Abstract
Cells with high CD44 but low CD24 expression (CD44high/CD24−/low) and high aldehyde dehydrogenase activity (ALDHbr) are widely considered to be drivers of metastasis, therapy resistance and tumor recurrence in breast cancer. However, the role of the CD44high/CD24−/low and ALDHbr phenotypes in identifying tumorigenic cells in breast cancer remains controversial due to the discrepancy in their distribution and tumorigenic potential in intrinsic breast cancer subtypes. In this study, we analyzed the cells expressing these markers in six different breast cancer cell lines representing major breast cancer subtypes (T47D, MCF-7, BT-474, AU-565, Hs578T and MDA-MB-231). CD44high/CD24−/low, ALDHbr and CD44−/low/CD24−/low cell populations were isolated by flow cytometry and analyzed for hallmark stem cell characteristics of differentiation, migration, invasiveness and metastasis using in vitro and in vivo techniques. Our results demonstrate that the CD44−/low/CD24−/low cell population, which is enriched in luminal cell lines (T47D, MCF-7 and BT-474), possesses metastatic and tumorigenic properties. We also show that, contrary to previous claims, the expression of the ALDH1 isoform ALDH1A1 does not affect the tumorigenic potential of cell lines with high ALDH activity (BT-474 and AU-565). Further transcriptomic and clinical studies are needed to determine the potential of these markers as early diagnostic tools and treatment targets.
Keywords: breast cancer; ALDH1A1; CD44; CD24; metastasis, ALDH
1. Introduction
Breast cancer is the most prevalent type of malignancy in women worldwide, leading to thousands of deaths every year. Based on the presence or absence of commonly evaluated hormone receptors—estrogen (ER), progesterone (PR) and human epidermal growth factor 2 (HER2)—breast cancer is divided into four main subtypes: luminal A (ER+/PR+/HER2−), luminal B (ER+/PR+/HER2+), HER2 overexpressing or HER2-OE (ER−/PR+/HER2+) and triple-negative/basal-like type (ER−/ER−/HER2−) [1].
According to the cancer stem cell (CSC) hypothesis, irrespective of the molecular subtypes, breast cancer cells can evade traditional therapy and are able to enter a metastatic state because of a small population of tumorigenic cells that possess a stem/progenitor cell-like capacity of self-renewal and differentiation. These tumorigenic cells are also referred to as breast cancer stem cells (BCSCs). This tumorigenic subpopulation is considered to be responsible for intratumoral heterogeneity, drug resistance and tumor recurrence [2]. CSC identification has been largely driven by a combination of different cellular markers that are considered to be CSC specific. Indeed, this hypothesis is supported by data obtained for solid tumors from a number of carcinomas and sarcomas, including brain [3], liver [4], oral [5], colon [6], osteosarcoma [7], rhabdomyosarcoma [8], and Ewing’s sarcoma [9].
For breast cancer, Al−Hajji et al. (2003) [10] showed that cells which overexpress the transmembrane glycoprotein CD44 and underexpress the cell membrane sialoglycoprotein CD24 (CD44high/CD24−/low), possess tumorigenic properties. Since that discovery, the CD44high/CD24−/low subpopulation has been found in other carcinomas such as squamous cell carcinoma of the oral cavity and head/neck region [11].
Aldehyde dehydrogenase (ALDH) is another biomarker that has been widely used for characterizing cell stemness [12]. The ALDH superfamily is associated with alcohol metabolism, retinoic acid (RA) metabolism and protection from reactive oxygen species [13]. ALDH-dependent RA signaling is implicated in morphogenesis, cell development and stem cell regulation [12]. Cells with increased ALDH activity (ALDHbr) have tumorigenic behavior in breast cancer [14]. A few studies have also suggested that a very small subpopulation of cells that overexpress both CD44 and ALDH (CD44high/CD24−/low/ALDHbr) may be potent initiators of breast tumorigenesis [15,16].
Recent studies, however, have shown that the phenotypes identified by these markers do not universally correlate with tumorigenic cells in different molecular subtypes. Studies indicate that the ALDHbr subpopulation is pronounced in the HER2-OE, whereas the CD44high/CD24−/low subpopulation is more apparent in the triple-negative/basal-like subtype [17]. The luminal subtypes, which constitute the majority of breast cancer cases, are predominantly CD44low/CD24–/low/ALDH–/low. It remains unclear whether CD44high or ALDHbr cells play a tumorigenic role in these breast cancer subtypes [17,18]. In contrast, CD44–/low/CD24high cells have been observed in the majority of invasive breast carcinomas [19]. Moreover, in breast cancers with high ALDH activity, the expression of the ALDH1 isoform ALDH1A1 did not correlate with high metastasis or poor survival [20]. Some studies have also suggested that tumor cell metastasis may not depend exclusively on tumor cells that express these stemness-related markers [21]. More importantly, from a clinical perspective, ALDH1A1 overexpression does not significantly correlate with a poor clinical outcome in different cancers [21,22,23]. Overall, these studies present a contrasting picture of the correlation between the expression level of these markers and tumorigenicity in breast cancer cells.
Considering the conflicting nature of data regarding BCSC phenotypes, it is important to systematically investigate the expression of CD44, CD24 and ALDH1A1 in different breast cancer subtypes to understand whether the expression of these biomarkers indeed correlates with tumorigenic potential. Specifically, in luminal breast cancers, which constitute 70% of all breast cancers, it is critical to assess the tumorigenicity and BCSC-like properties of CD44low/CD24–/low/ALDH– cells, because this subpopulation reflects the major luminal breast cancer phenotype.
In this study, we performed a systematic comparison of six breast cancer cell lines representing different molecular subtypes, including luminal A (T47D and MCF-7), luminal B (BT-474), HER2-OE (AU-565) and triple-negative/basal-like subtype (MDA-MB-231 and Hs578T) for CD44, CD24 expression and ALDH activity. We first categorized the cells into different subpopulations according to biomarker expression and then carried out a systematic and comparative analysis of these subpopulations for hallmark stem cell characteristics of differentiation, migration, invasiveness, and metastatic potential using established in vitro and in vivo techniques. We also analyzed the expression of genes that are considered to be hallmarks of metastasis, pluripotency and proliferation. Finally, we assessed the expression of ALDH1 isotypes ALDH1A1 and ALDH1A3 in these phenotypes to ascertain the role of ALDH1 in BCSC function.
2. Results
2.1. Luminal Subtypes Have a Predominantly CD44−/low/CD24−/low Phenotype
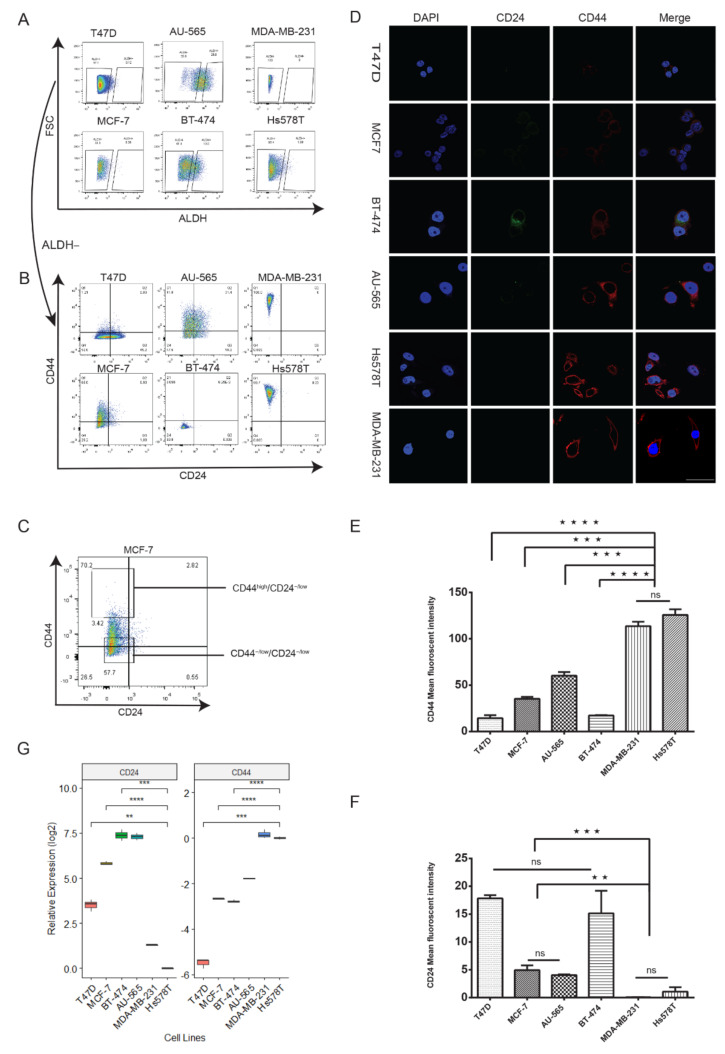
To understand the relation of CD44, CD24 and ALDH1 expression with tumorigenic cells in breast cancer, we chose six cell lines to represent the major molecular subtypes of breast cancer: MCF-7 and T47D (luminal A), BT-474 (luminal B, HER2-OE), AU-565 (HER2-OE), Hs578T and MDA-MB-231 (triple-negative/basal like). Recent research has suggested that a high CD44/CD24 ratio and high ALDH expression correlate positively with proliferation and tumorigenesis [14,24,25,26]. Using flow cytometry, we investigated the expression of CD44, CD24 and ALDH activity in these cell lines. We analyzed ALDH activity by treating the cell lines with ALDEFLUOR and calculating the percentage of ALDEFLUOR-stained cells (ALDHbr). ALDHbr cells were the highest in HER2-OE cell lines BT-474 (20%) and AU-565 (26%); the prevalence was <1% in all other lines (Figure 1A). However, we did not find ALDHbr cells in MDA-MB-231. Consistent with previous research [21,27], our results revealed that triple-negative cell lines had a primarily CD44high/CD24−/low/ALDH− profile (CD44high/CD24−/low), whereas luminal A cell lines MCF-7 and T47D had a CD44−/low/CD24−/low/ALDH− profile (CD44−/low/CD24−/low). Luminal B and HER2-OE cell lines BT-474 and AU-565 had a mixed profile of CD44−/low/CD24−/low and CD44−/low/CD24−/low/ALDHbr (ALDHbr) cells, with CD44−/low/CD24−/low constituting the majority of cells (Figure 1B, Supplementary Figure S1A). Fluorescence and real-time quantitative reverse transcription PCR (qRT-PCR) analysis of CD44 and CD24 in the cell lines showed a similar expression profile (Figure 1D–G), with triple-negative cell lines significantly overexpressing CD44. Interestingly, MCF-7 showed a lower expression of CD44 compared to AU-565 (Figure 1E, G) which is considered to have comparatively lower malignancy [28]. Taken together, these results show that luminal A breast cancer cell lines (T47D and MCF-7) were predominantly CD44−/low/CD24−/low and ALDH−. Cell lines overexpressing HER2 (BT-474 and AU-565) had a higher proportion of ALDHbr cells and triple-negative cell lines (Hs578T and MDA-MB-231) were almost exclusively CD44high/CD24−/low.
