Abstract
Background and Purpose
Lung oedema in association with suppressed fibrinolysis is a hallmark of lung injury. Here, we have tested whether plasmin cleaves epithelial sodium channels (ENaC) to resolve lung oedema fluid.
Experimental Approach
Human lungs and airway acid‐instilled mice were used for analysing fluid resolution. In silico prediction, mutagenesis, Xenopus oocytes, immunoblotting, voltage clamp, mass spectrometry, and protein docking were combined for identifying plasmin cleavage sites.
Key Results
Plasmin improved lung fluid resolution in both human lungs ex vivo and injured mice. Plasmin activated αβγENaC channels in oocytes in a time‐dependent manner. Deletion of four consensus proteolysis tracts (αΔ432‐444, γΔ131‐138, γΔ178‐193, and γΔ410‐422) eliminated plasmin‐induced activation significantly. Further, immunoblotting assays identified 7 cleavage sites (K126, R135, K136, R153, K168, R178, K179) for plasmin to trim both furin‐cleaved C‐terminal fragments and full‐length human γENaC proteins. In addition, 9 new sites (R122, R137, R138, K150, K170, R172, R180, K181, K189) in synthesized peptides were found to be cleaved by plasmin. These cleavage sites were located in the finger and the thumb, particularly the GRIP domain of human ENaC 3D model composed of two proteolytic centres for plasmin. Novel uncleaved sites beyond the GRIP domain in both α and γ subunits were identified to interrupt the plasmin cleavage‐induced conformational change in ENaC channel complexes. Additionally, plasmin could regulate ENaC activity via the G protein signal.
Conclusion and Implications
Plasmin can cleave ENaC to improve blood‐gas exchange by resolving oedema fluid and could be a potent therapy for oedematous lungs.
Abbreviations
- AFC
alveolar fluid clearance
- ARDS
acute respiratory distress syndrome
- AT2
alveolar type II epithelial cells
- ENaCs
epithelial sodium channels
- GRIP
gating relief of inhibition by proteolysis
- PAI‐1
plasminogen activator inhibitor‐1
- uPA
urokinase plasminogen activator
What is already known
Plasmin cleaves epithelial sodium channels (ENaC) proteolytically to activate channel function.
What this study adds
Plasmin improves AFC via cleaving 16 sites in two proteolytic centres in human γENaC subunits.
What is the clinical significance
Activation of ENaC by plasmin may improve resolution of lung oedema in ARDS lungs.
1. INTRODUCTION
One of the most common complications of sepsis is the acute respiratory distress syndrome (ARDS) with a hallmark of dysfunctional blood–gas barrier, occurring in nearly 200,000 patients per year with a mortality above 30% (Ware & Matthay, 2000). ARDS is characterized by hypoxaemia with bilateral infiltrates of the lungs with non‐cardiac origin (Ranieri et al., 2012; Sartori & Matthay, 2002). Prolonged suppression of normal fibrinolytic activity, paralleled by a sustained increase in plasminogen activator inhibitor‐1 (PAI‐1), has been consistently observed in clinical and preclinical studies of septic ARDS (Bertozzi et al., 1990). Fibrinolytic activity is further impaired following the development of sepsis with multiorgan failure (Asakura et al., 2001). An elevated plasma PAI‐1 concentration is associated with an adverse outcome in sepsis (Hermans et al., 1999). Caged plasminogen activators lose their ability to produce plasmin, a key molecule of the plasminogen/plasmin system that sustains normal fibrinolysis. The reduced fibrinolytic activity was also reported in bronchoalveolar fluid and pleural effusion of septic illnesses (Idell, James, & Coalson, 1992). Concurrently, increased permeability pulmonary oedema often develops. Accumulation of alveolar oedema fluid mainly results from an increase in lung endothelial and epithelial permeability that cannot be compensated by transepithelial fluid re‐absorption (Matthay, Folkesson, & Clerici, 2002), causing what is termed clinically ARDS (Matthay, Ware, & Zimmerman, 2012).
Epithelial sodium channels (ENaCs) located at the apical membrane regulate alveolar fluid reabsorption across the tight alveolar epithelium (Ji et al., 2012). Reduced ENaC expression and activity are described in both injured human lungs ex vivo and in the animal models of lung injury (Matthay et al., 2002). Decreased alveolar fluid clearance (AFC) is confirmed in mice with deficient scnn1 genes. Plasminogen activators may decrease the severity of lung injury and pleural effusion (Karandashova et al., 2013) and may reduce the risk of death from ARDS and multiorgan failure from sepsis (Hardaway et al., 2001; Lim & Chin, 1999). Recently, our meta‐analysis reports that fibrinolytic therapy might reduce lung injury, facilitate oedema fluid resolution, and reduce histological lung injury score in animal models of acute lung injury (Liu et al., 2018). However, whether exogenously administered plasmin enhances the removal of oedema fluid via ENaC and related mechanisms is not clear. We recently identified human ENaC as a novel target of urokinase (Ji et al., 2015). Based on the lesser specificity of plasmin substrates in amino acid sequence, it is a challenge to identify all of cleavage sites for plasmin, in particular for in vivo lifetime exposure to plasmin. On the other hand, whether or not plasmin could enhance reduced transepithelial fluid transport and restore impaired AFC in vivo has not been studied. Therefore, we aimed to test the potential beneficial effects of plasmin delivered intratracheally on AFC and determined proteolysis as post‐translational mechanisms.
2. METHODS
2.1. Animals and HCl instillation
All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of the University of Texas Health Science Center at Tyler. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010) and with the recommendations made by the British Journal of Pharmacology. Healthy, 8‐ to 12‐week‐old, male or female C57 BL/6 mice (Jackson Lab, Cat#: 000664. RRID:IMSR_JAX:000664) were used. . C57 BL/6 mice provide well‐characterized animal models for ARDS with pathogenesis similar to that in humans. Animals were kept under pathogen‐free conditions. To create an in vivo model of lung injury, mice were anaesthetized with ketamine (1.7 mg·ml‐1) and xylazine (20 mg·ml‐1), at 5ml kg‐1 by i.p. injection. No additional analgesia was applied. Anaesthetized mice were given 0.1‐M HCl in 2.5 μl·g−1, intratracheally, followed by 30 μl·g−1 of air (Nagase et al., 2000; Patel, Wilson, & Takata, 2012). Control mice were given the same volume of 0.9% NaCl and air.
AFC in mice
The clearance rate of alveolar fluid was measured in vivo as previously described (Han et al., 2010; Patel et al., 2012). Briefly, mice, 4 hr after the instillation of HCl, were anaesthetized with ketamine (1.7 mg·ml−1) and xylazine (20 mg·ml−1), given i.p. at 5 ml kg−1. Then the mice were placed on a continuous positive airway pressure system delivering 100% O2 at 8 cmH2O. All animals were maintained at a temperature of 37°C with a heating pad and ultrared bulb. An isosmotic instillate containing 5% bovine serum albumin was prepared with 0.9% NaCl as base solution. Control animals were given 50‐μl base solution intratracheally. Plasmin (one group with 60 μg·ml−1 and another group with 100 μg·ml−1), a mixture of plasmin and equal amount of α2‐antiplasmin (Pl + AP), and plasmin (60 μg·ml−1) in the presence of amiloride (1 mM) were intratracheally delivered. To maximize the collection of instilled BSA solution, the diaphragm was dissected after 30‐min of ventilation. Methodological concerns were addressed by using amiloride to inhibit water movement to confirm that measurements accurately reflected AFC. We assume that the concentration of installed BSA proteins is not altered significantly by the presence of excess alveolar fluid and/or proteins caused by HCl‐induced damage. HCl may impair the blood–gas barrier function and causes a significant leakage of murine serum proteins into the air space. The endogenous BSA may lead to an overestimation of AFC rates. This potential bias was corrected by measuring murine albumin (Cat#: EMA3201‐1, AssayPro, St. Charles, MO) and bovine albumin (Cat#: E10‐113, Bethyl Laboratories, Montgomery, TX) separately using specific ELISAs following the manufacturer's instructions (Wolk et al., 2008). Final AFC values were corrected by removing murine albumin present in the aspirate. In addition, we used an aliquot of the instillate that had been delivered into the lungs and removed within 5 min, rather than freshly prepared naive instillate per se. Any pre‐existing oedema or leaking murine plasma proteins would be excluded for it dilutes both the control at time point 0 min (P i) and final samples (P f). The calculation of AFC was as described for human lungs below.
2.2. Ex vivo human lung lobe studies
Discarded human lung lobes more than 5 cm away from tumour were collected 24 hr post‐surgery with an approved exempt from the IRB committee of the China Medical University and in accordance with the Declaration of Helsinki. The left and right lobes were chosen randomly upon availability. Lung lobes were ventilated with 100% O2 via a volume‐controlled ventilator (model 683, Harvard Apparatus) and were passively rewarmed at 37°C for 1 hr without perfusion in an incubator prior to measuring AFC. Plasmin (60 μg·ml−1) was added to the instillate (20 ml) and intrabronchially delivered. The instilled alveolar fluid was aspirated by applying gentle suction to the tracheal catheter with a 1‐ml syringe. The BSA content of the alveolar fluid was measured with a 96‐well microplate reader. AFC was calculated as follows: AFC = (V i − V f )/V i × 100, where V i and V f denote the volume of the instilled and recovered alveolar fluid respectively. V f was obtained as V f = (V i × P i )/P f, where P i and P f represent protein concentration of instilled and collected fluid respectively.
2.3. Primary type 2 alveolar epithelial (AT2) cell culture and bioelectrical measurements
Mouse AT2 cells were isolated from both WT and urokinase plasminogen activator (uPA) knockout (plau −/−) strains of C57BL/6 mice (Cat#: 000664 and 002509 respectively, Jackson Laboratory, USA) with a modified protocol as described (Demaio et al., 2009; Zhao et al., 2019). Briefly, lungs of killed mice were removed and incubated in dispase for 45 min and then were gently teased and incubated in DMEM/F‐12 + 0.01% DNase I for 10 min. Cells were passed through a serial of Nitex filters (100, 40, 30, and 10 microns; Corning, USA) and centrifuged at 300× g for 10 min. Resuspended cells were biotinylated with CD16/32 (0.65 μg·10−6cells), CD45 (1.5 μg·10−6cells), and Ter/119 (10 μl) antibodies and then incubated with streptavidin‐coated magnetic particles (2.5 μg·10−6cells). Fibroblasts were removed by a 2‐hr incubation in a mouse IgG coated plastic culture dish. To authenticate freshly isolated AT2 cells, only cells meeting the following criteria were used: (a) viability >90% as assessed by Trypan Blue extrusion and (b) purity >95% as analysed by the PAP stain, pro‐SPC immunofluorescent recognition, or FACS. Mouse laminin 1 precoated transwells (Corning Costar, Cat#: 3413) were seeded with AT2 cells (106 cells cm−2). Culture medium was replaced after 72 hr and then every 48 hr. Transepithelial resistance (R TE) and potential difference (V TE) were measured using an epithelial voltohmmeter (World Precision Instrument, RRID:SCR_008593). Equivalent short‐circuit current (IEQ) values were calculated as the ratio of V TE/R TE.
2.4. Construction of ENaC mutants
Deletion and site‐directed mutants were generated in human ENaC cDNAs cloned into a pGEM HE vector using the QuikChange II Site‐Directed Mutagenesis kit (Stratagene; Ji & Benos, 2004; Molina et al., 2011). cRNAs of human α, β, and γ ENaC subunits were prepared as described previously (Ji, Parker, Langloh, Fuller, & Benos, 2001). HA and V5 tags were introduced to the N‐ and C‐termini of α and γ ENaC subunits respectively.
