Abstract
Breast cancer is the most common malignancy, and metastasis is the main cause of cancer-associated mortality in women worldwide. Transforming growth factor-β (TGF-β) signaling, an inducer of epithelial-to-mesenchymal transition (EMT), plays an important role in breast cancer metastasis. Abnormal expression of miR-543 is associated with tumorigenesis and progression of various human cancers; however, the knowledge about the role of miR-543 in breast cancer metastasis is still unknown. In this study, we demonstrated that miR-543 inhibits the EMT-like phenotype and TGF-β-induced breast cancer metastasis both in vitro and in vivo by targeting ZNF281. ZNF281 transactivates the EMT-related transcription factor ZEB1 and Snail. Furthermore, both ZEB1 and Snail can transcriptionally suppress miR-543 expression. Taken together, our data uncover the ZNF281-miR-543 feedback loop and provide a mechanism to extend the understanding of TGF-β network complexity.
Keywords: miR-543, ZNF281, transforming growth factor-β, epithelial-to-mesenchymal transition, metastasis, breast cancer
Graphical Abstract
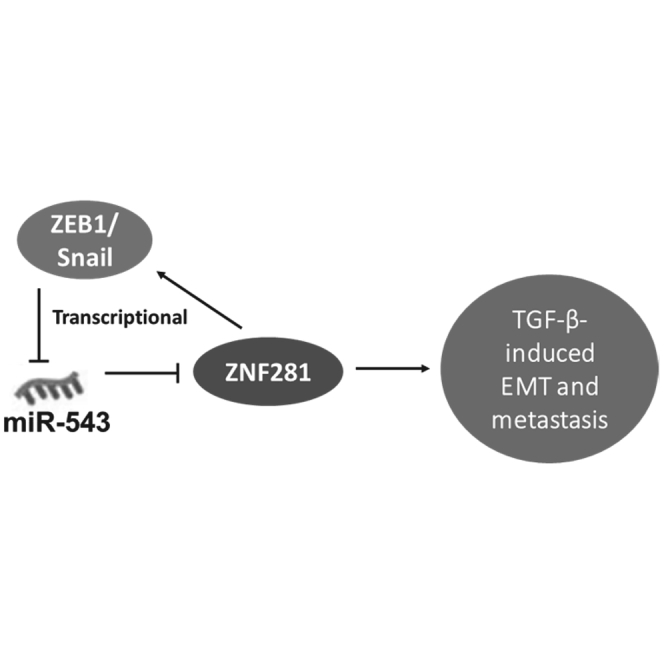
miR-543 suppresses breast cancer metastasis and EMT-like phenotype by targeting ZNF281. ZNF281 functions as an inducer of EMT and transactivates the expression of ZEB1 and Snail. Furthermore, miR-543 is a transcriptional target of ZEB1 and Snail. The ZNF281-miR-543 feedback loop regulates transforming growth factor-β-induced breast cancer metastasis.
Introduction
Breast cancer is the most common malignancy and the second cause of cancer-associated mortality in women worldwide.1 Although earlier diagnosis and effective treatment have improved the outcome of patients with breast cancer, distant metastasis remains the main cause of death. Therefore, understanding the mechanisms of breast cancer metastasis will be beneficial to develop effective anti-cancer drugs and improve breast cancer prognosis.
Metastasis is a multi-step process by which cancer cells spread from the primary tumor to colonize distant sites. Epithelial-to-mesenchymal transition (EMT), originally characterized in an embryo developmental program, was adopted by cancer cells during the metastatic cascade to gain migratory ability and reach distant organs, losing epithelial cell adhesion and cell-cell contacts, while undergoing cell shape remodeling and cytoskeleton rearrangement.2 EMT has been enthusiastically proposed as an essential step for metastasis. Most breast cancers are carcinomas of epithelial origin, in which the progression to malignancy appears to exploit a pathological EMT process.3
MicroRNAs (miRNAs), a group of endogenous non-coding and single-stranded small RNAs having a length of 21–23 nt, mainly bind to the 3ʹ untranslated region (UTR) of their target mRNAs, thereby inducing target mRNA translational repression or degradation.4 miR-543 is located in the imprinted DLK1-DIO3 region comprising a regulatory network that restrains the EMT program.5 To date, many studies have demonstrated that altered expression of miR-543 is associated with tumorigenesis and progression of various human cancers, including lung cancer,6,7 gastric cancer,8, 9, 10 colorectal cancer,11, 12, 13 prostate cancer,14 hepatocellular carcinoma,15 clear cell renal cell carcinoma,16 osteosarcoma,17 and glioma.18 miR-543 has been reported to function as an oncogene and promotes cell proliferation, migration, and invasion and induces an EMT-like phenotype in several types of cancers, including gastric cancer, colorectal cancer, hepatocellular carcinoma, prostate cancer, lung cancer, and clear cell renal cell carcinoma.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 Conversely, miR-543 exerts an opposing role in osteosarcoma, glioma, endometrial cancer, and breast cancer.17, 18, 19, 20 miR-543 suppresses breast cancer proliferation and induces cell apoptosis by targeting Extracellular Signal-Regulated Kinase 2.20 However, knowledge about the role of miR-543 in breast cancer metastasis is still unknown.
The zinc-finger transcription factor ZNF281 has been described to act as a transcriptional repressor of Nanog and control cellular stemness.21,22 ZNF281 antagonizes the differentiation of murine cortical neurons and neuroblastoma cells, and contributes to the DNA damage response.23, 24, 25 SOX4, a master regulator of EMT, is overexpressed in many types of human cancers and influences cancer progression.26 SOX4 has been demonstrated to bind to the ZNF281 promoter and regulate its expression in prostate cancer by using a chromatin immunoprecipitation sequencing (ChIP-seq) analysis.27 Moreover, ZNF281 induces EMT in colorectal cancer by regulating the expression of several EMT-related genes.28,29 However, the role of ZNF281 in breast cancer progression remains largely unknown.
In the present study, we demonstrated that the expression of miR-543 is downregulated in breast cancer. miR-543 inhibits breast cancer metastasis and EMT-like phenotype by regulating transforming growth factor-β (TGF-β) signaling. ZNF281 is a target of miR-543 and regulated by a feedback loop involving ZEB1 and Snail. Together, our study reveals a novel mechanism of TGF-β signaling pathway in breast cancer metastasis.
