Abstract
Using a mini-library of 1062 lentiviral shRNAs targeting 40 nuclear hormone receptors and 70 of their co-regulators, we searched for potential therapeutic targets that would be important during in vivo tumor growth using a parallel in vitro and in vivo shRNA screening strategy in the non-small cell lung cancer (NSCLC) line NCI-H1819. We identified 21 genes essential for in vitro growth, and nine genes specifically required for tumor survival in vivo, but not in vitro: NCOR2, FOXA1, HDAC1, RXRA, RORB, RARB, MTA2, ETV4, and NR1H2. We focused on FOXA1, since it lies within the most frequently amplified genomic region in lung adenocarcinomas. We found that 14q-amplification in NSCLC cell lines was a biomarker for FOXA1 dependency for both in vivo xenograft growth and colony formation, but not mass culture growth in vitro. FOXA1 knockdown identified genes involved in electron transport among the most differentially regulated, indicating FOXA1 loss may lead to a decrease in cellular respiration. In support of this, FOXA1 amplification was correlated with increased sensitivity to the complex I inhibitor phenformin. Integrative ChipSeq analyses reveal that FOXA1 functions in this genetic context may be at least partially independent of NKX2-1. Our findings are consistent with a neomorphic function for amplified FOXA1, driving an oncogenic transcriptional program. These data provide new insight into the functional consequences of FOXA1 amplification in lung adenocarcinomas, and identify new transcriptional networks for exploration of therapeutic vulnerabilities in this patient population.
Keywords: Non-small cell lung cancer, FOXA1, NKX2-1, Pooled shRNA screens, Xenograft
Introduction
Accumulated genetic aberrations in cancer cells, including mutated driver oncogenes and tumor suppressor genes, are major causes of the malignant phenotype. There is a growing body of evidence to suggest that the acquisition of these alterations also creates novel functional dependencies (“synthetic lethalities”, “oncogene addiction”) that do not exist in normal tissues, providing tumor-specific therapeutic windows. Systematic genomic analyses can sometimes identify alterations that account for such oncogene addiction. However, functional studies are often required to identify non-mutated genes that are involved in synthetic lethal relationships.
In lung adenocarcinoma, mutations involving genes such as EGFR and EML4-ALK are known to be clinically actionable, and targeted therapies against these genes leads to dramatic clinical benefit [1], [2]. Despite identification of other putative oncogene addiction relationships via sequencing and copy number profiling, ∼70% of NSCLCs do not harbor a mutation that is currently actionable in the clinic [3], [4]. This results in the urgent need to discover acquired vulnerabilities which may be tractable from a pharmaceutical standpoint, in order to improve treatment outcomes for this disease.
Loss-of-function studies using pooled short hairpin RNA, and more recently, CRISPR-Cas9 screening is a powerful method by which new cancer targets can be identified. Large-scale screens using cohorts of human cancer cell lines have identified context-specific essential genes, including NKX2-1 in lung cancer [5], [6], [7], [8], [9], [10]. Other studies identified functional dependencies and/or drug sensitivities that would have remained masked without functional interrogation of specific pathways [11], [12], [13]. Most of these types of studies are conducted in 2D tissue culture, which has advantages of both scale and versatility, but also restricts the interrogation space to cell autonomous phenomena that are apparent under the relatively low selection pressure of nutrient- and oxygen-rich tissue culture conditions. By contrast, in vivo screens can expand this space to include pathways that are active in low-nutrient, low-oxygen environments, and/or interactions with the tumor microenvironment. Recent reports of adapting these negative selection screens to in vivo settings have demonstrated their utility in identifying new context-specific vulnerabilities [14], [15], [16], [17].
Nuclear hormone receptors (NHRs) comprise a superfamily of ligand-dependent transcription factors that respond to a variety of endocrine cues in order to regulate diverse cellular processes [18]. Their function is highly dependent on the activity of associated co-regulators, which include co-activators that cooperate with agonist-bound receptors to induce gene expression, and co-repressors which interact with antagonist-bound or unliganded receptors to repress gene expression [19], [20]. NHRs and their co-regulators are aberrantly regulated in many tumor types, the most well-known examples being estrogen receptors (ER) in estrogen-dependent breast cancers and androgen receptors (AR) in androgen-dependent prostate cancers. However, it is possible they could be dysregulated in other cancers as well. In fact, a large percentage of drugs currently approved by the FDA target nuclear hormone receptors, making these proteins attractive targets to explore for new cancer therapeutics [21].
