Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has become a major global challenge. The virus infects host cells using its spike glycoprotein (S‐protein) and has significantly higher infectivity and mortality rates among the aged population. Here, based on bioinformatic analysis, I provide evidence that some members of the upper respiratory tract (URT) commensal bacteria express viral S‐protein ‐binding proteins. Based on this analysis and available data showing a decline in the population of these bacteria in the elderly, I propose that some URT commensal bacteria hamper SARS‐CoV‐2 infectivity and that a decline in the population of these bacteria contributes to the severity of infection. Further studies should provide a better understanding of the interaction of URT bacteria and SARS‐CoV‐2, which may lead to new therapeutic approaches.
Keywords: ACE2
Abbreviations
ACE2, angiotensin‐converting enzyme 2
ACE2‐PD, ACE2 protease domain
COVID‐19, coronavirus disease 2019
HA, haemagglutinin
LRT, lower respiratory tract
MERS‐CoV, Middle Eastern respiratory syndrome coronavirus
NA, neuraminidase
RBD, receptor‐binding domain
SARS‐CoV, Severe acute respiratory syndrome coronavirus
SARS‐CoV‐2, Severe acute respiratory syndrome coronavirus 2
SPI, signal peptidase I
S‐protein, spike glycoprotein
URT, upper respiratory tract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which causes coronavirus disease 2019 (COVID‐19), has led to many fatalities among humans and huge economical loss around the globe. Thus, there is an urgent need for developing therapeutics to prevent or treat infected individuals. The genome of the virus shows more than 80% identity with severe acute respiratory syndrome coronavirus (SARS‐CoV) and bat coronavirus [1, 2]. The high level of identity suggests that SARS‐CoV‐2 potentially uses a similar mechanism as that of SARS‐CoV to infect host cells. It has been demonstrated that the trimeric spike glycoprotein (S‐protein) of SARS‐CoV is primed by cellular proteases and is cleaved to S1 and S2 subunits [3, 4, 5]. S1 protein interacts with the host cell receptor angiotensin‐converting enzyme 2 (ACE2) [6, 7], and S2 mediates fusion of viral particles to cellular membranes [8, 9]. A similar mechanism of viral entry to host cells for SARS‐CoV‐2 has been proposed based on biochemical studies [10]. It has been shown that the SARS‐CoV‐2 S‐protein initially interacts with the serine protease TMPRSS2 [10]. This interaction was shown to be essential for viral entry into cells and infectivity, and an inhibitor of TMPRSS2 blocked viral entry [10]. Based on these findings, it is suggested that the SARS‐CoV‐2 uses host TMPRSS2 for S‐protein priming. This leads to the interaction of the S‐protein receptor‐binding domain (RBD) with ACE2 on the surface of host cells [10]. Structural studies have revealed the interaction of RBD of the S1 subunit of SARS‐CoV‐2 with ACE2 protease domain (ACE2‐PD) [11, 12]. These findings have provided the knowledge for designing therapeutic molecules to block the viral entry process and developing vaccines.
Since the emergence of SARS‐CoV about 19 years ago, in 2002, efforts to develop a vaccine or an effective therapeutic based on the structure of viral S‐protein and the interaction of viral proteins with different host cell proteins have not been successful. Therefore, there is a need for considering alternative therapeutic interventions. A process, the understanding of which can help in developing new medications, is the interaction of the virus with the upper respiratory tract (URT) commensal microbiota. Emerging evidence has confirmed the interaction of different viruses with commensal microbiota and how this interaction regulates infection and immune response [13, 14, 15, 16]. More recently, it has been reported that there is an association between URT microbiota and SARS‐CoV‐2 infection [17]. It was found that a high population of certain group of bacteria including members of proteobacteria is associated with less severe SARS‐CoV‐2 infection. Interestingly, the occurrence of this population of bacteria decreased linearly with age, suggesting a correlation of these bacteria with susceptibility to COVID‐19 [17].
Here, I briefly review available evidence in support of the interaction of URT microbiota with influenza virus. I used bioinformatics to test potential interaction of SARS‐CoV‐2 with URT commensal bacteria. I demonstrate that some bacteria, whose populations in the URT are reduced due to ageing, do produce membrane or secretory proteins with putative‐binding domains for the S‐protein of SARS‐CoV‐2. Thus, I hypothesize that the composition of the URT commensal microbiota is essential for reducing the severity of the disease caused by SARS‐CoV‐2. This proposal offers new preventive therapeutic approaches to avoid severe disease, which will significantly reduce the burden on global economy and health.
