Abstract
The geometrical representation of muscles in computational models of the musculoskeletal system typically consists of a series of line segments. These muscle anatomies are based on measurements from a limited number of cadaveric studies that recently have been used as atlases for creating subject-specific models from medical images, so potentially restricting the options for personalisation and assessment of muscle geometrical models. To overcome this methodological limitation, we propose a novel, completely automated technique that, from a surface geometry of a skeletal muscle and its attachment areas, can generate an arbitrary number of lines of action (fibres) composed by a user-defined number of straight-line segments. These fibres can be included in standard musculoskeletal models and used in biomechanical simulations. This methodology was applied to the surfaces of four muscles surrounding the hip joint (iliacus, psoas, gluteus maximus and gluteus medius), segmented on magnetic resonance imaging scans from a cadaveric dataset, for which highly discretised muscle representations were created and used to simulate functional tasks. The fibres’ moment arms were validated against measurements and models of the same muscles from the literature with promising outcomes. The proposed approach is expected to improve the anatomical representation of skeletal muscles in personalised biomechanical models and finite element applications.
Electronic supplementary material
The online version of this article (10.1007/s10439-020-02490-4) contains supplementary material, which is available to authorized users.
Keywords: Skeletal muscle, Musculoskeletal geometry, Moment arms, Lower limb, Line of action
Introduction
Computational models of the musculoskeletal system have been used in a variety of contexts, from estimating contact forces on lower limb joints31,46 to simulating musculotendon contraction mechanisms in healthy and pathological individuals.5 Despite the popularity of these models, there are limited options to represent the muscle anatomy, and the available musculoskeletal models4,14,16,36 are built using geometrical data from few dissection studies.9,10,26,57 This limitation impacts the approaches to personalised medicine because musculotendon paths in subject-specific models are defined using registration methods35,47,53 or statistical shape models61 that map existing representations of the muscular system to personalised bone geometries. The modelling assumptions about muscle anatomy are therefore maintained across applications, potentially hindering personalization and predictive accuracy. Based on previous literature, the two main design aspects to consider when representing the anatomy of skeletal muscles in multibody models are (a) the number of elements included in the representation (muscle discretization level), also called “fibres” in the following, and (b) the geometrical complexity of each of these fibres’ path, determined by the number of straight line segments constituting the line of action.
The number of fibres required to ensure an accurate representation of a muscle’s mechanical action has been previously investigated based on the dimensionality of their attachment sites,54 with only one study focusing on the lower limb.52 In that study, where errors due to muscle discretization were computed at the bone attachments and not at joint centres as in musculoskeletal simulations, it was found that the required level of discretization depended on the individual anatomy and on the lower limb posture. In a sensitivity study, Xiao and Higginson59 observed that the number of fibres affected muscle force estimation, while Moissenet et al. reported more accurate estimates of contact force at the tibiofemoral joint37 and hip joint38 for higher muscle discretization. Recently, other studies confirmed that high discretization level of the muscle surrounding the hip joint is necessary to provide an accurate estimation of joint contact forces.32,58 The effect of muscle discretization on musculoskeletal force outputs remains however largely unexplored, due to the lack of methodologies to systematically vary the level of discretization of the muscle representation.
It is common practice to represent muscle geometry in musculoskeletal models by connecting the origin and attachment sites with a series of line segments, enhanced using via points and wrapping surfaces that prevent bone penetration and improve bio-fidelity resulting in series-of-line-segments paths3,20 (referred to as straight-lines approach in the following). From the mechanical point of view, this is a valid representation of a three-dimensional muscle only as long as the line segments pass through the centroids of the force distribution in the considered muscle sections.2 This approach is therefore reasonable for muscles presenting fusiform shapes and well-defined muscle attachments, but less appropriate for those with complex paths and large attachment areas. To overcome this limitation, Jansen and Davy24 proposed a geometrical representation based on the line connecting the muscle section centroids (centroidal approach), which required a line of action with around 15 line-segments. They found that estimated moment arms were larger compared to those of the straight-lines approach, with differences up to 50% on certain components of the generated muscle moment. Moreover, their reported moment arms correlated with muscle volume, so highlighting the importance of considering individual muscle morphology. The lines of action’s geometrical representation can influence the predictions of musculoskeletal models; Modenese et al.34 suggested that the straight-lines representation of muscles surrounding the hip joint was limiting the accuracy of hip contact force predictions, while non-negligible differences between modelled and experimentally measured musculotendon lengths, that can influence muscle force generation, were reported both for lower and upper limbs models.25,33 Despite these known limitations, no approach has been developed to generate lines of action of appropriate complexity based on muscle morphological data, e.g. segmented muscle surfaces, to include them in musculoskeletal models.
In previous studies, continuous models provided realistic anatomical representations of skeletal muscles. Blemker and Delp6 developed a finite element model of hip muscles by mapping templates of fibre arrangements on surface meshes. The muscle geometries in their study were segmented from magnetic resonance imaging (MRI) scans collected on a young individual, and the deformations predicted by their model were validated against additional segmentations of MRI scans for multiple hip joint positions. The model was computationally expensive (5–10 CPU hours in 2005) and, although it produced fibre paths from which lengths and moment arms could be computed, those results were not employed in a multibody musculoskeletal model. Oberhofer et al.41 used the free form deformation technique to simulate a gait cycle with a model including deformable lower limb muscles, the shape of which was also validated against MRI scans, but no quantitative biomechanical variables were discussed. Kohout et al. developed a technique to decompose a muscle surface mesh in an arbitrary number of fibres and used it to create a simulation of walking intended as a visual aid for clinicians27,28 but did not provide any quantity of biomechanical interest. Despite the minimum computational cost, this approach27 still required an underlying straight-lines musculoskeletal model for solving the fibre kinematics. Other approaches to produce fibres from muscle surfaces are available in the literature,12,23,30,42 but it is unclear how to couple them with a multibody model for further biomechanical analyses.