Results
miR-543 Inhibits Breast Cancer Metastasis
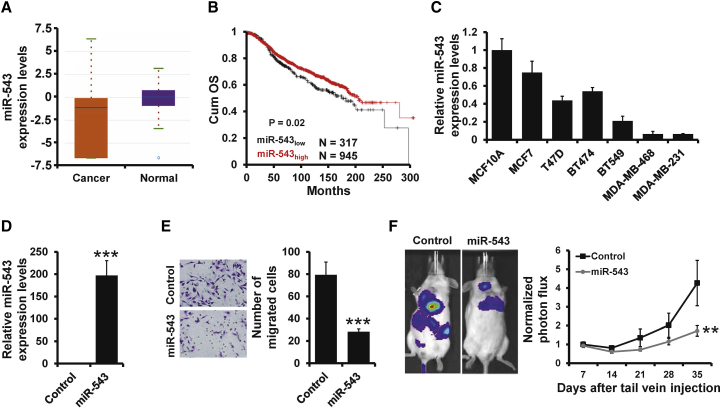
Although a previous study indicated that miR-543 suppresses breast cancer proliferation and induces cell apoptosis by regulation of the ERK/mitogen-activated protein kinase (MAPK) pathway,19 the role of miR-543 in breast cancer metastasis is still unknown. We first examined the expression of miR-543 in clinical breast cancer samples from The Cancer Genome Atlas (TCGA) database and observed that miR-543 expression was significantly decreased in breast cancer tissues compared with normal breast tissues (Figure 1A). Furthermore, we compared the overall survival in breast cancer patients with different miR-543 expression levels and found that patients with low miR-543 expression had a significantly poor outcome compared with those with high miR-543 expression (Figure 1B). We next determined the expression of miR-543 in breast cancer cell lines (MCF7, T47D, BT474, BT549, MDA-MB-468, and MDA-MB-231) and the normal breast epithelial cell line MCF10A by quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR). The expression of miR-543 was downregulated in all breast cancer cell lines as compared with MCF10A cells (Figure 1C). To further investigate the effect of miR-543 on breast cancer metastasis, we generated stable miR-543-expressing cells by lentiviral infection of MDA-MB-231-luciferase (MDA-MB-231-luc) cells (Figure 1D). Overexpression of miR-543 leads to an approximately 60%–70% reduction of cell invasion compared with the control group by Transwell analysis (Figure 1E). Next, we injected MDA-MB-231-luc cells stably expressing miR-543 and control cells via the tail vein into severe combined immunodeficiency (SCID) mice to evaluate the role of miR-543 in breast cancer metastasis. As expected, we observed that overexpression of miR-543 significantly decreases lung metastasis compared with those in control by bioluminescent imaging analysis (Figure 1F). Together, our results suggest that miR-543 inhibits breast cancer metastasis.
Figure 1.
miR-543 Inhibits Breast Cancer Metastasis
(A) The miR-543 expression in breast invasive carcinoma and normal tissues from TCGA database. (B) Kaplan-Meier analysis of the overall survival in patients with different miR-543 expression levels from TCGA database. (C) The expression of miR-543 in breast cancer cell lines and the normal breast cell line by qRT-PCR. (D) The expression of miR-543 in miR-543-overexpressed MDA-MB-231-luc and control cells by qRT-PCR. (E) Transwell analysis of cell invasion in cells as in (D). (F) Bioluminescent imaging and quantification plot of mice harboring lung metastases after tail vein injection of cells as in (D). ∗∗∗p < 0.001, ∗∗p < 0.01.
Knockdown of miR-543 Induces an EMT-like Phenotype
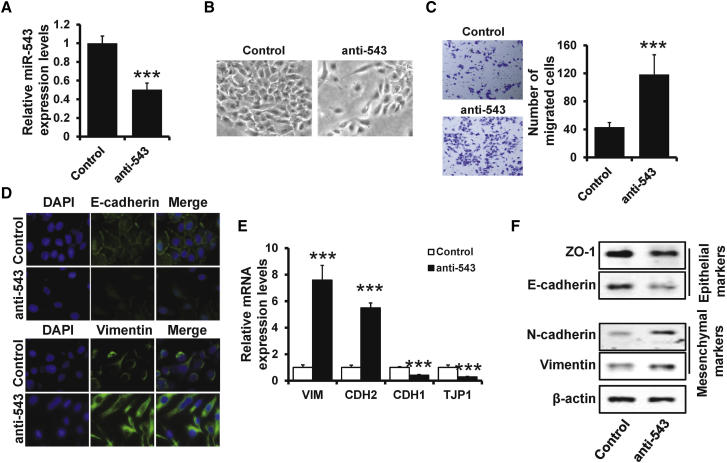
EMT, one of the main mechanisms in breast cancer metastasis, allows epithelial cells to acquire an invasive mesenchymal phenotype. We transfected miR-543 inhibitors into MCF10A cells (Figure 2A) and observed that miR-543-depleted MCF10A cells exhibited a fibroblast-like and spindle-shaped morphology, whereas the control cells retained their cobblestone-like morphology (Figure 2B). Furthermore, depletion of miR-543 inhibits the cell invasion by Transwell assay (Figure 2C). We next determined the expression of epithelial and mesenchymal markers by immunofluorescence (Figure 2D), qRT-PCR (Figure 2E), and western blotting (Figure 2F). miR-543-depleted MCF10A cells exhibited significant downregulation of epithelial markers ZO-1 and E-cadherin, but dramatic upregulation of the mesenchymal markers N-cadherin and Vimentin. Together, these results suggest that knockdown of miR-543 induces an EMT-like phenotype in breast cancer.
Figure 2.
Depletion of miR-543 Induces an EMT-like Phenotype
(A) The expression of miR-543 in MCF10A transfected with miR-543 inhibitors and control cells determined by qRT-PCR. (B) Morphology of cells as in (A). (C) Transwell analysis of cell invasion in cells as in (A). (D) Immunofluorescence analyses of epithelial marker E-cadherin and mesenchymal marker Vimentin in the cells as in (A). (E and F) mRNA (E) and protein (F) expression levels of epithelial markers E-cadherin and ZO-1 and mesenchymal markers N-cadherin and Vimentin in the cells described in (A) were evaluated by qRT-PCR and western blotting, respectively. ∗∗∗p < 0.001.
