Abstract
It is critical for the neuronal cell cycle to remain suppressed in terminally differentiated neurons as its activation results in aberrant cell cycle re-entry that causes neuronal apoptosis (CRNA), which has been observed in several neurodegenerative disorders like Alzheimer’s disease (AD). In the present study, we report that E3 ubiquitin ligase Itch is a major regulator of CRNA and elucidated the mechanism via which it is regulated in this process. Neurotoxic amyloid peptide Aβ42-treated neurons or neurons from an AD transgenic mouse model (TgAD) exhibited aberrant activation of the JNK pathway which resulted in the hyperphosphorylation of Itch. The phosphorylation of Itch primes it for autoubiquitination, which is necessary for its activation. These post-translational modifications of Itch facilitate its interaction with TAp73 resulting in its degradation. These series of events are critical for Itch-mediated CRNA and its phosphorylation and autoubiquitination site mutants reversed this process and were neuroprotective. These studies unravel a novel pathway via which neurodegeneration in AD and possibly other related disorders may be regulated by aberrant regulation of the neuronal cell cycle.
Subject terms: Cell death in the nervous system, Neurological disorders
Introduction
It is imperative for the cell cycle of terminally differentiated neurons to remain suppressed in order to maintain their viability. Neurotoxic conditions that include trophic factor withdrawal, encounter with misfolded proteins like beta amyloid peptide Aβ42 and DNA damaging agents are known to trigger aberrant re-entry into the cycle1,2. Instead of resulting in mitosis, it causes neuronal cell death3,4. One of the key features of CRNA is aberrant regulation of important cell cycle proteins which promote apoptosis. A series of events is triggered by neurotoxic insults resulting in an alarming increase in Cyclin D1 causing activation of CDK4/6, retinoblastoma phosphorylation and transcriptional activation of E2F4,5. As a result, neurons exhibit S-phase entry and DNA replication but most of these neurons undergo apoptosis3,6. Aberrant cell cycle re-entry and neuronal loss has been observed in several neurodegenerative disorders like Alzheimer’s disease (AD) in which neuronal loss and aberrant cell cycle re-entry are coincidental7–11.
Studies performed in vitro as well as in animal models of AD suggest that Aβ42 causes cell cycle re-entry and DNA replication that precedes cell death10,12,13. Cyclin D1 plays a critical role in this process as it is an upstream regulator of the cascade14,15 and its expression aberrantly increases and causes S-phase entry of neurons13. Previously, we demonstrated that hyper activation of the MEK-ERK pathway results in enhanced Cyclin D1 production13 and temporal down regulation of microRNA-34a, which targets Cyclin D1, resulted in aberrant increase in its expression12. TAp73 regulates expression of miR-34a which is important for neuronal differentiation and neurite outgrowth16. We demonstrated that TAp73 undergoes ubiquitination in response to Aβ42, which was the cause of its degradation12 and triggered aberrant cell cycle re-entry and apoptosis of neurons.
p53-family transcription factors p73 can be synthesized in at least seven isoforms of TAp73, which are mainly generated by alternative splicing at the 3′-end17. In addition, an alternate promoter and extra exon are used to generate N-terminal truncated versions of the full-length protein (ΔNp73)18. These truncated versions lack the N-terminal transactivation (TA) domain, which can block the function of full-length TAp7319,20. The relative expression of these two isoforms regulates cell fate21,22. Mice lacking either of these isoforms exhibit brain defects that include hippocampal dysgenesis, neurodegeneration, and genomic instability23,24.
We have identified Itch—a Nedd4 family E3 Ubiquitin ligase25,26 as a regulator of CRNA, which it achieves by promoting degradation of TAp73 in neurons. Thus far, Itch has mainly been implicated in chronic inflammation, T-cell response and other immunological functions. Itchy mice that have an inversion in Itch locus exhibit aberrant scratching and immune functions and inflammation27. Itch deficiency causes multi-system autoimmune disease28 and Itch-deficient mice exhibit chronic production of tumorigenic cytokines and exhibited higher propensity for tumor formation26,29,30. Several Itch targets have been identified in immune cells, which include c-jun, E3 ligase Cbl and TAp63, and TAp7325,31–33. Itch facilitates the degradation of p63 and p73 but not p53, which may affect tumor cell response29,30. RASSF5, a downstream effector of Ras involved in cell cycle arrest, is also targeted by Itch34. Itch is regulated by post-translation modifications like phosphorylation32,33, autoubiquitination35,36. Stress-induced JNK activation results in the phosphorylation of Itch in an N-terminal proline-rich region (PRR), which induces a conformational change facilitating its autoubiquitination at specific sites32,33.
Despite these studies, the role of Itch in neuronal development or neurodegeneration has remained almost unknown. In our quest to dissect mechanisms involved in TAp73 degradation during CRNA, we identified Itch as the candidate E3-ligase. We found that Aβ42 activates Itch in neurons by promoting its phosphorylation via the JNK pathway, which further facilitates its autoubiquitination at specific sites. These events prime Itch to interact with TAp73 in TgAD neurons resulting in degradation of the latter. As a result, neurons undergo aberrant cell cycle re-entry and apoptosis. We generated mutants of Itch defective in phosphorylation and autoubiquitination, which reversed the CRNA of TgAD neurons by preventing TAp73 degradation.
Results
Itch regulates the degradation of TAp73 mediated by Aβ42
TAp73 is critical for neuronal differentiation and survival16,37. Previously, we demonstrated that it is ubiquitinated and degraded in neurons upon treatment with neurotoxic Aβ4212, which promoted cell cycle re-entry and apoptosis. In order to get deeper insights into the underlying mechanisms, it was pertinent to identify the E3 Ub-ligase is involved in TAp73 degradation.
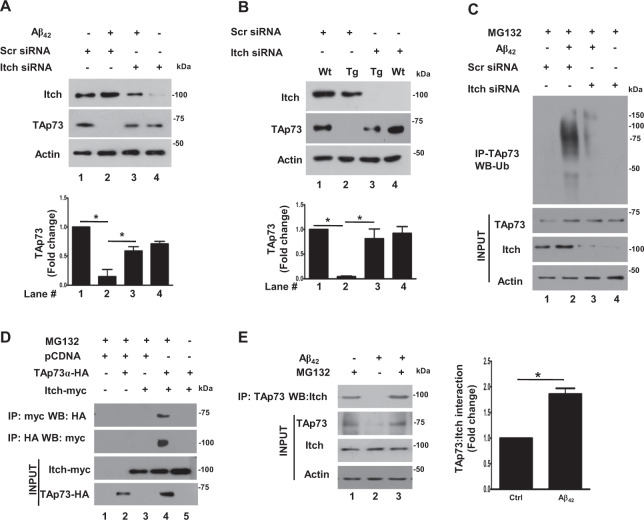
Previous studies performed in non-neuronal HEK293 or HeLa cells indicated that HECT-family ligase Itch is a major mediator of p73 degradation30. In order to explore if Itch is involved in TAp73 degradation in neurons in response to Aβ42, cortical neurons were treated with Aβ42 for 48 h as described previously12 in the presence or absence of siRNA against Itch. There was no major change in TAp73 under steady-state conditions. While TAp73 was degraded in response to Aβ42, Itch depletion almost completely reversed this process and TAp73 expression was retained (Fig. 1a). Similar experiments were also performed on cortical neurons derived from a mouse model for AD (TgAD) that overexpress mutant forms of amyloid precursor protein (APP) and Presnillin1 (PS1) associated with AD38,39, which results in enhanced Aβ42 production and CRNA has been observed in these animals12. The expression of TAp73 was barely detectable in cortical neurons from TgAD animals in contrast to WT. Strikingly, Itch siRNA, but not a control scrambled siRNA, caused a significant increase in TAp73 (Fig. 1b). The status of TAp73 ubiquitination was assessed by performing IP followed by Western blotting for ubiquitin. As reported previously12, Aβ42 caused TAp73 ubiquitination, which was markedly suppressed upon Itch depletion (Fig. 1c). Collectively, these results indicated that Itch mediates the ubiquitination and degradation of TAp73 in response to Aβ42. Next, the association of Itch with TAp73 was tested in neuronal cells, which has not been established as yet. To this end, TAp73 and Itch were overexpressed in neuronal PC12 cells followed by immunoprecipitation (IP). TAp73, co-immunoprecipitated with Itch and vice versa indicating that they interact with each other (Fig. 1d). The effect of Aβ42 on their association was tested in cortical neurons by performing co-IP experiments. While endogenous Itch and TAp73 interacted in untreated neurons, there was a significant increase upon Aβ42 treatment (Fig. 1e). These results explained enhanced degradation of TAp73 in an Itch-dependent manner (Fig. 1a) and established that Aβ42 promotes association of Itch with TAp73 leading to its ubiquitination and degradation.