2.5. Oocyte expression and voltage clamp studies
Xenopus oocytes were surgically removed from appropriately anaesthetized adult female Xenopus laevis (Xenopus Express, Cat#: HCG IMP FM, hCG injected Mature Female Xenopus laevis. RRID:SCR_016373) as described (Zhao et al., 2019). Briefly, the ovarian tissue was removed from frogs under anaesthesia with tricaine (0.1%) through a small incision in the lower abdomen. Ovarian lobes were removed and digested in OR‐2 calcium‐free medium (in mM: 82.5 NaCl, 2.5 KCl, 1.0 MgCl2, 1.0 Na2HPO4, and 10.0 HEPES, pH 7.5) with the addition of 2 mg·ml−1 collagenase (Roche, Indianapolis). To authenticate oocytes, only cells meeting the following criteria were used: (a) oocytes at the developmental stage VI, (b) rolling freely, (c) without leaking after cRNA microinjection, and (d) keep healthy through entire procedure. Defolliculated oocytes were randomly assigned to each group. Cells for ENaC expression were injected with ENaC cRNAs into the cytosol (25 ng per oocyte in 50 nl of RNase‐free water) and incubated in regular OR‐2 medium at 18°C. The two‐electrode voltage clamp technique was used to record whole‐cell currents 48 hr post injection. Oocytes were impaled with two electrodes filled with 3‐M KCl, having resistance of 0.5–2 MΩ. A TEV‐200A voltage clamp amplifier (Dagan) was used to clamp oocytes with concomitant recording of currents. Two reference electrodes were connected to the bath. The continuously perfused bathing solution was ND‐96 medium (in mM: 96.0 NaCl, 1.0 MgCl2, 1.8 CaCl2, 2.5 KCl, and 5.0 HEPES, pH 7.5). Whole‐cell currents were recorded as previously reported (Ji et al., 2006). Experiments were controlled by pCLAMP 10.7 software (Molecular Devices. RRID:SCR_011323), and currents were continuously monitored at an interval of 10 s. To analyse amiloride inhibition, we waited until the current level reduced and reach a plateau and last for at least 1 min. After amiloride application to the bath, data were sampled at the rate of 200 Hz and filtered at 50 Hz.
2.6. Biotinylation and immunoblotting
Biotinylation experiments were adapted from previous publications (Haerteis et al., 2012; Haerteis, Krueger, Korbmacher, & Rauh, 2009), using 20–40 oocytes per group. In some experiments, oocytes were preincubated either in ND‐96 solution or in low‐sodium solution (1‐mM NaCl and 96‐mM NMDG). Oocytes were incubated in freshly prepared biotinylation buffer (1.5 mg·ml−1 EZ‐Link Sulfo‐NHS‐SS‐Biotin in DPBS solution from Hyclone, pH 8.0) for 30 min at room temperature with gentle agitation. The biotinylation reaction was stopped by washing the oocytes three times for 5 min each with quenching buffer (in mM: 192 glycine and 25 Tris‐Cl, pH 7.5). Subsequently, the oocytes were incubated in ND‐96 solution or supplemented with 10 μg·ml−1 human plasmin for 60 min or designated periods for time‐dependent study. After washing the oocytes three times with ND‐96 solution, treated cells were lysed by passing them through a 27‐gauge needle in lysis buffer (in mM: 500 NaCl, 5 EDTA, 50 Tris, 1% Triton X‐100, 1% Igepal CA‐630, pH 7.4) and supplemented with Complete Mini EDTA‐free protease inhibitor cocktail (Roche, 04693159001) according to the manufacturer's instructions. The lysates were incubated in a shaker for 1 hr and centrifuged at 16,000× g for 15 min at 4°C. Supernatants were transferred to 1.5‐ml tubes (Eppendorf). Biotinylated proteins were precipitated with 50 μl of pre‐washed high‐capacity neutravidin agarose resin (Pierce, 29204). After overnight incubation at 4°C with overhead rotation, supernatants were removed, and beads were washed three times with lysis buffer containing protease inhibitors. 50 μl of 2× SDS‐PAGE sample buffer (Pierce, 39001) was added to the beads. Samples were boiled for 5 min at 95°C, centrifuged for 1 min and loaded on a 7.5% SDS‐PAGE gel. To detect small peptides by anti‐HA antibody, samples were run on a 16.5% Tris‐Tricine gel (BioRad, 4563063) for some experiments. To detect γ ENaC fragments, the membrane blots were blocked in 5% blocking buffer (5% non‐fat dry milk, Bio‐Rad, in TBST) for 1 hr at room temperature. Then, anti‐V5 or anti‐HA monoclonal antibodies were added to the samples (1:5,000 and 1:1,000 dilution, respectively). Our and other groups have demonstrated that non‐ENaC bands could be detected in parental oocytes or oocytes expressing ENaC constructs without these artificial flags (Carattino et al., 2014; Ji et al., 2015; Passero et al., 2008; Passero et al., 2012). HRP‐labelled secondary antibodies (Jackson ImmunoResearch, RRID:SCR_010488) were used (1:10,000). Chemiluminescence signals were detected using ECL Plus (Millipore). The immuno‐related procedures used comply with the recommendations made by the British Journal of Pharmacology.
2.7. In silico prediction of plasmin cleaved sites in α, β, γ, and δ ENaC subunits
Specific cleavage sites for human plasmin confirmed with phage substrate display and solution phase fluorogenic peptide microarray (Gosalia, Salisbury, Maly, Ellman, & Diamond, 2005; Hervio et al., 2000) were applied. Another substrate motif from P4 to P4′ was from the getMerops of the SitePrediction server (Verspurten, Gevaert, Declercq, & Vandenabeele, 2009). Default settings of the server were used. The predicted sites must meet these criteria: (a) The cleavage sites are located at the ectodomain of ENaC, (b) the size of predicted C‐terminal fragments is similar to that on Western blots or smaller considering potential multiple proteolysis, (c) the P1 protein must be either Arg (R) or Lys (K), (d) average score is >1, (e) specificity is >95%, and (f) predicted by applying both optimized substrate sequences.
2.8. MS analysis for plasmin cleaved fragments
Three pure peptides (with a purity >95%) with a continuous sequence from amino acid residue T121 to A190 of the human γ ENaC proteins were synthesized by the GenScript (NJ, USA). The N‐ and C‐termini were acetylated and amidated respectively. The sequence of each peptide is shown in Figure 8a. The synthesized peptides were dissolved in 400‐μl DPBS (1.0 mg·ml−1), and digested by adding 10 μg·ml−1 plasmin for 30 min at room temperature. Trifluoroacetic acid (0.1%) was used to inactivate the cleavage process. The samples were analysed by the University of Texas Southwest Medical Center Proteomics Core. Data were then analysed with the Proteoe Discoverer 2.2 software (Thermo Fisher, USA) based on a tryptic digestion with a maximum of six missed cleavages at amino acid residues R and K. F and V sites are searched by the strategy for non‐specific cleavage. The frequency of the fragments with cleaved end of a specific amino acid residue is the sum of peptide spectral matches for both termini.
FIGURE 8.
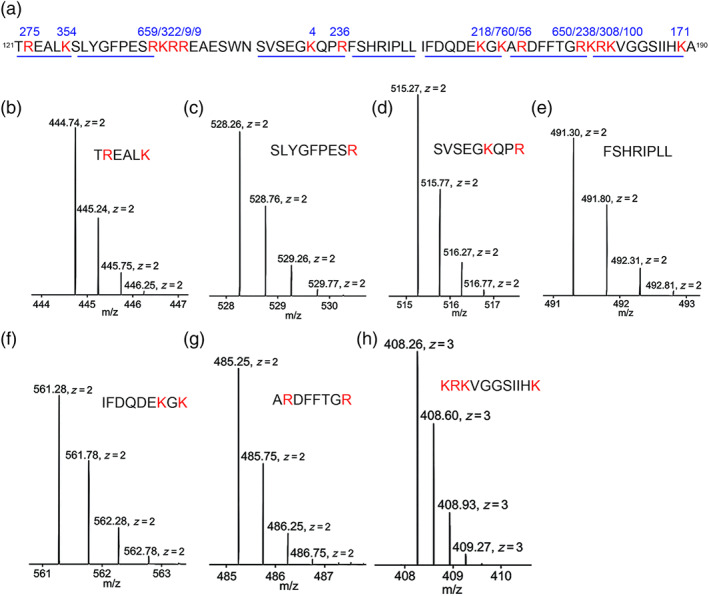
Validation of plasmin cleaved sites by MS. (a) Sequences of three synthesized peptides with identified plasmin cleavage sites (red font). Three peptides with a continuous sequence were separated by two gaps. The numbers above the cleaved sites are the frequency of the fragments with an end of this amino acid residue identified by MS. The underlining indicates the sequences of the fragments for panels b to h from left to right. (b–h) MS1 spectrum for the representing fragments. The size and charge are labelled for each isotope. Corresponding sequences of each fragment is shown as insets. n =1
2.9. Data and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology. Experimental procedures and statistical analysis were designed to be double‐blind. All results are presented as mean ± S.E. We designed the same sample size for the experiments performed in parallel. However, there was a loss due to the deaths of animals and cells before the completion of the experimental procedure. When there was a considerable diverse level in control groups between different sets of experiments, clearly due to reagents, experimenters or other experimental conditions, the data were normalized to the corresponding control group before data combination. ENaC activity is the difference between the total and amiloride‐resistant current fractions. Power analysis was performed for the sample size with the Origin Pro 2019 (OriginLab, RRID:SCR_014212). One‐way ANOVA was used to analyse the significance of the difference of the means. Bonferroni post hoc tests were conducted only if the F analysis was significant, and there was no variance inhomogeneity. P <.05 was considered significant.
2.10. Materials
Human recombined plasmin (Molecular Innovation, Novi, Michigan, USA. Cat#: HPLM), two‐chain urokinase plasminogen activator (uPA; Molecular Innovation, Novi, Michigan, USA. Cat#: UPA‐HTC), dispase (Corning, USA. Cat# 354235), DNase I (Sigma‐Aldrich, St. Louis, Missouri, USA. Cat#: DN25), bovine serum albumin (Sigma‐Aldrich, St. Louis, Missouri, USA. Cat#: A2153), biotin conjugated rat anti‐mouse CD16/32 (BD, Pharmingen, USA. Clone 2.4G2. Cat#: 553143. RRID:AB_394658), biotin conjugated rat anti‐mouse CD45 (BD, Pharmingen, USA. Clone 30‐F11. Cat#: 553078. RRID:AB_394608), biotin conjugated rat anti‐mouse Ter‐119/erythroid TER‐119 cells (BD, Pharmingen, USA. Cat# 553672. RRID:AB_394985), Dynabeads™ MyOne™ streptavidin T1 magnetic beads (Invitrogen, USA. Cat#: 65601), mouse IgG (Sigma‐Aldrich, St. Louis, Missouri, USA. Cat# I5381. RRID:AB_1163670), mouse laminin1 (Trevigen, USA. Cat#: 3401‐010‐02), tricaine‐S (MS‐222; Western Chemical, Inc. USA), EZ‐Link Sulfo‐NHS‐SS‐Biotin (Thermo Scientific, USA. Cat#: 21331), anti‐V5 mouse monoclonal IgG2a antibody (Invitrogen, USA. Cat#: R960‐25. RRID:AB_2556564), anti‐HA high affinity rat monoclonal antibody (Roche USA. Clone 3F10. Cat#: 11867423001. RRID:AB_390918).
2.11. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in the IUPHAR/BPS Guide to Pharmacology (RRID:SCR_013077; Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander, Christopoulos et al., 2019; Alexander, Fabbro et al., 2019; Alexander, Mathie et al., 2019).