The aim of this paper is to present an automated approach to generate muscle fibres based on surface meshes obtainable from medical images by segmentation. From each mesh, this method can produce an arbitrary number of fibres composed by a user-defined number of straight-line segments, usable as musculotendon actuators in musculoskeletal models and in biomechanical analyses. The approach is demonstrated on a hip joint model including highly discretised muscle representations of the surrounding muscles, for which moment arms will be computed using standard musculoskeletal approaches. The results will be assessed against a model with straight-lines muscle representations, created from the same anatomy, and results from previous literature, including a validated finite element model.6
Materials and Methods
Anatomical Dataset
A comprehensive anatomical dataset collected on a female cadaver (81 years old, 167 cm, 63 kg) was employed to create the musculoskeletal models used in this investigation (Fig. 1). The dataset, publicly available and known as LHDL dataset,56 was selected because it includes surface meshes of bones and muscles, segmented from computed tomography and MRI scans respectively, of quality similar to in vivo datasets. Muscle attachment areas were also identified and digitised during the dissection.55 The triangular muscle meshes were improved in a pre-processing step by removing non-manifold edges, duplicated vertices and degenerate triangles, followed by smoothing using MeshLab.13
Figure 1.
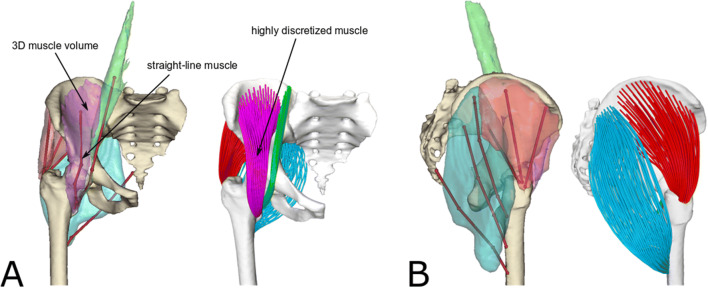
Frontal (a) and side view (b) of the bone and muscle geometries (iliacus: purple, psoas: green, gluteus maximus: cyan, gluteus medius: red) used for creating the musculoskeletal models. The straight-lines muscle representations and the segmented muscle surface meshes are shown together for comparison. The model with highly discretised muscle representations is shown on the right. All muscles were discretised using 100 fibres, each one consisting of a 15 line-segments polyline. Please note that although the gluteus maximus surface does not touch the femur, its insertion area lies on the bone.
Musculoskeletal Models
The geometries of the pelvis and right femur were employed in NMSBuilder51 to create a skeletal model of the right hip joint (Fig. 1), represented as a three-degrees of freedom ball-and-socket joint centred by fitting a sphere to the femoral head surface. This kinematic model was then exported in OpenSim 3.3 format15 and used as common baseline for both models described below.
Model with Highly Discretised Muscle Representations
Each muscle surface mesh was processed in two stages to generate a set of muscle fibres used in the simulations: (1) a muscle geometry decomposition step, performed in the scanning pose, in which the mesh is transformed in a user-defined number of fibres, and (2) a fibre kinematic step, in which the geometry of the fibres from the first step is updated to a new skeletal pose.
Muscle Geometry Decomposition
As the algorithm employed in the muscle geometry decomposition is described in details in a previous publication,28 only an overview of its main steps will be presented here.
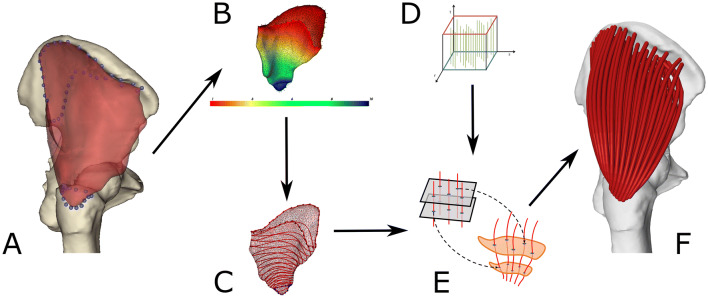
The required inputs of the method (Figs. 2a and 2d) are (1) a triangular surface mesh representing the muscle geometry, (2) a fibre template providing geometrical information about the internal fibre arrangement of the muscle and (3) the attachment areas of the muscle (origin and insertion), described as sets of landmarks fixed on the bone.
Figure 2.
Sequence of operations used for decomposing a muscle volume mesh in an arbitrary number of fibres of user-defined resolution. A muscle surface mesh, in red, and attachment areas, outlined by the blue markers, are taken as input (a). The attachment sites are mapped on the muscle mesh, from which their projected area is removed. A scalar field is defined on the resulting surface (b) and as many isolines as the desired fibre points (c) are identified. A muscle architecture template (d), containing the number of fibres desired from the decomposition, is then mapped to the muscle mesh using planes corresponding to the isolines (e), so generating fibres that can be imported in a standard OpenSim model (f). In this example, the gluteus medius is discretised in 100 fibres, each one consisting of 15 straight-line segments.
Firstly, the attachment areas are projected from the bones to the muscle mesh, outlining two areas that are subsequently removed, to produce a surface with two boundaries. A piece-wise linear scalar field, presenting minimum value on the origin boundary and maximum on the insertion one, is then computed over the vertices of the mesh (Fig. 2b).17 Contours corresponding to field isolines, i.e. lines connecting points where the field value is constant, are extracted for as many values as required by the user-specified number of straight-line segments in each fibre (Fig. 2c).
To represent muscle fibre architecture, templates consisting of unit space with an arbitrary (user-defined) number of fibres connecting two attachment areas were employed, similarly to Blemker and Delp.6 The fibres are expressed analytically by Bezier spline curves. In the decomposition step, an appropriate template is selected and evenly sliced using as many parallel planes as the specified fibre points. The resulting template sections are then mapped one-to-one to the contours of the scalar field isolines (Fig. 2e). As the position of the fibres is expressed relative to the contour of the template using generalized barycentric coordinates,22 they can be mapped on the muscle mesh using the same transformation. The fibres’ geometry is finalised with a step that ensures they connect with the attachments followed by a quadratic smoothing to eliminate noise (Fig. 2f).
