Abstract
Sweet pepper (Capsicum annuum L.) is a high value crop and one of the most widely grown vegetables belonging to the Solanaceae family. In addition to commercial varieties and F1 hybrids, a multitude of landraces are grown, whose genetic combination is the result of hundreds of years of random, environmental, and farmer selection. High genetic diversity exists in the landrace gene pool which however has scarcely been studied, thus bounding their cultivation. We re-sequenced four pepper inbred lines, within as many Italian landraces, which representative of as many fruit types: big sized blocky with sunken apex (‘Quadrato’) and protruding apex or heart shaped (‘Cuneo’), elongated (‘Corno’) and smaller sized sub-spherical (‘Tumaticot’). Each genomic sequence was obtained through Illumina platform at coverage ranging from 39 to 44×, and reconstructed at a chromosome scale. About 35.5k genes were predicted in each inbred line, of which 22,017 were shared among them and the reference genome (accession ‘CM334’). Distinctive variations in miRNAs, resistance gene analogues (RGAs) and susceptibility genes (S-genes) were detected. A detailed survey of the SNP/Indels occurring in genes affecting fruit size, shape and quality identified the highest frequencies of variation in regulatory regions. Many structural variations were identified as presence/absence variations (PAVs), notably in resistance gene analogues (RGAs) and in the capsanthin/capsorubin synthase (CCS) gene. The large allelic diversity observed in the four inbred lines suggests their potential use as a pre-breeding resource and represents a one-stop resource for C. annuum genomics and a key tool for dissecting the path from sequence variation to phenotype.
Subject terms: Genomics, Plant genetics
Introduction
The Capsicum genus (Solanaceae family) originated in the New World and includes 27 species of which five (C. annuum, C. chinense, C. frutescens, C. baccatum, and C. pubescens) have been domesticated1 and used worldwide as spices and vegetables. The most cultivated is C. annuum L. (2n = 2×= 24), which includes most of the Mexican chili peppers, hot peppers grown in Africa and Asia as well as numerous cultivars and landraces of sweet (non pungent) peppers cultivated in the temperate regions of Europe and North America1. China is at present the main worldwide pepper producer, with more than 17 M tons, followed by Mexico, Turkey and Indonesia. Italy, Spain and the Netherlands are the main European producers, and in several regions of the former, the heterogeneity of the land, climate and soil has favoured the cultivation of landraces. Each of them is the result of selection processes for adaptation to specific ecological niches, and differ in fruit shape, organoleptic properties and resistance against abiotic and biotic stresses2.
Molecular markers techniques and gene chip arrays have been applied in the last decades for pepper germplasm characterization3–10 with the goal to provide complementary descriptors to conventional morphological variation. However, triggered by advancements in sequencing technologies, the genome sequence of several crop is now available and the application of NGS technologies for resequencing represent the most powerful tool for exploring the DNA‐level diversity among members of a crop, with the ultimate goal to understand the molecular basis for phenotype–genotype relationships11. The first whole-genome sequences of C. annuum ('CM334') and C. chinense (PI159236) were released by Kim et al.12 in 2014. In the same year, Qin et al.13 published the genome sequences of C. annuum Zunla-1 and of Chiltepin (C. annuum var. glabriusculum). Both studies highlighted that the pepper genome size is ~3–3.5 Gb, it is characterized by a high percentage (over 80%) of repetitive elements, and includes 35 K genes. Afterwards, a resequencing effort14 made available the raw sequences of two C. annuum lines: ‘Dempsey’, a large bell-type genotype, and ‘Perennial’, a genotype with small elongated fruits. A few years later, an improved version of the reference genome of both ‘CM334’ and C. chinense ‘PI159236’ as well as the genome sequence of the domesticated C. baccatum were published and contributed to decipher the evolutionary relationships among the three species as well as to estimate the lineage-divergence times occurring in Capsicum15. Recently, Hulse-Kemp et al.16, by adopting the linked-read sequencing technology, obtained the genome sequence of an F1 hybrid from a cross between ‘CM334’ with a non-pungent blocky accession of C. annuum. In 2019, Du and co-worker17, to identifying a panel of 92 SNPs for population structure analysis, produced low-pass resequencing data from 35 different C. annuum lines.
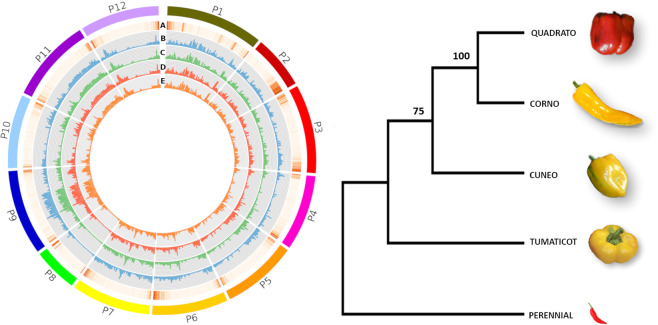
Here we report on the resequencing of four sweet pepper inbred lines, which we selected within four Italian landraces representative of as many fruit types, ranging from blocky to elongated and sub-spherical. The inbred lines produce sweet fruits with peculiar organoleptic and sensorial characteristics and of potential use for both fresh consumption and industrial transformation. The four genomes were reconstructed at a chromosomal scale and annotated. MiRNA loci as well as the number, position and phylogenetic relationships of putative resistance gene analogues (RGAs) and susceptibility genes (S-genes)18 were identified. Lastly, functionally characterised SNP/Indels and PAV in genes affecting berry size, shape and pigmentation were spotted.
Results and Discussion
Fruit morphological characterization
Four pepper lines, previously selected from landraces through a breeding program developed by DISAFA (www.disafa.unito.it), in collaboration with AGRION (www.agrion.it), were under study: ‘Cuneo’, ‘Quadrato di Carmagnola’ (later called ‘Quadrato’), ‘Corno di Carmagnola’ (later called ‘Corno’) and ‘Tumaticot’. The landrace ‘Cuneo’ produces red or yellow hearth shaped berries, mostly tri –lobed and, owning a fleshy and crunchy pericarp whose thickness ranges from 8 to 10 mm, and with a protruding apex characterized by a typical brownish anthocyanic spot, commonly called ‘moustache’. The landrace ‘Quadrato’ produces tetra-lobated berries with a sunken apex, a diameter ranging from 12 to 18 cm and whose pericarp is fleshy and aromatic. The landrace ‘Corno’ produces elongated fruits characterized by thinner pericarp which maintain its red or yellow color also after cooking, making them particularly suitable for the processing industry. ‘Tumaticot’, a landrace grown in the Piedmont region (North-West Italy) since the early ‘30 s, is characterized by smaller fruits, with a flattened shape and a flat or slightly sunken apex. The fruits are tri or tetra-lobated, with a color nuance from red to yellow and a pericarp thickness of 7–11 mm.
The average fruit weight and related components (diameter, length, fruit shape, thickness of the pericarp, number of lodges) of the inbred lines in study are reported in Table 1. Data for ‘Perennial’ genotype and ‘CM334’ were added to the analysis as references. The two blocky types ‘Cuneo’ and ‘Quadrato’ produce berries with the greatest average weight (272.7 and 270.5 g respectively), followed by ‘Tumaticot’, which produces heavier fruits (238.1 g) than ‘Corno’ (130.2 g), due to its thicker pericarp (Table 1). The latter is characterized by the longest berries, up to 18 cm (ratio lenght/diameter: 2.82), while ‘Quadrato’ is the only one producing red berries (Table 1).
Table 1.
Characteristics of the berries of Piedmontese inbred lines, ‘CM334’ and Perennial.
| Genotype | Berry colour | Average berry weight (gr) | Fruit Length (cm) | Fruit Diameter (cm) | Ratio L/D | Pericarp thickness (mm) | N° locules |
|---|---|---|---|---|---|---|---|
| Cuneo* | Yellow | 272.7 ± 11.28 | 8.7 ± 0.27 | 9.9 ± 0.15 | 0.80 ± 0.01 | 6.8 ± 0.20 | 3.3 ± 0.33 |
| Corno* | Yellow | 130.2 ± 11.55 | 18.2 ± 1.02 | 5.8 ± 0.31 | 2.82 ± 0.24 | 3.3 ± 0.11 | 3.0 ± 0.11 |
| Quadrato* | Red | 270.5 ± 222.72 | 9.4 ± 0.48 | 9.3 ± 0.50 | 1.01 ± 0.03 | 5.4 ± 0.42 | 3.7 ± 0.17 |
| Tumaticot** | Yellow | 238.1 ± 75.20 | 4.9 ± 0.10 | 10.3 ± 0.10 | 0.48 ± 0.28 | 5.3 ± 0.10 | 2.8 ± 0.12 |
| CM334*** | Red | 6.2 ± 1.10 | 5.9 ± 0.50 | 1.9 ± 0.20 | 3.17 ± 0.27 | 2.5 ± 0.20 | 2.3 ± 0.46 |
| Perennial**** | Red | 1.0 ± 0.00 | 3.0 ± 0.3 | 1.0 ± 0.01 | 3.00 ± 0.30 | — | 2.0 ± 0.00 |
Genome assembly and reconstruction
Genome sequencing of the four C. annuum genotypes yielded 3.315 million raw pair-end reads (in 1.658 million clusters, Table 2). For each genotype an average of 829 million sequences (150 bp) were produced for a total of ~497.4 Gb, reduced to 465.7 Gb after removal of low-quality bases. The sequencing coverage was 41.46X on average, ranging from 38.76X in ‘Corno’ to 43.54X in ‘Tumaticot’ (Table 2). The sequence data have been deposited into NCBI Short Read Archive with specific submission identifiers (from SAMN14253609 to SAMN14253612), under the Bioproject PRJNA609444. Analyzed data are available in the www.pepper-genomics.unito.it portal.