3. RESULTS
3.1. Plasmin increases AFC in healthy human lungs ex vivo
The beneficial effects of plasminogen activators on respiratory dysfunction in sepsis imply that plasmin might improve AFC via the ENaC pathway (Chen et al., 2014; Liu et al., 2018). To address this issue, the effects of plasmin on ENaC‐mediated fluid resolution were measured in ventilated ex vivo human lung lobes (Figure 1a). Plasmin significantly increased the overall AFC value in 1 hr. Approximately 75% of the AFC rate was ENaC‐dependent, the fraction that was inhibited by amiloride. As reflected by the proportion of amiloride‐inhibitable AFC, plasmin stimulated ENaC activity up to three times (Figure 1b).
FIGURE 1.
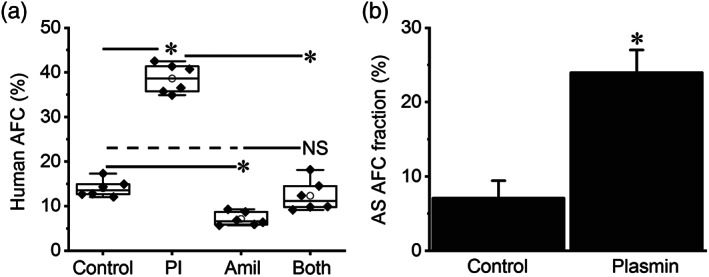
Intratracheally administered plasmin up‐regulates alveolar fluid clearance in normal human lungs ex vivo. (a) Total 60‐min alveolar fluid clearance (AFC60) in the presence and absence (control) of plasmin (Pl, 60 μg·ml−1, 0.7 μM) and/or amiloride (Amil, 1 mM). Adjacent human lung lobes within one resected sample were selected, and AFC values were measured in parallel for both control and treated groups. Data are presented as medians, means, 25% and 75% percentiles, and SEM; n = 6 per group. * P < .05, significantly different as indicated; NS, not significantly different from both control and Amil. (b) Amiloride sensitive (AS) fraction of AFC was computed as ENaC contribution in human lung lobes instilled with PBS only (control) and plasmin. * P < .05, significantly different from control; n = 6 per group
3.2. Plasmin increases alveolar oedema fluid clearance in the mouse model of injured lungs
We postulated that plasmin could restore the impaired ENaC function in a mouse model of gastric acid aspiration (Nagase et al., 2000). Intratracheal administration of two boluses of plasmin 1 hr after the instillation of HCl significantly reduced lung water content of injured mouse lungs (Figure 2a). In contrast, deactivation of the catalytic activity of plasmin with specific α2‐antiplasmin or amiloride did not alter the wet/dry ratio significantly. Correspondingly, in vivo AFC rate was restored close to a near normal level by proteolytically active plasmin but not by the mixture of plasmin with either α2‐antiplasmin or amiloride (Figure 2b). Together with the results in ex vivo human lungs, these data suggest that plasmin may resolve oedema fluid via the ENaC pathway in injured lungs.
FIGURE 2.
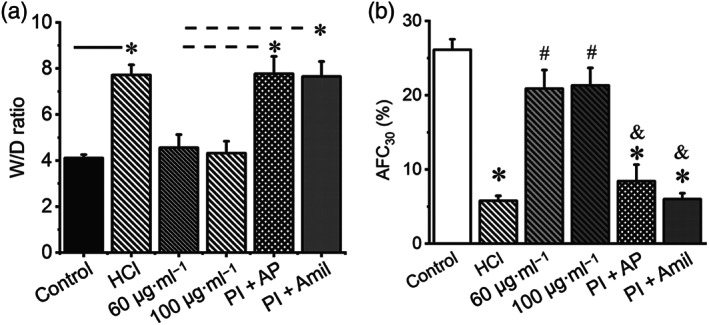
Plasmin stimulates alveolar fluid clearance in mouse lungs with acid aspiration injury. (a) Lung water content (wet/dry ratio). Mice were intratracheally instilled with 1‐N HCl (50 μl) as a model of gastric acid aspiration induced lung injury. Control animals were intratracheally instilled with 50‐μl PBS. Plasmin (60 and 100 μg·ml−1) or a mixture of plasmin and equal amount of α2‐antiplasmin (Pl + AP) were intratracheally instilled, 4 hr post HCl instillation. Lungs were dissected and weighed before and after drying. * P < .05, significantly different as indicated. Dashed line indicates the difference from more than one group. (b) Plasmin restores in vivo 30 min (AFC30) in mice injured by HCl. n = 20. * P < .05, significantly different from control. # P < .05, significantly different from HCl group. & P < .05, significantly different from plasmin only groups
3.3. Plasmin stimulates deficient ENaC activity in primary AT2 monolayers
Plasmin exhibited multifaceted effects on membrane‐arched receptors and enzymic substrates in epithelial and non‐epithelial cells (Castellino & Ploplis, 2005; Schaller & Gerber, 2011). To further confirm the stimulatory effects of plasmin on ENaC function located at the apical membrane of alveolar epithelial cells, we applied plasmin to polarized primary AT2 cell monolayers. As shown in Figure 3a,b, a significant suppression of ENaC activity, accompanied by an increment in transepithelial resistance, was seen in monolayers of AT2 cells from plau −/− mice, an in vitro model of lung injury mimicking injured lungs lacking fibrinolytic activity. The reduced ENaC function was increased by adding plasmin to the medium (Figure 3c). Combined with our previous studies in plau −/− mice and other groups' observations on the regulation of ENaC by plasmin (Chen et al., 2014; Haerteis et al., 2012; Ji et al., 2015; Passero et al., 2008), we decided to examine the post‐translational mechanisms underlying this action of plasmin, using human αβγENaCs heterologously expressed in Xenopus oocytes.
FIGURE 3.
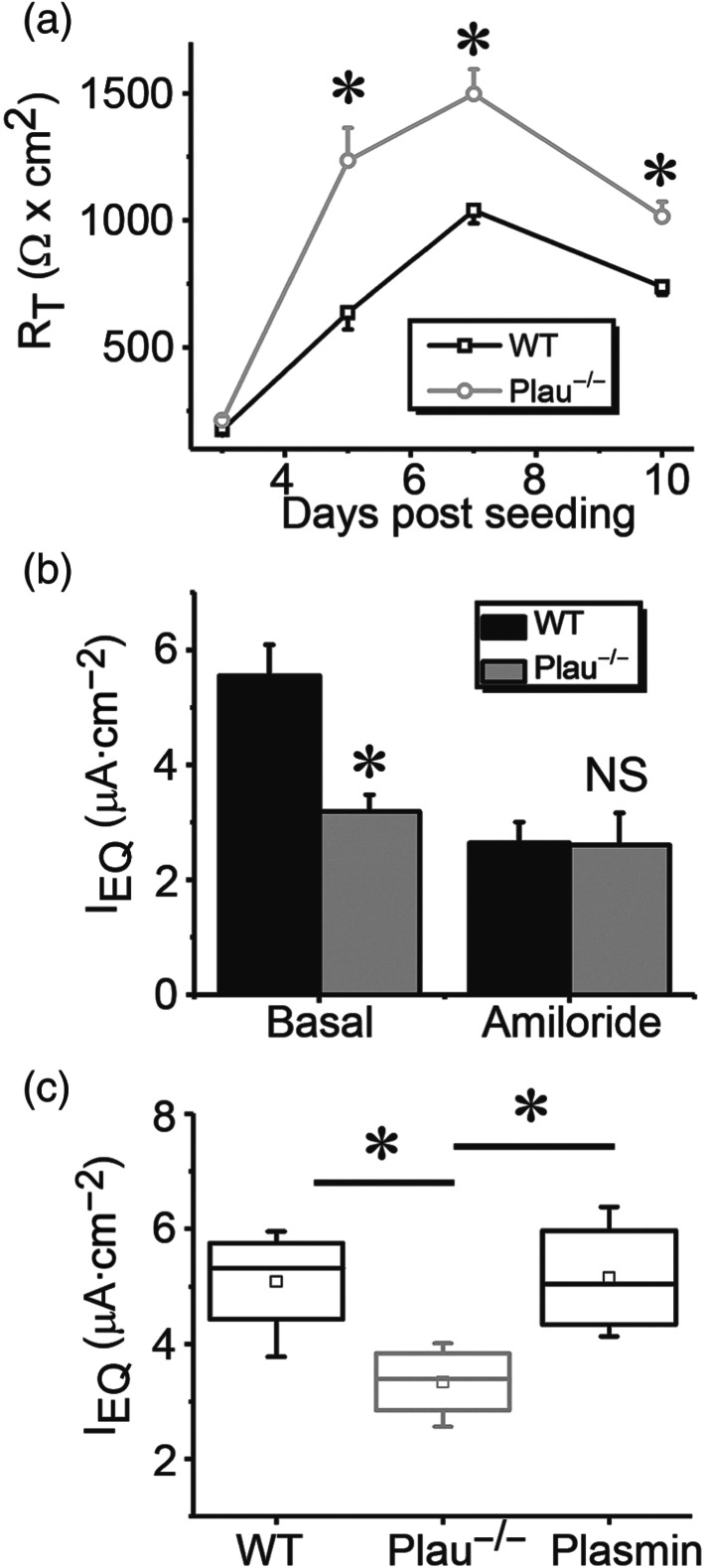
Plasmin restores deficient ENaC activity in monolayers of alveolar type 2 (AT2) epithelial cells from uPA deficient mice. (a) Transepithelial resistance in WT and plau −/− monolayers up to 10 days post seeding. * P < .05, significantly different from WT cells. n = 6. (b) Equivalent short‐circuit (IEQ) in polarized AT2 monolayers cultured at the air‐liquid mode. IEQ = ET/RT. ET is the transepithelial potential difference, and RT is the resistance across the monolayer read by an EVOM meter. Data are presented as means ±SEM; n = 6.. * P < .05, significantly different from WT group; NS, not significant. (c) Effects of plasmin on IEQ in polarized AT2 monolayers. Data were presented as medians, means, 25% and 75% percentiles, and SEM for wild type (WT; n = 5), plau knockout (plau −/− ; n = 7), and addition of plasmin (20 μg·ml−1; n = 6 ). * P < .05, significantly different as indicated
3.4. γENaC is targeted by plasmin
Plasmin is generated from plasminogen, a substrate of both tissue‐type and urokinase‐like plasminogen activators (tPA and uPA, respectively). To examine the effects of plasmin on ENaC activity, we incubated cells heterologously expressing human αβγENaC with plasmin for 1 hr with uPA as a positive control (Ji et al., 2015; Figure 4a). Plasmin activated ENaC currents approximately 7‐fold and this increase was greater than that following incubation with tc‐uPA. In sharp contrast, incubation with a mixture of plasmin and α2‐antiplasmin did not increase ENaC current. Plasmin activated human heterologous αβγENaC activity in a time‐dependent manner. The channel activity was activated maximally in 1 hr (Figure 4b). Subsequently, ENaC function declined but still maintained at a level significantly greater than controls for 8 hr. To identify which subunit (α, β or γ) was targeted by plasmin, the stimulatory effects of plasmin on ENaCs composed of α subunit alone, α + β subunits, and α + γ subunits were measured, using αβγ channels as a positive control (Figure 4c). Both current amplitude and fold of αγENaC was significantly increased by plasmin, (Figure 4d). whereas the currents of ENaCs consisting of α subunit only or α + β subunits were not significantly altered by plasmin. These results suggested that the γ subunit was the target of plasmin.
FIGURE 4.