ZNF281 Is a Direct Target of miR-543
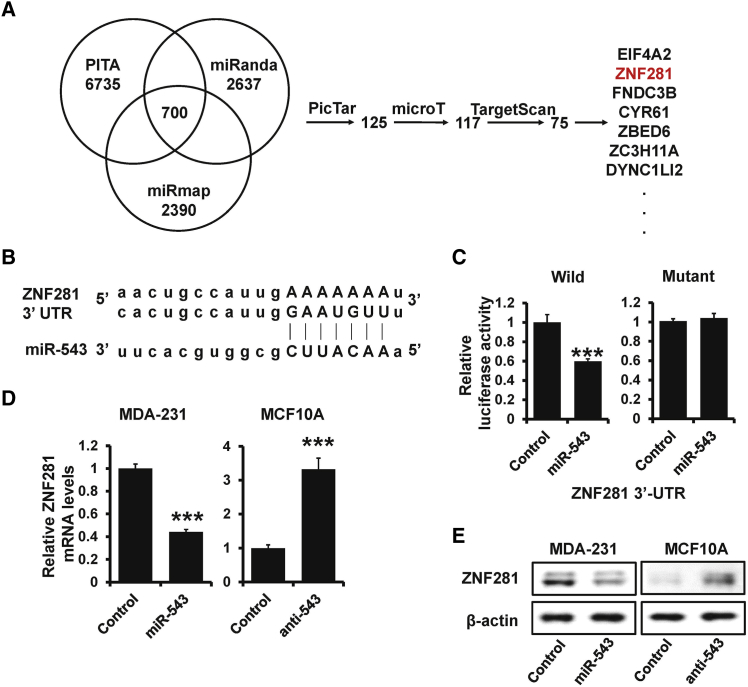
To elucidate the biological mechanisms underlying the role of miR-543 in breast cancer metastasis, we investigated the potential targets of miR-543 by using starBase v.3.0. We found several candidate genes by using multiple target-predicting programs (Figure 3A). EIF4A2 has been identified to be an oncogene and regulate chemosensitivity in breast cancer;30 however, the role of ZNF281 in breast cancer development and progression is still unknown. A target prediction program, TargetScan, was applied to identify ZNF281 as a putative miR-543 target (Figure 3B). To further confirm this regulation, we cloned ZNF281 and the miR-543 binding site mutated 3′ UTR into the psiCHEK2 luciferase reporter plasmid. Overexpression of miR-543 reduced the luciferase activity by about 40% compared with the control (Figure 3C, left), whereas mutation to the putative miR-543 binding site abolished the decreased luciferase activity induced by miR-543 overexpression (Figure 3C, right). Moreover, the expression of ZNF281 was decreased in miR-543-overexpressed MDA-MB-231 cells and was increased in miR-543-depleted MCF10A cells compared with that in control cells by qRT-PCR (Figure 3D) and western blotting (Figure 3E). Thus, these results indicate that ZNF281 is a direct target of miR-543.
Figure 3.
ZNF281 Is a Target of miR-543
(A) The potential target genes of miR-543 analyzed by starBase v.3.0. (B) The predicted binding of miR-543 with ZNF281 3ʹ UTR. (C) A dual-luciferase reporter assay was performed to validate the interaction between miR-543 and ZNF281. (D and E) The expression levels of ZNF281 in miR-543-overexpressed MDA-MB-231 (left) or miR-543-depleted MCF10A cells (right), as well as the control cells by qRT-PCR (D) and western blotting (E). ∗∗∗p < 0.001.
miR-543 Inhibits Breast Cancer Metastasis by Regulation of ZNF281
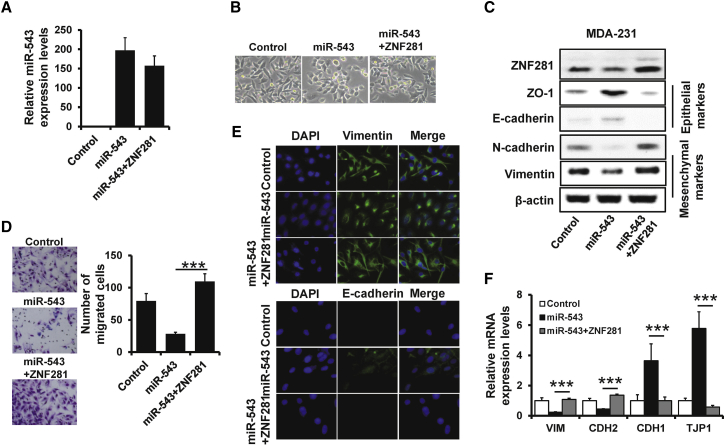
We next investigated whether miR-543 regulates breast cancer metastasis and EMT phenotype by regulating ZNF281 expression. We transfected pcDNA3-ZNF281 expression plasmid into miR-543-overexpressed MDA-MB-231 cells (Figure 4A) and observed that the expression of ZNF281 was restored in ZNF281-transfected miR-543-overexpressed MDA-MB-231 (Figure 4C). Overexpression of ZNF281 could induce a fibroblast-like and spindle-shaped morphology in miR-543-overexpressed MDA-MB-231 cells, whereas miR-543-overexpressed MDA-MB-231 cells retained their cobblestone-like morphology (Figure 4B). Moreover, upregulation of ZNF281 in miR-543-overexpressed MDA-MB-231 resulted in increased invasion in Transwell assay (Figure 4D). Furthermore, ZNF281-restored miR-543-overexpressed MDA-MB-231 cells exhibited significant downregulation of ZO-1 and E-cadherin but dramatic upregulation of N-cadherin and Vimentin, as determined by western blotting (Figure 4C), immunofluorescence (Figure 4E), and qRT-PCR (Figure 4F). Collectively, these results indicate that miR-543 inhibits breast cancer metastasis and EMT-like phenotype by regulation of ZNF281 expression.
Figure 4.
miR-543 Inhibits Breast Cancer Metastasis by Regulation of ZNF281
(A) The expression of miR-543 in miR-543- or ZNF281/miR-543-overexpressed MDA-MB-231, as well as the control cells determined by qRT-PCR. (B) Morphology of cells as in (A). (C) The protein expression levels of epithelial markers E-cadherin and ZO-1, mesenchymal markers N-cadherin and Vimentin, and ZNF281 in the cells described in (A) were evaluated by western blotting. (D) Transwell analysis of cell invasion in cells as in (A). (E) Immunofluorescence analyses of epithelial marker E-cadherin and mesenchymal marker Vimentin in the cells as in (A). (F) The mRNA expression levels of epithelial markers E-cadherin and ZO-1 and mesenchymal markers N-cadherin and Vimentin in the cells described in (A) were evaluated by qRT-PCR. ∗∗∗p < 0.001.