Figure 1.
CD44, CD24 and ALDH expression in breast cancer cell lines is subtype dependent (A–B) Flow cytometry analysis of ALDH, CD44 and CD24 expression in different molecular subtypes of breast cancer. Cells were treated with ALDEFLUOR reagent and double stained with anti-CD44-APC (allophycocyanin) and anti-CD24-PerCP-eFluor 710. The accuracy of the double immunostaining was confirmed by comparison with single immunostaining. FSC—forward scatter. ALDHbr cells were the highest in HER2-OE cell lines, CD44−/low/CD24−/low in luminal and CD44high/CD24−/low in triple-negative cell lines (Supplementary Figure S1). (C) Representative gating for the selection of CD44high/CD24−/low and CD44−/low/CD24−/low cells. (D) Representative immunofluorescence images showing the expression of CD24 (green), CD44 (red) and DAPI (blue) in breast cancer cell lines. Cells were stained with primary anti-CD24 and anti-CD44 antibodies. Scale bar = 100 μm. (E–F) Comparison of average fluorescence intensities of CD44 and CD24. (G) The relative mRNA expression of CD24 and CD44 by qRT-PCR shows that luminal cell lines have a low expression of both CD44 and CD24. All mRNA expression compared to Hs578T. Data represent the mean ± SD of three independent experiments; ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
2.2. Lumina CD44−/low/CD24−/low Cells Exhibit Properties of Self-Renewal and Differentiation
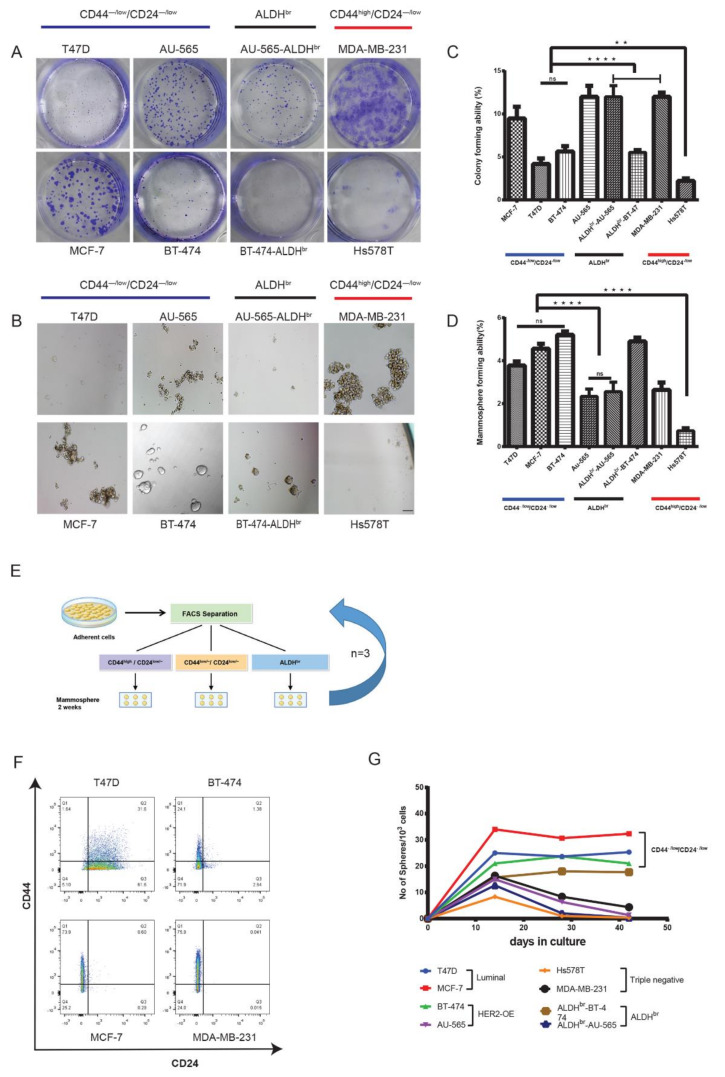
Because the luminal and HER2-OE cell lines had a predominantly CD44−/low/CD24−/low phenotype, we compared them with CD44high/CD24−/low and ALDHbr cells with respect to tumorigenicity and stem cell-like characteristics in vitro and in vivo. A number of studies have reported that cells with the CD44high/CD24−/low and ALDHbr phenotypes have greater tumorigenic potential than the CD44−/low/CD24−/low phenotype [24,29]. To verify this assertion, we carried out sphere and colony formation assays. These assays have been widely applied to assess key stem cell characteristics of cell survival and the ability of a cell to undergo “unlimited” cell division in vitro [30,31,32]. We seeded the CD44−/low/CD24−/low, CD44high/CD24−/low and ALDHbr subpopulations separated by flow cytometry at densities of 100, 500 and 1000 cells/well (in triplicate for each group) as monolayers and non-attached multicellular spheroids (mammospheres). CD44high/CD24−/low cells from the MDA-MB-231 cell line had the highest colony forming ability (CFA), whereas CD44high/CD24−/low cells from Hs578T had the lowest CFA. CD44−/low/CD24−/low cells from luminal A and B cell lines showed significantly higher CFA than CD44high/CD24−/low cells from Hs578T (Figure 2A,C). Mammosphere assay too showed a similar trend, with CD44high/CD24−/low cells from Hs578T showing the lowest mammosphere-forming ability. ALDHbr cells from HER2-OE cell lines had a similar colony and mammosphere-forming ability to that of CD44−/low/CD24−/low cells from their respective cell lines. However, both the CD44−/low/CD24−/low and ALDHbr subpopulations from the HER2-OE cell line AU-565 had high colony forming but low mammosphere-forming ability (Figure 2B,D). Interestingly, only luminal cell lines (T47D, MCF-7 and BT-474) formed tightly bound or fused spheres (Supplementary Figure S2A).
Figure 2.
Luminal CD44−/low/CD24−/low cells propagate as colonies and form long-term viable spheres. (A,C) Representative images and comparison of clonogenic assay and mammosphere assay (B,D) of the flow sorted CD44−/low/CD24−/low, CD44high/CD24−/low and ALDHbr subpopulations from breast cancer cell lines. Spheres were counted after 7 days in culture (Supplementary Figure S2). (E) Schematic representation of long-term mammosphere assay. FACS-separated populations were seeded at a density of 2 × 104 cells/mL in ultra-low-attachment plates and maintained as spheres. The whole procedure was repeated every 14 days, until 42 days (three generations). (F) FACS analysis of CD44−/low/CD24−/low, ALDHbr and CD44high/CD24−/low mammospheres after three generations (42 days) shows a decrease in CD44 expression in mammospheres of CD44high/CD24−/low origin from the MDA-MB-231 cell line. AU-565 and Hs578T cell lines could not form viable mammospheres after 7–12 days in culture. (G) Graph showing the number of viable mammospheres before each FACS separation. Spheres were counted under a microscope at 10× magnification. Data represent the mean ± SD of three independent experiments; ** p < 0.01 and **** p < 0. 0001. Scale bar = 200 μm.
Cells with tumorigenic or stem cell-like properties can survive and differentiate through multiple generations in a non-adherent environment [33]. Therefore, we compared the ability of CD44−/low/CD24−/low, ALDHbr and CD44high/CD24−/low cells to retain their diversity and viability upon serial passage as mammospheres. While the viability of CD44high/CD24−/low cells from MDA-MB-231 dropped substantially, CD44−/low/CD24−/low cells from luminal cell lines (T47D, MCF-7 and BT-474) maintained viable mammospheres through three passages over 42 days (Figure 2G). After 7–10 days, the Hs578T and AU-565 cell lines could not be propagated further owing to low viability. The proportion of cells with the CD44−/low/CD24−/low phenotype remained consistent in luminal cell lines (Figure 2F). As described previously, mammosphere assay enriches for tumorigenic cells [19], we therefore expected an increase in the proportion of the CD44high/CD24−/low and/or ALDHbr phenotypes. Instead, the proportion of CD44high/CD24–/low cells and the number of mammospheres in MDA-MB-231 decreased after the second passage (Figure 2F,G). A similar decrease in the proportion of CD44high/CD24−/low cells and mammosphere-forming ability in MDA-MB-231 was observed by Wang et al. (2014) [34], indicating a decrease in the proliferative ability of MDA-MB-231-derived cells in long-term sphere culture. Taken together, these results show that ALDHbr cells from the HER2-OE cell line Au-565 and CD44high/CD24–/low cells from triple-negative cell lines exhibit a decrease in survival and proliferative ability in sphere culture. Cells with CD44−/low/CD24−/low phenotype from luminal cell lines exhibit consistent survival and retains the ability to differentiate and maintain subpopulation diversity, which are hallmark stem cell characteristics of differentiation and long-term survival.
