Abstract
There is a rapidly expanding literature on the in vitro antiviral activity of drugs that may be repurposed for therapy or chemoprophylaxis against severe acute respiratory syndrome‐coronavirus 2 (SARS‐CoV‐2). However, this has not been accompanied by a comprehensive evaluation of the target plasma and lung concentrations of these drugs following approved dosing in humans. Accordingly, concentration 90% (EC90) values recalculated from in vitro anti‐SARS‐CoV‐2 activity data was expressed as a ratio to the achievable maximum plasma concentration (Cmax) at an approved dose in humans (Cmax/EC90 ratio). Only 14 of the 56 analyzed drugs achieved a Cmax/EC90 ratio above 1. A more in‐depth assessment demonstrated that only nitazoxanide, nelfinavir, tipranavir (ritonavir‐boosted), and sulfadoxine achieved plasma concentrations above their reported anti‐SARS‐CoV‐2 activity across their entire approved dosing interval. An unbound lung to plasma tissue partition coefficient (K p U lung) was also simulated to derive a lung Cmax/half‐maximal effective concentration (EC50) as a better indicator of potential human efficacy. Hydroxychloroquine, chloroquine, mefloquine, atazanavir (ritonavir‐boosted), tipranavir (ritonavir‐boosted), ivermectin, azithromycin, and lopinavir (ritonavir‐boosted) were all predicted to achieve lung concentrations over 10‐fold higher than their reported EC50. Nitazoxanide and sulfadoxine also exceeded their reported EC50 by 7.8‐fold and 1.5‐fold in lung, respectively. This analysis may be used to select potential candidates for further clinical testing, while deprioritizing compounds unlikely to attain target concentrations for antiviral activity. Future studies should focus on EC90 values and discuss findings in the context of achievable exposures in humans, especially within target compartments, such as the lungs, in order to maximize the potential for success of proposed human clinical trials.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Coronavirus disease 2019 (COVID‐19) is an acute infectious respiratory disease caused by infection with the coronavirus subtype severe acute respiratory syndrome‐coronavirus 2 (SARS‐CoV‐2), first detected in Wuhan, China, in December 2019. There are currently no available treatments or chemopreventative options, but several are being explored preclinically and clinically. Most publications reporting in vitro activity have focused on 50% maximum effective concentrations and not considered the achievable concentrations in plasma or relevant compartments for COVID‐19, which may be an insufficiently robust indicator of antiviral activity because of marked differences in the slope of the concentration‐response curve between drugs.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ This paper describes a comprehensive analysis of literature reported anti‐SARS‐CoV‐2 activity for approved medicines in the context of their known pharmacokinetic exposure. A combination of physiochemical and pharmacological parameters was used to predict the accumulation of these drugs within lung tissues using a widely accepted modeling approach. Plasma and lung pharmacokinetic parameters were then used to rank the reported molecules according to whether they would provide therapeutic or chemopreventative exposures with the plasma or lung tissue.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ Of the identified molecules with reported anti‐SARS‐CoV‐2 activity, the overwhelming majority are not expected to reach active concentrations within the key target compartments. However, a number of candidates were identified that are expected to exceed the concentrations necessary to provide viral suppression at doses approved for use in humans.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ This paper identifies key drug repurposing opportunities and dramatically highlights the importance of considering pharmacokinetic exposure when interpreting the emerging candidacy of drugs for COVID‐19 treatment and prevention.
Coronavirus disease 2019 (COVID‐19) is a respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection. Fever, a persistent cough, and respiratory symptoms are common, with some patients reporting vomiting, nausea, abdominal pains, and diarrhea. 1 To date, no specific treatment is available, and this has resulted in significant morbidity and mortality globally. According to the International Clinical Trials Registry Platform search portal, 927 clinical trials for COVID‐19 have been registered. 2 This rapidly expanding pandemic warrants the urgent development of strategies, particularly to protect people at high risk of infection. Repurposing currently available drugs that have been utilized clinically with a known safety profile is the quickest way to address this serious unmet clinical need. Antiviral drugs are urgently required for treatment of patients with mild/moderate disease to prevent the worsening of symptoms and reduce the burden upon healthcare systems. However, a different approach is likely to be needed for patients that are already in a critical state, due to the immune dysregulation, which is so apparent in severe cases. 3
Previous investigations have shown that the entry by SARS‐CoV‐2 occurs via the angiotensin converting enzyme 2 (ACE2) receptor. 4 A study on normal lung tissue showed that 83% of ACE2‐expressing cells were alveolar epithelial type II cells, 5 highlighting the lungs as the primary target organ that facilitate viral invasion and replication. Furthermore, the ACE2 receptor is also highly expressed in gastrointestinal epithelial cells, with SARS‐CoV‐2 RNA observed to be present in stool specimens of patients during infection. 1 , 6 A recent retrospective analysis of 85 patients with laboratory‐confirmed COVID‐19 also indicated that SARS‐CoV‐2 infects human kidney tubules and induces acute tubular damage in some patients. 7 Furthermore, 2–11% of patients with COVID‐19 exhibit liver comorbidities. 8 Of note is an observation of SARS and Middle East respiratory syndrome having a tropism to the gastrointestinal tract 9 and causing liver impairment in addition to respiratory disease. The genomic similarity between SARS‐CoV‐2 and SARS‐CoV (79.6% sequence identity) would imply that the current virus would act in a similar manner and be present within the body systemically. 10 , 11 , 12 Therefore, treatment options that provide therapeutic concentrations of drug(s) within the systemic circulation and other affected organs are likely to be required.
In the absence of a vaccine, antiviral drugs could also be deployed as chemoprophylaxis to protect against infection and would present an essential tool for protecting healthcare staff and other key workers, as well as household contacts of those already infected. For chemoprevention, drugs will need to penetrate into the multiple sites where SARS‐CoV‐2 infection occurs, and do so in sufficient concentrations to inhibit viral replication. 13 This may include the mucous membranes present in the nasal cavity and throat, the ocular surface, tears, and the upper respiratory tract/lungs. 14 , 15 However, therapeutic concentrations may not be needed in the systemic circulation for chemoprophylaxis, but this is yet to be determined. Although difficult and scarcely studied, work in animals has shown that the size of the inoculum of other respiratory viruses, such as influenza, is associated with the severity of the resultant disease. 16 , 17 Reports with SARS‐CoV‐2 indicate that higher viral loads are indicative of poorer prognosis and correlate with the severity of symptoms, with viral load in severe cases reported to be 60 times higher than that of mild cases. 18 , 19 In light of this, even if a chemoprophylactic drug reduced inoculum size without completely blocking transmission, major benefits for morbidity and mortality may still be achievable.
Many ongoing global research efforts are focused on screening the activity of existing compounds in vitro in order to identify candidates to repurpose for SARS‐CoV‐2. However, current data have not yet been systematically analyzed in the context of the plasma and target site exposures that are achievable after administration of the approved doses to humans. The purpose of this work was to evaluate the existing in vitro anti‐SARS‐CoV‐2 data to determine and prioritize drugs capable of reaching antiviral concentrations within the blood plasma. Accepted physiologically‐based pharmacokinetic equations were also used to predict the expected concentration in the lungs, 20 , 21 , 22 in order to assess the potential of these drugs for therapy in this key disease site and the potential for chemoprevention.
METHODS
Candidate analysis
To identify compounds and their relevant potency and pharmacokinetic data, we performed a literature search on PubMed, Google Scholar, BioRxiv, MedRxiv, and ChemRxiv. The following search terms were used for in vitro activity data—(COVID‐19 OR SARS‐CoV‐2) AND (half‐maximal effective concentration (EC50) OR half‐maximal inhibitory concentration OR antiviral). For pharmacokinetic data (Cmax OR pharmacokinetics) was used along with the drug name for drugs with reported anti‐SARS‐CoV‐2 activity (up to April 13, 2020). Further clinical pharmacokinetic data were obtained from the US Food and Drug Administration (FDA), the European Medicines Agency (EMA), and through publications available online. Inhaled medications were excluded from all analyses because the purpose was to assess systemically administered medicines.
Lung accumulation prediction
An indication of the degree to which candidate drugs are expected to accumulate in the lungs (a presumed site of primary efficacy and for prevention of SARS‐CoV‐2 infection) was provided by calculation of unbound lung to plasma tissue partition coefficient (K p U lung) according to the methodology of Rodgers and Rowland. 20 , 21 , 22 Equations therein were implemented in the R programming environment (version 3.6.3) and are replicated in the Supplementary Methods . Briefly, the physicochemical properties of the drug (pKa, log P, and classification as acid/base/neutral) and in vitro drug binding information (fraction unbound in plasma and blood to plasma ratio), in combination with tissue‐specific data (lipid content, volumes of intra/extracellular water, etc.) were used to predict tissue K p U values. Measured log P and pKa values were used where available but substituted with calculated values where necessary and all parameter values used for the calculations for each drug, and their references/sources, are provided in Table S1 . K p U lung values were converted to K p_lung by multiplying by the fraction unbound in plasma to allow estimation of lung exposure from in vivo measurements of plasma Cmax concentration. A similar analysis was conducted to assess the tissue distribution into other tissues. In the absence of observed tissue distribution data, the Rodgers and Rowland method is an accepted means to provide initial estimates of tissue partitioning for physiologically‐based pharmacokinetic modeling. However, there are known limits on accuracy with predicted K p U by the Rodgers and Rowland method generally reported to be within twofold to threefold of observed tissue K p U values. 20 , 21 , 22 This was confirmed for a limited number of drugs within the current dataset for which measure K p values for lungs were available from animal studies in the literature (see data analysis below).