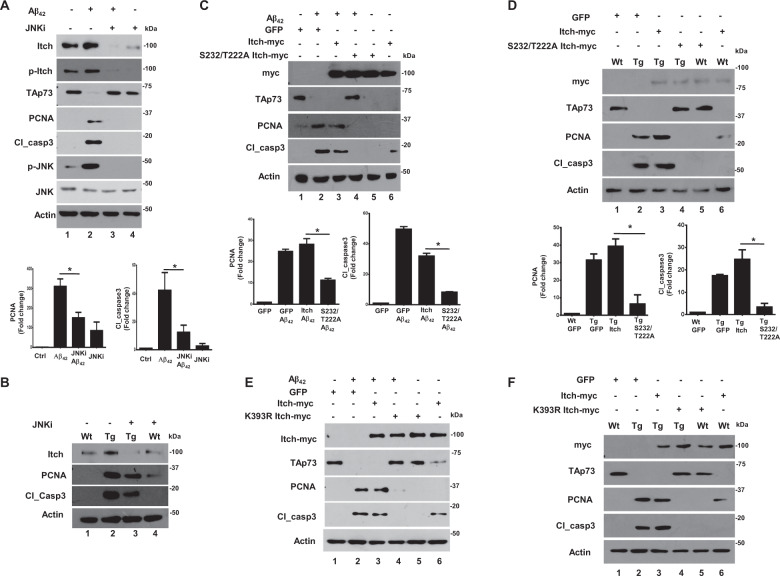
Fig. 1. Itch regulates Aβ42-mediated degradation of TAp73.
a Rat cortical neurons, which were transfected with siRNA against Itch or a scrambled siRNA, scr_siRNA (control) and were treated with Aβ42 for 48 h or left untreated. Subsequently, Western blotting was performed for assessing the expression of TAp73, Itch, or Actin (loading control). Itch depletion significantly prevented TAp73 degradation, which was quantified by densitometry of TAp73 bands (bottom panel), which was normalized with respect to Actin (mean ± SEM, ANOVA, p < 0.05, N = 3). b Cortical neurons were cultured from wild type (WT) or APP/PS1 transgenic (Tg) mice and transfected with Itch siRNA or scrambled scr_siRNA (control). After 48 h, cell lysate was prepared and Western blotting was performed for TAp73, Itch or Actin. Itch depletion significantly prevented TAp73 degradation that was quantified by densitometry of TAp73 bands, which was normalized with respect to Actin (mean ± SEM, ANOVA, p < 0.05, N = 3). c Rat cortical neurons, which were transfected with siRNA against Itch or a scrambled siRNA (control), were left untreated or treated with Aβ42 for 48 h in the presence of MG132. TAp73 was immunoprecipitated and the IP was used for Western blotting with anti-ubiquitin antibody. d Neuronal PC12 cells were transfected with expression plasmids for the overexpression of TAp73-HA or Itch-Myc or control vector (pcDNA) in presence or absence of MG132. After 48 h, cell lysates were prepared and used for immunoprecipitation using anti-HA or anti-Myc antibodies. HA-IP and Myc-IP were immunoblotted with anti-myc and anti-HA, respectively. Immunoblotting was also performed on total cell lysates (input). e Rat cortical neurons were treated with Aβ42 for 48 h in the presence or absence of proteasome inhibitor MG132. Subsequently, TAp73 was immunoprecipitated followed by Western blotting for Itch. Cell lysates were also immunoblotted to detect expression of these proteins. There was a significant increase in Itch co-immunoprecipitated with TAp73 upon Aβ42 treatment, which was also quantified by densitometry of bands corresponding to Itch (right panel, mean ± SEM, *p < 0.05, t-test, N = 3).
Itch promotes cell cycle re-entry and apoptosis of neurons
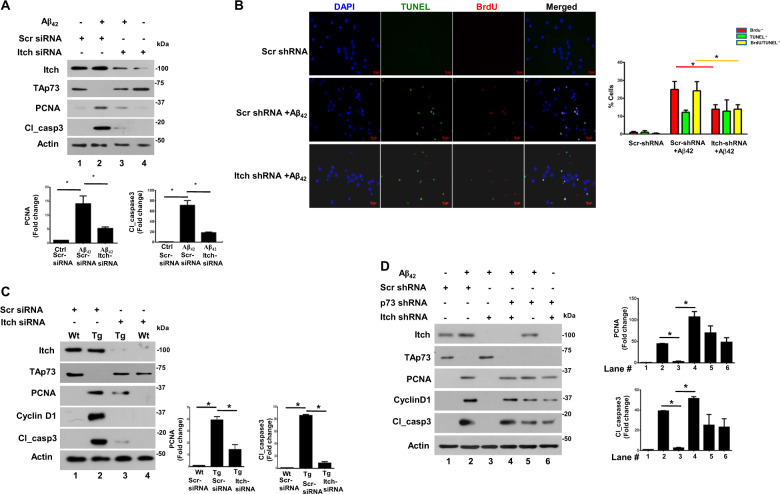
Next, we evaluated the role of Itch in Aβ42-induced cell cycle re-entry and apoptosis of neurons. We used two previously reported approaches for this purpose: levels of PCNA that suggest S-phase entry and cleaved caspase 3 (cl_caspase3), which represents the active form of caspase 3 and is indicative of apoptosis, were determined by Western blotting; BrdU incorporation and TUNEL labeling was performed to detect DNA replication and apoptosis at the single cell level, respectively12,13. PCNA is present at very low or almost undetectable levels in cortical neurons from E18 embryos suggesting that these cells are terminally differentiated and have exited the cell cycle (Fig. 2a). While Aβ42 treatment resulted in a significant increase in both these proteins, which was indicative of CRNA as reported previously12. The knockdown of Itch significantly reduced both PCNA and cl_casapse3 suggesting that it regulates CRNA (Fig. 2a). A significant increase in BrdU+/TUNEL+ neurons was observed upon Aβ42 treatment and most BrdU+ neurons were also TUNEL+, which indicated that those neurons that re-entered the cell cycle also underwent apoptosis and there was a population, which was only TUNEL+ but did not exhibit BrdU incorporation, that represented “conventional” apoptosis independent of cell cycle re-entry as described previously12,13. However, the knockdown of Itch caused a significant decrease in BrdU+/TUNEL+ cells whereas TUNEL+ cells were almost unaltered (Fig. 2b) suggesting that mainly those neurons that underwent apoptosis as a result of cell cycle re-entry were rescued by Itch depletion. These data suggested that Aβ42-mediated Itch regulation results in aberrant cell cycle re-entry and neuronal apoptosis. Next, studies were also carried out in neurons from TgAD animals. The knockdown of Itch in neurons from TgAD caused a significant decrease in PCNA and cleaved caspase3, which were elevated in these cells in comparison to WT neurons (Fig. 2c).