We have previously shown that NHRs have variable expression in lung tumors, including differences between tumor and normal lung tissues, and that the NHR expression patterns in NSCLC provided information on patient survival after surgical resection [22]. This prompted us to interrogate NHR and co-regulator gene sets for their roles in lung tumorigenesis. To start this effort, we used an NHR/CoReg mini-library of shRNAs to perform a parallel in vitro and in vivo drop out screen in a genomically well characterized lung adenocarcinoma line (NCI-H1819). By using both in vitro and in vivo selection in parallel, we aimed to discover novel tumor vulnerabilities that were not previously identified by standard 2D tissue culture screening methods. We found nine genes whose shRNA dropout occurred in vivo but not in vitro, including FOXA1. We go on to show that the FOXA1 gene is required for growth in lung adenocarcinoma cells harboring amplification on chromosome 14q, while expression and cistromic analyses revealed that co-amplification of FOXA1 with NKX2-1 drives a neomorphic transcriptional program in the 14q-amplified context which supports malignant growth.
Methods
Short hairpin library targeting NHRs and co-regulators
Mini-library screens were performed using a custom shRNA library (human MISSION lentiviral shRNA library, Sigma), originally developed by The RNAi Consortium (TRC) and based in the pLKO1 vector. A lentiviral shRNA mini-library comprised of both TRC1 and TRC2 vectors targeting 40 nuclear hormone receptors, 72 co-regulators and associated transcription factor genes, and several control genes was selected (142 genes, 1062 shRNAs total). The arrayed glycerol stocks and pooled, high titer virus for the mini-library was obtained commercially (Sigma). A list of genes included in the library and number of shRNA clones per gene is included in Supplementary Table 1.
Parallel in vitro and in vivo mini-library screen
NCI-H1819 cells (2.3 × 106) were infected with the library pool at low MOI (≤0.5), in the presence of 4.0 µg/mL polybrene (3 replicates). Average library coverage was estimated to be ∼800x across replicates. Transduced cells were selected in 1.0 µg/mL puromycin for four days, and then expanded for ∼3–4 population doublings (PDs). At this point a reference sample was collected (t = 0), and the remaining cells were either seeded into T175 flasks (1.5 × 106 cells per replicate) or resuspended in PBS and injected subcutaneously into female NOD-SCID mice (5 × 106 cells per injection). Cells maintained in vitro were cultured for 20 population doublings, and reference samples were harvested at incremental population doubling points (T = 2, T = 3), until PD20 (T = 4). Tumors were allowed to grow to a minimum volume of 300 mm3 before harvesting.
Preparation of TRC shRNAs for sequencing
Genomic DNA was extracted from cell pellets and tumors via PureGene Genomic DNA Purification Kit (Gentra). Short hairpins were PCR-amplified from gDNA using a common reverse primer and indexed forward primers, each containing the appropriate Illumina adapter sequences. The analysis described in this study used primers which amplified TRC1 shRNAs. To adequately cover the library, ∼6 µg gDNA from each sample were used as input into each PCR and spread across multiple 50 µL reactions (∼0.5 µg template/reaction). PCR reaction conditions were as follows: hot-start for 95 °C for 15 min, then 94 °C for 30 s, 66 °C for 45 sec, 72 °C for 60 s, repeat for 33 cycles, and a final extension at 72 °C for 5 min. PCR products were concentrated and gel purified, quantified by Picogreen assay, and sequenced on an Illumina HiSeq 2500 using a custom primer. Sequences for all primers are listed in Supplementary Table 2.
Analysis of shRNA abundance
Sequencing reads were mapped back to the shRNA library using Bowtie 2.0 via a local installation of Galaxy [23], [24], [25]. Short hairpins which were not detected in all three replicates at T = 0 were filtered out of all analyses. Relative abundance of each shRNA was calculated as a percentage of total mapped reads. Three different analyses were then calculated: log2 fold change in shRNA abundance at T = 4 vs. T = 0 (log2 T4/T0), tumors vs. T = 0 (log2 tumors/T0), and tumors vs. T = 4 (log2 tumors/T4).
Scoring of candidate genes
To score candidate genes, log2 fold changes for each shRNA were converted to a ranked integer list and the cumulative distribution of all these values for all shRNAs against a particular gene was compared to the background distribution in a one-sided ranked Kolmogorov–Smirnov test. A ranked KS test allows for the preferential ranking of sets that are separated from the background at the tails of the distribution.