The interaction of bacteria and influenza virus
The primary source of the interaction of viral particles and bacteria is the viral particle surface proteins. For the purpose of this work, which is respiratory viruses, I focus on evidence in support of the interaction of influenza virus surface glycoproteins and bacteria. The viral particles consist of two major surface glycoproteins: haemagglutinin (HA) and neuraminidase (NA) [18, 19]. Available data suggest that the interactions of URT bacteria with HA or NA either promote viral/bacterial infection or suppress viral infection (Fig. 1) [13, 15].
Fig. 1.
Interaction of influenza virus with commensal bacteria. (A) Influenza virus NA cleaves sialic acid from mucins allowing some of the pathogenic bacteria trapped in the URT to be released to the lungs and cause secondary infection. (B) Influenza virus NA cleaves the sialic acid from cell surface exposing the receptors that can be identified by pathogenic bacteria. (C) The commensal bacteria in the URT release matrix proteins, which trap viral particles and abolish viral infectivity rate.
The NA protein is shown to have a role at different stages of viral life cycle [20], which can induce bacterial infections. Firstly, enzymatic activity of NA cleaves the host cell surface sialic acids helping receptor recognition by HA and viral entry [21, 22]. Secondly, it is suggested that the cleavage of sialic acid by enzymatic activity of NA allows the newly synthesized viral particles to be detached from cells and released [22]. Finally, the NA sialidase activity is shown to cleave the sialic acid from mucin, the glycoprotein constituent of the mucus. This activity of NA allows the virus to abolish the ability of URT to trap viral particles [23]. The cleavage of sialic acid from the surface of host cells and mucins can promote subsequent bacterial infection in at least two different ways (Fig. 1). Removal of sialic acid from mucins will scrap their ability to trap inhaled bacteria in the URT, which leads to the accumulation of bacteria in lung and secondary infections (Fig. 1A). Alternatively, cleavage of sialic acid from cell surface will expose cryptic receptors that can be identified by inhaled pathogenic bacteria [24] (Fig. 1B). On the other hand, it is shown that entry and fusion of virus to host cells require the activity of the host cell surface proteases to cleave HA to two subunits HA1 and HA2 [25]. This cleavage is essential for viral infectivity because it exposes the fusion peptide, which initiates viral fusion and entry to cells [25]. It is shown that protease of some of the nasal bacteria including those of Staphylococcus, a Gram‐positive genus of bacteria, can enhance infectivity by proteolytic activation of HA [26]. While most available data suggest that the interaction of viral and URT bacteria promotes infectivity of one and/or the other, there is evidence that the interactions of viral and URT bacteria can suppress influenza virus infectivity in different ways. Firstly, the Gram‐positive Enterococcus faecium bacterium is shown to directly trap viral particles preventing viral infection in vitro [27]. Secondly, it was found that the LPS derived from commensal microbiota binds to the viral particles and changes their morphology. Consequently, it can decrease stability of the virus and potentially its infectivity [28]. Finally, the commensal bacteria Staphylococcus epidermidis, a Gram‐positive bacterium, produces an extracellular matrix‐binding protein, which stably binds to viral particles and blocks viral infection [29] (Fig. 1C).
As discussed above, there is emerging evidence suggesting suppression of influenza virus infection by bacteria. Additionally, it is shown that a peptidoglycan produced by the Gram‐positive bacterium Bacillus subtilis, a member of phylum Firmicutes, can abrogate infectivity of SARS‐CoV and Middle Eastern respiratory syndrome coronavirus (MERS‐CoV) [30]. Thus, although the interaction of SARS‐CoV‐2 and URT commensal microbiota should yet be investigated, it is reasonable to assume such an interaction plays a role in pathogenesis of the virus.
The composition of URT commensal microbiota changes due to ageing
To investigate whether available data support a role of URT microbiota in the SARS‐CoV‐2 infectivity, I evaluated available literature to test whether there is a correlation between the composition of URT bacteria and the higher infectivity and mortality rates in elderly. Different phyla of bacteria have been identified in URT [31, 32, 33, 34, 35, 36]. In general, it is shown that members of the bacterial phylum Firmicutes, most of which are Gram‐positive like Staphylococcus aureus and S. epidermidis, are predominant [33, 36]. Other prevalent bacterial phyla are Actinobacteria and Proteobacteria, which are Gram‐positive and Gram‐negative, respectively [33, 36]. Based on available data, it appears that the population of some bacterial phyla decreases due to ageing, while those of some other phyla increases [36]. Specifically, it appears that the overall population of Proteobacteria in the URT decreases upon ageing [36]. A decline in population of Proteobacteria was also reported due to smoking [34]. On the other hand, it is observed that the severity of SARS‐CoV‐2 infection and its mortality rate are higher in aged population [37] and may increase due to smoking [38]. It is shown that SARS‐CoV‐19 can actively replicate in the URT tissue [39]. Therefore, it is possible that a decrease in the population of members of Proteobacteria increases the susceptibility of URT or lower respiratory tract (LRT) to viral infection and replication leading to severe disease.
