Abstract
Respiratory syncytial virus (RSV) is the most important cause of respiratory tract illness especially in young infants that develop severe disease requiring hospitalization, and accounting for 74,000–126,000 admissions in the United States (Rezaee et al., 2017; Resch, 2017). Observations of neonatal and infant T cells suggest that they may express different immune markers compared to T-cells from older children. Flow cytometry analysis of cellular responses using “conventional” anti-viral markers (IL2, IFN-γ, TNF, IL10 and IL4) upon RSV-peptide stimulation detected an overall low RSV response in peripheral blood. Therefore we sought an unbiased approach to identify RSV-specific immune markers using RNA-sequencing upon stimulation of infant PBMCs with overlapping peptides representing RSV antigens. To understand the cellular response using transcriptional signatures, transcription factors and cell-type specific signatures were used to investigate breadth of response across peptides. Unexpected from the ICS data, M peptide induced a response equivalent to the F-peptide and was characterized by activation of GATA2, 3, STAT3 and IRF1. This along with upregulation of several unconventional T cell signatures was only observed upon M-peptide stimulation. Moreover, signatures of natural RSV infections were identified from the data available in the public domain to investigate similarities between transcriptional signatures from PBMCs and upon peptide stimulation. This analysis also suggested activation of T cell response upon M-peptide stimulation. Hence, based on transcriptional response, markers were chosen to validate the role of M-peptide in activation of T cells. Indeed, CD4+CXCL9+ cells were identified upon M-peptide stimulation by flow cytometry. Future work using additional markers identified in this study could reveal additional unconventional T cells responding to RSV infections in infants. In conclusion, T cell responses to RSV in infants may not follow the canonical Th1/Th2 patterns of effector responses but include additional functions that may be unique to the neonatal period and correlate with clinical outcomes.
Keywords: RSV, PBMC, Transcriptomics, Peptide stimulation
1. Introduction
Respiratory syncytial virus (RSV), the most important cause of respiratory tract illness in infants and young children, infects 50%–70% of infants during the first year of life [1]. Although most infections are relatively mild, 1%–3% of infected infants require hospitalization, accounting for 74,000–126,000 admissions of infants aged < 1 year annually in the United States [2], [3]. Additionally, RSV-related emergency department visits for infants aged ≤ 1 year of age range from 39 to 69 per 1000, and RSV-related office visits are 3 times as many [4].
Host immune responses are thought to influence disease manifestations during primary RSV infection [5], [6], [7], [8]. Higher levels of maternally derived transplacental antibodies have been associated with reduced illness severity and an increase in the age of acquisition of primary infection [9]. Prophylactic treatment of high-risk infants with RSV-specific polyclonal or monoclonal antibody can reduce hospitalization by half [10], [11]. RSV is generally non-cytopathic, and virus clearance is considered to be primarily mediated by cytotoxic CD8+ T cells [12], [13], [14]. However, animal models suggest that an exuberant CD8+ T cell response is associated with increases in pathogenicity and severity of disease [15]. Some studies in both animal models and in human infants have also associated a Th2 skewed CD4+ T cell response during primary infection with greater disease severity [16], [17].
T cell specificity and function against RSV can be measured in a variety of ways including proliferations assays, Enzyme Linked Immunosorbent Assay (ELISA), Enzyme Linked Immune Spot Assay (ELISpot), and intracellular cytokine staining followed by flow cytometry [18]. Typically, the T cells in these assays are stimulated with the virus or sets of overlapping peptides representing the individual proteins. The number of functions that can be measured in any one of these assays is limited, with flow cytometry and bead-based ELISA assays providing the most potential variety of analytes that can be measured. The infant immune system, and the T cells in particular, exhibit functions different than that of an older child or an adult [19]. For example, both CD4+ and CD8+ T cells in the neonate produce Interleukin 8 (IL8) as a “signature” cytokine [20]. Among these T cells, the frequency of CD31+ cells at birth is associated with reduced frequency and severity of respiratory infection over the first year of life [20]. Measuring the canonical Th1/Th2 effector functions such as IL2, IL4, IFN-γ, and TNF-α in the neonate would thus potentially miss the intermediate cellular phenotypes that could lead to different functionality. Hence an unbiased RNA sequencing (RNAseq) approach was utilized in this study for a comprehensive analysis of all possible effector functions. The comparison of gene-signatures of natural infection to the response to RSV-peptides reveals the role of M-peptides in generating a physiologically relevant response. The novel markers identified in the study were validated by flow cytometry revealing induction of CD4+CXCL9+ cells upon the M-peptide stimulation. Additional follow-up studies should reveal the function of CD4+CXCL9+ cells in RSV-pathogenesis and may aid in the development of novel approaches for vaccination.
2. Methods
Subjects and Assays: The Research Subject Review Boards of the University of Rochester Medical Center (URMC) and Rochester General Hospital (RGH) approved the study and a parent provided written informed consent was obtained for each subject. RSV-infected infants <10 months of age and undergoing their first RSV infection were selected for this analysis as part of the Assessing and Predicting Infant RSV Effects and Severity (AsPIRES) study of RSV pathogenesis [21]. Peripheral blood mononuclear cell (PBMC) samples were collected at three visits: v11 (acute), v12 (12–15 days after illness onset), and v13 (20–35 days after illness onset) from 75 infants, of who 28 had mild and 47 severe illness based on the global respiratory severity scores (GRSS [22]) (Table 1). An initial flow cytometry experiment was conducted on these PBMC samples for quantifying peptide-specific cytokine secretion in both CD4+ T and CD8+ T lymphocytes. RNAseq transcriptomics analysis was performed on an independent group of 10 subjects with mild and severe disease (Table 2). Finally, a follow-up flow cytometry experiment on a limited number of subjects was conducted to confirm some of the RNAseq results.
Table 1.
Demographic characteristics of infants participated in immune phenotyping study.
| Clinical variable | Mild disease (n = 28) | Severe disease (n = 47) | P value* |
|---|---|---|---|
| Gestational age, wk; mean ± SE | 39.07 ± 0.28 | 38.63 ± 0.20 | 0.22 |
| Birth weight, kg; mean ± SE | 3.42 ± 0.14 | 3.36 ± 0.09 | 0.71 |
| Gender, male; no. (%) | 14 (50) | 21 (45) | 0.81 |
| Race; no. (%) | |||
| Caucasian only | 17 (61) | 30 (64) | |
| African American only | 7 (25) | 9 (19) | 0.83 |
| Other | 4 (14) | 8 (17) | |
| Age, mo.; mean ± SE | 2.70 ± 0.56 | 2.68 ± 0.31 | 0.97 |
| Days of illness; mean ± SE | |||
| Exposure to tobacco smoke; no. (%) | 4 (14) | 18 (38) | 0.04 |
| RSV group A, B; no. each group | 15, 12 | 30, 16 | 0.46 |
P-values calculated by t-test for all except for male, race, smoking and RSV strain fisher’s exact test was used.
Table 2.
Demographic characteristics of infants participated in RNA-sequencing study.
| Clinical variable | Mild disease (n = 5) | Severe disease (n = 5) | P value* |
|---|---|---|---|
| Gestational age, wk; mean ± SE | 39.20 ± 0.66 | 37.80 ± 0.80 | 0.22 |
| Birth weight, kg; mean ± SE | 3.33 ± 0.11 | 3.42 ± 0.13 | 0.60 |
| Gender, male; no. (%) | 2 (40) | 2 (40) | 0.81 |
| Race; no. (%) | |||
| Caucasian only | 3 (60) | 4 (80) | |
| African American only | 2 (40) | 1 (20) | 1.00 |
| Other | 0 (0) | 0 (0) | |
| Age, mo.; mean ± SE | 3.34 ± 1.16 | 3.18 ± 1.44 | 0.93 |
| Days of illness; mean ± SE | |||
| Exposure to tobacco smoke; no. (%) | 3 (60) | 2 (40) | 0.36 |
| RSV group A, B; no. each group | 2, 3 | 4, 1 | 0.52 |
P-values calculated by t-test for all except for male, race, smoking and RSV strain fisher’s exact test was used.