Data analysis and interpretation
Because in the majority of papers only an EC50 value was available, concentration‐response data were digitized using the Web Plot Digitizer software. Graphs were then replotted in SigmaPlot version 14.0 (Systat Software) and curves were fitted to confirm EC50 values and determine effective concentration 90% (EC90) values. A Cmax/EC50 and Cmax/EC90 ratio was then calculated for each drug for which previous evidence of clinical use in humans and availability of human pharmacokinetic data were available. Lung and other tissue K p U values were used in combination with reported Cmax values to derive an estimate of lung exposure at Cmax for each drug. For a subset of molecules, the absence of available physicochemical or plasma protein binding parameters prohibited derivation of a K p U estimate. For the remaining drugs, a lung (or other tissue) Cmax/EC50 and lung Cmax/EC90 were calculated. Published plasma concentration‐time data for the most promising candidates were then digitized (where available) and replotted to visually represent human pharmacokinetics relative to the calculated EC50 and EC90 data. Equivalence between values for the predicted lung K p and those observed in vivo was undertaken for drugs with available animal lung and plasma concentration data. For this analysis, animal lung concentration data were available for anidulafungin (rat), bazedoxifene (rat), chloroquine (3 albino rat studies), favipiravir (monkey), hydroxychloroquine (2 albino rat studies), nitazoxanide (mouse), tamoxifen (rat), cyclosporine (rat), ritonavir (rat), azithromycin (mouse), dolutegravir (mouse), gilteritinib (albino rat), and lopinavir (rat). 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 Agreement between the predicted and measured K p was assessed by simple linear regression and by constructing Bland–Altman plots, the limits of agreement (mean ± 2 SD) were included in these plots as previously described. 33
RESULTS
Identified papers and methods
We identified 14 key studies that detailed the antiviral activity of 71 compounds. 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 The majority of the in vitro SARS‐CoV‐2 infection experiments were performed in Vero E6 cells (ATCC 1586) maintained in either Dulbecco’s Modified Eagle’s Medium or Minimum Essential Medium. Other studies utilized Vero‐hSLAM cells, Vero E6 cells expressing TMPRSS2, and the CACO‐2 cell line to cultivate the virus. The following SARS‐CoV‐2 strains were used across studies; WA‐1 strain—BEI #NR‐52281; Brazil/RJ‐314/2020; C‐Tan‐nCoV Wuhan strain 01; Wuhan/WIV04/2019; USA‐WA1/2020; nCoV‐2019BetaCoV/Wuhan/WIV04/2019; BetaCoV/Hong Kong/VM20001061/2020; Australia/VIC01/2020; βCoV/KOR/KCDC03/2020, and BavPat1/2020. Cells across all studies were infected with the virus with a multiplicity of infection of 0.002, 0.01, 0.0125, 0.02, 0.05, and 0.1. Drugs were added at concentrations varying between 0.01 and 500 μM. A summary of the differences in methodologies between studies reporting SARS‐CoV‐2 antiviral activity is presented in Table S2 . A ranking of included drugs based just on their EC50 and recalculated EC90 is presented in Figure S1 .
Identification of candidates achieving plasma concentrations expected to exert antiviral activity (Cmax/EC50 ratio)
Seventeen molecules had a reported Cmax value greater than at least one of the reported EC50 values against SARS‐CoV‐2 and these were nelfinavir, chloroquine, remdesivir, lopinavir (ritonavir boosted), eltrombopag, hydroxychloroquine, atazanavir (ritonavir boosted), indomethacin, favipiravir, sulfadoxine, niclosamide, mefloquine, tipranavir (ritonavir boosted), ritonavir, merimepodib, anidulafungin, and nitazoxanide. However, it should be noted that for amodiaquine, atazanavir, chloroquine, hydroxychloroquine, lopinavir, mefloquine, nelfinavir, remdesivir, and toremifene, more than one EC50 value had been reported across the available literature and these were not always in agreement (Figure 1a ). Moreover, this variability in reported EC50 values sometimes resulted in Cmax/EC50 ratios giving a different estimation of the likely value of the molecule. Meaning that for the same drug, the Cmax/EC50 ratio could be above or below 1 (Figure 1b ). For amodiaquine and toremifene, all reported EC50 values were below their reported Cmax and only for nelfinavir was the reported Cmax value expected to exceed both reported EC50 values. For atazanavir, chloroquine, hydroxychloroquine, lopinavir, mefloquine, and remdesivir, some EC50 values were above the Cmax whereas others were below. This observation dramatically highlights the sensitivity of the current analysis to the reported antiviral activity data, and this should be taken into account when interpreting the data presented hereafter.
Figure 1.
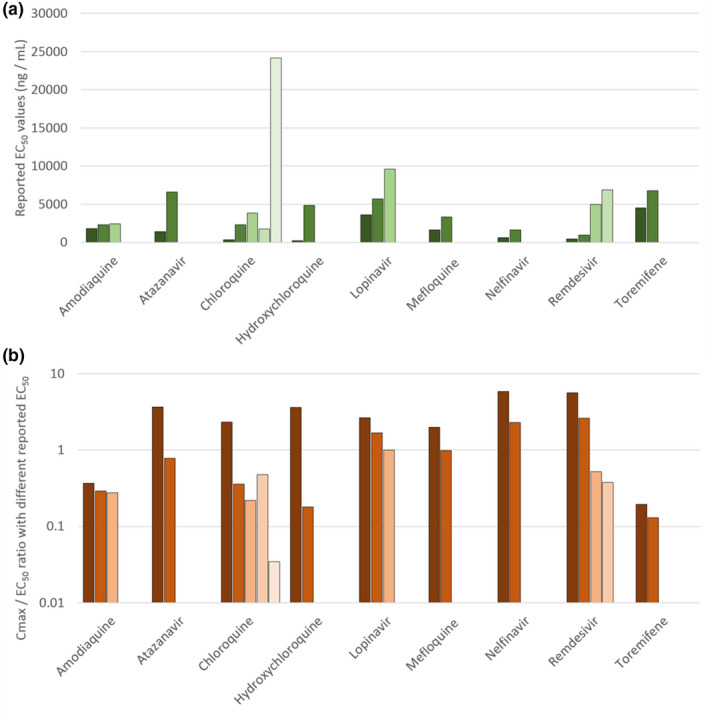
Assessment of the variation in reported half‐maximal effective concentration (EC50) values for severe acute respiratory syndrome‐coronavirus 2 across the drugs for which more than one value was available in the literature (a). The consequences of this variability in reported EC50 in terms of the peak plasma concentration (Cmax)/EC50 ratio is also provided (b). Amodiaquine and toremifene were estimated to exhibit subtherapeutic pharmacokinetics irrespective of which EC50 value was used. Similarly, nelfinavir was estimated to have Cmax value higher than its EC50 irrespective of which EC50 was used in the analysis. For the other drugs, interpretation was highly dependent upon which reported EC50 was utilised and this underscores the caution that should be taken in interpreting the available data.
Identification of candidates achieving plasma concentrations exceeding the SARS‐CoV‐2 EC90 (Cmax/EC90 ratio)
For 56 of the reported antiviral activities, data covering a sufficient concentration range were available for digitization and subsequent calculation of an EC90 value. For the remainder, it was not possible to calculate an EC90. Drugs with an available EC90 were ranked according to their Cmax/EC90 ratio (Figure 2 ). Drugs with a value above 1.0 achieved plasma concentrations above the concentrations reported to inhibit 90% of SARS‐CoV‐2 replication. Only eltrombopag, favipiravir, remdesivir, nelfinavir, niclosamide, nitazoxanide, and tipranavir were estimated to exceed at least one of their reported EC90 by twofold or more at Cmax. Anidulafungin, lopinavir, chloroquine, and ritonavir were also reported to exceed at least one of their reported EC90 values at Cmax but by less than twofold. It was not possible to calculate an EC90 value for sulfadoxine or indomethacin.
Figure 2.
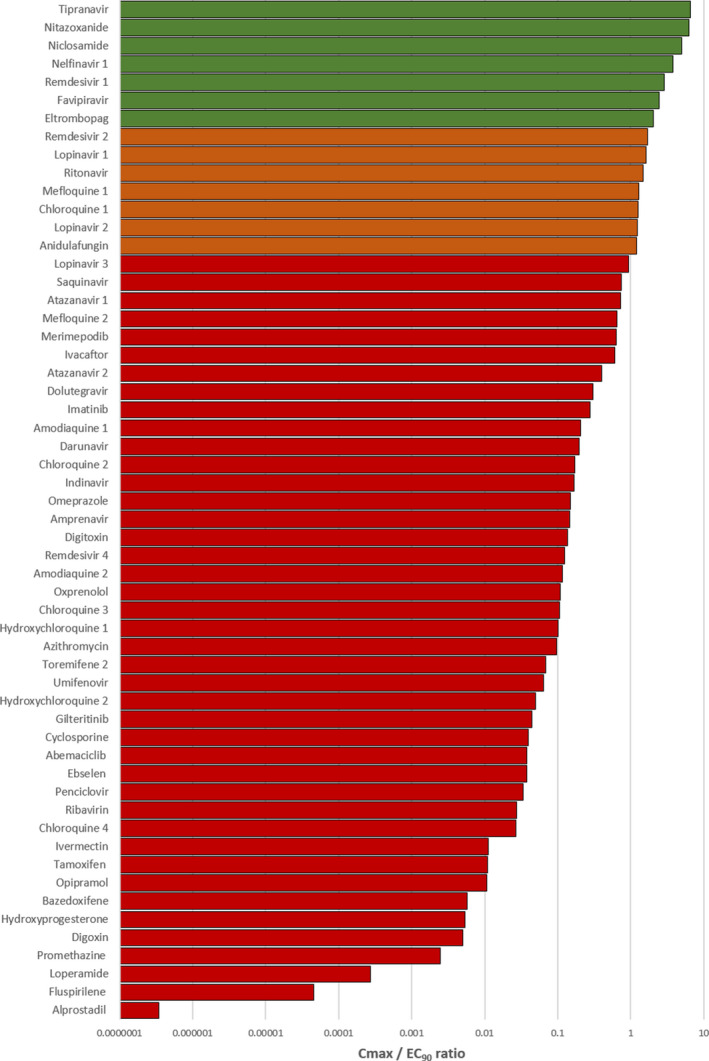
A bar chart displaying peak plasma concentration (Cmax)/effective concentration 90% (EC90) ratio for compounds studied for in vitro antiviral activity against severe acute respiratory syndrome‐coronavirus 2 for which data were available to recalculate an EC90. Drugs with a ratio below 1 were deemed not to provide plasma concentrations at their approved doses to exert sufficient systemic antiviral activity. Those drugs with a ratio above 1 (shown in orange) were deemed to have potential to provide plasma concentrations sufficient to exert at least some antiviral activity for at least some of their dosing interval at their approved dose. Drugs shown in green were predicted to exceed plasma concentrations over their EC90 by more than twofold.
