Abstract
The SARS-CoV-2 coronavirus, the etiologic agent responsible for COVID-19 coronavirus disease, is a global threat. To better understand viral tropism, we assessed the RNA expression of the coronavirus receptor, ACE2, as well as the viral S protein priming protease TMPRSS2 thought to govern viral entry in single-cell RNA-sequencing (scRNA-seq) datasets from healthy individuals generated by the Human Cell Atlas consortium. We found that ACE2, as well as the protease TMPRSS2, are differentially expressed in respiratory and gut epithelial cells. In-depth analysis of epithelial cells in the respiratory tree reveals that nasal epithelial cells, specifically goblet/secretory cells and ciliated cells, display the highest ACE2 expression of all the epithelial cells analyzed. The skewed expression of viral receptors/entry-associated proteins towards the upper airway may be correlated with enhanced transmissivity. Finally, we showed that many of the top genes associated with ACE2 airway epithelial expression are innate immune-associated, antiviral genes, highly enriched in the nasal epithelial cells. This association with immune pathways might have clinical implications for the course of infection and viral pathology, and highlights the specific significance of nasal epithelia in viral infection. Our findings underscore the importance of the availability of the Human Cell Atlas as a reference dataset. In this instance, analysis of the compendium of data points to a particularly relevant role for nasal goblet and ciliated cells as early viral targets and potential reservoirs of SARS-CoV-2 infection. This, in turn, serves as a biological framework for dissecting viral transmission and developing clinical strategies for prevention and therapy.
Introduction
In December 2019, a cluster of atypical pneumonia associated with a novel coronavirus was detected in Wuhan, China1. This coronavirus disease, termed COVID-19, was caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; previously termed 2019-nCoV)2. The virus has since spread worldwide, emerging as a serious global health concern in early 20203,4. Human-to-human transmission of the virus has been reported in several instances5–7 and is thought to have occurred since mid-December 20198. As of early March 2020, there were more than 100,000 confirmed COVID-19 cases4.
Patients with suspected COVID-19 have been treated in the Wuhan Jin Yintan Hospital since Dec 31st, 20199. In a meta-analysis of 50,466 hospitalized patients with COVID-19 from 10 studies, most patients were from China and the average age in the included studies ranged from 41 to 56 years old10. The prevalence rates of fever, cough, and muscle soreness or fatigue were 89.1%, 72.2%, and 42.5%. Critical illness requiring admission to an intensive care unit occurred in 18.1% of patients, and 14.8% developed acute respiratory distress syndrome (ARDS)10. Acute renal injury and septic shock have been observed in 4% and 5% of patients hospitalized with COVID-19, respectively1,9. Chest imaging demonstrated bilateral pneumonia involvement in more than 80% of cases1,9,11. Ground-glass opacities were the most common radiologic finding on chest computed tomography (CT)11,12. Abnormalities on CT were also observed preceding symptom onset in patients exposed to infected individuals, with an incidence of 93%10,11.
Pathological evaluation of a patient who died of severe disease revealed diffuse alveolar damage consistent with ARDS13. Currently, the estimated mortality rate is 3.4%14. These clinical data underscore the severity of this infection. The involvement of both lungs in most of the cases suggests viral dissemination after initial infection.
Viral RNA was detected in the upper airways from symptomatic patients, with higher viral loads observed in nasal swabs compared to those obtained from the throat15. Similar viral loads were observed in an asymptomatic patient15, indicating that the nasal epithelium is an important portal for initial infection, and may serve as a key reservoir for viral spread across the respiratory mucosa and an important locus mediating viral transmission. Identification of the cells hosting viral entry and permitting viral replication as well as those contributing to inflammation and disease pathology is essential to improve diagnostic and therapeutic interventions.
Cellular entry of coronaviruses depends on the binding of the spike (S) protein to a specific cellular receptor and subsequent S protein priming by cellular proteases. Similar to severe acute respiratory syndrome-associated coronavirus (SARS-CoV)16,17, the SARS-CoV-2 employs angiotensin-converting enzyme-2 (ACE2) as a receptor for cellular entry. In addition, studies have shown that the serine protease TMPRSS2 can prime S protein15,18 although other proteases like cathepsin B/L can also be involved18. For SARS, the binding affinity between the S protein and the ACE2 receptor was found to be a major determinant of viral replication rates and disease severity19. The SARS-CoV-2 has been shown to infect and replicate in Vero cells, a Cercopithecus aethiops (old world monkey) kidney epithelial cell line, and huh7 cells, a human hepatocarcinoma cell line15. The BHK21 cell line has been shown to facilitate viral entry by the SARS-CoV-2 S protein only when engineered to express the ACE2 receptor ectopically18. In addition, viral entry was found to depend on TMPRSS2 activity, although cathepsin B/L activity might substitute for the loss of TMPRSS218. The in vivo expression of ACE2 and TMPRSS2 (as well as other candidate proteases) by cells of the upper and lower airways and alveoli must be defined.