Table 2.
Summary of results from sequencing data for each of the four genotypes (before and after the clean-up phase).
| Genotype | duplicated sequences (%) | GC (%) | Length of sequences (bp) | Total sequences (M) | Coverage | Read Mapping (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre-cleaning | final | Pre-cleaning | Final | Pre-cleaning | Final | Pre-cleaning | Final | |||
| Corno | 10.55% | 9.97% | 35.00% | 35.67% | 150 | 141.33 | 870.8 | 866.7 | 43.54 × | 99.10 |
| Cuneo | 8.28% | 8.19% | 35.00% | 35.00% | 150 | 141.3 | 775.2 | 763.8 | 38.76 × | 98.11 |
| Quadrato | 9.77% | 9.67% | 35.00% | 35.67% | 150 | 141.33 | 816 | 812.5 | 40.80 × | 99.07 |
| Tumaticot | 10.94% | 10.37% | 35.00% | 35.67% | 150 | 141.67 | 854.4 | 850.4 | 42.72 × | 99.08 |
| Total | — | — | — | — | — | — | 3316.4 | 3293.4 | — | — |
| Average | 9.88% | 9.55% | 35.00% | 35.75% | 150 | 141.41 | 829.1 | 823.35 | 41.46 × | 98.84 |
Genome reconstruction, hampered by both the large genome size and its richness in repeated elements (~ 80%), was based on a combination of a de novo assembly procedure to generate contigs/scaffolds and an iterative read mapping strategy against the pepper reference genome to integrate contigs/scaffolds into pseudomolecules. In respect to a de novo assembly, this approach required lower sequencing depth and avoided the construction of multiple libraries.
An extensive k-mer survey (Table S1) identified the k-mer length of 63 as the one granting optimal contiguity metrics, and was adopted for the final assembly of all the genotypes (Table 3). In order to close the gaps emerging during the scaffolding process by SOAPdenovo, we applied Gapcloser, and lowered, in average, the number of unidentified nucleotides (Ns) in the assemblies from 5.83 to 0.04% (Table 3). The obtained assemblies were successfully reconstructed in 12 pseudomolecules, corresponding to the haploid chromosome number of the species. A variable number of scaffolds were anchored (range 4,062–4,688), while the unanchored fraction, of each genome, was attributed to chr. 0 (range 110–115 Mb).
Table 3.
Statistics of the four assembled genomes (k = 63), relative to the different steps of the assembly procedure (de novo, Gapclosing, filtering and reference-guided).
| Genotype | Metrics | De novo | Gap closed | Filtered > 500 bp | Reference guided |
|---|---|---|---|---|---|
| Corno | Number of scaffolds | 1,994,391 | 1,994,391 | 313,692 | 4,688 |
| Total size of scaffolds (bp) | 3,238,887,426 | 3,156,174,802 | 2,888,201,005 | 2,952,523,284 | |
| Longest scaffold | 977,366 | 967,969 | 967,969 | 304,682,413 | |
| Shortest scaffold | 100 | 100 | 500 | 800 | |
| Average size (scaffolds, bp) | 1,624 | 1,583 | 9,207 | 629,804 | |
| N50 | 47,527 | 47,339 | 55,304 | 244,870,051 | |
| L50 | 15,812 | 15,493 | 12,873 | 6 | |
| % of Ns in scaffolds | 6.63 | 0.04 | 0.03 | 2.97 | |
| Number of genes | 35,484 | — | — | 29,204 | |
| Cuneo | Number of scaffolds | 1,209,809 | 1,209,809 | 183,951 | 4,062 |
| Total size of scaffolds (bp) | 3,015,917,026 | 3,003,730,531 | 2,856,195,490 | 2,888,888,923 | |
| Longest scaffold | 785,116 | 783,752 | 783,752 | 298,303,901 | |
| Shortest scaffold | 100 | 100 | 500 | 800 | |
| Average size (scaffolds, bp) | 2,493 | 2,483 | 15,527 | 711,199 | |
| N50 | 53,466 | 53,354 | 57,217 | 239,928,576 | |
| L50 | 14,568 | 14,539 | 13,205 | 6 | |
| % of Ns in scaffolds | 3.92 | 0.04 | 0.03 | 1.76 | |
| Number of genes | 35,518 | — | — | 29,300 | |
| Quadrato | Number of scaffolds | 1,773,028 | 1,773,028 | 289,008 | 4,688 |
| Total size of scaffolds (bp) | 3,196,719,955 | 3,111,823,022 | 2,880,333,039 | 2,938,129,169 | |
| Longest scaffold | 684,241 | 678,098 | 678,098 | 303,210,577 | |
| Shortest scaffold | 100 | 100 | 500 | 800 | |
| Average size (scaffolds, bp) | 1,803 | 1,755 | 9,966 | 626,734 | |
| N50 | 46,314 | 46,168 | 52,573 | 243,527,465 | |
| L50 | 16,181 | 15,845 | 13,497 | 6 | |
| % of Ns in scaffolds | 6.68 | 0.04 | 0.03 | 2.74 | |
| Number of genes | 35,538 | — | — | 29,288 | |
| Tumaticot | Number of scaffolds | 1,191,858 | 1,191,858 | 173,746 | 4,322 |
| Total size of scaffolds (bp) | 3,088,650,784 | 3,016,051,866 | 2,871,230,234 | 2,901,582,212 | |
| Longest scaffold | 788,112 | 780,295 | 780,295 | 300,041,623 | |
| Shortest scaffold | 100 | 100 | 500 | 800 | |
| Average size (scaffolds, bp) | 2,591 | 2,531 | 16,525 | 671,352 | |
| N50 | 58,072 | 57,584 | 61,963 | 240,199,676 | |
| L50 | 13,578 | 13,360 | 12,147 | 6 | |
| % of Ns in scaffolds | 6.09 | 0.03 | 0.02 | 1.64 | |
| Number of genes | 35,723 | — | — | 29,465 |
1The reference guided assembly contained also chromosome zero.
Genome annotation and OrthoMCL analysis
Globally, ~80% of the resequenced genomes was masked, in line with recent findings15,19. The four assembled genomes were then structurally annotated with the Maker-P suite and the total number of genes identified was on average 35.5k (Fig. 1, AED ~ 0.35). The lowest number of genes (Table 3) was detected in ‘Corno’ (35.484) while the highest in ‘Tumaticot’ (35.723). No correlation between size of the assembled genomes and the number of predicted genes was highlighted; this suggests that the different genome sizes are attributable to non-coding regions or structural variants.
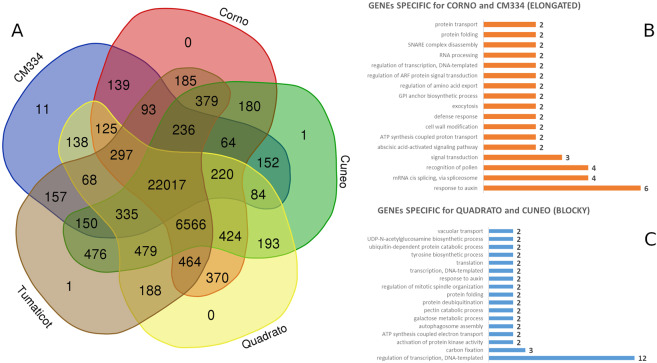
Figure 1.
(A) Venn diagram showing the grouping of the four proteomes and the one of the reference ‘CM334’ based on Orthofinder. (B) GO distribution related to the genes present in the 139 orthogroups specific for the elongated types; (C) GO distribution related to the genes present in the 193 orthogroups specific for the blocky types.
The functional annotation produced about 35.5k proteins, in line with what previously detected within the Solanaceae family (34.9k in ‘CM334’, 35.34k in ‘Zunla-1’, 34.5k in ‘Chiltepin’, 33.7k in tomato, 35,0k in eggplant and 38.5k in potato). Proteomes were validated using BUSCO; overall, more than 91% of 1,614 expected embryophyta genes were identified in our genome annotations as the complete and partial BUSCO profiles (Table S2).
The function attributed to each predicted protein was based on the results of BlastP (SwissProt) and the InterProScan domain inspection. InterProScan analyses highlighted about 77% of the predicted proteins with at least one IPR domain. Among the top 20 SUPERFAMILY domains, listed in Table S3, the most abundant in all the genomes was SSF52540 (P-loop containing nucleoside triphosphate hydrolase), which is involved in several UniPathways, including chlorophyll or coenzyme A biosynthesis. The other most abundant Superfamilies were: SSF56112 (protein Kinase-like domain), which acts on signalling and regulatory processes in the eukaryotic cell, SSF52058 (Leucine-rich repeat domain, L domain-like), which is related to resistance to pathogens and SSF48371 (Armadillo-type fold), which is involved, inter alia, in defense response and translation factor activity.
Clustering the proteomes (169,465 sequences) of the four genomes and the reference ‘CM334’ with Orthofinder produced a set of 34,192 gene families (plus 8,682 unassigned genes, Fig. 1A), of which 22,017 (including 123,662 genes) were shared. The proteome of ‘CM334’ highlighted 11 genome-specific orthogroups (118 genes), while just one specific orthogroup was found in the ‘Cuneo’ and ‘Tumaticot’ genomes and none in ‘Corno’ and ‘Quadrato’. A total of 193 orthogroups, that include 391 genes among which the transcription factors MED28 and NAP2, were in common between the two blocky types ‘Quadrato’ and ‘Cuneo’ (Fig. 1B). MED proteins in plants perform different functions ranging from development regulation20 to biotic and abiotic stresses response21 while NAP2 is involved in delayed leaf senescence and enhanced fruit yield and sugar content22. A total of 139 orthogroups, that include 344 genes, were also shared between the long-shaped peppers ‘CM334’ and ‘Corno’. The common genes comprise SAUR (Small Auxin UP RNAs) genes, which are known to promote primary growth (lengthening) by inducing cell elongation, increasing the rate of cell division as well as cell wall expansion23. In any case, a more in-depth functional characterization of these genes is needed.