ZNF281 Transactivates the Snail and ZEB1 Expression
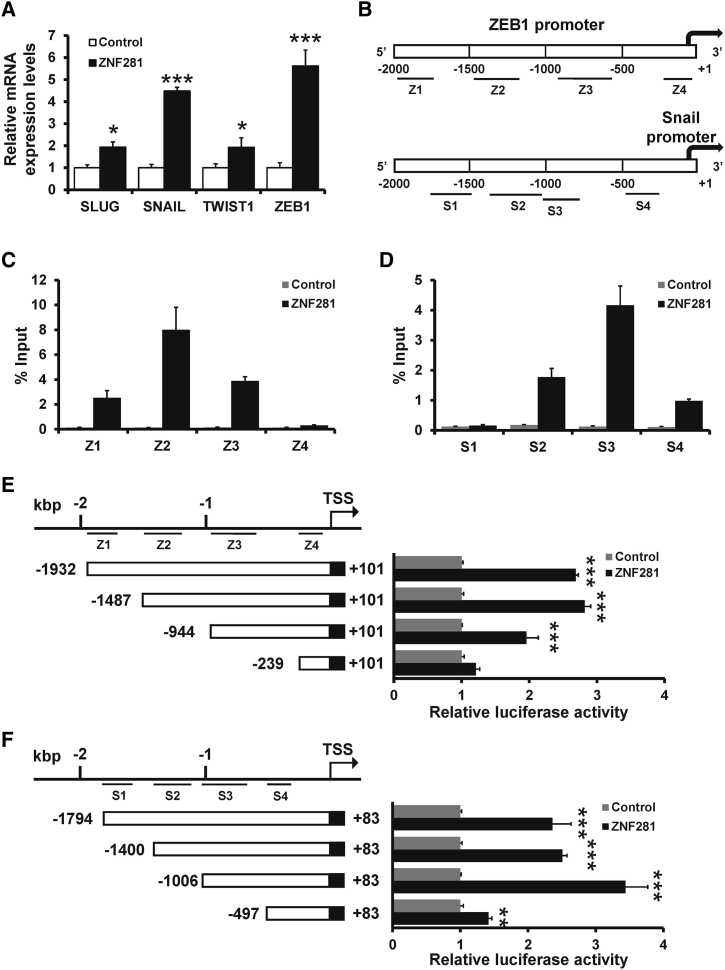
We next determined whether ZNF281 induced an EMT-like phenotype by regulation of EMT-related transcription factors. We transfected pcDNA3-ZNF281-hemagglutinin (HA) into MCF10A cells and observed that the expression of Snail and ZEB1 was increased more than 4-fold in ZNF281-transfected MCF10A cells compared with the control group (Figure 5A). ZNF281 is known to bind in GC-rich DNA sequences, and similar sequences are present in both Snail and ZEB1 promoter.31 Thus, we speculated that ZNF281 induces the Snail and ZEB1 expression by direct binding of the Snail and ZEB1 promoter (Figure 5B). The occupancy by ZNF281 was detected in the ZEB1 (Figure 5B) and Snail (Figure 5C) promoter region (−2,000 to +1) by a HA-specific ChIP analysis. ZNF281 was able to bind to both the ZEB1 promoter region (−2,000 to −500; Figure 5C) and the Snail promoter region (−1,500 to +1; Figure 5D). A series of deletion constructs of the ZEB1 and Snail promoter was used to further determine the regions involved in the regulation of ZNF281 by a dual-luciferase reporter assay. The activation of the ZEB1 promoter was most dominant for the −1,487 to +101 reporter (Figure 5E), which was in line with the dominant occupancy of ZNF281 to a region −1,500 to −1,000 bp (Z2) upstream of the transcription start site (TSS). In addition, consistent with the ChIP analysis, the activation of Snail promoter was most dependent on the −1,006 to +83 region (S3) (Figure 5F). Taken together, ZNF281 transactivates the Snail and ZEB1 expression.
Figure 5.
ZNF281 Transactivates the Snail and ZEB1 Expression
(A) The expression of EMT-related factors in ZNF281-overexpressed MCF10A and control cells. (B) Schematic depiction of the ZEB1 and SNAIL promoter. Black bars indicated primers for ChIP analysis. (C and D) ChIP analysis of interaction between ZNF281 and the ZEB1 (C) or Snail (D) promoter in ZNF281-transfected MCF10A and control cells using anti-HA or anti-IgG antibodies. Results represent the percentage of input chromatin. (E and F) Dual-luciferase analysis of ZEB1 (E) or Snail (F) promoter activity in ZNF281-transfected 293FT and control cells. ∗∗∗p < 0.001, ∗p < 0.01.
The Existence of a ZNF281-Snail/ZEB1-miR-543 Feedback Loop
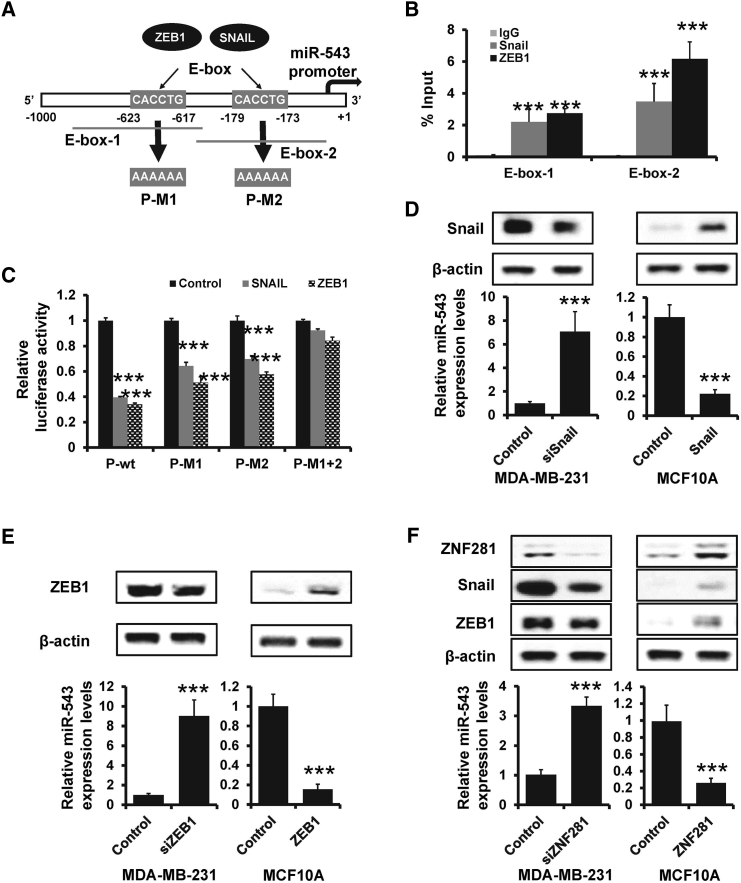
By analyzing the promoter region of miR-543, we found two E-boxes (CACCTG) on the miR-543 promoter region, which represent putative ZEB1 and Snail binding sites (Figure 6A). Next, we used ChIP assay to immunoprecipitate Snail or ZEB1 and used primers to amplify the E-box1 or E-box2 region of the miR-543 promoter region. As shown in Figure 6B, both Snail and ZEB1 could bind directly to the miR-543 promoter region (Figure 6B). To further determine whether miR-543 is mediated by ZEB1 or Snail via E-boxes, a region encompassing 1 kb upstream of the miR-543 TSS was subcloned into pGL3 luciferase reporter plasmid. As shown in Figure 6C, both Snail and ZEB1 could reduce the wild-type reporter luciferase activity, whereas mutation of E-box-1 or E-box-2 decreased the responsiveness to Snail or ZEB1. In addition, a reporter combined with mutation of E-box-1 and E-box-2 resulted in complete loss of responsiveness to both Snail and ZEB1 (Figure 6C). Furthermore, depletion of Snail or ZEB1 enhances the miR-543 expression, whereas ectopic Snail or ZEB1 reduced the miR-543 expression at both mRNA and protein levels in MDA-BM-231 or MCF7, respectively (Figures 6D and 6E). Moreover, we observed a decreased expression of Snail and ZEB1 and an increased miR-543 expression in ZNF281-depleted MDA-MB-231 cells (Figure 6F, left). Conversely, ectopic expression of ZNF281 enhanced the expression of Snail and ZEB1, and reduced the expression of miR-543 in MCF10A cells (Figure 6F, right). Together, these results identify the existence of a ZNF281-Snail/ZEB1-miR-543 feedback loop in breast cancer metastasis.