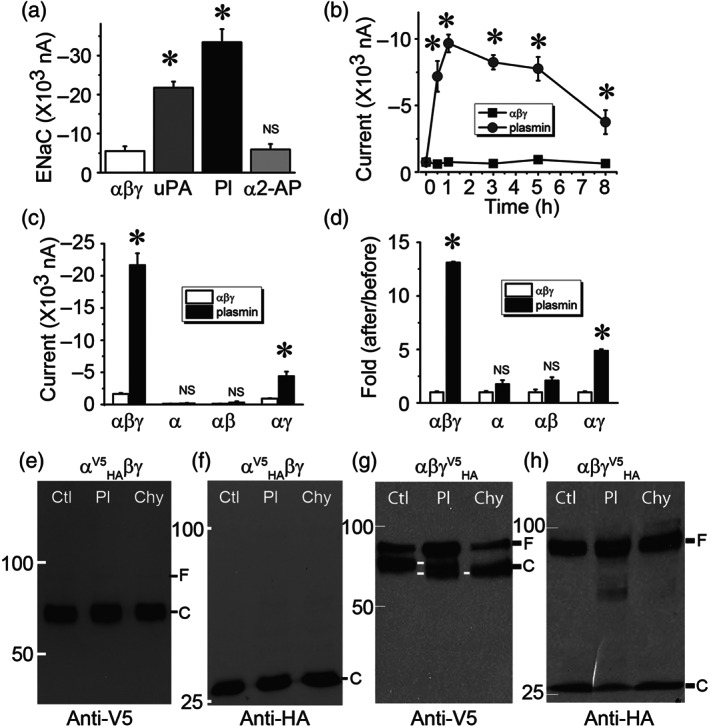
Plasmin specifically activates human αβγ‐ENaCs expressed in Xenopus oocytes. (a) Amiloride‐sensitive sodium currents (ENaC) were digitally sampled at the membrane potential of −100 mV in cells heterologously expressing human αβγ‐ENaCs, incubated with two‐chain uPA (10 μg·ml−1, 30 min at room temperature), plasmin (Pl, 10 μg·ml−1), and a mixture of plasmin (Pl, 10 μg·ml−1) and α2‐antiplasmin (α2‐AP, 10 μg·ml−1). n = 20 per group, * P < .05, significantly different from controls in the absence of fibrinolytic molecules; NS, not significant. (b) Time course of ENaC activation by plasmin. ENaC activity in the set of oocytes was recorded at designated time points after exposure to plasmin. n = 6 per group. * P < .05, significantly different from the corresponding time point of controls. (c) Effects of plasmin on the current levels of α, α + β (αβ), and α + γ (αγ) channels; n = 12 for α alone, n = 10 for αβ, and n = 12 for αγ group. * P < .05, significantly different from the corresponding controls in the absence of plasmin; NS, not significant. (d) Normalized incremental fold of ENaC activity. Ratio of current amplitudes recorded after over before addition of plasmin or perfusate was computed as fold of increase in channel activity; . * P < .05, significantly different from controls; NS, not significant.. (e,f) Detection of full‐length (F) and cleavage (C) of α ENaC by plasmin. HA (attached to the N‐terminal tail) and V5 (attached to the C‐terminal tail) tagged α subunit (αV5HA) was co‐expressed with βγ subunits complementarily. Biotinylated plasma membrane proteins were run on a 7.5% SDS‐PAGE gel and probed with anti‐V5 (e) and anti‐HA (f) monoclonal antibodies. Lanes from left to right were loaded with plasma membrane proteins of cells without plasmin treatment (Ctl), pretreated with plasmin (Pl, 10 μg·ml−1 for 30 min at room temperature), and chymotrypsin (Chy, 10 μg·ml−1) as a positive control. (g,h) Cleavage of HA and V5 tagged γ ENaC (γV5HA) by plasmin as recognized by anti‐V5 (g) and anti‐HA (h) antibodies. Cleaved bands are labelled with white lines at the same level. These experiments were repeated at least four times with similar observations
Further, with chymotrypsin as a positive control, we examined potential proteolytic cleavage of α and γ ENaC subunits with immunoblotting assays (Figure 4e–h). Both N‐ and C‐terminal fragments were detected by anti‐HA and anti‐V5 monoclonal antibodies respectively. As reported previously (Carattino et al., 2006), the full‐length αENaC subunit (90 kDa) was cleaved by endogenous proteases (i.e., furin) into two fragments in the control group (Figure 4e,f). Neither plasmin nor chymotrypsin altered furin‐cleaved V5‐ (64 kDa) and HA‐tagged fragments (26 kDa). In contrast, plasmin and chymotrypsin reduced the furin‐cleaved C‐terminal fragment (80 kDa) of full‐length γENaC proteins (90 kDa) to 70 kDa (Figure 4g). The N‐terminal fragment (26 kDa) as recognized by anti‐HA antibody was not affected by either plasmin or chymotrypsin (Figure 4h). Consistent with our previous studies, serine proteases are unable to cleave full‐length ENaC (Ji et al., 2015). These functional and immunoblotting data indicate that plasmin may specifically activate human αβγ channels by targeting γ subunits proteolytically and that plasmin as well as chymotrypsin may not be able to cleave the HA‐tagged N‐terminal fragments of both α and γ subunits cut off by furin.
3.5. Plasmin‐mediated regulation of deletion and site‐directed mutants missing putative cleavage domains
Three consensus domains at the extracellular loop for proteases to cleave ENaC proteins have been proposed (Kleyman, Carattino, & Hughey, 2009; Planes & Caughey, 2007; Rossier & Stutts, 2009). We attempted to find potential putative tracts responsible for plasmin‐mediated cleavage in both α and γ subunits. As shown in Figure 5a, removal of the first (aa173–178) and second (aa201–204) putative cleavage domains in the finger of α ENaC slightly but not significantly reduced and augmented the stimulation of plasmin on ENaC function respectively. By comparison, deletion of the third consensus cleavage tract (aa432–444) in the thumb markedly eliminated plasmin‐induced up‐regulation of the channel activity (Table 1 and Figure 5a). Similarly, all of three deletion mutants of γ ENaC subunit almost lost their responses to plasmin (Figure 5b). In contrast, plasmin‐cleaved C‐terminal peptides with variant sizes were seen for these γ deletion mutants (Figure 5c–f). Intriguingly, this process might not be furin cleavage‐dependent because plasmin was still able to cleave γΔ131–138, which lost furin cleavage sites‐R135K136 (Figure 5d). Plasmin was reported to potentially cleave both human and mouse γENaC subunits (Haerteis et al., 2012; Passero et al., 2008). Mutation of mouse γ K194 significantly suppressed acute activation of heterologous mouse αβγ ENaCs in minutes (Passero et al., 2008). Substitution of the corresponding site in human γ ENaC subunit (γK189A) with alanine, however, could not prevent the stimulatory effects of plasmin on human αβγK189A channels (Figure 5g). Instead, both the domains for mouse plasmin‐ and prostasin‐induced cleavage in human γ ENaC subunit termed γ5A (RKRK178–181AAAA + K189A) were crucial for plasmin to acutely activate human αβγ function (Haerteis et al., 2012). Surprisingly, plasmin still increased the current amplitude of this αβγ5A channels approximately nine and 20 times in 1 and 24 hr respectively (Figure 5h). Meanwhile, the plasmin‐cleaved C‐terminal fragments were recognized by anti‐V5 antibody (Figure 5i), suggesting additional new cleavage sites exist beyond these five amino acid residues for long‐term exposure to plasmin.
FIGURE 5.
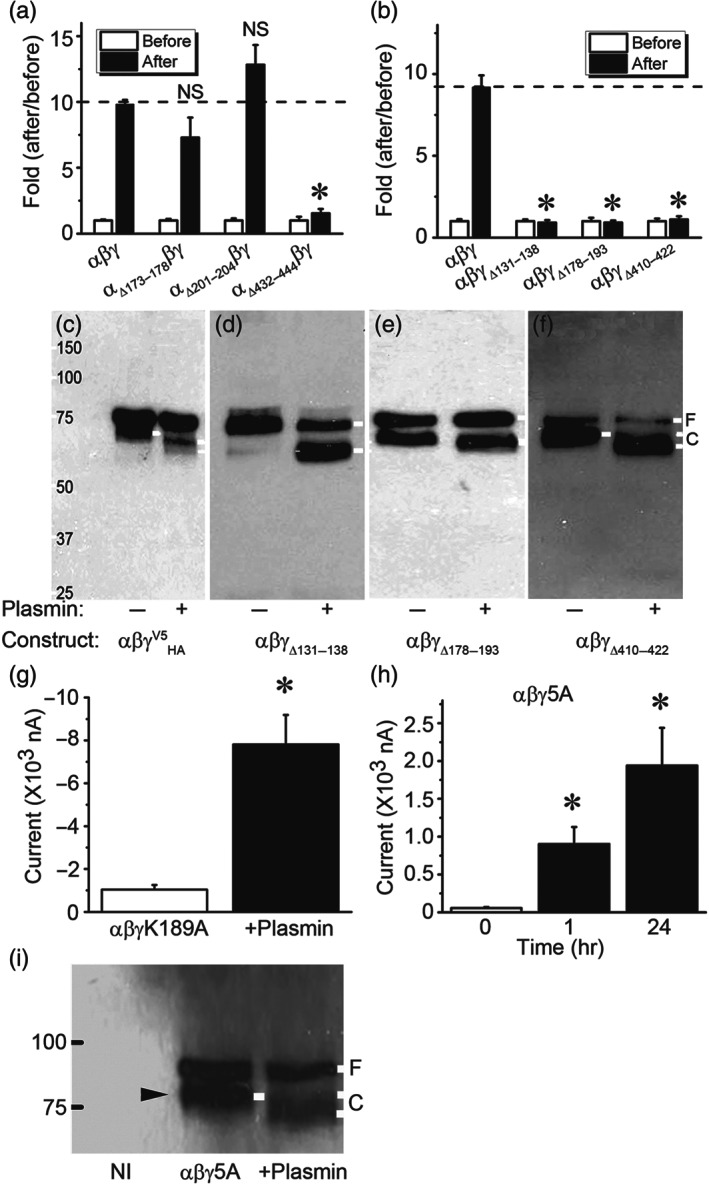
Effects of plasmin on α and γ‐ENaC mutants missing consensus cleavage domains. (a) Basal ENaC currents were recorded in oocytes expressing αβγ‐ENaC and three deletion mutants lack of putative cleavage domains, namely, αΔ173–178βγ (n = 24), αΔ201–204βγ (n = 26) and αΔ432–444βγ (n = 26). The activated currents were collected again 1 hr post incubation with plasmin (10 μg·ml−1, 30 min at room temperature) at the room temperature. The activated current levels were normalized to that basal currents. * P < .05, significantly different from αβγ‐ENaC; NS, not significant. n = 19–26. (b) Activation of three γ deletion mutants of αβγΔ131–138, αβγΔ178–193, and αβγΔ410–422 by plasmin. * P < .05, significantly different from αβγ‐ENaC; NS, not significant. (c–f) Immunoblotting detection of cleavage in full‐length and three deletion γ mutants. The most left lane is protein marker. Untreated cells were controls (−). Endogenous furin cleaved band in the absence of plasmin and plasmin‐cleaved band in the presence of plasmin (+) were labelled with white lines. These blots represent four experiments with similar observations. (g) Plasmin activates the activity of mouse mutant (αβγK189A) for plasmin cleavage site. Whole‐cell currents were digitized and then repeated 1 hr post incubation with plasmin (10 μg·ml−1). n = 8. * P < .05, significant effect of plasmin or time. (h) Plasmin stimulates the activity of a human mutant (αβγ5A) for plasmin cleavage (K189 and 178RKRK181 were substituted with alanine). Measurements of whole‐cell currents were performed 1 and 24 hr after exposure to plasmin. n = 22, * P < .05 significantly different from before addition of plasmin. (i) Western blot assay of biotinylated plasma membrane proteins an anti‐V5 monoclonal antibody. Plasma membrane proteins from noninjected oocytes (NI), cells expressing αβγ ENaC (Control), and cells pretreated with plasmin (Plasmin) were loaded on a 7.5% SDS‐PAGE gel. Furin and plasmin‐cleaved proteins are marked with black and white rightward arrowhead respectively. Similar results were observed in four blots
TABLE 1.