Previously, gene expression of ACE2 and TMPRSS2 has been reported to occur largely in type-2 alveolar (AT-2) epithelial cells15, which are central to SARS-CoV pathogenesis. A study reported that ACE2 expression is absent from the upper airways20. The rapid spread of the SARS-CoV-2 suggests efficient human-to-human transmission which would, in turn, seem to supersede the odds of dependency on alveolar epithelial cells as the primary point of entry and viral replication8,21,22. Indeed, protein expression, based on immunohistochemistry, of ACE2 and TMPRSS2 has been reported in both nasal and bronchial epithelium23. To clarify the expression patterns of ACE2 and TMPRSS2 and analyze the expression of the other potential genes associated with SARS-CoV-2 pathogens at cellular resolution, we interrogated single-cell transcriptome expression data from published scRNA-seq datasets from healthy donors generated by the Human Cell Atlas consortium24.
Results
ACE2 and TMPRSS2 are enriched in nasal tissues and enterocytes
We investigated the gene expression of ACE2 in multiple scRNA-seq datasets from different tissues, including those of the respiratory tree25, ileum26, colon27, liver28, placenta/decidua29, kidney30, testis31, pancreas32, and prostate gland33. While scRNA-seq is a comprehensive assay, we note that some studies may still miss specific cell types, due to either their rarity, challenges associated with their isolation, or analysis methodology that was used. Thus, while positive (presence) results are highly reliable, absence should be interpreted with care.
The expression of ACE2, in general, is relatively low in all of the datasets analyzed. Consistent with independent analyses34, we found that ACE2 is expressed in lung, airways, ileum, colon, and kidney (Fig. 1a; first column). It is worth noting that TMPRSS2, the primary protease important for viral entry, is highly expressed with a broader be a limiting factor for viral entry at the initial stage of infection. When taking into account distribution (Fig. 1a; second column), suggesting that ACE2, rather than TMPRSS2, may the expression of both genes, the cells found in mucosal epithelia in the respiratory tree, ileum, and colon are ACE2+ (Fig. 1a; third column), consistent with viral transmission by respiratory droplets, and the potential of fecal-oral transmission35. We also assessed ACE2 and TMPRSS2 expression in developmental datasets from fetal liver, fetal thymus, fetal skin, fetal bone marrow and fetal yolk sac36,37 and found little to no expression of ACE2 with no co-expression with TMPRSS2 (data not shown) even if single ACE2 expression is noticeable in certain cell types in placenta/decidua (Fig. 1a). While we cannot rule out the possibility that the virus uses alternative proteases for entry in such contexts, or that lung fetal tissue expresses the relevant genes, these results are at least consistent with early reports that fail to detect evidence of intrauterine infection through vertical transmission in women who develop COVID-19 pneumonia in late pregnancy38. If future epidemiologic data are consistent with a lack vertical viral transmission, these findings may form the basis of an explanatory model for the clinical finding. However, if future evidence for vertical transmission emerges, additional scRNA-seq data can be collected and further scrutinized for the presence of rare co-expressers or alternative receptors or proteases.
Fig. 1|. Expression of ACE2 and TMPRSS2 across different tissues and its enrichment in nasal epithelial cells.
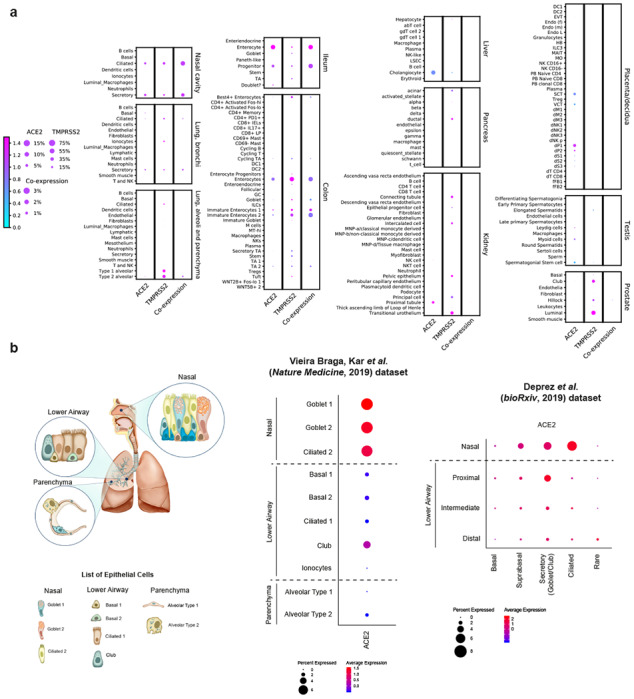
a, RNA expression of SARS-CoV-2 entry receptor ACE2 (first column), entry-associated protease TMPRSS2 (second column), and their co-expression (third column) from multiple published scRNA-seq datasets. Raw expression values were normalized, log transformed and summarized by published cell clustering where available, or reproduced clustering annotated using marker genes and cell type nomenclature from the respective studies. The size of the dots indicates the proportion of cells in the respective cell type having greater-than-zero expression of ACE2 (first column), TMPRSS2 (second column) or both (third column), while the colour indicates the mean expression of ACE2 (first and third columns) or TMPRSS2 (second column). b, Schematic illustration depicts the major anatomical regions in the human respiratory tree demonstrated in this study: nasal, lower airway, and lung parenchyma (left panel). Expression of ACE2 is from airway epithelial cell datasets: Vieira Braga, Kar et al. 2019 (middle panel) and Deprez et al. 2019 (right panel). The datasets were retrieved from existing sources, and the cell clustering and nomenclature were retained based on the respective studies. For gene expression results in the dot plots: the dot size represents the proportion of cells within the respective cell type expressing the gene and the dot color represents the average gene expression level within the particular cell type.
