Abstract
Background: Programmed cell death 1 ligand-1 (PD-L1) is an immune checkpoint molecule that acts to protect cancer cells from immune surveillance and is considered as a prognostic biomarker in several cancers, but the prognostic value of PD-L1 in bone and soft tissue sarcomas remains inconclusive. In the present meta-analysis, the clinicopathological and prognostic value of PD-L1 in sarcomas was evaluated.
Method: We performed a systemic and comprehensive meta-analysis by searching the PubMed, Medline, Cochrane Library, EMBASE, and Web of Science databases up to October 31, 2019. Eligible articles were incorporated, and pooled hazard ratios (HRs) and odds ratios (ORs) with their 95% confidence intervals (CIs) were used to estimate the outcomes.
Results: Thirty-six articles containing 39 independent studies with 3,680 bone and soft tissue sarcoma patients were included in our meta-analysis. The pooled results showed that PD-L1 overexpression could predict poor overall survival (HR 1.45, 95% CI 1.11–1.90, P < 0.01), metastasis-free survival (HR 1.58, 95% CI 1.14–2.19, P < 0.01), and event-free survival (HR 2.82, 95% CI 1.69–4.71, P < 0.01) in sarcomas. Furthermore, PD-L1 overexpression was correlated with a higher rate of tumor metastasis (OR 2.95, 95% CI 1.32–6.60, P < 0.01), a more advanced tumor grade (OR 3.63, 95% CI 2.55–5.16, P < 0.01), and more T lymphocyte infiltration (OR 5.55, 95% CI 2.86–10.76, P < 0.01). No obvious publication bias was observed, and the sensitivity analysis showed that our results were robust.
Conclusion: The results of our meta-analysis indicate that high PD-L1 expression might serve as a valuable and predictive biomarker for adverse clinicopathological features and poor prognosis in patients with sarcoma.
Keywords: programmed cell death 1 ligand-1, sarcoma, meta-analysis, prognosis, overall survival
Introduction
Bone and soft tissue sarcomas are a group of heterogeneous and rare neoplasms with more than 50 distinct histologic subtypes that originate from mesenchymal tissues, and these sarcomas account for <1% of adult cancers and 7–15% of pediatric malignancies (1). In recent years, due to the improvement of surgical techniques and the effective combining of chemotherapy or radiotherapy with surgery, a satisfactory but stagnant survival of over 60% has been achieved (2–4). Moreover, the response of sarcomas to conventional chemotherapy and radiotherapy varies, and for those patients with recurrent and metastatic disease, their survival remains unsatisfactory, with an average survival period of <1 year (5–7). Thus, novel treatment options for these patients are of utmost urgency.
Immunotherapy is considered as one of the foremost options for cancer treatment and has recently achieved tremendous success (8, 9). In the immune system, immune tolerance is an important process that inhibits the role of innate immunity in the cancer microenvironment. One of the immune tolerance mechanisms is the immune checkpoint mechanism. Immune checkpoint molecules, such as cytotoxic T lymphocyte antigen 4, lymphocyte activation gene 3, programmed cell death 1 (PD-1), and programmed cell death 1 ligand-1 (PD-L1), are expressed in the microenvironment of cancers (10), and suppressive signals are transmitted to T cells to prevent excessive immune responses. Thus, immune checkpoint inhibitors are emerging and becoming a prevailing form of systemic therapy regarded to “release the brakes” on the immune system. In malignant tumors, PD-L1 is often expressed on the surface of tumor cells and helps promote tumor evasion from the immune system by activating PD-1 and inhibiting the function of T cells. Immune checkpoint inhibitors targeting the PD-L1/PD-1 immune checkpoint axis have shown considerable success in cancers (11) but not in bone and soft tissue sarcomas (12–14). To investigate the underlying mechanism of PD-L1 in the progression of sarcomas and assess the clinical value of PD-L1, the correlation between PD-L1 expression and the clinical outcomes of sarcoma patients was assessed. However, more studies are emerging to show controversial results in the past several years (15–17). In addition, although several meta-analyses have been conducted to assess the prognostic value of PD-L1 in sarcomas, the results have been inconclusive (18, 19) due to the inconsistent results of PD-L1 assessment assays and the limited number of included studies.
In the present meta-analysis, the prognostic significance of PD-L1 expression was systemically and comprehensively evaluated with 39 independent studies of 3,680 bone and soft tissue sarcoma patients to investigate whether PD-L1 could serve as a prognostic biomarker in sarcomas.
Materials and Methods
Search Strategies
A systemic and comprehensive literature search was conducted in five databases with no language or publication date limitations: PubMed, Medline, Cochrane Library, EMBASE, and Web of Science. Articles published before October 2019 were included in the meta-analysis, and the last search was conducted on October 31, 2019. The following keywords were used to search the relevant literature: “sarcoma” or “bone sarcoma” or “soft tissue sarcoma” or “osteosarcoma” or “Ewing sarcoma” or “chondrosarcoma” or “myosarcoma” or “fibrosarcoma” or “synovial sarcoma” or “malignant fibrous histiocytoma” or “liposarcoma” or “epithelioid sarcoma” or “spindle cell sarcoma” or “carcinosarcoma” or “leiomyosarcoma” or “angiosarcoma” or “hemangiosarcoma” or “lymphangiosarcoma” or “malignant lymphangioendothelioma” and “PD-L1” or “B7-H1” or “CD274” or “programmed cell death 1 ligand 1 protein.” Moreover, we manually searched the references in each study to avoid missing potentially relevant data.
Inclusion and Exclusion Criteria
The eligible studies included in the meta-analysis had to be in accordance with the following criteria: (1) studies on patients with pathologically confirmed bone and/or soft tissue sarcomas, (2) studies focusing on the correlations between PD-L1 expression and the survival and/or clinicopathological outcome(s), and (3) studies utilizing defined cutoff values to classify the PD-L1 expression as “high” and “low” or “positive” and “negative.” Studies were excluded if they (1) were reviews, case reports, letters, or conference abstracts, (2) investigated cell experiments and/or animal experiments, (3) were not related to PD-L1 expression, or (4) comprised overlapping patients and/or insufficient data. The eligibility of the included studies was determined by two authors independently. Any discrepancy was resolved by a consensus after a discussion.
Data Extraction and Quality Assessment
The data of interest were extracted, including (1) basic characteristics (first author, publication year, tumor type, number and source of patients, presence of positive/high PD-L1 expression, cutoff value, assay method, biomarkers, and follow-up duration), (2) hazard ratios (HRs) with 95% confidence intervals (CIs) that either were given directly or could be calculated from published and available data by using the Cox regression method with a standard procedure (SPSS version 16.0) or Tierney's methods (20), and (3) data to estimate odds ratios (ORs) with 95% CIs for the correlations between PD-L1 and clinicopathological outcome(s). The HRs from the multivariate analysis were extracted if the HRs from both the univariate and the multivariate analysis were available.
The Newcastle–Ottawa Scale (NOS, https://www.ohri.ca/programs/clinical/epidemiology/oxford.asp) system was used to assess the methodological quality of each study, and the included studies were assessed for risk of bias by using the Quality and Prognosis Studies (QUIPS) method (21). The risk of bias was assessed for the following domains: study participation, study attrition, prognostic factor measurement, outcome measurement, study confounding, and statistical analysis. The score was classified and reported as low, moderate, or high. The assessment was performed by two authors independently. If there was any disagreement, it was resolved by a consensus after a discussion.
Statistical Analysis
Estimates of HRs, ORs, and their 95% CIs were used to assess the pooled survival related to and the clinicopathological significance of PD-L1 expression. Cochran's Q test and Higgins I2 statistic were employed to assess the statistical heterogeneity among studies. A fixed-effects model was appropriately used if there was no significant heterogeneity (P > 0.10 or I2 <50%); otherwise, a random-effects model was used, and a subgroup meta-analysis was conducted to identify the underlying heterogeneity when at least five studies were included. Publication bias was assessed by using a funnel plot analysis, Begg's tests, and Egger's tests, and a sensitivity analysis was used to assess the stability of the pooled results when at least five studies were included. P < 0.05 was considered as statistically significant. All analyses were conducted by using Stata version 14.0 (Stata Corporation, College Station, TX, USA).