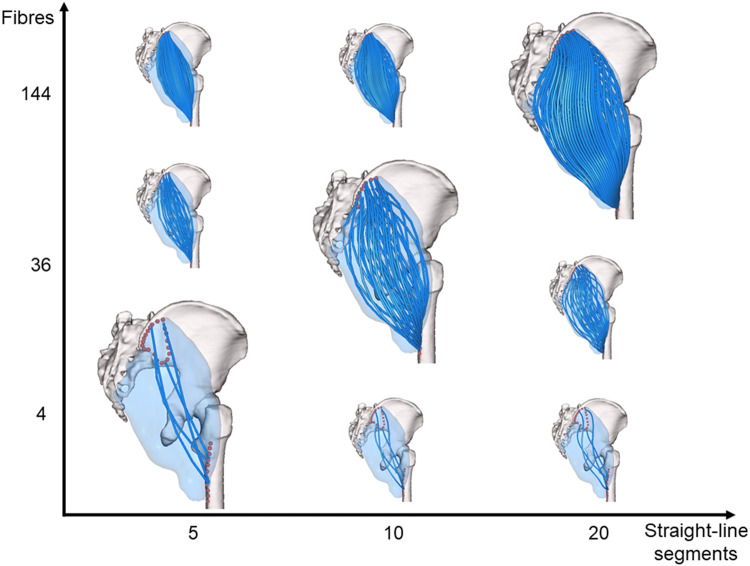
The muscle geometry representation resulting from this workflow can be customized by the user by choosing the total number of fibres and straight-line segments per fibre (Fig. 3).
Figure 3.
Muscle representations obtainable for gluteus maximus by combining different levels of muscle discretization (from 4 to 144 fibres) and numbers of line segments in the fibres (from 5 to 20 line-segments per fibre).
The surface meshes of gluteus maximus, gluteus medius, iliacus and psoas were decomposed in highly discretised models (100 fibres) using a template with parallel fibres composed by 15 line-segments, similarly to the centroidal paths of Jensen and Davy24 (Fig. 1). This was done in the MuscleWrapping software, part of a larger LHPBuilder application developed within the VPHOP project1, now concluded. In this explorative study, the pelvic attachment area of iliacus was used as origin also for the psoas muscle to avoid modelling its multiple origins on the lumbar spine. These muscles were selected because of their complex geometry and for consistency with Blemker and Delp.6
Fibre Kinematics
The algorithm solving the kinematics of the produced fibres is based on binding the points of the fibres to the bones using an automated procedure. As justified in details in the supplementary materials, every fibre point was associated with its two nearest bones, and its kinematic position calculated as a linear combination of the transformations of its rest-pose position with respect to these bones as:
where is the number of points in the fibres, and are the rotational and translational transformations of the j-th nearest bone and is a blending weight determining the bone influence, with . Although other options exists (see supplementary materials), the weights in this study were computed from the relative position of the i-th fibre point on the fibre (measured from the fibre origin ) using a quadratic function :
where , , and are muscle-specific parameters that determine how quickly the influence of an attachment bone diminishes along the fibre length. The first and the last fibre point positions are governed by the pelvis and the femur only, i.e., and , implying that and , thus leading to a formula with just one muscle-specific parameter to specify. In the current simulations, the value of was determined for each muscle by trial and error aiming to visually minimize the muscle-bone penetrations throughtout the flexion/extension range of motion (psoas: − 0.042, iliacus: − 0.024, gluteus maximus: − 0.042, gluteus medius: 0.0).
Model with Straight-Lines Muscle Representations
Musculotendon paths of iliacus, psoas, gluteus medius and gluteus maximus were defined as in Modenese et al.35 using the straight-lines muscle representation of the popular model gait239216 as reference atlas, with minor manual adjustments to take advantage of the available muscle geometries (Fig. 1). Consistently with the gait2392 model, iliacus and psoas were modelled using a single line of action, while gluteus medius and gluteus maximus were discretized using three lines of action each.
Simulations and Validations of Muscle Moment Arms
Simulations of hip extension/flexion (− 10° to 60°), hip ab/adduction (− 40° to 40°) and hip internal/external rotation (− 30° to 30°), performed in steps of 2°, were generated using both models.
The geometry of the fibres in the model with highly discretised muscles was updated at each frame of the kinematics using the application programming interface (API) of OpenSim v3.315 from MATLAB R2017b. The length of each fibre was then computed through the same API, interpolated with a 4th order polynomial function and used to calculate the moment arm of the i-th fiber with respect to the j-th coordinate using the tendon excursion method1:
The model with straight-lines muscles, instead, was imported in OpenSim 3.315 and its standard MuscleAnalysis tool was used to compute the moment arms49 for the same hip joint tasks. This approach was preferred to the tendon excursion method because it is more accurate for lines of action including conditional via points, for which the moment arm can change non smoothly (iliacus and psoas at around 40° flexion).
The moment arms computed with the highly discretised muscles were compared at each hip joint pose against those of the model with straight-lines muscles and data available from previous literature,3,18,40 including the results of the validated finite element model of Blemker and Delp,6 which were digitised using Graph Grabber v2.0 (https://www.quintessa.org). The percentage of poses for which the moment arms were in agreement, i.e. for which the range calculated with the highly discretized muscles model included the values from the other model or measurement, was calculated and reported. The peak and mean values of moment arms across the entire range of motion from the highly discretised muscles, together with those from the straight-line muscles and an estimation from Blemker and Delp6 were also reported for each task.
Results
For all muscles, the decomposition step took around 20 ms, while the simulations with highly discretised muscle representations took around one minute on a Z640 Dell Workstation (RAM: 64 GB, CPU: E5-2630 2.40 GHz).