Fig. 2. Itch regulates cell cycle-related neuronal apoptosis (CRNA).
a Rat cortical neurons were transfected with siRNA against Itch or a scrambled control followed by treatment with Aβ42 for 48 h. Subsequently, Western blotting was performed for assessing the expression of indicated proteins. Itch depletion significantly prevented TAp73 degradation and blocked the increase in PCNA and cl_caspase3. Bottom panel: Densitometry of PCNA and cl_caspase 3 bands was performed, and their level was normalized with respect to Actin. Fold change in these proteins with respect to untreated neurons transfected with control siRNA was determined (mean ± SEM, *p < 0.05 by ANOVA, N = 3). b Rat cortical neurons were transfected with Itch shRNA or control scr_shRNA and treated with Aβ42 followed by incubation with BrdU. Immunofluorescence and TUNEL assay were performed to detect BrdU incorporation (red) or apoptosis (green), respectively. Right panel, % cells that were BrdU+ (red), TUNEL+ (green), or were BrdU+/TUNEL+ (yellow) were determined (mean ± SEM, *p < 0.05, t-test, N = 3). c Cortical neurons were cultured from wild type (WT) or APP/PS1 Transgenic (Tg) mice and transfected with Itch siRNA or scrambled scr_siRNA (control). After 48 h, cell lysate was prepared and Western blotting was performed for indicated proteins. Right panel, Densitometry of PCNA and cl_caspase 3 bands was performed, and their expression was normalized with respect to actin. Fold change in these proteins with respect to WT neurons transfected with control siRNA was determined (mean ± SEM, *p < 0.01 by ANOVA, N = 3). d Rat cortical neurons were transfected with shRNA against Itch or a scrambled control. In addition, in some cases shRNA against TAp73 was also transduced using lentivirus. Following treatment with Aβ42 for 48 h, Western blotting was performed for assessing the expression of indicated proteins. Itch depletion significantly prevented TAp73 degradation and blocked increase in PCNA and cl_caspase3. TAp73 knockdown in Itch-depleted cells restored cl_caspase3 and PCNA expression. Right panel, Densitometry of PCNA and cl_caspase3 bands was performed, and their expression was normalized with respect to Actin. Fold change in these proteins with respect to scr-shRNA-transfected cells was determined (mean ± SEM, *p < 0.05, t-test, N = 3).
As reported above (Fig. 1a), Itch is involved in TAp73 degradation in response to Aβ42. Therefore, in order to investigate if Itch-mediated degradation of TAp73 caused this process; TAp73 was knocked down in combination with Itch depletion. Itch shRNA prevented the TAp73 degradation and CRNA as evidenced by suppressed PCNA and cleaved caspase3 levels (Fig. 2d lane 3 vs. lane 2). Simultaneous addition of TAp73 shRNA reversed this process as levels of both these proteins increased (lane 4 vs. lane 3). Based on these results, it is reasonable to state that Itch can cause CRNA by promoting degradation of TAp73.
Phosphorylation of Itch by the JNK pathway is critical for its autoubiquitination and interaction with TAp73
Next, we sought the mechanisms via which Itch is regulated in AD models. We did not find a major increase in the expression of Itch mRNA upon Aβ42 treatment of cortical neurons or in TgAD mouse neurons (Supplementary Fig. S1). There was only a modest increase in Itch protein levels (Figs. 1a and 2a, c and d). Therefore, we speculated the role of post-translational modifications in regulation of Itch as previous reports had indicated that it can be regulated by post-translational events like autoubiquitination and phosphorylation32,33,40.
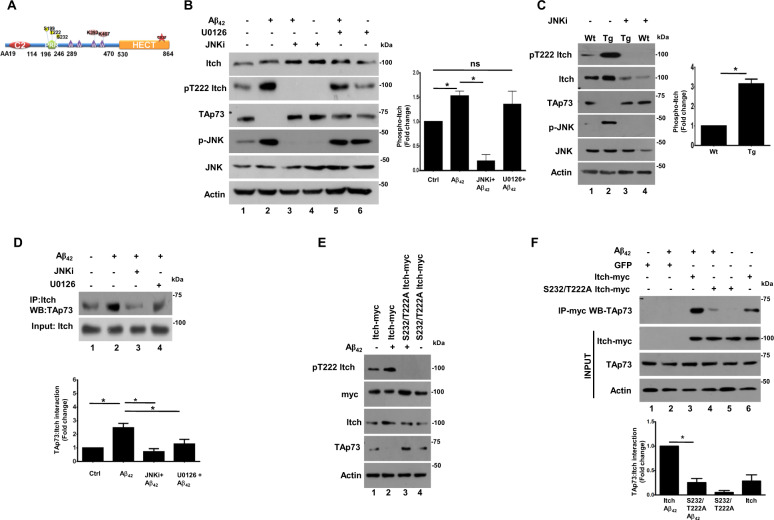
Previous reports also indicated that a PRR of Itch (Fig. 3a) is susceptible to phosphorylation by kinases like JNK at S199, T222, S232 which in turn is critical for its activation32,33. These sites are proline directed (SP/TP) which are typically targeted by MAP kinases, we speculated a role of MAP kinases like JNK and previous studies have shown that in immune cells JNK can phosphorylate these sites32. Therefore, first the role of JNK and ERK in phosphorylation of T222 was evaluated using a specific phospho antibody, which was commercially available. While Itch was phosphorylated at this site in neurons in steady-state conditions, Aβ42 caused a significant increase in phosphorylation of T222 (Fig. 3b, lane 1 vs. lane 2). The addition of JNK inhibitor SP600125 (JNKi) caused a dramatic decrease in the phosphorylation of Itch at T222 (lane 3) whereas MEK-ERK inhibitor U0126 did not cause much change (lane 5). Importantly, JNKi also prevented TAp73 degradation by Aβ42 suggesting Itch phosphorylation at this site by JNK in response to Aβ42. The phosphorylation of Itch at T222 was also higher in neurons derived from TgAD mice; JNKi suppressed the phosphorylation and also prevented TAp73 degradation (Fig. 3c). Further, the role of JNK-induced phosphorylation of Itch and TAp73 interaction was assessed by co-immunoprecipitation (co-IP), which revealed that Aβ42-induced interaction of Itch and TAp73 was significantly attenuated by JNK inhibition in both Aβ42-treated cortical neurons (Fig. 3d) as well as neurons from TgAD animals (not shown here). JNKi also prevented interaction between JNK and Itch (Supplementary Fig. S4B). These data indicated that JNK pathway triggers phosphorylation of Itch which promotes interaction with TAp73. As mentioned above, we have previously demonstrated that aberrant activation of the MEK-ERK pathway in TgAD or Aβ42-treated neurons results in TAp73 ubiquitination-degradation12, which was also observed in present studies (Fig. 3b, lane 5 vs. lane 2). Therefore, we tested if this pathway influences TAp73–Itch interaction and co-IP in the presence of U0126 revealed a significant decrease in interaction of these proteins in Aβ42-treated neurons (Fig. 3d, lane 4 vs. lane 2). Since MEK-ERK pathway does not seem to contribute to Itch phosphorylation at T222 (Fig. 3b), which is critical for its activation (see below), it is likely that this pathway influences phosphorylation of TAp73 to promote its interaction with Itch, although it needs experimental demonstration.