Cell culture
All cell lines were obtained from Hamon Center libraries and detailed mutation (whole exome DNAseq) and mRNA expression (RNAseq) datasets for the lung cancer lines reported in this paper are available at dbGaP (Accession phs001823.v1.p1) at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001823.v1.p1.) Lung cancer cell lines were maintained in RPMI-1650 supplemented with 5% fetal bovine serum (Sigma) (R5 media) and passaged using standard trypsinization methods. Immortalized human bronchial epithelial cell strains (HBEC)s [18] were maintained in KSFM supplemented with bovine pituitary extract and EGF (Life Technologies) and passaged using trypsin followed by an equal volume of Trypsin Neutralizing Solution (Lifeline Cell Technologies). Cell counts were performed on a Beckman Coulter Z2 Particle Count and Size Analyzer. Cell line identity was confirmed by DNA fingerprinting (PowerPlex 1.2 Kit, Promega) and mycoplasma-free status was verified by PCR (e-Myco Kit, Boca Scientific). All cell culture experiments were maintained in a humidified incubator at 37 °C, in 5% CO2.
Generation of stable cell lines
Lentivirus for individual short hairpin constructs was prepared by transfection of 293 T cells with packaging plasmids (pCMV-dR8.91 and pMD2-VSV-G) and the shRNA construct of interest using FuGene transfection reagent (Promega). Virus was harvested in R5 media over the course of 3 days, pooled, and then filtered through a 0.45 µm filter prior to use. Cells were infected at MOI ≥1 (pLKO1 constructs) or MOI ∼0.5 (TRIPZ constructs) in the presence of 4 µg/mL polybrene, followed by selection in 1.0–2.0 µg/mL puromycin for at least 3 days. Growth assays, colony formation assays, and harvesting for molecular profiling were typically performed 1–2 weeks after initial infection. TRC1 pLKO1 shRNA constructs used for validation experiments were clones TRCN0000014879 (shFOXA1-1), TRCN0000014880 (shFOXA1-2), and TRCN0000014882 (shFOXA1-3), where TRCN0000014882 was the clone used in subsequent experiments. Doxycycline-inducible short hairpin RNA plasmids (TRIPZ Human FOXA1 shRNA) were purchased from Dharmacon (#1: V2THS_16780, #2: V2THS_16814, #3: V3THS_414341). Short hairpin non-targeting control (shNTC)-TRIPZ plasmid was used as control.
Cell proliferation, clonogenicity, and drug response assays
For MTS cell proliferation assays, cells were seeded in 100 µL media at 1000–4000 cells/well in triplicate, in three 96-well plates. At the desired time points, 20 µL MTS reagent (Promega) was added to each well and the plates were incubated at 37 °C for 1–3 h. Absorbance readings were taken at Day 1 to assess for equal seeding density, and at Day 5 or Day 7 as an end point. Proliferation rate was estimated using the ratio of the test sample absorbance to control sample absorbance at the end time point. For clonogenicity assays using pLKO constructs, cells were plated at a density of 1000–2000 cells per well in 6-well dishes and allowed to incubate for 1–3 weeks before staining with crystal violet and counting colonies. For clonogenicity assays using dox-inducible TRIPZ-constructs, cells were serially diluted to appropriate concentrations and plated into 6-well plate (H1819:500 cells, H3132 and H1781:2000 cells/well) in triplicate allowed to attach for 24 h, and then 0.5 μg/mL of doxycycline was added every 48 h in DOX+ conditions. Drug response assays were performed as previously described [26].
Mouse xenograft experiments
Cells were counted and resuspended in PBS at a concentration of 1 injection per 200 µL and transported to the mouse facility on ice. 1–10 × 106 cells in 200 µL PBS were injected subcutaneously in the right flank of female NOD-SCID mice using 27-gauge needles and monitored for tumor formation for up to six months. All animal care was in accordance with institutional guidelines and approved IACUC protocols.