Prediction and annotation of miRNA
A search against miRBase (release 21) led to the prediction of non-redundant miRNAs, ranging from 169 in ‘Tumaticot’ and ‘Corno’ (within 213 and 210 genomic regions respectively) to 175 in ‘Cuneo’ (in 219 genomic regions) and 176 in ‘Quadrato’ (in 222 genomic regions) (Table S4), belonging to 62 (61 for ‘Tumaticot’) miRNA families. The same pipeline was applied to the recently improved ‘CM334’ genome, and resulted in the identification of 170 miRNAs, belonging to 59 families, in 215 genomic regions. It is well known that some miRNA families target known transcription factors related to plant development, morphology, flowering time, as well as response to stress24–26. Examples include miR164 and NAC-like proteins following drought or salinity stress27, miR160 and ARF (Auxin Response Factor), which control auxin-regulated transcription24, miR156 and SQUAMOSA promoter binding-like proteins which regulates the juvenile-to-adult vegetative transition and the vegetative-to-reproductive transition24, miR172 and AP2-like proteins24, which regulate plant development and response to stress. The search for genes targeted by the identified miRNAs (psRNATarget search) identified between 736 (‘Corno’) and 761 (‘Tumaticot’) putative miRNA:mRNA duplexes, involving 125–135 unique miRNAs and 275–584 unique transcripts. Almost 87% of genes encoding predicted target transcripts have functional InterPro annotations. The total number of miRNA families involved in miRNA:mRNA interactions varied according to the genotype, ranging from 53 in ‘Tumaticot’ to 58 in ‘Quadrato’ with miR2673, miR172 and miR395 being the top ranked family for all the four genotypes (Table S5). The same search in ‘CM334’ identified 635 putative miRNAs:mRNA duplexes, involving 1119 unique miRNAs and 412 unique transcripts (92% of genes having an InterPro annotation). The putative miRNA-target gene enrichment analysis in each genotype revealed significant enrichment for some GO terms (Table S6). Some of them appeared shared among all genotypes (Table S6), including GO:0009808/GO:0046274 (lignin metabolic/catabolic process) and GO:0003677 (DNA binding).
Resistance genes
Many plant-pathogen interactions are determined by the presence of resistance (R) genes/alleles, which enable plants to recognize pathogens effectors and subsequently activate effector-triggered immunity (ETI)28, followed by a defense response often leading to cell death or a hypersensitive response (HR)29. Most intracellular immune receptors in plants belong to the nucleotide-binding site and leucine-rich repeat (NLR, also known as NB-LRR) superfamily30,31. The NLR family proteins include two classes on the basis of the presence of a toll and interleukin-1 receptor domain in the N-terminus (TIR-NLR or TNL) or its absence (non-TIR-NLR or non-TNL). Some non-TNL proteins have a coiled-coil motif consisting of CC-NLR (CNL).
The RGAugury pipeline detected between 925 and 943 resistance gene analogues (RGAs), in the four genomes, representing about 2.5% of the total number of predicted genes, while up to 1,600 were found in ‘CM334’. The majority of RGAs were receptor like kinases (RLKs), followed by receptor like proteins (RLP) and nucleotide binding site leucine rich repeat (NBS-LRR), while only few RGAs contain at least one NB-ARC domain (Table 4). Few TNLs in the genomes of the four inbred lines were found, in line with results obtained by32, and Barchi et al.19 in pepper and other Solanaceae species such as eggplant, tomato and potato. Indeed, Kim et al.33 highlighted that some Asterids contain functional TNLs, whereas others do not, resulting in the identification of only 19 and 13 full length CNLs in sunflower and lettuce respectively, but no full length TNLs. Recently, Acquadro et al.34 reported that in the C. cardunculus genome, the RGAs belong almost exclusively to the RLK/RLP families, while no TNLs and few CNLs were identified. This species-specific RGAs distribution was also observed in Brassica oleracea, B. rapa, Arabidopsis and Teobroma cacao, where the number of TNL was higher than CNL, while an opposite situation was found for Populus trichocarpa, Vitis vinifera and Medicago truncatula35.
Table 4.
RGA proteins in the four pepper resequenced genotypes.
| Genotype/Species | NBS | CNL | TNL | CN | TN | NL | TX | Others | RLP | RLK | TM-CC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cuneo | 37 | 92 | 21 | 6 | 8 | 129 | 8 | 4 | 142 | 445 | 42 |
| Tumaticot | 30 | 98 | 20 | 7 | 9 | 122 | 6 | 5 | 150 | 450 | 46 |
| Quadrato | 38 | 89 | 18 | 6 | 9 | 121 | 9 | 5 | 137 | 454 | 42 |
| Cuneo | 32 | 90 | 23 | 4 | 9 | 116 | 6 | 5 | 145 | 455 | 40 |
| CM334 | 194 | 150 | 21 | 57 | 12 | 299 | 9 | 7 | 185 | 634 | 40 |
| Heinz 1706 (Tomato) | 60 | 66 | 22 | 12 | 9 | 81 | 7 | 1 | 72 | 481 | 47 |
Data on RGA in the reference ‘CM334’ genotype as well as in in tomato 'Heinz 1706' are also reported.
We found out clustering of RLKs, RLPs, NBS-encoding and TM-CC genes in some chromosome of the four inbred lines (Table S7), in agreement with classical genetics and analysis from large scale sequencing data in plant genomes36. The chr. 3 was the richest in RGAs followed by 12, 1 and 2, while chr. 8 was the poorest. Otherwise, ‘CM334’ was rich in NBS genes mainly in chr. 9, 6 and 5. The majority of RLK genes were found on chr. 2, 3, 7 and 4 while the majority of NBS on chr. 1, 9 and 11.
The alignments of the amino acid sequences and subsequent IQ-TREE analyses generated phylogenetic trees for each RGA class (Figure S1). As expected, resistance genes and orthologs clusters together, although in some taxa one or more orthologs were absent. Interestingly, in ‘CM334’ genome, 105 NBS genes were found to cluster into two main groups while no orthologs were identified in the genomes of the four inbred lines. Besides ‘CM334’ contained two clusters of 124 NL and 74 RLK specific gene. To explore the evolutionary relationships among pepper NLRs, a phylogenetic tree was constructed using the CNL and TNL proteins identified together with 39 known plant resistance (R) proteins. As expected, the TNL and CNL clades branched out, although five predicted CNL genes were nested within the TNL clade. The CNL genes were splitted in 13 subgroups, being G4 and G10 the largest. Subgroups G1 and G3 contain only genes of ‘CM334’ while all the others included RGAs identified in the four inbred lines as well. We could hypothesize that the specific CM334 clusters could be somehow related to their high levels of resistance to diverse pathogens, including Phytophthora capsici, pepper mottle virus and root-knot nematodes12,37,38, confirming an unequal gene duplication event among subgroups not only at the species level32, but also at genotypic level.
CNG-G13 was reported to be particular expanded in potato. Our findings seem to support this hypothesis, as just one gene from ‘CM334’ was found in this CNL subgroup. Interestingly, several putative RGAs were identified and showed missing domains, in line with what was reported by several authors39–48. Besides genes belonging to types TN, TX and CN, which might serve as reservoirs for diversity or serve to guard other NLRs from genetic aberration32, several NL genes (NBS-LRR lacking coiled-coil or Toll/Interleukin-1 receptor) were found, as observed in P. trichocarpa, V. vinifera and M. truncatula35 or Arachis spp.49. It was also reported that maintaining many NBS/resistance genes has potential fitness costs50,51 and it has been suggested that microRNAs are exploited by plants to regulate NBS gene expression52–56. Indeed, we found that 19 (in ‘Cuneo’) to 34 (in ‘CM334’) identified RGAs of the CNL, TNL, RLP and RLK classes (Table S7) were putatively targeted by a miRNA, suggesting that this mechanism could also be present in pepper.
Susceptibility genes
Typically, phytopathogens exploit plants’ susceptibility genes (S-genes) to facilitate their proliferation18, and their disruption may interfere with the compatibility between the host and the pathogens and consequently provide broad-spectrum and durable disease resistance57.
In the genome of the four inbred lines and the reference ‘CM334’, we explored the presence and variants of 11 S-genes (Table 5), which are involved in: (i) basic compatibility, which assists in host recognition and penetration, e.g. MLO; (ii) sustained compatibility, which is required for pathogen proliferation and spread, e.g. CESA3 and (iii) negative regulation of immune signals e.g. DMR158. PMR genes are involved in cell wall biology where they mediate structure formation and pectin accumulation59. Among them, the PMR5 family was the most represented, totalling from 67 to 69 gene members in the four inbred lines and 72 in CM334’, which were scattered along the genome. Less represented were the families of PMR6, whose number ranged from 25 to 27, and PMR4, with 13 genes in ‘CM334’, 12 in ‘Quadrato’ and ‘Tumaticot’, 11 in ‘Corno’ and 10 in ‘Cuneo’ (Table 5). MLO is one of the best-known S genes, which is required for powdery mildew penetration in epidermal cells. It represents a prominent example of robustness in durable pathogen-resistance programs and is conserved throughout monocots and dicots. We identified several MLO genes ranging from 16 in ‘Corno’ to 18 in ‘Cuneo’ (Table 5). MLO-like genes were found scattered along the genome with 4–5 loci on chr. 2, and 3 in chr. 8. The phylogenetic tree of the MLO gene family in the four pepper inbred lines and ‘CM334’ (Fig. 2) highlighted, as expected, the clustering of MLO orthologs.
