann F. Evidence for cohesin sliding along budding yeast chromosomes. Open Biol. 2016;6 doi: 10.1098/rsob.150178. 150178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bresson S, Tuck A, Staneva D, Tollervey D. Nuclear RNA Decay Pathways Aid Rapid Remodeling of Gene Expression in Yeast. Molecular cell. 2017;65:787–800.e5. doi: 10.1016/j.molcel.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 32.Perea-Resa C, Blower MD. Centromere Biology: Transcription Goes on Stage. Molecular and cellular biology. 2018;38:e00263–18. doi: 10.1128/MCB.00263-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stempor P. Package ‘seqplots’. 2015 [Google Scholar]
- 34.Belton JM, Dekker J. Hi-C in Budding Yeast. Cold Spring Harb Protoc. 2015;2015 doi: 10.1101/pdb.prot085209. pdb.prot085209. [DOI] [PubMed] [Google Scholar]
- 35.Schalbetter SA, Fudenberg G, Baxter J, Pollard KS, Neale MJ. Principles of Meiotic Chromosome Assembly. bioRxiv. 2018 doi: 10.1101/442038. 442038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome biology. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerpedjiev P, et al. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome biology. 2018;19:125. doi: 10.1186/s13059-018-1486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.