Figure 6.
The Feedback Loop of ZNF281-Snail/ZEB1-miR-543
(A) Scheme of the miR-543 promoter and ZEB1/Snail binding sites. Black bars indicated primers for ChIP analysis. (B) ChIP analysis of interaction between ZEB1/Snail and the miR-543 promoter in MDA-MB-231 cells using anti-ZEB1, anti-Snail, or anti-IgG antibodies. Results represent the percentage of input chromatin. (C) Dual-luciferase analysis of indicated miR-543 promoter activity in ZEB1 or Snail-transfected 293FT and control cells. (D) The expression of Snail and miR-543 in Snail-depleted MDA-MB-231 (left), Snail-overexpressed MCF10A (right), as well as the control cells determined by western blotting and qRT-PCR, respectively. (E) The expression of ZEB1 and miR-543 in ZEB1-depleted MDA-MB-231 (left), ZEB1-overexpressed MCF10A (right), as well as the control cells determined by western blotting and qRT-PCR, respectively. (F) The expression of ZNF281, Snail, ZEB1, and miR-543 in ZNF281-depleted MDA-MB-231 (left), ZNF281-overexpressed MCF10A (right), as well as the control cells determined by western blotting and qRT-PCR, respectively. ∗∗∗p < 0.001. P-M1, E-box-1 mutated; P-M2, E-box-2 mutated; P-M1+2, both E-box-1 and -2 mutated; P-wt, wild-type.
Regulation of ZNF281-miR-543 Feedback Loop on TGF-β-Induced EMT
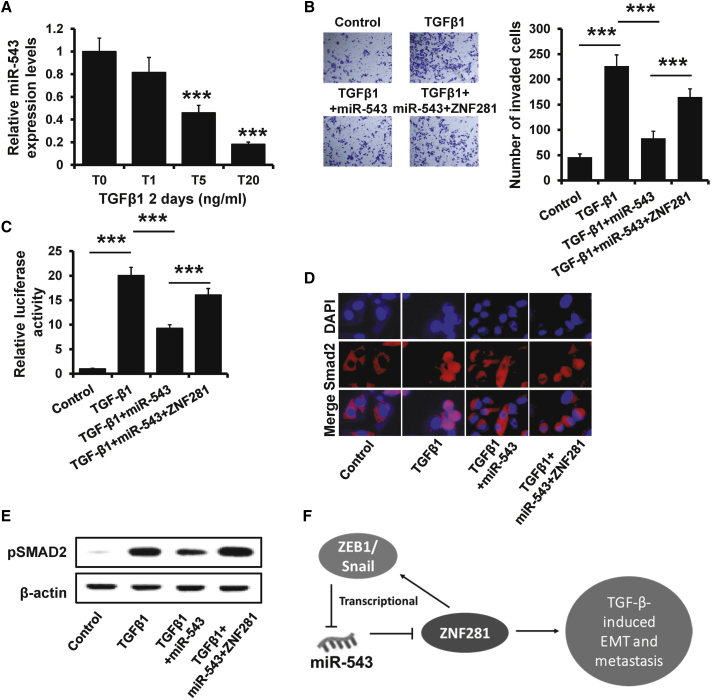
Both Snail and ZEB1 are master regulators in TGF-β-induced EMT during cancer metastasis.32 Thus, we investigated whether the ZNF281-miR-543 feedback loop regulates TGF-β-induced EMT in breast cancer metastasis. We observed that miR-543 expression was reduced in MCF10A cells after treatment with TGF-β1 for 2 days (Figure 7A). As shown in Figure 7B, the invasive ability of MCF10A cells was dramatically decreased in TGF-β1-treated MCF10A cells after transfection of miR-543 mimics. In contrast, restoration of ZNF281 abolished the miR-543-induced invasive reduction in TGF-β1-treated MCF10A cells (Figure 7B). To further explore the role of ZNF281-miR-543 feedback loop in TGF-β signaling, we detected the luciferase activity of SMAD reporter in miR-543-overexpressed MCF10A cells with or without ZNF281 overexpression after treatment with TGF-β1. The luciferase activity was significantly decreased in miR-543-transfected MCF10A cells compared with that in control cells after treatment with TGF-β1, but this effect was reversed after ZNF281 overexpression (Figure 7C). Moreover, the nuclear localization of SMAD2 and the expression of pSMAD2 were also increased in TGF-β1-treated and miR-543-overexpressed MCF10A cells after ZNF281 overexpression (Figures 7D and 7E). Together, these results indicate that the ZNF281-miR-543 feedback loop is involved in TGF-β-induced EMT.
Figure 7.
ZNF281-miR-543 Feedback Loop Is Involved in TGF-β-Induced EMT
(A) The expression of miR-543 in MCF10A cells after treatment with TGF-β1 at indicated concentration. (B) Transwell analysis of miR-543-overexpressed MCF10A or control cells with TGF-β1 and/or ZNF281 overexpression. (C) Luciferase reporter analysis of TGF-β signaling activity in cells as in (B). (D) Localization of SMAD2 in cells as in (B) as determined by immunofluorescence staining. (E) The expression of pSMAD2 in cells as in (B) by western blotting. (F) A model for the ZNF281-miR-543 feedback loop in TGF-β-induced breast cancer metastasis.
Discussion
In the present study, we could show that miR-543 functions as a tumor suppressor and an EMT inhibitor in breast cancer metastasis. ZNF281 was identified to be a target of miR-543 and a transcriptional activator on regulation of EMT-related factors Snail and ZEB1 expression. Furthermore, miR-543 is transcriptionally suppressed by both Snail and ZEB1. Ectopic expression of ZNF281 reverses the TGF-β-induced EMT in breast cancer. Therefore, our results revealed that the ZNF281-miR-543 feedback loop is involved in breast cancer metastasis by regulating TGF-β-induced EMT.