Specimen Collection: RSV-infected infants were evaluated by two members of the study team (a physician and a project nurse). Demographic data, illness symptoms, findings on physical examination, and results of standard laboratory and chest radiograph results, when available, were recorded at each visit. At each visit 2–3 ml of heparinized blood was collected and processed within 4 h of collection. PBMCs were isolated by centrifugation on a Ficoll-Hypaque gradient (Sigma-Aldrich, St. Louis, MO) at 1500 × g for 30 min at 20 °C, and the buffy coat layer washed 3 times by resuspension in phosphate buffered saline and centrifugation at 300 × g for 10 min at 4 °C. Viable cell numbers were assessed by trypan blue exclusion and cells were suspended in 90% fetal calf serum plus 10% DMSO, frozen, and stored in liquid nitrogen.
In vitro stimulation: For the initial flow cytometry and RNAseq assays, cryopreserved PBMC were rapidly thawed in a 37 °C water bath and washed two times (300 × g for 10 min) in complete (10% fetal calf serum) RPMI (Cellgro, Manassas, VA) supplemented with 1x penicillin/streptomycin (GIBCO, Carlsbad, CA), and 10 µg/ml DNaseI (Sigma-Aldrich, St. Louis, MO) before an overnight rest at 37 °C in 5% CO2. Following the rest, cell viability was tested by trypan blue dye exclusion, and samples with > 80% viability were plated into 96-well V-bottom plates (BD, Franklin Lakes, NJ) at 1–2 × 106 cells/well. Cells were stimulated with the following antigens: peptide (18-mers overlapping by 3) pools representing the RSV full-length F, G, M and N proteins (stock in DMSO; final dilution in complete RPMI); The number of peptides in each pool were: F-39, G-20, M-17, and N-28. The optimal concentration (1, 5, or 10 µg/ml) for each peptide pool was empirically determined using healthy adult PBMC that demonstrated robust responses to RSV. DMSO was used as the negative control, and Staphylococcal enterotoxin B (SEB) (1 µg/ml, Sigma- Aldrich, St. Louis, MO) was used as the positive control. Cells were cultured for 2 h at which point GolgiPlug (BD Biosciences) and 2 μM monensin (Sigma) were added; cells were further cultured for an additional 8 h. Following stimulation, cells were either processed for flow cytometry or in the case of the RNAseq assay, immediately lysed and stored in RNA-stabilization buffer (RNA Protect).
For the follow-up flow cytometry assay, cell stimulation was performed as above with the following exceptions: peptide pools G and N were omitted; RSV whole-virus (from cultured supernatant) was included.
Flow cytometry: For the initial flow cytometry assay (Fig. 1; Supplemental Table 1a, Panel 1), stimulated cells were washed once in PBS, stained for 30 min. on ice with an amine-reactive dye (LIVE/DEAD, Invitrogen), washed 1x in staining buffer (PBS + 2% fetal calf serum; filtered), and stained for 30 min. on ice with the following surface antibodies: CD4, CD8α, CD45RA, and CD25. Following the surface stain, cells were washed 2x in staining buffer, fixed-permeabilized in Fixation/Permeabilization buffer (eBioscience; ‘FOXP3 kit’) for 30 min. on ice, washed 2x in permeabilization buffer (eBioscience) and stained for 30 min. on ice with the following intracellular/nuclear antibodies: CD3, CD69, FOXP3, IFNɣ, IL-2, IL-4, IL-10 (biotin), IL17A, Ki67, and TNFα. Following the intracellular/nuclear stain, cells were washed 2x in permeabilization buffer, stained for 30 min. on ice with a streptavidin-secondary, washed 2x in permeabilization buffer, 1x in staining buffer, and finally resuspended in staining buffer and stored at 4 °C until acquisition on a BD LSR II.
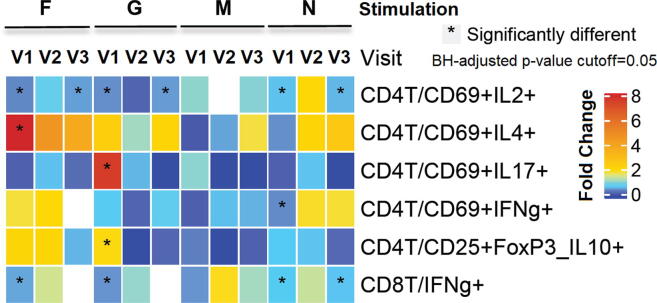
Fig. 1.
Peptide induced changes in composition of cell populations: Heatmap showing fold change of subsets of CD4+ and CD8+ T cells induced by RSV-peptide re-stimulation (first horizontal bar) compared to the DMSO treatment across the three visits: V1, V2, and V3. The proportions of the cell subsets were calculated using the number of CD4+ or CD8+ T cells as parents. The colors depict fold difference calculated by dividing the proportion of the cell subsets observed upon stimulation with the proportion observed upon DMSO stimulation. Note that the values <1 correspond to a smaller value than DMSO. Fold difference that is between 0.9 and 1.1 is in white, indicating a minor change from DMSO. A generalized linear mixed effect regression analysis model is applied to generate p-values after Benjamini-Hochberg (BH) correction to indicate the significance of the difference with BH-adjusted p-value cutoff = 0.05. * indicates a significant change with p-value < 0.05.
For the follow-up flow cytometry assay (Fig. 5; Supplemental Table1b, Panel 2), the staining was performed as above with the following exceptions: fixation and permeabilization buffers were substituted with Cytofix/Cytoperm and 1x PermWash buffers (BD Biosciences; ‘Cytofix/Cytoperm’ kit); surface antibodies included: CD4, CD8α, CD14, CD31, CD196, CD45RA, and TCR γ/δ; intracellular antibodies included: CD3, CD69, CXCL9, IFNɣ, IL-6, IL-8, IL-10, and Ki67.
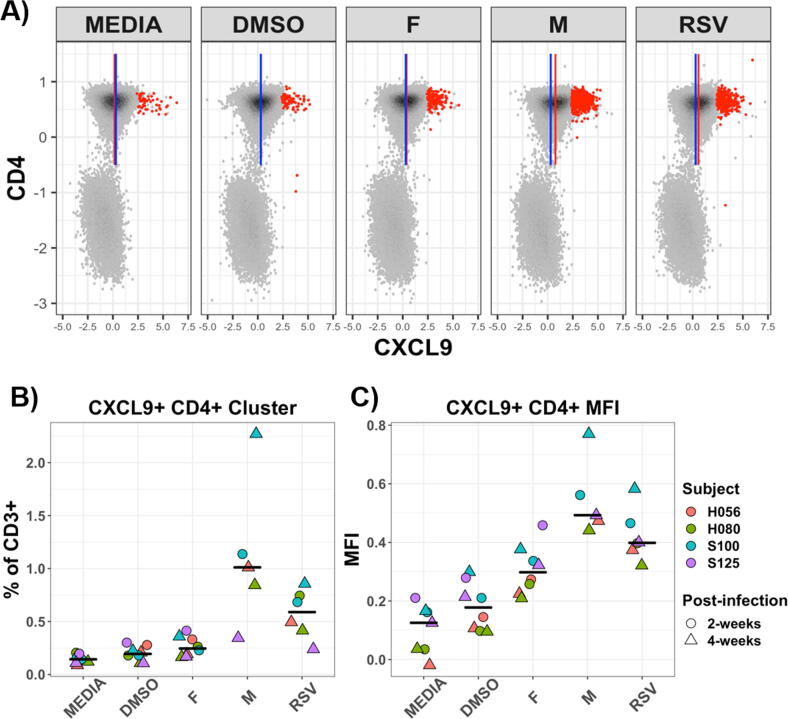
Fig. 5.