Characteristics of ENaC mutants
| Mutant (sequence) | 2D/3D location | In silico prediction | IB | TEVC (nA): Mutant vs. αβγ or controls | Protein docking | MS |
|---|---|---|---|---|---|---|
| αΔ173–178 (GSRSRR) | Before P1 after α1/finger | ‐ | ‐ | −2,092.63 ± 249.48 vs. −2,558.39 ± 177.42 | ‐ | ‐ |
| αΔ201–204 (RRAR) | P2, between α1 and α2/finger | ‐ | ‐ | −743.87 ± 110.93 vs. −2,558.39 ± 177.42 | ‐ | ‐ |
| αΔ432–444 (AYIFYPRPQNVEY) | Between α4 and α5/thumb | ‐ | Yes | −1,420.24 ± 399.51 vs. −2,558.39 ± 177.42 | ‐ | ‐ |
| γΔ131–138 (FPESRKRR) | Before P1 after α1, with furin site/finger | Yes | Yes | −916.10 ± 95.47 vs. −2,525.94 ± 200.39 | ‐ | R137, R138 |
| γΔ178–193 (RKRKV GGSIIHKASNV) | P2+, between α1 and α2, /finger | Yes | Yes | −214.09 ± 43.84 vs. −2,525.94 ± 200.39 | R178, R180 | R178, K179, R180, K181, K189 |
| γΔ410–422 (AQYSQP LPPAANYCNYQQH PNWM) | After α4 and part of α5, /thumb | ‐ | Yes | −253.81 ± 39.54 vs. −2,525.94 ± 200.39 | ‐ | ‐ |
| γΔ131–138 + 3A (K168A, R178A, K179A) | Before P1 after α1 + P2, /finger | Yes | Yes | −8,733.92 ± 875.22 vs. −870.30 ± 197.97 | R135, R137, K178 | R137, R138, K168, R178, K179 |
| γΔ131–138 + K168A | Before P1 and P2, /finger | Yes | Yes | −910.22 ± 147.01 vs. −1,120.30 ± 135.13 | R135, R137 | R137, R138, K168 |
| γΔ131–138 + R178A | Before P1 after α1 + P2, /finger | Yes | Yes | −1,033.91 ± 231.72 vs. −1,120.30 ± 135.13 | R135, R137, K178 | R137, R138, R178 |
| γΔ131–138 + K179A | Before P1 after α1 + P2, finger | Yes | Yes | −441.37 ± 121.09 vs. −1,120.30 ± 135.13 | R135, R137 | R137, R138, K179 |
| γK126A | Within α1/finger | Yes | Yes | ‐ | ‐ | Yes |
| γR135A | After α1/finger | Yes | Yes | −2,617.14 ± 589.75 vs. −2,573.49 ± 511.56 | Yes | Yes |
| γK136A | After α1/finger | Yes | Yes | −2,398.07 ± 341.08 vs. −2,573.49 ± 511.56 | ‐ | Yes |
| γR153A | Before P1/finger | Yes | Yes | ‐ | ‐ | Yes |
| γK168A | Before P2/finger | Yes | Yes | ‐ | ‐ | Yes |
| γK170A | Before P2/finger | Yes | ‐ | ‐ | ‐ | Yes |
| γR178A | Within P2/finger | Yes | Yes | ‐ | Yes | Yes |
| γK179A | Within P2/finger | Yes | Yes | ‐ | ‐ | Yes |
| γR180A | Within P2/finger | Yes | ‐ | Yes | Yes | |
| γΔ178–193 + 4A (K126A, R135A, K136A, R153A) | P2 + α1 (with furin site) /finger | Yes | Yes | −320.15 ± 113.04 vs. −299.47 ± 152.83 | R135, R137, R178, R180 | K126, R135, K136, R153, R178, K179, R180, K181, K189 |
| γΔ178–181 + K189A (5A) | P2+/finger | Yes | Yes | −968.10 ± 470.85 vs. −1,295 ± 266.52 | R178, R180 | R178, K179, R180, K181, K189 |
Note. All of deleted and single or multiple directed mutants are included for predicted location in the 2D/3D cryo‐EM model of truncated human ENaC (Noreng et al., 2018) and experimental evidence.
Abbreviations: 2D/3D, two or three‐dimensional structure; IB, immunoblotting; TEVC, two‐electrode voltage clamp Yes, positively detected/tested. “‐” means not available.
3.6. Identification of novel critical domains for activation of ENaC by plasmin
To identify substrate‐like motifs in the γENaC proteins, we carried out in silico predictions with the SitePrediction server with two different strategies (Figure 6a). Because plasmin cleaved both full‐length and furin‐catalysed proteins as shown in Figure 5d, we simplified our study by using the furin‐site deletion mutant, γΔ131–138 as a base construct to prepare a new triple mutant, namely, γΔ131–138 + 3A (K168A, R178A, and K179A). These three predicted sites were top ranked by the SitePrediction server. As shown in Figure 6b, the activity of this mutant was not activated by plasmin in both 1 and 24 hr. The cleaved C‐terminal fragments were not recognized compared with that of αβγΔ131–138 (Figure 6c). Moreover, three single point mutants were made: γΔ131–138 + K168A, γΔ131–138 + R178A, and γΔ131–138 + K179A. The activity of these site‐directed mutants was no longer increased by plasmin (Figure 6d). Immunoblotting assays showed a clear reduction in the fraction of cleaved proteins accompanied by increased full‐length bands (Figure 6e). Similarly, the densitometry ratio of cleaved bands over full‐length bands was markedly reduced for these single site‐directed mutants (Figure 6f).
FIGURE 6.
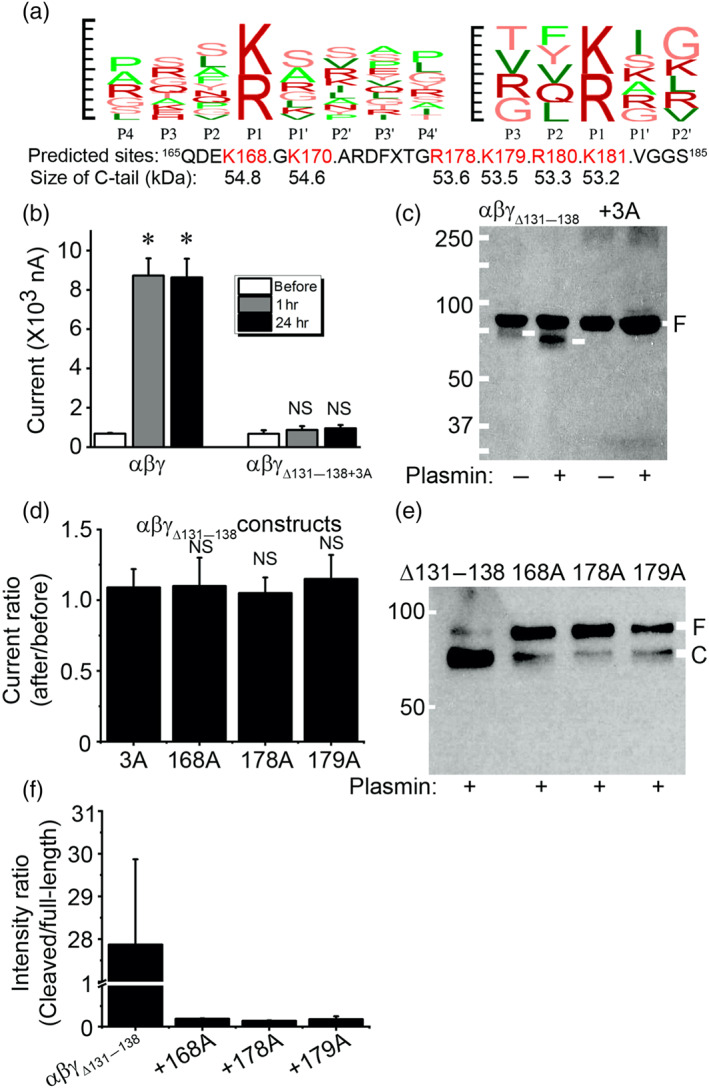
Potential proteolysis domains for plasmin beyond the putative cleaving motif (131–138) in γ ENaC subunit. (a) In silico prediction with the database for plasmin‐specific motifs. Left, a combination (logo) of reported 92 substrate motifs for plasmin from P4 to P4′. Right, a logo of input substrate sequences for plasmin from P3 to P2′. Top 10 predicted sites and the size of C‐terminal fragments by both strategies are listed below. Default set‐up was used for running the SitePrediction server (Gosalia et al., 2005; Hervio et al., 2000). Penalty: 0.1; sort order: average score. Similarity score: 100; specificity, >95%. (b) Effects of plasmin on αβγΔ131–138 + 3A (K168A, R178A, and K179A) channel activity in 1 and 24 hr. n = 18. * P < .05, significantly different from before; NS, not significant. (c) Immunoblotting assays of αβγΔ131–138 and αβγΔ131–138 + 3A channel proteins. These results are a representative blot of three experiments. (d) Regulation of single point mutants (γK168A, γR178A, and γK179A) derived from αβγΔ131–138 by plasmin. n = 12. NS, not significantly different from αβγΔ131–138 + 3A (3A) construct. (e) Detection of plasmin‐cleaved fragments of three single point mutants (from left to right are αβγΔ131–138, αβγΔ131–138 + K168A, αβγΔ131–138 + R178A, and αβγΔ131–138 + K179A). (f) Cleavage efficacy (cleaved band/uncleaved band). n = 3
The different size of cleaved band of γΔ178–193 mutant by plasmin indicates additional cleavage sites may exist. Based on the prediction (Figure 7a), we constructed a new mutant termed γΔ178–193 + 4A (K126A, R135A, K136A, and R153A). The activity of this mutant was not activated by plasmin (Figure 7b; Table 1). Moreover, the “full‐length” proteins of this mutant were increased significantly compared with the γΔ178–193 mutant (Figure 7c). This was further confirmed by the densitometrical ratio of cleaved/full‐length proteins (Figure 7d), suggesting that plasmin could cleave multiple sites in the finger domain of γENaC (Figure 7e).
FIGURE 7.