Proteobacteria secrete homologues of TMPRSS2
Next, I investigated if members of Proteobacteria express proteins with the potential ability to interact with SARS‐CoV‐2 S‐protein. I first tested if these bacteria express a homologue of human TMPRSS2, whose peptidase activity is essential for viral entry. I used blast analysis and identified peptidase S1‐domain‐containing proteins and serine peptidases in members of Proteobacteria. Multiple sequence analysis (Fig. S1) revealed that the proposed catalytic triad of TMPRSS2 [40], which consists of His296, Asp345, and Ser441, and the amino acid residues around this catalytic triad are highly conserved among all proteins (Fig. 2A). Using SignalP5.0 server [41], I found (with more than 70% probability) that the putative bacterial peptidases contain a secretory signal peptide [Sec/signal peptidase I (SPI)] at the N‐terminus. The SPI is responsible for the release of signal peptide, which is essential for transport of protein to the outer membrane and its secretion in Gram‐negative bacteria [42]. To test whether the highly conserved His, Asp, and Ser in bacterial peptidases form a catalytic triad, I used Phyre2 server [43] and predicted (> 90% accuracy for 90% of the amino acid residues) the structure of a peptidase S1‐domain‐containing protein from Gammaprotiobacteria bacterium (Gene ID: A3E01_06460) (Fig. 2B). The results show that the highly conserved His, Asp, and Ser form a catalytic triad.
Fig. 2.
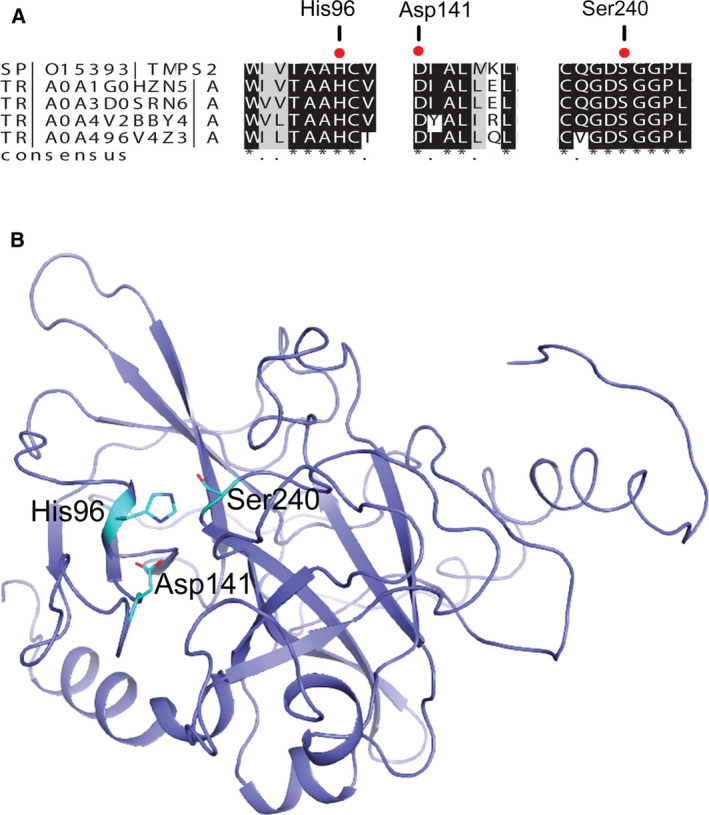
Members of Proteobacteria express homologues of TMPRSS2. (A) Multiple sequence alignment shows that the catalytic triad of human TMPRSS2, which consists of His, Asp, and Ser, is highly conserved. The amino acid numbering is for peptidase S1‐domain‐containing protein from Gammaproteobacteria bacterium (ID: A0A1G0HZN5). From top to bottom: human TMPRSS2; peptidase S1‐domain‐containing protein from G. bacterium; peptidase S1‐domain‐containing protein from G. bacterium; serine protease from Proteobacteria bacterium; and peptidase S1‐domain‐containing protein from G. bacterium. (B) The predicted structure of a peptidase S1‐domain‐containing protein from G. bacterium (ID: A0A1G0HZN5) confirms that the highly conserved His96, Asp141 and Ser240 form the catalytic site.