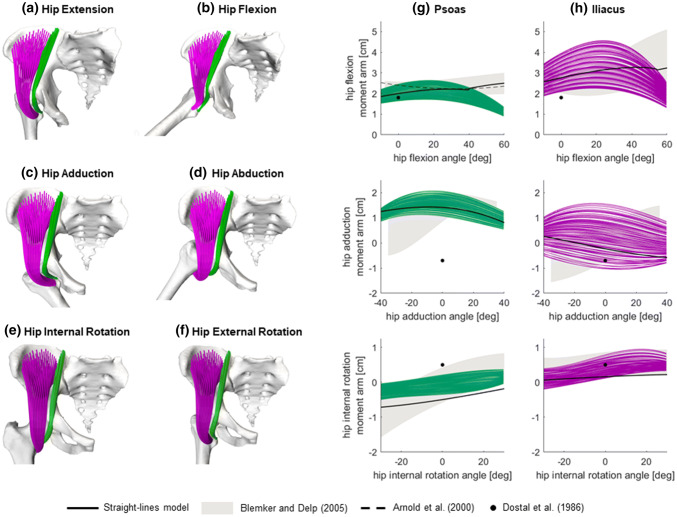
Overall, the results of the simulations were visually realistic for psoas and iliacus (Figs. 4a–4f), with the exception of flexion angles larger than 40 degrees (Fig. 4b), for which some fibres, especially of psoas, were penetrating the pelvis ridge geometry. On average, the moment arms from the straight-lines muscles were within the range estimated by the highly discretized muscles in 57% of the hip joint poses for psoas and 83% for iliacus (Table 1). Compared to Blemker and Delp,6 some differences in trend were observed for flexion angles larger than 35 degrees for both muscles, and when simulating ab/adduction for iliacus (Figs. 3g, 3h), leading to slightly lower level of agreement (psoas: 50%, iliacus: 77%).
Figure 4.
Results of the simulations of hip functional tasks for the iliacus and psoas muscles (in purple and green respectively). The resulting geometries of the highly discretised muscle models are shown on the left (a–f), and the correspondent moment arms are presented, for all fibres, on the right (g–h). The resulting moment arms are compared against the moment arms of the model with straight-lines muscles and other studies from the literature (see legend).
Table 1.
Comparison of the moment arms calculated with highly discretized muscles against estimations from the straight-lines muscles and previous data available in the literature
| Considered model or study | Description of metrics | Hip task | Psoas | Iliacus | Gluteus maximus | Gluteus medius |
|---|---|---|---|---|---|---|
| Straight-lines muscles | Percentage of poses in which moment arms are within the range estimated by the model with highly discretized muscles. | Flexion/extension | 72% | 90% | 91%a | 99%a |
| Adduction/abduction | 100% | 100% | 77%a | 79%a | ||
| Internal/external rotation | 0% | 60% | 78%a | 85%a | ||
| Mean | 57% | 83% | 82% | 88% | ||
| Blemker and Delp6 | Percentage of area outlined by results from Blemker and Delp6 that is overlapping with the area outlined by the highly-discretized moment arms results | Flexion/extension | 62% | 71% | 65% | 81% |
| Adduction/abduction | 44% | 79% | 86% | 54% | ||
| Internal/external rotation | 44% | 79% | 86% | 95% | ||
| Mean | 50% | 77% | 79% | 77% | ||
| Dostal et al.18 | Reported values fall within the range estimated by the model with highly discretized muscles (Y/N: yes/no) | Flexion/extension | Y | N | Y | Y (72%b) |
| Adduction/abduction | N | Y | Y | Y | ||
| Internal/external rotation | N | Y | Y | Y | ||
| Nemeth and Ohlsen (1985) | Reported values fall within the range estimated by the model with highly discretized muscles (Y/N: yes/no) | Flexion/extension | – | – | Y (91%b) | – |
| Adduction/abduction | – | – | Y | Y | ||
| Arnold et al.3 | Percentage of poses in which moment arms are within the range estimated by the model with highly discretized muscles | Flexion/extension | 61% | – | – | – |
aAveraged across the three fibres representing this muscle in the straight-lines model
bPercentage estimated as for the model with straight-lines muscles
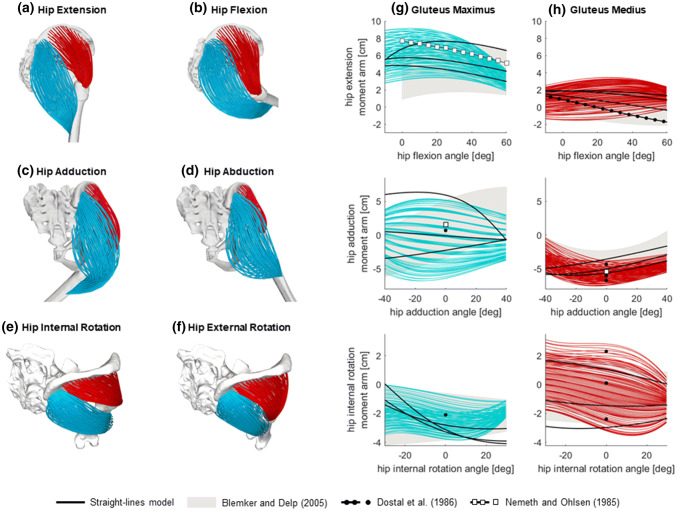
The simulations of gluteus maximus and gluteus medius also generated visually satisfactory geometries (Figs. 5a–5f), with fibres of the former muscle minimally penetrating the ischium and sacrum bones towards the extreme joint angles of hip flexion and adduction respectively, e.g. Figs. 5c. The moment arms of the straight-lines muscles model were consistent with those of the highly discretized model on average in 82% of the considered poses for gluteus maximus and 88% for gluteus medius (Figs. 5g, 5h, Table 1). The comparison with Blemker and Delp6 also suggested a remarkable similarity of the estimated moment arms (gluteus maximus: 79%, gluteus medius: 77%).
Figure 5.
Results of the simulations of hip functional tasks for the gluteus maximus and gluteus medius muscles (in cyan and red respectively). The resulting geometries of the highly discretised muscle models are shown on the left (A-F), and the corresponding moment arms are presented, for all fibres, on the right (g–h). The resulting moment arms are compared against the moment arms of the model with straight-lines muscles and other studies from the literature (see legend).
Moment arms from a physical model18 and medical images40 were also in strong agreement with those of the highly discretized muscles (in 9 out of 12 comparisons and three out of three comparisons respectively, see Table 1). Reasonable consistency, on 61% of the considered hip flexion angles, was also found with the cadaveric measurements of Arnold et al.3
The upper and lower bounds of the moment arms computed from the OpenSim models, and those from the digitised results of Blemker and Delp,6 are reported for all muscles in Table 2. The peaks and means of moment arms were consistent among the three models for the majority of the considered hip joint poses.