Results
Study Selection
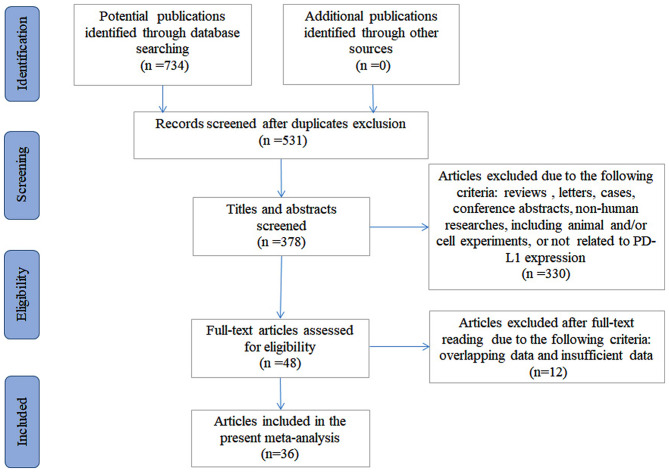
There were 734 articles identified initially from the systemic and comprehensive search. After the duplicates were excluded, 531 articles were subjected to further screening. After viewing the title and the abstract, 378 articles remained. Three hundred thirty articles were omitted due to irrelevance. After a full-text review, 12 articles were excluded, including one with overlapping data and 11 with insufficient data. In total, 36 articles containing 39 independent studies with 3,680 bone and soft tissue sarcoma patients were included (15–17, 22–54), among which 31 articles were used to evaluate the prognostic significance of PD-L1 expression and 27 articles were used to evaluate the clinicopathological significance. The selection process is shown in Figure 1.
Figure 1.
Flow diagram of the study selection process.
Study Characteristics and Quality Assessment
The summarized study characteristics are shown in Table 1. Briefly, the publication year of the eligible studies, written in English, ranged from 2013 to 2019. The patients were collected internationally from Asian (n = 17) to non-Asian (n = 23) populations. The number of patients ranged from 11 to 492. A variety of sarcomas were investigated in the studies, including osteosarcoma (n = 11), soft tissue sarcoma (STS, n = 9), unclassified sarcoma (n = 4), angiosarcoma (n = 4), leiomyosarcoma (n = 3), liposarcoma (n = 2), chondrosarcoma (n = 2), Ewing's sarcoma (n = 1), myeloid sarcoma (n = 1), pleomorphic sarcoma (n = 1), and synovial sarcoma (n = 1). Immunohistochemistry (IHC), quantitative polymerase chain reaction (qPCR), or RNA sequencing methodologies were employed to evaluate the PD-L1 expression. The cutoff values were available in studies using IHC, and the positivity ranged from 5.90 to 75.90%. Moreover, data from studies were extracted to analyze the HRs of overall survival (OS, n = 30), as a primary endpoint, and other survival outcomes, including recurrence-free survival (RFS, n = 4), metastasis-free survival (MFS, n = 3), event-free survival (EFS, n = 3), disease-free survival (DFS, n = 5), and progression-free survival (PFS, n = 4). The follow-up duration ranged from 1 to 366 months based on the available data. The correlation between PD-L1 expression and clinicopathological outcomes was assessed with data from 29 studies, including patient age and sex, tumor size, depth and localization, tumor grade and stage, metastasis, recurrence, previous chemotherapy/radiotherapy, chemotherapy response, tumor-infiltrating lymphocytes (TILs), and programmed cell death 1 (PD1) positivity.
Table 1.
Characteristics of the included studies.
| Study | References | Patients | Tumor type | Positive (%) | Source | Cutoff value | Assay method | Outcome | Biomarker* | HR source (M/U) | Follow-up (m) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | (17) | 29 | Angiosarcoma | 22 (75.90) | Japan | >5% of cells | IHC | OS, CPF | PD-L1, PD1 | SC (U) | NA |
| 2 | (23) | 56 | Liposarcoma | 13 (23.20) | China | IRS >0 | IHC | CPF | PD-L1, PD1, CD4, CD8, FoxP3, and CD20 | NA | NA |
| 3 | (22) | 19 | Osteosarcoma | 8 (42.10) | Japan | NA | qPCR | OS, MFS | PD-L1, GZMB, PRF, and IFNγ | SC (U) | 8.2–237.4 |
| 4 | (15) | 25 | Angiosarcoma | 5 (19.00) | USA | ≥5% of cells | IHC | OS | PD-L1, VEGF, and VEGFR | Directly (M) | NA |
| 5 | (16) | 128 | STS | 23 (40.40) | Germany | ≥0% of TCs | IHC | OS, DFS | PD-L1, PD-1, CD3, and CD8 | Directly (M) | 2–222 |
| 6 | (33) | 11 | Leiomyosarcoma | 4 (36.40) | Israel | >1% of TCs | IHC | OS,CPF | MMR proteins, PD-L1, PD-1, CD3, and CD8 | Cox-regression (M) | 4–120 |
| 7 | (32) | 131 | MS | 10 (10.20) | Japan | ≥1% of TCs | IHC | OS, PFS | PD-L1, PD1, TP53, and CXCR4 | SC (U) | 8 (1–100) |
| 8 | (31) | 106 | Leiomyosarcoma | 32 (30.20) | Netherlands | ≥1% of cells | IHC | OS, DFS, CPF | CD163, CD3, PD-L1/PD-L2, and HLA-I | SC (U) | NA |
| 9 | (30) | 370 | Ewing's sarcoma | 71 (19.20) | Spain | >5% of TCs | IHC | OS,PFS, CPF | PD-L1, PD-1, and CD8 | P value (U) | NA |
| 10 | (29) | 36 | SS | 9 (47.40) | Japan | NA | qPCR | OS, PFS | CD4, CD8, FOXP3, CD163, HLA, PD-L1, and PD-L2 | SC(U) | 113 (6–366) |
| 11 | Cohort a (28) | 32 | Liposarcoma | 7 (21.90) | Korea | ≥1% of TCs | IHC | OS, RFS, CPF | PD-L1 | SC(U) | 19 |
| 12 | Cohort b (28) | 60 | PS | 12 (20.00) | Korea | ≥1% of TCs | IHC | OS, RFS, CPF | PD-L1 | SC (U) | 49 |
| 13 | (27) | 46 | STS | 21 (45.70) | USA | >1% of cells | IHC | MFS,CPF | PD-L1 | SC (U) | 69.8 |
| 14 | (26) | 59 | Chondrosarcoma | 40 (67.80) | China | ≥1% of cells | IHC | RFS, CPF | PD-L1, PD-L2, Ki67, and TP53 | Directly (M) | 20 (1–118) |
| 15 | (25) | 92 | Osteosarcoma | 65 (70.70) | China | ≥1% of cells | IHC | CPF | PD-L1 | NA | NA |
| 16 | (24) | 234 | Sarcomas | 45 (20.50) | China/USA | ≥10% of cells | IHC | OS, CPF | PD-L1, PD-L2, and PD-1 | SC (U) | NA |
| 17 | (44) | 492 | STS | 196 (39.80) | France | NA | RNA seq | MFS,CPF | PD-L1 | Directly (M) | 0–120 |
| 18 | (43) | 24 | Angiosarcoma | 16 (66.70) | Italy | ≥5% of TCs | IHC | OS, DFS, CPF | PD-L1 | SC (U) | NA |
| 19 | Cohort a (42) | 256 | STS | 34 (21.10) | Germany | NA | RNA seq | OS | CD3D, CD4/CD8, PD-L1, and CD3Z | Directly (U) | NA |
| 20 | Cohort b (42) | 103 | Leiomyosarcoma | 16 (16.00) | Germany | NA | RNA seq | OS | CD3D, CD4, CD68, CD4/CD8, PD-L1, and CD3Z | Directly (U) | NA |
| 21 | (41) | 13 | Osteosarcoma | 9 (69.20) | Brazil | IRS > 2 | IHC | OS, CPF | HLA-G, HLA-E, and PD-L1 | Cox-regression (M) | 32 (2–156) |
| 22 | (40) | 72 | Osteosarcoma | 30 (41.70) | USA | Score ≥ 2 | IHC | OS, CPF | PD-L1 | Directly (M) | NA |
| 23 | (39) | 86 | Osteosarcoma | 12 (14.00) | Italy | >5% of cells | IHC | OS | CD8, Tia1,CD3, FOXP3, PD-1, PD-L1, Argase-1, CD303, CD68, and CD163 | Directly (M) | 96 (12–156) |
| 24 | (38) | 81 | STS | 48 (59.00) | USA | IHC score >0 | IHC | CPF | PD-L1 and PD-1 | NA | NA |
| 25 | (37) | 163 | STS | 19 (11.70) | China | >1% of TCs | IHC | OS, DFS, CPF | PD-L1 and FOXP3 | Directly (U) | 75 (1–176) |
| 26 | (36) | 22 | Osteosarcoma | 4 (18.20) | Netherlands | ≥1% of cells | IHC | OS, DFS, CPF | HLA-I, CD3, CD8, and PD-L1 | SC (U) | 56 (14–117) |
| 27 | (35) | 208 | Sarcomas | 18 (8.65) | Netherlands | ≥10% of TCs | IHC | OS | PD-1, PD-L1, and CD8 | SC (U) | NA |
| 28 | (34) | 93 | Osteosarcoma | 33 (35.50) | China | ≥10% of cells | IHC | OS, CPF | VEGFR2 and PD-L1 | SC (U) | NA |
| 29 | (49) | 106 | Angiosarcoma | 32 (30.20) | Japan | >5% of cells | IHC | OS, CPF | PD-L1 and PD1 | Directly (M) | 20 (3–100) |
| 30 | (48) | 82 | STS | 35 (42.70) | Korea | Score ≥ 2 | IHC | OS,RFS,CPF | PD-L1 | Directly (M)/SC (U) | 34 (4–85) |
| 31 | Cohort a (47) | 51 | Osteosarcoma | 3 (5.90) | USA | ≥1% of cells | IHC | EFS, CPF | PD-L1 | SC (U) | 84 (4–150) |
| 32 | Cohort b (47) | 41 | Osteosarcoma | 12 (29.30) | USA | ≥1% of cells | IHC | EFS, CPF | PD-L1, CD68, and CD1a | SC (U) | 54 (15–100) |
| 33 | (46) | 22 | Chondrosarcoma | 9 (41.00) | Netherlands | ≥1% of cells | IHC | OS | PD-L1 | SC (U) | 54 (15–100) |
| 34 | (45) | 66 | Sarcomas | 20 (30.30) | Turkey | >5% of cells | IHC | OS,PFS, CPF | PD-1 and PD-L1 | Cox-regression (M) | 30 (4–310) |
| 35 | (52) | 59 | Sarcomas | 36 (59.30) | UK | >5% of TCs | IHC | CPF | PD-L1 and CD8/PD1 | NA | 33 (3–200) |
| 36 | (51) | 47 | STS | 4 (8.50) | USA | >1% of TCs | IHC | OS,CPF | CD3, CD4, CD8, FOXP3, PD-L1, and PD1 | P value (U) | NA |
| 37 | (50) | 16 | Osteosarcoma | 12 (75.00) | USA | >1 cell /HPF | IHC | CPF | PD-L1 and PD1 | NA | NA |
| 38 | (53) | 38 | Osteosarcoma | 9 (27.00) | USA | Score >2-log | qPCR | OS,CPF | PD-L1 | SC (U) | 36 (1–200) |
| 39 | (54) | 105 | STS | 68 (64.80) | Korea | Score ≥8 | IHC | OS, EFS, CPF | PD1 and PD-L1 | Directly (M) | 35 (1–175) |
Analyzed for survival and/or clinicopathological outcomes.