Nasal goblet and ciliated cells display the highest expression of ACE2 within the larger population of respiratory epithelial cells
To further characterize specific epithelial cell types expressing ACE2, we evaluated the expression of ACE2 within lung/airway epithelia from a previous study25. We found that, despite a low level of expression overall, ACE2 is expressed in multiple epithelial cell types across the airway, as well as in AT-2 cells in the parenchyma, consistent with previous studies20,39. Importantly, nasal epithelial cells, including previously described two clusters of goblet cells and one cluster of ciliated cells, have the highest expression among all investigated cells in the respiratory tree (Fig. 1b; left panel). We confirmed enriched ACE2 expression in nasal epithelial cells from a second scRNA-seq study, which, in addition to nasal brushing samples seen in the earlier dataset, included nasal biopsies40. The results were consistent: we found the highest expression of ACE2 in nasal secretory cells (equivalent to the two goblet cell clusters in the previous dataset) and ciliated cells (Fig. 1b; right panel).
In addition, scRNA-seq data from an in vitro 3D epithelial regeneration system from nasal epithelial cells41 corroborated the expression of ACE2 in goblet/secretory cells and ciliated cells in these air-liquid interface (ALI) cultures (Extended Data Fig. 1). Of note, the differentiating cells in ALI acquire progressively more ACE2 and, unlike their corresponding progenitors, they have large luminal surfaces in the mature differentiated epithelium where viral entry is likely to occur (Extended Data Fig. 1). These results also suggest that such in vitro culture system is biologically relevant to the study of viral pathogenesis.
We also investigated the expression of known proteases associated with the entry of SARS-CoV and SARS-CoV-2. TMPRSS2, which was shown to be important for SARS-CoV/SARS-CoV-2 viral entry and SARS-CoV transmission,16–18 is expressed in a subset of ACE2+ cells (Extended Data Fig. 2), suggesting that the virus might use alternative pathways for entry. In fact, it was previously shown that SARS-CoV-2 could enter TMPRSS2− cells using cathepsin B/L18. Indeed, we found that they are much more promiscuously expressed than TMPRSS2, especially cathepsin B, which is expressed in more than 70%-90% of ACE2+ cells (Extended Data Fig. 2). However, whether cathepsin B/L can functionally replace TMPRSS2 has not been empirically determined. In the case of SARS-CoV, TMPRSS2 activity is documented to be important for viral transmission42,43.
Respiratory expression of viral receptor/entry-associated genes and implications for viral transmissivity
We next asked whether the enriched expression of viral receptors and entry-associated molecules in the nasal region/upper airway could be relevant to viral transmissivity. Here, we assessed the expression of viral receptor genes that are used by other coronaviruses and influenza viruses, including ANPEP (used by HCoV-22944) and DPP4 (used by MERS CoV45), as well as the enzymes ST6GAL1 and ST3GAL4 in the lung epithelial cell datasets. The latter genes are enzymes which are important for the synthesis of viral receptors used by influenza viruses: α(2,6)-linked sialic acid and α(2,3)-linked sialic acid46. Notably, the distribution of receptor/receptor-associated enzymes appears to coincide with viral transmissivity patterns based on a comparison to the basic reproduction number (R0), which estimates the number of people who can get infected from a single infected person; and the infection will be able to start spreading in a population when R0 > 1. The skewed distribution of the receptors/enzymes towards the upper airway is observed in viruses with relatively higher R0/infectivity, including those of SARS-CoV/SARS-CoV-2 (R0 ~ 1.4-5.08,21,22), influenza (mean R0 ~1.347) and HCoV-229E (unidentified R0; associated with common cold48). This distribution is in distinct contrast with that of DPP4, the receptor for MERS-CoV (R0 ~0.3-0.8), a coronavirus with limited human-to-human transmission49, with the skewed expression towards lower airway/lung parenchyma (Fig. 2a). Therefore, our data highlight the possibility that viral transmissivity is dependent on receptor accessibility based on spatial distribution along the respiratory tract.
Fig. 2|. Respiratory expression of viral receptor/entry-associated genes and implications for viral transmissivity and genes associated with ACE2 expression.