2.3. Luminal CD44−/low/CD24−/low Cells Express Key Pluripotency Regulators
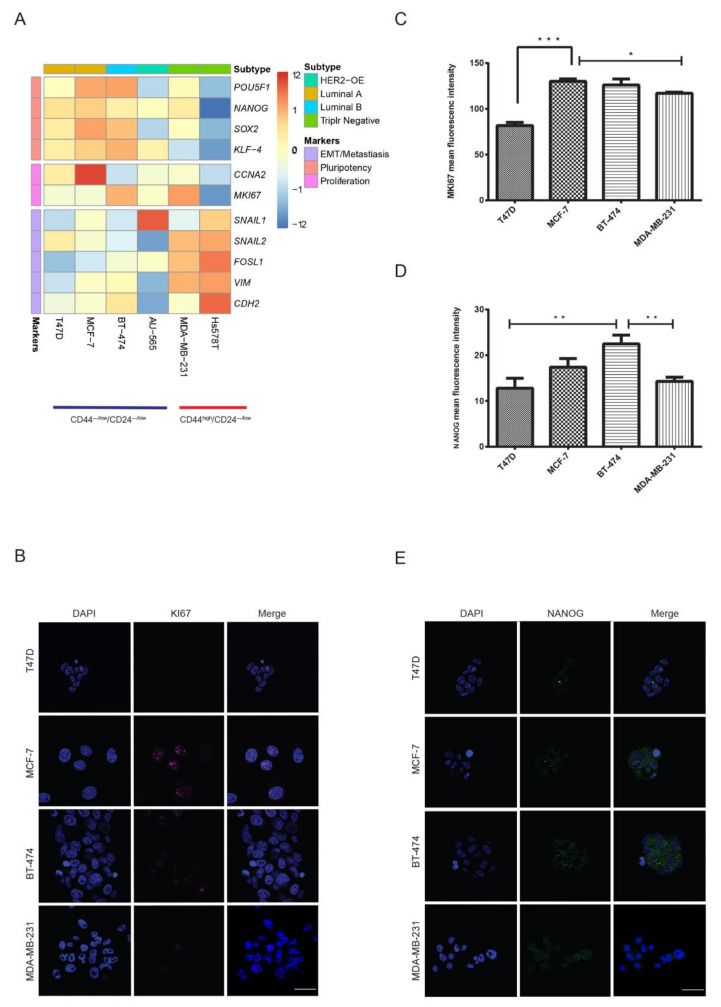
The lack of correlation between these markers and proliferative and survival capacity in the cell populations indicated differences in the expression of genes responsible for these characteristics. Therefore, we next compared the expression of key markers genes for pluripotency (POU5F1, NANOG, SOX2, KLF-4), proliferation (CCNA2, MKI67) and epithelial-mesenchymal transition(EMT)/metastasis (SNAIL1, SNAIL2, FOSL1, vimentin (VIM), CDH2) between the CD44−/low/CD24−/low (T47D, MCF-7 and BT-474, AU-565) and CD44high/CD24−/low subpopulations (Hs578T and MDA-MB-231) using qRT-PCR. There was a clear distinction between the mRNA expression profiles of these genes between CD44high/CD24−/low and CD44−/low/CD24−/low cells. Broadly, CD44−/low/CD24−/low cells from luminal cell lines (T47D, MCF-7 and BT-474) overexpressed pluripotency markers, whereas CD44high/CD24−/low cells from triple-negative cell lines (Hs578T and MDA-MB-231) showed the overexpression of EMT/metastasis markers (Figure 3A). Specifically, the expression of NANOG and SOX2, which are demonstrated to drive tumorigenesis and metastasis in breast cancer [35,36], was 5-fold higher in CD44−/low/CD24−/low cells from luminal cell lines compared to CD44high/CD24−/low cells from Hs578T. However, CD44high/CD24−/low cells from MDA-MB-231 too showed high expression of NANOG and SOX2. Conversely, FOSL1 and VIM, which are ubiquitously expressed in normal mesenchymal cells and overexpressed in many cancers [37], had a more than 5- to 10-fold higher expression in CD44high/CD24−/low cells from triple-negative/basal-like cell lines. However, compared to CD44−/low/CD24−/low cells from luminal cell lines, CD44−/low/CD24−/low cells from HER2-OE cell line AU-565 had a comparatively lower expression of pluripotency markers but a 2-fold higher expression of SNAIL1, indicating its divergence from luminal cell lines. Surprisingly, within triple-negative cell lines, the proliferation marker MKI67 was highly expressed in CD44high/CD24−/low cells from MDA-MB-231 (4-fold higher than that of Hs578T). CD44−/low/CD24−/low cells from luminal cell lines overall had a 2- to 3-fold higher expression of CCNA2 and MKI67 than that of CD44high/CD24−/low cells from Hs578T (Supplementary Figure S3A). Of all the subpopulations, CD44−/low/CD24−/low cells from AU-565 had the lowest expression of both proliferation and pluripotency markers. On the other hand, compared to CD44high/CD24−/low cells from Hs578T, CD44high/CD24−/low cells from MDA-MB-231 had a higher expression of both pluripotency and proliferation markers.
Figure 3.
Luminal CD44−/low/CD24−/low cells show a similar expression profile of pluripotency markers. Comparison of mRNA expression of markers for pluripotency, EMT/metastasis and proliferation for CD44high/CD24−/low and CD44−/low/CD24−/low cells, performed using qRT-PCR. (A) Heat map of genes with opposite expression patterns between CD44high/CD24−/low and CD44−/low/CD24−/low for pluripotency and EMT. Each row represents an RNA transcript; each column represents a cell line. CD44−/low/CD24−/low cells from luminal cell lines overexpressed pluripotency markers (Supplementary Figure S3). Representative image and analysis of immunofluorescence for MKI67 (B,C) and NANOG (D,E) expression in mammospheres from CD44−/low/CD24−/low and CD44high/CD24−/low cells. Flow cytometry sorted cell populations were maintained as spheres for 14 days and stained with DAPI (blue), NANOG (green) and MKI67 (magenta). Spheres were observed under fluorescence microscope. Data represent the mean ± SD (n = 3) of three independent experiments; * p < 0.05, ** p < 0.01 and *** p < 0.001. Scale bar = 100 μm.
Our mRNA expression results showed that the CD44−/low/CD24−/low subpopulation from luminal cell lines and CD44high/CD24−/low subpopulation from highly tumorigenic cell line MDA-MB-231 had similar expression of pluripotency markers and proliferation marker MKI67. We further compared the protein expression of NANOG and MKI67 in mammospheres generated by CD44−/low/CD24−/low cells from luminal cell lines with that of mammospheres from CD44high/CD24−/low cells of MDA-MB-231. Mammospheres of CD44−/low/CD24−/low origin from all luminal cell lines expressed NANOG equivalent to or higher than mammospheres of CD44high/CD24−/low origin. BT-474 mammospheres showed significantly higher expression than MDA-MB-231 (p < 0.01) (Figure 3D,E). Except for T47D, MKI67 expression in luminal mammospheres was similar to mammospheres from MAD-MB-231 (Figure 3B,C). These results demonstrated that the CD44−/low/CD24−/low subpopulation in luminal cell lines may indeed be comprised of cells with pluripotent potential. On the other hand, the overall low expression of proliferation and pluripotency markers in CD44high/CD24−/low cells from the Hs578T cell line revealed a stark difference in the expression of these markers within triple-negative cell lines. The overexpression of mesenchymal markers such as vimentin correlated more positively with the CD44high/CD24−/low phenotype in triple-negative cell lines.
2.4. ALDH1A3 Is Overexpressed in Breast Cancer Subtypes
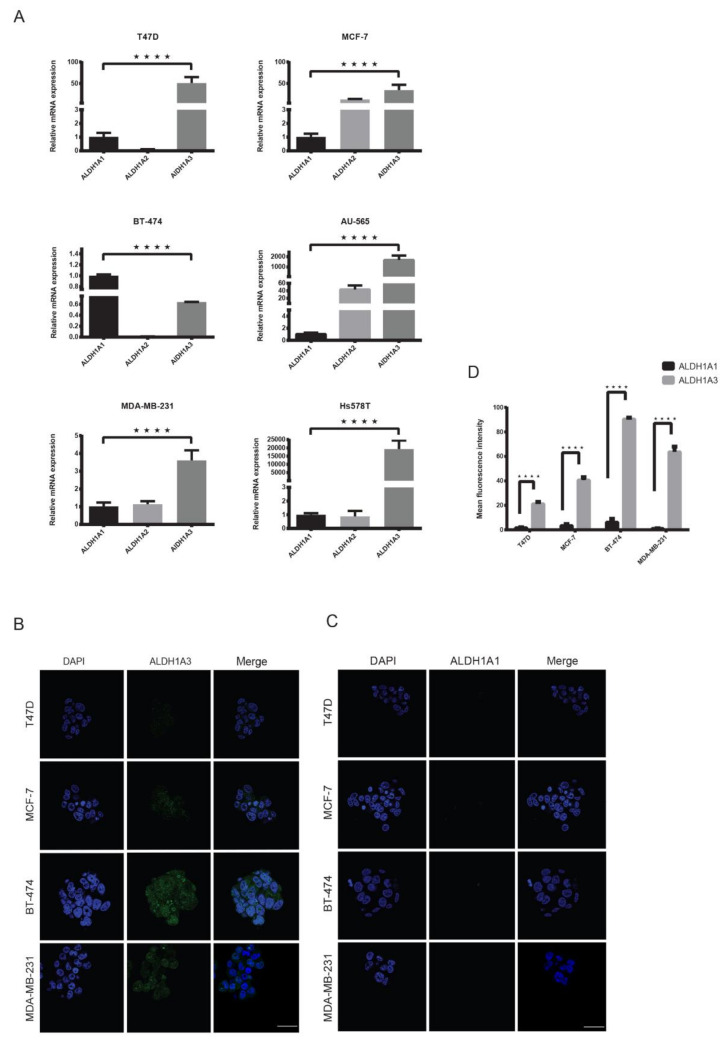
It has been hypothesized that the ALDH1 isoform ALDH1A1 is responsible for the metastatic property in breast cancer cells with high ALDH activity [25,38]. However, its role as a potential CSC remains controversial [12,22]. In our mammosphere experiments, ALDHbr cells did not show high viability and sphere-forming ability. Therefore, to verify this hypothesis, we first compared the expression of ALDH1 isoforms (A1, A2 and A3) in all breast cancer cell lines using qRT-PCR. We found that with the exception of BT-474, ALDH1A1 expression was very low in all cell lines, whereas ALDH1A3 was overexpressed in all lines (p < 0.0001) (Figure 4A). To further verify whether the mRNA expression translates to protein expression, we performed an immunofluorescence analysis of ALDH1A1 and ALDH1A3 in long-term mammospheres of luminal cell lines and compared this with the highly tumorigenic MDA-MB-231 cell line. The results showed high ALDH1A3 expression in all cell lines including BT-474 (p < 0.0001) (Figure 4B–D), which is surprising as it showed a higher expression of ALDH1A1 in qRT-PCR experiments. Finally, to analyze the claim that ALDHbr cells are key tumorigenic cells in HER2-OE breast cancer [14,26], we compared the mRNA expression of the aforementioned key markers genes for pluripotency, proliferation and EMT/metastasis between the CD44−/low/CD24−/low and ALDHbr subpopulations from the HER2-OE cell lines BT-474 and AU-565. The expression of these genes did not differ significantly between the CD44−/low/CD24−/low and ALDHbr subpopulations. Overall, these markers were marginally overexpressed in the CD44−/low/CD24−/low subpopulation, showing that ALDHbr cells may not correlate with EMT, pluripotency or proliferation in HER2-OE cell lines. (Supplementary Figure S3B). Our results support previous studies, which have shown that ALDH1A3 is highly expressed in ALDHbr cells and that ALDH1A1 expression may not be directly related to proliferation and metastasis in breast cancer [23,39].
Figure 4.