HR, hazard ratio; IHC, immunohistochemistry; qPCR, quantitative polymerase chain reaction; OS, overall survival; EFS, event-free survival; MFS, metastasis-free survival; PFS, progression-free survival; RFS, recurrence-free survival; DFS, disease-free survival; CPF, clinicopathological feature; STS, soft tissue sarcomas; MS, myeloid sarcoma; SS, synovial sarcoma; PS, pleomorphic sarcoma; SC, survival curve; IRS, immunoreactive score; NA, not available; TC, tumor cell; HPF, high-power field; U, univariate; M, multivariate; PD-L1, programmed cell death 1 ligand-1; PD-L2, programmed cell death 1 ligand-2; PD1, programmed cell death 1; VEGF, vascular endothelial growth factor; VEGFR, vascular endothelial growth factor receptor; MMR, mismatch repair; HLA, human leukocyte antigen; CXCR4, CXC chemokine receptor 4; TP53, tumor suppressor p53; GZMB, granzyme B; PRF, perforin; IFNγ, interferon-γ; FOXP3, forkhead box P3; Tia1, cytotoxic granule-associated RNA binding protein 1.
The included 39 studies were critically appraised for methodological quality and risk of bias by using the NOS system and the QUIPS criteria, respectively. The results showed that all of the studies were of high methodological quality, with scores that ranged from six to nine stars. Most of the studies were retrospective studies and were assessed as having a low or moderate risk of bias. Almost no study provided information about patients lost to follow-up. Due to a lack of sufficient data on survival outcome(s) or study confounding, studies with only clinicopathological features were scored with a high risk of bias. Some studies were scored with a high or moderate risk of study participation due to missing details and the limited number of patients. A few studies were scored with a high or moderate risk of study confounding because of insufficient survival analyses for some important factors. The results are shown in Supplementary Table 1.
Prognostic Value of PD-L1 Expression in Sarcomas
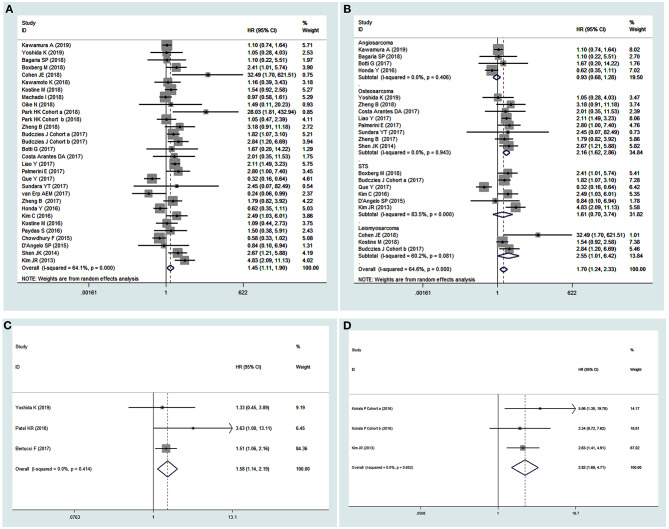
A random-effects model was used in the analysis for OS as obvious heterogeneity was observed (I2 = 64.10%, P = 0.00). The results showed that elevated PD-L1 expression was correlated with poor OS (HR 1.45, 95% CI 1.11–1.90, P < 0.01, Figure 2A). Moreover, to investigate the underlying source of heterogeneity, subgroup analyses by tumor type, publication date, sample size and source, analysis model of effects, PD-L1 positivity, assay method, and PD-L1 expression region (in the tumor or the tumor microenvironment) were conducted. The results showed that none of these factors were the underlying source of heterogeneity, and the overall value of PD-L1 did not change (Table 2). Moreover, as various tumor subtypes were observed in the present analysis, a stratified analysis by tumor subtype was conducted, in which no less than three studies were included. A random-effects model was used (I2 = 64.60%, P = 0.00), and the results showed that PD-L1 overexpression could predict poor OS specifically in osteosarcoma (P < 0.01) and leiomyosarcoma (P < 0.05, Figure 2B). Thus, it was indicated that PD-L1 was a predictor for poor OS in sarcoma patients.
Figure 2.
Forest plots of pooled HR for pooled overall survival (OS) (A) and OS stratified by tumor subtypes (B), metastasis-free survival (C), and event-free survival (D).
Table 2.
Summary of correlation between PD-L1 expression and survival outcomes.
| Subgroup | Study (n) | HR (95% CI) | P | Heterogeneity | Effects model | |
|---|---|---|---|---|---|---|
| I2 (%) | P | |||||
| OS | 30 | 1.45 (1.11–1.90) | <0.01* | 64.10 | 0.00 | Random effects |
| Tumor type | ||||||
| Bone sarcoma | 10 | 1.72 (1.30–2.28) | <0.01* | 14.30 | 0.31 | |
| Non-bone sarcoma | 20 | 1.33 (0.92–1.93) | 0.14 | 70.40 | 0.00 | |
| Publication (year) | ||||||
| ≥2018 | 12 | 1.41 (1.02–1.94) | <0.05* | 31.70 | 0.14 | |
| <2018 | 18 | 1.40 (0.94–2.09) | 0.10 | 73.60 | 0.00 | |
| Sample source | ||||||
| Asian | 14 | 1.51 (0.93–2.44) | 0.10 | 71.90 | 0.00 | |
| Non-Asian | 16 | 1.48 (1.08–2.01) | <0.05* | 52.60 | 0.01 | |
| Sample size (n) | ||||||
| ≥85 | 12 | 1.32 (0.87–2.01) | 0.19 | 78.50 | 0.00 | |
| <85 | 18 | 1.55 (1.09–2.21) | 0.01* | 42.40 | 0.03 | |
| Analysis model | ||||||
| Univariate | 21 | 1.27 (0.93–1.72) | 0.13 | 59.50 | 0.00 | |
| Multivariate | 9 | 2.01 (1.17–3.45) | <0.05* | 66.90 | 0.00 | |
| Positivity (%) | ||||||
| ≥30 | 15 | 1.62 (1.13–2.33) | <0.01* | 65.90 | 0.00 | |
| <30 | 15 | 1.28 (0.83–1.96) | 0.26 | 63.70 | 0.00 | |
| Assay method | ||||||
| IHC | 26 | 1.36 (1.00–1.86) | <0.05* | 67.10 | 0.00 | |
| Non-IHC | 5 | 2.06 (1.42–2.98) | <0.01* | 0.00 | 0.00 | |
| Expression region | ||||||
| Tumor cell | 14 | 1.32 (0.772–2.27) | 0.31 | 74.70 | 0.00 | |
| Non-tumor cell | 16 | 1.59 (1.24–2.03) | <0.01* | 34.80 | 0.08 | |
| RFS | 4 | 1.42 (0.92–2.21) | 0.12 | 0.00 | 0.45 | Fixed effects |
| MFS | 3 | 1.58 (1.14–2.19) | <0.01* | 0.00 | 0.41 | Fixed effects |
| EFS | 3 | 2.82 (1.69–4.71) | <0.01* | 0.00 | 0.65 | Fixed effects |
| DFS | 5 | 1.17 (0.53–2.58) | 0.70 | 74.5 | 0.00 | Random effects |
| Publication date | ||||||
| ≥2018 | 2 | 1.39 (0.66–2.91) | 0.39 | 47.30 | 0.17 | |
| <2018 | 3 | 1.09 (0.27–4.46) | 0.90 | 75.80 | 0.12 | |
| Sample size | ||||||
| ≥100 | 3 | 0.90 (0.35–2.34) | 0.83 | 83.20 | 0.00 | |
| <100 | 2 | 2.29 (0.77–6.76) | 0.14 | 0.00 | 0.84 | |
| Tumor type | ||||||
| STS | 2 | 0.89 (0.16–4.80) | 0.89 | 90.40 | 0.00 | |
| Non-STS | 3 | 1.24 (0.72–2.14) | 0.44 | 0.00 | 0.43 | |
| Positivity (%) | ||||||
| ≥35 | 2 | 2.27 (1.01–5.09) | 0.05 | 0.00 | 0.84 | |
| <35 | 3 | 0.82 (0.34–2.01) | 0.66 | 76.00 | 0.02 | |
| Cutoff value | ||||||
| ≥1% of tumor cells | 3 | 2.27 (1.01–5.09) | 0.66 | 0.00 | 0.84 | |
| Non-≥1% of tumor cells | 2 | 0.82 (0.34–2.01) | <0.05* | 76.00 | 0.02 | |
| Expression region | ||||||
| Tumor cell | 3 | 1.16 (0.28–4.75) | 0.84 | 84.10 | 0.00 | |
| Non-tumor cell | 2 | 1.16 (0.66–2.04) | 0.62 | 0.00 | 0.33 | |
| PFS | 4 | 1.00 (0.65–1.53) | 0.99 | 19.8 | 0.29 | Fixed effects |
Significant difference.