Fig. 3. Aβ42-induced phosphorylation of Itch via JNK is critical for its interaction with TAp73.
a Schematic illustrating domain architecture of Itch. In addition to HECT and WW domains, Itch has a proline-rich region (PRR), which is susceptible to phosphorylation at S199, T222, S232. The lysines K383 and K407 implicated in auto-ubiquitination and catalytic C832 are also indicated. b Rat cortical neurons were treated with Aβ42 for 48 h in the presence or absence of JNK inhibitor (JNKi) or MEK inhibitor (U0126). Cell lysates were prepared and immunoblotted with antibodies specific to Itch phosphorylated at T222, TAp73, and against total JNK and Itch. The level of Itch phosphorylation was quantitated by performing densitometry of phospho-Itch band, which was normalized with respect to total Itch and fold change in phospho-Itch was determined (right panel, mean ± SEM, *p < 0.05, t-test, N = 4). c WT and TgAD mouse cortical neurons were treated with JNK inhibitor (JNKi) for 48 h. Western blotting was performed for phospho-Itch, phospho-JNK and TAp73 as described in panel b. Actin was used as a loading control. The level of Itch phosphorylation was quantitated by performing densitometry of phospho-Itch band as described in panel b (right panel, mean ± SEM, *p < 0.05, t-test, N = 3). d Rat cortical neurons were treated with Aβ42 for 48 h in the presence or absence of JNKi or U0126 in the presence of MG132. Subsequently, Itch was immunoprecipitated followed by Western blotting for TAp73. Cell lysates were also immunoblotted to determine levels of Itch. The increase in Itch co-immunoprecipitated with TAp73 upon Aβ42 treatment was significantly reduced upon JNKi or U0126 treatment, which was quantified by densitometry of bands corresponding to TAp73 in Itch-IP (bottom panel, mean ± SEM, *p < 0.05, t-test, N = 3). e Rat cortical neurons were infected with adenovirus to overexpress Itch or its S232/T222A mutant or GFP (control). Subsequently, neurons were treated with Aβ42 for 48 h and Western blotting was performed by using antibodies against indicated proteins. S232/T222A mutant was not phosphorylated and it prevented the degradation of TAp73 in Aβ42-treated cells. f Rat cortical neurons were infected with adenovirus to express Itch or its S232/T222A mutant or GFP (control) in the presence of MG132. Subsequently, Aβ42 treatment was given for 48 h followed by immunoprecipitation using anti-myc antibody to IP Itch followed by Western blotting for TAp73. The interaction of S232/T222A mutant was significantly reduced to TAp73, which was also quantified by densitometry of bands corresponding to TAp73 in Itch-IP (bottom panel, mean ± SEM, *p < 0.05, t-test, N = 3).
To further investigate the role of Itch phosphorylation sites, T222 and a neighboring proline-directed site S232, which is also a putative JNK target was mutated to A and this phosphodeficient mutant (T222A/S232A) was overexpressed in cortical neurons using adenovirus. Expectedly, the phospho-T222-Itch antibody did not recognize the T222A/S232A mutant (Fig. 3e). Interestingly, the TAp73 degradation, which was observed in Aβ42-treated WT Itch overexpressing neurons (Fig. 3e, lane 2) was almost completely reversed in Itch phospho-deficient mutant overexpressing cells (lane 3). The role of S232/T222 phosphorylation in Itch–TAp73 interaction was also investigated. The interaction of Itch with TAp73 in the presence of Aβ42 was significantly impaired upon mutation of T222A/S232A (Fig. 3f). Similar results were obtained in TgAD neurons as both Itch phosphorylation and TAp73 degradation were attenuated upon overexpression of this phospho-deficient mutant (Supplementary Fig. S3). Collectively, these data established that phosphorylation of Itch at T222 and S232 is critical for TAp73 degradation.
The phosphorylation of Itch promotes its autoubiquitination
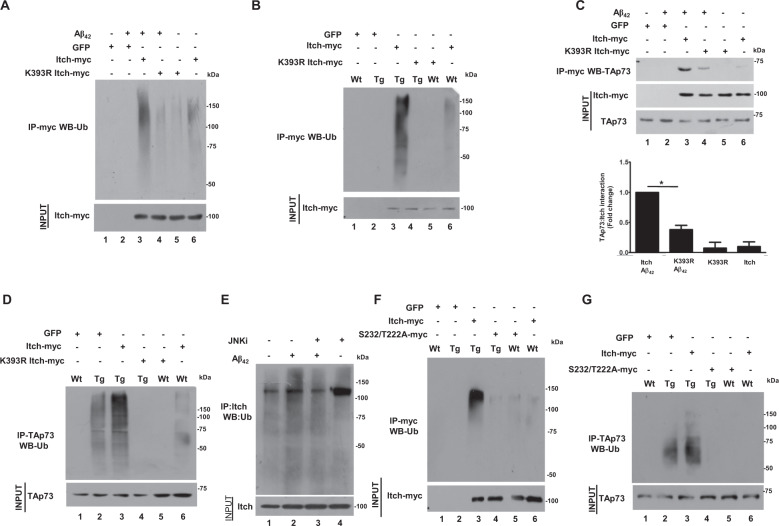
Previous studies have indicated that Itch needs to be autoubiquitinated to interact with some of its targets32,33,41. However, it is unknown if autoubiquitination is needed for interaction with TAp73 and also sites for autoubiquitination that are responsible for substrate interaction have not been established. Previous proteomics studies have revealed that Itch is ubiquitinated at several sites and most commonly at K393 and K40742. Therefore, K393 and K407 mutations were generated and first overexpressed in neuronal PC12 cells. In addition, the catalytic cysteine (C832) which is critical for ubiquitin-ligase activity was also mutated, which resulted in complete abrogation of Itch autoubiquitination (Supplementary Fig. S2A). While there was a dramatic decrease in autoubiquitination of K393R mutant of Itch upon Aβ42 treatment, K407R did not reveal a significant change (Supplementary Fig. S2A). These results indicated that K393 may be the major site for autoubiquitination and critical for TAp73 interaction. Therefore, for further studies, adenovirus was prepared to overexpress K393R mutant in neurons. Aβ42 caused a dramatic increase in Itch autoubiquitination, which was significantly reduced in K393R mutant (Fig. 4a). Itch autoubiquitination was observed in TgAD neurons and was impaired upon K393R mutation (Fig. 4b). We also assessed if autoubiquitination of Itch influences its interaction with TAp73 by performing co-IP, which revealed a significant reduction in Itch–TAp73 interaction upon K393R mutation in response to Aβ42 (Fig. 4c). Similar results were obtained in TgAD neurons as K393R-Itch exhibited reduced interaction with TAp73 (Supplementary Fig. S5). Importantly, TAp73 ubiquitination, which is stimulated in TgAD (Fig. 4d) or Aβ42-treated neurons by Itch was significantly reduced by the K393R mutant (Supplementary Fig. S6). These results established that Itch autoubiquitination promotes TAp73 interaction under neurotoxic conditions, which facilitates degradation of the later.