Detailed interrogation of the plasma pharmacokinetics in relation to reported anti‐SARS‐CoV‐2 activity
For drugs with Cmax concentrations above at least one of their reported EC90 values that are not already in clinical trials for COVID‐19, a detailed evaluation of concentrations across their approved dosing interval was undertaken. For this, published pharmacokinetics data were digitized and replotted relative to the calculated EC50 and EC90 data for SARS‐CoV‐2 (Figure 3 ). For tipranavir (ritonavir boosted), nelfinavir, sulfadoxine, and nitazoxanide, plasma concentrations after administration of the approved dose remained above SARS‐CoV‐2 effective concentrations across the entire dosing interval. For anidulafungin, eltrombopag, lopinavir (ritonavir boosted), mefloquine, and chloroquine, Cmax values were above EC90 at 2, 6, 8, and 24 hours postdose, respectively, but concentrations would be expected to dip below the EC50 at 3, 8, 10, 72, and 120 hours postdose, respectively, when given at approved doses and schedules. An overview of these drugs is presented in Table 1 .
Figure 3.
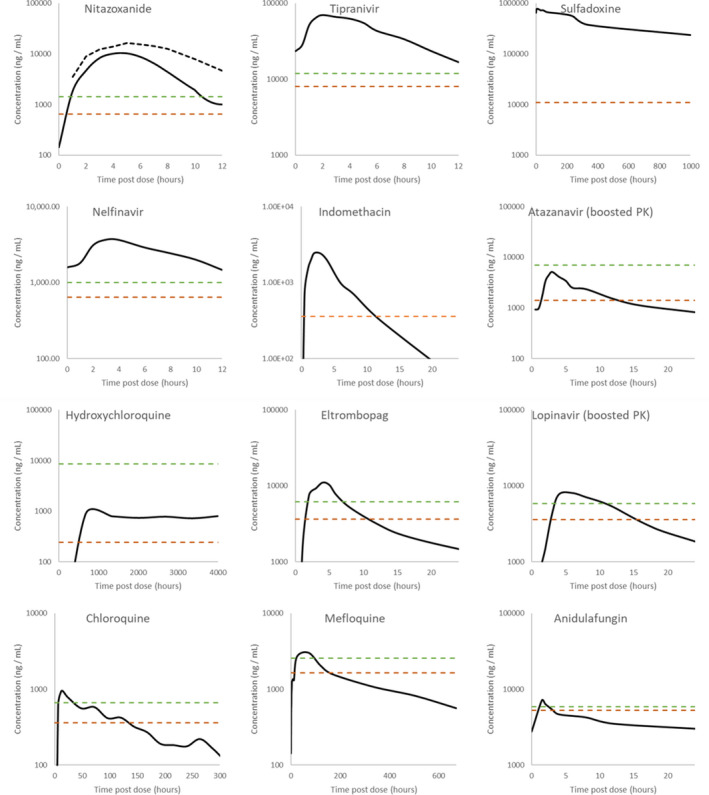
Digitized pharmacokinetic (PK) interrogation of all drugs calculated to have a peak plasma concentration (Cmax)/half‐maximal effective concentration (EC50) ratio above 1. The lowest reported severe acute respiratory syndrome‐coronavirus 2 EC50 (dashed orange lines) and associated recalculated effective concentration 90% (EC90; dashed green lines) are also highlighted. References for the utilized data are nitazoxanide 500 mg b.i.d. and 1,000 mg b.i.d., 95 tipranavir 500 mg b.i.d. with 200 mg ritonavir, 96 sulfadoxine 1,500 mg with 75 mg pyrimethamine, 82 nelfinavir 1,250 mg b.i.d., 97 indomethacin 50 mg t.i.d., 98 atazanavir 300 mg q.d. with 100 mg ritonavir, 99 hydroxychloroquine 2,000 mg hydroxychloroquine sulfate/1,550 mg base administered over 3 days, 100 eltrombopag 75 mg single dose, 101 lopinavir 400 mg with 100 mg ritonavir, 102 chloroquine 1,500 mg administered over 3 days, 103 mefloquine 1,200 mg over 3 days, 104 and anidulafungin 100 mg q.d. 105 Robust PK data were unavailable for niclosamide 500 mg, ritonavir 600 mg, and merimepodib 300 mg in order to conduct this digitized interrogation of these molecules.
Table 1.
Summary of the top leads identified
| Drug | Cmax:EC50 | Cmax:EC90 | Approval | Indications | Route of administration | Dosage | Ref |
|---|---|---|---|---|---|---|---|
|
Atazanavir and Ritonavir REYATAZ (Bristol‐Myers Squibb) |
3.643 | 0.728 |
EMA FDA |
HIV‐1 | Oral | 300/100 mg | 106, 107 |
|
Anidulafungin Eraxis/Ecalta (Pfizer) |
1.323 | 1.192 |
EMA FDA |
Invasive fungal infections | Intravenous infusion | 200 mg q.d. + 100 mg q.d. | 108 |
|
Chloroquine Aralen (Sanofi Aventis) |
2.318 | 1.261 | FDA |
Malaria Extraintestinal amebiasis |
Oral | 1,500 mg | 109 |
|
Eltrombopag Promacta/Revolade (Novartis) |
3.416 | 2.029 |
EMA FDA |
Primary immune thrombocytopenia Acquired severe aplastic anemia |
Oral | 75 mg q.d. | 110 |
|
Favipiravir Avigan (Fujifilm Toyama Chemical Co) |
6.326 | 2.469 | PMDA ‐ Japan | Influenza | Oral | 600 mg b.i.d. | 111 |
|
Hydroxychloroquine Plaquenil (Sanofi Aventis) |
3.598 | 0.101 |
EMA FDA |
Malaria | Oral | 400 mg | 112 |
|
Indomethacin Indocin (Merck & Co) |
5.366 | ‐ |
EMA FDA |
Rheumatoid arthritis | Oral | 50 mg t.i.d. | 113 |
|
Lopinavir and Ritonavir Kaletra (AbbVie) |
2.660/ 1.671 | 1.630/ 1.240 |
EMA FDA |
HIV‐1 | Oral | 400/100 mg b.i.d. | 114 |
|
Mefloquine Lariam (Roche) |
1.350 | 1.284 |
EMA FDA |
Malaria | Oral | 250 mg | 115 |
|
Merimepodib (Vertex Pharmaceuticals) |
1.629 | 0.638 | Not clinically approved | HCV | Oral | 300 mg t.i.d. | 116 |
|
Nelfinavir VIRACEPT (Roche) |
5.849/2.287 | 3.755 |
EMA FDA |
HIV‐1 | Oral | 1,250 mg b.i.d. | 117 |
|
Niclosamide Yomesan (Bayer) |
8.286 | 4.936 |
EMA FDA |
Infestation with tapeworms | Oral | 2,000 mg | 118 |
|
Nitazoxanide Alinia (Romark Pharmaceuticals) |
13.823 | 6.315 | FDA | Diarrhea caused by Giardia lamblia or Cryptosporidium parvum | Oral | 1,000–2,000 mg b.i.d. | 119 |
|
Remdesivir (Gilead) |
5.603/2.614 | 3.755/1.712 | Not clinically approved a | Ebola | Intravenous | 200 mg + 100 mg | 38 |
|
Ritonavir Norvir (AbbVie) |
1.800 |
EMA FDA |
HIV‐1 | Oral | 600 mg | 120 | |
|
Sulfadoxine and pyrimethamine Fansidar (Roche) |
6.577 | FDA ‐ discontinued | Malaria | Oral | 1,500/75 mg | 121 | |
|
Tipranavir and Ritonavir Aptivus (Boehringer Ingelheim Pharmaceuticals, Inc.) |
9.647 | 6.559 |
EMA FDA |
HIV‐1 | Oral | 500/200 mg b.i.d. | 122 |
Cmax, peak plasma concentration; EC50, half‐maximal effective concentration; EC90, effective concentration 90%; EMA, European Medicines Agency; FDA, US Food and Drug Administration; HCV, hepatitis C virus; PMDA, Pharmaceuticals and Medical Devices Agency.
Compassionate use program.
Simulated exposure relative to reported anti‐SARS‐CoV‐2 activity in lung and other tissues
Lung K p U was simulated for all molecules for which the necessary physicochemical properties and in vitro drug binding information were available. Regression and Bland–Altman plots were first used to assess the agreement between predicted lung K p and that observed in previously published animal studies for drugs with available prior data. Good agreement was observed across the available drugs with the exception of chloroquine. An r 2 = 0.86 was observed in linear regression when chloroquine was excluded, but decreased to r 2 = 0.22 when included (Figure S2a ). Similarly, good agreement between measured and predicted K p was observed by Bland–Altman analysis for all data points with the exception of one chloroquine measurement (Figure S2b ). K p U lung was then used along with fraction unbound in plasma (fu) and plasma Cmax values to calculate a predicted Cmax/EC50 (Figure 4 ) and Cmax/EC90 in the lungs (data not shown). Tissue Cmax/EC50 ratios are also shown for other tissues in Figure 5 . For four drugs, ebselen, merimepodib, niclosamide, and remdesivir, the f u data were unavailable. For six other drugs, benztropine, indinavir, loperamide, nelfinavir, saquinavir, and toremifene, the blood to plasma ratios were unavailable. For a further four drugs, camostat, emetine, fluspirilene, and umifenovir, both f u and blood to plasma ratios were unavailable. Therefore, these drugs were excluded from the analysis. A total of 18 drugs with available data were predicted to give concentrations in the lungs above at least one of their reported EC50 against SARS‐CoV‐2 (Figure 4 ) and eight of these were predicted to exceed their EC50 by > 10‐fold. The rank order of lung Cmax/EC90 ratio was chloroquine > atazanavir (ritonavir boosted) > tipranavir (ritonavir boosted) > hydroxychloroquine> mefloquine > ivermectin > lopinavir (ritonavir boosted) > azithromycin > nitazoxanide > ritonavir > gilteritinib > amodiaquine> imatinib > oxprenolol (data excluded due to this analysis only being possible for 33 of the 56 drugs).
Figure 4.