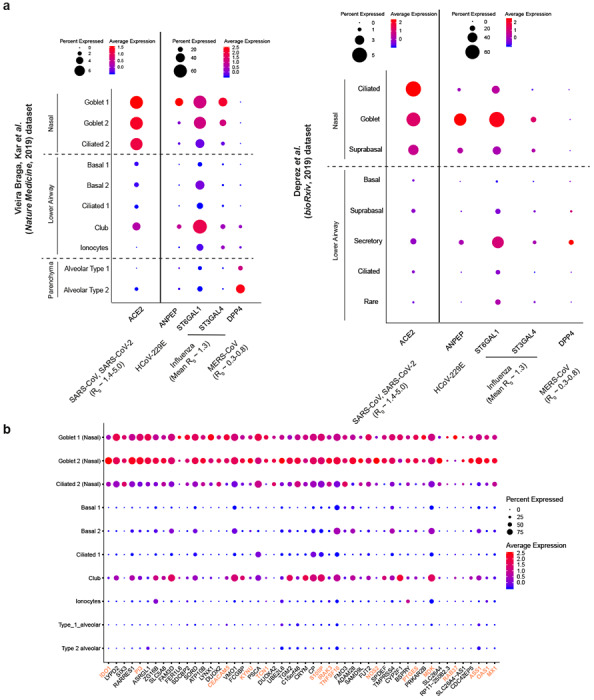
a, Expression of ACE2 (an entry receptor for SARS-CoV and SARS-CoV-2), ANPEP (an entry receptor for HCoV-229E), ST6GAL1/ST3GAL4 (enzymes important for synthesis of influenza entry receptors), and DPP4 (an entry receptor for MERS-CoV) from the airway epithelial datasets: Vieira Braga, Kar et al. 2019 (left panel) and Deprez et al. 2019 (right panel). The basic reproductive number (R0) for respective viruses, if available, are shown. b, Respiratory epithelial expression of the top 50 genes correlated with ACE2 expression based on Spearman correlation analysis (with Benjamini-Hochberg-adjusted p-values) on genes associated with ACE2 across all cells within the Vieira Braga, Kar et al. lung epithelial dataset. The colored gene names represent genes that are immune-associated (GO:0002376: immune system process or GO:0002526: acute inflammatory response). For gene expression results in the dot plots: the dot size represents the proportion of cells within the respective cell type expressing the gene and the color represents the average gene expression level within the particular cell type.
Expression of genes associated with ACE2 expression: innate immunity and carbohydrate metabolism
To gain more insight into the expression patterns of genes associated with ACE2, we performed Spearman correlation analysis with Benjamini-Hochberg-adjusted p-values on genes associated with ACE2 across all cells within the lung epithelial cell dataset25. While the correlation coefficients are relatively low (< 0.11), likely due to low expression of ACE2, the expression pattern of the top 50 ACE2-correlated genes (all with adjusted p-value close to 0; ranked by correlation coefficients) across the respiratory tree is similar to that of ACE2, with a skewed expression toward upper airway (Fig. 2b). To our surprise, while some of the genes are associated with carbohydrate metabolism (possibly due to the role of goblet cells in mucin synthesis), a number of genes associated with immune functions including innate and antiviral immune functions, are over-represented in the rank list, including IDO1, IRAK3, NOS2, TNFSF10, OAS1, and MX1 (Fig. 2b and Supplementary Table 1). These genes have the highest expression in nasal goblet 2 cells (Fig. 2b), consistent with the phenotype previously described25. Nonetheless, nasal goblet 1 and nasal ciliated 2 cells also significantly express these genes, but less so elsewhere (Fig. 2b). Given their environmental exposure and the high expression of receptor/receptor-associated enzymes (Fig. 2a), it is plausible that the nasal epithelial cells were conditioned and primed to express these immune-associated genes to prevent viral susceptibility. This association with innate immune pathways not only highlights the importance of host-microbe dynamics in nasal epithelia, but it may also have implications for subsequent viral pathogenesis and immune-associated protection/pathology.
Discussion
In this study, we explored multiple scRNA-seq datasets generated within the HCA consortium, and found that SARS-CoV-2 entry receptor ACE2 is more highly expressed (and co-expressed with viral entry-associated protease TMPRSS2) in nasal epithelial cells, specifically goblet and ciliated cells. This finding implicates these cells as loci of original infection and possible reservoirs for dissemination within a given patient and from person to person. Importantly, viral infection itself could drastically change the gene expression landscape in the nose and other tissues later on.
The up-regulation of innate immune genes, in association with ACE2, in highly-exposed nasal epithelial cells could be the result of their responsiveness to persistent environmental challenges, including viral infection. It would be of great interest to further investigate how other genetic, demographic, and environmental factors might affect this poised state in these cells and whether such state could influence the susceptibility to infection due to its association with viral receptor expression. Future meta-analysis of HCA data can help further assess some of these aspects.