Table 2.
Peak values (minimum and maximum) and mean values with standard deviations (sd) of moment arms calculated across the considered ranges of motion for the highly discretized muscles, the straight-lines muscles and the results extrapolated from Figs. 6 and 7 of Blemker and Delp6 using Graph Grabber 2.0
| Hip task | Model | Muscle moment arms (cm) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Psoas | Iliacus | Gluteus maximus | Gluteus medius | ||||||||||
| Min | Max | Mean (sd) | Min | Max | Mean (sd) | Min | Max | Mean (sd) | Min | Max | Mean (sd) | ||
| Flexion(+) /extension (−) | Highly discretized muscles | 0.9 | 2.6 | 1.9 (0.3) | 1.3 | 4.6 | 3.0 (0.4) | − 9.2 | − 1.9 | − 5.7 (0.9) | − 3.5 | 1.5 | − 1.5 (0.3) |
| Straight-lines muscles | 1.8 | 2.5 | 2.2 (0.2) | 2.6 | 3.3 | 3.1 (0.2) | − 7.7 | − 2.9 | − 5.5 (1.5) | − 1.9 | 0.4 | − 1.3 (0.6) | |
| Blemker and Delp (2005) | 1.7 | 3.0 | 2.3 (0.1) | 1.9 | 5.1 | 3.0 (0.4) | − 7.2 | − 0.9 | − 4.2 (0.3) | − 1.7 | 2.1 | − 0.0 (0.3) | |
| Adduction (+)/abduction (−) | Highly discretized muscles | 0.5 | 2.1 | 1.4 (0.2) | − 1.0 | 1.6 | 0.2 (0.2) | − 6.8 | 5.4 | − 0.6 (0.3) | − 7.5 | − 0.4 | − 5.1 (0.6) |
| Straight-lines muscles | 0.8 | 1.4 | 1.3 (0.2) | − 0.6 | 0.3 | − 0.2 (0.3) | − 3.5 | 6.4 | 0.8 (3.6) | − 5.9 | − 1.6 | − 4.5 (1.0) | |
| Blemker and Delp 6 | − 0.5 | 2.0 | 1.2 (0.5) | − 1.5 | 1.5 | − 0.0 (0.5) | − 6.1 | 7.1 | 0.8 (1.9) | − 5.4 | 0.6 | − 3.1 (0.7) | |
| Internal (+)/external (−) rotation | Highly discretized muscles | − 0.5 | 0.4 | − 0.1 (0.1) | − 0.2 | 0.9 | 0.3 (0.1) | − 3.9 | 0.0 | − 2.3 (0.3) | − 3.5 | 3.1 | − 0.4 (0.5) |
| Straight-lines muscles | − 0.7 | − 0.2 | − 0.5 (0.2) | 0.1 | 0.2 | 0.2 (0.0) | − 4.1 | 0.1 | − 2.8 (0.2) | − 3.0 | 1.7 | − 1.1 (1.9) | |
| Blemker and Delp 6 | − 1.8 | 0.8 | − 0.2 (0.4) | − 0.1 | 0.9 | 0.5 (0.1) | − 4.2 | − 0.1 | − 2.2 (0.0) | − 2.7 | 2.2 | − 0.4 (0.2) | |
Discussion
The aim of this investigation was to present an automated technique to create complex, three-dimensional representations of skeletal muscles from their surface meshes that can be used in standard musculoskeletal models and demonstrate its use on a hip joint model. The proposed approach overcomes the traditional dualism of straight-lines versus more geometrically complex muscle representations such as centroidal lines, because paths of varying complexity can be automatically generated based on the user’s specifications. At the same time, the muscle discretization level can be altered in a systematic, reproducible way while still ensuring anatomical accuracy of the fibre set (Fig. 3).
In the absence of additional MRI scans for validating the highly discretised muscle geometries in various poses, we created a musculoskeletal model with straight-lines muscles, as this is the state of the art for representing musculotendon anatomies in multibody approaches. Results were also evaluated against a validated finite element model6 and previous measurements of moment arms from medical images and physical models.3,18,40 The agreement with the straight-lines muscles model and previous studies was generally very positive, especially for the hip extensors (Figs. 5g, 5h). For psoas and iliacus, however, we observed deviations from the straight-lines model for hip flexions larger than 40 and 55 degrees respectively. At those angles, conditional via points in the straight-lines muscles became inactive, similarly to the reference model gait2392, letting the moment arms increase consistently with experimental measurements3 and the results of the validated finite element model.6 Conversely, in the current formulation of the proposed technique the fibre points behaved essentially like via points with position regulated by a weight function but nevertheless active throughout the motion, so creating a sort of “adhesion” to the femur at high hip flexion (see Fig. 3b, in the hip joint area) that affected the moment arms estimation. We plan to improve this limitation of the methodology by implementing a position-based dynamics system39 that will have the further benefit of detecting muscle-bone contacts and preventing the occasional muscle-bone penetrations we observed. Further differences from Blemker and Delp6 could be attributed to various causes, including the identification of the muscle attachments, the muscle morphology in the elderly specimen (54 years older than their participant) and different segmentation of the psoas muscle, which presents in their study a planar cut at the level of the sacrum. Our results, however, compared overall positively against the validation datasets, suggesting that the new technique provides realistic muscle fibre configurations, especially within the range of motion typical of walking48 (hip extension/flexion: − 10° to 40°, ab/adduction: − 10° to 10° and axial rotation: − 7° to 7°).