CXCL9 expression following stimulation with RSV peptide or whole virus: Four subjects (matched study visit 12 and 13; 2-weeks and 4-weeks post-RSV infection, respectively) PBMC were re-stimulated with RSV specific peptides or whole virus. (A) Representative bivariate plots (single subject; pre-gated on CD3+ T cells) showing expression of CXCL9 and CD4 as measured by flow cytometry (arcsinh transformed/scaled; 50,000 events each). Blue line indicates CD4+CXCL9 median fluorescence intensity (MFI) for the DMSO control; red line indicates MFI for the respectively labeled stimulation condition. Red dot overlays indicate meta-clustered cell events as assigned by FlowSOM (clustering algorithm). Frequency of the CXCL9+CD4+ cluster (B) and MFI of CXCL9+ on bulk CD4+ (C) are shown; black line indicates respective median value. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Identification of Predefined Cell Populations: The flow cytometry experiment using Panel 1 generated 656 FCS3.0 files. To improve reproducibility of the analysis and reduce human bias in the manual gating procedure, a computational analysis approach called DAFi [23] was applied to identify the following 10 predefined cell populations: 9 subsets in CD4+ T lymphocytes: CD69+IFNg+, CD69+IL2+, CD69+IL4+, CD69+IL10+, CD69+IL17+, CD69+Ki67+, CD69+TNFa+, CD69+FoxP3+, CD69+FoxP3+IL10+, and one subset in CD8+ T lymphocytes: IFNɣ+. Details of the DAFi analysis of Panel 1 are provided in the Supplementary Text 1. A generalized linear mixed model (GLMM) was applied to test the effects of the 7 stimulations (DMSO, N1, N5, F1, F5, G5, and M10), three visits (v11, v12, and v13), and two conditions (mild and severe RSV infection) on the cell population percentages identified by the DAFi method. Specifically, we applied the approach in [24], which used binomial distribution with GLMM taking the cell count relative to the parent population (to represent proportions of cell populations) as the dependent variable. A multiple comparison correction across the cell populations was done using Benjamini-Hochberg correction to control the false discovery rate (FDR) at 0.05 level. Details of the statistical comparison can be found in the Supplemental Methods.
The Panel 2 flow cytometry experiment generated 34 FCS files (4 subjects, 2 visits (V12, V13 – some visits/stimulations missing due to limited subjects/volumes), and 5 stimulations. Because of the relatively small number of samples, we applied the unsupervised FlowSOM [25] analysis to identify the CD4+CXCL9+ cells and compare them across samples. The advantages of FlowSOM include the use of a self-organizing map to visualize the high-dimensional flow data for identification of the cell populations. It supports unsupervised meta-clustering for cross-sample comparison that suits well to the size of the Panel 2 FCM data. Default parameters of the FlowSOM algorithm were used in our analysis.
Library Construction and Sequencing: RNA sequencing (RNA-seq) was performed as previously described, starting with 1 ng of RNA, and using the SMARter Ultra Low amplification kit (Clontech, Mountain View, CA) [26]. Libraries were constructed using the NexteraXT library kit (Illumina, San Diego, CA) and sequenced on the Illumina HiSeq2500 to generate ∼ 20 million 100-bp single end reads per sample. Pre-analysis and data processing was performed as previously described [26].
RNAseq analysis: The sequencing data was normalized for sequencing depth and gene length using Fragments Per Kilobase of transcript per Million (FPKM) transformation. 10 samples with low mean correlations (<0.5) with other samples were removed from subsequent analyses. A non-specific filtering strategy was used to select 12,149 genes with relatively large and meaningful expression values. We then applied a linear mixed effects regression analysis to test the effects of 5 stimulations, three visits, and two conditions. We first tested whether there are overall expression differences by ANOVA F-test for each gene. Among those genes with significant overall difference, we performed post hoc analyses to test for the specific effect of stimulations, visits, and conditions. Differentially expressed genes were defined by application of Benjamini-Hochberg multiple testing procedure to control FDR at 0.05 and absolute value of fold change greater than one for each stimulation compared to DMSO. Gene-sets (GSs), pathways from KEGG and MSiGDB were used in enrichment analysis performed using Qgen function from QuSAGE [27]. The multivariate pathway analysis was performed using GRSS and visit numbers as covariates. Transcription factor analysis was performed using binding sites obtained from JASPER as described previously [28]. Hypergeometric test was performed to identify enriched binding sites [29], [30].
Meta-analysis of transcriptomic data available in public domain: The raw data from GSE34205 [31] and GSE69606 [32] series were downloaded and quantile normalized using limma package [33] and were combined as described previously [34]. GSE34205 transcriptomic data was collected from 51 RSV infected and 10 healthy infants [31]. GSE69606 transcriptomic data was collected from 9 mild, 9 moderate and 8 severely RSV infected infants [32]. GSs were identified using Fuzzy-C-Means algorithm as described previously [35]. Ward’s minimum variance method was used to estimate the initial centers for Fuzzy-C-Means which produced stable and consistent clusters. Ward’s method (based on analysis of variance) minimized the total within-cluster variance and maximized between-clusters variance. Cluster membership was evaluated by calculating the total sum of squared deviations from the mean of a cluster. At the initial step, all clusters were singletons (each cluster containing a single gene), which were merged in each next step so that the merging contributed least to the variance criterion. This distance measure called the Ward distance was defined by:
where a and b denote two specific clusters, and denote the number of data points in the two clusters. and denote the cluster centroids and ‖‖ is the Euclidean norm. Clustering was performed using Cluster package in R (ref). 13 GSs were robustly inferred. The smallest GS had 81 genes and largest had 1045 genes.
Data availability: All data including phenotypic data is available in dbGAP under phs001201 accession number with links to raw flow cytometry (fcs) files available at IMMPORT and RNA-sequencing files available at Sequence Read Archive (SRA).
3. Results
3.1. Cytokine-secreting CD4+ and CD8+ cellular response to RSV peptide stimulations measured by flow cytometry
Both the quantitative DAFi analysis and visual examination of the dot plots of the flow cytometry data from Panel 1 show very low frequency responses for the cytokines IL2, IL4, IL10, IL17α and IFN-γ in both CD4+ and CD8+ T cells. Also, the M proteins did not seem to generate detectable responses for the cytokines tested. However, as Fig. 1 shows, we found significant fold change of proportions of cell populations from DMSO for a) F peptide stimulated CD4+CD69+ IL2+ and IL4+, as well as CD8+IFNγ+; and b) G peptide stimulated CD4+CD69+IL2+, IL17+, and CD4+CD25+FoxP3+IL10+, as well as CD8+IFN γ+ populations. Overall, the result from the flow cytometry data analysis suggests that F and G peptide stimulation induces T cell proliferation and activation. However, most of the responses in peripheral blood were very low. Some of our other studies in infants suggest unconventional T cell cytokine responses (i.e. IL8) [20]. Therefore, to discover novel immune markers and model comprehensive responses generated by peptide stimulations, we performed an unbiased analysis by RNA-sequencing following RSV peptide stimulation.
3.2. Gene expression changes upon stimulation of infant PBMCs with RSV peptides
RNA-sequencing was performed on up to 30 samples from 10 subjects resulting in 138 different conditions. Re-stimulation with peptides revealed large variation in response to F-peptides, and the least variation across subjects in response to M-peptide (Fig. 4A). The linear mixed effect model was developed on a filtered set of 12,149 genes to find differentially expressed genes upon peptide re-stimulations. M peptide re-stimulation led to the most number of differentially expressed genes (416 in total, with 81 upregulated and 335 downregulated), followed by F peptide re-stimulation (180 in total with 77 upregulated and 103 downregulated) and N peptide (6 in total with 2 upregulated and 6 downregulated) (Supplementary Table 2). Since we were interested in evaluating cellular responses upon peptide stimulation, transcription factor analysis (Fig. 4B) of differentially expressed genes was performed. T helper cell associated regulators were significantly enriched upon M peptide stimulation, namely GATA3, GATA2, IRF1, and STAT3. GATA3 and 2 were also involved in F-peptide dependent upregulation. Thus, transcriptional signatures suggest an activation of T helper cells in response to M peptide re-stimulation.