Dysregulation of miRNAs has been reported to be involved in almost every cellular process during carcinogenesis and progression in various types of cancer, including breast cancer.33 The abnormal expression and function of miR-543 was observed in various types of human cancers. Furthermore, miR-543 may play an opposite role in different tumor environments. miR-543 promotes tumor growth and metastasis in lung cancer, gastric cancer, and colorectal cancer, whereas miR-543 functions as a tumor suppressor in glioma and osteosarcoma.6, 7, 8, 9, 10, 11, 12, 13,17,18 Although it has been indicated that miR-543 expression is decreased in breast cancer, and ectopic expression of miR-543 leads to cell-cycle arrest and suppresses breast cancer proliferation, the effect of miR-543 on breast cancer metastasis is still unknown.19 Our results indicated that miR-543 suppresses breast cancer metastasis both in vitro and in vivo. Metastasis is a multi-step process by which cancer cells spread from the primary tumor to colonize distant sites, and EMT has been shown to play important roles in these steps to promote metastasis.34 During EMT, epithelial cells lose their cell polarity and cell adhesion, and acquire an invasive mesenchymal phenotype. An increased expression of mesenchymal markers Vimentin and N-cadherin and a decreased expression of epithelial markers E-cadherin and ZO-1 are often observed during the EMT process.35 Here, we found that miR-543 reduced the expression of Vimentin and N-cadherin, and induced the expression of E-cadherin and ZO-1, whereas depletion of miR-543 yielded the opposite effects, suggesting that miR-543 is an EMT suppressor in breast cancer metastasis.
ZNF281, a transcription factor that contains four Krüppel-type zinc-finger domains, regulates the expression of several genes dependent on the cellular context by transcriptional activation and repression.21,28 ZNF281 has been identified to be an oncogene and promotes cancer progression in pancreatic cancer and colon cancer by activation of Wnt/β-Catenin signaling and EMT-like phenotype.28,29,36 Moreover, ZNF281 inhibits neuronal differentiation and is a prognostic marker for neuroblastoma.23 Previous studies indicated that ZNF281 is regulated by miR-1 and miR-34a in muscle differentiation and colon cancer metastasis, respectively.28,37 In the present study, we demonstrated that ZNF281 is a target of miR-543 and reverses the miR-543-induced inhibition of EMT and breast cancer metastasis. As a transcription factor, ZNF281 can regulate the expression of target genes by binding to GC-rich sequences in their promoter region. We found that similar GC-rich sequences are present on the promoter region of EMT-related factors ZEB1 and Snail. Moreover, ZN281 transactivates the ZEB1 and Snail expression, suggesting that ZNF281 is an EMT-promoting transcription factor in breast cancer metastasis.
Both ZEB1 and Snail are zinc-finger transcription factors involved in the regulation of cell differentiation fate and determination.32 The zinc-finger domain is highly conserved and allows DNA occupancy at the E-box in the promoter region to regulate the expression of target genes. Aberrant expression of ZEB1 and Snail has been observed in many human cancers, including breast cancer.38,39 Here, we found that ZEB1 and Snail function as a transcriptional repressor, which depends on the E-box binding sites on the miR-543 promoter region. Increasing evidence supports an extensive cross-talk between miRNA and EMT-related transcription factors during breast cancer metastasis.40 Our results further extend this network by adding reciprocal connections between ZEB1, Snail, ZNF281, and miR-543.
TGF-β signaling, a critical player in embryonic development, is closely involved in the EMT and plays a key role in breast cancer metastasis. It is driven by a set of EMT-related transcription factors, including ZEB1 and Snail, which function as inducers of mesenchymal markers, such as Vimentin and N-cadherin, and repressors of E-cadherin and ZO-1. The TGF-β signaling pathway regulates these transcription factors, which confers TGF-β a potent inducer of EMT.41 In this study, we observed a decreased expression of miR-543 after treatment with TGF-β1. Overexpression of miR-543 reverses the TGF-β1-induced EMT-like phenotype and breast cancer metastasis, whereas ectopic expression of ZNF281 abolishes these effects, suggesting that the ZNF281-miR-543 feedback loop regulates breast cancer metastasis by regulation of TGF-β signaling.
Conclusions
In summary, we demonstrated that miR-543 is a tumor suppressor in breast cancer metastasis. miR-543 suppresses breast cancer metastasis and EMT-like phenotype by targeting ZNF281. ZNF281 functions as an inducer of EMT and transactivates the expression of ZEB1 and Snail. Furthermore, miR-543 is a transcriptional target of ZEB1 and Snail. Based on the findings from our observation, we propose a model that highlights the role of miR-543 in regulating TGF-β signaling during breast cancer metastasis (Figure 7F). The uncovering of this ZNF281-miR-543 feedback loop will extend our understanding of TGF-β network complexity.
Materials and Methods
Cell Culture
Normal breast epithelial cell line MCF10A, breast cancer cell lines BT549, MDA-MB-231, MDA-MB-468, T47D, and MCF7, and human embryonic kidney 293FT cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured as previously described.42
Plasmids, siRNA, miRNA, and Antibodies
The antibodies, plasmids, miRNA, and small interfering RNA (siRNA) are described in the Supplemental Materials and Methods.
Transfection and Generation of Stable miR-543-Overexpressed Cell Line
For transient transfection, miRNA or plasmids were transfected into different cell lines using FuGENE HD Transfection Reagent (Promega, Madison, WI, USA) or TransFast Transfection Reagent (Promega) according to the manufacturer’s recommendations. To generate stable miR-543-overexpressed cells, we used the lentiviruses (RiboBio, Shanghai, China) to infect MDA-MB-231 cells according to the manufacturer’s recommendations.
Western Blotting and Immunofluorescence
Standard procedures for western blotting and immunofluorescence are described in the Supplemental Materials and Methods.
RNA Extraction and qRT-PCR
Total RNA was extracted from cultured cells using a mirVana PARISTM Kit (Life Technologies) according to the manufacturer’s instructions. TaqMan qRT-PCR was performed to detect the expression of mature miRNAs using a TaqMan miRNA Reverse Transcription Kit, has-RNU6B (U6, ABI Assay ID: 001093), and miR-190 (ABI Assay ID: 002376) according to the manufacturer’s instructions (Life Technologies). qPCR was performed with GoTaq qPCR Master Mix (Promega). The cycle threshold (Ct) values of each gene were averaged from triplicate reactions. The gene expression was determined by 2−ΔCt method. The primers of qPCR are listed in Table S1.
Luciferase Reporter Assays
For the luciferase reporter assay, 293FT cells were seeded in a 12-well plate at 5 × 104 cells per well and transfected with FuGENE HD Transfection Reagent (Promega) for 48 h with 200 ng of the indicated firefly luciferase reporter plasmid, 200 ng pcDNA3-ZNF281/ZEB/Snail, and 20 ng Renilla reporter as a normalization control. Firefly and Renilla luciferase activities were determined by a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s recommendation.