The presented methodology presents some limitations. First, the approach requires muscle attachment areas, which are normally not available for in vivo datasets. They can, however, be estimated using statistical shape approaches19,61 or mapped from existing atlases, like the dataset provided with this paper, using registration techniques.43 Second, personalized surface meshes of muscles are also required by this technique, and they are currently time-consuming to segment from medical images. However, semi-automated segmentation procedures are becoming available both in commercial29 and open-source software,60 and, depending on the intended application, it might not be necessary to represent all the muscles as highly discretised, but only those of interest, e.g. presenting abnormal volumes that could affect moment arms.21 Third, only a template with parallel fibres was used in the current investigation, but the methodology is straightforward to extend27 to fibre templates proposed in previous studies.6,7
In conclusion, we believe that the proposed approach is a promising and fully automated solution to provide subject-specific representations of muscle geometries usable in multibody models that could benefit multiple applications in biomechanics. For example, sensitivity studies on muscle discretization similar to Valente et al.52 and their extension to muscle forces and joint reactions will be enabled. Future studies will investigate aspects of the methodology that require clarification before adoption in automated workflows for musculoskeletal and finite element simulations, such as sensitivity analyses with respect to the desirable number of straight-line segments in muscle fibres and to the uncertainty in muscle attachment areas identification, and in more advanced applications like the use of the computed moment arms and muscle lengths for muscle force estimation in dynamic simulations. Finite element models will particularly benefit from this technique because highly discretised muscle forces can be easily distributed on attachment areas, so avoiding stress concentrations on a small set of attachment nodes45 while still applying equilibrated force sets provided by the multibody systems.44,50 The muscle decomposition could also inform finite element models of skeletal muscles about fibre arrangements, or be replaced by experimentally derived fibre arrangements, e.g. from diffusion tensor imaging,8,11 to use in kinematic simulations.
The anatomical dataset and the OpenSim models used in this study are freely available for download at https://github.com/ComputationalBiomechanics/3d-muscles and https://simtk.org/projects/3d-muscles.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by an Imperial College Research Fellowship granted by Imperial College London to LM and by the Czech Ministry of Education, Youth and Sports, Project PUNTIS (LO1506) to JK.
Conflict of interest
The authors declare that they do not have any financial or personal relationship with other people or organizations that could have inappropriately influenced this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.An KN, Takahashi K, Harrigan TP, Chao EY. Determination of muscle orientations and moment arms. J. Biomech. Eng. 1984;106:280–282. doi: 10.1115/1.3138494. [DOI] [PubMed] [Google Scholar]
- 2.Andrews JG, Hay JG. Biomechanical considerations in the modeling of muscle function. Acta Morphologica Neerlando-Scandinavica. 1983;21:199–223. [PubMed] [Google Scholar]
- 3.Arnold AS, Salinas S, Asakawa DJ, Delp SL. Accuracy of muscle moment arms estimated from MRI-based musculoskeletal models of the lower extremity. Comput. Aided Surg. 2000;5:108–119. doi: 10.1002/1097-0150(2000)5:2<108::AID-IGS5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Arnold E, Ward S, Lieber R, Delp S. A model of the lower limb for analysis of human movement. Ann. Biomed. Eng. 2010;38:269–279. doi: 10.1007/s10439-009-9852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barber L, Carty C, Modenese L, Walsh J, Boyd R, Lichtwark G. Medial gastrocnemius and soleus muscle-tendon unit, fascicle, and tendon interaction during walking in children with cerebral palsy. Dev. Med. Child Neurol. 2017;59:843–851. doi: 10.1111/dmcn.13427. [DOI] [PubMed] [Google Scholar]
- 6.Blemker SS, Delp SL. Three-dimensional representation of complex muscle architectures and geometries. Ann. Biomed. Eng. 2005;33:661–673. doi: 10.1007/s10439-005-1433-7. [DOI] [PubMed] [Google Scholar]
- 7.Blemker SS, Delp SL. Rectus femoris and vastus intermedius fiber excursions predicted by three-dimensional muscle models. J. Biomech. 2006;39:1383–1391. doi: 10.1016/j.jbiomech.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Bolsterlee B, Finni T, D’Souza A, Eguchi J, Clarke EC, Herbert RD. Three-dimensional architecture of the whole human soleus muscle in vivo. PeerJ. 2018;6:e4610. doi: 10.7717/peerj.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brand RA, Crowninshield RD, Wittstock CE, Pedersen DR, van Clark CR, Krieken FM. A model of lower extremity muscular anatomy. J. Biomech. Eng. 1982;104:304–310. doi: 10.1115/1.3138363. [DOI] [PubMed] [Google Scholar]
- 10.Carbone V, Fluit R, Pellikaan P, van der Krogt M, Janssen D, Damsgaard M, Vigneron L, Feilkas T, Koopman H, Verdonschot N. TLEM 2.0–A comprehensive musculoskeletal geometry dataset for subject-specific modeling of lower extremity. J. Biomech. 2015;48:734–741. doi: 10.1016/j.jbiomech.2014.12.034. [DOI] [PubMed] [Google Scholar]
- 11.Charles JP, Moon C-H, Anderst WJ. Determining subject-specific lower-limb muscle architecture data for musculoskeletal models using diffusion tensor imaging. J. Biomech. Eng. 2019;141:060905. doi: 10.1115/1.4040946. [DOI] [PubMed] [Google Scholar]
- 12.Choi HF, Blemker SS. Skeletal muscle fascicle arrangements can be reconstructed using a laplacian vector field simulation. PLoS ONE. 2013;8:e77576. doi: 10.1371/journal.pone.0077576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cignoni, P., Callieri, M., Corsini, M., Dellepiane, M., Ganovelli, F. Ranzuglia, G. Meshlab: an open-source mesh processing tool. In: Eurographics Italian chapter conference, 2008.