Quantitative RT-PCR
Total RNA was isolated from cell pellets using the RNeasy Plus Mini Kit (QIAGEN), and cDNAs were generated using the iScript cDNA Synthesis Kit (Bio-Rad). Gene-specific TaqMan probes (Applied Biosystems) were utilized for quantitative analyses of mRNA transcript levels. Probes used in this study were: FOXA1 (Hs04187555_m1), GAPDH (Hs02758991_g1), IGFBP3 (Hs00365742_g1), THBS1 (Hs00962908_m1). The GAPDH gene was used as an internal reference to normalize input cDNA. PCR reactions were performed in triplicate, run on an ABI 7300 Real-time PCR System, and analyzed with the SDS software (Applied Biosystems). The delta-delta-CT method was used to calculate relative mRNA expression levels.
Western blotting
For pLKO-shRNA validation experiments in H1819, cell pellets were lysed in 0.1% SDS Lysis Buffer (50 mM NaCl; 1 mM EDTA; 2.5 mM Tris, pH 7.4; 0.1% SDS; 1.0% NP-40) and protein concentration was quantified using Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad). Proteins were separated by SDS-PAGE and electrotransferred to nitrocellulose membranes (Millipore). Membranes were blocked for 1 h at room temperature (RT) in a 5% milk in TBST (Tris Buffered Saline with 1% Tween-20). For profiling of baseline FOXA1 expression in cell line panels, cells were grown to ∼80% confluency and harvested in RIPA buffer with protease inhibitor cocktail and Phosphatase Inhibitor cocktail (Roche). For TRIPZ-shRNA validation experiments, cells were harvested in 4× Loading Buffer (250 mM Tris, pH 6.8; 8% SDS; 20% sucrose; 0.08 mg/mL bromophenol blue). Protein concentration for these experiments was then quantified by Pierce BCA protein assay (ThermoFisher). After SDS-PAGE, proteins were electrotransferred to nitrocellulose, and membranes were blocked in 5% BSA in TBST. Membranes were incubated with primary antibody overnight at 4 °C, and detected using horseradish peroxidase-conjugated secondary antibody followed by chemiluminescence. Primary antibodies used in this study are as follows: FOXA1 (Abcam, ab23738), E-cadherin (BD Biosciences, BD610181), vimentin (BD Biosciences, BD550513), GAPDH (GeneTex, GTX627408), and vinculin (Sigma V9131).
Copy number analyses
To obtain copy number data on cell lines, genomic DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen) and quantified using UV spectroscopy. Whole-genome SNP array profiling of 75 NSCLC cell lines was performed at MD Anderson (J. Heymach) using the Illumina Human 1M-Duo DNA Analysis BeadChip (Illumina). Raw data was normalized using BeadStudio software, and Log R Ratios were then aggregated by gene using their mean values. The R package DNAcopy was used for segmentation into DNA regions of copy number alterations.
To obtain copy number data on primary lung adenocarcinomas in the MD Anderson Cancer Center “PROSPECT” NSCLC tumor panel (see below), snap-frozen tumor tissue was processed on a cryostat to generate multiple 5-micron thick shavings. Genomic DNA was extracted using the Qiagen QIAamp DNA Mini Kit (Qiagen) and quantified by Quant-iT PicoGreen dsDNA Assay kit (Life Technologies). Tumor DNA was labeled with Cy5 (red) and normal reference (Human Genomic DNA, Female, Promega, Inc., Cat #G1521) was labeled with Cy3 (green). Labeled DNA was hybridized on Agilent Human Genome CGH Microarray 244A using the Oligonucleotide Array-Based CGH for Genomic DNA Analysis protocol (Agilent) and scanned on an Agilent G2505C scanner. Normalized and background-corrected intensities were used to calculate log2 ratios (Cy5/Cy3) of test/reference for each probe. The array CGH data has been publicly deposited in the GEO database as GSE74948.
Tumor cohorts
The Profiling of Resistance patterns and Oncogenic Signaling Pathways in Evaluation of Cancers of the Thorax (PROSPECT) cohort, consists of surgically resected tumors with both neoadjuvant and adjuvant treatment, representing 183 lung adenocarcinomas and 80 squamous cell carcinomas with detailed clinical annotations on treatment history [27]. Genome-wide Illumina Human WG-6V3 microarray expression profiles from this cohort were obtained as previously described [27], and had been deposited in the GEO database as GSE42127. RNA-seq expression profiles for lung adenocarcinomas in The Cancer Genome Atlas (TCGA) cohort (n = 126 lung adenocarcinomas) were accessed from the publicly available database [3].