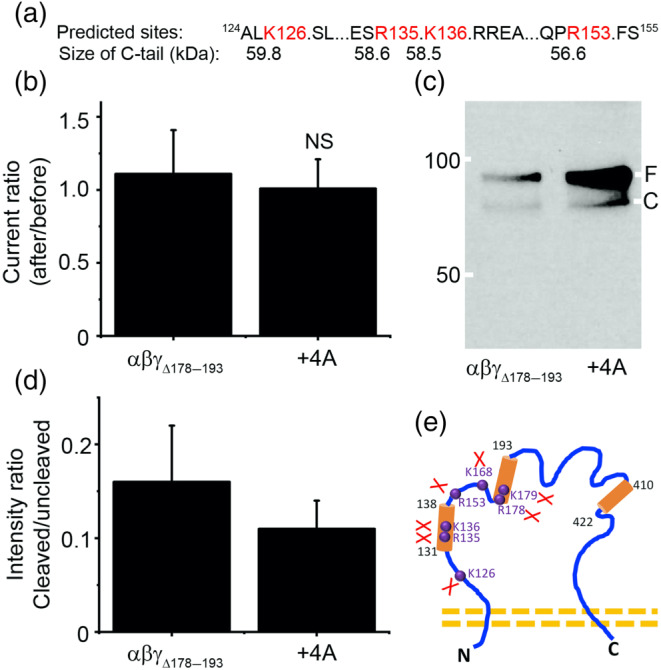
Prediction and validation of putative plasmin cleaved sites preceding γK168. (a) In silico predicted four potential cleavage sites (K126, R135, K136, and R153) and the peptide size of C‐terminal fragments. (b) Effects of plasmin on the channel activity of αβγΔ178–193 (n = 14) and a mutant combining γΔ178–193 and four predicted sites (+4A) (n = 9) that were replaced with alanine. NS, not significantly different from αβγΔ178–193 mutant. (c) Immunoblotting assays. This blot represents four experiments with similar results. (d) Contribution of four predicted cleavage sites. n = 4. (e) Summary of newly identified and validated cleavage sites for plasmin in γ‐ENaC subunit. Three putative proteolytic regions for truncation mutants are marked with cylinders; four identified amino acid residues are labelled in purple font and balls
3.7. Validation of plasmin cleavage sites by MS
To corroborate identified cleavage sites, three synthesized peptides identical to the sequence of γENaC from 121T to A190 amino acid residues were treated with plasmin and analysed by LC–MS. In addition to confirming the identified critical residues, nine more cleavage sites were detected (Figure 8a and Table 1). The frequency for the fragments with either the R or K amino acid residue as the C‐terminal tail or the next amino acid residue as the N‐terminal tail was computed, the confirmed seven cleavage sites identified by mutagenesis and confirmed by MS occurred more than 200 times. In comparison, the frequency of most MS discovered new cleavage sites was less than 100 except R122, K170, R180, and K189 (Figure 8a). A representative MS1 spectrum of seven fragments was shown in Figure 8b–h. In general, four to five isotopes with variant intensity/counts for the same fragment were seen. The corresponding MS/MS spectrum for these fragments is included in Figures S1–S7.
3.8. Identified cleavage sites for plasmin compose two proteolytic centres
To address the hypothesis that the identified 16 sites may be composed of cleavage centres spatially separated in the 3D structure (Figure 9a), we labelled three deletion mutants in key domain organization scheme (Figure 9b) and gating relief of inhibition by proteolysis (GRIP) domain (Figure 9c) based on the cryo‐EM model (Noreng, Bharadwaj, Posert, Yoshioka, & Baconguis, 2018). Apparently, one centre was made up of six cleavage sites at the end of the α1 helix and another with cleavage sites located in the antiparallel P1 and P2 strands. On the other hand, we employed a protein docking approach to substantiate this possibility and potential interactions between individual amino acid residues. We generated a homology model of human γENaC with the I‐TASSER server (Figure S8a; Noreng et al., 2018; Roy, Kucukural, & Zhang, 2010). There were three highly accessible clusters of positively charged residues in the GRIP domain of the γ ENaC (Figure S8b). As positive control, Figure S9c,d shows interactions between plasmin and two arginine residues (R17 and R19) of textilinin‐1 (pdbID: 3uir). Using the interacting residues in plasmin, we found the best docking result between 178RKRK in γ ENaC and D735 and G764 of plasmin (Figure S8e,f). Further, interactions between 135RKRR of r RNaC and plasmin were seen (Figure S8g,h). Together, two spatial centres for the cleavage by plasmin were identified in the vicinity of α1 terminal and antiparallel P1/P2 strands.
FIGURE 9.
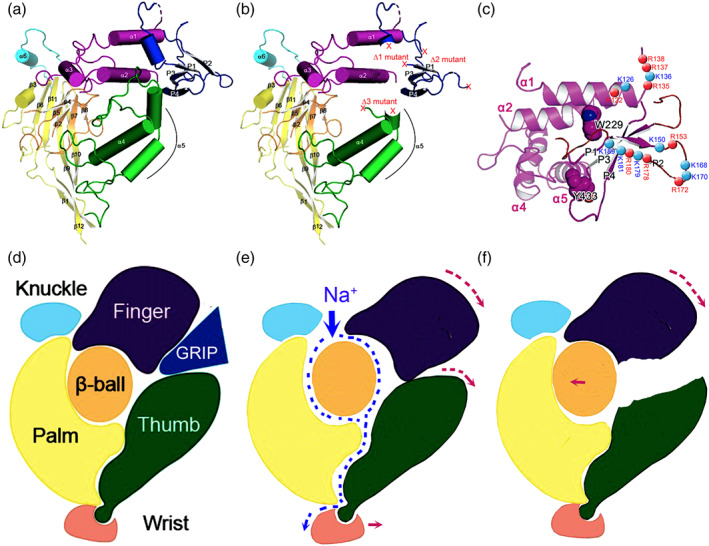
Location of deletion and single or multiple point mutants in 3D model and gating mechanism. (a) 3D structure of truncated human ENaC subunits, adapted from Noreng et al. (2018). (b) Location of deleted regions (Δ1, Δ 2, and Δ 3) for both α and γ‐ENaC subunits. (c) Positions of identified amino acid residues for plasmin cleavage in human γ subunit. (d–f) Schematic mechanism for plasmin to cleave and gate ENaC protein complexes. Plasmin cleaves full‐length ENaC (d) to remove the GRIP domain (e). The finger domain will subsequently fall down to hit the thumb to increase the width of channel pore, allowing Na+ ions to go through channel pore faster as shown by increased currents. However, for some mutants, for example, Δ3 deletion mutants of α and γ subunits, the link between the finger and the thumb disappears so that downward movement of the finger cannot reach the thumb to enlarge the channel pore (f)
3.9. G protein pathway may be involved
ENaC is regulated by G proteins, which are downstream molecules of the plasmin signal pathway (Greenlee et al., 2013). Because the effects of plasmin lasted up to 24 hr, plasmin could regulate ENaC via activation of the G protein signal. To exclude this possibility, Pertussis toxin was used to specifically inactivate Gi/o activity. Activation of ENaC currents by uPA and plasmin was not affected significantly (Figure S9a). However, the increase of plasmin‐activated current was significantly reduced, compared with controls and uPA groups (Figure S9b). These data suggest that plasmin but not uPA could activate ENaC by activating the G protein signal pathway as a long‐term effect.
4. DISCUSSION
Given the pharmacological features of plasmin and depressed fibrinolytic activity in ARDS, we set out to address the hypothesis that plasmin could improve AFC by cleaving ENaC proteolytically. We here provide preclinical evidence showing that intratracheally administered plasmin augmented AFC in both normal and acid aspiration injured lungs. For the first time, we found that plasmin catalysed both full‐length and furin‐trimmed human γENaC proteins. Plasmin may be a newly identified pharmacological intervention for removing oedema fluid from the air spaces in ARDS.
We identified novel cleavage sites for plasmin beyond three putative regions described previously (Kleyman et al., 2009; Planes & Caughey, 2007; Rossier & Stutts, 2009; Table 1). The GRIP domain is composed of four β strands (referred to as P1–4 segments; Figure 9a,d; Noreng et al., 2018). Plasmin cleaved 16 sites, at least two of them are out of this GRIP domain but within the α1 helix of the finger. The antiparallel arrangement of P1 strand, P2 strand, and the α1 helix place all of plasmin cleavage sites in close proximity to facilitate proteolytic efficiency of plasmin by forming two proteolytic centres. Plasmin is the most powerful serine protease and shares cleavage sites with prostasin, trypsin I and IV, uPA, elastases, chymotrypsin, and furin.
One of our novel observations is that long‐term exposure of human ENaC proteins to plasmin leads to the cleavage of full‐length γ ENaC. The current concept is that serine proteases cannot cleave full‐length ENaC but can cleave furin‐cut C‐terminal fragments because the action of furin exposes hidden catalytic triads for chymotrypsin and other proteases (Kleyman et al., 2009; Rossier & Stutts, 2009). Similar to furin, plasmin may cleave at the furin sites and expose additional proteolytic sites for itself. Our findings that long‐term exposure to plasmin hydrolyses full‐length ENaC proteins has altered the concept that only furin‐cleaved ENaC proteins are accessible for external proteinases. This may explain why plasmin activated ENaC to a greater extent than tc‐uPA. Additionally, the finding that plasmin, but not tc‐uPA, activated the G‐protein pathway may also facilitate ENaC activation.
The differences between immunoblotting and functional assays in the third deletion mutants (i.e., αΔ432–444 and γΔ410–422) could be due to the dis‐association of the finger and the thumb. Removal of the GRIP domain by proteolysis results in a downward movement of the finger, subsequently hitting the thumb to enlarge the channel pore and gate. Eventually, the channel activity is up regulated maximally (Figure 9e). Even though cleavage of the GRIP domain by plasmin, as shown on Western blots, still allows the finger to move down, truncation of the thumb of these deletion mutants would not be hit by the moving finger, and thus would not enlarge the channel pore and function. In addition, deletion of these putative proteolytic regions may alter the formation of the channel pore. This hypothesis is compatible with the first 3D model of the human ENaC proteins (Noreng et al., 2018).
Our in silico prediction excludes the cleavage of human α, δ, and β subunits by plasmin. To date, only α and γ‐ENaC subunits are cleaved by tested proteases. Although the δ subunit affects the cleavage of these two subunits, cleavage of δ‐ENaC has not been reported. The cleavage of β‐ENaC by serine proteases is rare and weak (Garcia‐Caballero, Dang, He, & Stutts, 2008; Jovov, Berdiev, Fuller, Ji, & Benos, 2002). Based on our in silico prediction and functional results, plasmin is much less likely to cleave β subunits than the γ subunits.
The greater real size of C‐terminal fragments on blots compared with the prediction by the server could be due to post‐translational modifications. Similar to our observations, up to 110 kDa of αβγ‐ENaC proteins were described, which was even larger than that detected. Recently, we carried out MS for the two bands of δ‐ENaC on western blots. Both the band with a predicted size (75 kDa) and a larger band (110 kDa) were confirmed to have δ‐ENaC sequences (Zhao et al., 2019). Therefore, we deduce that cleavage of ENaC by plasmin could facilitate post‐translational modifications.
We could not exclude the involvement of the protease‐activated receptors, PAR1, 2, and 4 in the process of fluid resolution in human and mouse lungs (Bock et al., 2015; Carmo et al., 2014; Mannaioni et al., 2008; Quinton, Kim, Derian, Jin, & Kunapuli, 2004). However, activation of lung epithelial PAR isoforms by plasmin has not been reported.
Under physiological conditions, plasmin in the bronchoalveolar fluid (1.5 μmol·L−1) may activate ENaC indirectly through cleavage of sc‐uPA to produce active tc‐uPA. However, this possibility could be ruled out in diseased lungs. In the bronchoalveolar lavage fluid of ARDS, antiplasmin molecules, including antithrombin, α1‐antitrypsin, α2‐antiplasmin, and α2‐macroglobulin, are markedly elevated. For example, α1‐antitrypsin concentration increased 60‐fold in diseased lungs (Wewers, Herzyk, & Gadek, 1988). These high levels of plasmin inhibitors are likely to prevent the enzymic activity of any plasmin in the bronchoalveolar fluid. On the other hand, increased urokinase inhibitors, PAI‐1, would form complexes with uPA and eliminate physical interactions between uPA and plasmin. Therefore, a dose above physiological concentration is needed for treating arterial occlusive diseases and macular oedema.