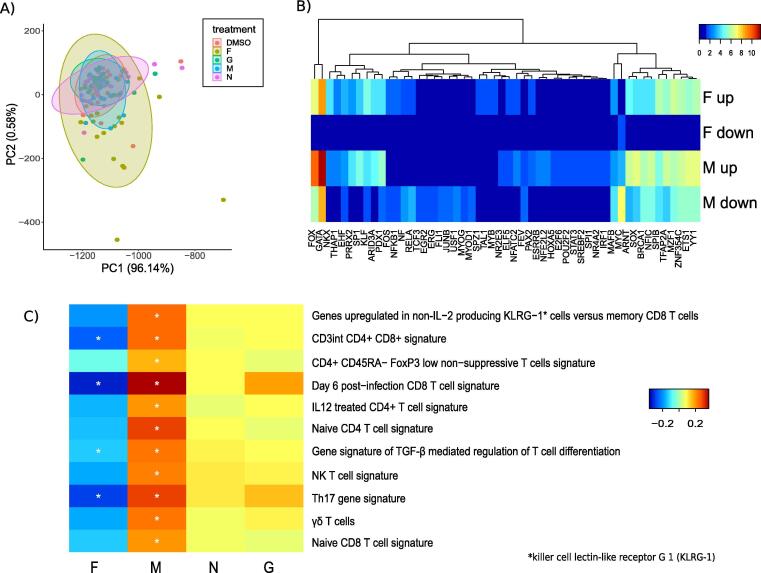
Fig. 4.
Transcriptional response of PBMCs to RSV peptide stimulation: A) Principle Component Analysis (PCA) representing each sample upon re-stimulation with DMSO, F, M, N and G, B) Enrichment of transcription factor binding sites in differentially expressed genes upon re-stimulation with F and M peptide. The colors represent –log10(P-value), calculated by hypergeometric test, C) Multivariate enrichment analysis of gene signatures of T cell subtypes performed using Qgen using visit number and GRSS as covariates. The colors blue to red indicate activities ranging from low to high. Stars indicate significance * p < 0.05. The names of the transcription factor binding sites in B and gene signatures of T cell subtypes in C have been modified for ease of understanding. The original names corresponding to those names have been provided in Supplementary Table 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To investigate immune cell subsets induced by the peptide re-stimulations, we performed enrichment analysis using T cell subset specific signatures from MSigDB [36]. These gene signatures define characteristic T cell responses upon treatment with various cytokines and stimulation conditions, (Fig. 4C). In the multivariate analysis, visit and severity score were used as covariates to test the response to each peptide re-stimulation compared to DMSO, whereas in the univariate models gene-sets responsive to each peptide re-stimulation were evaluated at different visits. F peptide re-stimulation produced significant downregulation of all T cell gene signatures. Interestingly, in consensus with the observed enrichment of the transcription factors, M peptide re-stimulation significantly upregulated signatures of naïve and effector CD4+ and CD8+ subsets. Additionally, enrichment of gene signatures associated with CD4+CD8+ double positive thymocytes, possibly reflect that many neonatal T cells are recent thymic emigrants [20]. TGF-β mediated T cell differentiation and FOXP3low non-suppressive CD4+ T cells were also enriched. Thus, the results suggest elicitation of effector CD8+ and CD4+ response upon M-peptide re-stimulation.
3.3. M peptide re-stimulation upregulates a physiologically relevant response
To compare transcriptional responses upon peptide re-stimulation with the natural infant immune response upon infection, we performed a meta-analysis of publicly available gene expression measurements from PBMCs of 77 RSV infected infants with varying severity and 27 healthy infants in the age range of 1 to 10 months. The genes were grouped into 13 RSV response clusters using robust fuzzy-C-means clustering as described before (Fig. 2A) [35]. The clusters grouped the functionally relevant genes, for example cluster-4 contained anti-viral signaling and cluster-1 grouped the genes involved in T cell signaling. Evaluation of cell specific gene signatures revealed that cluster 12 was enriched in naïve and effector CD4+ and CD8+ subset specific gene signatures.
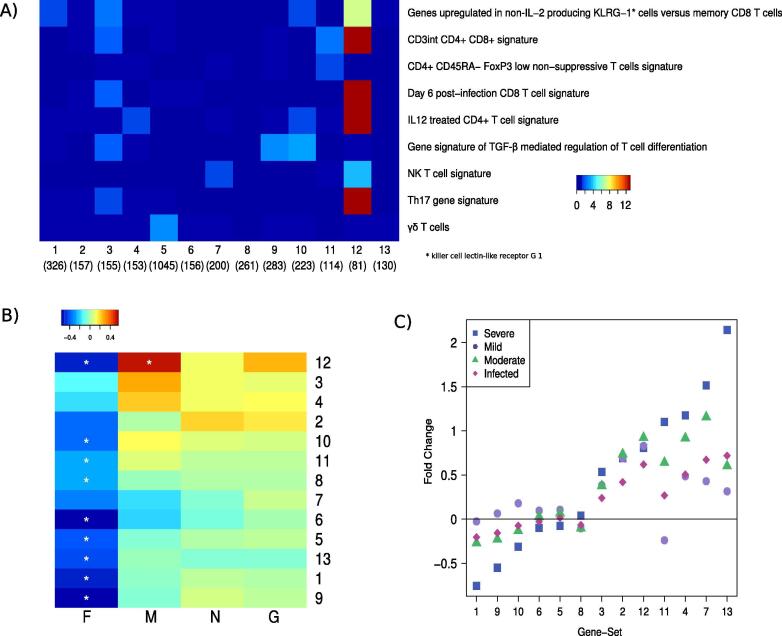
Fig. 2.
Evaluation of activities of infant RSV gene signatures in RSV-peptide stimulation study: A) Enrichment of T cell associated gene-signatures (y-axis) in 13 RSV response clusters (x-axis) identified using previously published transcriptomic data measuring response to RSV infections in infant PBMCs [22], [23]. Colors represent –log10(p-value) calculated by hypergeometric test, blue represents < 1.3 (non-significant), B) Activities of the 13 clusters upon re-stimulation of PBMCs with RSV-peptides (x-axis) and C) in transcriptomic data obtained from [22], [23] using Qgen where visit number and GRSS are covariates. These previously published studies stratified the subjects into mild (red circles, data from 9 subjects), moderate (green triangles, data from 9 subjects), severe (blue diamonds, data from 8 subjects) and infected (black rectangles, data from 51 subjects). The names of gene signatures of T cell subtypes in A have been modified for ease of understanding. The original names corresponding to those names have been provided in Supplementary Table 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We then asked whether any of the 13 functionally related modules of infant RSV response are elicited upon RSV-peptide re-stimulation. F peptide downregulated all of the 13 clusters while M peptide upregulated cluster-12 in multivariate analysis and both clusters-12 and 3 in the univariate analysis (Fig. 2B). Notably, cluster-12 was upregulated in previously published data from mild, moderate and severely infected infants [31], [32] (Fig. 2C). Several genes from cluster-12 such as SLC1A4, a protein involved in transport of cysteine/cysteine across membranes, are strongly activated during T cell activation and proliferation [37]. Other genes such as CD59 and BIRC5 (downstream target of STAT3) are also linked to CD4+ T cells [38], [39]. To further test if genes in cluster-12 are related to CD4+ T cells, we evaluated transcriptomic measurements of CD4+ T cells from RSV infected infants using public data sets (“infected” Fig. 2C) [26]. Interestingly, 7 genes from cluster-12 were overlapping with CD4+ genes associated with severe response (worst SaO2 response) indicating a CD4 related response to M peptide re-stimulation. These results suggest cellular proliferation and activation of CD4+ T cells in response to M antigen re-stimulation.