Microarray and RNA-Seq analyses
RNA for NSCLC cell lines analyzed by microarray was prepared using the RNeasy Mini Plus kit (QIAGEN). Microarray expression profiling on lung cancer cell lines was performed using Illumina HumanWG-6V3 Expression BeadChips (Illumina Inc., San Diego, CA). The data have been deposited as part of GEO dataset GSE32036. Bead-level data were obtained and pre-processed using the R package mbcb for background correction and probe summarization [28]. Pre-processed data were then quantile-normalized and log-transformed. Class comparison was performed using MATRIX v1.508 software. Two-sample t-tests were used to assess statistical significance. Samples derived from squamous cell carcinomas and ASCL1-positive tumors were omitted from all NSCLC cell line expression analyses. Unsupervised clustering analyses across the panel were performed using a filtered gene list, obtained first by filtering out low expressing genes (median expression value of 5 or less), and then using the top 20% variant genes. Centered Pearson correlation was used with average linkage clustering to cluster the cell line panel by the remaining genes.
RNA for cell lines analyzed by RNA-seq were prepared using either the RNeasy Mini or RNeasy Mini Plus kits (QIAGEN). Samples prepared using the RNeasy Mini kit were additionally treated with Dnase I (Life Technologies) prior to library preparation. Sequencing libraries were prepared and sequenced on a HiSeq 2500 machine according to manufacturer’s guidelines (Illumina). Reads were mapped to the human genome with Tophat [29] and differentially expressed genes were identified using Cuffdiff [30].
ChIP
Three or two 15 cm dishes of H1819 or H1781, respectively, were grown to 70–80% confluency prior to chromatin immunoprecipitation. Cross-linking was performed in 1% formaldehyde in RPMI for 10 min at room temperature, followed by reaction termination with 125 mM glycine for 5 min. Cells were harvested with a scraper in 5 mL ice-cold PBS supplemented with Complete Mini, EDTA-free Protease Inhibitor Cocktail (Roche), pelleted at 720g for 10 min at 4 °C. Cross-linked cells were resuspended in 1 mL sonication buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) and 4 °C with a Bioruptor (Diagenode) (30 s ON and 30 s OFF at highest power for 15 min). Insoluble debris was pelleted at 20,000g for 10 min, and supernatant was stored at −80 °C. GammaBind G Sepharose beads (GE Healthcare) were prepared in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl, pH 8.1, plus protease inhibitors) and blocked in 50 mg/mL BSA (Ambion) for 1 h at 4 °C. Sheared chromatin was diluted 1:10 in ChIP dilution buffer and incubated with 100 µL blocked beads for 1 h at 4 °C for pre-clearing, and supernatant was then incubated with 5 µg FOXA1 antibody (Abcam, ab23738) overnight at 4 °C. After incubation, the chromatin/antibody mixture was incubated with 80 µL blocked beads for 1 h at 4 °C. Beads were then serially washed for 10 min at 4 °C in washing buffer 1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 150 mM NaCl, pH 8.1, plus protease inhibitors), washing buffer 2 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 500 mM NaCl, pH 8.1, plus protease inhibitors), washing buffer 3 (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1), and Tris-EDTA twice (10 mM Tris-HCl, 1 mM disodium EDTA, pH 8.0 – Sigma-Aldrich). Chromatin-antibody complexes were eluted were eluted twice in 100 µL ChIP elution buffer (100 mM NaCO3, 1% SDS) for 15 min at 65 °C, adjusted to 200 mM NaCl, and incubated overnight at 65 °C. Following RNase and proteinase K treatment, ChIPed DNA was purified using a PCR purification kit (Qiagen) and eluted in 30 × 2 µL EB using a Min-Elute column. Libraries were then prepared using a TruSeq ChIP Sample Prep Kit (Illumina) and sequenced according to manufacturer’s guidelines (Illumina).
Chip-Seq read alignment and peak calling
Reads were aligned to human reference genome (hg19) using bowtie2 (v.2.1.0) [31]. A maximum of two mismatches were allowed during the read alignment. Low-quality reads and duplicate reads were removed from aligned files using “samtools view-bh-F 0 × 04 -q 10” (v1.3) [32] and “Picard MarkDuplicates.jar” (v. 1.131) commands (http://broadinstitute.github.io/picard). Chip-seq signal enriched regions were identified using the “findPeaks” module available in HOMER software (v.4.7) [33].