- 14.De Pieri E, Lund ME, Gopalakrishnan A, Rasmussen KP, Lunn DE, Ferguson SJ. Refining muscle geometry and wrapping in the TLEM 2 model for improved hip contact force prediction. PLoS ONE. 2018;13:e0204109. doi: 10.1371/journal.pone.0204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delp SL, Anderson FC, Arnold AS, Loan P, Habib A, John CT, Guendelman E, Thelen DG. OpenSim: open-source software to create and analyze dynamic simulations of movement. IEEE Trans. Biomed. Eng. 2007;54:1940–1950. doi: 10.1109/TBME.2007.901024. [DOI] [PubMed] [Google Scholar]
- 16.Delp SL, Loan JP, Hoy MG, Zajac FE, Topp EL, Rosen JM. An interactive graphics-based model of the lower extremity to study orthopaedic surgical procedures. IEEE Trans. Biomed. Eng. 1990;37:757–767. doi: 10.1109/10.102791. [DOI] [PubMed] [Google Scholar]
- 17.Dong S, Kircher S, Garland M. Harmonic functions for quadrilateral remeshing of arbitrary manifolds. Comput. Aided Geometr. Des. 2005;22:392–423. [Google Scholar]
- 18.Dostal WF, Soderberg GL, Andrews JG. Actions of hip muscles. Phys. Ther. 1986;66:351–361. doi: 10.1093/ptj/66.3.351. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda N, Otake Y, Takao M, Yokota F, Ogawa T, Uemura K, Nakaya R, Tamura K, Grupp RB, Farvardin A, Armand M, Sugano N, Sato Y. Estimation of attachment regions of hip muscles in CT image using muscle attachment probabilistic atlas constructed from measurements in eight cadavers. Int. J. Comput. Assist. Radiol. Surg. 2017;12:733–742. doi: 10.1007/s11548-016-1519-8. [DOI] [PubMed] [Google Scholar]
- 20.Garner BA, Pandy MG. The obstacle-set method for representing muscle paths in musculoskeletal models. Comput. Methods Biomech. Biomed. Eng. 2000;3:1–30. doi: 10.1080/10255840008915251. [DOI] [PubMed] [Google Scholar]
- 21.Handsfield GG, Meyer CH, Abel MF, Blemker SS. Heterogeneity of muscle sizes in the lower limbs of children with cerebral palsy. Muscle Nerve. 2016;53:933–945. doi: 10.1002/mus.24972. [DOI] [PubMed] [Google Scholar]
- 22.Hormann K, Floater MS. Mean value coordinates for arbitrary planar polygons. ACM Trans. Graph. (TOG) 2006;25:1424–1441. [Google Scholar]
- 23.Inouye, J., G. Handsfield, anD S. Blemker. Fiber tractography for finite-element modeling of transversely isotropic biological tissues of arbitrary shape using computational fluid dynamics. In: Proceedings of the conference on summer computer simulation. Society for Computer Simulation International, 2015
- 24.Jensen RH, Davy DT. An investigation of muscle lines of action about the hip: A centroid line approach vs the straight line approach. J. Biomech. 1975;8:103–110. doi: 10.1016/0021-9290(75)90090-1. [DOI] [PubMed] [Google Scholar]
- 25.Klein Breteler MD, Spoor CW, Van der Helm FCT. Measuring muscle and joint geometry parameters of a shoulder for modeling purposes. J. Biomech. 1999;32:1191. doi: 10.1016/s0021-9290(99)00122-0. [DOI] [PubMed] [Google Scholar]
- 26.Klein Horsman MD, Koopman HF, van der Helm FC, Prose LP, Veeger HE. Morphological muscle and joint parameters for musculoskeletal modelling of the lower extremity. Clin. Biomech. 2007;22:239–247. doi: 10.1016/j.clinbiomech.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Kohout J, Clapworthy GJ, Zhao Y, Tao Y, Gonzalez-Garcia G, Dong F, Wei H, Kohoutová E. Patient-specific fibre-based models of muscle wrapping. Interface Focus. 2013;3:20120062. doi: 10.1098/rsfs.2012.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohout J, Kukačka M. Real-time modelling of fibrous muscle. Comput. Graph. Forum. 2014 doi: 10.1111/cgf.12354. [DOI] [Google Scholar]
- 29.Kolk S, Klawer E, Schepers J, Weerdesteyn V, Visser EP, Verdonschot N. Muscle activity during walking measured using 3D MRI segmentations and [18F]-fluorodeoxyglucose in combination with positron emission tomography. Med. Sci. Sports Exerc. 2015;47:1896–1905. doi: 10.1249/MSS.0000000000000607. [DOI] [PubMed] [Google Scholar]
- 30.Lee D, Glueck M, Khan A, Fiume E, Jackson K. A survey of modeling and simulation of skeletal muscle. ACM Trans. Graph. 2010;28:162. [Google Scholar]
- 31.Marra MA, Vanheule V, Fluit R, Koopman BH, Rasmussen J, Verdonschot N. A subject-specific musculoskeletal modeling framework to predict in vivo mechanics of total knee arthroplasty. J. Biomech. Eng. 2015;137:020904. doi: 10.1115/1.4029258. [DOI] [PubMed] [Google Scholar]
- 32.Mathai B, Gupta S. Numerical predictions of hip joint and muscle forces during daily activities: a comparison of musculoskeletal models. Proc. Inst. Mech. Eng. 2019;233:636–647. doi: 10.1177/0954411919840524. [DOI] [PubMed] [Google Scholar]
- 33.Modenese L, Ceseracciu E, Reggiani M, Lloyd DG. Estimation of musculotendon parameters for scaled and subject specific musculoskeletal models using an optimization technique. J. Biomech. 2016;49:141–148. doi: 10.1016/j.jbiomech.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Modenese L, Gopalakrishnan A, Phillips ATM. Application of a falsification strategy to a musculoskeletal model of the lower limb and accuracy of the predicted hip contact force vector. J. Biomech. 2013;46:1193–1200. doi: 10.1016/j.jbiomech.2012.11.045. [DOI] [PubMed] [Google Scholar]
- 35.Modenese L, Montefiori E, Wang A, Wesarg S, Viceconti M, Mazzà C. Investigation of the dependence of joint contact forces on musculotendon parameters using a codified workflow for image-based modelling. J. Biomech. 2018;73:108–118. doi: 10.1016/j.jbiomech.2018.03.039. [DOI] [PubMed] [Google Scholar]
- 36.Modenese L, Phillips ATM, Bull AMJ. An open source lower limb model: Hip joint validation. J. Biomech. 2011;44:2185–2193. doi: 10.1016/j.jbiomech.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 37.Moissenet F, Chèze L, Dumas R. Influence of the level of muscular redundancy on the validity of a musculoskeletal model. J. Biomech. Eng. 2015;138:021019. doi: 10.1115/1.4032127. [DOI] [PubMed] [Google Scholar]
- 38.Moissenet F, Giroux M, Chèze L, Dumas R. Validity of a musculoskeletal model using two different geometries for estimating hip contact forces during normal walking. Comput. Methods Biomech. Biomed. Eng. 2015;18:1–2. doi: 10.1080/10255842.2015.1069596. [DOI] [PubMed] [Google Scholar]
- 39.Müller M, Heidelberger B, Hennix M, Ratcliff J. Position based dynamics. J. Vis. Commun. Image Represent. 2007;18:109–118. [Google Scholar]
- 40.Németh G, Ohlsén H. In vivo moment arm lengths for hip extensor muscles at different angles of hip flexion. J. Biomech. 1985;18:129–140. doi: 10.1016/0021-9290(85)90005-3. [DOI] [PubMed] [Google Scholar]
- 41.Oberhofer K, Mithraratne K, Stott N, Anderson I. Anatomically-based musculoskeletal modeling: prediction and validation of muscle deformation during walking. Visual Comput. 2009;25:843–851. [Google Scholar]
- 42.Otake, Y., M. Takao, N. Fukuda, S. Takagi, N. Yamamura, N. Sugano, and Y. Sato. Registration-based patient-specific musculoskeletal modeling using high fidelity cadaveric template model. In: Medical image computing and computer assisted intervention – MICCAI 2018. Cham: Springer International Publishing, 2018.