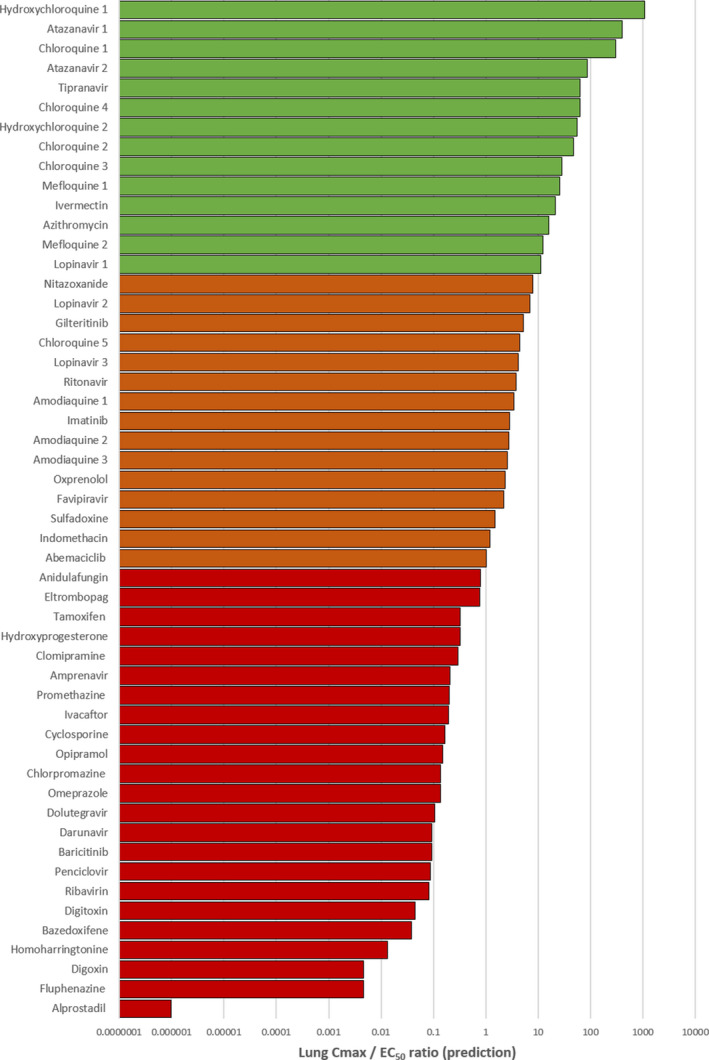
A bar chart displaying the simulated lung peak plasma concentration (Cmax)/half‐maximal effective concentration (EC50). Drugs with a ratio below 1 were deemed not to provide lung concentrations at their approved doses to exert sufficient pulmonary antiviral activity for treatment or prevention strategies. Those drugs with a ratio above 1 (shown in orange) were estimated to provide lung concentrations sufficient to exert at least some antiviral activity at their approved dose. Drugs shown in green were predicted to exceed lung concentrations over their EC50 by > 10‐fold.
Figure 5.
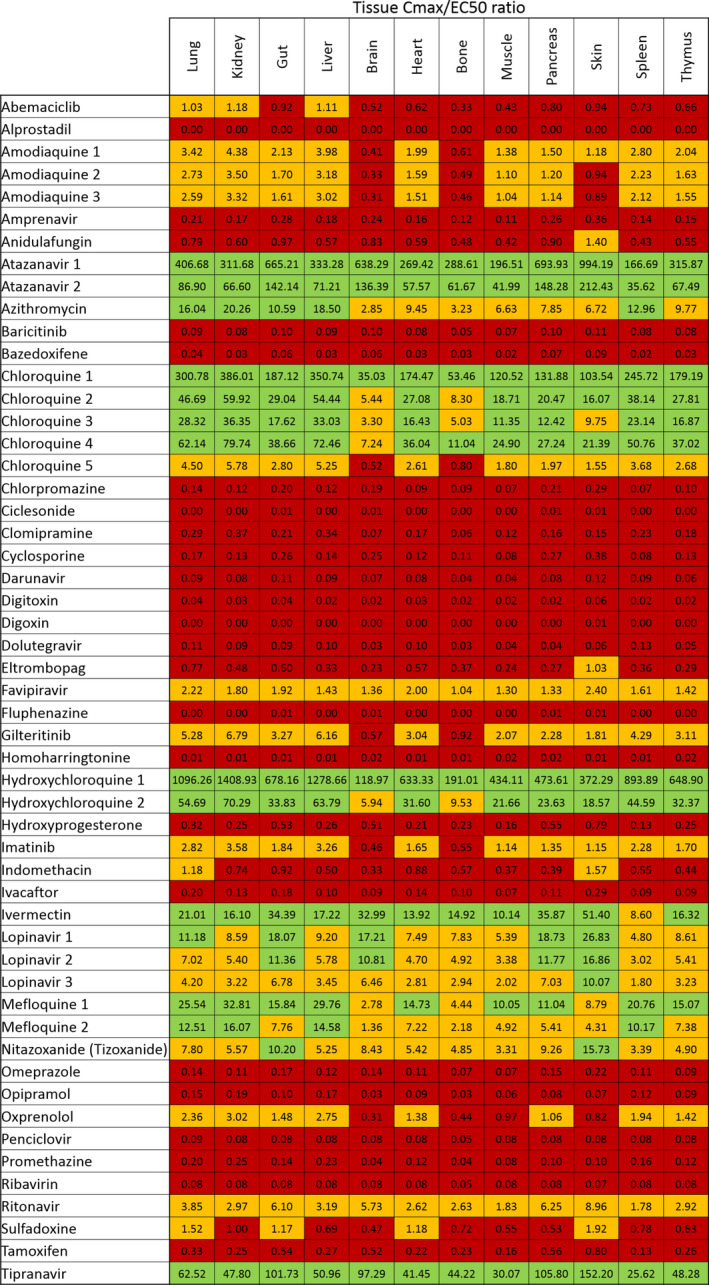
A heatmap displaying the simulated tissue peak plasma concentration (Cmax)/half‐maximal effective concentration (EC50) values for all drugs with available data. Those drugs with a ratio above 1 (shown in orange) were estimated to provide tissue concentrations sufficient to exert at least some antiviral activity at their approved dose. Drugs shown in green were predicted to exceed tissue concentrations over their EC50 by > 10‐fold.
DISCUSSION
The systematic development of mechanism‐based inhibitors for key targets involved in viral replication or pathogenesis is likely to result in highly effective and safe medicines in the coming years. However, the repurposing of already approved medicines in antiviral treatment or chemoprevention strategies is undoubtedly the fastest way to bring forward therapeutic options against the urgent unmet need posed by SARS‐CoV‐2. A range of different drugs and drug classes have been demonstrated to display varying degrees of antiviral activity against SARS‐CoV‐2 in vitro, and many of these drugs are already licensed for use in humans for a range of indications. However, currently, the data emerging from global screening efforts are not being routinely benchmarked and prioritized against achievable concentrations after administration of doses proven to have acceptable safety profiles in humans.
The current analysis indicates that only 12 drugs with reported antiviral activity are likely to achieve plasma exposures above that required for antiviral activity for at least some of their dosing intervals. Notably, neither chloroquine, hydroxychloroquine, nor lopinavir/ritonavir exhibited a sustained plasma concentration above their reported SARS‐CoV‐2 EC90 across their reported dosing intervals. Ultimately, the implications of this for therapy will depend upon whether systemic suppression is a prerequisite for a reduction in morbidity or mortality, but this does raise some concern for ongoing trials with these drugs (chloroquine: NCT04323527 and NCT04333628; hydroxychloroquine: NCT04316377, NCT04333225, and NCT04307693; and lopinavir/ritonavir: NCT04331834, NCT04255017, and NCT04315948). However, the predicted lung accumulation rather than plasma exposure may provide some therapy advantage and/or give more reassurance for ongoing chemoprevention trials.
At least 7 of the 13 candidates achieving Cmax above one of their reported EC50 and derived EC90 are already in clinical evaluation for treatment of SARS‐CoV‐2. These include remdesivir (NCT04292730, NCT04292899, NCT04257656, NCT04252664, and NCT04315948), favipiravir (NCT04310228 and NCT04319900), niclosamide (NCT04345419), mefloquine (NCT04347031), lopinavir/ritonavir, and chloroquine. No robust antiviral activity data were found for galidesivir on which to conduct an analysis but it is also under clinical investigation (NCT03800173). A recent trial for favipiravir demonstrated some success with an improvement over arbidol from 56–71% (P = 0.02) in patients without risk factors (but not critical cases or patients with hypertension and/or diabetes). 49 The results of compassionate use of remdesivir in severely ill patients was also recently reported, and if confirmed in ongoing randomized, placebo‐controlled trials, will serve as a further validation of the other candidates presented here. 50 Of particular interest, nitazoxanide, tipranavir, sulfadoxine, and nelfinavir may be expected to sustain their plasma pharmacokinetic exposure above their lowest reported EC50 and derived EC90 (where available) for the duration of their approved dose and dosing interval.