Motif discovery and motif enrichment analysis
De novo motif discovery was performed using the “findMotifsGenome” module available in HOMER software (v.4.7) [33]. We used 200 bp around the peak summit to identify primary binding motif and other potential DNA-binding co-factor motifs. We analyzed the enrichment of known transcription factor motifs in JASPAR by determining the frequency of known motifs in TF bound regions and in 100,000 random sets of the same sample size by using Motif Scanner [34]. The background model selected for this study is the 3rd-order Markov model designed using the human promoter sequences in eukaryotic promoter databases (EPD) [34]. The motif enrichment score was calculated as the ratio of the motif frequency in TF binding region set and the mean motif frequency in 100,000 random sets. The Z value and statistical significance (p-value) of the enrichment score was calculated based on the variance and the mean obtained from the 100,000 random simulations.
FOXA1-bound regions were identified as genomic regions with a significant read enrichment and binding peak profile in the FOXA1 reads over the input reads by using the Model-based Analysis of ChIP-Seq (MACS) software tool (v.1.4.2) [25]. Using an in-house script, all genes whose transcription start sites (TSSs) were within ±10 kb distance with respect to peak summit were called as target genes. If no gene was identified with in ±10 kb distance, the nearest gene was considered as the putative target. De novo motif discovery analysis for shared FOXA1-bound regions was performed with the Hypergeometric Optimization of Motif EnRichment (HOMER) software tool [33].
For analyses of publicly available FOXA1 and NKX2-1 ChIP-Seq data, GEO dataset GSE39998 was downloaded from the GEO database, and dataset SRP045118 was downloaded from the NCBI Sequence Read Archive. All datasets were processed as described above.
Immunohistochemistry
We determined the FOXA1 expression of 217 formalin-fixed paraffin embedded (FFPE) surgical resected primary non-small cell lung carcinoma (NSCLC) placed in tissue microarrays (TMAs). All lung cancer tissues were evaluated and underwent surgical resection at The University of Texas MD Anderson Cancer Center. Informed consent was obtained from all patients under a protocol approved by the Institutional Review Board (IRB).
The slides were stained with FOXA1 antibody (Abcam, ab23738) in a Leica Bond Max automated stainer (Leica Biosystems, Nussloch, GmbH). The tissue sections were deparaffinized and rehydrated following the Leica Bond protocol. Antigen retrieval was performed with Bond Solution #1 (Leica Biosystems, equivalent to citrate buffer pH 6.0) for 20 min; the primary antibody dilution employed was 1:500 for 20 min at room temperature. The primary antibody was detected using the Bond Polymer Refine kit (Leica Biosystems) with diaminobenzidine (DAB) as chromogen. The slides were counterstained with hematoxylin, dehydrated and coverslipped.
Immunohistochemical (IHC) staining for FOXA1 was performed on TMA samples as follows: 5 µm-thick formalin-fixed, paraffin-embedded tissue sections were deparaffined, hydrated, heated in a Biocare decloaker for 30 min pretreated with Target Retrieval Solution (Dako), and washed in Tris buffer. Peroxide blocking was performed with 3% H2O2 in methanol at room temperature for 15 min, followed by 35 min incubations in Tris-buffered saline containing 15% FBS. Slides were incubated with the primary antibody against FOXA1 (Abcam, ab23738, 1:500) at room temperature for 65 min, washed with Tris-buffered saline, followed by incubation with Envision Dual Link + Polymer-Labeled System (Dako) for 30 min. Staining was developed with chromogen substrate (Dako) for 5 min and then counterstained with hematoxylin, dehydrated, and mounted. FOXA1 expression was quantitated using a four-value intensity score (0, 1, 2, and 3) and the percent of IHC+ tumor cells (0–100%). An H-score was calculated by adding the product of the percentage cells stained at a given staining intensity (0–100) and the staining intensity (0–3), as previously described [35].