3.4. Markers of infant immune responses
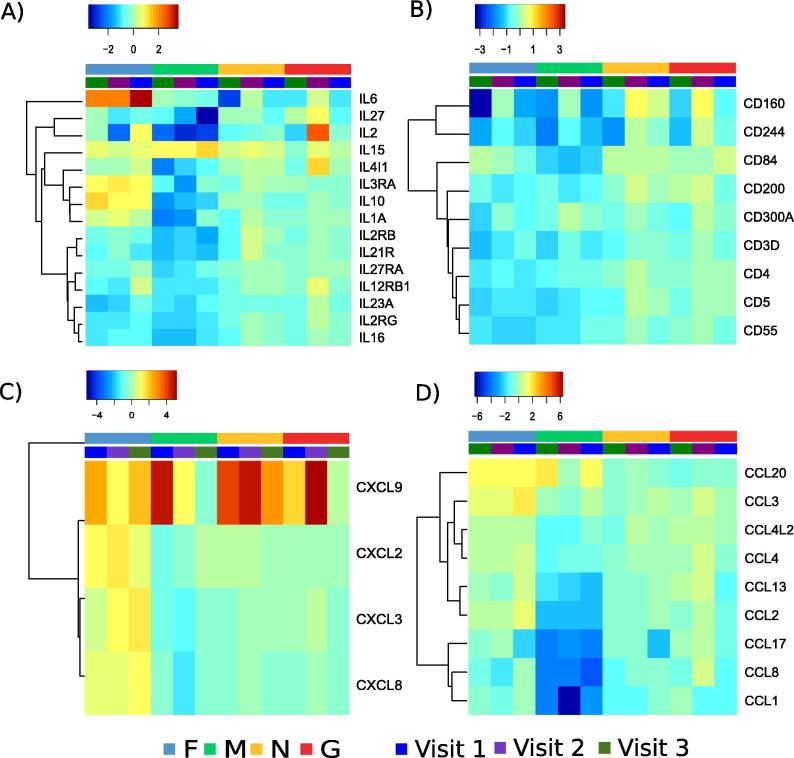
To further characterize the cell-types responding to peptide re-stimulation, genes expressing cluster of differentiation (CD) markers, interleukins, chemokine receptors and chemokines were analyzed. F-peptides stimulated expression of IL6, IL3RA, IL10 and IL1A (Fig. 3A), CCL3, CXCL8, CXCL3 and CXCL2 (Fig. 3C). CXCL8 (IL8) is a signature cytokine of neonatal T cells [27]. CXCL9 is a T cell chemoattractant induced by IFNγ which was upregulated by all the peptide re-stimulations, especially at the early visits (Fig. 3C). CCL20 was upregulated by both F and M peptide. Most of the CD genes were downregulated upon peptide re-stimulation, but CD160, an indicator of cytolytic activity by NK and CD8 T cells was slightly upregulated upon N and G peptide re-stimulation. CD8 and CD69 were slightly increased at the convalescent visit. F peptide re-stimulation led to increased levels of IL6, IL3RA, IL10 and IL1A (Fig. 3). M-peptide re-stimulation increased expression of IL-15 reaching its highest level on visit 3. G peptide increases IL2 levels at the convalescent visit as observed by original flow experiment. Thus, the expression of surface markers along with previous results indicating proliferation and differentiation of CD4 and CD8 cells in response to M peptide suggest new markers that could facilitate characterization of T cells in response to RSV. To test whether the T cells expressing any of the markers identified above exist in vivo we performed intracellular staining using TCRγ, CD8A, CD196, CXCL9 and CD4. Fig. 5 shows that CD4+CXCL9+ cells were significantly enriched upon M peptide and RSV re-stimulation. Thus, transcriptional profiling of peptide-stimulated PBMCs have revealed novel markers for investigating infant immune response to RSV infection. We did not see strong induction of Th2-associated cytokines, instead observing a mostly Th1-associated expression profile that fits with the CXCL9 expression.
Fig. 3.
Immune markers identified by transcriptomics study: All interleukins, chemokines and cluster differentiation genes were examined upon four re-stimulations by RSV peptides (F in blue, M in green, N in yellow and G in red). Colors blue to red represent low to high fold change compared to DMSO treatment. The visits 1, 2 and 3 are represented by blue, purple and green colors. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Based on results using small animal models of RSV infection it has been proposed that the nature of the immune response is pivotal in determining the outcome of the infection [40], [41]. However, a full understanding of the cellular immune mechanisms as they relate to both successful recovery from infection and potentially severe disease in infants has been elusive. This is especially relevant for normal, full-term, healthy infants, the population that comprises the majority of infants brought to medical attention for RSV disease. Most studies investigating RSV disease pathogenesis have measured the presence and levels of various inflammatory proteins, such as cytokines and chemokines in blood or respiratory secretions, or their in vitro production by peripheral blood mononuclear cells, often with disparate results [35]. We recently employed an unbiased transcriptomic approach using RNAseq to study in vivo peripheral CD4 T cell gene expression responses in young infants during active RSV infection [26]. Those results suggested that CD4 T cells from hospitalized infants with severe illness had more evident gene expression patterns consistent with a Th2 bias when compared to mildly ill outpatients, though the majority of those cells were naïve T cells and the expression observed is hypothesized to be an imprint on those cells likely coming from localized cytokine production in the airways or lymphatic tissues [26].
Here we examined the T cell response in infants with primary RSV disease using peptide re-stimulation with flow cytometry and identified an activated IL2+CD4+ T cell response to F stimulation and IL17+ and IL10+CD4+ T cell response to G peptide re-stimulation. However, these responses were limited and overall low. The flow cytometry investigation is dependent on the use of specific pre-chosen markers, and there were concerns that we may have missed key cytokine responses. IL8 (CXCL8) for example is a signature cytokine produced by CD31+ neonatal T cells [20]. Hence we utilized unbiased transcriptional profiling to study the responses induced by peptide re-stimulation in RSV infected infants.
Transcriptional profiling revealed that M peptide re-stimulations led to activation of naïve and effector T cell subset specific signatures and transcription factors involved in differentiation of T cell subtypes. Particularly, IRF1 and STAT3 enriched upon M peptide stimulation have been shown to regulate T cell lineages [42]. STAT3 also has potent anti-inflammatory effects and regulates critical cellular processes such as cell growth, apoptosis and transcription of inflammatory genes. Recent studies suggest that IL6 driven ability of CD4+ T cells to promote B cell activation is dependent on STAT3 [42]. STAT3 is also involved in regulating cytotoxic function of CD4+ and CD8+ [43]. Previous studies have reported deficits in T cell function in IRF-1-/- mice including low levels of CD8+ T cells in peripheral lymphoid tissue [44], [45]. GATA3 enriched in F and M response, is a transcriptional activator that is required for T-helper 2 (Th2) differentiation. It also plays a role in differentiation of Th9 cells, among other functions in T cell developmental stages [46]. Thus evaluation of transcriptional responses suggested that M-peptide might be responsible for inducing T cell response and that it is different from a “conventional” anti-viral response characterized by (IL2, IFN-γ, TNF, IL10 and IL4). Surprisingly, cytokines, cluster differentiation genes and chemokines were not very prominently expressed upon M-peptide stimulation as shown in Fig. 3. Although F-peptide upregulated markers of infant T cells such as CXCL8 and IL10, it led to downregulation of CD4 and CD8 subset-specific gene-sets. Thus, transcriptional analysis highlighted the proliferation of T cells in response to M peptide.