Transwell
Matrigel-coated Transwell was used to evaluate the invasion ability of breast cancer cells in vitro. A total of 5 × 104 cells were seeded to the upper chamber, and medium containing 20% FBS was added into the lower chamber. Twenty-four hours later, the migrant cells that had attached to the lower surface were fixed with 20% methanol and stained for 20 min with crystal violet. The membranes were then carved and embedded under coverslips with the cells on the top. The number of migrating cells was counted under a microscope in five predetermined fields.
ChIP Analysis
ChIP analysis was performed according to the protocol of Upstate Biotechnology as previously described.42 To calculate the binding to the DNA region analyzed, we expressed the enrichment using the specific antibody compared with the isotype control as %input. The sequences of oligonucleotides used as ChIP primers are listed in Table S2.
Xenograft
Female SCID mice (6–7 weeks old) were injected with the stable miR-543-overexpressed MDA-MB-231 cells or the control cells (2 × 106 cells) at lateral tail vein intravenously. The formation of metastasis was observed and assessed by bioluminescence imaging using a Xenogen IVIS 200 Imaging System (Caliper Life Sciences, Hopkinton, MA, USA) at multiple time points (days 7, 14, 21, 28, and 35). All of the animal protocols were reviewed and approved by the Animal Ethics Committee of Tianjin Medical University Cancer Institute & Hospital and were performed according to the guidelines for the welfare and use of animals in cancer research and national law.
Statistical Analysis
Data are presented as mean ± standard deviation. The Student’s t test (two-tailed) was used to determine the differences between the experimental and control groups. The level of significance was set to p < 0.05; asterisks generally indicate: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. All calculations were performed with the SPSS for Windows statistical software package (SPSS, Chicago, IL, USA).
Author Contributions
Y.Y. and X.-C.C. designed the study. W.J., Q.M., and X.-Y.L. performed the experiments. W.J. and Y.Y. wrote and revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant 81502518) and the Natural Science Foundation of Tianjin City (grant 17JCQNJC10400).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.05.020.
Contributor Information
Xu-Chen Cao, Email: caoxuchen@tmu.edu.cn.
Yue Yu, Email: yuyue@tmu.edu.cn.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Punzi S., Balestrieri C., D’Alesio C., Bossi D., Dellino G.I., Gatti E., Pruneri G., Criscitiello C., Lovati G., Meliksetyan M. WDR5 inhibition halts metastasis dissemination by repressing the mesenchymal phenotype of breast cancer cells. Breast Cancer Res. 2019;21:123. doi: 10.1186/s13058-019-1216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y., Chen L., Hong D., Chen Z., Zhang J., Fu L., Pan D., Zhang Y., Xu Y., Gan S. Baicalein inhibits fibronectin-induced epithelial-mesenchymal transition by decreasing activation and upregulation of calpain-2. Cell Death Dis. 2019;10:341. doi: 10.1038/s41419-019-1572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haga C.L., Phinney D.G. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J. Biol. Chem. 2012;287:42695–42707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S., Jiang W., Zhang X., Lu Z., Geng Q., Wang W., Li N., Cai X. LINC-PINT alleviates lung cancer progression via sponging miR-543 and inducing PTEN. Cancer Med. 2020;9:1999–2009. doi: 10.1002/cam4.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J., Chen Y., Li X., Miao H., Li R., Chen D., Wen Z. THUMPD3-AS1 Is Correlated With Non-Small Cell Lung Cancer And Regulates Self-Renewal Through miR-543 And ONECUT2. OncoTargets Ther. 2019;12:9849–9860. doi: 10.2147/OTT.S227995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y., Yang Z., Zhang T., Shen L., Li Y., Ding S. SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer. Cell Death Dis. 2019;10:625. doi: 10.1038/s41419-019-1859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J., Wang F., Wang X., He Z., Zhu X. miRNA-543 promotes cell migration and invasion by targeting SPOP in gastric cancer. OncoTargets Ther. 2018;11:5075–5082. doi: 10.2147/OTT.S161316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Dong G., Wang B., Gao W., Yang Q. miR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem. Biophys. Res. Commun. 2016;469:15–21. doi: 10.1016/j.bbrc.2015.11.062. [DOI] [PubMed] [Google Scholar]
- 11.Liang Y., Zhu D., Zhu L., Hou Y., Hou L., Huang X., Li L., Wang Y., Li L., Zou H. Dichloroacetate Overcomes Oxaliplatin Chemoresistance in Colorectal Cancer through the miR-543/PTEN/Akt/mTOR Pathway. J. Cancer. 2019;10:6037–6047. doi: 10.7150/jca.34650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G., Zhou J., Dong M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci. Rep. 2019;39 doi: 10.1042/BSR20190249. BSR20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J., Zhou J., Dong M., Sheng W. Dysregulation of MicroRNA-543 expression in colorectal cancer promotes tumor migration and invasion. Mol. Carcinog. 2017;56:250–257. doi: 10.1002/mc.22489. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y.C., He W.Y., Dong C.H., Pei L., Ma Y.L. lncRNA HCG11 regulates cell progression by targeting miR-543 and regulating AKT/mTOR pathway in prostate cancer. Cell Biol. Int. 2019;43:1453–1462. doi: 10.1002/cbin.11194. [DOI] [PubMed] [Google Scholar]
- 15.Yu L., Zhou L., Cheng Y., Sun L., Fan J., Liang J., Guo M., Liu N., Zhu L. MicroRNA-543 acts as an oncogene by targeting PAQR3 in hepatocellular carcinoma. Am. J. Cancer Res. 2014;4:897–906. [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F., Ma J., Tang Q., Zhang W., Fu Q., Sun J., Wang H., Song B. MicroRNA-543 promotes the proliferation and invasion of clear cell renal cell carcinoma cells by targeting Krüppel-like factor 6. Biomed. Pharmacother. 2018;97:616–623. doi: 10.1016/j.biopha.2017.10.136. [DOI] [PubMed] [Google Scholar]
