Abstract
BACKGROUND
Osteoarthritis (OA) is a chronic complex multifactorial joint disease, and a major degenerative form of arthritis. Existing studies on the association between polymorphisms of the IL-17 gene and the risk of OA in different populations have yielded conflicting findings.
AIM
To investigate the association between polymorphisms of the IL-17 gene and the risk of OA.
METHODS
We conducted a meta-analysis by systematically searching databases, including PubMed, EMBASE, MEDLINE, Cochrane Library, and Google Scholar to evaluate this association by calculating pooled odds ratios with 95% confidence intervals. Moreover, subgroup analyses stratified by ethnicity and OA type were also conducted.
RESULTS
In a total of 6 citations involving 8 studies (2131 cases and 2299 controls), 4 single nucleotide polymorphisms were identified. Of these 4 polymorphisms, 2 (rs2275913, rs763780) were common in five case-control studies. Together, the pooled results revealed that the A allele and genotype AA/GA of the rs2275913 polymorphism, and the C allele and genotype CC of the rs763780 polymorphism in the IL-17 gene increased the risk of OA. Furthermore, stratification analyses by ethnicity and OA type showed that the rs2275913 polymorphism increased the risk of OA among Asians and in knee/hip OA, respectively. In addition, stratification analyses also revealed that the rs763780 polymorphism increased OA risk among both Asians and Caucasians in knee/hip OA.
CONCLUSION
The rs763780 polymorphism of the IL-17F gene increased the risk of OA, whereas the rs2275913 polymorphism of the IL-17A gene increased the risk of OA only among Asians. Due to the limitations of this study, these findings should be validated in future studies.
Keywords: Interleukin-17, Polymorphism, Osteoarthritis, Meta-analysis, Odds ratio, Confidence interval
Core tip: Osteoarthritis (OA) is the combined result of complex pathogenic factors, including mechanical, biochemical, environmental, endocrine, metabolic, and genetic factors, which account for nearly 50% of the risk of OA development. Although the pathogenesis and etiology of OA are not known, it is likely that interleukin-17 (IL-17) might play an important role in OA development. Existing studies on the association between polymorphisms of the IL-17 gene and the risk of OA in different populations have yielded conflicting findings. We meta-analyzed relevant articles to pool available data and investigated whether IL-17 gene polymorphisms were associated with OA susceptibility.
INTRODUCTION
Osteoarthritis (OA) is a common form of arthritis that can cause progressive loss of joint function[1]. OA is characterized by softening, splitting, and fragmentation of articular cartilage, which is usually accompanied by subchondral bone sclerosis, bone cysts, and bony outgrowths at the joint margins[2]. OA is the combined result of complex pathogenic factors, including mechanical, biochemical, environmental, endocrine, metabolic, and genetic factors, which account for nearly 50% of the risk of OA development[3-6]. Some previous genome-wide association studies[7,8] have suggested that polymorphisms in some genes may affect OA pathogenesis. Genetic markers, in combination with imaging (such as X-ray, magnetic resonance imaging and ultrasound[9,10]) and biochemical markers, have the potential to assist in identifying and diagnosing OA in the early stage[11].
One of the most important family of genes associated with OA is the inflammatory cytokines gene family[12]. Inflammatory cytokines [such as interleukin-1β (IL-1β), IL-6, tumor necrosis factor-α, matrix metalloprotein-13] and moderate physical activity together play a possible biological role in the development of OA[13,14]. IL-17 involves five confirmed receptors and six members (IL-17A-F)[15,16]. Recent evidence has shown that IL-17 was detectable in serum samples and synovial fluid from OA subjects, and a direct correlation between the IL-17 concentration and the severity of OA was observed[17,18]. IL-17 induces the release of chemokines by chondrocytes and synovial fibroblasts, which contributed to synovial infiltration and cartilage collapse in OA[19]. Although the pathogenesis and etiology of OA are not known, it is likely that IL-17 might play an important role in OA development.
To date, several studies[12,20-24] have explored the relationship between polymorphisms of the IL-17 gene and OA susceptibility. The association between IL-17 gene single nucleotide polymorphisms (SNPs) and OA susceptibility may provide novel research directions for OA studies[2]. However, the results of previous studies are inconclusive and conflicting due to clinical heterogeneity, different ethnic populations and small sample sizes. Meta-analysis is a statistical system for assembling results from different studies to produce a single approximate of the major effect with enhanced precision, especially when the results from single case-control studies are incomprehensive and conflicting. Therefore, in the present study we aimed to pool available data and investigated whether IL-17 gene polymorphisms were associated with OA susceptibility.
MATERIALS AND METHODS
The current meta-analysis was performed by following the PRISMA statement[25].
Search strategy
We systematically conducted a literature search using the following electronic databases: PubMed, EMBASE, MEDLINE, Cochrane Library, and Google Scholar to identify epidemiological studies published up to September 2019 to retrieve genetic association studies on OA. The terms “Interleukin-17”, “IL-17”, “SNP”, “polymorphism”, “variant”, “osteoarthritis” and “OA” were used to identify all publications reporting IL-17 gene polymorphisms and OA risk. No language or other restrictions were placed on the search. Eligible studies were retrieved and cautiously evaluated. Furthermore, the reference lists of all related citations were screened to identify any missing studies.
Inclusion and exclusion criteria
Articles were filtered by two independent reviewers (Yang HY and Liu YZ) to assess the appropriateness of the articles selected using a standardized protocol and data collection form. The inclusion criteria were as follows: (1) Case-control studies on humans; (2) Comparison between OA patients and controls; (3) Studies evaluating the association between IL-17 gene polymorphisms and OA susceptibility; and (4) Studies with sufficient genetic frequency for extraction. The exclusion criteria were as follows: (1) Studies with lack of information for data extraction; (2) Non-human studies, abstracts only, comments, reviews, editorials or letters, mechanistic studies, and studies missing controls; and (3) Duplicate or overlapping publications. All questionable publications were discussed and addressed by consensus.
Data extraction and quality assessment
From each eligible study, two reviewers (Yang HY and Liu YZ) independently extracted the following data: Authors, publication date, country, ethnicity, sample size, type of OA, source of controls, allele, and genotype frequency distribution. In addition, the two reviewers independently assessed the methodological quality of the included studies according to the Newcastle-Ottawa Scale (NOS)[26]. NOS criteria were scored based on three aspects: (1) Subject selection, 0-4; (2) Comparability of subjects, 0-2; and (3) Exposure, 0-3. The NOS score ranged from 0 (lowest) to 9 (highest). The Hardy-Weinberg equilibrium (HWE) in controls was tested with Pearson’s χ2 test (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl).