ICS data characterizing “conventional” anti-viral response measured by (IL2, IFN-γ, TNF, IL10 and IL4) corroborated with transcriptional observation of IL2 activation by G-peptide and IL4 activation by F peptide. While G peptide induced activation of surface markers transcriptional response was minimal. Further ICS analysis was designed to test several makers including CCL20, CXCL9 and IL15. Unfortunately, only antibodies against CXCL9 worked in our experiment validating activation of CD4+CXCL9+ cells upon M-peptide stimulation. CXCL9 is an IFN γ–inducible CD8+ and CD4+ T cell chemoattractant [47]. CXCL9 has also been shown to support CD4 T cell accumulation in lymph node and is thought to be an important benchmark for vaccine efficacy and perhaps immunotherapy [48]. While we did find weak signatures of CD8+IFNγ+ and CD4+IFNγ+ T cells in peripheral blood, it is more likely the CXCL9+ cells had received IFN γ signal in the lymphoid or infected tissues. The transcriptional analysis also suggest additional markers for future work. For example, IL15+ cells in response to M-peptide stimulation might play interesting role in neonatal immune memory [49]. Furthermore, transcriptional profiling also suggests response of F-specific CCL20+ and IL8+ T cells.
In conclusion, our study supports that infant immune response is different compared to what we know about involvement of immune cells in RSV pathogenesis. Novel immune markers defining cellular phenotypes are required to probe the infant immune response and we suggest combination of transcriptional profiling along with flow cytometry as a tool to investigate infant immune response.
CRediT authorship contribution statement
Juilee Thakar, David J. Topham and Edward E. Walsh conceptualized the study; Edward E. Walsh and Mary T. Caserta Supervised enrollment and sample collection; Thomas J. Mariani and David J. Topham Methodology for transcriptomics, Formal analysis; ChinYi Chu and Christopher Slaunwhite Software, Formal Analysis, Validation; Juilee Thakar, Xing Qiu and Lauren Benoodt Supervision of transcriptomics, Formal Analysis, Validation; Lauren Benoodt and Lu Wang performed Formal Analysis of the transcriptomic data; Xing Qiu and Richard H. Scheuermann Supervised flow cytometry data analysis; Xing Qiu, Ivan Chang, Aishwarya Mandava, David Roumanes and Nathan Laniewski performed flow cytometry Methodology; Juilee Thakar, David J. Topham and Xing Qiu performed writing-original draft, Writing-review and editing; David J. Topham and Ann R. Falsey obtained funding.
Funding source
This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, Division of Microbiology and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272201200005C.
Ethics statement
The study was approved by the Institutional Review Boards of the University of Rochester study id RSRB00042632. All the data were de-identified.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jvacx.2020.100065.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Genes responding to F and M peptide stimulation: Heatmaps showing fold change of the differentially expressed genes upon F (A) and M (B) peptide stimulation compared to DMSO stimulation. The color from blue to red shows the z-score of the log2 fold change.
Voom normalized expression of CXCL9 and IFN-γ expression. A) Highlighting the difference between subjects with mild (black diamonds) and severe (red triangles) infections. B) Differences in expression colored based on stimulation with F (black circles), M (red squared), N (green diamonds), or G (blue triangles) peptides.
Flow panels.
Information on gene-sets.
Details of DAFi analysis.
References
- 1.Hall C.B. Respiratory syncytial virus and parainfluenza virus. New Engl J Med. 2001;344(25):1917–1928. doi: 10.1056/NEJM200106213442507. Epub 2001/06/23. doi: 10.1056/NEJM200106213442507. PubMed PMID: 11419430. [DOI] [PubMed] [Google Scholar]
- 2.Hall C.B., Weinberg G.A., Blumkin A.K., Edwards K.M., Staat M.A., Schultz A.F., Poehling K.A., Szilagyi P.G., Griffin M.R., Williams J.V., Zhu Y., Grijalva C.G., Prill M.M., Iwane M.K. Respiratory syncytial virus-associated hospitalizations among children less than 24 months of age. Pediatrics. 2013;132(2):e341–e348. doi: 10.1542/peds.2013-0303. Epub 2013/07/24. doi: 10.1542/peds.2013-0303. PubMed PMID: 23878043. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Hong, Thompson William W., Viboud Cecile G., Ringholz Corinne M., Cheng Po-Yung, Steiner Claudia, Abedi Glen R., Anderson Larry J., Brammer Lynnette, Shay David K. Hospitalizations associated with influenza and respiratory syncytial virus in the United States, 1993–2008. Clin Infect Dis. 2012;54(10):1427–1436. doi: 10.1093/cid/cis211. Epub 2012/04/13. doi: 10.1093/cid/cis211. PubMed PMID: 22495079; PubMed Central PMCID: PMCPMC3334364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall Caroline Breese, Weinberg Geoffrey A., Iwane Marika K., Blumkin Aaron K., Edwards Kathryn M., Staat Mary A., Auinger Peggy, Griffin Marie R., Poehling Katherine A., Erdman Dean, Grijalva Carlos G., Zhu Yuwei, Szilagyi Peter. The burden of respiratory syncytial virus infection in young children. N Engl J Med. 2009;360(6):588–598. doi: 10.1056/NEJMoa0804877. Epub 2009/02/07. doi: 10.1056/NEJMoa0804877. PubMed PMID: 19196675; PubMed Central PMCID: PMCPMC4829966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins PL, Fearns R, Graham BS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol. 2013;372:3-38. Epub 2013/12/24. doi: 10.1007/978-3-642-38919-1_1. PubMed PMID: 24362682; PubMed Central PMCID: PMCPMC4794264. [DOI] [PMC free article] [PubMed]
- 6.Mella Cesar, Suarez-Arrabal M.Carmen, Lopez Santiago, Stephens Julie, Fernandez Soledad, Hall Mark W., Ramilo Octavio, Mejias Asuncion. Innate immune dysfunction is associated with enhanced disease severity in infants with severe respiratory syncytial virus bronchiolitis. J Infect Dis. 2013;207(4):564–573. doi: 10.1093/infdis/jis721. Epub 2012/12/04. doi: 10.1093/infdis/jis721. PubMed PMID: 23204162; PubMed Central PMCID: PMCPMC3611762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Openshaw Peter J., Chiu Christopher. Protective and dysregulated T cell immunity in RSV infection. Curr Opin Virol. 2013;3(4):468–474. doi: 10.1016/j.coviro.2013.05.005. Epub 2013/06/29. doi: 10.1016/j.coviro.2013.05.005. PubMed PMID: 23806514; PubMed Central PMCID: PMCPMC4295022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell C.D., Unger S.A., Walton M., Schwarze J. The human immune response to respiratory syncytial virus infection. Clin Microbiol Rev. 2017;30(2):481–502. doi: 10.1128/CMR.00090-16. doi: 10.1128/CMR.00090-16. PubMed PMID: 28179378; PubMed Central PMCID: PMCPMC5355638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowe J.E., Jr. Influence of maternal antibodies on neonatal immunization against respiratory viruses. Clin Infect Dis. 2001;33(10):1720–1727. doi: 10.1086/322971. Epub 2001/10/12. doi: 10.1086/322971. PubMed PMID: 11595986. [DOI] [PubMed] [Google Scholar]