Table 5.
Number of the S-genes and their variant proportion in the four genotypes.
| S-Gene | Cuneo | Quadrato | Tumaticot | Corno | CM334 | Polymorphic genes | Number of variants | ratio (SNP/genes) |
|---|---|---|---|---|---|---|---|---|
| PMR4 | 10 | 12 | 12 | 11 | 13 | 5 | 61 | 5.26 |
| PMR5 | 69 | 68 | 68 | 67 | 72 | 30 | 71 | 1.03 |
| PMR6 | 26 | 27 | 27 | 25 | 26 | 7 | 68 | 2.60 |
| DMR1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2.00 |
| DMR6 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 0.50 |
| DND | 22 | 23 | 23 | 23 | 24 | 9 | 30 | 1.30 |
| MLO | 18 | 17 | 18 | 16 | 17 | 7 | 18 | 1.05 |
| CPR5 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2.00 |
| CESA3 | 35 | 34 | 36 | 37 | 32 | 13 | 52 | 1.49 |
| BIK1 | 25 | 22 | 26 | 25 | 30 | 10 | 42 | 1.64 |
| SR1 | 9 | 8 | 7 | 8 | 10 | 5 | 15 | 1.79 |
PMR4/5/6 = powdery mildew resistance 4/5/6; DMR1 = downy mildew resistance 1 (homoserine kinase); DND = defence no death (cyclic nucleotide-gated ion channel); MLO = mildew resistance locus O; CPR5 = constitutive expressor of pathogenesis-related gene 5; CESA3 = cellulose synthase 3; BIK1 = Botrytis–induced kinase 1; SR1 = signal responsive 1 (calmodulin binding transcription factor).
Figure 2.
Pepper statistics on S-Genes. Left side: Number of genes, polymorphic genes, variants in S-genes. Right side: Phylogenetic tree of proteins belonging to MLO-like genes from the four genotypes analyzed and considering ‘CM334’ different plant orthologous proteins; CaMLO-1 and 2 groups are highlighted in yellow and orange, respectively; bootstrap values were inferred from 100 replicates.
DND are cyclic nucleotide–gated calcium channels genes, which seem to specifically suppress the HR. We identified many DND genes ranging from 22 in ‘Cuneo’ to 24 in ‘CM334’ (Table 5), scattered along all the chromosomes except chr. 4. CESA3 is involved in the pathway of plant cellulose biosynthesis and the homozygous recessive mutant alleles can confer resistance to multiple pathogens, as a result of an increase in planta of abscissic acid, jasmonic acid and ethylene levels57. Its copies ranged from 34 to 37 in the four pepper inbred lines and were 32 in ‘CM334’. BIK 1 encodes a receptor-like cytoplasmic kinase that mediates PTI signalling from multiple pathogen-associated molecular pattern receptors60. Its copies ranged from 22 in the ‘Quadrato’ to 30 in the reference ‘CM334’. SR1 is a transcription factor which binds to the promoter of EDS1, a key regulator of plant defense responses, and represses its expression. Thus, its loss-of-function was found to display enhanced disease resistance61. Ten copies of the genes were found in ‘CM334’ and from 7 to 9 in the genome of the four inbred lines. The less represented S-genes were DMR6, which has been used to trigger broad-spectrum resistance against multiple pathogens and was present in two copies in all the genomes, and the DMR1 and CPR5 genes. The formers have been described to confer resistance to downy mildew in Arabidopsis and tomato by accumulating elevated levels of homoserine59, while the latter to be involved in cell proliferation and cell death control62.
S-genes were analyzed for the presence of functional SNPs (Fig. 2) and high impact variants in both homozygous and heterozygous state (Table S9). The number of deleterious SNPs fixed in homozygosity was two in the CESA3, PMR5 and MLO genes and one in the DND1 and SR1 genes. Their effect on plant pathogen resistance deserves to be tested in the inbred lines in study. Furthermore, we detected a number of deleterious SNPs in heterozygosity, i.e. two in the PMR5 genes and one in PMR4, PMR6 and BIK1 genes. Their effect should be tested after plant selfing with the goal to assess their effect when recessive S-genes are fixed in homozygosity.
SNP/Indel mining and annotation
The sequence reads of the four inbred lines were aligned back to the reference genome (‘CM334’) and their mapping rate was on average 98,84% (Table 2). The number of SNP/Indel variants detected ranged from about 16.33 to 18,08 M, for a total of ~19,63 M non-redundant SNPs. The majority of variants (13,07 M, 66,57%) were shared among the four genotypes, attributable to their high diversity in respect to ‘CM334’, which can be considered as a precursor of modern sweet peppers17. The SNP frequency observed at whole genome level was quite similar among the four inbred lines, ranging from 1 SNP/Indel every ~168,5 to 1 every 179,3 bp (~0.56–0.59%, Table 6). Some biases were instead recorded in the 12 chromosomes (Table S10), as can be seen from the Circos (Fig. 3). Chr. 9 resulted the most polymorphic in the four inbred lines (one polimorphism every 83.5 bp on average), while chr. 8 was the least polymorphic (one variation every 414 bp). Most of the SNP/Indels identified in the four inbred lines were highly homozygous, as the rate of heterozygosity ranged from 0,098% in ‘Tumaticot’ to 0,196% in ‘Quadrato’ and ‘Corno’ (Table 6), coherently with their inbreeding history5. The majority of variants in homozygous state was observed in ‘Cuneo’ and ‘Tumaticot’: this, together with their low heterozygosis level (~0.098% and 0.112%; Table 6), suggest a higher genetic stability compared to the other two inbred lines, confirming a more effective selection carried out over the time by farmers5,63. The identified SNP/Indels were used to estimate the genetic relatedness among the four inbred lines (Fig. 3). They did not group according to the fruit shape, as ‘Quadrato’ and ‘Corno’ showed to be closer each other in respect to ‘Cuneo’ and ‘Tumaticot’. This might be explained because ‘Quadrato’ and ‘Corno’ are currently cultivated on larger areas more suited to horticulture and over time have been subject to a greater selection for traits of agronomic interest. Vice versa, ‘Cuneo’ and ‘Tumaticot’ are widespread in more limited and foothills areas and therefore adapted to more niche soil and climatic conditions.
Table 6.
SNP/Indel (DP > 15) statistics identified in the four pepper genotypes with their relative frequency for each analysed genotype.
| Genotype | unfiltered SNP/Indel | SNP/Indel | SNP | Indel | N°SNP/Indel (%) | SNP/Indel in 1000 bp | 1 SNP/Indel every (bp) | N° SNP/Indel Homozygosis | SNP/Indel Heterozygosis | Heteroz. (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Cuneo | 17,678,065 | 16,649,886 | 16,033,452 | 616,434 | 0.586% | 5.86 | 170.6 | 13,228,374 | 3,421,512 | 0.112% |
| Quadrato | 18,972,695 | 18,019,108 | 17,347,941 | 671,167 | 0.593% | 5.93 | 168.5 | 12,019,779 | 5,999,329 | 0.196% |
| Corno | 18,941,981 | 18,076,252 | 17,389,251 | 687,001 | 0.585% | 5.85 | 171.0 | 12,082,809 | 5,993,443 | 0.196% |
| Tumaticot | 17,224,938 | 16,330,222 | 15,701,405 | 628,817 | 0.558% | 5.58 | 179.3 | 13,311,815 | 3,018,407 | 0.098% |
The reference genome (‘CM334’) considered (without N) is of length equal to 3.065.158.452 bp.
Figure 3.
Left side: Diagram showing gene density and SNP distributions: (A) Heat map of gene density in the reference genome; 1Mbp histograms of SNP density for ‘Cuneo’ (trace B) ‘Tumaticot’ (trace C), ‘Quadrato’ (trace D), ‘Corno’ (trace E). Right side: UPGMA-based dendrogram of the four genotypes taking into account genomic SNPs; ‘Perennial’ was used as outgroup and bootstrap values were inferred from 100 replicates.
About 80% of the SNP/Indel variants were found in intergenic region and just at most 1.8% in exons. The frequency of SNP/Indels in intronic region was 6%, while upstream and downstream gene variants were 12% and 9%, respectively (Table S11). Following the SnpEff analysis focused on coding regions, the majority (63.8%) of variations were non-synonymous (missense), followed by synonymous (silent), while just 2.4% were nonsense. By considering homozygous or heterozygous variants (Table S12), no significant differences in number, among inbred lines, were observed for high effect SNPs in homozygous state (4.65 K on average), while the heterozygous ones widely varied, ranging from about 1.98 K in ‘Tumaticot’ to 3.47 K in ‘Quadrato’ (Table S13). An analogous trend was also observed for the missense mutations, whose total number varied from about 65k in ‘Tumaticot’ to about 84 K in ‘Quadrato’. (Table S13).
Variants in fruit related genes
The size of the fruit is a key trait for breeding, which is controlled by several genes and multiple interacting biosynthetic pathways64. In pepper, a large number of QTLs affecting fruit size and shape have been identified so far in different genetic backgrounds14,37, and most of them locate on chr.s 2 and 4. Hill et al.64 detected in pepper a QTL (Chaim2.2), including both the regulatory genes CaOVATE and CaWUS, which explain up to 17% of the variation in the berry diameter, while in tomato, Frary et al.65 demonstrated that the fw2.2 gene explains about 30% of the fruit weight variation.