All in all, our findings may have significant implications for understanding viral transmissivity, considering that the primary viral transmission is through respiratory droplets. Moreover, as SARS-CoV-2 is an enveloped virus, its release does not require cell lysis. Thus, the virus might exploit existing secretory pathways in nasal goblet cells for low-level, continuous-release at the early stage with no overt pathology. These discoveries could have clinical implications with respect to targeting nasal epithelial cells, especially nasal goblet cells, beyond the current usage of face masks, providing a candidate clinical option for transmission prevention and/or early-stage intervention.
Finally, it is worth highlighting that this is the first collaborative effort by a Human Cell Atlas Biological Network (the Lung), and illustrates the opportunities from integrative analyses of Human Cell Atlas data, with future examples of consortium work expected soon.
Methods
The datasets were retrieved from existing sources based on previously published data as specifically specified in the reference. We retained the cell clustering when available or reprocessed using scanpy50 and harmony51, and annotated the clusters with marker genes and cell type nomenclature based on the respective studies. Illustration of the results was generated using scanpy50 and Seurat52.
Extended Data
Extend Data Fig. 1|. Gene expression of ACE2 in an in vitro 3D air-liquid interface (ALI) system.
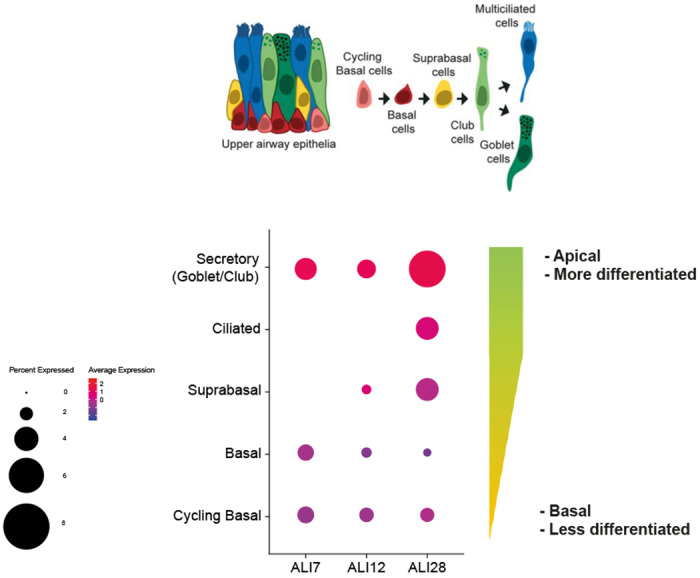
Epithelial regeneration system from nasal epithelial cells was used for in vitro cultures on successive days (7, 12 and 28), resulting in different epithelial cell types along differentiation trajectory characterized in Ruiz García et al. 2019. The cultures were differentiated in Pneumacult media. Schematic illustration depicts the respective cell types in the differentiation trajectory, and the dot plot illustrates the cultured cell types along the differentiation pseudotime, along with their respective location within the epithelial layers. For gene expression results in the dot plot: the dot size represents the proportion of cells within the respective cell type expressing the gene and the dot color represents the average gene expression level within the particular cell type.
Extended Data Fig. 2|.
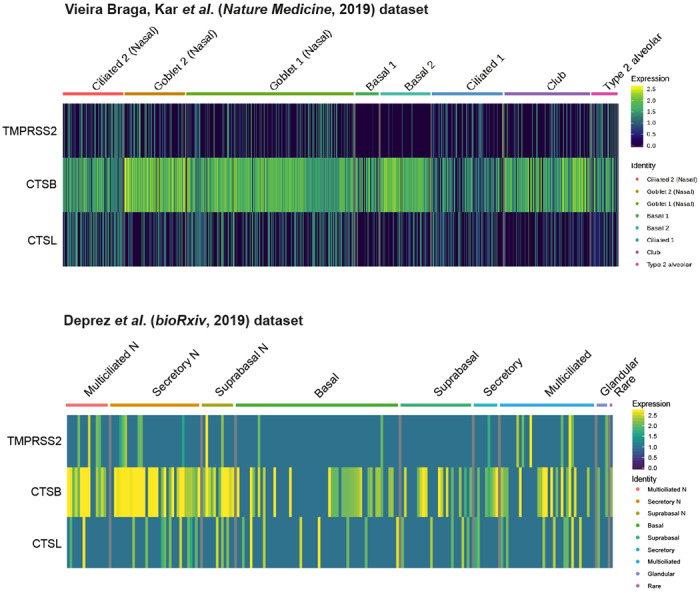
Expression and co-expression of SARS-CoV-2 entry-associated proteases in ACE2+ airway epithelial cells: TMPRSS2, CTSB, and CTSL in ACE2+ cells from the Vieira Braga, Kar et al. (top) and Deprez et al. (bottom) airway epithelial datasets. The color represents the expression level at the single-cell resolution and the cells are grouped based on the cell types specified.