High expression of ALDH1A3 in breast cancer cell lines. (A) Relative mRNA expression analysis of ALDH1A1, ALDH1A2 and ALDH1A3 in breast cancer cell lines using qRT-PCR showing a significantly higher expression of ALDH1A3 in all cell lines except BT-474. mRNA expression was normalized to ALDH1A1. (B,C) Representative image and analysis of immunofluorescence for ALDH1A3 and ALDH1A1 expression in mammospheres showing a significantly higher expression of ALDH1A3. Mammospheres formed after 7 days were separately stained with DAPI (blue), anti-ALDH1A1 (green) and anti-ALDH1A3 (green) antibodies. (D) Comparison of A1 and A3 immunofluorescence. Scale bar = 100 μm. Data represent the mean ± SD of three independent experiments; **** p < 0.0001.
2.5. Luminal CD44−/low/CD24−/low Cells Show Strong Tumorigenic and Metastatic Behavior
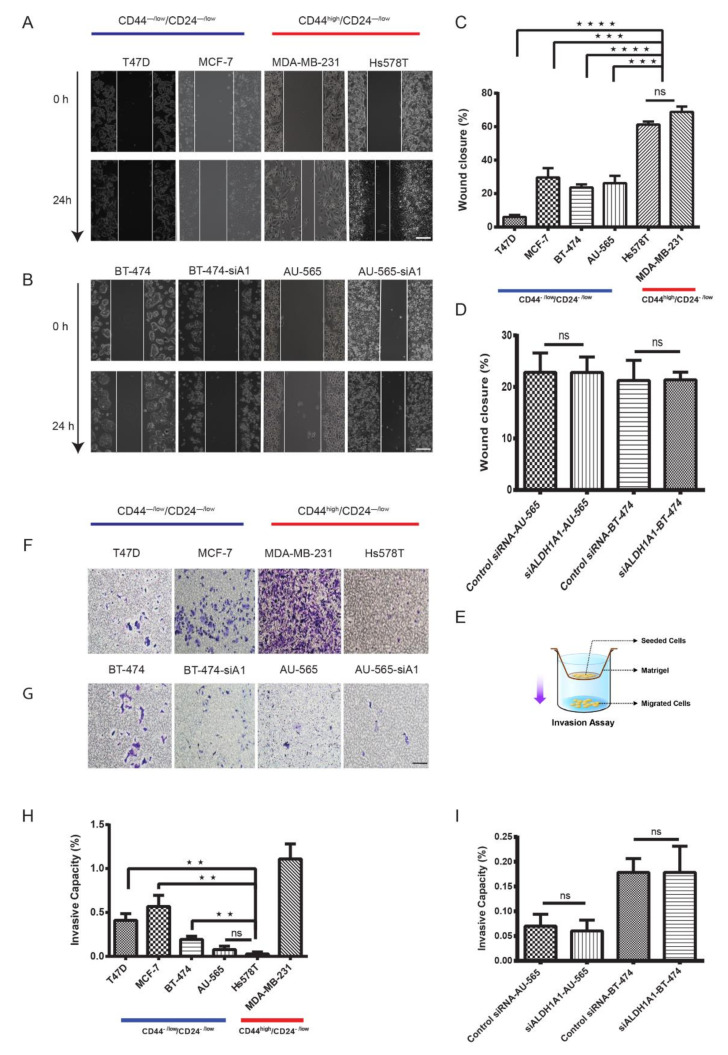
The ability of cancer cells to proliferate, migrate and invade other tissues is a key aspect of metastasis which leads to mortality in cancer patients [40]. To investigate migratory and invasive properties in the CD44−/low/CD24−/low subpopulation, we performed transwell invasion and wound healing assays. To further assess the metastatic potential of these cells, we performed xenotransplantation of CD44−/low/CD24−/low cells into the mammary fat pad of 6-week-old female NOD-scid IL2rγ (NSG) mice. Expectedly, the CD44high/CD24−/low subpopulation from the highly invasive MDA-MB-231 cell line had the highest number of cells with random migration/invasion. However, CD44−/low/CD24−/low cells from luminal cell lines MCF-7, T47D and BT-474 had a significantly higher number of (3–5 times) invasive cells than the CD44high/CD24−/low subpopulation from Hs578T and the CD44−/low/CD24−/low subpopulation from AU-565 (Figure 5F–H). The wound healing results, however, did not mirror those of the transwell assay—CD44high/CD24−/low cells from the triple-negative cell lines MDA-MB-231 and Hs578T had the highest rate of migration over a period of 24 h, whereas CD44−/low/CD24−/low cells from luminal cell lines had a significantly lower rate of migration (Figure 5A–C).
Figure 5.
Luminal CD44−/low/CD24−/low cells show low migratory but high invasive properties. Representative images and (A,B) comparison of the wound closure rate (C) of different populations show that CD44high/CD24−/low cells from triple-negative cell lines had a high rate of cell migration. (E) Schematic representation of the transwell invasion assay. The 1 × 104 cells in serum-free medium were seeded in matrigel. Cells migrating to lower chambers filled with complete medium were counted after 24 h under a microscope. (F,G) Representative images of migrated cells. (H) Comparison of invasive capacity showing low invasive properties of CD44high/CD24−/low cells and CD44−/low/CD24−/low cells from the Hs578T and AU-565 cell lines, respectively. (D,I) Comparison of siALDH1A1 with control in HER2-OE cell lines for wound healing and transwell invasion assays. Data represent the mean ± SD (n = 3); ** p < 0.01, *** p < 0.001, and **** p < 0.0001. Scale bar = 200 μm.
Our analysis of ALDH1 isotypes showed a low expression of ALDH1A1 in breast cancer cell lines. This led us to suspect that ALDH1A1, which is believed to be the key promoter of tumorigenicity in HER2-OE breast cancer [14,26], may not play a key role in tumorigenicity. We knocked down ALDH1A1 using siRNA in HER2-OE cell lines BT-474 and AU-565 and further compared its effect with normal cells in both the AU-565 and BT-474 cell lines. Transwell invasion and wound healing assays show that, as suspected, the suppression of ALDH1A1 activity in these two cell lines did not significantly affect their proliferation or migration (Figure 5D,I).
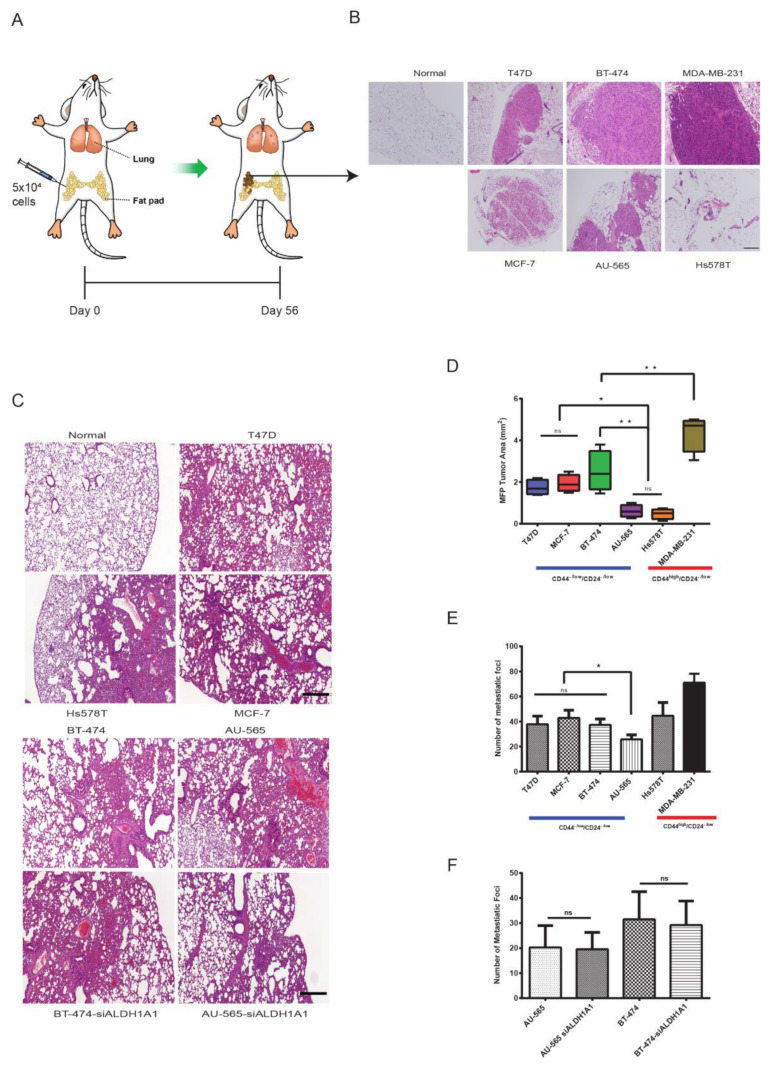
Finally, we compared the tumorigenic and metastatic potential of these populations in xenotransplanted 6-week-old female NSG mice. FACS-isolated 5 × 104 cells were injected in the mammary fat pad (MFP) at the 4th or 5th nipple site orthotopically and, after 56 days (8 weeks), lungs and the MFP were harvested. Locally, at the site of injection, MDA-MB-231 CD44high/CD24−/low xenotransplants formed the largest tumors. However, luminal CD44−/low/CD24−/low xenotransplants formed larger tumors than Hs578T (p < 0.05) (Figure 6B,D). In terms of lung metastasis, CD44−/low/CD24−/low xenotransplants from luminal cell lines formed significantly more metastatic foci (≥200 µm) compared to CD44−/low/CD24−/low xenotransplants from AU-565 (p < 0.05). CD44high/CD24−/low xenotransplants from the triple-negative MDA-MB-231 cell line showed a higher number of metastatic foci compared to CD44−/low/CD24−/low xenotransplants from luminal cell lines, while CD44high/CD24−/low xenotransplants from Hs578T had only a marginally higher number of metastatic foci compared to CD44−/low/CD24−/low xenotransplants from the luminal cell lines T47D and BT-474 (Figure 6C,E). Knockdown of ALDH1A1 did not affect the metastatic efficiency of ALDHbr cells in HER2-OE cell lines (Figure 6C,F).
Figure 6.
Luminal CD44−/low/CD24−/low cells cause metastasis in NSG mice. (A) Schematic representation of the xenotransplant experiment. Female NSG mice were injected with 5 × 104 cells from CD44high/CD24−/low, CD44−/low/CD24−/low and siALDH1 populations. The mammary fat pad (MFP) and lungs were compared for tumorigenic and metastatic potential after 8 weeks of incubation. (B) Representative images of H&E staining of the site of injection in the MFP. (C) Representative images of H&E staining of lung metastasis. (D) Analysis of tumor area in the MFP of NSG mice. (E) Analysis of lung tumor foci. (F) Comparison of lung tumor metastatic foci between non-si and siALDH1A1 in HER2-OE cell lines. Data represent the mean ± SD of three independent experiments; * p < 0.05 and ** p < 0.01. Scale bar = 200 μm.