HR, hazards ratio; CI, confidence interval; OS, overall survival; RFS, recurrence-free survival; MFS, metastasis-free survival; EFS, event-free survival; DFS, disease-free survival; PFS, progression-free survival; STS, soft tissue sarcoma; IHC, immunohistochemistry.
Other extracted important survival outcomes in the present analysis, including RFS, MFS, PFS, EFS, and DFS, are survival parameters related to disease according to their definition, but these data had limited numbers and thus insufficient effect size. In our analysis, the results showed that an elevated PD-L1 expression predicted poor MFS (HR 1.58, 95% CI 1.14–2.19, P < 0.01, Figure 2C) and EFS (HR 2.82, 95% CI 1.69–4.71, P < 0.01, Figure 2D) but was not associated with other survival outcomes (Table 2). As obvious heterogeneity was observed (I2 = 74.50%, P = 0.003) in the DFS analysis, a random-effects model was used, and subgroup analyses were conducted. The results of the subgroup analyses showed that the heterogeneity was not reduced by stratification of the data by publication date, tumor type, sample size, PD-L1 positivity, cutoff value, and PD-L1 expression region. However, the pooled HR did not change, indicating that the result was stable. In addition, although stratified analyses by tumor type (bone sarcoma vs. non-bone sarcoma) were conducted for other survival outcomes, no conclusive and reliable results were observed due to the limited number of included studies, as shown in Supplementary Table 2.
Clinicopathological Value of PD-L1 Expression in Sarcomas
The clinicopathological value of PD-L1 expression is summarized in Table 3. Briefly, 29 studies were included, and comprehensive meta-analyses were conducted to determine the correlation between the PD-L1 expression and patient age (n = 4) and sex (n = 21), tumor size (n = 6), depth (n = 3) and localization (n = 8), tumor grade (n = 15) and stage (n = 8), metastasis (n = 11), recurrence (n = 7), previous chemotherapy/radiotherapy (n = 10 and n = 7, respectively), chemotherapy response (n = 5), and TILs (n = 12). A random-effects model or a fixed-effects model was used based on the results of the heterogeneity analysis for each parameter, as shown in Table 3. PD-L1 overexpression was positively correlated with a higher rate of metastasis (OR 2.95, 95% CI 1.32–6.60, P < 0.01), a higher tumor grade (OR 3.63, 95% CI 2.55–5.16, P < 0.01), and more TILs (OR 5.55, 95% CI 2.86–10.76, P < 0.01) but was not correlated with other clinicopathological outcomes. For the TIL subset analysis, the PD-L1 expression was not correlated with CD4-positive TILs (OR 2.45, 95% CI 0.69–8.73, P = 0.17). Moreover, a stratified analysis by bone sarcoma vs. non-bone sarcoma was conducted for each outcome to clarify whether the correlation was tumor type dependent. The results showed that PD-L1 was specifically correlated with metastasis and CD8+ TILs in bone sarcomas and CD3+ TILs in non-bone sarcomas. In addition, PD-L1 overexpression was correlated with a good response to chemotherapy in bone sarcomas but with a poor response in non-bone sarcomas (Supplementary Table 3). For the outcomes with obvious heterogeneity, subgroup analyses were conducted. The results showed that some factors might be the underlying source of heterogeneity in some of the clinicopathological outcomes, as shown in Supplementary Table 4.
Table 3.
Summary of correlation between PD-L1 expression and clinicopathological outcomes.
| Category | Study (n) | Heterogeneity | Effects model | OR (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| I2 (%) | P | |||||
| <60 years/≥60 years | 4 | 0.00 | 0.64 | Fixed | 0.65 (0.39–1.10) | 0.11 |
| Male/female | 21 | 19.00 | 0.21 | Fixed | 1.07 (0.88–1.30) | 0.50 |
| Size 5 cm/ ≤ 5 cm >10 cm/ ≤ 10 cm |
6 3 3 |
54.40 57.20 54.40 |
0.05 0.10 0.05 |
Random Random Random |
1.14 (0.54–2.42) 0.86 (0.29–2.50) 1.54 (0.48–4.97) |
0.74 0.78 0.47 |
| Depth | 3 | 65.70 | 0.05 | Random | 1.60 (0.63–4.06) | 0.33 |
| Extremity/non-extremity | 8 | 52.30 | 0.04 | Random | 1.01 (0.58–1.75) | 0.99 |
| Stage (high/low) | 8 | 58.50 | 0.02 | Random | 1.94 (0.70–5.39) | 0.20 |
| Grade (high/low) | 15 | 17.2 | 0.26 | Fixed | 3.63 (2.55–5.16) | <0.01* |
| Radiotherapy (Y/N) | 7 | 66.60 | 0.01 | Random | 1.36 (0.58–3.20) | 0.48 |
| Chemotherapy (Y/N) | 10 | 0.00 | 0.95 | Fixed | 1.06 (0.70–1.59) | 0.79 |
| Met/non-met | 11 | 75.80 | 0.00 | Random | 2.95 (1.32–6.60) | <0.01* |
| Rec/non-rec | 7 | 51.30 | 0.06 | Random | 2.08 (0.87–4.96) | 0.10 |
| CR (good/poor) | 5 | 86.70 | 0.00 | Random | 0.55 (0.09–3.53) | 0.53 |
| TIL (Y/N) | 12 | 68.90 | 0.00 | Random | 5.55 (2.86–10.76) | <0.01* |
| PD1 + TIL (Y/N) | 5 | 0.00 | 0.41 | Fixed | 3.21 (1.85–5.58) | <0.01* |
| CD3 + TIL (Y/N) | 5 | 0.00 | 0.51 | Fixed | 3.12 (1.49–6.54) | <0.01* |
| CD4 + TIL (Y/N) | 2 | 0.00 | 0.94 | Fixed | 2.45 (0.69–8.73) | 0.17 |
| CD8 + TIL (Y/N) | 7 | 70.10 | 0.00 | Random | 6.90 (2.62–18.20) | <0.01* |
| FOXP3 + TIL (Y/N) | 2 | 0.00 | 0.62 | Fixed | 14.28 (4.47–45.65) | <0.01* |
Statistically significant.
OR, odds ratio; CI, confidence interval; Met, metastasis; Rec, recurrence; CR, chemotherapy response; TIL, tumor-infiltrating lymphocyte; PD1, programmed cell death 1.
Publication Bias and Sensitivity Analysis
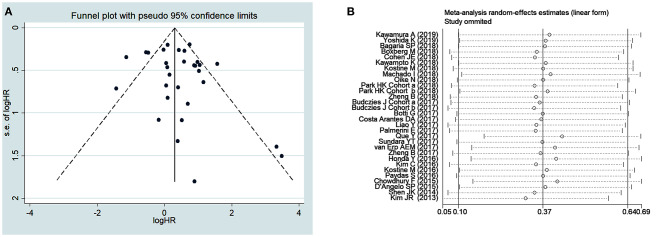
Publication bias was assessed by using funnel plot analysis (Figure 3) and Begg's and Egger's tests (Table 4). For survival outcomes, no obvious publication bias was observed in the evaluation of OS (Figure 3A) or RFS, MFS, EFS, and DFS (Table 4), but publication bias was noted for PFS (P = 0.04 in Egger's tests). For clinicopathological outcomes, Egger's tests showed that there might be a publication bias in the analysis of PD1+ TILs (P = 0.03). No obvious publication bias was observed in any other analysis of clinicopathological outcomes.