- 10.Resch Bernhard. Product review on the monoclonal antibody palivizumab for prevention of respiratory syncytial virus infection. Hum Vaccines Immunotherapeutics. 2017;13(9):2138–2149. doi: 10.1080/21645515.2017.1337614. doi: 10.1080/21645515.2017.1337614. PubMed PMID: 28605249; PubMed Central PMCID: PMCPMC5612471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezaee Fariba, Linfield Debra T., Harford Terri J., Piedimonte Giovanni. Ongoing developments in RSV prophylaxis: a clinician’s analysis. Curr Opin Virol. 2017;24:70–78. doi: 10.1016/j.coviro.2017.03.015. Epub 2017/05/14. doi: 10.1016/j.coviro.2017.03.015. PubMed PMID: 28500974; PubMed Central PMCID: PMCPMC5541395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson Teresa R., Hong Seokmann, Van Kaer Luc, Koezuka Yasuhiko, Graham Barney S. NK T cells contribute to expansion of CD8+ T cells and amplification of antiviral immune responses to respiratory syncytial virus. JVI. 2002;76(9):4294–4303. doi: 10.1128/JVI.76.9.4294-4303.2002. Epub 2002/04/05. doi: 10.1128/jvi.76.9.4294-4303.2002. PubMed PMID: 11932395; PubMed Central PMCID: PMCPMC155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostler T, Davidson W, Ehl S. Virus clearance and immunopathology by CD8(+) T cells during infection with respiratory syncytial virus are mediated by IFN-gamma. Eur J Immunol 2002;32(8):2117-23. Epub 2002/09/05. doi: 10.1002/1521-4141(200208)32:8<2117::AID-IMMU2117>3.0.CO;2-C. PubMed PMID: 12209623. [DOI] [PubMed]
- 14.Rutigliano John A., Johnson Teresa R., Hollinger Tonya N., Fischer Julie E., Aung Sandra, Graham Barney S. Treatment with anti-LFA-1 delays the CD8+ cytotoxic-T-lymphocyte response and viral clearance in mice with primary respiratory syncytial virus infection. JVI. 2004;78(6):3014–3023. doi: 10.1128/JVI.78.6.3014-3023.2004. Epub 2004/03/03. doi: 10.1128/jvi.78.6.3014-3023.2004. PubMed PMID: 14990720; PubMed Central PMCID: PMCPMC353752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh K.B., Teijaro J.R., Brock L.G., Fremgen D.M., Collins P.L., Rosen H., Oldstone M.B.A. Animal model of respiratory syncytial virus: CD8+ T cells cause a cytokine storm that is chemically tractable by sphingosine-1-phosphate 1 receptor agonist therapy. J Virol. 2014;88(11):6281–6293. doi: 10.1128/JVI.00464-14. Epub 2014/03/29. doi: 10.1128/JVI.00464-14. PubMed PMID: 24672024; PubMed Central PMCID: PMCPMC4093868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin Ling, Qiu Ke-zi, Hu Cheng-ping, Wu Guo-jun, Wang Li-li, Tan Yu-rong. Bronchial epithelial cells promote the differentiation of Th2 lymphocytes in airway microenvironment through jagged/notch-1 signaling after RSV infection. Int Arch Allergy Immunol. 2019;179(1):43–52. doi: 10.1159/000495581. Epub 2019/04/04. doi: 10.1159/000495581. PubMed PMID: 30943513. [DOI] [PubMed] [Google Scholar]
- 17.Shrestha B, You D, Saravia J, Siefker DT, Jaligama S, Lee GI, et al. IL-4Ralpha on dendritic cells in neonates and Th2 immunopathology in respiratory syncytial virus infection. J Leukoc Biol. 2017;102(1):153-61. Epub 2017/04/09. doi: 10.1189/jlb.4A1216-536R. PubMed PMID: 28389622; PubMed Central PMCID: PMCPMC5470835. [DOI] [PMC free article] [PubMed]
- 18.Karlsson A.C., Martin J.N., Younger S.R., Bredt B.M., Epling L., Ronquillo R. Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J Immunol Methods. 2003;283(1–2):141–153. doi: 10.1016/j.jim.2003.09.001. Epub 2003/12/09 PubMed PMID: 14659906. [DOI] [PubMed] [Google Scholar]
- 19.Heinonen Santtu, Rodriguez-Fernandez Rosa, Diaz Alejandro, Oliva Rodriguez-Pastor Silvia, Ramilo Octavio, Mejias Asuncion. Infant immune response to respiratory viral infections. Immunol Allergy Clinics North America. 2019;39(3):361–376. doi: 10.1016/j.iac.2019.03.005. Epub 2019/07/10. doi: 10.1016/j.iac.2019.03.005. PubMed PMID: 31284926; PubMed Central PMCID: PMCPMC6625527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheible Kristin M., Emo Jason, Laniewski Nathan, Baran Andrea M., Peterson Derick R., Holden-Wiltse Jeanne, Bandyopadhyay Sanjukta, Straw Andrew G., Huyck Heidie, Ashton John M., Tripi Kelly Schooping, Arul Karan, Werner Elizabeth, Scalise Tanya, Maffett Deanna, Caserta Mary, Ryan Rita M., Reynolds Anne Marie, Ren Clement L., Topham David J., Mariani Thomas J., Pryhuber Gloria S. T cell developmental arrest in former premature infants increases risk of respiratory morbidity later in infancy. JCI Insight. 2018;3(4) doi: 10.1172/jci.insight.96724. Epub 2018/02/23. doi: 10.1172/jci.insight.96724. PubMed PMID: 29467329; PubMed Central PMCID: PMCPMC5916253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh Edward E., Mariani Thomas J., Chu ChinYi, Grier Alex, Gill Steven R., Qiu Xing, Wang Lu, Holden-Wiltse Jeanne, Corbett Anthony, Thakar Juilee, Benoodt Lauren, McCall Matthew N., Topham David J., Falsey Ann R., Caserta Mary T. Aims, study design, and enrollment results from the assessing predictors of infant respiratory syncytial virus effects and severity study. JMIR Res Protoc. 2019;8(6):e12907. doi: 10.2196/12907. Epub 2019/06/15. doi: 10.2196/12907. PubMed PMID: 31199303; PubMed Central PMCID: PMCPMC6595944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caserta M.T., Qiu X., Tesini B., Wang L., Murphy A., Corbett A. Development of a global respiratory severity score for respiratory syncytial virus infection in infants. J Infect Dis. 2017;215(5):750–756. doi: 10.1093/infdis/jiw624. doi: 10.1093/infdis/jiw624. PubMed PMID: 28011907; PubMed Central PMCID: PMCPMC5388274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Alexandra J., Chang Ivan, Burel Julie G., Lindestam Arlehamn Cecilia S., Mandava Aishwarya, Weiskopf Daniela, Peters Bjoern, Sette Alessandro, Scheuermann Richard H., Qian Yu. DAFi: A directed recursive data filtering and clustering approach for improving and interpreting data clustering identification of cell populations from polychromatic flow cytometry data: DAFi. Cytometry. 2018;93(6):597–610. doi: 10.1002/cyto.a.23371. Epub 2018/04/18. doi: 10.1002/cyto.a.23371. PubMed PMID: 29665244; PubMed Central PMCID: PMCPMC6030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowicka Malgorzata, Krieg Carsten, Weber Lukas M., Hartmann Felix J., Guglietta Silvia, Becher Burkhard, Levesque Mitchell P., Robinson Mark D. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 2017;6:748. doi: 10.12688/f1000research.11622.1. doi: 10.12688/f1000research.11622.2. PubMed PMID: 28663787; PubMed Central PMCID: PMCPMC5473464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry Part A: J Int Soc Anal Cytology 2015;87(7):636-45. Epub 2015/01/13. doi: 10.1002/cyto.a.226PubMed PMID: 25573116. [DOI] [PubMed]