We analysed variants in 55 genes belonging to categories/pathways controlling berry size and shape (Table S14), and also included in the analysis orthologs of tomato genes involved in the domestication syndrome66, i.e.: fw2.2/CNR controlling fruit weight, OVATE and SUN controlling fruit shape, fas/CLV3 and lc/WUSCHEL controlling fruit size and shape. On the whole, most of the genes affecting fruit shape and size (e.g.: WUS, CLV3, SUN,) were found to harbour more mutations than the ones influencing berry weight (e.g.: fw2.2 and fw3.2).
By using ‘CM334’ as reference, the inbred lines in study contained 35 SNPs per gene on average, of which most resided in regulatory regions, but no deleterious variants were spotted (Figure S3). Half of the genes (Figure S2) exhibited more than 20 SNPs in upstream regions (cis-regulatory regions), with a potential role in affecting agronomic traits67.
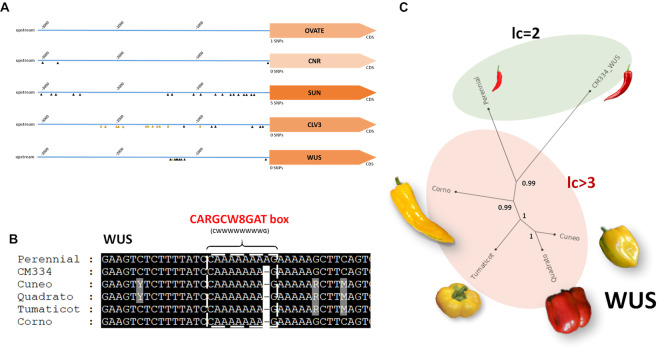
Promoters of the four inbred lines (Fig. 4) were compared with the ones of ‘CM334’ and ‘Perennial’, which produce very small berries (Table 1). In the 3 kb upstream (Figure S4) region of SUN, WUS and CLV3, consensus variants were spotted in the small-fruited ‘CM334’ and ‘Perennial’ as well as in our four inbred lines, as disclosed by the genetic trees (Fig. 4). In SUN and CLV3 the variants were widespread in the 3 kb upstream the genes, while in WUS was highlighted a cluster of 18 SNP in a small region (130–140 bp). Due to the absence of some cis-regulatory elements (File S1), those mutations might impact the WUS and CLV3 promoter with phenotipic consequences. As example, a mutated CARGCW8GAT box, a variant of CArG motif with an extended A/T-rich sequence68, was observed in the WUS promoter of the four inbred lines in study as well as in ‘CM334’ (Fig. 4). Many variants in CLV3 were found conserved between ‘CM334’ and’Perennial’ in the 1500–2200 bp upstream the gene start codon (Figure S4) eliminating some cis-element (CCAAT box, MYb1 box, File S1). Although not functionally validated, the mutations in the WUS and CLV3 promoters of the four inbred lines (Fig. 4) might affect CLV3 expression and its interaction with WUS in the classical CLAVATA-WUSCHEL stem cell circuit (CLV-WUS) controlling meristem size, as already observed in tomato69,70, in which they led to an increased fruit size during domestication. Mutations in WUS and CLV promoters might be related to the number of fruit lodges, which ranges from 3 to 4 in the four piedmontese inbred lines, while is two in ‘CM334’ (2.28) and ‘Perennial’. However, more complex allelic epistatic interactions as well as other genes might be involved in determining fruit shape, as SNPs in promoter region of many other genes involved in organ size/development were detected (Figure S2). Currently, the effect of mutations in gene regulatory regions is based on the expensive and laborious selection of rare mutations occurring in these regions. However, hereafter, the application of the CRISPR/Cas9 technology on promoters of WUS and CLV may contribute to shedding light on the impact of the detected variants by modelling different cis-regulatory alleles and providing quantitative variations67.
Figure 4.
(A) SNP distribution in upstream regions of OVATE, CNR, SUN, CLV3 and WUS genes. Triangles show detected SNPs. Orange triangles represent conserved SNPs in elongated vs blocky genotypes. (B) Example of one altered cis-element in the WUS gene. (C) Genetic tree constructed using information on SNPs present in the WUS promoter, with bootstrap values (100 iterations). “lc” notation means “locule average number”.
Detection of presence/absence variants (PAVs)
Gene presence–absence variations (PAVs) within a species can contribute to trait variation and recently Ou et al.71 published the PAN-genome of cultivated pepper, based on a wide set of cultivars belonging to the four Capsicum cultivated species, and applied a gene PAV approach for performing phylogenetic analysis and a genome-wide association study (GWAS)71. In our study, of the 875 PAV loci identified, most (654, i.e. about 75.0%) were present only in the ‘CM334’ genome. Among the other 221 PAV loci detected, 78 were absent in one, 43 in two and 100 in three inbred lines (Table S15). Of the 100 PAV loci exclusive to a genotype, 39 were spotted in ‘Cuneo’, 42 in ‘Tumaticot’, 11 in ‘Corno’ and 8 in ‘Quadrato’ (Table S16). Some of the PAVs related to RGAs seem justify some discrepancies detected in the RGA orthologous clusters (missing of some genes among genotypes, Figure S1), as well as some genotype-specific clusterization of NBS-LRRs, which likely occurred via segmental and/or tandem duplications (Table S6). Among the PAVs missing in a inbred line, two small clusters were detected, of which one chr. 11 (249,24 Mb − 249,25 Mb and 257,90 Mb 257,94 Mb) of ‘Cuneo’ and one in chr. 7 (0,77 Mb - 1,61 Mb) of ‘Tumaticot’ carrying three transcriptional factors (SAP4 Zinc finger A20 and AN1 domain-like) and four resistance RGAs. Among PAVs unique to a genotype, three small clusters were observed: two in ‘Cuneo’ located in chr. 2 (133,78 Mb - 133,81 Mb), and containing three RGAs in a 10k wide region and on chr. 6 (4,48 Mb-4,74 Mb); the third was located on chr. 9 (268,65 Mb-269,32 Mb) of ‘Tumaticot’ carrying four resistance gene analogues (RGAs, Figure S5).
The GO enrichment analysis for the five clusters showed just one over-representation (“nucleotide binding proteins”, GO:0000166) related to the PAVs exclusively present in ‘Tumaticot’.
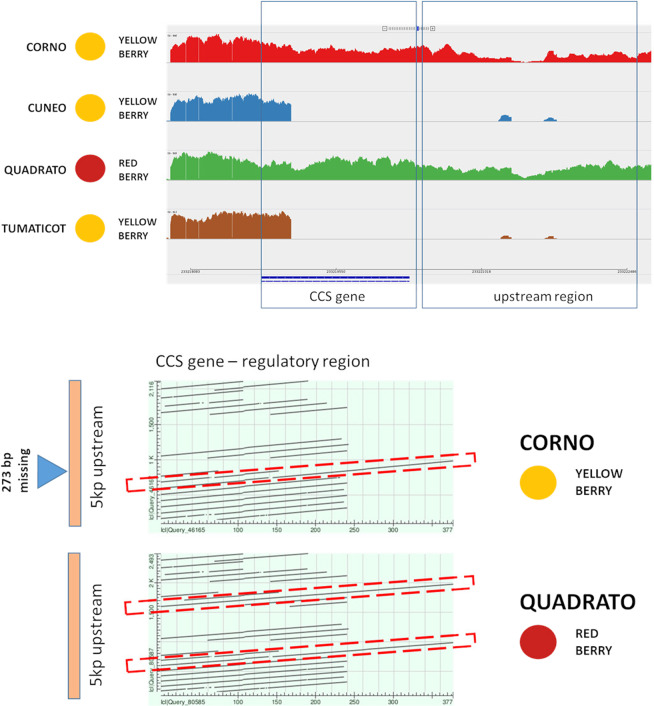
In pepper, capsanthin/capsorubin content is regulated by the capsanthin/capsorubin synthase (CCS) gene activity, and other three genes (PSY, LCYB, CRTZ) are responsible for red/orange/yellow pigmentation72. Three of the four pepper inbred lines in study (‘Cuneo’, ‘Corno’, and ‘Tumaticot’) produce yellow fruits while one (Quadrato) red fruits (Table 1). Since no deleterious SNPs were observed in the four genes involved in the berry pigmentation, we perfomed comparative analyses in order to detect possible PAVs. In two of the yellow-fruited inbred lines (‘Cuneo’ and ‘Tumaticot’, Fig. 5) missing reads in the CCS gene surrounding the coding sequence and its promoter, and leading to a trunked protein, were spotted (Fig. 5). This was not so in the red fruited ‘Quadrato’, but also in the yellow-fruited ‘Corno’, although the latter lacks of a 378 bp region located in the CCS promoter (1200–1300 bp upstream the start codon, Fig. 5). This region was scanned and many regulatory motifs were identified (e.g.: CAATBOX 1-like and myb-like cis-elements, Table S17). Moreover, some differences were also present in the common distal box, such as a triple CAAAT box in ‘Quadrato’, in respect of a double CAAAT box in ‘Corno’, the latter due to a 15 bp deletion. It is known that multiple repeats in a promoter segment can cause transcription factor autoregulation, and in red apples the number of such modules correlates with increased transactivation by a MYB10 protein73. A similar behaviour was reported in C. annuum, where the CCS promoter presents a 3-modular redundant structure72, in which a single repeat ensures CCS gene expression. By considering the 5 kb region up-stream the CCS gene in ‘Quadrato’ and ‘Corno’, the former (red) showed the presence of 2 modules (2 ×378 bp), while the latter (yellow) exhibited only one distal module, with a 15 bp deletion depleting one CCAT box (Fig. 5). As previously reported the transcriptional regulation of CCS expression is complex74, and we hypothesize that the CCS function in ‘Corno’ is thus compromised by its incomplete promoter structure.
Figure 5.