Fig. 4. Aβ42 promotes autoubiquitination of Itch, which is dependent on its phosphorylation and is critical for interaction with TAp73.
a Rat cortical neurons were infected with adenovirus to express Itch or its K393R mutant or GFP (control) in the presence of MG132. Subsequently, neurons were treated with Aβ42 for 48 h followed by immunoprecipitation using anti-myc antibody to IP Itch, which was immunoblotted for ubiquitin. A significant decrease in ubiquitination of the K393R mutant was observed. b Neurons from WT or TgAD mice were infected with adenovirus to express Itch or K393R mutant or GFP (control) in the presence of MG132. Subsequently, immunoprecipitation was performed using anti-myc antibody to IP Itch, which was immunoblotted for ubiquitin. A significant decrease in ubiquitination of the K393R mutant was observed in TgAD neurons. c Rat cortical neurons were infected with adenovirus to express Itch or K393R mutant or GFP (control) in the presence of MG132. Subsequently, neurons were treated with Aβ42 for 48 h followed by immunoprecipitation using anti-myc antibody to IP Itch followed by Western blotting for TAp73. Densitometry of bands corresponding to TAp73 in Itch-IP was performed for quantitation of interaction and fold change was determined with respect to wild type Itch transduced neurons (bottom panel, mean ± SEM, *p < 0.01, t-test, N = 3). d Neurons from WT or TgAD mice were infected with adenovirus to express Itch or K393R mutant or GFP (control) in the presence of MG132 as described in panel a. Immunoprecipitation was performed using TAp73 antibody, which was immunoblotted for ubiquitin. A significant decrease in ubiquitination of TAp73 was observed in K393R mutant expressing TgAD neurons. Since the same cell lysate was used for co-immunoprecipitation of Itch-myc and TAp73 described in Supplementary Fig. S5A, the input TAp73 Western blot here has also been used for Supplementary Fig. S5A. e Rat cortical neurons were treated with Aβ42 for 48 h in the presence or absence of JNK inhibitor followed by immunoprecipitation using anti-Itch antibody, Itch-IP was immunoblotted for ubiquitin. A significant decrease in ubiquitination in JNKi-treated cells was observed. f Neurons from WT or TgAD mice were infected with adenovirus to express Itch or its S232/T222A mutant or GFP (control) in presence of MG132. Anti-myc antibody was used to IP Itch-myc, which was immunoblotted for ubiquitin. A significant decrease in ubiquitination of the mutant was observed in TgAD neurons. g Neurons from WT or TgAD mice were infected with adenovirus to express Itch or S232/T222A mutant or GFP (control) in the presence of MG132. Subsequently, immunoprecipitation was performed using anti-TAp73 antibody, which was immunoblotted for ubiquitin. A significant decrease in ubiquitination of TAp73 with mutant overexpression was observed in TgAD neurons.
Given that both phosphorylation and autoubiquitination of Itch regulates its interaction with TAp73, we probed if there is a link between these two processes. First, the role of JNK pathway, which regulates Itch phosphorylation, in autoubiquitination was tested. Aβ42 induced the autoubiquitination of Itch was significantly reduced in the presence of JNK inhibitor (Fig. 4e). Similarly, treatment of neurons from TgAD animals that exhibited enhanced Itch autoubiquitination was significantly reduced by JNK inhibitor (Supplementary Fig. S4A). Next, the ubiquitination of phospho-deficient mutants was compared to WT Itch. While WT Itch autoubiquitination was significantly enhanced upon Aβ42 treatment of cortical neurons (Supplementary Fig. S7A) or TgAD neurons (Fig. 4f), a marked reduction was observed when S232/T222 was mutated to alanine. Concurrently, a significant reversal of TAp73 ubiquitination was also observed in TgAD neurons by this mutant (Fig. 4g), which was also the case in Aβ42-treated neurons (Supplementary Fig. S7B). Collectively these studies revealed that the ubiquitination of Itch, which is critical for its interaction with TAp73, is regulated by JNK-mediated phosphorylation of S232/T222. Upon autoubiquitination, Itch interacts with TAp73 resulting in its ubiquitination and degradation.
Itch phosphorylation and ubiquitination promotes CRNA
Having established that Itch is regulated by phosphorylation and autoubiquitination by Aβ42, it was pertinent to study if these events are critical for CRNA. Since Itch phosphorylation at T222/S232 was mediated by the JNK pathway (Fig. 3b, c), we first tested if this pathway is critical for CRNA. Aβ42 treatment resulted in aberrant activation of the JNK pathway, which correlated with high PCNA and cl_caspase3 expression. The treatment with JNKi caused a significant reduction in these proteins, which indicated reversal in CRNA and corroborated well with reduced phosphorylation of Itch (Fig. 5a). Similar experiments were also performed on cortical neurons derived from TgAD animals, which expressed higher PCNA and cl_caspase3 in comparison to WT neurons. The inhibition of JNK also caused a significant reversal of CRNA in TgAD cells (Fig. 5b). These results confirmed that Aβ42-induced aberrant activation of JNK pathway causes Itch hyperphosphorylation and CRNA. Given that this phosphorylation occurs at T222 and S232, we next tested direct effects of the phosphorylation on CRNA by using S232/T222A mutant. The overexpression of WT Itch and/or Aβ42 treatment causes significant degradation of TAp73 (Fig. 5c, lane 1 vs. lanes 2 and 3) accompanied by CRNA as indicated by increase in cl_caspase3 and PCNA expression. The S232/T222A mutant not only prevented TAp73 degradation but also significantly reverted CRNA (lane 4). Similar observations were obtained in TgAD mouse neurons as significant reversal in CRNA (Fig. 5d, lane 4 vs. lane 3) was caused by the S232/T222A mutant.
Fig. 5. Post-translational modifications of Itch promotes CRNA.
a Rat cortical neurons were treated with Aβ42 for 48 h in the presence or absence of JNKi followed by Western blotting to detect indicated proteins. JNKi prevented PCNA and cl_caspase3 expression, which was quantitated by densitometry (bottom panel, mean ± SEM, *p < 0.05, t-test, N = 3). b Cortical neurons from WT and TgAD animals were treated with JNKi for 48 h followed by Western blotting. A significant decrease in cl_caspase3 and PCNA was observed in JNKi-treated TgAD neurons. c Rat cortical neurons were infected with adenovirus to express Itch or its S232/T222A mutant or GFP (control). Subsequently, neurons were treated with Aβ42 for 48 h followed by Western blotting for indicated proteins. The levels of PCNA and cl_caspase3 were quantified by densitometry (bottom panel, mean ± SEM, *p < 0.05, ANOVA, N = 3). d Cortical neurons from WT and TgAD animals were infected with adenovirus to express Itch, S232/T222A mutant, or GFP. Subsequently, Western blotting was performed for indicated proteins. Mutant overexpression prevented cl_caspase3 and PCNA expression in TgAD neurons. The levels of PCNA and cl_caspase3 were quantified by densitometry (bottom panel, mean ± SEM, *p < 0.05, ANOVA, N = 3). e Rat cortical neurons were infected with adenovirus to express Itch or its K393R mutant or GFP (control). Subsequently, neurons were treated with Aβ42 for 48 h followed by Western blotting for indicated proteins. The levels of PCNA and cl_caspase3 were quantified by densitometry (Supplementary Fig. S8A). f Cortical neurons from WT and TgAD animals were infected with adenovirus to express Itch or its K393R mutant or only GFP. Subsequently, Western blotting was performed for indicated proteins. K393R-Itch overexpression prevented cl_caspase3 and PCNA expression in TgAD neurons. The levels of PCNA and cl_caspase3 were quantified by densitometry (Suppementary Fig. S8B).
Next, the role of Itch autoubiquitination at K393, which is critical for interaction with TAp73 on CRNA was tested. To this end, K393R mutant was used which when overexpressed in cortical neurons prevented TAp73 ubiquitination (Fig. 4d) and degradation (Fig. 5e). Strikingly, this mutant also dramatically reduced PCNA and cl_caspase3 in Aβ42-treated cells (Fig. 5e, Supplementary Fig. S8A). These findings were further established in experiments performed on TgAD neurons: K393R mutant almost completely abolished CRNA in these cells (Fig. 5f, lane 4 vs. lane 3 and 2, Supplementary Fig. S8B). Similar results were obtained in neuronal PC12 cells (Supplementary Fig. S2B).