Figure 3.
Funnel plots for publication bias (A) and sensitivity analysis (B) of overall survival.
Table 4.
Begg's test and Egger's test for publication bias.
| Analysis value | Study (n) | Begg's test | Egger's test | ||
|---|---|---|---|---|---|
| z | P | t | P | ||
| OS | 30 | 0.82 | 0.41 | 1.10 | 0.28 |
| RFS | 4 | 1.02 | 0.31 | 1.18 | 0.36 |
| MFS | 3 | 1.04 | 0.30 | 0.73 | 0.60 |
| EFS | 3 | 1.04 | 0.30 | 0.69 | 0.62 |
| DFS | 5 | 0.73 | 0.46 | 1.98 | 0.14 |
| PFS | 4 | 1.70 | 0.09 | 5.06 | 0.04* |
| Age | 4 | 0.34 | 0.73 | 2.78 | 0.11 |
| Gender | 21 | 0.27 | 0.79 | −0.41 | 0.69 |
| Size | 6 | 0.00 | 1.00 | 0.00 | 1.00 |
| Radiotherapy | 7 | 0.00 | 1.00 | 0.56 | 0.60 |
| Chemotherapy | 10 | 1.07 | 0.28 | −0.76 | 0.47 |
| Localization | 8 | 0.37 | 0.71 | −0.32 | 0.76 |
| Depth | 3 | 1.04 | 0.30 | −3.64 | 0.17 |
| Grade | 15 | 0.89 | 0.37 | 1.39 | 0.19 |
| Stage | 8 | 0.62 | 0.54 | −0.08 | 0.94 |
| Metastasis | 11 | 0.00 | 1.00 | 0.41 | 0.69 |
| Recurrence | 7 | 1.50 | 0.13 | 0.70 | 0.52 |
| CR | 5 | 0.24 | 0.81 | −0.09 | 0.93 |
| TIL (Y:N) | 12 | 0.07 | 0.95 | −0.27 | 0.79 |
| PD1 + TIL (Y:N) | 5 | 1.22 | 0.22 | 4.10 | 0.03* |
| CD3 + TIL (Y:N) | 5 | −0.24 | 1.00 | 1.44 | 0.25 |
| CD4 + TIL (Y:N) | 2 | 0.00 | 1.00 | – | – |
| CD8 + TIL (Y:N) | 7 | 0.00 | 1.00 | −0.31 | 0.77 |
| FOXP3 + TIL (Y:N) | 2 | 0.00 | 1.00 | – | – |
Significant difference.
OS, overall survival; RFS, recurrence-free survival; MFS, metastasis-free survival; EFS, event-free survival; DFS, disease-free survival; PFS, progression-free survival; CR, chemotherapy response; TIL, tumor-infiltrating lymphocyte; PD1, programmed cell death 1.
A sensitivity analysis was performed to assess the effects of each individual study on the pooled HRs and ORs of our analysis. The results of the sensitivity analysis, which was performed by omitting one study at a time, showed that no unique study significantly affected the pooled HRs for OS (Figure 3B) and other outcomes (Supplementary Figures 1–14), indicating that the results of the present meta-analysis were stable and credible.
Discussion
PD-L1, one of the immune checkpoint molecules, can inhibit T cell activation and proliferation by binding with PD-1 and protect tumor cells from the host immunologic surveillance system. Immunotherapy, by administration of immune checkpoint inhibitors, has been considered as the foremost method in individualized medicine due to its tremendous success in cancer treatment. Clinical trials have shown satisfactory results by using anti-PD-1/PD-L1 blockade in some cancers (11). However, the results in bone and soft tissue sarcomas are unsatisfactory (14). Recent studies have indicated that the high expression of PD-L1 is associated with poor prognosis in some cancers (55, 56). Moreover, several meta-analyses have evaluated the clinical significance of PD-L1 expression in patients with sarcomas, but the results have been inconclusive (18, 19). In the present meta-analysis, 39 independent studies (including 3,680 patients) were included, and the pooled results showed that PD-L1 overexpression could predict poor OS, MFS, and EFS. Furthermore, an elevated PD-L1 expression was correlated with a higher tumor metastasis rate, a more advanced tumor grade, and more T lymphocyte infiltration.
In terms of survival outcomes, the results showed that PD-L1 overexpression could predict poor OS and EFS, which was similar to the findings of previous reports (18, 19). As obvious heterogeneity was observed for OS, a random-effects model was used to reduce heterogeneity, and subgroup analyses were conducted. Although the results did not identify the underlying source of heterogeneity, the pooled results did not change in each subgroup analysis. Moreover, a stratified analysis by tumor subtype showed that PD-L1 overexpression could specifically predict poor OS in osteosarcoma and leiomyosarcoma. There was no obvious publication bias, and the sensitivity analysis indicated that the results were stable. Therefore, our results were robust and reliable. Some other factors should be considered to explain the underlying heterogeneity. There are more than 50 different subtypes of sarcoma, and more than 10 subtypes were included in the present analysis, which introduced heterogeneity into the whole meta-analysis. Moreover, differences in the detection techniques, such as antibody selection and dilution, and the antigen retrieval methods may have affected the sensitivity of detection, and the positivity of the PD-L1 expression might have been affected by the different cutoff values. In addition, the region used to define the PD-L1 expression was different among the included studies. These might be other explanations for the heterogeneity.
It was initially reported that PD-L1 overexpression predicted poor MFS but was not associated with DFS, RFS, or PFS in patients with sarcoma. A previous meta-analysis showed a similar result that the PD1/PD-L1 expression was associated with lymph node metastasis in patients with osteosarcoma (57). PD-L1 is widely considered to function in the tumor immunologic surveillance system; however, the tumor-intrinsic roles of PD-L1 in regulating the ability of human cancers to disseminate and to metastasize are currently not understood. PD-L1 is suggested to be involved in cancer metastasis via several mechanisms. PD-L1 is shown to promote epithelial-to-mesenchymal transition and is involved in tumor proliferation, migration, and invasion (58, 59). Moreover, cancer stem cells (CSCs) are considered to be involved in tumor metastasis (60), and PD-L1 is suggested to sustain stemness by promoting OCT4 and Nanog expression in CSCs (61) and promoting tumor-initiating cell generation and virulence in cancer (62). Thus, it is suggested that PD-L1 is involved in the possible immune evasion mechanism employed by CSCs during metastasis (63). In addition, several signaling pathways have been suggested to modulate the role of PD-L1 in cancer initiation, metastasis, and chemoresistance, including the MAPK/ERK pathway, the PI3K/AKT pathway, and the RAS/MEK pathway (64, 65). Sarcomas are a group of malignancies with a high propensity for metastasis, and the prognosis of metastatic patients remains poor. Further studies are encouraged to investigate the mechanism by which PD-L1 is involved in sarcoma metastasis. However, as there were only three studies in the pooled analysis for MFS, the results should be interpreted with caution.
The results from the analyses of clinicopathological outcomes were similar to those from previous reports (18, 19). An elevated PD-L1 expression was correlated with a higher rate of metastasis, a higher tumor grade, and more T lymphocyte infiltration. In the analysis for tumor grade, the result was different from a previous report (19). In other cancers, the correlation between PD-L1 expression and tumor grade remains controversial (55, 66), which might arise from the inherent differences between cancers. Moreover, the prognosis of patients with high-grade sarcomas is not satisfactory. It was suggested that PD-L1 overexpression might predict the presence of advanced sarcoma. In addition, our results might be more conclusive and reliable than those from other meta-analyses because more studies were included (n = 15). Moreover, although the results showed that PD-L1 expression was not correlated with chemotherapy response, stratified analyses by tumor type showed that PD-L1 overexpression might predict good response to chemotherapy in bone sarcomas but poor response in non-bone sarcomas, and this difference might be due to the difference in chemotherapy sensitivity between these two types of sarcoma.
Another major result in our analysis was that high PD-L1 expression was correlated with more TILs, which was similar to the findings of previous analyses in sarcomas and some other cancers (19, 67, 68). However, in the analyses of T cell subtype, the results showed that elevated PD-L1 expression was mainly correlated with PD1+ T cells, CD3+ T cells, and CD8+ T cells but not with CD4+ T cells. In the process of tumor immune tolerance, the PD1/PD-L1 axis acts to suppress the T cell response. PD-L1 is expressed on tumor cells and binds with PD-1, which is expressed on tumor-infiltrating CD8+ T cells as well as CD4+ T cells and other inflammatory cells. Additionally, PD-L1 makes tumor cells less susceptible to specific CD8+ T cells (69). Therefore, the killing effect of T cells on tumor cells is inhibited (70, 71). Moreover, PD-L1 expression can be modulated by tumor cells. One feasible pathway is induced by IFNγ production and subsequent IFNGR/JAK/STAT signaling in tumor cells, which depends on TILs (72, 73). Thus, the correlation between PD-L1 overexpression and increased tumor-infiltrating T cells in sarcomas might indicate a higher capacity of sarcomas to evade immune surveillance. However, inhibitors targeting the PD1/PD-L1 axis have not resulted in satisfactory results in sarcomas. The combination of immunotherapy with other therapeutics is postulated to show improved responses over checkpoint inhibitor monotherapies (74, 75), and investigations to explore the underlying mechanisms by which PD-L1 is involved in sarcoma are encouraged.