Statistical analysis
Quantitative meta-analysis was employed using STATA 11.0 software (STATA Corporation, College Station, TX, United States). To assess the association between IL-17 gene polymorphisms and the risk of OA, pooled odds ratios (ORs) with 95% confidence intervals (CIs) were calculated. Due to a lack of original data (sex and age), crude ORs were calculated, and five genetic models were included. P < 0.05 was considered statistically significant. Heterogeneity across studies was assessed by Q statistics with its P value and I2 statistics[27,28]. If I2 > 50% and P < 0.10, a random effects model was used in the calculations; otherwise, a fixed effects model was applied. Subgroup analyses were carried based on ethnicity and type of OA. Moreover, the Begg’s test and Egger’s linear regression analysis were applied to prevent publication bias[29]. To evaluate the precision and consistency of the primary meta-analysis, sensitivity analysis was performed to verify the effects associated with any individual study. Furthermore, false-positive report probability (FPRP) was conducted to evaluate the significant findings and rule out any false associations due to multiple tests[30]. Generally, the FPRP value for a given association between IL-17 gene polymorphisms and OA risk was calculated with different prior probability. An association with a FPRP value < 0.2 at a prior probability of 0.1 indicated a significant relationship.
RESULTS
Characteristics of the included studies
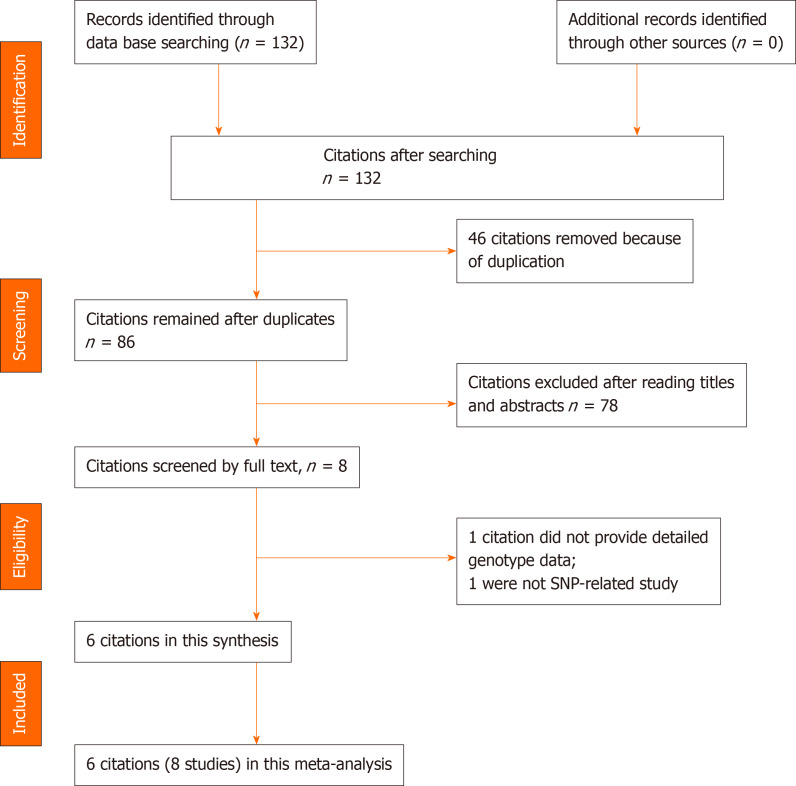
In this study, the literature search yielded 132 citations, among which 46 duplicates were removed. Then, 78 of the 86 remaining citations were excluded after reviewing the titles and abstracts. The remaining 8 citations were sent for full text review, which resulted in exclusion of 1 citation due to the lack of detailed genotype data and 1 non-SNP study. Finally, 6 citations (2131 cases and 2299 controls) involving 8 studies and 4 SNPs were included in this study. The year of publication ranged between 2014 and 2019. In four of the included citations[20-22,24], the association between IL-17 gene polymorphisms in an Asian population were investigated, and in two included citations[12,23] focus was on the Caucasian population. In all 6 citations[12,20-24], the association between rs2275913/rs763780 polymorphisms of the IL-17 gene and risk of OA were studied. Detailed characteristics of the included citations are shown in Tables 1 and 2. A flowchart of reviews, showing the detailed selection process, is illustrated in Figure 1. The NOS scores ranged from 6 to 7 stars.
Table 1.
Characteristics of included studies
| Ref. | Yr | Nationality | Osteoarthritis type |
Sample size (female / male) |
Age (mean) |
Study single nucleotide polymorphisms | Genotype method |
Newcastle-Ottawa Scale |
Hardy-Weinberg equilibrium | ||||
| Case | Control | Case | Control | I | II | III | |||||||
| Zhang et al[20] | 2019 | China | Knee | 122 (90/32) | 124 (88/36) | 66 | 66 | rs2275913 | PCR | 3 | 0 | 3 | Yes |
| rs763780 | PCR | 3 | 0 | 3 | Yes | ||||||||
| Zhang et al[20] | 2019 | China | Knee | 76 (52/24) | 68 (49/19) | 62 | 61 | rs2275913 | PCR | 3 | 0 | 3 | Yes |
| rs763780 | PCR | 3 | 0 | 3 | Yes | ||||||||
| Jiang et al[21] | 2019 | China | NA | 410 (271/139) | 507 (348/159) | 57.35 | 57.88 | rs2275913 | PCR-RFLP | 3 | 1 | 3 | Yes |
| rs763780 | PCR-RFLP | 3 | 1 | 3 | Yes | ||||||||
| Bai et al[22] | 2019 | China | Knee | 594 (393/201) | 576 (402/174) | 59.5 | 58.3 | rs2275913 | PCR | 3 | 1 | 3 | Yes |
| rs763780 | PCR | 3 | 1 | 3 | Yes | ||||||||
| Bafrani et al[12] | 2019 | Iran | Knee | 127 (69/58) | 127 (73/54) | 68.06 | 66.84 | rs2275913 | PCR-RFLP | 3 | 1 | 3 | Yes |
| rs763780 | PCR-RFLP | 3 | 1 | 3 | Yes | ||||||||
| rs2397084 | PCR-RFLP | 3 | 1 | 3 | Yes | ||||||||
| Vrgoc et al[23] | 2018 | Croatia | Hip | 260 (175/85) | 597 (152/445) | 67.82 | 42.64 | rs2275913 | TaqMan | 3 | 1 | 3 | Yes |
| rs763780 | TaqMan | 3 | 1 | 3 | Yes | ||||||||
| rs1889570 | TaqMan | 3 | 1 | 3 | Yes | ||||||||
| Vrgoc et al[23] | 2018 | Croatia | Knee | 240 (174/66) | 597 (152/445) | 69.74 | 42.64 | rs2275913 | TaqMan | 3 | 1 | 3 | Yes |
| rs763780 | TaqMan | 3 | 1 | 3 | Yes | ||||||||
| rs1889570 | TaqMan | 3 | 1 | 3 | Yes | ||||||||
| Han et al[24] | 2014 | SouthKorea | Knee | 302 (245/57) | 300 (136/164) | 60 | 51 | rs2275913 | PCR-SSCP | 3 | 0 | 3 | No |
| rs763780 | PCR-SSCP | 3 | 0 | 3 | No | ||||||||
Newcastle-Ottawa Scale is available from http://www.ohri.ca/programs/clinical epidemiology/oxford.asp. I: Selection; II: Comparability; III: Exposure; NA: Not available; PCR: Polymerase chain reaction; RFLP: Restriction fragment length polymorphism; SSCP: Single-strand conformation polymorphism.
Table 2.