Nitazoxanide is an antiprotozoal drug that has previously been demonstrated to display broad antiviral activity against human and animal coronaviruses 51 as well as various strains of influenza. 52 , 53 Importantly, nitazoxanide is rapidly metabolized to tizoxanide in humans and this active metabolite is being investigated against SARS‐CoV‐2 (NCT04341493 and NCT04343248). Tizoxanide has been reported to exhibit similar activities to nitazoxanide for other viruses as well as other pathogens. 52 , 54 , 55 The mechanism of antiviral action is not fully understood for nitazoxanide, but it has been reported to affect viral genome synthesis, prevent viral entry, and interfere with the N‐glycosylation and maturation of the influenza hemagglutinin. 56 , 57 , 58 , 59 Notably, the SARS‐CoV‐2 spike protein is also highly N‐glycosylated. 60 This drug has also been shown to elicit an innate immune response that potentiates the production of type 1 interferons. 56 , 61 and a phase IIb/III clinical trial demonstrated a reduction in symptoms and viral shedding in patients with uncomplicated influenza. 53 The safety of nitazoxanide is well understood, but it has not been fully investigated during renal or hepatic impairment. The antiviral activity of nitazoxanide for SARS‐CoV‐2 requires further study but the existing data for this drug are encouraging. Niclosamide is another antiprotozoal drug that exhibits broad antiviral activity due to its ability to perturb the pH‐dependent membrane fusion required for virus entry, 62 but it was reported to have no impact upon the attachment and entry of SARS‐CoV‐2. 63 For MERS‐CoV, niclosamide was observed to inhibit SKP2 activity impairing viral replication. 64 Niclosamide has been reported to be well‐tolerated and does not influence vital organ functions. 65 However, it has low aqueous solubility and poor oral bioavailability, 66 and, despite a higher reported SARS‐CoV‐2 potency 39 than nitazoxanide, 38 the Cmax/EC90 ratio was slightly lower. There is a paucity of published pharmacokinetic data for niclosamide and this prohibited a thorough investigation of exposures in relation to activity over its entire dosing interval. Both nitazoxanide and niclosamide have also been reported to be potent antagonists of TMEM16A, calcium‐activated chloride channels that modulate bronchodilation. 67
Tipranavir and nelfinavir are HIV protease inhibitors 68 and both drugs ranked highly in terms of their Cmax/EC90 ratio. Moreover, a more in‐depth analysis demonstrated that the concentrations across the dosing interval for both these drugs remained above the calculated EC90 values at approved doses and schedules. Unlike nelfinavir, tipranavir has to be co‐administered with a low dose of ritonavir to boost its pharmacokinetics via CYP3A4 inhibition. 69 Because ritonavir itself has been reported to exert anti‐SARS‐CoV‐2 activity, this could be advantageous, but would need to be balanced against the much higher risk of drug‐drug interactions that could negatively impact patient management. The implications of drug interactions have already been raised for this reason with lopinavir/ritonavir use for COVID‐19 70 and are likely to be exacerbated with the higher ritonavir dose needed for tipranavir. Moreover, tipranavir has a black box warning from the FDA for fatal and nonfatal intracranial hemorrhage as well as severe hepatotoxicity. 71 , 72 , 73 The major route of metabolic clearance for nelfinavir is via CYP2C19 and this pathway generates the M8 metabolite that retains activity against the HIV protease. 74 No data are available for inhibition of SARS‐CoV‐2 replication by the M8 metabolite but, if active, this could provide an advantage for nelfinavir over tipranavir for COVID‐19. Conversely, although the analysis of pharmacokinetics relative to potency of these molecules against SARS‐CoV‐2 is encouraging, it should be noted that the reported in vitro activity for HIV 68 , 75 is far higher than that against SARS‐CoV‐2 and both drugs are highly protein bound. 76 , 77 Given that tipranavir and nelfinavir are associated with long‐term toxicities, 68 , 78 , 79 , 80 there will be concern over giving even short‐term exposure for COVID‐19.
Sulfadoxine is another antimalarial drug that is usually administered in combination with pyrimethamine as a folic acid antagonist combination. 81 Sulfadoxine inhibits the activity of dihydropteroate synthase within the malaria parasite, but its mechanism of action for SARS‐CoV‐2 is unclear. It should also be noted that the authors can find no data describing antiviral activity of this drug against other viruses. In addition, the concentrations used in the in vitro activity used in this analysis 37 were not high enough to reach or calculate an EC90 value. Therefore, like other molecules described in this paper, in vitro anti‐SARS‐CoV‐2 activity should be repeated. Notwithstanding, sulfadoxine plasma concentrations far above the reported EC50 are maintained in patients receiving a single 1,500 mg dose (with 75 mg pyrimethamine) for over 40 days. 82 Compared with some other reported molecules, sulfadoxine is not expected to have as high an accumulation in the lungs, but concentrations higher than its EC50 are estimated from the analysis of its lung K p U. Therefore, if the reported antiviral activity is confirmed, this drug may offer opportunities for therapy and/or chemoprophylaxis.
Indomethacin is a nonsteroidal anti‐inflammatory drug that is indicated for rheumatoid arthritis, ankylosing spondylitis, osteoarthritis, acute painful shoulder, or acute gouty arthritis. The recommended dose for acute gouty arthritis is 50 mg 3 times a day and the pharmacokinetic exposure for this is shown in Figure 3 relative to the reported EC50. The indomethacin mechanism of action for SARS‐CoV‐2 remains elusive, but it was shown to inhibit translation of the vesicular stomatitis virus by activating protein kinase R leading to the phosphorylation of eukaryotic initiation factor‐2 α‐subunit. 83 This abrogated viral protein translation, leading to a dramatic inhibition of viral replication and infectious viral particle production. The reported in vitro antiviral activity data for indomethacin were insufficient to calculate an EC90 and this activity requires confirmation in other studies. 40 Furthermore, the drug has a black box warning for serious cardiovascular and gastrointestinal events from the FDA so its use should be managed with caution. 84
Considering that most of the impact of severe disease occurs in the lungs and that this tissue may be a key site for transmission, the potential of candidate drugs to accumulate in lung tissue was considered. The lung K p predictions were validated across 13 drugs for which previously reported animal plasma and lung concentrations were available, and showed good agreement for all agents other than chloroquine. The poor fit for chloroquine does highlight that the predictions may not be accurate for all of the drugs listed and this should be considered in interpretation. Notwithstanding, the analysis of predicted lung Cmax/EC50 ratio revealed more candidates expected to exceed the concentrations needed for antiviral activity in this tissue. Hydroxychloroquine, chloroquine, mefloquine, atazanavir (ritonavir boosted), tipranavir (ritonavir boosted), ivermectin, and lopinavir were all predicted to achieve lung concentrations over 10‐fold higher than their reported EC50. All of these drugs were also predicted to exceed their EC90 in the lungs by at least 3.4‐fold (data not shown). The lung prediction was not possible for nelfinavir because insufficient data were available to calculate K p U lung, but nitazoxanide and sulfadoxine were also predicted to exceed their reported EC50 by 7.8‐fold and 1.5‐fold in the lungs, respectively. Nitazoxanide was predicted to exceed its EC90 by 3.6‐fold in the lungs but an EC90 was not calculable from the available data for sulfadoxine.
Predictions for Cmax/EC50 ratio were also made for other tissues, and were generally in agreement with observations in the lungs with some important exceptions. Gliteritinib, amodiaquine, imatinib, indomethacin, oxprenolol, and sulfadoxine were predicted to be subtherapeutic in the brain and bones, with indomethacin and sulfadoxine being predicted to be subtherapeutic across most of the tissues in which Cmax was estimated.
During inflammation or injury, changes to the vascular microenvironment could have a profound effect on the ability of these drugs to accumulate in lung cells. Due to the recruitment of neutrophils and leaky endothelial cells, 85 the lung inflammatory microenvironment is characterized by increased body temperature, excessive enzymatic activity, and, most importantly, by a low interstitial pH. 86 In the case of chloroquine and hydroxychloroquine, these diprotic weak bases are exquisitely dependent on a pH gradient to drive lysosomal uptake as a mechanism of lung accumulation. It has been demonstrated that cellular chloroquine uptake is diminished 100‐fold for every pH unit of external acidification. 87 This situation is likely to deteriorate further on mechanical ventilation, which also induces acidification of the lung tissue, independently of inflammation. 88 , 89 Therefore, the benefits of lung accumulation for many of these drugs may be lost during treatment of severe SARS‐CoV‐2 infection. Conversely, mefloquine is monoprotic and more lipophilic than chloroquine, which may make it much less reliant on the pH gradient to drive cellular accumulation in the lungs. It is likely that the charged form of the drug is sufficiently lipophilic to allow movement across biological membranes along a concentration gradient. 90 Only two studies have described mefloquine uptake into cells, one study suggested that mefloquine uptake is not energy dependent and the other suggested that mefloquine uptake is mediated by secondary active transport, rather than passive proton trapping. 91 , 92 Mefloquine is known to cause severe psychiatric side effects in some patients and so use of this drug should be managed with care. 93 Therefore, mefloquine may offer opportunities for treatment during severe disease that are not available with other drugs currently being tested for COVID‐19 therapy. If the high lung exposures are proven empirically for the drugs on this list, then some may also prove to be valuable for chemoprevention strategies.
Limitations of this analysis
This study represents the first holistic view of drugs with reported activity against SARS‐CoV‐2 in the context of their achievable pharmacokinetic exposure in humans. Although the analysis does provide a basis to rationally selected candidates for further analysis, there are some important limitations. First, Cmax was the only pharmacokinetic parameter that was universally available for all of the candidate drugs, but minimum plasma concentration (Cmin) values are generally accepted as a better marker of efficacy because they represent the lowest plasma concentration over the dosing interval. However, Cmax was only used to assess whether plasma concentration would exceed those required at any point in the dosing interval, and this was followed by a more in‐depth analysis of the most promising candidates.
Second, an EC50 value only equates to a concentration required to suppress 50% of the virus, and data were unavailable to calculate EC90 values for some of the drugs. EC90 values are a preferred marker of activity because the slope of the concentration‐response curve can vary substantially between different molecules and between different mechanisms of action. Although EC90 values were not calculable for all drugs, the authors deemed it appropriate to deprioritize molecules not achieving EC50 at Cmax in this analysis. Third, the reported antiviral activities were conducted under different conditions (Table S2 ) and in several cases varied between the same molecule assessed in different studies (Figure 1 ). In addition, some of the studied drugs (e.g., nitazoxanide and amodiaquine) are rapidly metabolized such that the major species systemically is a metabolite that has not been investigated for anti‐SARS‐CoV‐2 activity. No mitigation strategy was possible for these limitations and the data should be interpreted in the context that the quality of the available data may profoundly impact the conclusions. In vitro activity should be confirmed for the promising candidates and/or relevant metabolites.
Fourth, plasma protein binding can be an important factor in determining whether sufficient free drug concentrations are available to exert antiviral activity 94 and insufficient data were available across the dataset to determine protein binding‐adjusted EC90 values. This is important because, for highly protein bound drugs, the antiviral activity in plasma may be lower than reported in in vitro activity because protein concentrations used in culture media are lower than those in plasma. Fifth, robust pharmacokinetic data were not available for all the molecules and subtle differences have been reported in the pharmacokinetics in different studies. Where possible, this analysis utilized the pharmacokinetics described at the highest doses approved for other indications and checked them to ensure that profound differences were not evident between different studies. However, in some cases, higher doses and/or more frequent dosing has been investigated for some of the drugs mentioned so higher exposures may be available for some drugs with off‐label dosing. Sixth, the digitized pharmacokinetic plots presented in this paper represent the mean or median profiles depending on what was presented in the original papers. Many of the drugs presented are known to exhibit high interindividual variability that is not captured within the presented analysis and it is possible that even for promising candidates, a significant proportion of patients may have subtherapeutic concentrations despite population mean/median being higher than the Cmax. Advanced pharmacokinetic modeling approaches will be needed to unpick the exposure‐response relationship and these studies are currently underway by the authors.