Several limitations in our analysis should be acknowledged. First, there might be some potential sources of bias. The HRs and their corresponding 95% CIs were extracted indirectly or directly from the studies, including from multivariate and univariate analyses, which might lead to statistical bias. Moreover, most studies included in this meta-analysis were retrospective, investigating patients with osteosarcoma and STS. A limited number of studies of other rare sarcomas were included, which might lead to subject selection bias. In addition, some other underlying sources of publication bias should be considered, such as unpublished negative results (76) or studies that could not be included due to language limitations or insufficient data (77). Second, due to the limited number of studies included for some outcomes, the results should be interpreted with caution. Third, a lack of uniformity in the definitions of some clinicopathological outcomes was observed, such as tumor stage, size and depth, and patient age, which might be due to the authors' preferences or the characteristics of certain sarcomas, and the results should be interpreted with caution. In addition, although the onset of sarcomas is age dependent and the immune context might play a different role in young patients than in elderly patients, we could not obtain a conclusive result regarding whether PD-L1 expression was correlated with patient age due to the limited number of studies and the lack of uniformity in age classification. In addition, PD-L1 expression has been considered to correlate with tumor response to anti-PD-1/PD-L1 agents in melanoma patients (65), but no study investigated the correlation between PD-L1 expression and administration of PD-L1 inhibitors, which may be a very important factor for evaluating their efficacy. Because sufficient data could not be extracted, the correlation between PD-L1 expression and PD1-positive tumors was not assessed. Sarcomas are a group of rare malignancies with low morbidity, and there were not sufficient numbers of studies for some tumor subtypes; therefore, more investigations are encouraged to clarify the significance of PD-L1 for those rare tumor subtypes.
Moreover, the assessment of PD-L1 expression through IHC is advocated by many studies, and a variety of antibodies and cutoff values for positive PD-L1 labeling are used. Other assay methods, such as qPCR and RNA sequencing, are also used to assess PD-L1 expression. To avoid selection bias and missing important data, we included studies using IHC and other assay methods. The results of the subgroup analysis by assay method showed that high PD-L1 expression predicted poor OS at both the protein level (HR 1.36, 95% CI 1.00–1.86, P < 0.01) and the mRNA level (HR 2.06, 95% CI 1.42–2.98, P < 0.01), and the assay method was not an underlying source of heterogeneity. As different cutoff values for PD-L1 positivity might affect the reliability of the evaluation, a subgroup analysis by cutoff value for IHC data was conducted. The results showed that PD-L1 overexpression could not predict poor OS with a defined cutoff value of ≥1% of cells, ≥5% of cells, or ≥10% of cells (Supplementary Figure 15), suggesting that the cutoff value could impact the prognostic value of PD-L1. It has been reported that different IHC methodologies for PD-L1 assessment provide different results (78); therefore, other approaches should be employed to improve the evaluation of PD-L1 expression in clinical practice (79), including in studies of patients with sarcomas.
Conclusions
In this meta-analysis, the results showed that PD-L1 overexpression could predict poor survival and was correlated with adverse tumor status in sarcoma patients. The results suggest that PD-L1 is a valuable prognostic biomarker in bone and soft tissue sarcomas, although more well-designed prospective studies with appropriate multivariate analyses are needed to validate our results.
Data Availability Statement
All datasets analyzed for this study are included in the article/Supplementary Material.
Author Contributions
FW and ZZ conceived and designed the project. FW, TY, and HZ analyzed and interpreted the data. FW, TY, and ZZ wrote the paper. All the authors have read and approved the final version of the manuscript and participated in acquiring the data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00749/full#supplementary-material
References
- 1.Burningham Z, Hashibe M, Spector L, Schiffman JD. The epidemiology of sarcoma. Clin Sarcoma Res. (2012) 2:14. 10.1186/2045-3329-2-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu CY, Yen CC, Chen WM, Chen TH, Chen PC, Wu HT, et al. Soft tissue sarcoma of extremities: the prognostic significance of adequate surgical margins in primary operation and reoperation after recurrence. Ann Surg Oncol. (2010) 17:2102–11. 10.1245/s10434-010-0997-0 [DOI] [PubMed] [Google Scholar]
- 3.Friebele JC, Peck J, Pan X, Abdel-Rasoul M, Mayerson JL. Osteosarcoma: a meta-analysis and review of the literature. Am J Orthop. (2015) 44:547–53. [PubMed] [Google Scholar]
- 4.Lahat G, Tuvin D, Wei C, Anaya DA, Bekele BN, Lazar AJ, et al. New perspectives for staging and prognosis in soft tissue sarcoma. Ann Surg Oncol. (2008) 15:2739–48. 10.1245/s10434-008-9970-6 [DOI] [PubMed] [Google Scholar]
- 5.Italiano A, Mathoulin-Pelissier S, Cesne AL, Terrier P, Bonvalot S, Collin F, et al. Trends in survival for patients with metastatic soft-tissue sarcoma. Cancer. (2011) 117:1049–54. 10.1002/cncr.25538 [DOI] [PubMed] [Google Scholar]
- 6.Hagleitner MM, de Bont ES, Te Loo DM. Survival trends and long-term toxicity in pediatric patients with osteosarcoma. Sarcoma. (2012) 2012:636405. 10.1155/2012/636405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukushi S, Nishida Y, Urakawa H, Kozawa E, Ishiguro N. Prognostic significance of histological invasion in high grade soft tissue sarcomas. Springerplus. (2014) 3:544. 10.1186/2193-1801-3-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maccio A, Madeddu C. Blocking inflammation to improve immunotherapy of advanced cancer. Immunol. (2020) 159:357–64. 10.1111/imm.13164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taefehshokr N, Baradaran B, Baghbanzadeh A, Taefehshokr S. Promising approaches in cancer immunotherapy. Immunobiology. (2019) 225:151875. 10.1016/j.imbio.2019.11.010 [DOI] [PubMed] [Google Scholar]
- 10.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. (2012) 12:252–64. 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romano E, Romero P. The therapeutic promise of disrupting the PD-1/PD-L1 immune checkpoint in cancer: unleashing the CD8 T cell mediated anti-tumor activity results in significant, unprecedented clinical efficacy in various solid tumors. J Immunother Cancer. (2015) 3:15. 10.1186/s40425-015-0059-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K, Hasegawa K, Katsumata N, Matsumoto K, Mukai H, Takahashi S, et al. Efficacy and safety of nivolumab in Japanese patients with uterine cervical cancer, uterine corpus cancer, or soft tissue sarcoma: multicenter, open-label phase 2 trial. Cancer Sci. (2019) 110:2894–904. 10.1111/cas.14148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groisberg R, Hong DS, Behrang A, Hess K, Janku F, Piha-Paul S, et al. Characteristics and outcomes of patients with advanced sarcoma enrolled in early phase immunotherapy trials. J Immunother Cancer. (2017) 5:100. 10.1186/s40425-017-0301-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chawla SP, van Tine BA, Pollack S, Ganjoo KN, Elias AD, Riedel RF, et al. A phase II randomized study of CMB305 and atezolizumab versus atezolizumab in NY-ESO-1+ soft tissue sarcoma: analysis of immunogenicity, tumor control, and patient survival. J Clin Oncol. (2019) 37(Suppl. 15):11011. 10.1200/JCO.2019.37.15_suppl.11011 [DOI] [Google Scholar]