Genotype distributions of IL-17 gene polymorphisms in the included studies
| Ref. | Source of control | Ethnicity |
Allele |
Case |
Control |
Association with osteoarthritis | |||||
| 1 | 0 | 11 | 10 | 00 | 11 | 10 | 00 | ||||
| rs2275913 | |||||||||||
| Zhang et al[20], 2019 | HB | Asian | G | A | 42 | 53 | 27 | 49 | 63 | 12 | AA genotype/A allele increased OA risk |
| Zhang et al[20], 2019 | HB | Asian | 23 | 32 | 21 | 26 | 28 | 14 | Not related | ||
| Jiang et al[21], 2019 | HB | Asian | 204 | 170 | 34 | 289 | 194 | 23 | AA genotype/A allele increased OA risk | ||
| Bai et al[22], 2019 | HB | Asian | 189 | 271 | 134 | 207 | 265 | 104 | AA genotype/A allele increased OA risk | ||
| Bafrani et al[12], 2019 | HB | Caucasians | 83 | 35 | 9 | 69 | 51 | 7 | GA genotype decreased OA risk | ||
| Vrgoc et al[23], 2018 | HB | Caucasians | 78 | 71 | 23 | 190 | 172 | 45 | Not related | ||
| Vrgoc et al[23], 2018 | HB | Caucasians | 76 | 85 | 25 | 190 | 172 | 45 | Not related | ||
| Han et al[24], 2014 | HB | Asian | 52 | 109 | 141 | 97 | 106 | 97 | GG genotype decreased OA risk | ||
| rs763780 | |||||||||||
| Zhang et al[20], 2019 | HB | Asian | T | C | 91 | 30 | 1 | 98 | 24 | 2 | Not related |
| Zhang et al[20], 2019 | HB | Asian | 59 | 16 | 1 | 54 | 13 | 1 | Not related | ||
| Jiang et al[21], 2019 | HB | Asian | 356 | 49 | 4 | 423 | 80 | 2 | Not related | ||
| Bai et al[22], 2019 | HB | Asian | 380 | 188 | 26 | 411 | 155 | 10 | CC/TC genotype/C allele increased OA risk | ||
| Bafrani et al[12], 2019 | HB | Caucasians | 98 | 26 | 3 | 112 | 13 | 2 | TC genotype/C allele increased OA risk | ||
| Vrgoc et al[23], 2018 | HB | Caucasians | 198 | 34 | 0 | 493 | 34 | 1 | C allele increased OA risk | ||
| Vrgoc et al[23], 2018 | HB | Caucasians | 195 | 14 | 1 | 493 | 34 | 1 | Not related | ||
| Han et al[24], 2014 | HB | Asian | 226 | 59 | 17 | 236 | 56 | 8 | Not related | ||
| rs2397084 | |||||||||||
| Bafrani et al[12], 2019 | HB | Caucasians | T | C | 111 | 16 | 0 | 109 | 18 | 0 | Not related |
| rs1889570 | |||||||||||
| Vrgoc et al[23], 2018 | HB | Caucasians | G | A | 57 | 129 | 58 | 49 | 91 | 46 | Not related |
| Vrgoc et al[23], 2018 | HB | Caucasians | 68 | 111 | 51 | 49 | 91 | 46 | Not related | ||
HB: Hospital-based; OA: Osteoarthritis.
Figure 1.
Flowchart of the literature search and selection for this study.
Association between rs2275913 polymorphism and OA susceptibility
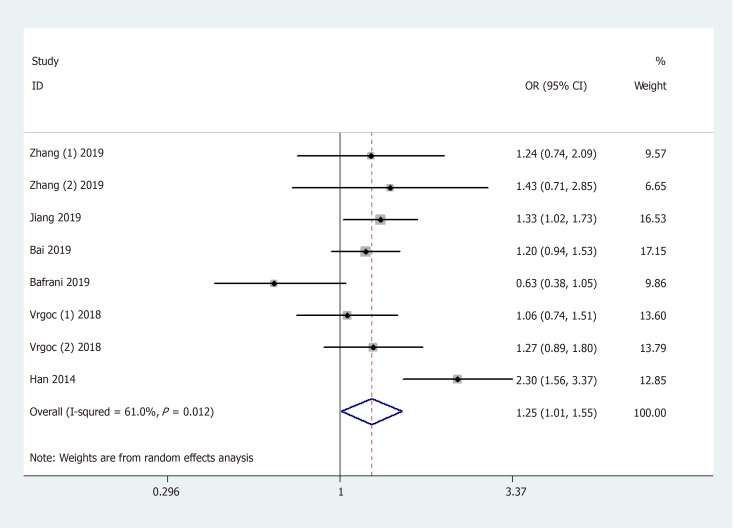
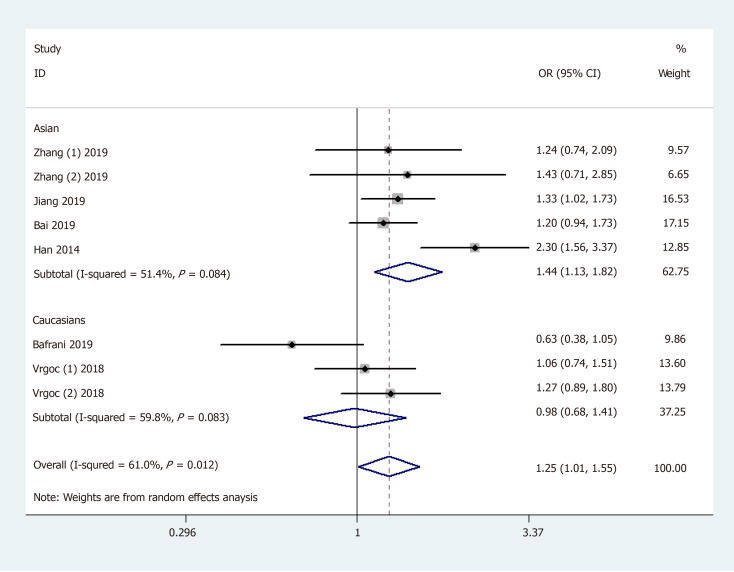
General analysis showed that the polymorphism rs2275913 of the IL-17A gene increased OA risk [OR and 95%CI: 1.26 (1.08, 1.47) in A vs G; 1.25 (1.01, 1.55) in AA + GA vs GG; 1.53 (1.30, 1.81) in AA vs GA + GG; 1.71 (1.42, 2.06) in AA vs GG; and 1.15 (1.01, 1.32) in GA vs GG, Table 3 and Figure 2]. Stratification analysis by ethnicity showed that the rs2275913 polymorphism increased the risk of OA among Asians [OR and 95%CI: 1.40 (1.18, 1.67) in A vs G; 1.44 (1.13, 1.82) in AA + GA vs GG; 1.62 (1.34, 1.96) in AA vs GA + GG; 1.89 (1.52, 2.34) in AA vs GG; and 1.25 (1.06, 1.46) in GA vs GG, Table 3 and Figure 3); however, this was not observed in Caucasians. Subgroup analysis by OA type revealed that the rs2275913 polymorphism increased the risk of both knee and hip OA. A change in conclusion was not observed after eliminating a study[24] that did not meet the HWE.