Proteobacteria secret homologues of the ACE2‐PD
Next, I used amino acid blast analysis to identify bacterial homologues of human ACE2. The results revealed the presence of peptidyl peptidases in members of Proteobacteria phylum with high similarity to the ACE2 peptidase domain (ACE2‐PD). Multiple sequence alignment suggests that the bacterial peptidyl peptidases have 30–50% identity at the amino acid level with human ACE2‐PD (Fig. S2). I used the SignalP5.0 server [41] and tested if bacterial ACE2‐PD‐like peptidyl peptidase has a secretory signal peptide. The results revealed (with more than 99% probability) the presence of the secretory lipoprotein signal peptide [secretory (Sec)/signal peptidase II (SPII)] in the N‐terminus. This signal peptide plays a role in secretion of lipoproteins in Gram‐negative bacterium [42, 44]. Subsequently, I predicted the structure of a peptidyl peptidase from Proteobacteria bacterium (Gene ID: DIU56_02030). The structure of protein (97% of the amino acid residues, except the first 27 amino acid residues at the N‐terminus) was predicted with more than 90% accuracy using Phyre 2 server [43] (Fig. 3A). The predicted amino acid residues that may interact with SARS‐CoV‐2 S‐protein RBD are shown (Fig. 3A). I aligned the predicted structure of bacterial peptidyl peptidase with that of human ACE2‐PD (Fig. 3B), as determined previously using electron microscopy [11]. The results revealed potential interaction between bacterial ACE2‐PD‐like peptidyl peptidase and the RBD of S‐protein. Although these bioinformatics data are limited and further experimental data are required, the results suggest that secretory peptidases homologue of TMPRSS2 and ACE2‐PD are produced by Proteobacteria and may interact with the RBD of the SARS‐CoV‐2 S‐protein. This interaction can potentially block viral infectivity like the effect observed by soluble form of human ACE2 and a wide range of orthologues of ACE2 from different species. It is shown that a soluble form of human ACE2 [45] and 17 orthologues of ACE2 from different species [46] can inhibit SARS‐CoV‐2 infection.
Fig. 3.
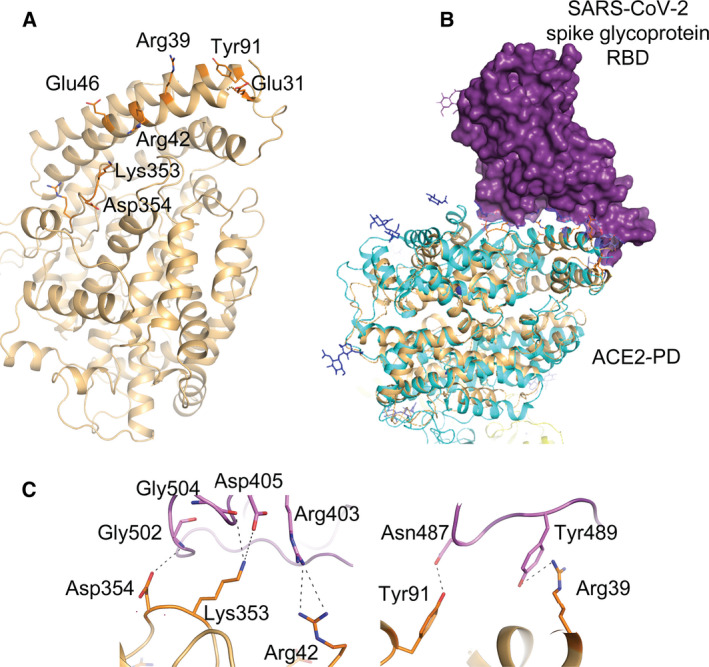
Members of proteobacteria express a peptidyl peptidase with similarity to the PD of human ACE2. (A) The predicted structure of peptidyl peptidase from Proteobacteria bacterium. The structure was predicted using Phyre2 server. The amino acid residues of the putative‐binding site of the S‐protein are numbered. (B) Alignment of the predicted structure of the bacterial ACE2‐PD‐like peptidyl peptidase (ochre) with the 3D structure of ACE2‐PD (cyan) in complex with the RBD of the SARS‐CoV‐2 S‐protein (purple). (C) Possible interactions between amino acid residues of bacterial ACE2‐PD‐like peptidyl peptidase (orange) and those of RBD of the SARS‐CoV‐2 (Pink).