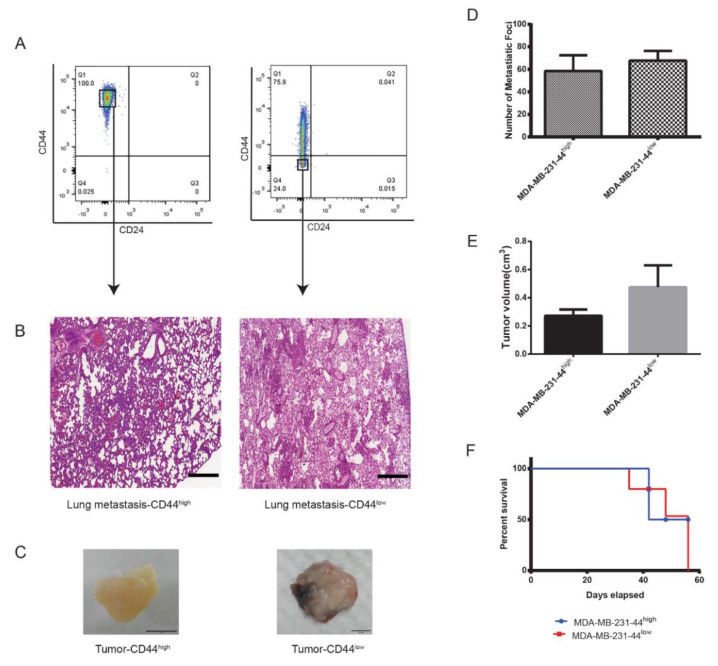
The large difference in tumorigenic behavior within triple-negative cell lines Hs578T and MDA-MB-231 led us to speculate that CD44high/CD24−/low cells may not always correlate to highly tumorigenic and invasive behavior in triple-negative cell lines. To further confirm this, we xenotransplanted 1000 cells of the CD44−/low/CD24−/low phenotype isolated from the mammosphere of MDA-MB-231 in the mammary fat pad of NSG mice. Both the CD44high/CD24−/low and CD44−/low/CD24−/low populations had comparable metastatic efficiency with the CD44−/low population forming slightly larger tumors (Figure 7C–E). Further, mice injected with the CD44−/low/CD24−/low population showed higher mortality than mice injected with CD44high/CD24−/low cells (Figure 7G). Overall, MDA-MB-231-derived CD44−/low/CD24−/low cells showed high malignancy, forming a large number of lung metastatic foci, and shortened disease-free survival.
Figure 7.
High tumorigenicity in MDA-MB-231 is not CD44 dependent. Analysis of the CD44high/CD24−/low and CD44−/low/CD24−/low populations in MDA-MB-231. Cells from short-term (7-days) and long-term (21 days) mammosphere culture were isolated by FACS (A) and injected in the MFP of female NSG mice as described earlier. (B) Representative images of H&E-stained lung metastasis. (C) Representative images of solid tumors. Scale bar = 0.5 cm. Comparison of lung metastatic foci (D), tumor volume (E), and survival rate (F). Data represent the mean ± SD of three independent experiments. Scale bar = 200 μm.
These results demonstrated that, while CD44high/CD24–/low cells from both the triple-negative cell lines had a significantly higher migration rate than CD44−/low/CD24−/low cells from luminal cell lines, the CD44high/CD24−/low cells from Hs578T were only weakly invasive, less so than CD44−/low/CD24−/low cells from luminal cell lines. ALDHbr cells did not show a significant difference in proliferative or migratory potential from CD44−/low/CD24−/low cells in their respective cell lines. Consistently, the mouse xenotransplant study revealed a similar pattern, which showed that CD44−/low/CD24−/low xenotransplants from luminal cell lines had higher tumorigenicity than CD44−/low/CD24−/low xenotransplants from AU-565 and CD44high/CD24−/low xenotransplants from Hs578T. However, in terms of lung metastasis, only CD44−/low/CD24−/low xenotransplants from AU-565 showed significantly lower metastasis than CD44−/low/CD24−/low xenotransplants from luminal cell lines. The overall weak tumorigenic and metastatic behavior of the AU-565 subpopulations is in keeping with the in vitro and gene expression results. CD44high/CD24−/low xenotransplants from the MDA-MB-231 cell line, had the highest tumorigenicity as well as metastatic potential. However, within the MAD-MB-231 cell line, mammosphere-isolated CD44−/low/CD24−/low xenotransplants showed marginally higher tumorigenicity and metastatic potential than CD44high/CD24−/low xenotransplants. Although various reports have concluded that CD44 expression is critically associated with high tumorigenicity, a direct correlation is lacking [41]. In our experiments, the CD44high/CD24−/low phenotype in triple-negative cell lines seems to be positively correlated with proliferation and migration. Moreover, CD44−/low/CD24−/low cells from luminal cell lines show low proliferative capacity but high tumorigenic and metastatic properties. Overall, CD44−/low/CD24−/low cells from luminal breast cancer cell lines seem to behave akin to pluripotent quiescent, slow-cycling cells.
2.6. CD44−/low/CD24−/low Xenotransplants Express NANOG and ALDH1A3 at Metastatic Site
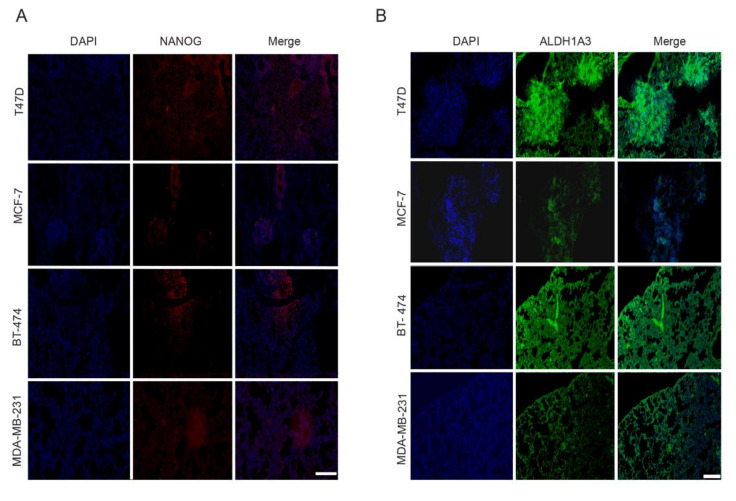
There is increasing evidence that the gene expression profile of CSCs or tumorigenic cells depends on the changes in their microenvironment and thus is dynamic [42,43]. This aspect may complicate the identification of CSC-specific biomarkers. This dynamism would be reflected in the differential gene expression observed from the primary tumor site to the site of metastasis in CSCs. To assess this dynamic, we analyzed the expression of NANOG and ALDH1A3 in lung metastatic sites by immunohistofluorescence. These genes are highly expressed in luminal CD44−/low/CD24−/low and MDA-MB-231 cells, as demonstrated by our qRT-PCR and immunofluorescence analysis. Our analysis shows that both NANOG and ALDH1A3 were consistently expressed at the metastatic site (Figure 8A,B) in all luminal xenotransplants. Mice xenotransplanted with CD44high/CD24−/low cells from the triple-negative MDA-MB-231 cell line showed a similar expression of both NANOG and ALDH1A3. Overall, our results show that the expression of NANOG and ALDH1A3 remains stable during metastasis, probably not affected by change in the tumor microenvironment.
Figure 8.
Luminal CD44−/low/CD24−/low cells express NANOG and ALDH1A3 at the metastasis site. Representative images of double immunohistostained metastatic sections of lungs from NSG mice. (A) DAPI (blue), anti-NANOG (red) (B) DAPI (blue), anti-ALDH1A3 (green). Scale bar = 200 μm.
3. Discussion
Identification of tumorigenic cells specific for luminal breast cancer, which represents more than 70% of all breast cancers, will significantly improve understanding of tumor microenvironment and signaling events that are involved in discrete steps of breast cancer progression, including metastasis. This insight is critical because, despite improved prognosis of breast cancer due to the availability of targeted agents, drug resistance and tumor recurrence still remain major concerns. Here, we show that, in luminal breast cancer cell lines, cells with the CD44−/low/CD24−/low phenotype display features typically associated with cells with tumorigenic and metastatic property.
Previous studies have shown cells with the CD44high/CD24−/low and ALDHbr phenotypes to be highly tumorigenic, potential BCSCs [12,24,25,44,45]. Cells with high ALDH activity are considered to possess stem cell-like properties in both normal and malignant cells [13,46] and ALDH1A1 expression has been shown to correlate with higher metastasis and worse prognosis in clinical samples of breast cancer [23,47]. However, a direct association between these phenotypes and invasion, homing and proliferation at sites of metastasis is lacking [21]. MCF-7, which is a luminal A, ER-positive cell line, is one such example to the contrary. The CD44−/low/CD24−/low subpopulation from MCF-7 causes osteosclerotic bone lesions on intracardiac injection in nude mice, showing high invasiveness and metastatic potential [48]. By comparing and analyzing the tumorigenic and metastatic properties of these two phenotypes with the CD44−/low/CD24−/low phenotype, we found that tumorigenic cells in luminal cell lines typically had a CD44−/low/CD24−/low phenotype, whereas the CD44high/CD24−/low phenotype was associated with triple-negative/basal-like cell lines. Similar to previous studies [29], we found that ALDH expression was the strongest in HER2-OE cell lines, while it was very low in luminal and triple-negative/basal-like cell lines. Contrary to previous reports, we did not find differences in the tumorigenic ability of ALDHbr cells vis-a-vis CD44−/low/CD24−/low cells. In terms of invasiveness, metastasis and tumorigenicity, CD44−/low/CD24−/low cells from MDA-MB-231 had the highest tumorigenic and invasive capacity, whereas these properties were the weakest in CD44high/CD24−/low and ALDHbr cells from the Hs578T and AU-565 cell lines, respectively. These results suggest that the CD44high/CD24−/low and ALDHbr phenotypes may not universally relate to high tumorigenicity or invasiveness in breast cancer subtypes. Our results are more supportive of the argument put forth in a few previous studies [49,50] that these phenotypes could be markers for HER2-OE and triple-negative/basal-like breast cancer.