Present studies delineate a novel pathway triggered by neurotoxic amyloid peptide Aβ42, which regulates post-translational regulation of E3 ligase Itch. Aberrant activation of the JNK pathway causes hyperphosphorylation of Itch at S232/T222, which possibly causes a conformational change conducive for its autoubiquitination at K393. As a result, these modifications of Itch promote its interaction with substrates like TAp73 and facilitate its degradation. As a consequence of these events, neurons re-enter the cell cycle and undergo apoptosis, which may contribute to AD pathology (Fig. 6).
Fig. 6. Regulation of Itch in Aβ42-induced CRNA.
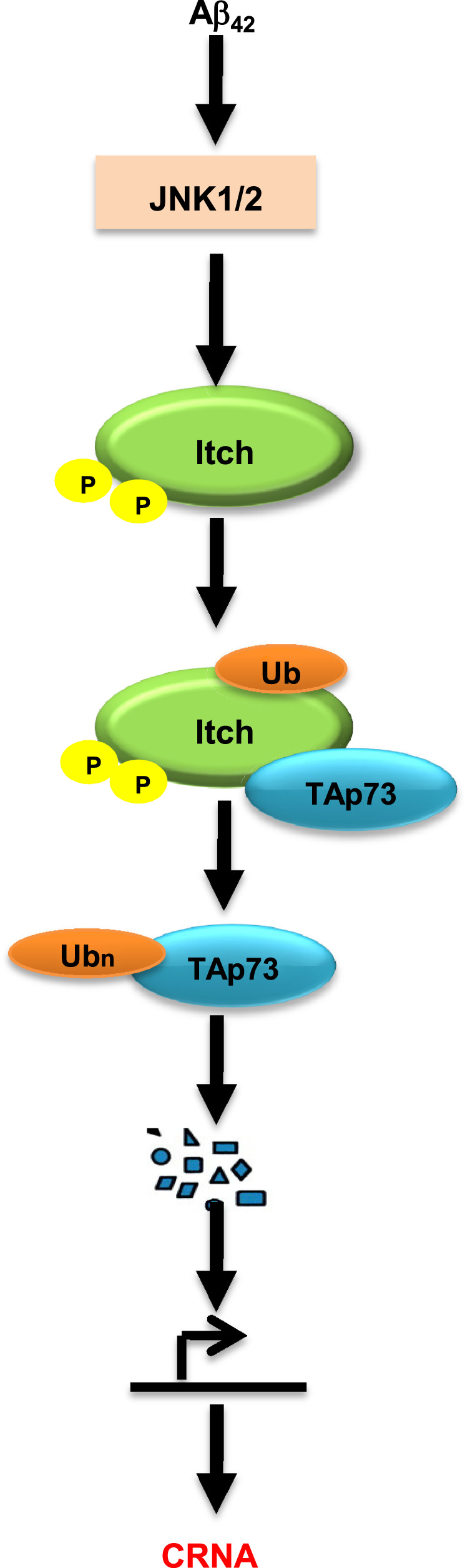
Aβ42 stimulates aberrant activation of the JNK pathway, which results in hyperphosphorylation of Itch at T222 and S232 by JNK (Fig. 3). The phosphorylation at these sites promotes its autoubiquitination of Itch at K393 (Fig. 4e and f). These post-translational modifications induce TAp73–Itch interaction (Figs. 3d, e and 4c), which results in TAp73 ubiquitination (Fig. 4d, g) and degradation (Fig. 5c–f) and may affect the transcription of TAp73 target genes. Collectively, these events result in cell cycle re-entry and neuronal apoptosis (CRNA; Figs. 2 and 5).
Discussion
Terminally differentiated neurons exit the cell cycle and remain in this state for the remainder of their life, which is critical to maintain their differentiated or arrested state. The reactivation of cell cycle in response to Αβ42 results in their apoptosis. We have demonstrated that the aberrant activation of E3 ubiquitin ligase, Itch deregulates the neuronal cell cycle (Fig. 2), which is expressed in neurons in the steady-state conditions. Aβ42 does not cause a significant change in its transcripts (Supplementary Fig. S1) and only a modest change at the protein level (Fig. 1a, b). Its depletion under physiological or steady-state conditions did not cause any neuronal apoptosis and activation of the cell cycle (Fig. 2a, c). These observations seem to suggest that Itch may not have a major physiological function in neurons at least in neuronal survival. In addition, studies on Itchy mice (Itch−/−) did not reveal any defects in brain development27. There is only one report in which over expression of constitutively active Itch mutants in the brain resulted in enhanced neuronal migration36.
Itch is a multi-domain protein with a WW domain, a C2 domain and a PRR in addition to its catalytic HECT domain (Fig. 3a). The WW domain is typically critical for interaction with other proteins43 and present studies indicate that the post-translational regulation of Itch in WW domain and the PRR via autoubiquitination and phosphorylation aberrantly activates it in neurons. We established that indeed Itch exhibits significantly enhanced interaction with TAp73 in the presence of Aβ42. A previous study also shows that Itch and TAp73 interact which leads to degradation of the later but these studies were performed in cancer and dividing cells and Itch regulation by post-translational modifications was not studied in this report30. While a PRR of TAp73 was implicated in their interaction, the underlying mechanism by which Itch regulates TAp73 degradation had remained almost unknown. Itch is regulated by phosphorylation in dividing cells: For instance, ATM phosphorylates it at S161 which causes degradation of c-FLIP-L and c-Jun44; phosphorylation at Y371 within the Itch WW domain induced by Src kinase Fyn upon T-cell receptor (TCR) stimulation alters the binding affinity between Itch and JunB and reduces JunB degradation45. In addition, JNK1 phosphorylates it at three sites (S199, T222, and S232) within PRR in T-cells32, leading to conformational changes that disrupt the self-inhibitory intramolecular interaction between the WW and the HECT domains and increase the catalytic activity of Itch.
Our studies demonstrate that Aβ42-induced JNK pathway activation causes aberrant phosphorylation of Itch at S232 and T222, which enhances its interaction with TAp73. While aberrant activation of the JNK pathway contributes to neuronal apoptosis in response to Aβ4246–48, its regulation of cell cycle machinery has remained largely unknown. We found that JNK pathway contributes to the process of CRNA, as its inhibition reverts this process by preventing phosphorylation of Itch at S232/T222. Interestingly, closely related MEK-ERK pathway did not seem to influence Itch phosphorylation at T222 (Fig. 3b). However, this pathway contributed to the interaction of TAp73 with Itch (Fig. 3d). It is reasonable to suggest that it may directly or indirectly promote phosphorylation of TAp73, which may contribute to its ubiquitination by Itch. MEK-ERK pathway was previously reported to cause TAp73 ubiquitination and degradation12. Therefore, it will be interesting to study if TAp73 phosphorylation also contributes to its interaction with Itch in response to Aβ42.
We also found that the inhibition of the JNK pathway prevented autoubiquitination of Itch, which is significantly enhanced in response to Aβ42 (Fig. 4e). Furthermore, phospho-deficient mutant S232/T222A exhibited dramatic reduction in autoubiquitination (Fig. 4f). The phosphorylation of the PRR region has been suggested to introduce conformational changes that may release Itch from its auto-inhibited state32. Our present findings corroborate well with these studies as autoubiquitination was dependent on the phosphorylation of this region. K393 was identified as the primary site as its mutation almost completely abolished autoubiquitination. Importantly, the phospho-deficient mutant T222A/S232A as well as K393R mutant prevented Itch–TAp73 interaction (Figs. 4c, 3f) as well as ubiquitination of TAp73 (Fig. 4d, g).
These results highlighted that the Itch regulation by Aβ42 is a tightly regulated process which is dependent on cellular machinery like the activation of a JNK pathway. While TAp73 has also been shown to be a Itch target in dividing cells30, these studies were performed in HEK293 or HeLa cells that are very distinct from neurons. Moreover, the mechanism via which Itch is regulated was not dissected in these studies as they were more focussed on p73.