Top side: Presence absence variation in the Locus Y (CCS) is shown in the four resequenced pepper genotypes showing the CCS gene (in blue, bottom) and 5 kb upstream regions. Bottom side: 5Kb upstream region of the CCS gene. Pairwise blast of the 378 bp in the ‘Corno’ versus the 5 kb region of ‘Corno’ (top part) and ‘Quadrato’ (bottom part).
Conclusions
In spite of the high organoleptic and sensorial quality of their products, landraces of horticultural species have been displaced over time from market-driven production due to their lower yields, poorer pest and disease resistance, and often limited postharvest shelf-life in respect to bred commercial varieties and F1 hybrids. It is thus crucial to characterize the genetic variation within and between landraces of vegetables, with the goal to actualize their agronomic performances and make them fitting with current agriculture and consumption standards.
Recent advances in high-throughput sequencing technologies makes easily available big amounts of data, which integrated with phenotypic information, enable the identification of genes affecting key agronomic traits. We performed the genome resequencing of four pepper inbred lines selected within as many Italian landraces producing different berry types, ranging from blocky to elongated or sub-spherical. Our resequencing data revealed large genetic variations among them and demonstrate that resequencing provides an efficient way for gathering a large amounts of genomic information, although further analyses and functional studies will serve for practical applications in marker-assisted selection programs.
The identified RGAs provide tools for designing diagnostic markers and identifying quantitative trait loci (QTL) or markers associated with plant disease resistance. The identified susceptibility genes (S-genes), which favour the plant colonization by pathogens, represent ideal target for genomic editing, with the goal to disrupt their function and confer durable resistance to diseases. In addition, the spotted genes related to fruit quality traits may represent target for pepper breeding as well as for understanding the genomic features that distinguish modern from traditional varieties.
Materials and Methods
DNA extraction
Seeds were germinated and plantlets cultivated for four weeks in a growth chamber in conditions of darkness/light (8/16 hours) at 25 °C. Subsequently, total genomic DNA was extracted from fresh leaves of each genotype, using the DNA Mini Plant kit (Qiagen, Hilden, Germany). RNAse A was used to remove RNA contamination. DNA quality was checked by 1% (w/v) agarose gel electrophoresis, and its quantity was assessed by Qubit 2.0 based on Qubit dsDNA HS Assay (Thermo Fisher Scientific, Waltham, MA, USA).
Sequencing, genome assembly and reference-guided reconstruction
A total amount of 1 μg of DNA was used for the construction of four short insert (length 350 bp) genomic libraries (Novogene, Hong Kong), which were sequenced using an Illumina sequencer (Illumina Inc., San Diego, CA, USA) with paired-end chemistry (2×150 bp). Raw reads were cleaned with Scythe (v0.994, https://github.com/vsbuffalo/scythe) for removing contaminant residual adapters and Sickle (v1.33, https://github.com/najoshi/sickle), which allows to remove reads with poor quality ends (Q < 30). A two-step approach was adopted for assembly. The first one (de novo), which generated contigs/scaffolds, was performed with SoapDenovo v2.0475 using specific assembly parameters (avg_ins=300; max_rd_len=150; reverse_seq=0; asm_flags=3; rd_len_cutoff=100; rank=1; pair_num_cutoff=3; map_len=32). The second one (reference-guided), which integrates the contigs/scaffolds into large pseudomolecules corresponding to the chromosomes, was performed using the Chromosomer (v 0.1.4a) tool76, with default parameters. In the first one, a k-mer set (wide range: 51, 61, 71, 81 and 91) was preliminarily tested to identify the best k-mer range. Then, a second series of k-mers (e.g.: narrow range, 53–69), which revealed the best assembly performances, was scanned. Metrics for assessing the quality of a genome assembly (e.g.: N50, contig/scaffold number/size/length, genome length) were assessed using the perl script Assemblathon_stats.pl (https://github.com/ucdavis-bioinformatics/assemblathon2-analysis). GapCloser v1.12 (https://sourceforge.net/projects/soapdenovo2/files/GapCloser) was used to fill in the gap emerging in the assembly/scaffolding process. Finally, only the contigs/scaffolds with a length exceeding 500 bp were taken into consideration for genome reconstruction. The genome reconstruction of each genotype was performed using the Chromosomer76 pipeline taking into consideration the scaffolds/contig previously obtained and the sequence of the Capsicum annuum genome (‘CM334’)15 as a guide. Chromosomer uses two parameters influencing the assembly process. The first represents the alignment score threshold (r = 1), which is used to distinguish between anchored and unlocated fragments. If the ratio of the scores of the two fragment alignments with the highest score exceeds the threshold, the fragment is considered anchored, otherwise it will not be positionable and excluded from subsequent analyzes. The alignment score threshold must be a positive number greater than one. The second parameter is the average size of the sequenced fragments, which is used to insert gaps in regions not covered by fragments to be anchored. Raw data from ‘CM334’ and ‘Perennial’ genotypes were downloaded from NCBI (PRJNA223222 and PRJNA298503).
Structural and functional annotation
Each genotype was masked using RepeatMasker77 v4.1.0 using a combination of homology-based and de novo approaches. A species specific repeats library was constructed following the Repeat Library Construction Advanced pipeline (http://weatherby.genetics.utah.edu/MAKER/wiki/index.php/Repeat_Library_Construction-Advanced) which requires the use of mite hunter78, LTRdigest79, LTR_harvest80 (available in genome tools81 v1.5.10) and Repeatmodeler82 v1.0.11. The new library was then combined with Repbase-viridiplantae83 to identify TEs. TEs were classified into two main classes (typical of plant genomes): Class I (retrotransposon elements) and Class II (DNA transposons). Gene prediction was performed using Maker-P84 v2.31.08. Augustus85 v3.3.2 Hidden Markov Models and SNAP86 gene prediction algorithms were combined with transcripts and proteins alignments as evidence to support prediction. All predicted gene models were filtered and only the ones with an AED ≤ 0.35 were maintained. AED measures the concordance of a gene predicted with aligned transcripts, mRNA-seq and protein homology data. AED scores range from 0 and 1, where 0 indicates perfect concordance between evidence and gene prediction, while 1 absence of concordance.
For each predicted gene, the gene function was assigned by a BlastP87 search against the Uniprot/Swissprot Viridiplantae database88, using the default parameters, with the exception of the e-value (<1e-5).
To measure the quality and completeness of the predicted proteomes, a quantitative assessment was carried out based on evolutionary informed expectations of gene content known as Benchmarking Universal Single-Copy Orthologs (BUSCO89 v3.0.2., Embryophyta odb 10).
The sequences of the predicted proteins were also noted using InterproScan590 compared to all the available databases (ProSitePro les-20.119, PANTHER-10.0, Coils-2.2.1, PIRSF-3.01, Hamap-201511.02, Pfam-29.0, ProSitePatterns - 20.119, SUPERFAMILY-1.75, ProDom-2006.1, SMART-7.1, Gene3D-3.5.0 and TIGRFAM-15.0)91–101. Data obtained from the four proteomes were illustrated in a Venn diagram constructed with Interactivenn102. Then, GOfeat103 was used to identify the enrichment of GO terms for specific gene clusters. Some candidate genes were specifically analyzed in the four resequenced pepper inbred lines. To the scope, protein sequences of the orthologous genes (from pepper, tomato, Arabidopsis) and involved in specific traits/pathways (e.g.: fruit shape, colour, S-genes) were downloaded from NCBI, and used to retrieve, via BlastP, putative orthologous proteins in the proteomes of ‘Corno’, ‘Quadrato’, ‘Tumaticot’ and ‘Cuneo’.
Prediction and annotation of miRNA
The MIReNA v2.0 software104 was used for the identification of high confidence miRNA-coding sequences (miRBase release 21105) in each pseudomolecule and chr. 0 of all the analyzed inbred lines. A homology search was conducted with known miRNAs from a group of plants and algae species, which included: Solanum lycopersicum, Solanum tuberosum, Nicotiana tabacum, Vitis vinifera, Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Medicago trunculata, Zea mays, Picea abies, Triticum aestivum, Physcomitrella patens, Chlamydomonas reinhardtii. MIReNA was run with default parameters and the maximum number of allowed mismatches between known miRNAs and putative miRNAs was set to 10. For each inbred line, miRNA identified were named based on the miRNA family with the addition of the name of the genotype (‘Corno’, ‘Cuneo’, ‘Quadrato’, ‘Tumaticot’). psRNATarget106 (2017 update) was applied to identify the targets of the identified pepper miRNA on the predicted CDSs. Results were parsed to keep only those targets having expectation <of 2.5. GO term enrichment of target sequences for each line was carried out with AGRIGOv2107 to find out a representative subset of the GO terms previously identified with the Interproscan pipeline. AGRIGOv2 cross comparison of SEA (SEACOMPARE) was used to identify common and different enrichment GO terms between the genotypes showing GO terms enrichment.