Table 3.
Meta-analysis of the association between IL-17 gene polymorphisms and osteoarthritis risk
| SNP | Comparison | Category | Category | Studies | Odds ratio (95% confidence interval) | P value | I2 |
| rs2275913 | A vs G | Total (random model) | 8 | 1.26 (1.08, 1.47) | 0.003 | 61.6% | |
| Allele model | Ethnicity | Asian | 5 | 1.40 (1.18, 1.67) | < 0.001 | 56.1% | |
| Caucasian | 3 | 1.05 (0.84, 1.31) | 0.680 | 38.8% | |||
| OA type | Knee | 6 | 1.28 (1.04, 1.59) | 0.023 | 69.9% | ||
| Hip | 1 | 1.09 (0.83, 1.42) | 0.545 | - | |||
| AA + GA vs GG | Total (random model) | 8 | 1.25 (1.01, 1.55) | 0.039 | 61.0% | ||
| Dominant model | Ethnicity | Asian | 5 | 1.44 (1.13, 1.82) | 0.003 | 51.4% | |
| Caucasian | 3 | 0.98 (0.68, 1.41) | 0.926 | 59.8% | |||
| OA type | Knee | 6 | 1.27 (0.93, 1.73) | 0.130 | 70.2% | ||
| Hip | 1 | 1.06 (0.74, 1.51) | 0.769 | - | |||
| AA vs GA + GG | Total (fixed model) | 8 | 1.53 (1.30, 1.81) | < 0.001 | 0.0% | ||
| Recessive model | Ethnicity | Asian | 5 | 1.62 (1.34, 1.96) | < 0.001 | 13.3% | |
| Caucasian | 3 | 1.25 (0.88, 1.78) | 0.209 | 0.0% | |||
| OA type | Knee | 6 | 1.53 (1.27, 1.84) | < 0.001 | 0.0% | ||
| Hip | 1 | 1.24 (0.73, 2.13) | 0.430 | - | |||
| AA vs GG | Total (fixed model) | 8 | 1.71 (1.42, 2.06) | < 0.001 | 30.2% | ||
| Homozygote model | Ethnicity | Asian | 5 | 1.89 (1.52, 2.34) | < 0.001 | 40.8% | |
| Caucasian | 3 | 1.28 (0.88, 1.86) | 0.192 | 0.0% | |||
| OA type | Knee | 6 | 1.74 (1.41, 2.15) | < 0.001 | 39.8% | ||
| Hip | 1 | 1.25 (0.71, 2.20) | 0.449 | - | |||
| GA vs GG | Total (fixed model) | 8 | 1.15 (1.01, 1.32) | 0.034 | 47.7% | ||
| Heterozygote model | Ethnicity | Asian | 5 | 1.25 (1.06, 1.46) | 0.007 | 23.7% | |
| Caucasian | 3 | 0.98 (0.77, 1.24) | 0.846 | 63.1% | |||
| OA type | Knee | 6 | 1.15 (0.98, 1.36) | 0.090 | 60.3% | ||
| Hip | 1 | 1.01 (0.69, 1.47) | 0.977 | - | |||
| rs763780 | C vs T | Total (random model) | 8 | 1.32 (1.06,1.64) | 0.013 | 51.5% | |
| Allele model | Ethnicity | Asian | 5 | 1.19 (0.95, 1.48) | 0.132 | 42.6% | |
| Caucasian | 3 | 1.75 (1.16, 2.66) | 0.008 | 39.3% | |||
| OA type | Knee | 6 | 1.37 (1.17, 1.60) | < 0.001 | 0.0% | ||
| Hip | 1 | 2.24 (1.38, 3.63) | 0.001 | - | |||
| CC + TC vs TT | Total (random model) | 8 | 1.32 (1.02, 1.72) | 0.033 | 58.4% | ||
| Dominant model | Ethnicity | Asian | 5 | 1.15 (0.90, 1.48) | 0.274 | 44.0% | |
| Caucasian | 3 | 1.82 (1.11, 3.00) | 0.018 | 51.9% | |||
| OA type | Knee | 6 | 1.35 (1.14, 1.61) | 0.001 | 0.0% | ||
| Hip | 1 | 2.42 (1.47, 3.99) | 0.001 | - | |||
| CC vs TC + TT | Total (fixed model) | 8 | 2.08 (1.30, 3.32) | 0.002 | 0.0% | ||
| Recessive model | Ethnicity | Asian | 5 | 2.17 (1.32, 3.59) | 0.002 | 0.0% | |
| Caucasian | 3 | 1.48 (0.38, 5.67) | 0.570 | 0.0% | |||
| OA type | Knee | 6 | 2.10 (1.28, 3.45) | 0.003 | 0.0% | ||
| Hip | 1 | 0.76 (0.03, 3.32) | 0.864 | - | |||
| CC vs TT | Total (fixed model) | 8 | 2.19 (1.37, 3.51) | 0.001 | 0.0% | ||
| Homozygote model | Ethnicity | Asian | 5 | 2.28 (1.37, 3.77) | 0.001 | 0.0% | |
| Caucasian | 3 | 1.62 (0.42, 6.23) | 0.481 | 0.0% | |||
| OA type | Knee | 6 | 2.23 (1.35, 3.67) | 0.002 | 0.0% | ||
| Hip | 1 | 0.83 (0.03, 20.43) | 0.909 | - | |||
| TC vs TT | Total (random model) | 8 | 1.29 (0.97, 1.71) | 0.078 | 62.6% | ||
| Heterozygote model | Ethnicity | Asian | 5 | 1.09 (0.85, 1.40) | 0.505 | 40.5% | |
| Caucasian | 3 | 1.84 (1.06, 3.17) | 0.030 | 57.6% | |||
| OA type | Knee | 6 | 1.28 (1.07, 1.54) | 0.008 | 0.0% | ||
| Hip | 1 | 2.49 (1.51, 4.12) | < 0.001 | - |
OA: Osteoarthritis; SNP: Single nucleotide polymorphism.
Figure 2.
Forest plot shows odds ratios for the associations between rs2275913 polymorphism and osteoarthritis risk (AA + GA vs GG).
Figure 3.
Stratification analysis by ethnicity shows odds ratio for the association between rs2275913 polymorphism and osteoarthritis risk (AA + GA vs GG).
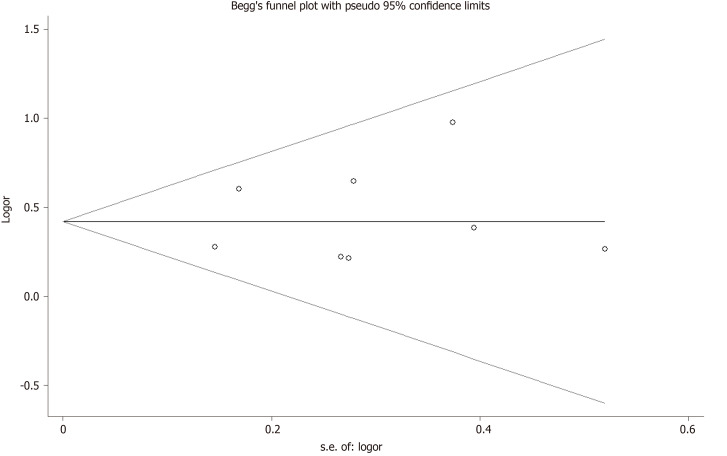
Sensitivity analysis was used to determine the pooled ORs regarding the effects of this SNP on OA risk. Taken together, the results indicated that our data were stable and credible. Neither Egger’s nor Begg's tests revealed obvious publication bias for the rs2275913 polymorphism (Figure 4).