Proteobacteria express membrane or secretory glycan‐binding proteins
Another possible interaction can occur between secreted lectins from bacteria and glycans on the surface of the viral S‐protein. To test this possibility, I first investigated if there is a C‐type‐like lectin present in any member of the URT commensal bacteria. I tested the presence of this specific lectin because it is known that in humans, it can recognize different glycans and activates the immune response [47]. Additionally, it has been shown that in response to MERS‐CoV infection, the C‐type receptor pathway is activated, suggesting interaction of this lectin with the virus S‐protein [48]. Therefore, using the blast analysis of the amino acid sequence, I investigated the presence of C‐type‐like lectins and found that different members of Proteobacteria and Gammaproteobacteria express putative C‐type‐like lectins (Table 1). Additionally, bioinformatics analysis led to the identification of putative lectin leg‐B domain‐containing and ricin B‐type lectin domain‐containing proteins. All these proteins are either localized in the membrane or have a secretory signal peptide, with more than 98% probability as predicted using SignalP 5.0 server [41].
Table 1.
Bioinformatics analyses suggest the presence of glycoprotein‐binding proteins in members of the URT commensal bacteria. The proteins were identified using blast analysis, and the presence of secretory signal peptide was determined, with more than 98% probability, using SignalP‐5.0 server [41].
| Lectin domain | Bacteria | Gene ID | Localization |
|---|---|---|---|
| C‐type | Gammaproteobacteria bacterium | CMQ88_02965 | Transmembrane |
| C‐type | Gammaproteobacteria bacterium | A9Q81_23845 | Signal peptide (Sec/SPI) |
| C‐type | Proteobacteria bacterium | CMK59_07605 | Lipoprotein signal peptide (Sec/SPII) |
| C‐type | Rhodobacteraceae bacterium | CML44_00865 | Signal peptide (Sec/SPI) |
| Ricin B‐type | Gammaproteobacteria bacterium | DEQ32_10900 | Signal peptide (Sec/SPI) |
| Ricin B‐type | Alphaproteobacteria bacterium | DEP10_04400 | Signal peptide (Sec/SPI) |
| LegB | Gammaproteobacteria bacterium | CBC55_05095 | Transmembrane |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Discussion and Hypothesis
Systematic analysis of published data between December 2019 and March 2020 suggests that although children are as susceptible as the elderly in getting infected with SARS‐CoV‐2, they show milder symptoms and less severe disease [49, 50, 51]. Additionally, PCR analysis of samples obtained from patients in different age groups suggests very similar viral loads among infected individuals. However, the number of patients increases significantly with increasing age [52], which is consistent with the general observation of a less severe disease in children. It has been reported that the expression of host cell entry proteins like ACE2 and TMPRSS2 does not increase with age and cannot explain a more severe disease in the elderly [53]. Different risk factors have been proposed to contribute to the increase in severity of the disease due to ageing [53, 54, 55]. Some of these risk factors are higher expression of genes associated with cell adhesion and oxytocin signalling, lower expression of genes involved in mitochondrial translation and mitosis, decrease in total alveolar macrophages, immunosenescence and metabolic diseases. These studies highlight the importance of understanding different mechanisms by which children are protected against severe disease.
A very recent study of healthy and infected individuals in the Netherlands suggests that the composition of URT microbiota is associated with SARS‐CoV‐2 infection and that this association is age‐dependent [17]. Based on these data and the reported data with other respiratory viruses, it is evident that the interaction of commensal bacteria with viral particles or host cells plays an important role in the pathogenesis of the virus [13, 14, 15, 16]. Bacterial components may interact with virus particles and/or the virus‐infected eukaryotic cells displaying viral proteins on their surface. While in many cases it is reported that such interactions can promote bacterial infection, there is evidence demonstrating that the commensal bacteria can hamper viral infectivity. Therefore, I investigated the possibility that a decline in the population of some URT commensal bacteria due to ageing increases the SARS‐CoV‐2 infectivity and mortality rates among the aged population. Firstly, my bioinformatic analysis to identify bacterial proteins with potential ability to interact with SARS‐CoV‐2 S‐protein revealed two possible groups of proteins from members of Proteobacteria: (a) secretory peptidases with similarity to human TMPRSS2 and the ACE2‐PD and (b) transmembrane or secretory lectins. Secondly, available data regarding the change in population of different URT commensal bacteria suggest that generally the population of Proteobacteria decreases due to ageing. Finally, it is known that the SARS‐CoV‐2 infectivity and mortality levels increase in elderly. Based on these data together, I put forward the following hypothesis: a decline in the population of certain URT commensal bacteria, such as members of Proteobacteria, abolishes the ability of the upper respiratory system to trap viral particles (Fig. 4). Consequently, the rate of infectivity and replication of the virus increase, which is a positive contributing factor to the severity of the COVID‐19 disease. It should be noted that other alternative mechanisms are possible. For example, it is conceivable that some metabolites or peptides released by URT bacteria bind to the viral S‐protein and block its interaction with host cell receptors. Alternatively, the interaction of viral particles with a cell‐wall protein of a bacteria leads to translocation and accumulation of the bacteria in the LRT and formation of secondary bacterial infection.