- 15.Bagaria SP, Gatalica Z, Maney T, Serie D, Parasramka M, Attia S, et al. Association between programmed death-ligand 1 expression and the vascular endothelial growth factor pathway in Angiosarcoma. Front Oncol. (2018) 8:71. 10.3389/fonc.2018.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boxberg M, Steiger K, Lenze U, Rechl H, von Eisenhart-Rothe R, Wortler K, et al. PD-L1 and PD-1 and characterization of tumor-infiltrating lymphocytes in high grade sarcomas of soft tissue - prognostic implications and rationale for immunotherapy. Oncoimmunology. (2018) 7:e1389366. 10.1080/2162402X.2017.1389366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamura A, Kawamura T, Riddell M, Hikita T, Yanagi T. Regulation of programmed cell death ligand 1 expression by atypical protein kinase C lambda/iota in cutaneous angiosarcoma. Cancer Sci. (2019) 110:1780–9. 10.1111/cas.13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng C, You W, Wan P, Jiang X, Chen J, Zheng Y, et al. Clinicopathological and prognostic significance of PD-L1 expression in sarcoma: a systematic review and meta-analysis. Medicine. (2018) 97:e11004. 10.1097/MD.0000000000011004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Z, Jin Z, Zhang M, Tang Y, Yang G, Yuan X, et al. Prognostic value of programmed death-ligand 1 in sarcoma: a meta-analysis. Oncotarget. (2017) 8:59570–80. 10.18632/oncotarget.19168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. (2007) 8:16. 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden JA, Van Der Windt DA, Cartwright JL, Côté P, Bombardier C. Assessing bias in studies of prognostic factors. Ann Intern Med. (2013) 158:280–6. 10.7326/0003-4819-158-4-201302190-00009 [DOI] [PubMed] [Google Scholar]
- 22.Yoshida K, Okamoto M, Sasaki J, Kuroda C, Ishida H, Ueda K, et al. Clinical outcome of osteosarcoma and its correlation with programmed death-ligand 1 and T cell activation markers. Onco Targets Ther. (2019) 12:2513–8. 10.2147/OTT.S198421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L, Wang Z, Cui C, Guan X, Dong B, Zhao M, et al. Comprehensive immune characterization and T-cell receptor repertoire heterogeneity of retroperitoneal liposarcoma. Cancer Sci. (2019) 110:3038–48. 10.1111/cas.14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng B, Ren T, Huang Y, Sun K, Wang S, Bao X, et al. PD-1 axis expression in musculoskeletal tumors and antitumor effect of nivolumab in osteosarcoma model of humanized mouse. J Hematol Oncol. (2018) 11:16 10.1186/s13045-018-0580-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Zhu G, Yang X, Zeng K, Liu F, Sun J. Expression of PD-L1 associated with Ki-67 and chemotherapy response but not p53 in osteosarcoma. Int J Clin Exp Med. (2018) 11:7. [Google Scholar]
- 26.Yang X, Zhu G, Yang Z, Zeng K, Liu F, Sun J. Expression of PD-L1/PD-L2 is associated with high proliferation index of Ki-67 but not with TP53 overexpression in chondrosarcoma. Int J Biol Markers. (2018) 33:507–13. 10.1177/1724600818774464 [DOI] [PubMed] [Google Scholar]
- 27.Patel KR, Martinez A, Stahl JM, Logan SJ, Perricone AJ, Ferris MJ, et al. Increase in PD-L1 expression after pre-operative radiotherapy for soft tissue sarcoma. Oncoimmunology. (2018) 7:e1442168. 10.1080/2162402X.2018.1442168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park HK, Kim M, Sung M, Lee SE, Kim YJ, Choi YL. Status of programmed death-ligand 1 expression in sarcomas. J Transl Med. (2018) 16:303. 10.1186/s12967-018-1658-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oike N, Kawashima H, Ogose A, Hotta T, Hatano H, Ariizumi T, et al. Prognostic impact of the tumor immune microenvironment in synovial sarcoma. Cancer Sci. (2018) 109:3043–54. 10.1111/cas.13769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machado I, Lopez-Guerrero JA, Scotlandi K, Picci P, Llombart-Bosch A. Immunohistochemical analysis and prognostic significance of PD-L1, PD-1, and CD8+ tumor-infiltrating lymphocytes in Ewing's sarcoma family of tumors (ESFT). Virchows Arch. (2018) 472:815–24. 10.1007/s00428-018-2316-2 [DOI] [PubMed] [Google Scholar]
- 31.Kostine M, Briaire-de Bruijn IH, Cleven AHG, Vervat C, Corver WE, Schilham MW, et al. Increased infiltration of M2-macrophages, T-cells and PD-L1 expression in high grade leiomyosarcomas supports immunotherapeutic strategies. Oncoimmunology. (2018) 7:e1386828. 10.1080/2162402X.2017.1386828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawamoto K, Miyoshi H, Suzuki T, Kiyasu J, Yokoyama S, Sasaki Y, et al. Expression of programmed death ligand 1 is associated with poor prognosis in myeloid sarcoma patients. Hematol Oncol. (2018) 36:591–9. 10.1002/hon.2506 [DOI] [PubMed] [Google Scholar]
- 33.Cohen JE, Eleyan F, Zick A, Peretz T, Katz D. Intratumoral immune-biomarkers and mismatch repair status in leiyomyosarcoma -potential predictive markers for adjuvant treatment: a pilot study. Oncotarget. (2018) 9:30847–54. 10.18632/oncotarget.25747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng B, Ren T, Huang Y, Guo W. Apatinib inhibits migration and invasion as well as PD-L1 expression in osteosarcoma by targeting STAT3. Biochem Biophys Res Commun. (2018) 495:1695–701. 10.1016/j.bbrc.2017.12.032 [DOI] [PubMed] [Google Scholar]
- 35.van Erp AEM, Versleijen-Jonkers YMH, Hillebrandt-Roeffen MHS, van Houdt L, Gorris MAJ, van Dam LS, et al. Expression and clinical association of programmed cell death-1, programmed death-ligand-1 and CD8(+) lymphocytes in primary sarcomas is subtype dependent. Oncotarget. (2017) 8:71371–84. 10.18632/oncotarget.19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundara YT, Kostine M, Cleven AH, Bovee JV, Schilham MW, Cleton-Jansen AM. Increased PD-L1 and T-cell infiltration in the presence of HLA class I expression in metastatic high-grade osteosarcoma: a rationale for T-cell-based immunotherapy. Cancer Immunol Immunother. (2017) 66:119–28. 10.1007/s00262-016-1925-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Que Y, Xiao W, Guan YX, Liang Y, Yan SM, Chen HY, et al. PD-L1 Expression is associated with FOXP3+ regulatory T-cell infiltration of soft tissue sarcoma and poor patient prognosis. J Cancer. (2017) 8:2018–25. 10.7150/jca.18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollack SM, He Q, Yearley JH, Emerson R, Vignali M, Zhang Y, et al. T-cell infiltration and clonality correlate with programmed cell death protein 1 and programmed death-ligand 1 expression in patients with soft tissue sarcomas. Cancer. (2017) 123:3291–304. 10.1002/cncr.30726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmerini E, Agostinelli C, Picci P, Pileri S, Marafioti T, Lollini PL, et al. Tumoral immune-infiltrate (IF), PD-L1 expression and role of CD8/TIA-1 lymphocytes in localized osteosarcoma patients treated within protocol ISG-OS1. Oncotarget. (2017) 8:111836–46. 10.18632/oncotarget.22912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao Y, Chen L, Feng Y, Shen J, Gao Y, Cote G, et al. Targeting programmed cell death ligand 1 by CRISPR/Cas9 in osteosarcoma cells. Oncotarget. (2017) 8:30276–87. 10.18632/oncotarget.16326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa Arantes DA, Goncalves AS, Jham BC, Duarte ECB, de Paula EC, de Paula HM, et al. Evaluation of HLA-G, HLA-E, and PD-L1 proteins in oral osteosarcomas. Oral Surg Oral Med Oral Pathol Oral Radiol. (2017) 123:e188–96. 10.1016/j.oooo.2016.12.002 [DOI] [PubMed] [Google Scholar]
- 42.Budczies J, Mechtersheimer G, Denkert C, Klauschen F, Mughal SS. PD-L1 (CD274) copy number gain, expression, and immune cell infiltration as candidate predictors for response to immune checkpoint inhibitors in soft-tissue sarcoma. Oncoimmunology. (2017) 6:e1279777. 10.1080/2162402X.2017.1279777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Botti G, Scognamiglio G, Marra L, Pizzolorusso A, Di Bonito M, De Cecio R, et al. Programmed death ligand 1 (PD-L1) expression in primary angiosarcoma. J Cancer. (2017) 8:3166–72. 10.7150/jca.19060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertucci F, Finetti P, Perrot D, Leroux A, Collin F, Le Cesne A, et al. PDL1 expression is a poor-prognosis factor in soft-tissue sarcomas. Oncoimmunology. (2017) 6:e1278100. 10.1080/2162402X.2016.1278100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paydas S, Bagir EK, Deveci MA, Gonlusen G. Clinical and prognostic significance of PD-1 and PD-L1 expression in sarcomas. Med Oncol. (2016) 33:93. 10.1007/s12032-016-0807-z [DOI] [PubMed] [Google Scholar]
- 46.Kostine M, Cleven AH, de Miranda NF, Italiano A, Cleton-Jansen AM, Bovee JV. Analysis of PD-L1, T-cell infiltrate and HLA expression in chondrosarcoma indicates potential for response to immunotherapy specifically in the dedifferentiated subtype. Mod Pathol. (2016) 29:1028–37. 10.1038/modpathol.2016.108 [DOI] [PubMed] [Google Scholar]