Figure 4.
Begg's tests between rs2275913 polymorphism and osteoarthritis (AA vs GA + GG).
Association between rs763780 polymorphisms and OA susceptibility
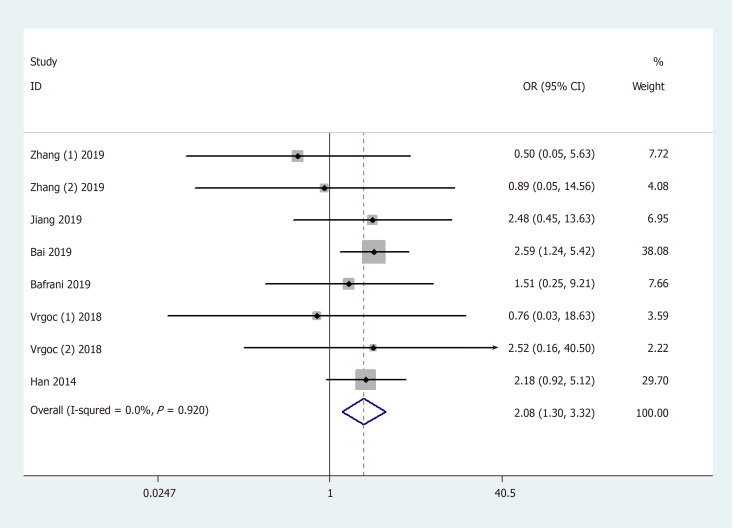
Results of the pooled analysis on the association between IL-17F gene rs763780 polymorphism and OA risk are shown in Table 3 and Figure 5 [OR and 95%CI: 1.32 (1.06, 1.64) in C vs T; 1.32 (1.02, 1.72) in CC + TC vs TT; 2.08 (1.30, 3.32) in CC vs TC + TT; and 2.19 (1.37, 3.51) in CC vs TT]. Moreover, subgroup analysis by ethnicity indicated that rs763780 polymorphism increased OA risk among Caucasians in Allele/Dominant/Heterozygote models [OR and 95%CI: 1.75 (1.16, 2.66) in C vs T; 1.82 (1.11, 3.00) in CC + TC vs TT; and 1.84 (1.06, 3.17) in TC vs TT], whereas among Asians in Recessive/Homozygote models [OR and 95%CI: 2.17 (1.32, 3.59) in CC vs TC + TT; and 2.28 (1.37, 3.77) in CC vs TT]. Stratification analysis by OA type revealed that the rs763780 polymorphism increased the risk of both knee and hip OA. A change in conclusion was not observed after eliminating a study[24] that did not meet the HWE. Both sensitivity analysis and the publication bias test revealed that our data were stable and credible (data not shown).
Figure 5.
Forest plot shows odds ratios for the associations between rs763780 polymorphism and osteoarthritis risk (CC vs TC + TT).
FPRP analysis result
For all statistically significant results, the FPRP values are summarized in Table 4. For a prior probability of 0.1 and crude ORs, the FPRP analysis indicated that the significant association between IL-17A rs2275913 polymorphism and OA risk was significant for all subjects (allele: FPRP = 0.052, recessive: FPRP = 0.009, and homozygous comparison: FPRP = 0.009), Asians (allele: FPRP = 0.013, dominant: FPRP = 0.048, recessive: FPRP = 0.009, homozygous: FPRP = 0.009, and heterozygous comparison: FPRP = 0.103), and knee OA (recessive comparison: FPRP = 0.010). Similarly, the association of the IL-17F rs763780 polymorphism with OA risk also deserved attention in the overall population, especially among Caucasians and individuals with knee/hip OA. In our study, greater values were observed of FPRP > 0.2 for other positive findings between IL-17 gene polymorphisms and OA risk, which may be attributed to the limited sample size. For future studies, additional validation will be needed using studies with a larger sample size.
Table 4.
False-positive report probability values for associations between IL-17 gene polymorphisms and risk of osteoarthritis
| Variables | Odds ratio (95%CI) | P value | Power |
Prior probability |
||||
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| rs2275913 | ||||||||
| A vs G | ||||||||
| All | 1.26 (1.08, 1.47) | 0.003 | 0.488 | 0.018 | 0.052 | 0.378 | 0.860 | 0.984 |
| Asian | 1.40 (1.18, 1.67) | < 0.001 | 0.673 | 0.004 | 0.013 | 0.128 | 0.597 | 0.937 |
| Knee OA | 1.28 (1.04, 1.59) | 0.023 | 0.483 | 0.125 | 0.300 | 0.825 | 0.979 | 0.998 |
| AA + GA vs GG | ||||||||
| All | 1.25 (1.01, 1.55) | 0.039 | 0.488 | 0.194 | 0.419 | 0.888 | 0.988 | 0.999 |
| Asian | 1.44 (1.13, 1.82) | 0.003 | 0.533 | 0.017 | 0.048 | 0.358 | 0.849 | 0.983 |
| AA vs GA + GG | ||||||||
| All | 1.53 (1.30, 1.81) | < 0.001 | 0.952 | 0.003 | 0.009 | 0.094 | 0.512 | 0.913 |
| Asian | 1.62 (1.34, 1.96) | < 0.001 | 0.953 | 0.003 | 0.009 | 0.094 | 0.512 | 0.913 |
| Knee OA | 1.53 (1.27, 1.84) | < 0.001 | 0.890 | 0.003 | 0.010 | 0.100 | 0.529 | 0.918 |
| AA vs GG | ||||||||
| All | 1.71 (1.42, 2.06) | < 0.001 | 0.991 | 0.003 | 0.009 | 0.091 | 0.502 | 0.910 |
| Asian | 1.89 (1.52, 2.34) | < 0.001 | 0.995 | 0.003 | 0.009 | 0.091 | 0.501 | 0.910 |
| GA vs GG | ||||||||
| All | 1.15 (1.01, 1.32) | 0.034 | 0.447 | 0.186 | 0.406 | 0.883 | 0.987 | 0.999 |
| Asian | 1.25 (1.06, 1.46) | 0.007 | 0.548 | 0.037 | 0.103 | 0.559 | 0.927 | 0.992 |
| rs763780 | ||||||||
| C vs T | ||||||||
| All | 1.32 (1.06,1.64) | 0.013 | 0.509 | 0.071 | 0.187 | 0.717 | 0.962 | 0.996 |
| Caucasian | 1.75 (1.16, 2.66) | 0.008 | 0.487 | 0.047 | 0.129 | 0.619 | 0.943 | 0.994 |
| Knee OA | 1.37 (1.17, 1.60) | < 0.001 | 0.753 | 0.004 | 0.012 | 0.116 | 0.570 | 0.930 |
| Hip OA | 2.24 (1.38, 3.63) | 0.001 | 0.006 | 0.018 | 0.167 | 0.669 | 0.953 | 0.995 |
| CC + TC vs TT | ||||||||
| All | 1.32 (1.02, 1.72) | 0.033 | 0.470 | 0.174 | 0.387 | 0.874 | 0.986 | 0.999 |
| Caucasian | 1.82 (1.11, 3.00) | 0.018 | 0.099 | 0.247 | 0.783 | 0.973 | 0.973 | 0.997 |
| Knee OA | 1.35 (1.14, 1.61) | 0.001 | 0.520 | 0.006 | 0.017 | 0.160 | 0.658 | 0.951 |
| Hip OA | 2.42 (1.47, 3.99) | 0.001 | 0.569 | 0.005 | 0.016 | 0.148 | 0.637 | 0.946 |
| CC vs TC + TT | ||||||||
| All | 2.08 (1.30, 3.32) | 0.002 | 0.492 | 0.012 | 0.035 | 0.287 | 0.802 | 0.976 |
| Caucasian | 2.17 (1.32, 3.59) | 0.002 | 0.471 | 0.013 | 0.037 | 0.296 | 0.809 | 0.977 |
| Knee OA | 2.10 (1.28, 3.45) | 0.003 | 0.485 | 0.018 | 0.053 | 0.380 | 0.861 | 0.984 |
| CC vs TT | ||||||||
| All | 2.19 (1.37, 3.51) | 0.001 | 0.487 | 0.006 | 0.018 | 0.169 | 0.672 | 0.954 |
| Caucasian | 2.28 (1.37, 3.77) | 0.001 | 0.469 | 0.006 | 0.019 | 0.174 | 0.681 | 0.955 |
| Knee OA | 2.23 (1.35, 3.67) | 0.002 | 0.526 | 0.011 | 0.033 | 0.274 | 0.792 | 0.974 |
| TC vs TT | ||||||||
| Caucasian | 1.84 (1.06, 3.17) | 0.030 | 0.511 | 0.150 | 0.346 | 0.853 | 0.983 | 0.998 |
| Knee OA | 1.28 (1.07, 1.54) | 0.008 | 0.486 | 0.047 | 0.129 | 0.620 | 0.943 | 0.994 |