Fig. 4.
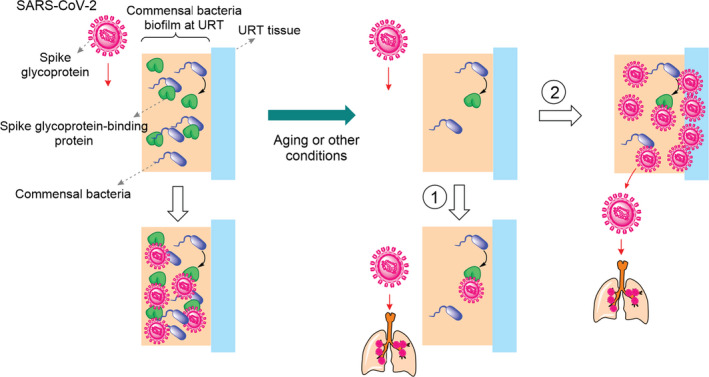
URT commensal bacteria can hamper SARS‐CoV‐2 replication and infection. In healthy individuals, who show no symptom of COVID‐19 disease, the S‐protein‐binding proteins expressed by URT commensal bacteria trap viral particles. Consequently, the viral replication rate abolishes. Due to ageing or other conditions, the population of the URT commensal bacteria, which produce the S‐protein ‐binding proteins, reduces. As a result, either viral particles easily reach LRT causing severe disease (path 1) or viral particles infect the URT tissue (path 2), which increases replication rate and viral load. The resulting viral particles reach the LRT and cause severe disease.
Future studies will provide a better understanding of the interaction between the URT commensal bacteria and SARS‐CoV‐2. These findings may lead to the discovery of new antiviral molecules or therapeutic approaches based on manipulating the URT bacteria, as it is suggested in the case of other diseases [56, 57, 58]. I speculate that it might be possible to manipulate the population of URT commensal microbiota or to identify bacterial S‐protein ‐binding proteins or metabolites to trap viral particles at the URT, block viral entry and reduce viral replication.
Supporting information
Fig. S1. Members of Proteobacteria express homologues of human TMPRSS2.
Fig. S2. Members of Proteobacteria express homologues of ACE2‐PD.
Acknowledgement
I am grateful to Professor Fraser Armstrong (University of Oxford) and Professor Wilfred Hagen (TU Delft) for generous support of my scientific endeavour. I thank the European Molecular Biology Organization (EMBO) for a long‐term fellowship (ALTF 157‐2015) and COST (European Cooperation in Science and Technology) Action CA15133 (ECOST‐STSM‐Request‐CA15133‐44200) for supporting my career.