Supplementary Material
Acknowledgements
We are grateful to Cori Bargmann, Jeremy Farrar, and Sarah Aldridge for stimulating discussions. We thank Jana Eliasova (scientific illustrator) for support with the design of figures.
Funding
This work was supported by a Seed Network grant from the Chan Zuckerberg Initiative to P.B., T.D., T.E.D., O.E., P.H., N.K., M.K., K.B.M., A.M., M.C.N., D.P., J.R., P.R.T., S.Q., A.R., O.R., H.B.S., D.S., A.T., J.W. and K.Z. and by the European Union’s H2020 research and innovation programme under grant agreement No 874656 (discovAIR) to P.B., A.B., M.K., S.L., J.L., K.B.M., M.C.N., K.S.P., C.S., H.B.S., J.S., F.T. and M.vd.B. W.S. acknowledges funding from the Newton Fund, Medical Research Council (MRC), The Thailand Research Fund (TRF), and Thailand’s National Science and Technology Development Agency (NSTDA). M.C.N acknowledge funding from GSK Ltd, Netherlands Lung Foundation project no. 5.1.14.020 and 4.1.18.226. T.D. acknowledges funding from HubMap consortium and Stanford Child Health Research Institute- Woods Family Faculty Scholarship. T.E.D. acknowledges funding from HubMap. P.H. acknowledges funding from LENDULET-BIOMAG Grant (2018-342) and the European Regional Development Funds (GINOP-2.3.2-15-2016-00006, GINOP-2.3.2-15-2016-00026, GINOP-2.3.2-15-2016-00037). N.K. acknowledges funding from NIH grants R01HL127349, U01HL145567 and an unrestricted grant from Three Lakes Foundation. M.K. acknowledges HHMI and Wall Center for Pulmonary Vascular Disease. H.L. coordinators for the HCA Lung Biological Network. acknowledges funding from National Research Foundation of Korea. K.M. acknowledges funding from Wellcome Trust. A.M. acknowledges funding from NIH grants HL135124, AG049665 and AI135964. M.Z.N. acknowledges funding from Rutherford Fund Fellowship allocated by the Medical Research Council and the UK Regenerative Medicine Platform (MR/5005579/1 to M.Z.N.). J.O.-M. acknowledges funding from Richard and Susan Smith Family Foundation. D.P. acknowledges funding from Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center. J.P. acknowledges funding from National Health and Medical Research Council. P.R.T. acknowledges funding from R01HL146557 from NHLBI/NIH. E.L.R. acknowledges funding from MRC MR/P009581/1 and MR/SO35907/1. A.R. and O. R. acknowledge HHMI, the Klarman Cell Observatory, and the Manton Foundation. K.S.-P. acknowledges NIHR Cambridge Biomedical Research Centre. C.S. acknowledges Swedish research Council, Swedish Cancer Society, and CPI. H.B.S. acknowledges German Center for Lung Research and Helmholtz Association. J.S. acknowledges Boehringer Ingelheim, by the German Research Foundation (DFG; EXC2151/1, ImmunoSensation2 - the immune sensory system, project number 390873048), project numbers 329123747, 347286815) and by the HGF grant sparse2big. A.K.S. acknowledges the Beckman Young Investigator Program, a Sloan Fellowship in Chemistry, the NIH (5U24AI118672), and the Bill and Melinda Gates Foundation. F.T. Theis the German Center for Lung Research. M.vd.B. acknowledges from Ministry of Economic Affairs and Climate Policy by means of the PPP. J.W. acknowledges NIH, U01 HL148856 LungMap Phase II.
Competing interests
N.K. was a consultant to Biogen Idec, Boehringer Ingelheim, Third Rock, Pliant, Samumed, NuMedii, Indaloo, Theravance, LifeMax, Three Lake Partners, Optikira in the last three years and received non-financial support from MiRagen. J.L. is a scientific consultant for 10X Genomics Inc. A.R. is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas, and an SAB member of ThermoFisher Scientific, Syros Pharmaceuticals, Asimov, and Neogene Therapeutics. O.R. is a co-inventor on patent applications filed by the Broad Institute to inventions relating to single cell genomics applications, such as in PCT/US2018/060860 and US Provisional Application No. 62/745,259. A.K.S. reports compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Cellarity, Cogen Therapeutics, Orche Bio, and Dahlia Biosciences. F.T. reports receiving consulting fees from Roche Diagnostics GmbH, and ownership interest in Cellarity Inc. S.A.T. was a consultant at Genentech, Biogen and Roche in the last three years.
This publication is part of the Human Cell Atlas - www.humancellatlas.org/publications.