| Hip | 2.49 (1.51, 1.71) | < 0.001 | ||||||
OA: Osteoarthritis.
DISCUSSION
Although the definite pathogenesis of OA remains unclear, genetic factors are considered to be strong determinants. IL-17 is a cytokine that is mainly synthesized by activated T cells, and its receptors are present in osteoblasts. IL-17 can enhance proinflammatory cytokines, including tumor necrosis factor-α, IL-1 and IL-6, that together play a key role in cartilage degradation and the inhibition of cartilage repair[31,32]. Furthermore, in patients with OA, IL-17 stimulates the release of vascular endothelial growth factor in synovial fibroblasts that was isolated from their joints[19]. Therefore, an increasing number of studies focused on the association between the IL-17 gene polymorphisms and OA susceptibility.
Han et al[24] first examined the alleles and genotypes of IL-17A rs2275913 and IL-17F rs763780 in a Korean population, and found a significant association between rs2275913 and the susceptibility of knee OA. In addition, Vrgoc et al[23] did not observe an association between IL-17A rs2275913 and the risk of hip or knee OA in a Croatian population. The C allele of the IL-17F gene rs763780 polymorphism increased the risk of hip OA, but not of knee OA. The results of a study by Bafrani et al[12] indicated a significant association between rs2275913‐GA genotype and a decrease in the risk of knee OA. However, the rs763780‐TC genotype and rs763780‐C allele were related to an increased risk of knee OA. Zhang et al[20] suggested that the genotype AA frequency of IL-17A (rs2275913) was significantly different between knee OA patients and the control group in a Chinese Han population, but not in a Tibetan population. Furthermore, Bai et al[22] indicated that the IL-17A rs2275913 polymorphism had a significant impact on the risk of knee OA. Additionally, the rs763780 C allele was found to be related to a greatly increased risk of developing knee OA. Jiang et al[21] suggested a significant association between the increased risk of OA and IL-17A rs2275913, but not of IL-17F rs763780. The discrepancy in the above-mentioned studies may be due to the following: First, the inclusion criteria differed among studies. For example, Bai et al[22] enrolled other types of arthritis or joint diseases, such as inflammatory arthritis, which were excluded by other citations. Moreover, Jiang et al[21] used clinical symptoms and radiological evidence of joints, whereas in the Han et al[24] study, OA cases who had undergone total knee arthroplasty were enrolled. Second, the allele frequencies of the rs2275913/rs763780 polymorphism in the cases were diverse. Third, the affected joint sites differed. Fourth, the genetic background of OA may vary among races. Finally, the difference in sample sizes may also account for this discrepancy.
Due to the limited sample sizes, previous single studies may have been underpowered and thereby may have presented conflicting findings, especially given the diverse inheritance of the heterogeneous and complex OA etiology, different ethnicities, clinical heterogeneity, and other causes. Therefore, we conducted this meta-analysis.
Our data showed that the rs2275913 and rs763780 polymorphisms increased the risk of OA. Stratification analyses by ethnicity and OA type showed that the rs2275913 polymorphism increased the risk of OA among Asians as well as in knee/hip OA, respectively. Stratification analyses also revealed that the rs763780 polymorphism increased OA risk among both Asians and Caucasians, as well as in knee/hip OA. There may be several possible reasons for the different findings regarding the rs2275913 polymorphism between Asians and Caucasians. First, genetic heterogeneity for OA exists in different populations. Second, these discrepancies may be explained by clinical heterogeneity. Third, the sample sizes of the Caucasian populations were not large enough to support a definitive conclusion. Additionally, different OA types and varying clinical parameters of different populations may also be potential reasons for the inconclusive findings. Furthermore, different characteristics of the OA groups (such as age and sex) and disease severity may also be possible reasons for the discrepancies observed. Finally, varying environmental factors may also have contributed, because the interaction between genetic factors and environmental factors can eventually lead to the development of OA. Gao et al[33] and Lee et al[34] also conducted meta-analyses to study the association between IL-17 gene rs2275913/rs763780 polymorphism and OA risk. We consider that our meta-analysis had several additional advantages. First, compared to the work by Lee et al[34], our meta-analysis of the rs3134069 polymorphism included one more case-controlled study[20]. Second, for rs2275913, Gao et al[33] regarded the G allele as a minor allele and then analyzed it in 5 genetic models. In fact, according to the dbSNP database, A allele is the true minor allele. Gao et al[33] misidentified the minor allele, which led to the wrong choice of genetic model. If the correct genetic model is selected, their results are consistent with our findings. This mistake was also found in the rs763780 polymorphism analysis. Third, FPRP was conducted in our meta-analysis to evaluate the significant findings and rule out any false associations due to multiple tests.