The role of ALDH isozyme ALDH1A1 in cancer has been a subject of intensive study, with a number of reports implicating that it is a key factor in maintaining stem cell-like properties in ALDHbr cells while some reports dispute this role [22,51,52]. We therefore determined the role of ALDH1A1 in ALDHbr cells by first measuring its mRNA and protein expression and then using siRNA to specifically knockdown ALDH1A1. At the mRNA level, except for BT-474, all cell lines showed a significantly high level of ALDH1A3 but not A1. Moreover, knockdown of ALDH1A1 did not show a significant difference in proliferation, migration or metastasis in ALDHbr cell lines. The CD44−/low/CD24−/low subpopulation showed self−renewal, proliferative and metastatic properties equivalent to the ALDHbr subpopulation in HER2-OE cell lines. Interestingly, ALDHbr cells from luminal A subtype did not show tumorigenic and metastatic properties and did not survive long-term sphere culture, indicating that ALDH activity may not a key marker for cancer progression in luminal A cell lines. Our results also show that ALDH1A3 is overexpressed in breast cancer cell lines of different subtypes. Further studies are required to ascertain whether the overexpression of ALDH1A3 plays a role in tumorigenesis and metastasis. Overall, our results show that ALDH1A1 expression is not consistent across breast cancer subtypes, and the presence of ALDHbr cells may not be a marker of strong pluripotency or metastatic behavior. The presence and proportion of ALDHbr cells in MDA-MB-231 is unclear. Some investigators have reported no presence of ALDHbr cells in MDA-MB-231, whereas others have reported the presence of a small CD44high/CD24−/ALDHbr subpopulation considered to be highly tumorigenic [17,21,25,26]. However, in our experiments, we could not find ALDHbr cells in the MDA-MB-231 cell line. We were not able ascertain whether CD44high/CD24−/low/ALDHbr cells did indeed have a comparatively higher tumorigenic potential than CD44high/CD24−/low cells. A more detailed study encompassing an in-depth mechanistic and transcriptomic experimental design is required in order to understand the role of ALDHbr cells in breast cancer.
Gene expression analysis of key EMT, metastasis and pluripotency markers has been widely implied in many studies to identify tumorigenic or stem cell-like cells in cancer [53]. Our qRT-PCR and immunofluorescence analysis of a number of these genes in CD44high/CD24−/low, CD44−/low/CD24−/low and ALDHbr cells shows that luminal CD44−/low/CD24−/low cells and CD44high/CD24−/low cells from the triple-negative MDA-MB-231 cell line overexpress stem cell marker genes NANOG, KLF4 and SOX2 compared to their counterparts from AU-565 and Hs578T—both of which showed significantly low invasiveness in the transwell assay. SNAIL2, VIM and FOSL1 genes, which are typically associated with EMT/metastasis and mesenchymal cells [54], had a higher expression in both the triple-negative cell lines. ALDHbr cells from both BT-474 and AU-565 cell lines showed expression profiles very similar to CD44−/low/CD24−/low cells of their respective cell lines. Expression analysis of proliferation marker MKI67 showed that except for T47D, MKI67 was overexpressed in CD44−/low/CD24−/low cells from luminal and CD44high/CD24−/low cells from MDA-MB-231 cell lines. The sharp difference in the invasive and metastatic behavior and expression profile of key pluripotency markers between CD44high/CD24−/low cells from the Hs578T and MDA-MB-231 cell lines does indicate an intrinsic variation in tumorigenic potential within cell lines with CD44high/CD24−/low phenotype. It would seem that the relatively high expression of pluripotency makers in luminal CD44−/low/CD24−/low cells is the reason for their long-term survival and tumorigenicity, perhaps also contributing to therapy resistance [55]. Further investigation of the tumorigenic potential of CD44−/low/CD24−/low cells in luminal breast cancer will be the key to understanding their tumorigenic potential and in developing biomarkers which identify the tumorigenic cells with more accuracy.
The importance of NANOG in the maintenance of pluripotency and proliferation is well documented [56,57]. It is therefore likely that upregulation of NANOG expression is critically related to tumorigenic cells in cancer [58]. Recently, Lu et al. [35] elucidated the role of NANOG in promoting breast cancer metastasis. Our comparison of NANOG expression in CD44−/low/CD24−/low cells from luminal cell lines and CD44high/CD24−/low cells from the highly invasive triple-negative MDA-MB-231 cell line showed similar expression levels, indicating their potential key role in maintaining pluripotency in these cell populations. Aberrations in developmentally conserved signaling pathways such as Janus kinase (JAK)/signal transducer activator of transcription protein (STAT) and activator of transcription 3 (STAT3) signaling pathways are deemed to be critical for the formation of CSCs [59]. In the JAK/STAT pathway, the phosphorylation of STAT3 results in enhancement of stem cell marker genes POU5F1, SOX2, KLF4 and NANOG, leading to tumorsphere formation and drug resistance [60,61]. It is therefore likely that the activation of the JAK/STAT pathway is the key factor for the tumorigenic behavior of CD44−/low/CD24−/low cells. Further, various other signaling pathways and signaling crosstalk could contribute to the formation of breast CSCs, and therefore further research into the activation of these signaling pathways, particularly the JAK/STAT pathway, is required in order to understand the genetic and molecular mechanism involved in the maintenance of tumorigenic populations in breast cancer.
As our results are based on representative cell lines, further research with primary and metastatic tumor-derived samples from both luminal and triple-negative breast cancer patients representing different age groups is critical in order to understand the tumorigenic role of these phenotypes in breast cancer.
4. Materials and Methods
4.1. Antibodies and Reagents
Supplementary Table S1 provides a list of all reagents and antibodies used.
4.2. Cell Culture
All cell lines were obtained from the Bioresource Collection and Research Center, Taipei, Taiwan. T47D, AU-565, MCF-7, BT-474, Hs578T and MDA-MB-231 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were maintained in a humidified atmosphere of 5% CO2/95% air at 37 °C. Cells were subcultivated using 0.25% trypsin and 5 mM Ethylenediaminetetraacetic acid (EDTA).
4.3. Mammosphere Culture
Cell lines were plated into ultra-low-attachment plates (Corning, NY, USA) at a density of 2 × 104 cells/mL and cultured in serum-free DMEM/F12 (1:1) medium supplemented with 20 ng/mL epidermal growth factor (EGF, PeproTech, St. Louis, MO, USA), 10 ng/mL basic fibroblast growth factor (b-FGF, PeproTech, St. Louis, MO, USA), ITS (insulin, transferrin and selenium, Sigma-Aldrich, St. Louis, MO, USA), and B27 (GIBCO, Waltham, MA, USA). A volume of 2 mL fresh mammosphere media was added every 2 to 3 days without decanting old media. Mammospheres were either disassociated using Accutase reagent or collected every 7, 14, 21 and 42 days for further analysis. Mammosphere-forming efficiency (MFE) was calculated using the formula: MFE = (number of mammospheres counted/number of cells plated) × 100.
4.4. ALDEFLUOR Assay
Breast cancer cells at the logarithmic growth phase were digested with 0.25% trypsin or Accutase (from Mammosphere) and washed three times with phosphate-buffered saline (PBS), followed by resuspension in 100 µl PBS. The ALDEFLUOR kit (StemCell Technologies, Durham, NC, USA) was used to analyze the population with high ALDH enzymatic activity. Briefly, the cells were incubated in the ALDEFLUOR assay buffer containing ALDH substrate BODIPY-aminoacetaldehyde (BAAA), 1 μmol per 1 × 106 cells, and incubated for 40 min at 37 °C. Each experiment included a negative control that contained 50 mM diethylaminobenzaldehyde (DEAB), which specifically inhibits ALDH activity.
4.5. Fluorescence-Activated Cell Sorting (FACS)
Immediately after ALDEFLOUR staining, the cells were stained with anti-CD44-APC (allophycocyanin) and anti-CD24-PerCP-eFluor 710, and separate aliquots of cells were single stained with anti-CD44-APC and anti-CD24-PerCP-eFluor 710 and used as controls. Aliquots were also treated with their isotype controls at 4 °C in the dark for 15 min. Dead cells were stained with 7-aminoactinomycin D (7-AAD). The samples were then washed by PBS three times and finally re-suspended in 500 μL PBS. Flow cytometry analysis was performed with a BD Facs Aria IIIu Flow Cytometer (BD Bioscience Franklin Lakes, NJ, USA).
4.6. Colony Formation Assay
After flow cytometry, separated cells were directly seeded on 24-well plates (Corning, NY, USA, #3526) supplemented with DMEM containing 10% fetal bovine serum. Cells were plated at different densities (100, 500 and 1000 cells/well) with 3 replicates of each density, and the plates were incubated at 37 °C in a 5% CO2 environment for 1–3 weeks depending on the cell line. The plates were then washed with 0.9% saline and fixed with 10% neutral buffered formalin solution for 15–30 min. The wells of each plate were stained with 5 mL 0.01% (w/v) crystal violet in dH2O for 30−60 min, washed with distilled water and counted using a microscope or stereomicroscope. Colonies containing more than 50 individual cells were counted and image captured. Colony forming efficiency (CFE) was calculated using the formula: CFE = (number of colonies counted/number of cells plated) × 100.
4.7. Transwell Migration Assay
The Transwell assay was applied using the transwell chamber with 8.0 μm pore polycarbonate membrane inserts (Corning, NY, USA, #3422). Flow cytometry isolated subpopulations from the aforementioned breast cancer cell lines were seeded into the upper chambers of the inserts (1 × 104 cells/chamber) in 200 μL serum-free DMEM/F-12 medium at 37 °C. Complete DMEM (600 μL) was added in the lower chambers. After 48 h of incubation, cells migrating to lower chambers were dyed with crystal violet and counted under a microscope.
4.8. Wound Healing Assay
FACS-sorted or siRNA-treated cells were plated onto a 6-well plate to create confluent monolayers. Then 2-well culture inserts (Ibidi, Martinsried, Planegg, Germany #80209) were used in petri plates (Corning, NY, USA, #353801) to create a uniform “scratch” area across each monolayer. The insert was removed, and 2 mL complete medium was added. The dishes were placed under a live cell-imaging phase-contrast microscope (Leica DMI 6000 B, Wetzlar, Germany) and images were acquired at 1 h intervals. For each image, the distance between either side of the scratch was measured at certain intervals (mm). By comparing the distances from time 0 to the last time point (24 h), the migration distance of each cell was obtained.