Seventh, the presented predictions for lung accumulation may offer a basis for ranking molecules for expected accumulation in that organ, but ultimate effectiveness of a chemoprophylactic approach will likely depend upon penetration into other critical matrices in the upper airways for which there are currently no robustly validated methods of prediction. In addition, although a generally accepted method for assessing K p U was used, the predictions were only validated for a subset of drugs for which previous animal lung accumulation data were available. In addition, the K p U method assumes all the processes are passive and perfusion limited, and the complexity of pulmonary tissue pharmacokinetics is not captured in this analysis. The lungs include different structures, including airways, bronchioles, and alveoli, with different blood flow perfusion and more detailed modeling validated through animal experiments will be required to capture this complexity.
Finally, this analysis assumes that drugs need to be active within the systemic compartment in order to have efficacy against SARS‐CoV‐2. Because current evidence suggests that the virus is widely disseminated throughout the body this is a logical assumption. However, ultimate efficacy of any drug can only be demonstrated with robust clinical trial designs.
SUMMARY
The current analysis reveals that many putative agents are never likely to achieve target concentrations necessary to adequately suppress SARS‐CoV‐2 under normal dosing conditions. It is critical that candidate medicines emerging from in vitro antiviral screening programs are considered in the context of their expected exposure in humans where possible. Clinical trials are extremely time‐consuming and expensive, and it is critical that only the best options are progressed for robust analysis as potential monotherapy or combination therapy or prevention options. Finally, it would be highly beneficial for activity data for SARS‐CoV‐2 to be performed with standardized protocol and with activity reported as EC90 values as a better marker of the concentrations required to suppress the virus to therapeutically relevant levels. Based upon the currently reported data, atazanavir, chloroquine, favipiravir, hydroxychloroquine, indomethacin, lopinavir, mefloquine, nitazoxanide, ritonavir, sulfadoxine, and tipranavir are predicted to have mean/median Cmax concentrations above their reported EC50 in both plasma and lungs. Anidulafungin, eltrombopag, merimepodib, nelfinavir, niclosamide, and remdesivir also had mean/media Cmax above available EC50 in plasma but a lung prediction was not possible. Only atazanavir, indomethacin, nelfinavir, nitazoxanide, sulfadoxine, and tipranavir were predicted to have mean/median plasma Cmax concentrations above their reported EC50 for the duration of their dosing interval, but full concentration‐time profiles were not available to make this judgment for favipiravir, niclosamide, and remdesivir.
Funding
The authors received no funding for the current work. A.O. acknowledges research funding from EPSRC (EP/R024804/1; EP/S012265/1), NIH (R01AI134091; R24AI118397), European Commission (761104), and Unitaid (project LONGEVITY). G.A.B. acknowledges support from the Medical Research Council (MR/S00467X/1). G.A. acknowledges funding from the MRC Skills Development Fellowship.
Conflict of Interest
D.J.B. has received honoraria or advisory board payments from AbbVie, Gilead, ViiV, Merck, Janssen, and educational grants from AbbVie, Gilead, ViiV, Merck, Janssen, and Novartis. A.O. and S.P.R. are Directors of Tandem Nano Ltd. A.O. has received research funding from ViiV, Merck, Janssen, and consultancy from Gilead, ViiV and Merck not related to the current paper. P.O.N. is currently engaged in a collaboration with Romark LLC but this interaction did not influence the prioritization or conclusions in the current paper. All other authors declared no competing interests for this work.
Author Contributions
All authors wrote the paper. A.O. designed the research. U.A., H.B., L.T., H.P., and A.O. performed the research. R.R., H.P., U.A., and A.O. analyzed the data.
Supporting information
Figure S1
Figure S2
Table S1
Table S2
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgments
The authors thank Nathan Morin from Alberta Health Services for being proactive in making them aware of previously published data for indomethacin. The authors also thank Articulate Science for publication support.
References
- 1. Gu, J. , Han, B. & Wang, J. COVID‐19: Gastrointestinal manifestations and potential fecal‐oral transmission. Gastroenterology 158, 1518–1519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organisation . COVID‐19 Trials ‐ International Clinical Trials Registry Platform (ICTRP) <https://www.who.int/ictrp/search/en/> (2020).
- 3. Qin, C. et al. Dysregulation of immune response in patients with COVID‐19 in Wuhan, China. Clin. Infect. Dis. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li, W. et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao, Y. , Zhao, Z. , Wang, Y. , Zhou, Y. , Ma, Y. & Zuo, W. Single‐cell RNA expression profiling of ACE2, the receptor of SARS‐CoV‐2. J bioRxiv. 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu, Y. et al. Prolonged presence of SARS‐CoV‐2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diao, B.H. et al. Kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) Infection. J medRxiv. 10.1101/2020.03.04.20031120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang, C. , Shi, L. & Wang, F.S. Liver injury in COVID‐19: management and challenges. Lancet Gastroenterol. Hepatol. 5, 428–430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo, Y.R. et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID‐19) outbreak ‐ an update on the status. Mil Med Res 7, 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoffmann, M. et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang, H. et al. The digestive system is a potential route of 2019‐nCov infection: a bioinformatics analysis based on single‐cell transcriptomes. J bioRxiv. 10.1101/2020.01.30.927806. [DOI] [Google Scholar]
- 12. Wong, S.H. , Lui, R.N. & Sung, J.J. COVID‐19 and the digestive system. J. Gastroenterol. Hepatol. 35, 744–748 (2020). [DOI] [PubMed] [Google Scholar]
- 13. Zhang, W. et al. Molecular and serological investigation of 2019‐nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 9, 386–389 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun, C.B. , Wang, Y.‐Y. , Liu, G.‐H. , Liu, Z . Role of the Eye in Transmitting Human Coronavirus: What We Know and What We Do Not Know. Frontiers in Public Health 8, (2020). 10.3389/fpubh.2020.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu, C.W. , Liu, X.F. & Jia, Z.F. 2019‐nCoV transmission through the ocular surface must not be ignored. Lancet 395, e39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith, C.A. , Kulkarni, U. , Chen, J. & Goldstein, D.R. Influenza virus inoculum volume is critical to elucidate age‐dependent mortality in mice. Aging Cell 18, e12893 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller, D.S. , Kok, T. & Li, P. The virus inoculum volume influences outcome of influenza A infection in mice. Lab. Anim. 47, 74–77 (2013). [DOI] [PubMed] [Google Scholar]
- 18. Chen, X. et al. Detectable serum SARS‐CoV‐2 viral load (RNAaemia) is closely associated with drastically elevated interleukin 6 (IL‐6) level in critically ill COVID‐19 patients. Clin. Infect. Dis. 10.1101/2020.02.29.20029520. [DOI] [PMC free article] [PubMed]
- 19. Liu, Y. et al. Viral dynamics in mild and severe cases of COVID‐19. Lancet Infect. Dis. 10.1016/S473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodgers, T. , Leahy, D. & Rowland, M. Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate‐to‐strong bases. J. Pharm. Sci. 94, 1259–1276 (2005). [DOI] [PubMed] [Google Scholar]
- 21. Rodgers, T. & Rowland, M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J. Pharm. Sci. 95, 1238–1257 (2006). [DOI] [PubMed] [Google Scholar]
- 22. Rodgers, T. & Rowland, M. Mechanistic approaches to volume of distribution predictions: understanding the processes. Pharm. Res, 24, 918–933 (2007). [DOI] [PubMed] [Google Scholar]
- 23. Damle, B. , Stogniew, M. & Dowell, J. Pharmacokinetics and tissue distribution of anidulafungin in rats. Antimicrob. Agents Chemother. 52, 2673–2676 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chandrasekaran, A. , Ahmad, S. , Shen, L. , DeMaio, W. , Hultin, T. & Scatina, J. Disposition of bazedoxifene in rats. Xenobiotica 40, 578–585 (2010). [DOI] [PubMed] [Google Scholar]
- 25. Browning, D.J. Pharmacology of chloroquine and hydroxychloroquine. In: Hydroxychloroquine Chloroquine Retinopathy. 35–63 (Springer, New York, NY, 2014). [Google Scholar]
- 26. McChesney, E.W. , Banks, W.F. Jr & Fabian, R.J. Tissue distribution of chloroquine, hydroxychloroquine, and desethylchloroquine in the rat. Toxicol. Appl. Pharmacol. 10, 501–513 (1967). [DOI] [PubMed] [Google Scholar]
- 27. Gupta, A. , Tulsankar, S.L. , Bhatta, R.S. & Misra, A. Pharmacokinetics, metabolism, and partial biodistribution of "pincer therapeutic" nitazoxanide in mice following pulmonary delivery of inhalable particles. Mol. Pharm. 14, 1204–1211 (2017). [DOI] [PubMed] [Google Scholar]
- 28. Lien, E.A. , Solheim, E. & Ueland, P.M. Distribution of tamoxifen and its metabolites in rat and human tissues during steady‐state treatment. Cancer Res. 51, 4837–4844 (1991). [PubMed] [Google Scholar]
- 29. Rivulgo, V. et al. Comparative plasma exposure and lung distribution of two human use commercial azithromycin formulations assessed in murine model: a preclinical study. Biomed. Res. Int. 2013, 392010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moss, L. , Wagner, D. , Kanaoka, E. , Olson, K. , Yueh, Y.L. & Bowers, G.D. The comparative disposition and metabolism of dolutegravir, a potent HIV‐1 integrase inhibitor, in mice, rats, and monkeys. Xenobiotica 45, 60–70 (2015). [DOI] [PubMed] [Google Scholar]
- 31. Kawai, R. , Mathew, D. , Tanaka, C. & Rowland, M. Physiologically based pharmacokinetics of cyclosporine A: extension to tissue distribution kinetics in rats and scale‐up to human. J. Pharmacol. Exp. Ther. 287, 457–468 (1998). [PubMed] [Google Scholar]
- 32. Pharmaceuticals and Medical Devices Agency (PMDA) . Report on the deliberation results ‐ Xospata tablets 40 mg <https://www.pmda.go.jp/drugs/2014/P201400148/800155000_22600AMX01325_I100_1.pdf> (2018). Accessed May 4, 2020.