Our study has several limitations that should be considered. First, due to limited data, we were unable to conduct stratification analyses of other potential factors, such as age, sex, and age at OA onset. Second, our results were based on unadjusted estimates of confounding factors, which might have affected the final results. Third, although funnel plots and Egger’s tests revealed no publication bias, selection bias could not be fully prevented, because only studies published in the English language were searched. Fourth, we were unable to assess potential gene-gene or gene-environment interactions due to the lack of relevant data. Fifth, in future studies, clinical cases should be investigated to support the analytical results observed in this study. Sixth, we can only infer but cannot conclude that the IL-17 gene rs2275913/rs763780 polymorphism is a susceptibility locus for other types of OA. Thus, further investigation into more types of OA is warranted. Finally, five genetic models of inheritance were used, thus, type I error may have arisen through lack of correction for multiple testing.
In conclusion, the present meta-analysis demonstrated that the rs763780 polymorphism of the IL-17F gene increased the risk of OA, whereas the rs2275913 polymorphism of the IL-17A gene only increased the risk of OA among Asians. Given the study limitations, further well-designed prospective studies with large sample sizes should be performed to validate these findings. In the future, it will become feasible to identify and diagnose OA in the early stage, as a result of genetic findings. Furthermore, the biological and functional relevance of these genetic findings is essential to help put the research into a clinical context to benefit people with OA.
ARTICLE HIGHLIGHTS
Research background
Osteoarthritis (OA) is the combined result of complex pathogenic factors, including mechanical, biochemical, environmental, endocrine, metabolic, and genetic factors, which account for nearly 50% of the risk of OA development. Although the pathogenesis and etiology of OA are not known, it is likely that interleukin-17 (IL-17) might play an important role in OA development.
Research motivation
To date, several studies have explored the relationship between polymorphisms of the IL-17 gene and OA susceptibility. The association between IL-17 gene single nucleotide polymorphisms and OA susceptibility may provide novel research directions for OA studies. However, the results of previous studies are inconclusive and conflicting due to clinical heterogeneity, different ethnic populations and small sample sizes.
Research objectives
We meta-analyzed relevant articles regarding the association between the polymorphisms of IL-17 gene and OA susceptibility.
Research methods
We systematically conducted the literature search using the following electronic databases: PubMed, EMBASE, MEDLINE, Cochrane Library, and Google Scholar to identify epidemiological studies published up to September 2019 to retrieve genetic association studies on OA. Pooled odds ratios with 95% confidence intervals were calculated. Subgroup analyses were carried based on ethnicity and type of OA. Furthermore, false-positive report probability was conducted to evaluate the significant findings and rule out any false associations due to multiple tests.
Research results
In a total of 6 citations involving 8 studies (2131 cases and 2299 controls), 4 single nucleotide polymorphisms were identified. Of these 4 polymorphisms, 2 (rs2275913, rs763780) were common in five case-control studies. Together, the pooled results revealed that the A allele and genotype AA/GA of the rs2275913 polymorphism, and the C allele and genotype CC of the rs763780 polymorphism in the IL-17 gene increased the risk of OA. Furthermore, stratification analyses by ethnicity and OA type showed that the rs2275913 polymorphism increased the risk of OA among Asians and in knee/hip OA, respectively. In addition, stratification analyses also revealed that the rs763780 polymorphism increased OA risk among both Asians and Caucasians in knee/hip OA.
Research conclusions
The rs763780 polymorphism of the IL-17F gene increased the risk of OA, whereas the rs2275913 polymorphism of the IL-17A gene increased the risk of OA only among Asians. Due to the limitations of this study, these findings should be validated in future studies.
Footnotes
Conflict-of-interest statement: The authors declare that they have no conflict of interest.
PRISMA 2009 Checklist statement: The guidelines of the PRISMA 2009 Statement have been adopted.
Manuscript source: Invited manuscript
Peer-review started: December 11, 2019
First decision: January 7, 2020
Article in press: April 24, 2020
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El-Razek AA, Musumeci G, Ünver B S-Editor: Dou Y L-Editor: Webster JR E-Editor: Xing YX
Contributor Information
Hao-Yu Yang, Department of Orthopedics, Wuxi 9th People’s Hospital affiliated to Soochow University, Wuxi 214000, Jiangsu Province, China.
Yu-Zhou Liu, Department of Orthopedics, Wuxi 9th People’s Hospital affiliated to Soochow University, Wuxi 214000, Jiangsu Province, China.
Xin-Die Zhou, Department of Orthopedics, The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, Changzhou 213003, Jiangsu Province, China.
Yong Huang, Department of Orthopedics, The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, Changzhou 213003, Jiangsu Province, China. huangyong@njmu.edu.cn.
Nan-Wei Xu, Department of Orthopedics, The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, Changzhou 213003, Jiangsu Province, China.
References
- 1.Valdes AM, Spector TD. Genetic epidemiology of hip and knee osteoarthritis. Nat Rev Rheumatol. 2011;7:23–32. doi: 10.1038/nrrheum.2010.191. [DOI] [PubMed] [Google Scholar]
- 2.Valdes AM, Spector TD. The contribution of genes to osteoarthritis. Rheum Dis Clin North Am. 2008;34:581–603. doi: 10.1016/j.rdc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Loughlin J. The genetic epidemiology of human primary osteoarthritis: current status. Expert Rev Mol Med. 2005;7:1–12. doi: 10.1017/S1462399405009257. [DOI] [PubMed] [Google Scholar]
- 4.Meulenbelt I. Osteoarthritis year 2011 in review: genetics. Osteoarthritis Cartilage. 2012;20:218–222. doi: 10.1016/j.joca.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Zou T, Yang L, Lee AM, Tan X, Wong E, Bai HX. Can genetics explain the higher risk of worsening knee pain in offspring of people with total knee replacement for severe primary knee osteoarthritis? Ann Rheum Dis. 2016;75:e45. doi: 10.1136/annrheumdis-2016-209380. [DOI] [PubMed] [Google Scholar]
- 6.Reynard LN. Analysis of genetics and DNA methylation in osteoarthritis: What have we learnt about the disease? Semin Cell Dev Biol. 2017;62:57–66. doi: 10.1016/j.semcdb.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 7.Pang H, Luo F, Dai F, Wu XH, Xu JZ. Genome-wide association study for osteoarthritis. Lancet. 2013;381:372–273; discussion 373. doi: 10.1016/S0140-6736(13)60167-1. [DOI] [PubMed] [Google Scholar]