- 17.Wang L.H., Tsai H.C., Cheng Y.C., Lin C.Y., Huang Y.L., Tsai C.H., Xu G.H., Wang S.W., Fong Y.C., Tang C.H. CTGF promotes osteosarcoma angiogenesis by regulating miR-543/angiopoietin 2 signaling. Cancer Lett. 2017;391:28–37. doi: 10.1016/j.canlet.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Xu L., Yu J., Wang Z., Zhu Q., Wang W., Lan Q. miR-543 functions as a tumor suppressor in glioma in vitro and in vivo. Oncol. Rep. 2017;38:725–734. doi: 10.3892/or.2017.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen P., Xu W., Luo Y., Zhang Y., He Y., Yang S., Yuan Z. MicroRNA 543 suppresses breast cancer cell proliferation, blocks cell cycle and induces cell apoptosis via direct targeting of ERK/MAPK. OncoTargets Ther. 2017;10:1423–1431. doi: 10.2147/OTT.S118366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bing L., Hong C., Li-Xin S., Wei G. MicroRNA-543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch. Gynecol. Obstet. 2014;290:533–541. doi: 10.1007/s00404-014-3219-3. [DOI] [PubMed] [Google Scholar]
- 21.Fidalgo M., Shekar P.C., Ang Y.S., Fujiwara Y., Orkin S.H., Wang J. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells. 2011;29:1705–1716. doi: 10.1002/stem.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fidalgo M., Faiola F., Pereira C.F., Ding J., Saunders A., Gingold J., Schaniel C., Lemischka I.R., Silva J.C., Wang J. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc. Natl. Acad. Sci. USA. 2012;109:16202–16207. doi: 10.1073/pnas.1208533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pieraccioli M., Nicolai S., Pitolli C., Agostini M., Antonov A., Malewicz M., Knight R.A., Raschellà G., Melino G. ZNF281 inhibits neuronal differentiation and is a prognostic marker for neuroblastoma. Proc. Natl. Acad. Sci. USA. 2018;115:7356–7361. doi: 10.1073/pnas.1801435115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pieraccioli M., Nicolai S., Antonov A., Somers J., Malewicz M., Melino G., Raschellà G. ZNF281 contributes to the DNA damage response by controlling the expression of XRCC2 and XRCC4. Oncogene. 2016;35:2592–2601. doi: 10.1038/onc.2015.320. [DOI] [PubMed] [Google Scholar]
- 25.Nicolai S., Mahen R., Raschellà G., Marini A., Pieraccioli M., Malewicz M., Venkitaraman A.R., Melino G. ZNF281 is recruited on DNA breaks to facilitate DNA repair by non-homologous end joining. Oncogene. 2020;39:754–766. doi: 10.1038/s41388-019-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiwari N., Tiwari V.K., Waldmeier L., Balwierz P.J., Arnold P., Pachkov M., Meyer-Schaller N., Schübeler D., van Nimwegen E., Christofori G. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 27.Scharer C.D., McCabe C.D., Ali-Seyed M., Berger M.F., Bulyk M.L., Moreno C.S. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn S., Jackstadt R., Siemens H., Hünten S., Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin C.J., Bu P.L., Zhang Q., Chen J.T., Li Q.Y., Liu J.T., Dong H.C., Ren X.Q. ZNF281 Regulates Cell Proliferation, Migration and Invasion in Colorectal Cancer through Wnt/β-Catenin Signaling. Cell. Physiol. Biochem. 2019;52:1503–1516. doi: 10.33594/000000104. [DOI] [PubMed] [Google Scholar]
- 30.Liu M., Gong C., Xu R., Chen Y., Wang X. MicroRNA-5195-3p enhances the chemosensitivity of triple-negative breast cancer to paclitaxel by downregulating EIF4A2. Cell. Mol. Biol. Lett. 2019;24:47. doi: 10.1186/s11658-019-0168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisowsky T., Polosa P.L., Sagliano A., Roberti M., Gadaleta M.N., Cantatore P. Identification of human GC-box-binding zinc finger protein, a new Krüppel-like zinc finger protein, by the yeast one-hybrid screening with a GC-rich target sequence. FEBS Lett. 1999;453:369–374. doi: 10.1016/s0014-5793(99)00754-1. [DOI] [PubMed] [Google Scholar]
- 32.Simeone P., Trerotola M., Franck J., Cardon T., Marchisio M., Fournier I., Salzet M., Maffia M., Vergara D. The multiverse nature of epithelial to mesenchymal transition. Semin. Cancer Biol. 2019;58:1–10. doi: 10.1016/j.semcancer.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Loh H.Y., Norman B.P., Lai K.S., Rahman N.M.A.N.A., Alitheen N.B.M., Osman M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019;20:4940. doi: 10.3390/ijms20194940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu B.W., Yu Z.H., Chen A.X., Chi J.R., Ge J., Yu Y., Cao X.C. Estrogen receptor-α-miR-1271-SNAI2 feedback loop regulates transforming growth factor-β-induced breast cancer progression. J. Exp. Clin. Cancer Res. 2019;38:109. doi: 10.1186/s13046-019-1112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji W., Diao Y.L., Qiu Y.R., Ge J., Cao X.C., Yu Y. LINC00665 promotes breast cancer progression through regulation of the miR-379-5p/LIN28B axis. Cell Death Dis. 2020;11:16. doi: 10.1038/s41419-019-2213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian Y., Li J., Xia S. ZNF281 Promotes Growth and Invasion of Pancreatic Cancer Cells by Activating Wnt/β-Catenin Signaling. Dig. Dis. Sci. 2017;62:2011–2020. doi: 10.1007/s10620-017-4611-1. [DOI] [PubMed] [Google Scholar]
- 37.Nicolai S., Pieraccioli M., Smirnov A., Pitolli C., Anemona L., Mauriello A., Candi E., Annicchiarico-Petruzzelli M., Shi Y., Wang Y. ZNF281/Zfp281 is a target of miR-1 and counteracts muscle differentiation. Mol. Oncol. 2020;14:294–308. doi: 10.1002/1878-0261.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Zhang Z., Zhang Q., Zhang Q., Sun P., Xiang R., Ren G., Yang S. ZEB1 confers chemotherapeutic resistance to breast cancer by activating ATM. Cell Death Dis. 2018;9:57. doi: 10.1038/s41419-017-0087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W.S., Chan S.H., Chang H.T., Li G.C., Tu Y.T., Tseng H.H., Fu T.Y., Chang H.Y., Liou H.H., Ger L.P., Tsai K.W. Isocitrate dehydrogenase 1-snail axis dysfunction significantly correlates with breast cancer prognosis and regulates cell invasion ability. Breast Cancer Res. 2018;20:25. doi: 10.1186/s13058-018-0953-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zare M., Bastami M., Solali S., Alivand M.R. Aberrant miRNA promoter methylation and EMT-involving miRNAs in breast cancer metastasis: Diagnosis and therapeutic implications. J. Cell. Physiol. 2018;233:3729–3744. doi: 10.1002/jcp.26116. [DOI] [PubMed] [Google Scholar]
- 41.Hao Y., Baker D., Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019;20:2767. doi: 10.3390/ijms20112767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Y., Luo W., Yang Z.J., Chi J.R., Li Y.R., Ding Y., Ge J., Wang X., Cao X.C. miR-190 suppresses breast cancer metastasis by regulation of TGF-β-induced epithelial-mesenchymal transition. Mol. Cancer. 2018;17:70. doi: 10.1186/s12943-018-0818-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.