Furthermore, to our knowledge, no neuronal substrates of Itch have been identified. Present findings suggest that Itch may regulate CRNA mainly by targeting TAp73, which was reflected when TAp73 was depleted in Itch-siRNA-transfected cells (Fig. 2d) as reversal of CRNA protection was observed. As reported in the past by us and others10,12,13, soluble Aβ42 oligomers generated as a result of miscleavage of APP promote aberrant cell cycle re-entry and apoptosis of neurons. It is likely that TAp73 is involved in transcription of genes that suppress the cell cycle progression. Therefore, Itch activation may serve as a key upstream event in this cascade, which prevents this process. Consistent with this, Itch depletion reinstated TAp73 and reversed CRNA (Fig. 2c and d).
Given that CRNA contributes to neuronal loss in AD7,49,50, strategies targeting neurons undergoing aberrant cell cycle may be useful. The fact that both K393R and S232/T222A Itch mutants were able to reverse the re-entry of neurons into the cell cycle and apoptosis suggests that these mutants may be used to reverse neurodegeneration or Itch may be targeted in AD neurons for therapeutic purposes.
Materials and methods
Information related to antibodies and other reagents is provided in Supplementary information.
Cell culture
Cortical neurons from Embryonic day 18 (E18) Sprague-Dawley rats or Embryonic day 16 (E16) APP/PS1 transgenic AD mice were isolated and cultured as previously published12,13. Briefly, E18 rat or E16 mouse embryos were dissected and cortical region of the brain was isolated and treated with Trypsin-DNAse followed by addition of serum-containing media (SCM) and centrifugation at 500 × g for 5 min at room temperature. Cell pellet was resuspended in SCM and plated on poly-l-lysine-coated six-well plates. After 12 h, cells were washed with Tyrode’s CMF PBS supplemented with glucose and NaHCO3 and were maintained in serum-free medium (SFM) containing B27 and N2 supplement (Gibco, Life technologies), 1× penicillin–streptomycin, L-glutamine, and glucose in 5% CO2, for 5 days. Typically, in vitro transfections or Aβ1-42 treatments were performed at DIV5.
PC12 (rat pheochromocytoma) cells (ATCC) were maintained in Dulbecco’s modified Eagles medium (DMEM) (Gibco, Life technologies) with 10% heat-inactivated horse serum (Gibco, Life technologies) and 5% heat-inactivated fetal bovine serum (FBS) (Gibco, Life technologies) and Antibiotic/Antimycotic (Gibco, Life technologies). PC12 cells were differentiated in DMEM containing 1% FBS and treated with 50 ng/ml of 2.5 s nerve growth factor (NGF) for 5 days.
HEK293T/A (human embryonic kidney) cells (ATCC) were maintained in DMEM with 10% FBS and 1× antibiotic/antimycotic at 37 °C in 5% CO2.
Transfection and treatment
Lipofectamine 2000 reagent (Invitrogen) was used for transfection of plasmid DNA and siRNA according to manufacturer’s instructions. Cortical neurons and differentiated PC12 cells were transfected with 1–3 μg of plasmid DNA or 100 pmoles of siRNA per well in a six-well plate in SFM without antibiotic. After 3–4 h of transfection, cultures were moved to medium with supplements and antibiotic. Various treatments were typically initiated after 48 h. Adenovirus for GFP (control), Itch or human TAp73α (gifted by Dr. Sanjeev Das, NII) was used to overexpress these proteins. 0.5 µM of soluble oligomers of Aβ1-42 (R-peptide) was used as described previously12,13,51 for 48 h. Typically, cells were treated with 20 μM SP600125/JNKi (Merck) for 48 h, 10 μM U0126 (V112A, Promega) for 48 h and 10 μM MG132 (474790, Merck) for 12 h.
Immunoblotting
Cells were washed with PBS and lysed using ice cold lysis buffer containing 100 mM Tris–HCl pH 7.4, 5 mM EDTA, 100 mM NaCl, 1% Triton x100, and 10% glycerol, 1 mM phenyl methane sulfonyl fluoride, 1 mM sodium orthovanadate, 20 mM β-glycero-phosphate, and 1x protease inhibitor cocktail was added before use. Immunoblotting was performed as described previously12 using primary antibodies and secondary antibody conjugated with horse radish peroxidase (HRP). Chemiluminescence reagent West Pico or West Dura (Pierce) was used for detection as per manufacturer’s instructions.
Immunoprecipitation
Typically, 50–100 μg of protein was incubated with 1 μg of desired antibody for 12 h at 4 °C with shaking in a 250 μl reaction volume. Subsequently, 50 μl of protein A + G Sepharose (Santa Cruz Biotechnology) beads were added to the antibody–protein complex and incubated on a shaker for 5–7 h at 4 °C. The resin was washed at 4 °C to remove unbound proteins and resuspended in lysis buffer and immunoblotting was performed as described above.
5-bromo-2′-deoxyuridine (BrdU) incorporation and TUNEL assay
BrdU labeling was performed to detect DNA replication. Anti-BrdU antibody (GE) was used to detect incorporated BrdU12,13. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay to detect cell death was performed by using Dead End fluorometric TUNEL system (G3250, Promega) as per manufacturer’s guidelines and Hoechst 33342 (Molecular Probes) was used to stain the nuclei. These two assays were performed simultaneously and labeled cells were visualized using a Zeiss AxioImager microscope and Axiovision software was used for image acquisition and processing images and population of cells positive for BrdU and/or TUNEL was determined.
Image and statistical analysis
Image J (NIH) software was for densitometry analysis of desired bands in Western blots. The band intensity of the loading control (Actin) was used for the normalization. Unless indicated otherwise, one-way analysis of variance (ANOVA) or t-test was used for statistical analysis (Graph Pad software Inc., USA). Data are represented as mean ± standard error of mean (SEM).
Animal ethics
All the experiments were designed and performed in accordance with the guidelines of Institutional Ethics Committee. Animal work has been approved by Institutional Animal Ethics Committee with IAEC serial #394/15 and 461/18.
Supplementary information
Acknowledgements
Studies were supported by a grant (SB/SO/BB/006/2014) from the Department of Biotechnology (DBT) and funds from NII core. P.S. is a recipient of J.C. Bose Fellowship; M.C. received Senior Research Fellowship from DBT. P.K.M. received DBT-RA fellowship. Assistance of Surbhi Jaiswal in some experiments is also acknowledged.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by R. Aqeilan
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-2647-1).