- 33. Bland, J.M. & Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1, 307–310 (1986). [PubMed] [Google Scholar]
- 34. Weston, S. , Haupt, R. , Logue, J. , Matthews, K. & Frieman, M.B. FDA approved drugs with broad anti‐coronaviral activity inhibit SARS‐CoV‐2 in vitro. J bioRxiv. 10.1101/2020.03.25.008482. [DOI] [Google Scholar]
- 35. Ge, Y. et al. A data‐driven drug repositioning framework discovered a potential therapeutic agent targeting COVID‐19. J bioRxiv. 10.1101/2020.03.11.986836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bojkova, D. et al. SARS‐CoV‐2 and SARS‐CoV differ in their cell tropism and drug sensitivity profiles. J bioRxiv . 10.1101/2020.04.03.024257. [DOI] [Google Scholar]
- 37. Touret, F. et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS‐CoV‐2 replication. J bioRxiv. 10.1101/2020.04.03.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang, M. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res. 30, 269–271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeon, S. et al. Identification of antiviral drug candidates against SARS‐CoV‐2 from FDA‐approved drugs. J bioRxiv. 10.1101/2020.03.20.999730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu, T. , Gao, X. , Wu, Z. , Selinger, D.W. & Zhou, Z. Indomethacin has a potent antiviral activity against SARS CoV‐2 in vitro and canine coronavirus in vivo. J bioRxiv. 10.1101/2020.04.01.017624. [DOI] [Google Scholar]
- 41. Fintelman‐Rodrigues, N. et al. Atazanavir inhibits SARS‐CoV‐2 replication and pro‐inflammatory cytokine production. J bioRxiv. 10.1101/2020.04.04.020925. [DOI] [Google Scholar]
- 42. Yamamoto, N. , Matsuyama, S. , Hoshino, T. & Yamamoto, N. Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. J bioRxiv. 10.1101/2020.04.06.026476. [DOI] [Google Scholar]
- 43. Bukreyeva, N. , Mantlo, E.K. , Sattler, R.A. , Huang, C. , Paessler, S. & Zeldis, J. The IMPDH inhibitor merimepodib suppresses SARS‐CoV‐2 replication in vitro. J bioRxiv. 10.1101/2020.04.07.028589. [DOI] [Google Scholar]
- 44. Jin, Z. et al. Structure of Mpro from COVID‐19 virus and discovery of its inhibitors. Nature. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 45. Yao, X. et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Clin. Infect. Dis. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Choy, K.‐T. et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS‐CoV‐2 replication in vitro. Antiviral. Res. 178, 104786 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Caly, L. , Druce, J.D. , Catton, M.G. , Jans, D.A. & Wagstaff, K.M. The FDA‐approved drug ivermectin inhibits the replication of SARS‐CoV‐2 in vitro. Antiviral Res. 178, 104787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu, Z. et al. Nelfinavir is active against SARS‐CoV‐2 in Vero E6. Cells. 10.26434/chemrxiv.12039888. [Google Scholar]
- 49. Chen, C. et al. Favipiravir versus Arbidol for COVID‐19: a randomized clinical trial. J. medRxiv. 10.1101/2020.03.17.20037432. [DOI]
- 50. Grein, J. et al. Compassionate use of remdesivir for patients with severe Covid‐19. N. Engl. J. Med. 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rossignol, J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect. Public Health 9, 227–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tilmanis, D. , van Baalen, C. , Oh, D.Y. , Rossignol, J.F. & Hurt, A.C. The susceptibility of circulating human influenza viruses to tizoxanide, the active metabolite of nitazoxanide. Antiviral Res. 147, 142–148 (2017). [DOI] [PubMed] [Google Scholar]
- 53. Haffizulla, J. et al. Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double‐blind, randomised, placebo‐controlled, phase 2b/3 trial. Lancet Infect Dis. 14, 609–618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gekonge, B. , Bardin, M.C. & Montaner, L.J. Short communication: Nitazoxanide inhibits HIV viral replication in monocyte‐derived macrophages. AIDS Res. Hum. Retroviruses 31, 237–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Trabattoni, D. et al. Thiazolides elicit anti‐viral innate immunity and reduce HIV replication. Sci. Rep. 6, 27148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rossignol, J.F. Nitazoxanide: a first‐in‐class broad‐spectrum antiviral agent. Antiviral Res. 110, 94–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rossignol, J.F. , La Frazia, S. , Chiappa, L. , Ciucci, A. & Santoro, M.G. Thiazolides, a new class of anti‐influenza molecules targeting viral hemagglutinin at the post‐translational level. J. Biol. Chem. 284, 29798–29808 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hickson, S.E. , Margineantu, D. , Hockenbery, D.M. , Simon, J.A. & Geballe, A.P. Inhibition of vaccinia virus replication by nitazoxanide. Virology 518, 398–405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang, Y.M. et al. Antiviral activities of niclosamide and nitazoxanide against chikungunya virus entry and transmission. Antiviral Res. 135, 81–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang, Y. et al. Site‐specific N‐glycosylation characterization of recombinant SARS‐CoV‐2 spike proteins using high‐resolution mass spectrometry. J bioRxiv. 10.1101/2020.03.28.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Clerici, M. , Trabattoni, D. , Pacei, M. , Biasin, M. & Rossignol, J.‐F. The anti‐infective nitazoxanide shows strong immumodulating effects. J. Immuno. 186, 155.21 (2011). [Google Scholar]
- 62. Jurgeit, A. , McDowell, R. , Moese, S. , Meldrum, E. , Schwendener, R. & Greber, U.F. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS Pathog. 8, e1002976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang, X.W. & Yap, Y.L. Old drugs as lead compounds for a new disease? Binding analysis of SARS coronavirus main proteinase with HIV, psychotic and parasite drugs. Bioorg. Med. Chem. 12, 2517–2521 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gassen, N.C. et al. SKP2 attenuates autophagy through Beclin1‐ubiquitination and its inhibition reduces MERS‐Coronavirus infection. Nat. Commun. 10, 5770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li, R. et al. Inhibition of STAT3 by niclosamide synergizes with erlotinib against head and neck cancer. PLoS One 8, e74670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lin, C.K. et al. Preclinical evaluation of a nanoformulated antihelminthic, niclosamide, in ovarian cancer. Oncotarget 7, 8993–9006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miner, K. et al. Drug repurposing: the anthelmintics niclosamide and nitazoxanide are potent TMEM16A antagonists that fully bronchodilate airways. Front. Pharmacol. 10, 51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lv, Z. , Chu, Y. & Wang, Y. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV AIDS (Auckl) 7, 95–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Streeck, H. & Rockstroh, J.K. Review of tipranavir in the treatment of drug‐resistant HIV. Ther. Clin. Risk Manag. 3, 641–651 (2007). [PMC free article] [PubMed] [Google Scholar]
- 70. Sanders, J.M. , Monogue, M.L. , Jodlowski, T.Z. , Cutrell, J.B. pharmacologic treatments for coronavirus disease 2019 (COVID‐19). JAMA. 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 71. Justice, A.C. et al. Drug toxicity, HIV progression, or comorbidity of aging: does tipranavir use increase the risk of intracranial hemorrhage? Clin. Infect. Dis. 47, 1226–1230 (2008). [DOI] [PubMed] [Google Scholar]
- 72. Flexner, C. , Bate, G. & Kirkpatrick, P. Tipranavir. Nat. Rev. Drug Discovery 4, 955–956 (2005). [DOI] [PubMed] [Google Scholar]
- 73. Chan‐Tack, K.M. , Struble, K.A. & Birnkrant, D.B. Intracranial hemorrhage and liver‐associated deaths associated with tipranavir/ritonavir: review of cases from the FDA's Adverse Event Reporting System. AIDS Patient Care STDS 22, 843–850 (2008). [DOI] [PubMed] [Google Scholar]
- 74. Hirani, V.N. , Raucy, J.L. & Lasker, J.M. Conversion of the HIV protease inhibitor nelfinavir to a bioactive metabolite by human liver CYP2C19. Drug Metab. Dispos. 32, 1462–1467 (2004). [DOI] [PubMed] [Google Scholar]
- 75. Zhang, K.E. et al. Circulating metabolites of the human immunodeficiency virus protease inhibitor nelfinavir in humans: structural identification, levels in plasma, and antiviral activities. Antimicrob. Agents Chemother. 45, 1086–1093 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Motoya, T. et al. Characterization of nelfinavir binding to plasma proteins and the lack of drug displacement interactions. HIV Med. 7, 122–128 (2006). [DOI] [PubMed] [Google Scholar]
- 77. King, J.R. & Acosta, E.P. Tipranavir. Clin. Pharmacokinet. 45, 665–682 (2006). [DOI] [PubMed] [Google Scholar]
- 78. Markowitz, M. et al. Long‐term efficacy and safety of tipranavir boosted with ritonavir in HIV‐1‐infected patients failing multiple protease inhibitor regimens: 80‐week data from a phase 2 study. J. Acquir. Immune Defic. Syndr. 45, 401–410 (2007). [DOI] [PubMed] [Google Scholar]
- 79. Justice, A.C. et al. Drug toxicity, HIV progression, or comorbidity of aging: does tipranavir use increase the risk of intracranial hemorrhage? Clin. Infect. Dis. 47, 1226–1230 (2008). [DOI] [PubMed] [Google Scholar]