- 43.Pellikaan P, van der Krogt MM, Carbone V, Fluit R, Vigneron LM, van Deun J, Verdonschot N, Koopman HFJM. Evaluation of a morphing based method to estimate muscle attachment sites of the lower extremity. J. Biomech. 2014;47:1144–1150. doi: 10.1016/j.jbiomech.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Phillips ATM, Villette CC, Modenese L. Femoral bone mesoscale structural architecture prediction using musculoskeletal and finite element modelling. Int. Biomech. 2015;2:43–61. [Google Scholar]
- 45.Polgar K, Gill H, Viceconti M, Murray D, O’Connor J. Strain distribution within the human femur due to physiological and simplified loading: finite element analysis using the muscle standardized femur model. Proc. Inst. Mech. Eng. 2003;217:173–189. doi: 10.1243/095441103765212677. [DOI] [PubMed] [Google Scholar]
- 46.Saxby DJ, Modenese L, Bryant AL, Gerus P, Killen B, Fortin K, Wrigley TV, Bennell KL, Cicuttini FM, Lloyd DG. Tibiofemoral contact forces during walking, running and sidestepping. Gait Posture. 2016;49:78–85. doi: 10.1016/j.gaitpost.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 47.Scheys L, Loeckx D, Spaepen A, Suetens P, Jonkers I. Atlas-based non-rigid image registration to automatically define line-of-action muscle models: a validation study. J. Biomech. 2009;42:565–572. doi: 10.1016/j.jbiomech.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz MH, Rozumalski A, Trost JP. The effect of walking speed on the gait of typically developing children. J. Biomech. 2008;41:1639–1650. doi: 10.1016/j.jbiomech.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 49.Sherman, M., A. Seth, and S. L. Delp. What is a moment arm? Calculating muscle effectiveness in biomechanical models using generalized coordinates. In: Proceedings of the ASME 2013 international design engineering technical conferences & computers and information in engineering conference, August 4–7, 2013, Portland, Oregon, USA, 2013. [DOI] [PMC free article] [PubMed]
- 50.Speirs AD, Heller MO, Duda GN, Taylor WR. Physiologically based boundary conditions in finite element modelling. J. Biomech. 2007;40:2318–2323. doi: 10.1016/j.jbiomech.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 51.Valente G, Crimi G, Vanella N, Schileo E, Taddei F. nmsBuilder: Freeware to create subject-specific musculoskeletal models for OpenSim. Comput. Methods Progr. Biomed. 2017;152:85–92. doi: 10.1016/j.cmpb.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Valente G, Martelli S, Taddei F, Farinella G, Viceconti M. Muscle discretization affects the loading transferred to bones in lower-limb musculoskeletal models. Proc. Inst. Mech. Eng. 2012;226:161–169. doi: 10.1177/0954411911425863. [DOI] [PubMed] [Google Scholar]
- 53.Valente G, Pitto L, Testi D, Seth A, Delp SL, Stagni R, Viceconti M, Taddei F. Are subject-specific musculoskeletal models robust to the uncertainties in parameter identification? PLoS ONE. 2014;9:e112625. doi: 10.1371/journal.pone.0112625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Helm FC, Veenbaas R. Modelling the mechanical effect of muscles with large attachment sites: application to the shoulder mechanism. J. Biomech. 1991;24:1151–1163. doi: 10.1016/0021-9290(91)90007-a. [DOI] [PubMed] [Google Scholar]
- 55.Van Sint Jan S. Introducing anatomical and physiological accuracy in computerized anthropometry for increasing the clinical usefulness of modeling systems. Crit. Rev. Phys. Rehabil. Med. 2005;17:249–274. [Google Scholar]
- 56.Viceconti M, Clapworthy G, Van Sint Jan S. The virtual physiological human - a European initiative for in silico human modelling. J. Physiol. Sci. 2008;58:441–446. doi: 10.2170/physiolsci.RP009908. [DOI] [PubMed] [Google Scholar]
- 57.Ward S, Eng C, Smallwood L, Lieber R. Are current measurements of lower extremity muscle architecture accurate? Clin. Orthop. Relat. Res. 2009;467:1074–1082. doi: 10.1007/s11999-008-0594-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weinhandl JT, Bennett HJ. Musculoskeletal model choice influences hip joint load estimations during gait. J. Biomech. 2019;91:124–132. doi: 10.1016/j.jbiomech.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 59.Xiao M, Higginson J. Sensitivity of estimated muscle force in forward simulation of normal walking. J. Appl. Biomech. 2010;26:142. doi: 10.1123/jab.26.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Fernandez J, Hislop-Jambrich J, Besier TF. Lower limb estimation from sparse landmarks using an articulated shape model. J. Biomech. 2016;49:3875–3881. doi: 10.1016/j.jbiomech.2016.10.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.