*The HCA Lung Biological Network are: Pascal Barbry1, Alvis Brazma2, Tushar Desai3, Thu Elizabeth Duong4, Oliver Eickelberg5, Muzlifah Haniffa6, Peter Horvath7, Naftali Kaminski8, Mark Krasnow9, Malte Kuhnemund10, Haeock Lee11, Sylvie Leroy12, Joakim Lundeberg13, Kerstin B. Meyer14, Alexander J. Misharin15, Martijn C. Nawijn16, Marko Z. Nikolic17, Jose Ordovas Montanes18, Dana Pe’er19, Joseph Powell20, Steve Quake21, Jay Rajagopal22, Purushothama Rao Tata23, Emma L. Rawlins24, Aviv Regev25, Orit Rozenblatt-Rosen26, Kourosh Saeb-Parsy27, Christos Samakovlis28, Herbert B. Schiller29, Joachim L. Schultze30, Alex K. Shalek31, Douglas Shepherd32, Xin Sun33, Sarah A. Teichmann34, Fabian Theis35, Alexander Tsankov36, Maarten van den Berge37, Jeffrey Whitsett38, and Kun Zhang39.
Affiliations
1Université Côte d’Azur, CNRS, IPMC, Sophia-Antipolis 06560, France
2European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SD, UK
3Department of Medicine and Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Stanford, CA 94116, USA
4Department of Pediatrics Division of Respiratory Medicine, University of California San Diego and Rady Children’s Hospital San Diego, San Diego, CA 92123, USA
5Division of Pulmonary Sciences and Critical Care Medicine, Department of Medicine, University of Colorado, Anschutz Medical Campus, Aurora, CO, US
6Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge CB10 1SA, UK; Biosciences Institute, Faculty of Medical Sciences, Newcastle University, Newcastle upon Tyne NE2 4HH, UK; Department of Dermatology and NIHR Newcastle Biomedical Research Centre, Newcastle Hospitals NHS Foundation Trust, Newcastle upon Tyne NE2 4LP, UK
7Synthetic and Systems Biology Unit, Biological Research Centre (BRC), Szeged, Hungary; Institute for Molecular Medicine Finland, University of Helsinki
8Pulmonary, Critical Care and Sleep Medicine, Yale University School of Medicine, New Haven, CT 06520, USA
9Department of Biochemistry and Wall Center for Pulmonary Vascular Disease, Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, CA 94305, USA
10Cartana AB, Nobels vag 16, 17165 Stockholm, Sweden
11Department of Biomedicine and Health Sciences, The Catholic University of Korea, Seoul, Korea
12Université Côte d’Azur, CHU de Nice, FHU OncoAge, Department of Pulmonary Medicine and Allergology, Nice, France; CNRS UMR 7275 - Institut de Pharmacologie Moléculaire et Cellulaire, Sophia Antipolis, France
13SciLifeLab, Department of Gene Technology, KTH Royal Institute of Technology, SE-100 44, Stockholm, Sweden
14Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SA, UK
15Division of Pulmonary and Critical Care Medicine, Northwestern University, Chicago, Illinois 60611, USA
16Department of Pathology and Medical Biology, University of Groningen, GRIAC Research Institute, University Medical Center Groningen, 9713 AV Groningen, Netherlands
17UCL Respiratory, Division of Medicine, University College London, WC1 E 6JF, London, UK
18Division of Gastroenterology Boston Children’s Hospital, Boston, MA 02115, USA; Program in Immunology, Harvard Medical School, Boston, MA 02115, USA; Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Harvard Stem Cell Institute, Cambridge, MA 02138, USA.
19Computational and Systems Biology Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York 10065, USA
20Garvan-Weizmann Centre for Cellular Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; UNSW Cellular Genomics Futures Institute, University of New South Wales, Sydney, NSW, Australia
21Depts of Bioengineering and Applied Physics, Stanford University, and the Chan Zuckerberg Biohub, Stanford University, Stanford, CA 94305, USA
22Harvard Stem Cell Institute, Cambridge, MA 02138, USA; Center for Regenerative Medicine, Massachusetts General Hospital, Boston, MA 02114, Boston
23Department of Cell Biology, Regeneration Next Initiative, Duke University School of Medicine, Durham, NC 27710, USA
24Wellcome Trust/CRUK Gurdon Institute and Department Physiology, Development and Neuroscience, University of Cambridge, Cambridge, CB2 1QN, UK
25Klarman Cell Observatory, Broad Institute of MIT and Harvard, Howard Hughes Medical Institute, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02142, USA
26Klarman Cell Observatory, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA
27Department of Surgery, University of Cambridge and NIHR Cambridge Biomedical Research Centre, CB2 0QQ, UK
28SciLifeLab, Department of Molecular Biosciences, Stockholm University, Stockholm Sweden; Cardiopulmonary Institute, Justus Liebig University, Giessen, Germany
29Comprehensive Pneumology Center (CPC) / Institute of Lung Biology and Disease (ILBD), Helmholtz Zentrum MGnchen, Member of the German Center for Lung Research (DZL), Munich, Germany
30Joachim L. Schultze, 1 Department for Genomics & Immunoregulation, LIMES-Institute, University of Bonn, 53115 Bonn, Germany; 2 PRECISE Platform for Single Cell Genomics & Epigenomics, German Center for Neurodegenerative Diseases and University of Bonn, Bonn, Germany
31Ragon Institute of MGH, MIT, and Harvard, Cambridge, MA, USA; Institute for Medical Engineering and Science (IMES), Koch Institute for Integrative Cancer Research, and Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA, USA; Broad Institute of MIT and Harvard, Cambridge, MA, USA
32Center for Biological Physics and Department of Physics, Arizona State University, Tempe, AZ 85287, USA
33Department of Pediatrics, Department of Biological Sciences, University of California SD, 9500 Gilman Dr. MC0766, San Diego, CA 92093, USA
34Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SA, UK; Theory of Condensed Matter Group, Cavendish Laboratory/Department of Physics, University of Cambridge, Cambridge CB3 0HE, UK
35Institute of Computational Biology, Helmholtz Zentrum München and Departments of Mathematics and Life Sciences, Technical University Munich, Germany
36Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
37Department of Pulmonary diseases and tuberculosis, University of Groningen, GRIAC Research Institute, University Medical Center Groningen, 9713 AV Groningen, Netherlands
38Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229, USA
39UCSD Department of Bioengineering, 9500 Gilman Drive, MC0412, PFBH402, La Jolla, CA 92093, USA
Pascal Barbry, Alexander Misharin, Martijn Nawijn and Jay Rajagopal serve as the coordinators for the HCA Lung Biological Network.