Edited by Urs Greber
References
- 1. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B and Sette A (2020) A Sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS‐CoV‐2. Cell Host Microbe 27, 671–680.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pohlmann S, Heurich A, Gierer S, Zmora P and Simmons G (2013) Proteolytic activation of the SARS‐coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res 100, 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Tsegaye TS, He Y, Gnirss K, Niemeyer D et al (2011) Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol 85, 4122–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M and Taguchi F (2010) Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84, 12658–12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC et al (2003) Antio‐tensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li F, Li W, Farzan M and Harrison SC (2005) Structure of SARS coronavirus spike receptor‐binding domain complexed with receptor. Science 309, 1864–1868. [DOI] [PubMed] [Google Scholar]
- 8. Petit CM, Melancon JM, Chouljenko VN, Colgrove R, Farzan M, Knipe DM and Kousoulas KG (2005) Genetic analysis of the SARS‐coronavirus spike glycoprotein functional domains involved in cell‐surface expression and cell‐to‐cell fusion. Virology 341, 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Supekar VM, Bruckmann C, Ingallinella P, Bianchi E, Pessi A and Carfi A (2004) Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc Natl Acad Sci USA 28, 17958–17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoffman M, Kleine‐Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A et al (2020) SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan R, Zhang Y, Li Y, Xia L, Guo Y and Zhou Q (2020) Structural basis for the recognition of SARS‐CoV‐2 by full‐length human ACE2. Science 367, 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L et al (2020) Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 13. McCullers JA (2014) The co‐pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 12, 252–262. [DOI] [PubMed] [Google Scholar]
- 14. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS and Iwasaki A (2011) Microbiota regulates immune defence against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA 108, 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li N, Ma W‐T, Pang M, Fan Q‐L and Hua J‐L (2019) The commensal microbiota and viral infection: a comprehensive review. Front Immunol 10, 1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neu U and Mainou BA (2020) Virus interactions with bacteria: partners in the infectious dance. PloS Pathog 16, e1008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Budding AE, Sieswerda E, Wintermans BB and Bos MP (2020) An age dependent pharyngeal microbiota signature associated with SARS‐CoV‐2 infection. Preprint with the Lancet. 10.2139/ssrn.3582780. [DOI] [Google Scholar]
- 18. Das K, Aramini JM, Ma L‐C, Krug RM and Arnold E (2010) Structures of influenza A proteins and insights into antiviral drug targets. Nat Struct Mol Biol 17, 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gamblin SJ and Skehel JJ (2010) Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem 285, 28403–28409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dou D, Revol R, Ostbye H, Wang H and Daniels R (2018) Influenza A virus cell entry, replication, virion assembly, and movement. Front Immunol 9, 1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagner R, Wolf T, Herwig A, Pleschka S and Klenk H‐D(2000) Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza growth: a study by reverse genetics. J Virol 74, 6316–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wen F and Wan X‐F (2019) Influenza neuraminidase: underrated role in receptor binding. Trends Microbiol 27, 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cohen M, Zhang X‐Q, Senaati HP, Chen H‐W, Varki NM, Schooley RT and Gagneux P (2013) Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 10, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peltola VT, Murti KG and McCullers JA (2005) Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J Infect Dis 192, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bertram S, Glowacka I, Steffen I, Kuhl A and Pohlmann S (2010) Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol 20, 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scheilblauer H, Reinacher M, Tashiro M and Rott R (1992) Interactions between bacteria and influenza A virus in the development of influenza pneumonia. J Infect Dis 166, 783–791. [DOI] [PubMed] [Google Scholar]
- 27. Wang Z, Chai W, Burwinkel M, Twardziok S, Wrede P, Palissa C, Esch B and Schmidt MFG (2013) Inhibitory influence of Enterococcus faecium on the propagation of swine influenza A virus in vitro . PLoS One 8, e53043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bandoro C and Runstadler JA (2017) Bacterial lipopolysaccharide destabilizes influenza viruses. mSphere 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen H‐W, Liu P‐F, Liu Y‐T, Kuo S, Zhang X‐Q, Schooley RT, Rohde H, Gallo RL and Huang C‐M (2016) Nasal commensal Staphylococcus epidermidis counteracts influenza virus. Sci Rep 2, 27870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson BA, Hage A, Kalveram B, Mears M, Plante JA, Rodriguez SE, Ding Z, Luo X, Bente D, Bradrick SS et al (2019) Peptidoglycan‐associated cyclic lipopeptide disrupts viral infectivity. J Virol 93, e01282–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahmed N, Mahmoud NF, Solyman S and Hanora A (2019) Human nasal microbiome as characterized by metagenomics differs markedly between rural and industrial communities in Egypt. Omi A J Integr Biol 23, 573–582. [DOI] [PubMed] [Google Scholar]