Results
Parallel in vitro and in vivo NHR and coregulator screen
In order to investigate the role of hormone receptor pathways in lung tumorigenesis, we assembled a mini-library of 1062 shRNAs against 40 hormone receptors and 72 co-regulators (125 genes targeted, including negative and positive controls), with an average coverage of ∼7.4 shRNAs/gene (gene list provided in Supplementary Table 1). A well-known limitation of xenograft experiments is the unknown tumor-initiating capacity of individual cell lines. The lung adenocarcinoma cell line H1819 was chosen for this study because it harbors a large (38%) ALDH-positive cell subpopulation, a marker we have previously shown to correlate with tumor-initiating capacity [36], [37]. Since ALDH-positive subpopulations in NSCLC cell lines are enriched for cells capable of tumor-initiation, we reasoned that cell lines with a large ALDH-positive population would be able to maintain representation of the library during xenograft growth in vivo, due to high engraftment efficiency. H1819 cells were transduced with the shRNA lentiviral library pool in triplicate biological replicates, followed by puromycin selection for cells with stably integrated shRNA (Fig. 1A). Cell counts of the replicate infected flasks were then compared with a non-infected flask seeded at the same time, confirming an infection efficiency of ∼30%, and an average library coverage of ∼863-fold. After a brief period of expansion, reference samples were collected for each replicate, and then the remaining cells in each replicate were split between either continued in vitro culture or subcutaneous injection into two NOD-SCID mice for each biological replicate. In order to maintain a high level of library representation in the tumors, 5 million cells were injected per mouse. For continued in vitro culture, cells were passaged for a total of ∼20 population doublings with reference samples collected at various time points, while tumors were allowed to reach a volume of ∼300 mm3 before harvesting. Tumors from the two mice in the third biological replicate were disparate in their growth rates, and thus were excluded from further downstream analysis, leaving tumors from four mice in the dropout analysis (Supplementary Fig. 1). Genomic DNA was harvested from the first reference time point (T = 0), PD20 time point (T = 4) and tumors. Short hairpin sequences were then amplified by PCR and quantified by deep sequencing.
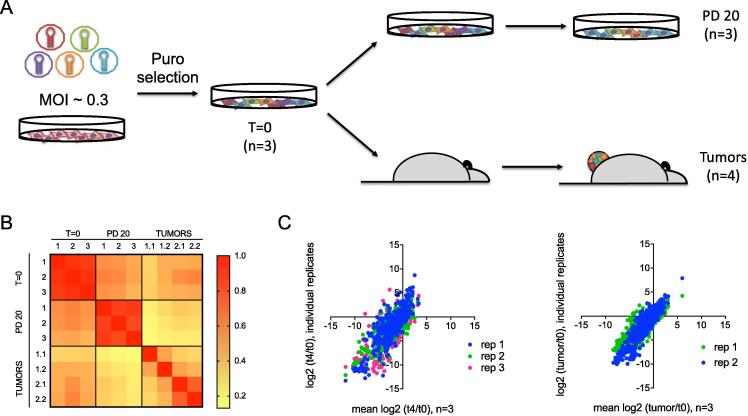
Fig. 1.
Overview of the parallel in vitro and in vivo screening strategy. (A) Experimental outline. H1819 cells were infected with a pooled shRNA library against nuclear hormone receptors and coregulators at low MOI, in three biological replicates. After puromycin selection and expansion, a reference sample was collected (t = 0), and the remaining cells were split into parallel in vitro and in vivo assays. Three biological replicates were maintained for in vitro samples. Two of these biological replicates were carried forward in vivo, using two mice for each technical replicate. Cells maintained in vitro were cultured for 20 population doublings, at which time a final reference sample was collected (t = 4; PD20). Cells in the in vivo arm were injected subcutaneously into NOD-SCID mice, and tumors were harvested after surpassing a minimum volume of 300 mm3. Short hairpins were PCR-amplified from gDNA and quantified by massively parallel sequencing. (B) Pearson correlation matrix of shRNA abundance detected in each time point sampled, demonstrating good correlation between experimental replicates. (C) Comparison of log2 fold changes observed between in vitro replicates and tumors. Plots of log2 fold changes in the in vitro screen (left) and log2 fold changes in the in vivo screen (right) confirm good correlation between experimental replicates.
The PCR primers used in this study detected >97% of TRC1 library constructs in all three T = 0 reference replicates (Supplementary Table 2), and >94% of these constructs were detected at all later time points, confirming good library representation throughout the screen. Subsequent analyses were restricted only to shRNAs detected in all three replicates at T = 0. Relative abundance of each shRNA was calculated as a percentage of total mapped reads (Supplementary Table 3). The abundance of individual shRNAs correlated very well across in vitro replicates (Fig. 1B). ShRNAs in tumors from the same replicate infection also showed a higher correlation than tumors from different replicates, however Spearman correlation coefficients for all replicates were still above 0.6. Thus, these data confirmed good reproducibility among our screening replicates.