4.9. Immunofluorescence, Immunohistochemistry, and Image Quantitative Analysis
For monolayers, cells were plated at a concentration of 5 × 104 cells/mL in the wells of 8-well chamber slides (Ibidi Martinsried, Planegg, Germany). The cells were fixed with 4% PFA for 20 min and permeabilized with PBS containing 0.2% (w/v) Triton X-100 on the following day. The cells were then blocked for 30 min at 37 °C with 5% bovine serum albumin (BSA) and incubated at 37 °C for 1 h with anti-CD44 primary antibody and anti-CD24-PE. Mammospheres acquired after 7 days in culture were incubated at 37 °C for 1 h with unconjugated primary antibodies for ALDH1A1, ALDH1A3, and NANOG and MKI67 (conjugated with Alexa Fluor 647). Cells stained with unconjugated primary antibodies were then washed with PBS and further stained for 1 h in dark at 37 °C. Cells incubated with primary antibodies were further stained with the appropriate secondary antibodies. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen, Waltham, MA, USA). The samples were then washed twice with PBS and mounted with anti-bleaching coverslips. For histochemical staining, tissue sections were stained with hematoxylin and eosin (H&E). For immunohistostaining, Paraffin-embedded sections were deparaffinized in xylene and rehydrated in a graded series of ethanol. Antigen enhancement was carried out by incubating the sections in citrate buffer, pH 6 (Dakocytomation, Copenhagen, Denmark). Double fluorescence staining for ALDH1A3 and NANOG was carried out using the aforementioned procedure. All the samples were examined and photographed using a single-photon laser confocal imaging system, Zeiss510 (Carl Zeiss, Oberkochen, Baden-Württemberg, Germany).
4.10. Mouse Xenotransplant
All the animal experiments were performed according to guidelines for the care and use of laboratory animals of the Animal Study Committee of the Institute of Biomedical Sciences, Academia Sinica, and were carried out according to the protocol approved by the Institutional Animal Care Facility (IACUC-14-9-733). Mice were maintained under specific pathogen-free conditions, and all the efforts were made to minimize animal suffering. Female NOD-scid IL2rγ (NSG) mice at 6 weeks of age were purchased from Animal Laboratory, Institute of Biomedical Sciences, Academia Sinica. After trypsinization, different subpopulations from breast cancer cell lines were isolated by flow cytometry. The cells were washed once with PBS, resuspended in culture medium at 5 × 104 cells per 100 µL, and injected in triplicate into mammary fat pads of female NSG mice. Mice with xenotransplants from ER+ cell lines (T47D, MCF-7 and BT-474) were supplemented with 0.18 mg 60-day release 17β-estradiol (Innovative Research of America, Sarasota, FL, USA, SE-121). The mice were monitored daily for 8 weeks. In the case of palpable tumors, size was measured with calipers after sacrifice. The tumor volume was determined by the formula: tumor volume = 1/2(length × width2). Lungs, the mammary fat pad and solid tissue (if applicable) were obtained from mice after euthanization, fixed in 4% paraformaldehyde (PFA) (Sigma, St. Louis, MO, USA) at 4 °C and processed for H&E and immunohistochemical staining by embedding in paraffin. Average lung metastatic foci were estimated by counting the number of metastases of size 200 µm or greater in each H&E-stained lung section.
4.11. Quantitative Reverse Transcription-PCR (qRT-PCR)
Total RNA was extracted according to the RNeasy microextraction kit (Qiagen, Hilden, Germany, #74004) from the FACS-isolated phenotypes. Extracted RNA was reverse-transcribed to generate first-strand cDNA (QuantiTect Reverse Transcription Kit, Qiagen, Hilden, Germany) for use in qPCR. qPCR was performed with Dnase-treated RNA using the Power SYBR Green Master Mix (Thermofisher Scientific, Waltham, MA, USA) on a real-time PCR system (Applied Biosystems, Waltham, MA, USA). qPCR primers were designed using PrimER-Blast software (http://www.ncbi.nlm.nih.gov/tools/primer-blast) and were based on previous studies. Supplementary Table S2 lists all primers used for this study.
4.12. RNA Silencing
Double-stranded ALDH1 (SMARTpool: ON-TARGETplus ALDH1 siRNA, L-008722-00-0005 5 nmol) siRNA and a scrambled control siRNA were purchased from Dharmacon (Lafayette, CO, USA). Cells at 50% confluence were transfected with ALDH1A1 siRNA in triplicate in 2 mL complete medium in 6-well plates. Transfections were performed with 50 nM siRNA using transfection reagent (DharmaFECT 1, T-2001-02, Dharmacon). The cells were then incubated at 37 °C in 5% CO2 for 48 h, harvested, and processed for further assays.
4.13. Statistical Analysis
GraphPad Software (version 8.0.0 for Windows, San Diego, CA, USA) and R package ggplot2 (https://ggplot2.tidyverse.org) were used to analyze data. Two-tailed student’s t-tests and one-way analysis of variance (ANOVA) were used to determine statistical differences. p-values <0.05 were considered statistically significant.
5. Conclusions
Tumorigenic cells for luminal breast cancers, which represent the majority of breast cancers, remain to be identified. In this report, we show that luminal breast cancer cell lines are enriched in CD44−/low/CD24−/low cells, which show characteristic properties of stemness and tumorigenicity, and we therefore conclude that this population contains tumorigenic and/or cancer progenitor cells.
Acknowledgments
The authors would like to thank the Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII108-113) for cell sorting and fluorescence microscopy service, Ms. Chi-Chun Huang, department of pathology and laboratory medicine, University of California, Davis, and Mr. Kolin Liao, Institute of Biomedical Sciences and Academia Sinica Pathology Core Facility, for their help and guidance in animal handling, xenotransplant and tissue preparation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/5/1239/s1, Figure S1: Gating strategy to isolate ALDHbr, CD44high/CD24−/low and CD44−/low/CD24−/low populations, Figure S2: Luminal CD44−/low/CD24−/low cells form tightly packed spheres, Figure S3: Relative expression of markers for pluripotency, proliferation and EMT/metastasis, Table S1: Antibodies and reagents used, Table S2: Primers used for qRT-PCR.
Author Contributions
Conceptualization, R.V.; Formal analysis, R.V.; Funding acquisition, C.-Y.S.; Methodology, S.-C.H.; Project administration, C.-Y.S.; Resources, W.C.C. and S.-C.H.; Supervision, C.-Y.S.; Visualization, R.V.; Writing–original draft, R.V.; Writing–review & editing, R.V. and W.C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Rouzier R., Perou C.M., Symmans W.F., Ibrahim N., Cristofanilli M., Anderson K., Hess K.R., Stec J., Ayers M., Wagner P., et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 2.Yu Z., Pestell T.G., Lisanti M.P., Pestell R.G. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012;44:2144–2151. doi: 10.1016/j.biocel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan P.M., Frame M. Identification of a novel cancer stem cell that might be important for glioma cell invasion. Lancet. 2014;383:S10. doi: 10.1016/S0140-6736(14)60273-7. [DOI] [Google Scholar]
- 4.Ma S., Chan K.-W., Hu L., Lee T.K.-W., Wo J.Y.-H., Ng I.O.-L., Zheng B.-J., Guan X.-Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Han J., Fujisawa T., Husain S.R., Puri R.K. Identification and characterization of cancer stem cells in human head and neck squamous cell carcinoma. BMC Cancer. 2014;14:173. doi: 10.1186/1471-2407-14-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weichert W., Denkert C., Burkhardt M., Gansukh T., Bellach J., Altevogt P., Dietel M., Kristiansen G. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin. Cancer Res. 2005;11:6574–6581. doi: 10.1158/1078-0432.CCR-05-0606. [DOI] [PubMed] [Google Scholar]
- 7.Tirino V., Desiderio V., d’Aquino R., De Francesco F., Pirozzi G., Graziano A., Galderisi U., Cavaliere C., De Rosa A., Papaccio G., et al. Detection and characterization of CD133+ cancer stem cells in human solid tumours. PLoS ONE. 2008;3:e3469. doi: 10.1371/annotation/6475ab87-5c24-4ee6-9989-0762186dd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pressey J.G., Haas M.C., Pressey C.S., Kelly V.M., Parker J.N., Gillespie G.Y., Friedman G.K. CD133 marks a myogenically primitive subpopulation in rhabdomyosarcoma cell lines that are relatively chemoresistant but sensitive to mutant HSV. Pediatric Blood Cancer. 2013;60:45–52. doi: 10.1002/pbc.24117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suvà M.-L., Riggi N., Stehle J.-C., Baumer K., Tercier S., Joseph J.-M., Suvà D., Clément V., Provero P., Cironi L., et al. Identification of Cancer Stem Cells in Ewing’s Sarcoma. Cancer Res. 2009;69:1776–1781. doi: 10.1158/0008-5472.CAN-08-2242. [DOI] [PubMed] [Google Scholar]
- 10.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S.J., Bourguignon L.Y.W. Role of hyaluronan-mediated CD44 signaling in head and neck squamous cell carcinoma progression and chemoresistance. Am. J. Pathol. 2011;178:956–963. doi: 10.1016/j.ajpath.2010.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomita H., Tanaka K., Tanaka T., Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7:11018–11032. doi: 10.18632/oncotarget.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma I., Allan A.L. The Role of Human Aldehyde Dehydrogenase in Normal and Cancer Stem Cells. Stem Cell Rev. Rep. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 14.Ginestier C., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C.G., Liu S., et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colacino J.A., Azizi E., Brooks M.D., Harouaka R., Fouladdel S., McDermott S.P., Lee M., Hill D., Madden J., Boerner J., et al. Heterogeneity of Human Breast Stem and Progenitor Cells as Revealed by Transcriptional Profiling. Stem Cell Rep. 2018 doi: 10.1016/j.stemcr.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S., Cong Y., Wang D., Sun Y., Deng L., Liu Y., Martin-Trevino R., Shang L., McDermott S.P., Landis M.D., et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of their Normal Counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryspayeva D.E., Smolanka I.I., Dudnichenko А.S., Lyashenko A.A., Grinevich Y.A., Gurianov V.G., Koshubarova M.V., Seleznev A.A. Are CD44+/CD24- cells the assumed cancer stem cells in breast cancer? Exp. Oncol. 2017;39:224–228. doi: 10.31768/2312-8852.2017.39(3):224-228. [DOI] [PubMed] [Google Scholar]
- 18.Significance of CD44 and CD24 as Cancer Stem Cell Markers: An Enduring Ambiguity. [(accessed on 26 April 2020)]; doi: 10.1155/2012/708036. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3369436/ [DOI] [PMC free article] [PubMed]