Resistance genes analogs (RGA)
Candidate resistance genes were identified using RGAugury108 for all the four inbred lines in study as well as for the reference line 'CM334'. The pipeline first identifies RGA-related protein domains and motifs, namely nucleotide binding site (NB-ARC), leucine rich repeat (LRR), transmembrane (TM), serine/threonine and tyrosine kinase (STTK), lysine motif (LysM), coiled-coil (CC) and Toll/Interleukin-1 receptor (TIR). RGA candidates were identified and classified into four major families based on the presence of combinations of these RGA domains and motifs: NBS-encoding, TM-CC, and membrane associated RLP and RLK. The NBS-encoding gene family members are further divided into several subgroups according to their domain architecture, namely NBS (NBS domain), CNL (CC-NBS-LRR domains), TNL (TIR-NBS-LRR), TN (TIR-NBS), CN (CC-NBS), NL (NBS-LRR), TX (TIR-unknown domain) and other. To compare the RGAs within and among the four inbred lines in study and the reference line ‘CM334’, we divided the RGAs identified, following RGAugury analyses, in 6 separated groups: RLP (Receptor Like Proteins), RLK (Receptor like Kinases), NBS, CNL, NL and TM-CC. Furthermore, NLR proteins (CNL and TNL) in Solanaceae together with known R proteins from Arabidopsis were used. The multiple alignments were performed using MAFFT109 v7.450 with the following parameters: –ep 0 –reorder –maxiterate 1000 –genafpair. Genetic relationships were described by constructing a phylogenetic tree by maximum likelihood by using the IQ-TREE software110 v.1.6.12; branch supports were obtained with the ultrafast bootstrap111 with 1000 replicates. Trees were visualized using interactive Tree of Life (iTOL) v3112. To identify the number of RGAs per chromosome across all the 5 genotypes, as well as the presence of clusters, coordinates of the genes belonging to the classes LP (Receptor Like Proteins), RLK (Receptor like Kinases), NBS, CNL, NL and TM-CC were extracted and BEDTools113 intersected using genome windows of 1 Mb to count the number of genes falling into these regions.
Susceptibility genes
A preliminary BLASTP analysis allowed to identify the possible orthologos for susceptibility genes, using information from several plant species (Table S8), considering as a preferential choice criterion the e-value (range 0–1e−10), the percentage of similarity and the query coverage. Since many genes were present in multi-gene family, filtering criteria were varied each time and previous functional annotations were used to filter out wrong candidates. Multiple sequence alignments and phylogenetic analyses were carried out using Clustal Omega114. Phylogenetic trees were generated for candidate S-genes families using the neighbor-joining (NJ) algorithm. A confidence level was established for each node by performing a bootstrap analysis with 100 iterations. Trees were plotted using Figtree graphical viewer115.
SNP calling and variant annotation
The sequences were mapped to the reference genome of the ‘CM334'15 line using the Burrows-Wheeler Aligner (BWA, v0.7.17) program and the ‘mem’ command with the default parameters116. BAM files were processed and use for the SNP calling using with Samtools (v1.6)117 mpileup with default parameters except for i) minimum mapping quality (Q = 20) and filtering out multimapping events (-q > 1). A vcf (variant call format) file was produced and was subsequently used to construct a tree diagram using Tassel v4.0118. SNP/Indels were counted and analyzed using custom bash scripts. The estimation of the heterozygous level of each genome was calculated by considering, per each inbred line, the ratio between the number of SNP/Indels (called in heterozygous state) and the size of the ‘CM334’ genome, deprived of Ns (3,065,158,452 bp). The identified genomic variants were analyzed with the SnpEff v4.3 program119, to infer their functional annotation and any potential deleterious effect on protein structure. The effect of each SNP/Indel was classified into four of classes of effects: 1) modifier effect, as variants located outside genes (non-transcribed regions or introns); 2) low effect, as synonymous variants in coding regions; 3) moderate effect, as variants altering the aminoacidic sequence and 4) high effect, as variants changing frameshift thereby introducing/eliminating stop codons or modifying splice sites. Upstream (3 kb) gene regions were searched for the presence/absence of cis-regulatory elements using PLACE (www.dna.affrc.go.jp/PLACE), a database which collect plant cis-acting regulatory DNA elements.
Identification and characterization of PAV genes
Samtools117 was used to generate a file containing the number of reads mapping on the reference genome. The number of reads that mapped at each gene location for every pepper inbred line were extracted and normalized by the total number of reads mapping to the whole reference genome (‘CM334’) for each inbred line. Parameters were set up to spot unmapped regions, as follows: samtools view -c -F 4 -q 1 file.sort.bam. To identify putative PAV genes, all genes with less than six mapped reads from at least one inbred line and more than 29 mapped reads from at least another inbred line were selected. The list of candidate PAV genes obtained were then described using the available ‘CM334’ functional and structural annotation.
Supplementary information
Acknowledgements
This research was funded by the ‘Cassa di Risparmio di Cuneo’ Foundation, under the research project: Resequencing of sweet pepper ecotypes for enhancing their quality and for their traceability (RISEPP).
Author contributions
S.L., E.P. and D.V. selected/provided plant materials and maintained all living materials. M.N., D.V., LB and A.A performed in field plant sampling and DNA extraction. A.A., L.B. and M.N performed the sequencing experiments, conducted the genome assembly/reconstruction, setup SNP mining pipelines. L.B. performed functional annotation, gene prediction analyses, conducted miRNAs and RGA analysis. A.A and L.B. analyzed/interpreted data. A.A. and L.B. drafted the paper and curated the supplementary materials. E.P., A.A. and S.L. planned the study, contributed to the data interpretation, and revised the manuscript. All the authors discussed the results and commented on the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-66053-2.
References
- 1.Pickersgill B. Genetic resources and breeding of Capsicum spp. Euphytica. 1997;96:129–133. [Google Scholar]
- 2.Portis E, Nervo G, Cavallanti F, Barchi L, Lanteri S. Multivariate analysis of genetic relationships between Italian pepper landraces. Crop Sci. 2006;6:2517–2525. [Google Scholar]
- 3.Lanteri S, Acquadro A, Quagliotti L, Portis E. RAPD and AFLP assessment of genetic variation in a landrace of pepper (Capsicum annuum L.), grown in North-West Italy. Genet. Resour. Crop Evol. 2003;50:723–735. [Google Scholar]
- 4.Portis E, et al. The design of Capsicum spp. SSR assays via analysis of in silico DNA sequence, and their potential utility for genetic mapping. Plant Sci. 2007;172:640–648. [Google Scholar]
- 5.Portis E, Acquadro A, Comino C, Lanteri S. Effect of farmers’ seed selection on genetic variation of a landrace population of pepper (Capsicum annuum L.), grown in North-West Italy. Genet. Resour. Crop Evol. 2004;51:581–590. [Google Scholar]
- 6.Akbar N, Habib A, Ghafoor S, Begum K, Gul S. Estimation of Genetic Diversity in Capsicum Germplasm Using Randomly Amplified Polymorphic DNA. Asian J. Agric. Sci. 2010;2(2):53–56, 2010. [Google Scholar]
- 7.Hill TA, et al. Characterization of Capsicum annuum genetic diversity and population structure based on parallel polymorphism discovery with a 30K Unigene pepper GeneChip. PLoS One. 2013;8:e56200. doi: 10.1371/journal.pone.0056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X. min et al. Genetic diversity of pepper (Capsicum spp.) germplasm resources in China reflects selection for cultivar types and spatial distribution. J. Integr. Agric. 2016;15:1991–2001. [Google Scholar]
- 9.Igwe DO, Afiukwa CA, Acquaah G, Ude GN. Genetic diversity and structure of Capsicum annuum as revealed by start codon targeted and directed amplified minisatellite DNA markers. Hereditas. 2019;156:1–13. doi: 10.1186/s41065-019-0108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzmán FA, Moore S, de Vicente MC, Jahn MM. Microsatellites to enhance characterization, conservation and breeding value of Capsicum germplasm. Genet. Resour. Crop Evol. 2019;67:569–585. [Google Scholar]
- 11.Jackson SA, Iwata A, Lee SH, Schmutz J, Shoemaker R. Sequencing crop genomes: Approaches and applications. New Phytologist. 2011;191:915–925. doi: 10.1111/j.1469-8137.2011.03804.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim SS-BS-Y, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 2014;46:270–8. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- 13.Qin C, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. 2014;111:5135–5140. doi: 10.1073/pnas.1400975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han K, et al. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum) DNA Res. 2016;23:81–91. doi: 10.1093/dnares/dsv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, et al. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 2017;18:210. doi: 10.1186/s13059-017-1341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hulse-Kemp AM, et al. Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 2018;5:4. doi: 10.1038/s41438-017-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du H, et al. Target sequencing reveals genetic diversity, population structure, core-SNP markers, and fruit shape-associated loci in pepper varieties. BMC Plant Biol. 2019;19:578. doi: 10.1186/s12870-019-2122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Schie CCN, Takken FLW. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014;52:551–581. doi: 10.1146/annurev-phyto-102313-045854. [DOI] [PubMed] [Google Scholar]
- 19.Barchi L, et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019;9:11769. doi: 10.1038/s41598-019-47985-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buendía-Monreal M, Gillmor CS. Mediator: A key regulator of plant development. Developmental Biology. 2016;419:7–18. doi: 10.1016/j.ydbio.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Mathur S, Vyas S, Kapoor S, Tyagi AK. The mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (Rice) during reproduction and abiotic stress. Plant Physiol. 2011;157:1609–1627. doi: 10.1104/pp.111.188300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X, et al. The NAC transcription factor SLNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol. 2018;177:1286–1302. doi: 10.1104/pp.18.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majda, M. & Robert, S. The role of auxin in cell wall expansion. International Journal of Molecular Sciences 19 (2018). [DOI] [PMC free article] [PubMed]
- 24.Rhoades MW, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 25.Spanudakis E, Jackson S. The role of microRNAs in the control of flowering time. J. Exp. Bot. 2014;65:365–380. doi: 10.1093/jxb/ert453. [DOI] [PubMed] [Google Scholar]
- 26.Sun X, et al. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.) BMC Genomics. 2015;16:197. doi: 10.1186/s12864-015-1416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang Y, Xie K, Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014;65:2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekhwal MK, et al. Disease resistance gene analogs (RGAs) in plants. International Journal of Molecular Sciences. 2015;16:19248–19290. doi: 10.3390/ijms160819248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaidi SS, Mukhtar MS, Mansoor S. Genome Editing: Targeting susceptibility genes for plant disease resistance. Trends in Biotechnology. 2018;36:898–906. doi: 10.1016/j.tibtech.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Eitas TK, Dangl JL. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Current Opinion in Plant Biology. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H-A, Yeom S-I. Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief. Funct. Genomics. 2015;14:233–242. doi: 10.1093/bfgp/elv012. [DOI] [PubMed] [Google Scholar]