References
- 1.Park DS, et al. Multiple pathways of neuronal death induced by DNA-damaging agents, NGF deprivation, and oxidative stress. J. Neurosci. 1998;18:830–840. doi: 10.1523/JNEUROSCI.18-03-00830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park DS, Levine B, Ferrari G, Greene LA. Cyclin dependent kinase inhibitors and dominant negative cyclin dependent kinase 4 and 6 promote survival of NGF-deprived sympathetic neurons. J. Neurosci. 1997;17:8975–8983. doi: 10.1523/JNEUROSCI.17-23-08975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat. Rev. Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 4.Giovanni A, Wirtz-Brugger F, Keramaris E, Slack R, Park DS. Involvement of cell cycle elements, cyclin-dependent kinases, pRb, and E2F x DP, in B-amyloid-induced neuronal death. J. Biol. Chem. 1999;274:19011–19016. doi: 10.1074/jbc.274.27.19011. [DOI] [PubMed] [Google Scholar]
- 5.Park DS, et al. Involvement of retinoblastoma family members and E2F/DP complexes in the death of neurons evoked by DNA damage. J. Neurosci. 2000;20:3104–3114. doi: 10.1523/JNEUROSCI.20-09-03104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrup K, Busser JC. The induction of multiple cell cycle events precedes target-related neuronal death. Development. 1995;121:2385–2395. doi: 10.1242/dev.121.8.2385. [DOI] [PubMed] [Google Scholar]
- 7.Busser J, Geldmacher DS, Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer’s disease brain. J. Neurosci. 1998;18:2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrup K. The involvement of cell cycle events in the pathogenesis of Alzheimer’s disease. Alzheimers Res. Ther. 2010;2:13. doi: 10.1186/alzrt37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Cheung T, Chen J, Herrup K. A comparative study of five mouse models of Alzheimer’s disease: cell cycle events reveal new insights into neurons at risk for death. Int. J. Alzheimers Dis. 2011;2011:171464. doi: 10.4061/2011/171464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varvel NH, et al. Abeta oligomers induce neuronal cell cycle events in Alzheimer’s disease. J. Neurosci. 2008;28:10786–10793. doi: 10.1523/JNEUROSCI.2441-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Herrup K. Cell division in the CNS: protective response or lethal event in post-mitotic neurons? Biochim. Biophys. Acta. 2007;1772:457–466. doi: 10.1016/j.bbadis.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modi PK, Jaiswal S, Sharma P. Regulation of neuronal cell cycle and apoptosis by microRNA 34a. Mol. Cell. Biol. 2016;36:84–94. doi: 10.1128/MCB.00589-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modi PK, Komaravelli N, Singh N, Sharma P. Interplay between MEK-ERK signaling, cyclin D1, and cyclin-dependent kinase 5 regulates cell cycle reentry and apoptosis of neurons. Mol. Biol. Cell. 2012;23:3722–3730. doi: 10.1091/mbc.E12-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman RS, Estus S, Johnson EM., Jr. Analysis of cell cycle-related gene expression in postmitotic neurons: selective induction of Cyclin D1 during programmed cell death. Neuron. 1994;12:343–355. doi: 10.1016/0896-6273(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 15.Malik B, et al. Loss of neuronal cell cycle control as a mechanism of neurodegeneration in the presenilin-1 Alzheimer’s disease brain. Cell Cycle. 2008;7:637–646. doi: 10.4161/cc.7.5.5427. [DOI] [PubMed] [Google Scholar]
- 16.Agostini M, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc. Natl Acad. Sci. USA. 2011;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene. 1999;18:4993–4998. doi: 10.1038/sj.onc.1202817. [DOI] [PubMed] [Google Scholar]
- 18.Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 19.Allocati N, Di Ilio C, De Laurenzi V. p63/p73 in the control of cell cycle and cell death. Exp. Cell Res. 2012;318:1285–1290. doi: 10.1016/j.yexcr.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Tomasini R, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, et al. TAp73 promotes cell survival upon genotoxic stress by inhibiting p53 activity. Oncotarget. 2014;5:8107–8122. doi: 10.18632/oncotarget.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conforti F, et al. Relative expression of TAp73 and DeltaNp73 isoforms. Aging (Albany, NY) 2012;4:202–205. doi: 10.18632/aging.100441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pozniak CD, et al. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 24.Pozniak CD, et al. p73 is required for survival and maintenance of CNS neurons. J. Neurosci. 2002;22:9800–9809. doi: 10.1523/JNEUROSCI.22-22-09800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou X, Levy-Cohen G, Blank M. Molecular functions of NEDD4 E3 ubiquitin ligases in cancer. Biochim. Biophys. Acta. 2015;1856:91–106. doi: 10.1016/j.bbcan.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Melino G, et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008;15:1103–1112. doi: 10.1038/cdd.2008.60. [DOI] [PubMed] [Google Scholar]
- 27.Perry WL, et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 28.Lohr NJ, et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am. J. Hum. Genet. 2010;86:447–453. doi: 10.1016/j.ajhg.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi M, et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl Acad. Sci. USA. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi M, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellomaria A, Barbato G, Melino G, Paci M, Melino S. Recognition mechanism of p63 by the E3 ligase Itch: novel strategy in the study and inhibition of this interaction. Cell Cycle. 2012;11:3638–3648. doi: 10.4161/cc.21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl Acad. Sci. USA. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao M, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 34.Suryaraja R, Anitha M, Anbarasu K, Kumari G, Mahalingam S. The E3 ubiquitin ligase Itch regulates tumor suppressor protein RASSF5/NORE1 stability in an acetylation-dependent manner. Cell Death Dis. 2013;4:e565. doi: 10.1038/cddis.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scialpi F, et al. Itch self-polyubiquitylation occurs through lysine-63 linkages. Biochem. Pharm. 2008;76:1515–1521. doi: 10.1016/j.bcp.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 36.Zhu K, et al. Allosteric auto-inhibition and activation of the Nedd4 family E3 ligase Itch. EMBO Rep. 2017;18:1618–1630. doi: 10.15252/embr.201744454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agostini M, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc. Natl Acad. Sci. USA. 2011;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jankowsky JL, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 39.Jankowsky JL, et al. Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol. Eng. 2001;17:157–165. doi: 10.1016/s1389-0344(01)00067-3. [DOI] [PubMed] [Google Scholar]
- 40.Perez JM, Chen Y, Xiao TS, Abbott DW. Phosphorylation of the E3 ubiquitin protein ligase ITCH diminishes binding to its cognate E2 ubiquitin ligase. J. Biol. Chem. 2018;293:1100–1105. doi: 10.1074/jbc.RA117.000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, et al. A tunable brake for HECT ubiquitin ligases. Mol. Cell. 2017;66:345–357 e346. doi: 10.1016/j.molcel.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y, et al. A data set of human endogenous protein ubiquitination sites. Mol. Cell. Proteom. 2011;10:M110 002089. doi: 10.1074/mcp.M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang SS, Hsu LJ, Chang NS. Functional role of WW domain-containing proteins in tumor biology and diseases: Insight into the role in ubiquitin-proteasome system. FASEB bioAdv. 2020;2:234–253. doi: 10.1096/fba.2019-00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santini S, et al. ATM kinase activity modulates ITCH E3-ubiquitin ligase activity. Oncogene. 2014;33:1113–1123. doi: 10.1038/onc.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang C, et al. Negative regulation of the E3 ubiquitin ligase itch via Fyn-mediated tyrosine phosphorylation. Mol. Cell. 2006;21:135–141. doi: 10.1016/j.molcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Shoji M, et al. JNK activation is associated with intracellular beta-amyloid accumulation. Brain Res. Mol. Brain Res. 2000;85:221–233. doi: 10.1016/s0169-328x(00)00245-x. [DOI] [PubMed] [Google Scholar]
- 47.Zhu X, et al. Differential activation of neuronal ERK, JNK/SAPK and p38 in Alzheimer disease: the ‘two hit’ hypothesis. Mech. Ageing Dev. 2001;123:39–46. doi: 10.1016/s0047-6374(01)00342-6. [DOI] [PubMed] [Google Scholar]
- 48.Zhu X, Ogawa O, Wang Y, Perry G, Smith MA. JKK1, an upstream activator of JNK/SAPK, is activated in Alzheimer’s disease. J. Neurochem. 2003;85:87–93. doi: 10.1046/j.1471-4159.2003.01645.x. [DOI] [PubMed] [Google Scholar]
- 49.Webber KM, et al. The cell cycle in Alzheimer disease: a unique target for neuropharmacology. Mech. Ageing Dev. 2005;126:1019–1025. doi: 10.1016/j.mad.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y, Mufson EJ, Herrup K. Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer’s disease. J. Neurosci. 2003;23:2557–2563. doi: 10.1523/JNEUROSCI.23-07-02557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stine WB, Jr, Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J. Biol. Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.