- 19.Rabinovich I., Sebastião A.P.M., Lima R.S., de Urban C.A., Schunemann E., Anselmi K.F., Elifio-Esposito S., de Noronha L., Moreno-Amaral A.N. Cancer stem cell markers ALDH1 and CD44+/CD24– phenotype and their prognosis impact in invasive ductal carcinoma. Eur. J. Histochem. 2018;62:231–237. doi: 10.4081/ejh.2018.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan E.Y., Thike A.A., Tan P.H. ALDH1 expression is enriched in breast cancers arising in young women but does not predict outcome. Br. J. Cancer. 2013;109:109–113. doi: 10.1038/bjc.2013.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Nenutil R., Appleyard M.V., Murray K., Boylan M., Thompson A.M., Coates P.J. Lack of correlation of stem cell markers in breast cancer stem cells. Br. J. Cancer. 2014;110:2063–2071. doi: 10.1038/bjc.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka K., Tomita H., Hisamatsu K., Nakashima T., Hatano Y., Sasaki Y., Osada S., Tanaka T., Miyazaki T., Yoshida K., et al. ALDH1A1-overexpressing cells are differentiated cells but not cancer stem or progenitor cells in human hepatocellular carcinoma. Oncotarget. 2015;6:24722–24732. doi: 10.18632/oncotarget.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croker A.K., Rodriguez-Torres M., Xia Y., Pardhan S., Sing Leong H., Lewis J.D., Allan A.L. Differential Functional Roles of ALDH1A1 and ALDH1A3 in Mediating Metastatic Behavior and Therapy Resistance of Human Breast Cancer Cells. Int. J. Mol. Sci. 2017;18:2039. doi: 10.3390/ijms18102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan W., Chen Y., Yao Y., Zhang H., Wang T. Increased invasion and tumorigenicity capacity of CD44+/CD24- breast cancer MCF7 cells in vitro and in nude mice. Cancer Cell Int. 2013;13:62. doi: 10.1186/1475-2867-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W., Ma H., Zhang J., Zhu L., Wang C., Yang Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 2017;7:13856. doi: 10.1038/s41598-017-14364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croker A.K., Goodale D., Chu J., Postenka C., Hedley B.D., Hess D.A., Allan A.L. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J. Cell. Mol. Med. 2009;13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricardo S., Vieira A.F., Gerhard R., Leitao D., Pinto R., Cameselle-Teijeiro J.F., Milanezi F., Schmitt F., Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011;64:937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- 28.Kenny P.A., Lee G.Y., Myers C.A., Neve R.M., Semeiks J.R., Spellman P.T., Lorenz K., Lee E.H., Barcellos-Hoff M.H., Petersen O.W., et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C., Luo Y., Liu X., Lu P., Zhao Z. Clinical Implications of CD44+/CD24− Tumor Cell Ratio in Breast Cancer. Cancer Biother. Radiopharm. 2012;27:324–328. doi: 10.1089/cbr.2011.1155. [DOI] [PubMed] [Google Scholar]
- 30.Shaheen S., Ahmed M., Lorenzi F., Nateri A.S. Spheroid-Formation (Colonosphere) Assay for in Vitro Assessment and Expansion of Stem Cells in Colon Cancer. Stem Cell Rev. 2016;12:492–499. doi: 10.1007/s12015-016-9664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manuel Iglesias J., Beloqui I., Garcia-Garcia F., Leis O., Vazquez-Martin A., Eguiara A., Cufi S., Pavon A., Menendez J.A., Dopazo J., et al. Mammosphere formation in breast carcinoma cell lines depends upon expression of E-cadherin. PLoS ONE. 2013;8:e77281. doi: 10.1371/journal.pone.0077281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajendran V., Jain M.V. Methods in Molecular Biology. Volume 1692. Humana Press Inc.; New York, NY, USA: 2018. In vitro tumorigenic assay: Colony forming assay for cancer stem cells; pp. 89–95. [DOI] [PubMed] [Google Scholar]
- 33.Dontu G., Abdallah W.M., Foley J.M., Jackson K.W., Clarke M.F., Kawamura M.J., Wicha M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R., Lv Q., Meng W., Tan Q., Zhang S., Mo X., Yang X. Comparison of mammosphere formation from breast cancer cell lines and primary breast tumors. J. Thorac. Dis. 2014;6:829–837. doi: 10.3978/j.issn.2072-1439.2014.03.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu X., Mazur S.J., Lin T., Appella E., Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2014;33:2655–2664. doi: 10.1038/onc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domenici G., Aurrekoetxea-Rodríguez I., Simões B.M., Rábano M., Lee S.Y., Millán J.S., Comaills V., Oliemuller E., López-Ruiz J.A., Zabalza I., et al. A Sox2–Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene. 2019;38:3151–3169. doi: 10.1038/s41388-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brzozowa M., Wyrobiec G., Kołodziej I., Sitarski M., Matysiak N., Reichman-Warmusz E., Żaba M., Wojnicz R. The aberrant overexpression of vimentin is linked to a more aggressive status in tumours of the gastrointestinal tract. Przeglad gastroenterologiczny. 2015;10:7–11. doi: 10.5114/pg.2014.47502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma F., Li H., Li Y., Ding X., Wang H., Fan Y., Lin C., Qian H., Xu B. Aldehyde dehydrogenase 1 (ALDH1) expression is an independent prognostic factor in triple negative breast cancer (TNBC) Medicine. 2017;96 doi: 10.1097/MD.0000000000006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcato P., Dean C.A., Pan D., Araslanova R., Gillis M., Joshi M., Helyer L., Pan L., Leidal A., Gujar S., et al. Aldehyde Dehydrogenase Activity of Breast Cancer Stem Cells Is Primarily Due To Isoform ALDH1A3 and Its Expression Is Predictive of Metastasis. Stem Cells. 2011;29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 40.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging Biological Principles of Metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann C., Jobs G., Thomas M., Burtscher H., Kubbies M. Established breast cancer stem cell markers do not correlate with in vivo tumorigenicity of tumor-initiating cells. Int. J. Oncol. 2012;41:1932–1942. doi: 10.3892/ijo.2012.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwamoto T., Niikura N., Ogiya R., Yasojima H., Watanabe K.I., Kanbayashi C., Tsuneizumi M., Matsui A., Fujisawa T., Iwasa T., et al. Distinct gene expression profiles between primary breast cancers and brain metastases from pair-matched samples. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-50099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed F., Haass N.K. Microenvironment-driven dynamic heterogeneity and phenotypic plasticity as a mechanism of melanoma therapy resistance. Front. Oncol. 2018;8:173. doi: 10.3389/fonc.2018.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McFarlane S., Coulter J.A., Tibbits P., O’Grady A., McFarlane C., Montgomery N., Hill A., McCarthy H.O., Young L.S., Kay E.W., et al. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget. 2015;6:11465–11476. doi: 10.18632/oncotarget.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheridan C., Kishimoto H., Fuchs R.K., Mehrotra S., Bhat-Nakshatri P., Turner C.H., Goulet R., Badve S., Nakshatri H. CD44+/CD24-breast cancer cells exhibit enhanced invasive properties: An early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreb J. Aldehyde Dehydrogenase as a Marker for Stem Cells. Curr. Stem Cell Res. Ther. 2008;3:237–246. doi: 10.2174/157488808786734006. [DOI] [PubMed] [Google Scholar]
- 47.Sjostrom M., Hartman L., Honeth G., Grabau D., Malmstrom P., Hegardt C., Ferno M., Niméus E. Stem cell biomarker ALDH1A1 in breast cancer shows an association with prognosis and clinicopathological variables that is highly cut-off dependent. J. Clin. Pathol. 2015;68:1012–1019. doi: 10.1136/jclinpath-2015-203092. [DOI] [PubMed] [Google Scholar]
- 48.Yi B., Williams P.J., Niewolna M., Wang Y., Yoneda T. Tumor-derived platelet-derived growth factor-BB plays a critical role in osteosclerotic bone metastasis in an animal model of human breast cancer. Cancer Res. 2002;62:917–923. [PubMed] [Google Scholar]
- 49.Honeth G., Bendahl P.-O., Ringnér M., Saal L.H., Gruvberger-Saal S.K., Lövgren K., Grabau D., Fernö M., Borg Å., Hegardt C. The CD44+/CD24-phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H., Wang L., Song Y., Wang S., Huang X., Xuan Q., Kang X., Zhang Q. CD44+/CD24- phenotype predicts a poor prognosis in triple-negative breast cancer. Oncol. Lett. 2017;14:5890–5898. doi: 10.3892/ol.2017.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen M.-H., Weng J.-J., Cheng C.-T., Wu R.-C., Huang S.-C., Wu C.-E., Chung Y.-H., Liu C.-Y., Chang M.-H., Chen M.-H., et al. Biology of Human Tumors ALDH1A3, the Major Aldehyde Dehydrogenase Isoform in Human Cholangiocarcinoma Cells, Affects Prognosis and Gemcitabine Resistance in Cholangiocarcinoma Patients. Clin. Cancer Res. 2016;22:4225–4235. doi: 10.1158/1078-0432.CCR-15-1800. [DOI] [PubMed] [Google Scholar]
- 52.Horimoto Y., Arakawa A., Sasahara N., Tanabe M., Sai S., Himuro T., Saito M. Combination of Cancer Stem Cell Markers CD44 and CD24 Is Superior to ALDH1 as a Prognostic Indicator in Breast Cancer Patients with Distant Metastases. PLoS ONE. 2016;11:e0165253. doi: 10.1371/journal.pone.0165253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zwaka T.P. Pluripotency network in embryonic stem cells: Maybe leibniz was right all along. Cell Stem Cell. 2012;11:441–442. doi: 10.1016/j.stem.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 54.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hepburn A.C., Steele R.E., Veeratterapillay R., Wilson L., Kounatidou E.E., Barnard A., Berry P., Cassidy J.R., Moad M., El-Sherif A., et al. The induction of core pluripotency master regulators in cancers defines poor clinical outcomes and treatment resistance. Oncogene. 2019;38:4412–4424. doi: 10.1038/s41388-019-0712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog Is the Gateway to the Pluripotent Ground State. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang D., Lu P., Zhang H., Luo M., Zhang X., Wei X., Gao J., Zhao Z., Liu C. Oct-4 and nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget. 2014;5:10803–10815. doi: 10.18632/oncotarget.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., He X., Semenza G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takebe N., Miele L., Harris P.J., Jeong W., Bando H., Kahn M., Yang S.X., Ivy S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L., Shi P., Zhao G., Xu J., Peng W., Zhang J., Zhang G., Wang X. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020;5:1–35. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuan I.I., Liang K.H., Wang Y.P., Kuo T.W., Meir Y.J.J., Wu S.C.Y., Yang S.C., Lu J., Wu H.C. EpEX/EpCAM and Oct4 or Klf4 alone are sufficient to generate induced pluripotent stem cells through STAT3 and HIF2α. Sci. Rep. 2017;7:41852. doi: 10.1038/srep41852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.