- 32.Seo, E., Kim, S., Yeom, S. I. & Choi, D. Genome-wide comparative analyses reveal the dynamic evolution of nucleotide-binding leucine-rich repeat gene family among solanaceae plants. Front. Plant Sci. 7 (2016). [DOI] [PMC free article] [PubMed]
- 33.Kim J, et al. A genome-wide comparison of NB-LRR type of resistance gene analogs (RGA) in the plant Kingdom. Mol. Cells. 2012;33:385–392. doi: 10.1007/s10059-012-0003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acquadro A, et al. Genome reconstruction in Cynara cardunculus taxa gains access to chromosome-scale DNA variation. Sci. Rep. 2017;7:5617. doi: 10.1038/s41598-017-05085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, J. et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics15 (2014). [DOI] [PMC free article] [PubMed]
- 36.Rody HVS, et al. Genome survey of resistance gene analogs in sugarcane: genomic features and differential expression of the innate immune system from a smut-resistant genotype. BMC Genomics. 2019;20:809. doi: 10.1186/s12864-019-6207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barchi L, et al. QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor. Appl. Genet. 2009;118:1157–1171. doi: 10.1007/s00122-009-0970-0. [DOI] [PubMed] [Google Scholar]
- 38.Bonnet J, et al. Are the polygenic architectures of resistance to Phytophthora capsici and P. parasitica independent in pepper? Theor. Appl. Genet. 2007;115:253–264. doi: 10.1007/s00122-007-0561-x. [DOI] [PubMed] [Google Scholar]
- 39.Xiao S, et al. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–20. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- 40.Gururani MA, et al. Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 2012;78:51–65. [Google Scholar]
- 41.Xiao S, et al. Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol. Biol. Evol. 2004;21:1661–1672. doi: 10.1093/molbev/msh165. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, et al. A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting. Plant Cell. 2013;25:4242–4261. doi: 10.1105/tpc.113.117226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Büschges R, et al. The marley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 44.Panstruga R. Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol. Biol. 2005;59:485–500. doi: 10.1007/s11103-005-0353-0. [DOI] [PubMed] [Google Scholar]
- 45.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Wretblad, S., Bohman, S. & Dixelius, C. Overexpression of a Brassica nigra cDNA gives enhanced resistance to Leptosphaeria maculans in B. napus, 10.1094/MPMI.2003.16.6.477 (2007). [DOI] [PubMed]
- 47.Brandwagt, B. F., Kneppers, T. J. A., Nijkamp, H. J. J. & Hille, J. Overexpression of the tomato Asc-1 gene mediates high insensitivity to AAL toxins and fumonisin B1 in tomato hairy roots and confers resistance to Alternaria alternata f. sp. lycopersici in Nicotiana umbratica plants, 10.1094/MPMI.2002.15.1.35 (2007). [DOI] [PubMed]
- 48.Cao A, et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. 2011;108:7727–7732. doi: 10.1073/pnas.1016981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song H, et al. Comparative analysis of NBS-LRR genes and their response to Aspergillus flavus in Arachis. PLoS One. 2017;12:e0171181. doi: 10.1371/journal.pone.0171181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- 51.Orgil U, Araki H, Tangchaiburana S, Berkey R, Xiao S. Intraspecific genetic variations, fitness cost and benefit of RPW8, a disease resistance locus in Arabidopsis thaliana. Genetics. 2007;176:2317–33. doi: 10.1534/genetics.107.070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eckardt NA. A microRNA cascade in plant defense. Plant Cell. 2012;24:840. doi: 10.1105/tpc.112.240311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shivaprasad PV, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–74. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–15. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kallman T, Chen J, Gyllenstrand N, Lagercrantz U. A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. PLANT Physiol. 2013;162:741–754. doi: 10.1104/pp.113.214643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun K, et al. Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 2016;25:731–742. doi: 10.1007/s11248-016-9964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saijo Y, Loo E. P. iian & Yasuda, S. Pattern recognition receptors and signaling in plant–microbe interactions. Plant J. 2018;93:592–613. doi: 10.1111/tpj.13808. [DOI] [PubMed] [Google Scholar]
- 59.Porterfield R, Meru G. Candidate Susceptibility Genes for powdery and downy mildew in watermelon and squash. J. Phylogenetics Evol. Biol. 2017;05:1–14. [Google Scholar]
- 60.Eckardt NA. BIK1 function in plant growth and defense signaling. Plant Cell. 2011;23:2806. doi: 10.1105/tpc.111.230811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nie H, et al. SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 2012;158:1847–1859. doi: 10.1104/pp.111.192310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirik V, et al. CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr. Biol. 2001;11:1891–1895. doi: 10.1016/s0960-9822(01)00590-5. [DOI] [PubMed] [Google Scholar]
- 63.Portis E, Baudino M, Magurno F, Lanteri S. Genetic structure and preservation strategies of autochthonous vegetable crop landraces of north-western Italy. Ann. Appl. Biol. 2012;160:76–85. [Google Scholar]
- 64.Hill, T. A. et al. Regions underlying population structure and the genomics of organ size determination in Capsicum annuum. Plant Genome 10 (2017). [DOI] [PubMed]
- 65.Frary AA, et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science (80-.). 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 66.van der Knaap, E. et al. What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5 (2014). [DOI] [PMC free article] [PubMed]
- 67.Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171:470–480.e8. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 68.Theißen G, Saedler H. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- 69.Somssich M, Je B, Il, Simon R, Jackson D. Clavata-Wuschel signaling in the shoot meristem. Development (Cambridge) 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- 70.Xu C, et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015;47:784–792. doi: 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- 71.Ou L, et al. Pan-genome of cultivated pepper (Capsicum) and its use in gene presence-absence variation analyses. New Phytol. 2018;220:360–363. doi: 10.1111/nph.15413. [DOI] [PubMed] [Google Scholar]
- 72.Tian S-L, Li Z, Li L, Shah SNM, Gong Z-H. Analysis of tandem repeat units of the promoter of capsanthin/capsorubin synthase (Ccs) gene in pepper fruit. Physiol. Mol. Biol. Plants. 2017;23:685–691. doi: 10.1007/s12298-017-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Espley RV, et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li, Z., Wang, S., Gui, X. L., Chang, X. B. & Gong, Z. H. A Further analysis of the relationship between yellow ripe-fruit color and the capsanthin-capsorubin synthase gene in pepper (Capsicum sp.) indicated a new mutant variant in c. annuum and a tandem repeat structure in promoter region. PLoS One, 10.1371/journal.pone.0061996 (2013). [DOI] [PMC free article] [PubMed]
- 75.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamazian G, et al. Chromosomer: a reference-based genome arrangement tool for producing draft chromosome sequences. Gigascience. 2016;5:38. doi: 10.1186/s13742-016-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smit, AFA, Hubley, R & Green, P. RepeatMasker Open-4.0.
- 78.Han Y, Wessler SR. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38:e199–e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steinbiss S, Willhoeft U, Gremme G, Kurtz S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009;37:7002–13. doi: 10.1093/nar/gkp759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gremme G, Steinbiss S, Kurtz S. Genome tools: A comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2013;10:645–656. doi: 10.1109/TCBB.2013.68. [DOI] [PubMed] [Google Scholar]
- 82.http://www.repeatmasker.org/RepeatModeler/. Available at: http://www.repeatmasker.org/RepeatModeler/.
- 83.Kohany O, Gentles AJ, Hankus L. & Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell MS, et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 2014;164:513–24. doi: 10.1104/pp.113.230144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–9. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204-212 (2014). [DOI] [PMC free article] [PubMed]
- 89.Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics btv351- (2015), 10.1093/bioinformatics/btv351 [DOI] [PubMed]
- 90.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sigrist CJA, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–7. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 94.Wu CH, et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–4. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lima, T. et al. HAMAP: A database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 37 (2009). [DOI] [PMC free article] [PubMed]
- 96.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Lima Morais DA, et al. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic Acids Res. 2011;39:D427–34. doi: 10.1093/nar/gkq1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bru C, et al. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–5. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–5. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lees J, et al. Gene3D: a domain-based resource for comparative genomics, functional annotation and protein network analysis. Nucleic Acids Res. 2012;40:D465–71. doi: 10.1093/nar/gkr1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haft DH, et al. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013;41:D387–95. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Araujo FA, Barh D, Silva A, Guimarães L, Ramos RTJ. GO FEAT: A rapid web-based functional annotation tool for genomic and transcriptomic data. Sci. Rep. 2018;8:1–4. doi: 10.1038/s41598-018-20211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mathelier A, Carbone A. MIReNA: finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics. 2010;26:2226–34. doi: 10.1093/bioinformatics/btq329. [DOI] [PubMed] [Google Scholar]
- 105.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dai X, Zhuang Z, Zhao P. X. psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tian T, et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li P, et al. RGAugury: A pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genomics. 2016;17:852. doi: 10.1186/s12864-016-3197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Minh, B. Q. et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol., 10.1093/molbev/msaa015 (2020). [DOI] [PMC free article] [PubMed]
- 111.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Madeira F, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.FigTree. Available at, http://tree.bio.ed.ac.uk/software/figtree/.
- 116.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. (2013).
- 117.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bradbury PJ, et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 119.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Portis E, Baudino M, Magurno F, Lanteri S. Genetic structure and preservation strategies of autochthonous vegetable crop landraces of north-western Italy. Ann. Appl. Biol. 2012;160:76–85. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.