In summary, this study for the first time demonstrates the beneficial effects of plasmin on AFC and provides novel mechanisms underlying the cleavage of human ENaCs at multiple sites by plasmin.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
R.Z.Z., D.B., and H.L.J. expressed ENaC in oocytes, performed voltage clamp recordings, analysed AFC in mice and analysed results. R.Z.Z. carried out mutagenesis, the immunoblotting assays, and sample preparation for MS. H.G.N. performed ex vivo AFC in human lungs. G.A. performed bioelectrical assays in mouse AT2 monolayers. Y.C. performed homology modelling and protein–protein docking. H.L.J. and M.A.M. were responsible for experimental design, data analysis, result assembly, and manuscript preparation.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis, Immunoblotting and Immunochemistry, and Animal Experimentation, and as recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Figure S1. Online supplementary figures are the representative MS/MS spectrum for the peptide fragments in Figure 8.
Figure S8. Homology model and docking results for plasmin cleavage centres. a. 3D model of the human γ ENaC generated by I‐TASSER. b. The GRIP domain (aa114–239) of the human γ ENaC model with three positively charged residue clusters facing outward and highly accessible. Four P strands are composed of following amino acids: P1, aa160–166; P2, aa182–190; P3, aa199–208; and P4, aa213–219. Three cleavage centres were highlighted. c. The interaction of plasmin (chain A with surface presentation) with textilinin‐1 (chain C) from the original crystal structure 3uir highlighted with two arginine residues in the substrate interacting with plasmin. d. Detailed interactions between two arginine residues and plasmin: Arg17 interacting with Asp735, Ser741, and Gly764; Arg19 interacting with Glu687 and Phe587. We used this as a docking guide as attractors (Asp735 and Gly764). The colour of the substrate in d, f, and h is set yellow for clarity. e. Docking result with 178RKRK in the γ ENaC as attractors to the plasmin, showing that two arginine residues Arg178 and Arg180 docked to the plasmin to the similar positions in c. f. The detailed interactions of Arg178 and Arg180 with the plasmin: Arg178 interacting with Asp735 and other residues (Tyr774, Gln738); Arg180 interacting with Glu623, His586. g. Docking result with 135RKRR in the γ ENaC as attractor to the plasmin active centre also resulted in two arginine residues (Arg137 and Arg135) interacting with the plasmin active centre. h. The detailed interactions of Arg137 and Arg135 with the plasmin residues as shown.
Figure S9. Plasmin regulates ENaC activity via G protein. a. ENaC activity in the absence (Control) of two‐chain urokinase plasminogen activator (tc‐uPA, 10 μg・ml‐1, 30 min at room temperature) and plasmin (10 μg・ml‐1, 30 min at room temperature) and presence of protease and Gi/o protein antagonist, Pertussis toxin (PTX, 2 μg・ml‐1, 24 hr prior to addition of proteases at 20oC). n = 12. NS, not significant; *P < 0.05 and **P < 0.01, respectively, correspondingly vs those before application of proteases. b. Normalized ENaC activity. ** P < 0.01 vs that in the absence of PTX. n = 12.
ACKNOWLEDGEMENTS
This study was supported by NIH grants HL87017, HL095435, and HL134828; AHA Award AHA14GRNT20130034 and AHA16GRNT30780002; and NSFC 81670010. The authors thank Ms. Yun Jiang (University of Texas Health Science Centre at Tyler) and Dr. Steven Idell (University of Texas Health Science Centre at Tyler) for their superb technical support and thoughtful discussion. The authors acknowledge Dr. Andrew Lemoff and Dr. Xuemei Luo of the UTSW Proteomics Core for their professional assistance with MS analysis.
Zhao R, Ali G, Nie H‐G, et al. Plasmin improves blood–gas barrier function in oedematous lungs by cleaving epithelial sodium channels. Br J Pharmacol. 2020;177:3091–3106. 10.1111/bph.15038
Present Address Deepa Bhattarai, Department of Paediatric Surgery, McGovern Medical School, University of Texas Health Science Centre at Houston, Texas, USA.
Xuefeng Su, Tasly Pharmaceutical Inc., Rockville, Maryland, USA.
Xiaoli Zhao, Division of Pharmacology, Physiology, and Biological Chemistry, National Institute of General Medical Sciences, Bethesda, Maryland, USA.
Contributor Information
Michael A. Matthay, Email: michael.matthay@ucsf.edu.
Hong‐Long Ji, Email: james.ji@uthct.edu.
REFERENCES
- Alexander, S. P. H. , Christopoulos, A. , Davenport, A. P. , Kelly, E. , Mathie, A. , Peters, J. A. , … Collaborators, C. G. T. P. (2019). The Concise Guide to PHARMACOLOGY 2019/20: G protein‐coupled receptors. British Journal of Pharmacology, 176, S21–S141. 10.1111/bph.14748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … Collaborators, C. G. T. P. (2019). The Concise Guide to PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology, 176, S297–S396. 10.1111/bph.14752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Mathie, A. , Peters, J. A. , Veale, E. L. , Striessnig, J. , Kelly, E. , … Collaborators, C. G. T. P. (2019). The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology, 176, S142–S228. 10.1111/bph.14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura, H. , Ontachi, Y. , Mizutani, T. , Kato, M. , Saito, M. , Kumabashiri, I. , … Nakao, S. (2001). An enhanced fibrinolysis prevents the development of multiple organ failure in disseminated intravascular coagulation in spite of much activation of blood coagulation. Critical Care Medicine, 29, 1164–1168. 10.1097/00003246-200106000-00015 [DOI] [PubMed] [Google Scholar]
- Bertozzi, P. , Astedt, B. , Zenzius, L. , Lynch, K. , LeMaire, F. , Zapol, W. , & Chapman HA Jr (1990). Depressed bronchoalveolar urokinase activity in patients with adult respiratory distress syndrome. The New England Journal of Medicine, 322, 890–897. 10.1056/NEJM199003293221304 [DOI] [PubMed] [Google Scholar]
- Bock, A. , Tucker, N. , Kelher, M. R. , Khan, S. Y. , Gonzalez, E. , Wohlauer, M. , … Silliman, C. C. (2015). α‐enolase causes proinflammatory activation of pulmonary microvascular endothelial cells and primes neutrophils through plasmin activation of protease‐activated receptor 2. Shock, 44, 137–142. 10.1097/SHK.0000000000000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattino, M. D. , Mueller, G. M. , Palmer, L. G. , Frindt, G. , Rued, A. C. , Hughey, R. P. , & Kleyman, T. R. (2014). Prostasin interacts with the epithelial Na+ channel and facilitates cleavage of the γ‐subunit by a second protease. American Journal of Physiology. Renal Physiology, 307, F1080–F1087. 10.1152/ajprenal.00157.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattino, M. D. , Sheng, S. , Bruns, J. B. , Pilewski, J. M. , Hughey, R. P. , & Kleyman, T. R. (2006). The epithelial Na+ channel is inhibited by a peptide derived from proteolytic processing of its α subunit. The Journal of Biological Chemistry, 281, 18901–18907. 10.1074/jbc.M604109200 [DOI] [PubMed] [Google Scholar]
- Carmo, A. A. , Costa, B. R. , Vago, J. P. , de Oliveira, L. C. , Tavares, L. P. , Nogueira, C. R. , et al. (2014). Plasmin induces in vivo monocyte recruitment through protease‐activated receptor‐1‐, MEK/ERK‐, and CCR2‐mediated signaling. Journal of Immunology, 193, 3654–3663. 10.4049/jimmunol.1400334 [DOI] [PubMed] [Google Scholar]
- Castellino, F. J. , & Ploplis, V. A. (2005). Structure and function of the plasminogen/plasmin system. Thrombosis and Haemostasis, 93, 647–654. 10.1160/TH04-12-0842 [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Zhao, R. , Zhao, M. , Liang, X. , Bhattarai, D. , Dhiman, R. , … Ji, H. L. (2014). Regulation of epithelial sodium channels in urokinase plasminogen activator deficiency. American Journal of Physiology. Lung Cellular and Molecular Physiology, 307, L609–L617. 10.1152/ajplung.00126.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaio, L. , Tseng, W. , Balverde, Z. , Alvarez, J. R. , Kim, K. J. , Kelley, D. G. , … Borok, Z. (2009). Characterization of mouse alveolar epithelial cell monolayers. American Journal of Physiology. Lung Cellular and Molecular Physiology, 296, L1051–L1058. 10.1152/ajplung.00021.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Caballero, A. , Dang, Y. , He, H. , & Stutts, M. J. (2008). ENaC proteolytic regulation by channel‐activating protease 2. The Journal of General Physiology, 132, 521–535. 10.1085/jgp.200810030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalia, D. N. , Salisbury, C. M. , Maly, D. J. , Ellman, J. A. , & Diamond, S. L. (2005). Profiling serine protease substrate specificity with solution phase fluorogenic peptide microarrays. Proteomics, 5, 1292–1298. 10.1002/pmic.200401011 [DOI] [PubMed] [Google Scholar]
- Greenlee, M. M. , Mitzelfelt, J. D. , Yu, L. , Yue, Q. , Duke, B. J. , Harrell, C. S. , … Eaton, D. C. (2013). Estradiol activates epithelial sodium channels in rat alveolar cells through the G protein‐coupled estrogen receptor. American Journal of Physiology. Lung Cellular and Molecular Physiology, 305, L878–L889. 10.1152/ajplung.00008.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerteis, S. , Krappitz, M. , Diakov, A. , Krappitz, A. , Rauh, R. , & Korbmacher, C. (2012). Plasmin and chymotrypsin have distinct preferences for channel activating cleavage sites in the γ subunit of the human epithelial sodium channel. The Journal of General Physiology, 140, 375–389. 10.1085/jgp.201110763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerteis, S. , Krueger, B. , Korbmacher, C. , & Rauh, R. (2009). The δ‐subunit of the epithelial sodium channel (ENaC) enhances channel activity and alters proteolytic ENaC activation. The Journal of Biological Chemistry, 284, 29024–29040. 10.1074/jbc.M109.018945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, D. Y. , Nie, H. G. , Gu, X. , Nayak, R. C. , Su, X. F. , Fu, J. , … Ji, H. L. (2010). K+ channel openers restore verapamil‐inhibited lung fluid resolution and transepithelial ion transport. Respiratory Research, 11, 65 10.1186/1465-9921-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardaway, R. M. , Harke, H. , Tyroch, A. H. , Williams, C. H. , Vazquez, Y. , & Krause, G. F. (2001). Treatment of severe acute respiratory distress syndrome: A final report on a phase I study. The American Surgeon, 67, 377–382. [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … NC‐IUPHAR . (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46, D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans, P. W. , Hibberd, M. L. , Booy, R. , Daramola, O. , Hazelzet, J. A. , de Groot, R. , et al. (1999). 4G/5G promoter polymorphism in the plasminogen‐activator‐inhibitor‐1 gene and outcome of meningococcal disease. Meningococcal Research Group. Lancet, 354, 556–560. [DOI] [PubMed] [Google Scholar]
- Hervio, L. S. , Coombs, G. S. , Bergstrom, R. C. , Trivedi, K. , Corey, D. R. , & Madison, E. L. (2000). Negative selectivity and the evolution of protease cascades: The specificity of plasmin for peptide and protein substrates. Chemistry & Biology, 7, 443–453. 10.1016/s1074-5521(00)00125-3 [DOI] [PubMed] [Google Scholar]