- 80. Unis, G. et al. Mitochondrial mechanisms of nelfinavir toxicity in human brain microvascular endothelial cells. The FASEB Journal 30, 953.4‐.4 (2016). [Google Scholar]
- 81. Lovegrove, F.E. & Kain, K.C. Chapter 6 ‐ malaria prevention. In: The Travel and Tropical Medicine Manual. 4th ed. (eds. Jong, E.C. & Sanford, C. ) 76–99 (W.B. Saunders, Edinburgh, 2008). [Google Scholar]
- 82. de Kock, M. et al. Pharmacokinetics of sulfadoxine and pyrimethamine for intermittent preventive treatment of malaria during pregnancy and after delivery. CPT Pharmacometrics Syst. Pharmacol. 6, 430–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Amici, C. , La Frazia, S. , Brunelli, C. , Balsamo, M. , Angelini, M. & Santoro, M.G. Inhibition of viral protein translation by indomethacin in vesicular stomatitis virus infection: role of eIF2alpha kinase PKR. Cell Microbiol. 17, 1391–1404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nalamachu, S. & Wortmann, R. Role of indomethacin in acute pain and inflammation management: a review of the literature. Postgrad. Med. 126, 92–97 (2014). [DOI] [PubMed] [Google Scholar]
- 85. Pober, J.S. & Sessa, W.C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7, 803–815 (2007). [DOI] [PubMed] [Google Scholar]
- 86. Zhang, R. et al. Tumor‐associated inflammatory microenvironment in non‐small cell lung cancer: correlation with FGFR1 and TLR4 expression via PI3K/Akt pathway. J. Cancer 10, 1004–1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Geary, T.G. , Divo, A.D. , Jensen, J.B. , Zangwill, M. & Ginsburg, H. Kinetic modelling of the response of Plasmodium falciparum to chloroquine and its experimental testing in vitro. Implications for mechanism of action of and resistance to the drug. Biochem. Pharmacol. 40, 685–691 (1990). [DOI] [PubMed] [Google Scholar]
- 88. Pugin, J. , Dunn‐Siegrist, I. , Dufour, J. , Tissieres, P. , Charles, P.E. & Comte, R. Cyclic stretch of human lung cells induces an acidification and promotes bacterial growth. Am. J. Respir. Cell Mol. Biol. 38, 362–370 (2008). [DOI] [PubMed] [Google Scholar]
- 89. Drachman, N. , Kadlecek, S. , Pourfathi, M. , Xin, Y. , Profka, H. & Rizi, R. In vivo pH mapping of injured lungs using hyperpolarized [1‐(13) C]pyruvate. Magn. Reson. Med. 78, 1121–1130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ginsburg, H. , Nissani, E. & Krugliak, M. Alkalinization of the food vacuole of malaria parasites by quinoline drugs and alkylamines is not correlated with their antimalarial activity. Biochem. Pharmacol. 38, 2645–2654 (1989). [DOI] [PubMed] [Google Scholar]
- 91. Vanderkooi, G. , Prapunwattana, P. & Yuthavong, Y. Evidence for electrogenic accumulation of mefloquine by malarial parasites. Biochem. Pharmacol. 37, 3623–3631 (1988). [DOI] [PubMed] [Google Scholar]
- 92. Fitch, C.D. , Chevli, R. & Gonzalez, Y. Chloroquine‐resistant Plasmodium falciparum: effect of substrate on chloroquine and amodiaquin accumulation. Antimicrob. Agents Chemother. 6, 757–762 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ritchie, E.C. , Block, J. & Nevin, R.L. Psychiatric side effects of mefloquine: applications to forensic psychiatry. J. Am. Acad. Psychiatry Law 41, 224–235 (2013). [PubMed] [Google Scholar]
- 94. Gonzalez, D. , Schmidt, S. & Derendorf, H. Importance of relating efficacy measures to unbound drug concentrations for anti‐infective agents. Clin. Microbiol. Rev. 26, 274–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fox, L.M. & Saravolatz, L.D. Nitazoxanide: a new thiazolide antiparasitic agent. Clin. Infect. Dis. 40, 1173–1180 (2005). [DOI] [PubMed] [Google Scholar]
- 96. la Porte, C.J. , Sabo, J.P. , Beique, L. & Cameron, D.W. Lack of effect of efavirenz on the pharmacokinetics of tipranavir‐ritonavir in healthy volunteers. Antimicrob. Agents Chemother. 53, 4840–4844 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kruse, G. et al. The steady‐state pharmacokinetics of nelfinavir in combination with tenofovir in HIV‐infected patients. Antivir. Ther. 10, 349–355 (2005). [PubMed] [Google Scholar]
- 98. Rainsford, K.D. et al. Effects of misoprostol on the pharmacokinetics of indomethacin in human volunteers. Clin. Pharmacol. Ther. 51, 415–421 (1992). [DOI] [PubMed] [Google Scholar]
- 99. Burger, D.M. , Agarwala, S. , Child, M. , Been‐Tiktak, A. , Wang, Y. & Bertz, R. Effect of rifampin on steady‐state pharmacokinetics of atazanavir with ritonavir in healthy volunteers. Antimicrobial Agents Chemother. 50, 3336–3342 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tett, S.E. , Cutler, D.J. , Beck, C. & Day, R.O. Concentration‐effect relationship of hydroxychloroquine in patients with rheumatoid arthritis–a prospective, dose ranging study. J. Rheumatol. 27, 1656–1660 (2000). [PubMed] [Google Scholar]
- 101. Shida, Y. , Takahashi, N. , Nohda, S. & Hirama, T. Pharmacokinetics and pharmacodynamics of eltrombopag in healthy Japanese males. Jpn. J. Clin. Pharmacol. Ther. 42, 11–20 (2011). [Google Scholar]
- 102. US Food and Drug Administration (FDA) . Abbott L. Clinical Pharmacology and Biopharmaceutics review of Kaletra oral solution (NDA#021251). <https://www.accessdata.fda.gov/drugsatfda_docs/nda/2000/21‐226_Kaletra_biopharmr_P1.pdf> (2020). Accessed April 13, 2020. [Google Scholar]
- 103. Na‐Bangchang, K. , Limpaibul, L. , Thanavibul, A. , Tan‐Ariya, P. & Karbwang, J. The pharmacokinetics of chloroquine in healthy Thai subjects and patients with Plasmodium vivax malaria. Br. J. Clin. Pharmacol. 38, 278–281 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Krudsood, S. et al. New fixed‐dose artesunate‐mefloquine formulation against multidrug‐resistant Plasmodium falciparum in adults: a comparative phase iib safety and pharmacokinetic study with standard‐dose nonfixed artesunate plus mefloquine. Antimicrob. Agents Chemother. 54, 3730–3737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Liu, P. , Ruhnke, M. , Meersseman, W. , Paiva, J.A. , Kantecki, M. & Damle, B. Pharmacokinetics of anidulafungin in critically ill patients with candidemia/invasive candidiasis. Antimicrob. Agents Chemother. 57, 1672–1676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Raja, A. , Lebbos, J. & Kirkpatrick, P. Atazanavir sulphate. Nat. Rev. Drug Discov. 2, 857–858 (2003). [DOI] [PubMed] [Google Scholar]
- 107. Harrison, T.S. & Scott, L.J. Atazanavir. Drugs 65, 2309–2336 (2005). [DOI] [PubMed] [Google Scholar]
- 108. Murdoch, D. & Plosker, G.L. Anidulafungin. Drugs 64, 2249–2258 (2004). [DOI] [PubMed] [Google Scholar]
- 109. Goel, P. & Gerriets, V. Chloroquine. In: StatPearls. (StatPearls Publishing, Treasure Island (FL). https://www.ncbi.nlm.nih.gov/books/NBK551512/ 2020. [PubMed]
- 110. Garnock‐Jones, K.P. Eltrombopag. Drugs 71, 1333–1353 (2011). [DOI] [PubMed] [Google Scholar]
- 111.Pharmaceuticals and Medical Devices Agency (PMDA) . Report on the Deliberation Results. <https://www.pmda.go.jp/files/000210319.pdf> (2014). [Google Scholar]
- 112. Ben‐Zvi, I. , Kivity, S. , Langevitz, P. & Shoenfeld, Y. Hydroxychloroquine: from malaria to autoimmunity. Clin. Rev. Allergy Immunol. 42, 145–153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yeh, K.C. Pharmacokinetic overview of indomethacin and sustained‐release indomethacin. Am. J. Med. 79, 3–12 (1985). [DOI] [PubMed] [Google Scholar]
- 114. Corbett, A.H. , Lim, M.L. & Kashuba, A.D. Kaletra (lopinavir/ritonavir). Ann. Pharmacother. 36, 1193–1203 (2002). [DOI] [PubMed] [Google Scholar]
- 115. Price, R.N. et al. Artesunate/mefloquine treatment of multi‐drug resistant falciparum malaria. Trans. R Soc. Trop. Med. Hyg. 91, 574–577 (1997). [DOI] [PubMed] [Google Scholar]
- 116. McHutchison, J.G. et al. A randomized, double‐blind, placebo‐controlled dose‐escalation trial of merimepodib (VX‐497) and interferon‐alpha in previously untreated patients with chronic hepatitis C. Antivir. Ther. 10, 635–643 (2005). [PubMed] [Google Scholar]
- 117. James, J.S. Nelfinavir (Viracept) approved: fourth protease inhibitor available. AIDS Treat. News 267, 1–2 (1997). [PubMed] [Google Scholar]
- 118. Chen, W. , Mook, R.A. , Premont, R.T. & Wang, J. Niclosamide: beyond an antihelminthic drug. Cell. Signal. 41, 89–96 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Anderson, V.R. & Curran, M.P. Nitazoxanide. Drugs 67, 1947–1967 (2007). [DOI] [PubMed] [Google Scholar]
- 120. Porche, D.J. Ritonavir (Norvir). J. Assoc. Nurses AIDS Care 8, 81–83 (1997). [DOI] [PubMed] [Google Scholar]
- 121. Miller, K.D. , Lobel, H.O. , Satriale, R.F. , Kuritsky, J.N. , Stern, R. & Campbell, C.C. Severe cutaneous reactions among American travelers using pyrimethamine‐sulfadoxine (Fansidar) for malaria prophylaxis. Am. J. Trop. Med. Hyg. 35, 451–458 (1986). [DOI] [PubMed] [Google Scholar]
- 122. Orman, J.S. & Perry, C.M. Tipranavir: a review of its use in the management of HIV infection. Drugs 68, 1435–1463 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Table S1
Table S2
Supplementary Material
Supplementary Material
Supplementary Material