- 26.Mariani T.J., Qiu X., Chu C., Wang L., Thakar J., Holden-Wiltse J. Association of dynamic changes in the CD4 T-cell transcriptome with disease severity during primary respiratory syncytial virus infection in young infants. J Infect Dis. 2017;216(8):1027–1037. doi: 10.1093/infdis/jix400. doi: 10.1093/infdis/jix400. PubMed PMID: 28962005; PubMed Central PMCID: PMCPMC5853440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaari Gur, Uduman Mohamed, Kleinstein Steven H. Quantifying selection in high-throughput immunoglobulin sequencing data sets. Nucleic Acids Res. 2012;40(17):e134. doi: 10.1093/nar/gks457. PubMed PMID: 22641856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Twisk D., Murphy S.P., Thakar J. Optimized logic-rules reveal IFN-gamma induced modes regulated by histone deacetylases and protein tyrosine phosphatases. Immunology. 2017 doi: 10.1111/imm.12707. PubMed PMID: 28054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakar J., Hartmann B.M., Marjanovic N., Sealfon S.C., Kleinstein S.H. Comparative analysis of anti-viral transcriptomics reveals novel effects of influenza immune antagonism. BMC Immunol. 2015;16(1) doi: 10.1186/s12865-015-0107-y. doi: 10.1186/s12865-015-0107-y. PubMed PMID: 26272204; PubMed Central PMCID: PMC4536893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaslavsky E., Nudelman G., Marquez S., Hershberg U., Hartmann B.M., Thakar J. Reconstruction of regulatory networks through temporal enrichment profiling and its application to H1N1 influenza viral infection. BMC Bioinf. 2013;14(Suppl 6):S1. doi: 10.1186/1471-2105-14-S6-S1. PubMed PMID: 23734902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis I., McNally B., Willette M., Peeples M.E., Chaussabel D., Durbin J.E., Ramilo O., Mejias A., Flano E. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol. 2012;86(10):5422–5436. doi: 10.1128/JVI.06757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand H.K., Ahout I.M.L., de Ridder D., van Diepen A., Li Y., Zaalberg M., Andeweg A., Roeleveld N., de Groot R., Warris A., Hermans P.W.M., Ferwerda G., Staal F.J.T., Schildgen Oliver. Olfactomedin 4 serves as a marker for disease severity in pediatric respiratory syncytial virus (RSV) infection. PLoS ONE. 2015;10(7):e0131927. doi: 10.1371/journal.pone.013192710.1371/journal.pone.0131927.g001. PubMed PMID: 26162090; PubMed Central PMCID: PMCPMC4498630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritchie Matthew E., Phipson Belinda, Wu Di, Hu Yifang, Law Charity W., Shi Wei, Smyth Gordon K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. Epub 2015/01/22. doi: 10.1093/nar/gkv007. PubMed PMID: 25605792; PubMed Central PMCID: PMCPMC4402510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katanic D., Khan A., Thakar J. PathCellNet: Cell-type specific pathogen-response network explorer. J Immunol Methods. 2016 doi: 10.1016/j.jim.2016.09.005. PubMed PMID: 27659011. [DOI] [PubMed] [Google Scholar]
- 35.Khan A., Katanic D., Thakar J. Meta-analysis of cell- specific transcriptomic data using fuzzy c-means clustering discovers versatile viral responsive genes. BMC Bioinf. 2017;18(1):295. doi: 10.1186/s12859-017-1669-x. doi: 10.1186/s12859-017-1669-x. PubMed PMID: 28587632; PubMed Central PMCID: PMCPMC5461682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. doi: 10.1093/bioinformatics/btr260. PubMed PMID: 21546393; PubMed Central PMCID: PMC3106198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levring T.B., Hansen A.K., Nielsen B.L., Kongsbak M., von Essen M.R., Woetmann A. Activated human CD4+ T cells express transporters for both cysteine and cystine. Sci Rep. 2012;2:266. doi: 10.1038/srep00266. doi: 10.1038/srep00266. PubMed PMID: 22355778; PubMed Central PMCID: PMCPMC3278673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aoki Y., Feldman G.M., Tosato G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood. 2003;101(4):1535–1542. doi: 10.1182/blood-2002-07-2130. PubMed PMID: 12393476. [DOI] [PubMed] [Google Scholar]
- 39.Sivasankar B., Longhi M.P., Gallagher K.M., Betts G.J., Morgan B.P., Godkin A.J. CD59 blockade enhances antigen-specific CD4+ T cell responses in humans: a new target for cancer immunotherapy? J Immunol. 2009;182(9):5203–5207. doi: 10.4049/jimmunol.0804243. doi: 10.4049/jimmunol.0804243. PubMed PMID: 19380765; PubMed Central PMCID: PMCPMC2704398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazur N.I., Higgins D., Nunes M.C., Melero J.A., Langedijk A.C., Horsley N. The respiratory syncytial virus vaccine landscape: lessons from the graveyard and promising candidates. Lancet Infect Dis. 2018;18(10):e295–e311. doi: 10.1016/S1473-3099(18)30292-5. Epub 2018/06/20. doi: 10.1016/S1473-3099(18)30292-5. PubMed PMID: 29914800. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt Alexander C., Johnson Teresa R., Openshaw Peter J.M., Braciale Thomas J., Falsey Ann R., Anderson Lawrence J., Wertz Gail W., Groothuis Jessie R., Prince Gregory A., Melero Jose A., Graham Barney S. Respiratory syncytial virus and other pneumoviruses: a review of the international symposium—RSV 2003. Virus Res. 2004;106(1):1–13. doi: 10.1016/j.virusres.2004.06.008. Epub 2004/11/04. doi: 10.1016/j.virusres.2004.06.008. PubMed PMID: 15522442. [DOI] [PubMed] [Google Scholar]
- 42.Eddahri F., Denanglaire S., Bureau F., Spolski R., Leonard W.J., Leo O. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. 2009;113(11):2426–2433. doi: 10.1182/blood-2008-04-154682. Epub 2008/11/21. doi: 10.1182/blood-2008-04-154682. PubMed PMID: 19020307; PubMed Central PMCID: PMCPMC2656270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciucci Thomas, Vacchio Melanie S., Bosselut Rémy. A STAT3-dependent transcriptional circuitry inhibits cytotoxic gene expression in T cells. Proc Natl Acad Sci USA. 2017;114(50):13236–13241. doi: 10.1073/pnas.1711160114. Epub 2017/11/29. doi: 10.1073/pnas.1711160114. PubMed PMID: 29180433; PubMed Central PMCID: PMCPMC5740647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuyama T., Kimura T., Kitagawa M., Pfeffer K., Kawakami T., Watanabe N. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75(1):83–97. Epub 1993/10/08 PubMed PMID: 8402903. [PubMed] [Google Scholar]
- 45.Penninger J.M., Sirard C., Mittrucker H.W., Chidgey A., Kozieradzki I., Nghiem M. The interferon regulatory transcription factor IRF-1 controls positive and negative selection of CD8+ thymocytes. Immunity. 1997;7(2):243–254. doi: 10.1016/s1074-7613(00)80527-0. Epub 1997/08/01 PubMed PMID: 9285409. [DOI] [PubMed] [Google Scholar]
- 46.Hosoya T., Maillard I., Engel J.D. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev. 2010;238(1):110–125. doi: 10.1111/j.1600-065X.2010.00954.x. doi: 10.1111/j.1600-065X.2010.00954.x. PubMed PMID: 20969588; PubMed Central PMCID: PMCPMC2965564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groom Joanna R., Luster Andrew D. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89(2):207–215. doi: 10.1038/icb.2010.158. Epub 2011/01/12. doi: 10.1038/icb.2010.158. PubMed PMID: 21221121; PubMed Central PMCID: PMCPMC3863330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinde Paurvi, Liu Wenhai, Ménoret Antoine, Luster Andrew D., Vella Anthony T. Optimal CD4 T cell priming after LPS-based adjuvanticity with CD134 costimulation relies on CXCL9 production. J Leukoc Biol. 2017;102(1):57–69. doi: 10.1189/jlb.1A0616-261RR. Epub 2017/04/23. doi: 10.1189/jlb.1A0616-261RR. PubMed PMID: 28432083; PubMed Central PMCID: PMCPMC5470836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schonland S.O., Zimmer J.K., Lopez-Benitez C.M., Widmann T., Ramin K.D., Goronzy J.J. Homeostatic control of T-cell generation in neonates. Blood. 2003;102(4):1428–1434. doi: 10.1182/blood-2002-11-3591. PubMed PMID: 12714521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes responding to F and M peptide stimulation: Heatmaps showing fold change of the differentially expressed genes upon F (A) and M (B) peptide stimulation compared to DMSO stimulation. The color from blue to red shows the z-score of the log2 fold change.
Voom normalized expression of CXCL9 and IFN-γ expression. A) Highlighting the difference between subjects with mild (black diamonds) and severe (red triangles) infections. B) Differences in expression colored based on stimulation with F (black circles), M (red squared), N (green diamonds), or G (blue triangles) peptides.
Flow panels.
Information on gene-sets.
Details of DAFi analysis.