- Idell, S. , James, K. K. , & Coalson, J. J. (1992). Fibrinolytic activity in bronchoalveolar lavage of baboons with diffuse alveolar damage: Trends in two forms of lung injury. Critical Care Medicine, 20, 1431–1440. 10.1097/00003246-199210000-00012 [DOI] [PubMed] [Google Scholar]
- Ji, H. L. , & Benos, D. J. (2004). Degenerin sites mediate proton activation of δβγ‐epithelial sodium channel. The Journal of Biological Chemistry, 279, 26939–26947. 10.1074/jbc.M401143200 [DOI] [PubMed] [Google Scholar]
- Ji, H. L. , Parker, S. , Langloh, A. L. , Fuller, C. M. , & Benos, D. J. (2001). Point mutations in the post‐M2 region of human α‐ENaC regulate cation selectivity. American Journal of Physiology. Cell Physiology, 281, C64–C74. 10.1152/ajpcell.2001.281.1.C64 [DOI] [PubMed] [Google Scholar]
- Ji, H. L. , Su, X. F. , Kedar, S. , Li, J. , Barbry, P. , Smith, P. R. , … Benos, D. J. (2006). δ‐subunit confers novel biophysical features to δβγ‐human ENaC via a physical interaction. The Journal of Biological Chemistry, 281, 8233–8241. 10.1074/jbc.M512293200 [DOI] [PubMed] [Google Scholar]
- Ji, H. L. , Zhao, R. , Komissarov, A. A. , Chang, Y. , Liu, Y. , & Matthay, M. A. (2015). Proteolytic regulation of epithelial sodium channels by urokinase plasminogen activator: Cutting edge and cleavage sites. The Journal of Biological Chemistry, 290, 5241–5255. 10.1074/jbc.M114.623496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H. L. , Zhao, R. Z. , Chen, Z. X. , Shetty, S. , Idell, S. , & Matalon, S. (2012). δ ENaC: A novel divergent amiloride‐inhibitable sodium channel. American Journal of Physiology. Lung Cellular and Molecular Physiology, 303, L1013–L1026. 10.1152/ajplung.00206.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovov, B. , Berdiev, B. K. , Fuller, C. M. , Ji, H. L. , & Benos, D. J. (2002). The serine protease trypsin cleaves C termini of β‐ and γ‐subunits of epithelial Na+ channels. The Journal of Biological Chemistry, 277, 4134–4140. 10.1074/jbc.M108354200 [DOI] [PubMed] [Google Scholar]
- Karandashova, S. , Florova, G. , Azghani, A. O. , Komissarov, A. A. , Koenig, K. , Tucker, T. A. , … Idell, S. (2013). Intrapleural adenoviral delivery of human plasminogen activator inhibitor‐1 exacerbates tetracycline‐induced pleural injury in rabbits. American Journal of Respiratory Cell and Molecular Biology, 48, 44–52. 10.1165/rcmb.2012-0183OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Animal research: Reporting in vivo experiments: The ARRIVE guidelines. British Journal of Pharmacology, 160, 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyman, T. R. , Carattino, M. D. , & Hughey, R. P. (2009). ENaC at the cutting edge: Regulation of epithelial sodium channels by proteases. The Journal of Biological Chemistry, 284, 20447–20451. 10.1074/jbc.R800083200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, T. K. , & Chin, N. K. (1999). Empirical treatment with fibrinolysis and early surgery reduces the duration of hospitalization in pleural sepsis. The European Respiratory Journal, 13, 514–518. 10.1183/09031936.99.13351499 [DOI] [PubMed] [Google Scholar]
- Liu, C. , Ma, Y. , Su, Z. , Zhao, R. , Zhao, X. , Nie, H. G. , … Ji, H. L. (2018). Meta‐analysis of preclinical studies of fibrinolytic therapy for acute lung injury. Frontiers in Immunology, 9, 1898 10.3389/fimmu.2018.01898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannaioni, G. , Orr, A. G. , Hamill, C. E. , Yuan, H. , Pedone, K. H. , McCoy, K. L. , … Traynelis, S. F. (2008). Plasmin potentiates synaptic N‐methyl‐D‐aspartate receptor function in hippocampal neurons through activation of protease‐activated receptor‐1. The Journal of Biological Chemistry, 283, 20600–20611. 10.1074/jbc.M803015200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay, M. A. , Folkesson, H. G. , & Clerici, C. (2002). Lung epithelial fluid transport and the resolution of pulmonary edema. Physiological Reviews, 82, 569–600. 10.1152/physrev.00003.2002 [DOI] [PubMed] [Google Scholar]
- Matthay, M. A. , Ware, L. B. , & Zimmerman, G. A. (2012). The acute respiratory distress syndrome. The Journal of Clinical Investigation, 122, 2731–2740. 10.1172/JCI60331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, R. , Han, D. Y. , Su, X. F. , Zhao, R. Z. , Zhao, M. , Sharp, G. M. , … Ji, H. L. (2011). Cpt‐cAMP activates human epithelial sodium channels via relieving self‐inhibition. Biochimica et Biophysica Acta, 1808, 1818–1826. 10.1016/j.bbamem.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase, T. , Uozumi, N. , Ishii, S. , Kume, K. , Izumi, T. , Ouchi, Y. , & Shimizu, T. (2000). Acute lung injury by sepsis and acid aspiration: a key role for cytosolic phospholipase A2. Nature Immunology, 1, 42–46. 10.1038/76897 [DOI] [PubMed] [Google Scholar]
- Noreng, S. , Bharadwaj, A. , Posert, R. , Yoshioka, C. , & Baconguis, I. (2018). Structure of the human epithelial sodium channel by cryo‐electron microscopy. eLife, 7, e39340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passero, C. J. , Mueller, G. M. , Myerburg, M. M. , Carattino, M. D. , Hughey, R. P. , & Kleyman, T. R. (2012). TMPRSS4‐dependent activation of the epithelial sodium channel requires cleavage of the γ‐subunit distal to the furin cleavage site. American Journal of Physiology. Renal Physiology, 302, F1–F8. 10.1152/ajprenal.00330.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passero, C. J. , Mueller, G. M. , Rondon‐Berrios, H. , Tofovic, S. P. , Hughey, R. P. , & Kleyman, T. R. (2008). Plasmin activates epithelial Na+ channels by cleaving the γ subunit. The Journal of Biological Chemistry, 283, 36586–36591. 10.1074/jbc.M805676200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, B. V. , Wilson, M. R. , & Takata, M. (2012). Resolution of acute lung injury and inflammation: A translational mouse model. The European Respiratory Journal, 39, 1162–1170. 10.1183/09031936.00093911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planes, C. , & Caughey, G. H. (2007). Regulation of the epithelial Na+ channel by peptidases. Current Topics in Developmental Biology, 78, 23–46. 10.1016/S0070-2153(06)78002-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinton, T. M. , Kim, S. , Derian, C. K. , Jin, J. , & Kunapuli, S. P. (2004). Plasmin‐mediated activation of platelets occurs by cleavage of protease‐activated receptor 4. The Journal of Biological Chemistry, 279, 18434–18439. 10.1074/jbc.M401431200 [DOI] [PubMed] [Google Scholar]
- Ranieri, V. M. , Rubenfeld, G. D. , Thompson, B. T. , Ferguson, N. D. , Caldwell, E. , Fan, E. , et al. (2012). Acute respiratory distress syndrome: The Berlin Definition. JAMA, 307, 2526–2533. 10.1001/jama.2012.5669 [DOI] [PubMed] [Google Scholar]
- Rossier, B. C. , & Stutts, M. J. (2009). Activation of the epithelial sodium channel (ENaC) by serine proteases. Annual Review of Physiology, 71, 361–379. 10.1146/annurev.physiol.010908.163108 [DOI] [PubMed] [Google Scholar]
- Roy, A. , Kucukural, A. , & Zhang, Y. (2010). I‐TASSER: A unified platform for automated protein structure and function prediction. Nature Protocols, 5, 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori, C. , & Matthay, M. A. (2002). Alveolar epithelial fluid transport in acute lung injury: New insights. The European Respiratory Journal, 20, 1299–1313. 10.1183/09031936.02.00401602 [DOI] [PubMed] [Google Scholar]
- Schaller, J. , & Gerber, S. S. (2011). The plasmin‐antiplasmin system: Structural and functional aspects. Cellular and Molecular Life Sciences, 68, 785–801. 10.1007/s00018-010-0566-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspurten, J. , Gevaert, K. , Declercq, W. , & Vandenabeele, P. (2009). SitePredicting the cleavage of proteinase substrates. Trends in Biochemical Sciences, 34, 319–323. 10.1016/j.tibs.2009.04.001 [DOI] [PubMed] [Google Scholar]
- Ware, L. B. , & Matthay, M. A. (2000). The acute respiratory distress syndrome. The New England Journal of Medicine, 342, 1334–1349. 10.1056/NEJM200005043421806 [DOI] [PubMed] [Google Scholar]
- Wewers, M. D. , Herzyk, D. J. , & Gadek, J. E. (1988). Alveolar fluid neutrophil elastase activity in the adult respiratory distress syndrome is complexed to α‐2‐macroglobulin. The Journal of Clinical Investigation, 82, 1260–1267. 10.1172/JCI113724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk, K. E. , Lazarowski, E. R. , Traylor, Z. P. , Yu, E. N. , Jewell, N. A. , Durbin, R. K. , … Davis, I. C. (2008). Influenza A virus inhibits alveolar fluid clearance in BALB/c mice. American Journal of Respiratory and Critical Care Medicine, 178, 969–976. 10.1164/rccm.200803-455OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R. , Ali, G. , Chang, J. , Komatsu, S. , Tsukasaki, Y. , Nie, H.‐G. , et al. (2019). Proliferative regulation of alveolar epithelial type 2 progenitor cells by human Scnn1d gene. Theranostics, 9, 8155–8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Online supplementary figures are the representative MS/MS spectrum for the peptide fragments in Figure 8.
Figure S8. Homology model and docking results for plasmin cleavage centres. a. 3D model of the human γ ENaC generated by I‐TASSER. b. The GRIP domain (aa114–239) of the human γ ENaC model with three positively charged residue clusters facing outward and highly accessible. Four P strands are composed of following amino acids: P1, aa160–166; P2, aa182–190; P3, aa199–208; and P4, aa213–219. Three cleavage centres were highlighted. c. The interaction of plasmin (chain A with surface presentation) with textilinin‐1 (chain C) from the original crystal structure 3uir highlighted with two arginine residues in the substrate interacting with plasmin. d. Detailed interactions between two arginine residues and plasmin: Arg17 interacting with Asp735, Ser741, and Gly764; Arg19 interacting with Glu687 and Phe587. We used this as a docking guide as attractors (Asp735 and Gly764). The colour of the substrate in d, f, and h is set yellow for clarity. e. Docking result with 178RKRK in the γ ENaC as attractors to the plasmin, showing that two arginine residues Arg178 and Arg180 docked to the plasmin to the similar positions in c. f. The detailed interactions of Arg178 and Arg180 with the plasmin: Arg178 interacting with Asp735 and other residues (Tyr774, Gln738); Arg180 interacting with Glu623, His586. g. Docking result with 135RKRR in the γ ENaC as attractor to the plasmin active centre also resulted in two arginine residues (Arg137 and Arg135) interacting with the plasmin active centre. h. The detailed interactions of Arg137 and Arg135 with the plasmin residues as shown.
Figure S9. Plasmin regulates ENaC activity via G protein. a. ENaC activity in the absence (Control) of two‐chain urokinase plasminogen activator (tc‐uPA, 10 μg・ml‐1, 30 min at room temperature) and plasmin (10 μg・ml‐1, 30 min at room temperature) and presence of protease and Gi/o protein antagonist, Pertussis toxin (PTX, 2 μg・ml‐1, 24 hr prior to addition of proteases at 20oC). n = 12. NS, not significant; *P < 0.05 and **P < 0.01, respectively, correspondingly vs those before application of proteases. b. Normalized ENaC activity. ** P < 0.01 vs that in the absence of PTX. n = 12.