- 8.arcOGEN Consortium ; arcOGEN Collaborators, Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, Lopes MC, Boraska V, Esko T, Evangelou E, Hoffman A, Houwing-Duistermaat JJ, Ingvarsson T, Jonsdottir I, Jonnson H, Kerkhof HJ, Kloppenburg M, Bos SD, Mangino M, Metrustry S, Slagboom PE, Thorleifsson G, Raine EV, Ratnayake M, Ricketts M, Beazley C, Blackburn H, Bumpstead S, Elliott KS, Hunt SE, Potter SC, Shin SY, Yadav VK, Zhai G, Sherburn K, Dixon K, Arden E, Aslam N, Battley PK, Carluke I, Doherty S, Gordon A, Joseph J, Keen R, Koller NC, Mitchell S, O'Neill F, Paling E, Reed MR, Rivadeneira F, Swift D, Walker K, Watkins B, Wheeler M, Birrell F, Ioannidis JP, Meulenbelt I, Metspalu A, Rai A, Salter D, Stefansson K, Stykarsdottir U, Uitterlinden AG, van Meurs JB, Chapman K, Deloukas P, Ollier WE, Wallis GA, Arden N, Carr A, Doherty M, McCaskie A, Willkinson JM, Ralston SH, Valdes AM, Spector TD, Loughlin J. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–823. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razek AA, El-Basyouni SR. Ultrasound of knee osteoarthritis: interobserver agreement and correlation with Western Ontario and McMaster Universities Osteoarthritis. Clin Rheumatol. 2016;35:997–1001. doi: 10.1007/s10067-015-2990-2. [DOI] [PubMed] [Google Scholar]
- 10.Razek AA, Fouda NS, Elmetwaley N, Elbogdady E. Sonography of the knee joint() J Ultrasound. 2009;12:53–60. doi: 10.1016/j.jus.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner S, Valdes A. The Genetics of Osteoarthritis: A Review. J Funct Morphol Kinesiol. 2016;1:140–153. [Google Scholar]
- 12.Bafrani HH, Ahmadi M, Jahantigh D, Karimian M. Association analysis of the common varieties of IL17A and IL17F genes with the risk of knee osteoarthritis. J Cell Biochem. 2019;120:18020–18030. doi: 10.1002/jcb.29105. [DOI] [PubMed] [Google Scholar]
- 13.Castrogiovanni P, Di Rosa M, Ravalli S, Castorina A, Guglielmino C, Imbesi R, Vecchio M, Drago F, Szychlinska MA, Musumeci G. Moderate Physical Activity as a Prevention Method for Knee Osteoarthritis and the Role of Synoviocytes as Biological Key. Int J Mol Sci. 2019:20. doi: 10.3390/ijms20030511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Rosa M, Castrogiovanni P, Musumeci G. The Synovium Theory: Can Exercise Prevent Knee Osteoarthritis? The Role of “Mechanokines”, A Possible Biological Key. J Funct Morphol Kinesiol. 2019;4:11. doi: 10.3390/jfmk4010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaguchi M, Adachi M, Oda N, Kokubu F, Huang SK. IL-17 cytokine family. J Allergy Clin Immunol. 2004;114:1265–1273; quiz 1274. doi: 10.1016/j.jaci.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Kolls JK, Lindén A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Neogi T, Zhang Y. Epidemiology of osteoarthritis. Rheum Dis Clin North Am. 2013;39:1–19. doi: 10.1016/j.rdc.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agarwal S, Misra R, Aggarwal A. Interleukin 17 levels are increased in juvenile idiopathic arthritis synovial fluid and induce synovial fibroblasts to produce proinflammatory cytokines and matrix metalloproteinases. J Rheumatol. 2008;35:515–519. [PubMed] [Google Scholar]
- 19.Honorati MC, Bovara M, Cattini L, Piacentini A, Facchini A. Contribution of interleukin 17 to human cartilage degradation and synovial inflammation in osteoarthritis. Osteoarthritis Cartilage. 2002;10:799–807. doi: 10.1053/joca.2002.0829. [DOI] [PubMed] [Google Scholar]
- 20.Zhang PL, Yang FM, Qiao ZZ, Liu J, Yang QY, Wang YJ, Qi M, Cui LN, Meng L, Li XP. [Association between interleukin-17A and 17F single nucleotide polymorphisms and knee osteoarthritis] Zhonghua Yi Xue Za Zhi. 2019;99:1870–1874. doi: 10.3760/cma.j.issn.0376-2491.2019.24.007. [DOI] [PubMed] [Google Scholar]
- 21.Jiang L, Zhou X, Xiong Y, Bao J, Xu K, Wu L. Association between interleukin-17A/F single nucleotide polymorphisms and susceptibility to osteoarthritis in a Chinese population. Medicine (Baltimore) 2019;98:e14944. doi: 10.1097/MD.0000000000014944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai Y, Gao S, Liu Y, Jin S, Zhang H, Su K. Correlation between Interleukin-17 gene polymorphism and osteoarthritis susceptibility in Han Chinese population. BMC Med Genet. 2019;20:20. doi: 10.1186/s12881-018-0736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vrgoc G, Vrbanec J, Eftedal RK, Dembic PL, Balen S, Dembic Z, Jotanovic Z. Interleukin-17 and Toll-like Receptor 10 genetic polymorphisms and susceptibility to large joint osteoarthritis. J Orthop Res. 2018;36:1684–1693. doi: 10.1002/jor.23823. [DOI] [PubMed] [Google Scholar]
- 24.Han L, Lee HS, Yoon JH, Choi WS, Park YG, Nam SW, Lee JY, Park WS. Association of IL-17A and IL-17F single nucleotide polymorphisms with susceptibility to osteoarthritis in a Korean population. Gene. 2014;533:119–122. doi: 10.1016/j.gene.2013.09.113. [DOI] [PubMed] [Google Scholar]
- 25.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8:336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 27.Melsen WG, Bootsma MC, Rovers MM, Bonten MJ. The effects of clinical and statistical heterogeneity on the predictive values of results from meta-analyses. Clin Microbiol Infect. 2014;20:123–129. doi: 10.1111/1469-0691.12494. [DOI] [PubMed] [Google Scholar]
- 28.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 30.He J, Wang MY, Qiu LX, Zhu ML, Shi TY, Zhou XY, Sun MH, Yang YJ, Wang JC, Jin L, Wang YN, Li J, Yu HP, Wei QY. Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population. Mol Carcinog. 2013;52 Suppl 1:E70–E79. doi: 10.1002/mc.22013. [DOI] [PubMed] [Google Scholar]
- 31.Tokuda H, Kanno Y, Ishisaki A, Takenaka M, Harada A, Kozawa O. Interleukin (IL)-17 enhances tumor necrosis factor-alpha-stimulated IL-6 synthesis via p38 mitogen-activated protein kinase in osteoblasts. J Cell Biochem. 2004;91:1053–1061. doi: 10.1002/jcb.20004. [DOI] [PubMed] [Google Scholar]
- 32.Katz Y, Nadiv O, Beer Y. Interleukin-17 enhances tumor necrosis factor alpha-induced synthesis of interleukins 1,6, and 8 in skin and synovial fibroblasts: a possible role as a "fine-tuning cytokine" in inflammation processes. Arthritis Rheum. 2001;44:2176–2184. doi: 10.1002/1529-0131(200109)44:9<2176::aid-art371>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Gao S, Mao C, Cheng J, Deng Q, Sheng W. Association of IL-17A-197G/A and IL-17F-7488T/C polymorphisms and osteoarthritis susceptibility: A meta-analysis. Int J Rheum Dis. 2020;23:37–46. doi: 10.1111/1756-185X.13737. [DOI] [PubMed] [Google Scholar]
- 34.Lee YH, Song GG. Association between IL-17 gene polymorphisms and circulating IL-17 levels in osteoarthritis: a meta-analysis. Z Rheumatol. 2019 doi: 10.1007/s00393-019-00720-2. [DOI] [PubMed] [Google Scholar]