Reference
- 1.Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Naming the coronavirus disease (COVID-19) and the virus that causes it. Vol. 2020 (2020). [Google Scholar]
- 3.Zhu N., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahase E. Covid-19: UK records first death, as world’s cases exceed 100 000. BMJ 368, m943 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Phan L.T., et al. Importation and Human-to-Human Transmission of a Novel Coronavirus in Vietnam. N Engl J Med 382, 872–874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothe C., et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395, 514–523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q., et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun P., et al. Clinical characteristics of 50466 hospitalized patients with 2019-nCoV infection. Journal of medical virology (2020). [Google Scholar]
- 11.Shi H., et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. The Lancet. Infectious diseases (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan W.J., et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet. Respiratory medicine (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. WHO Director-General’s opening remarks at the media briefing on COVID-19 - 3 March 2020. Vol. 2020 (2020). [Google Scholar]
- 15.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuyama S., et al. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. Journal of virology 84, 12658–12664 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M., et al. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv, 2020.2001.2031.929042 (2020). [Google Scholar]
- 19.Li W., et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. The EMBO journal 24, 1634–1643 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamming I., et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of pathology 203, 631–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallinga J. & Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. American journal of epidemiology 160, 509–516 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riou J. & Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 25(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertram S., et al. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PloS one 7, e35876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regev A., et al. The Human Cell Atlas. eLife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vieira Braga F.A., et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nature medicine 25, 1153–1163 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Martin J.C., et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 178, 1493–1508. e1420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smillie C.S., et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730. e722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacParland S.A., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nature communications 9, 4383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vento-Tormo R., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart B.J., et al. Spatiotemporal immune zonation of the human kidney. Science (New York, N.Y.) 365, 1461–1466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo J., et al. The adult human testis transcriptional cell atlas. Cell research 28, 1141–1157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron M., et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell systems 3, 346–360. e344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry G.H., et al. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell Rep 25, 3530–3542. e3535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi F., Qian S., Zhang S. & Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. bioRxiv, 2020.2002.2016.951913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerging microbes & infections 9, 386–389 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popescu D.M., et al. Decoding human fetal liver haematopoiesis. Nature 574, 365–371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J.E., et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (New York, N.Y.) 367(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H., et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. The Lancet (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y., et al. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv, 2020.2001.2026.919985 (2020). [Google Scholar]
- 40.Deprez M., et al. A single-cell atlas of the human healthy airways. bioRxiv, 2019.2012.2021.884759 (2019). [Google Scholar]
- 41.Ruiz Garcia S., et al. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development (Cambridge, England) 146(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y., et al. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral research 116, 76–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwata-Yoshikawa N., et al. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. Journal of virology 93(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeager C.L., et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357, 420–422 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raj V.S., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broszeit F., et al. N-Glycolylneuraminic Acid as a Receptor for Influenza A Viruses. Cell Rep 27, 3284–3294. e3286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coburn B.J., Wagner B.G. & Blower S. Modeling influenza epidemics and pandemics: insights into the future of swine flu (H1N1). BMC medicine 7, 30 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendley J.O., Fishburne H.B. & Gwaltney J.M. Jr. Coronavirus infections in working adults. Eightyear study with 229 E and OC 43. The American review of respiratory disease 105, 805–811 (1972). [DOI] [PubMed] [Google Scholar]
- 49.Killerby M.E., Biggs H.M., Midgley C.M., Gerber S.I. & Watson J.T. Middle East Respiratory Syndrome Coronavirus Transmission. Emerging infectious diseases 26, 191–198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf F.A., Angerer P. & Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korsunsky I., et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butler A., Hoffman P., Smibert P., Papalexi E. & Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