- 47.Koirala P, Roth ME, Gill J, Piperdi S, Chinai JM, Geller DS, et al. Immune infiltration and PD-L1 expression in the tumor microenvironment are prognostic in osteosarcoma. Sci Rep. (2016) 6:30093. 10.1038/srep30093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim C, Kim EK, Jung H, Chon HJ, Han JW, Shin KH, et al. Prognostic implications of PD-L1 expression in patients with soft tissue sarcoma. BMC Cancer. (2016) 16:434. 10.1186/s12885-016-2451-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honda Y, Otsuka A, Ono S, Yamamoto Y, Seidel JA, Morita S, et al. Infiltration of PD-1-positive cells in combination with tumor site PD-L1 expression is a positive prognostic factor in cutaneous angiosarcoma. Oncoimmunology. (2017) 6:e1253657. 10.1080/2162402X.2016.1253657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lussier DM, O'Neill L, Nieves LM, McAfee MS, Holechek SA, Collins AW, et al. Enhanced T-cell immunity to osteosarcoma through antibody blockade of PD-1/PD-L1 interactions. J Immunother. (2015) 38:96–106. 10.1097/CJI.0000000000000065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D'Angelo SP, Shoushtari AN, Agaram NP, Kuk D, Qin LX, Carvajal RD, et al. Prevalence of tumor-infiltrating lymphocytes and PD-L1 expression in the soft tissue sarcoma microenvironment. Hum Pathol. (2015) 46:357–65. 10.1016/j.humpath.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chowdhury F, Dunn S, Mitchell S, Mellows T, Ashton-Key M, Gray J. PD-L1 and CD8(+)PD1(+) lymphocytes exist as targets in the pediatric tumor microenvironment for immunomodulatory therapy. OncoImmunology. (2015) 4:e1029701 10.1080/2162402X.2015.1029701 [DOI] [Google Scholar]
- 53.Shen JK, Cote GM, Choy E, Yang P, Harmon D, Schwab J, et al. Programmed cell death ligand 1 expression in osteosarcoma. Cancer Immunol Res. (2014) 2:690–8. 10.1158/2326-6066.CIR-13-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JR, Moon YJ, Kwon KS, Bae JS, Wagle S, Kim KM, et al. Tumor infiltrating PD1-positive lymphocytes and the expression of PD-L1 predict poor prognosis of soft tissue sarcomas. PLoS ONE. (2013) 8:e82870. 10.1371/journal.pone.0082870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu L, Sun J, Wang L, Li Z, Wang L, Li Z. Prognostic and clinicopathological significance of PD-L1 in patients with bladder cancer: a meta-analysis. Front Pharmacol. (2019) 10:962. 10.3389/fphar.2019.00962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Xu Y, Wan B, Song Y, Zhan P, Hu Y, et al. The clinicopathological and prognostic significance of PD-L1 expression assessed by immunohistochemistry in lung cancer: a meta-analysis of 50 studies with 11,383 patients. Transl Lung Cancer Res. (2019) 8:429–49. 10.21037/tlcr.2019.08.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang X, Zhang W, Zhang Z, Shi D, Wu F, Zhong B, et al. Prognostic value of programmed cell death 1 ligand-1 (PD-L1) or PD-1 expression in patients with osteosarcoma: a meta-analysis. J Cancer. (2018) 9:2525–31. 10.7150/jca.25011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li J, Chen L, Xiong Y, Zheng X, Xie Q, Zhou Q, et al. Knockdown of PD-L1 in human gastric cancer cells inhibits tumor progression and improves the cytotoxic sensitivity to CIK therapy. Cell Physiol Biochem. (2017) 41:907–20. 10.1159/000460504 [DOI] [PubMed] [Google Scholar]
- 59.Chen L, Xiong Y, Li J, Zheng X, Zhou Q, Turner A, et al. PD-L1 expression promotes epithelial to mesenchymal transition in human esophageal cancer. Cell Physiol Biochem. (2017) 42:2267–80. 10.1159/000480000 [DOI] [PubMed] [Google Scholar]
- 60.Bighetti-Trevisan RL, Sousa LO. Cancer stem cells: powerful targets to improve current anticancer therapeutics. Stem Cells Int. (2019) 2019:9618065. 10.1155/2019/9618065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Almozyan S, Colak D, Mansour F, Alaiya A, Al-Harazi O, Qattan A, et al. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer. (2017) 141:1402–12. 10.1002/ijc.30834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta HB, Clark CA, Yuan B, Sareddy G, Pandeswara S, Padron AS, et al. Tumor cell-intrinsic PD-L1 promotes tumor-initiating cell generation and functions in melanoma and ovarian cancer. Signal Transduct Target Ther. (2016) 1:16030. 10.1038/sigtrans.2016.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong P, Xiong Y, Yue J, Hanley SJB, Watari H. Tumor-intrinsic PD-L1 signaling in cancer initiation, development and treatment: beyond immune evasion. Front Oncol. (2018) 8:386. 10.3389/fonc.2018.00386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu S, Chen S, Yuan W, Wang H, Chen K, Li D, et al. PD-1/PD-L1 interaction up-regulates MDR1/P-gp expression in breast cancer cells via PI3K/AKT and MAPK/ERK pathways. Oncotarget. (2017) 8:99901–12. 10.18632/oncotarget.21914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abdel-Rahman O. PD-L1 expression and outcome of advanced melanoma patients treated with anti-PD-1/PD-L1 agents: a meta-analysis. Immunotherapy. (2016) 8:1081–9. 10.2217/imt-2016-0025 [DOI] [PubMed] [Google Scholar]
- 66.Li S, Chen L, Jiang J. Role of programmed cell death ligand-1 expression on prognostic and overall survival of breast cancer: a systematic review and meta-analysis. Medicine. (2019) 98:e15201. 10.1097/MD.0000000000015201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang W, Ran R, Shao B, Li H. Prognostic and clinicopathological value of PD-L1 expression in primary breast cancer: a meta-analysis. Breast Cancer Res Treat. (2019) 178:17–33. 10.1007/s10549-019-05371-0 [DOI] [PubMed] [Google Scholar]
- 68.Kelly CM, Antonescu CR, Bowler T, Munhoz R, Chi P, Dickson MA, et al. Objective response rate among patients with locally advanced or metastatic sarcoma treated with talimogene laherparepvec in combination with pembrolizumab: a phase 2 clinical trial. JAMA Oncol. (2020) 6:402–8. 10.1001/jamaoncol.2019.6152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci USA. (2002) 99:12293–7. 10.1073/pnas.192461099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meng X, Huang Z, Teng F, Xing L, Yu J. Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat Rev. (2015) 41:868–76. 10.1016/j.ctrv.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 71.Schutz F, Stefanovic S, Mayer L, von Au A, Domschke C, Sohn C. PD-1/PD-L1 pathway in breast cancer. Oncol Res Treat. (2017) 40:294–7. 10.1159/000464353 [DOI] [PubMed] [Google Scholar]
- 72.Schalper KA. PD-L1 expression and tumor-infiltrating lymphocytes: revisiting the antitumor immune response potential in breast cancer. Oncoimmunology. (2014) 3:e29288. 10.4161/onci.29288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lastwika KJ, Wilson W, 3rd, Li QK, Norris J, Xu H, Ghazarian SR, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. (2016) 76:227–38. 10.1158/0008-5472.CAN-14-3362 [DOI] [PubMed] [Google Scholar]
- 74.Hall F, Villalobos V, Wilky B. Future directions in soft tissue sarcoma treatment. Curr Probl Cancer. (2019) 43:300–7. 10.1016/j.currproblcancer.2019.06.004 [DOI] [PubMed] [Google Scholar]
- 75.Thanindratarn P, Dean DC, Nelson SD, Hornicek FJ, Duan Z. Advances in immune checkpoint inhibitors for bone sarcoma therapy. J Bone Oncol. (2019) 15:100221. 10.1016/j.jbo.2019.100221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Torabi A, Amaya CN, Wians FH, Jr, Bryan BA. PD-1 and PD-L1 expression in bone and soft tissue sarcomas. Pathology. (2017) 49:506–13. 10.1016/j.pathol.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 77.Kim ST, Klempner SJ, Park SH, Park JO, Park YS, Lim HY, et al. Correlating programmed death ligand 1 (PD-L1) expression, mismatch repair deficiency, and outcomes across tumor types: implications for immunotherapy. Oncotarget. (2017) 8:77415–23. 10.18632/oncotarget.20492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sheffield BS, Fulton R, Kalloger SE, Milne K, Geller G, Jones M, et al. Investigation of PD-L1 biomarker testing methods for PD-1 axis inhibition in non-squamous non-small cell lung cancer. J Histochem Cytochem. (2016) 64:587–600. 10.1369/0022155416665338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ilie M, Hofman V, Dietel M, Soria JC, Hofman P. Assessment of the PD-L1 status by immunohistochemistry: challenges and perspectives for therapeutic strategies in lung cancer patients. Virchows Arch. (2016) 468:511–25. 10.1007/s00428-016-1910-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets analyzed for this study are included in the article/Supplementary Material.