- 32. Bassis CM, Tang AL, Young VB and Pynnonen MA (2014) The nasal cavity microbiota of healthy adults. Microbiome 2, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN and Pace NR (2010) The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 5, e10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kumpitsch C, Koskinen K, Schopf V and Moissl‐Eichinger C (2019) The microbiome of the upper respiratory tract in health and disease. BMC Biol 17, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bomar L, Brugger SD and Lemon KP (2018) Bacterial microbiota of the nasal passages across the span of human life. Curr Opin Microbiol 41, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schenck LP, Surette MG and Bowdish DM (2016) Composition and immunological significance of the upper respiratory tract microbiota. FEBS Lett 590, 3705–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dudley JP and Lee NT (2020) Disparities in age‐specific morbidity and mortality from SARS‐CoV‐2 in China and the Republic of Korea. Clin Infect Dis ciaa354 10.1093/cid/ciaa354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vardavas CI and Nikitara K (2020) COVID‐19 and smoking: a systematic review of the evidence. Tob Induc Dis 18, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Wollmar P, Rothe C et al (2020) Virological assessment of hospitalized patients with COVID‐2019. Nature 581, 465–469. [DOI] [PubMed] [Google Scholar]
- 40. Meng T, Cao H, Zhang H, Kang Z, Xu D, Gong H, Wang J, Li Z, Cui X, Xu H et al (2020) The insert sequence in SARS‐CoV‐2 enhances spike protein cleavage by TMPRSS. bioRxiv. 10.1101/2020.02.08.926006. “[PREPRINT]” [DOI] [Google Scholar]
- 41. Nielsen H, Tsirigos KD, Brunak S and von Heijne G (2019) A brief history of protein sorting prediction. Protein J 38, 200–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dalbey RE, Wang P and van Dijl JM (2012) Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol Mol Biol Rev 76, 311–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kelley LA, Mezulis S, Yates CM, Wass MN and Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pugsley AP (1993) The complete general secretory pathway in gram‐negative bacteria. Microbiol Rev 57, 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado del Pozo C, Prosper F et al (2020) Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell 181, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y, Wang H, Tang X, Ma D, Du C, Wang Y, Pan H, Zou Q, Zheng J, Xu L et al (2020) Potential host range of multiple SARS‐like coronaviruses and an improved ACE2‐Fc variant that is potent against both SARS‐CoV‐2 and SARS‐CoV‐1. bioRxiv. 10.1101/2020.04.10.032342. “[PREPRINT]” [DOI] [Google Scholar]
- 47. Geijtenbeek TBH and Gringhuis SI (2009) Signalling through C‐type lectin receptors: shaping immune responses. Nat Rev Immunol 9, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao X, Chu H, Wong BH‐Y, Chiu MC, Wang D, Li C, Liu X, Yang D, Poon VK‐M, Cai J et al (2019) Activation of C‐type lectin receptor and (RIG)‐I‐like receptors contributes to proinflammatory response in middle east respiratory syndrome coronavirus‐infected macrophages. J Infect Dis 221, 647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mehta N, Mytton O, Mullins E, Fowler T, Falconer C, Murphy O, Langenberg C, Jayatunga W, Eddy D and Nguyen‐Van‐Tam JS (2020) ARS‐CoV‐2 (COVID‐ 19): what do we know about children? A systematic review. Preprint with the Lancet. 10.2139/ssrn.3558015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee PI, Hu YL, Chen PY, Huang YC and Hsueh PR (2020) Are children less susceptible to COVID‐19? J Microbiol Immunol Infect. 53, 371–372. 10.1016/j.jmii.2020.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liguoro I, Pilotto C, Bonanni M, Ferrari ME, Pusiol A, Nocerino A, Vidal E and Cogo P (2020) SARS‐CoV‐2 infection in children and newborns: a systematic review. Res Sq. 10.1007/s00431-020-03684-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jones TC, Muhlemann B, Veith T, Zuchowski M, Hofmann J, Stein A, Edelmann A,Corman VM and Drosten C. An analysis of SARS‐CoV‐2 viral load by patient age. https://www.drperlmutter.com/wp‐content/uploads/2020/05/analysis‐of‐SARS‐CoV‐2‐viral‐load‐by‐patient‐age.pdf
- 53. Chow RD and Chen S (2020) The aging transcriptome and cellular landscape of the human lung in relation to SARS‐CoV‐2. Biorxiv. 10.1101/2020.04.07.030684. “[PREPRINT]” [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X et al (2020) Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet 395, 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stilhano RS, Costa AJ, Nishino MS, Shams S, Bartolomeo CS, Breithaupt‐Faloppa AC, Silva EA, Ramirez AL, Prado CM and Ureshino RP (2020) SARS‐CoV‐2 and the possible connection to ERs, ACE2 and RAGE: focus on susceptibility factors. Preprints. 10.20944/preprints202005.0178.v1. [DOI] [PMC free article] [PubMed]
- 56. Scott KP, Jean‐Michel A, Midtvedt T and van Hemert S (2015) Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health Dis 26, 25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jin D, Zhang H and Sun J (2014) Manipulation of microbiome, a promising therapy for inflammatory bowel diseases. J Clin Cell Immunol 5, 234. [Google Scholar]
- 58. Song W, Anselmo AC and Huang L (2019) Nanotechnology intervention of the microbiome for cancer therapy. Nat Nanotechnol 14, 1093–1103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Members of Proteobacteria express homologues of human TMPRSS2.
Fig. S2. Members of Proteobacteria express homologues of ACE2‐PD.