Identification of candidate genes
To assess the ability of our screen to detect biologically meaningful relationships, two initial analyses were calculated from the relative shRNA abundance: log2 fold change in shRNA abundance in vitro at PD 20 vs. T = 0 (log2 T4/T0), and in vivo in tumors vs. T = 0 (log2 tumors/T0). Fold changes between replicates correlated well (Fig. 1C). We then assessed the performance of several shRNAs which were included as either positive or negative controls. For positive controls, we included 3 shRNAs against UBB and 4 against UBC, as these are known to be essential genes and would be expected to lead to drop out early on in the screen. We also included shRNAs against ALDH1A3, as prior work from our lab has established that knockdown of this gene should eliminate in vivo clonogenicity of ALDH-positive cell lines, such as H1819 [35,36]. For negative controls, we included three independent shRNAs which should have had no available target in a human cell line: one non-targeting scrambled control, one shRNA targeting non-mammalian genes, and one against GFP. As expected, we observed that in the in vitro screen, the abundance of shRNAs targeting UBB, UBC, and ALDH1A3 were significantly depleted compared to the negative control shRNA set by t-test (p = 0.0443, 0.014, and 0.0261, respectively; Supplementary Fig. 2). Notably, shRNAs against some of the target genes in the library can also be used as controls. For example, CCND1 is classified as acommonly essential gene in Project DRIVE (depmap.org), and also showed significant depletion in the in vitro screen. Conversely, since H1819 harbors an inactivating mutation in SMARCA4 and does not express any of the protein, shRNAs against this gene would be expected to have no effect. Consistent with this, there was no significant difference between the abundance of these shRNAs compared to negative controls in the in vitro assay (Supplementary Fig. 2D).
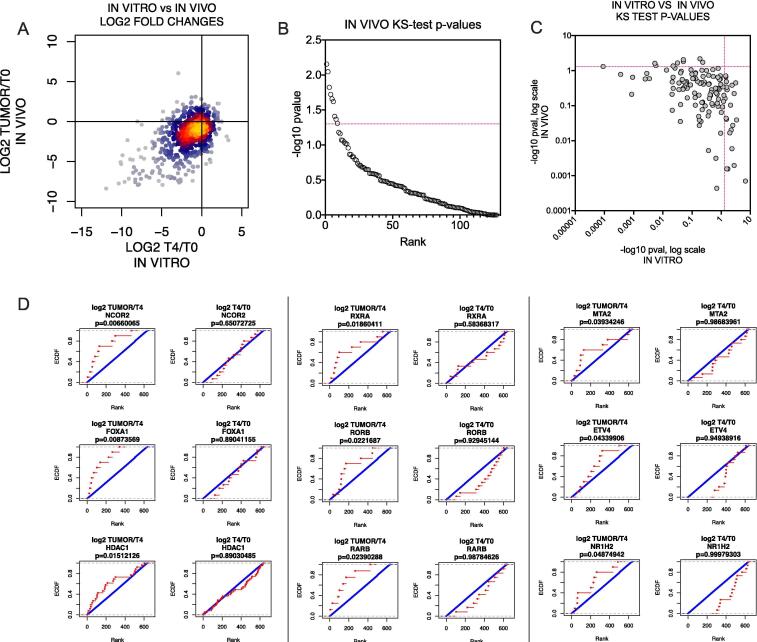
A direct comparison of log2 fold changes in vitro vs in vivo shows that most shRNAs behaved similarly in both conditions, but that a small subset of shRNAs were only depleted in vivo (Fig. 2A, lower right quadrant). In order to score hits from the two screens, we compared the distribution of log2 values for all shRNAs against each gene to all other shRNAs in each dataset. For the in vitro screen, the log2 T4/T0 values for shRNA abundance were used. For the in vivo screen, we calculated a log2 tumor/T4 value, which compares the abundance in the tumor to the last in vitro time point. Using these values, we performed a KS-test for each gene in each dataset to identify statistically significant drop-outs (p < 0.05). This approach identified 21 genes as being required for in vitro growth, and 9 genes required specifically for in vivo growth: NCOR2, FOXA1, HDAC1, RXRA, RORB, RARB, MTA2, ETV4, and NR1H2 (Fig. 2; Supplementary Fig. 3; Supplementary Table 4). Comparison of our data with Project Achilles data (CRISPR) available in DepMap revealed that many of the genes identified as required for in vitro growth were generally pan-essential across multiple tissue types, while genes identified as required for in vivo growth exhibited more selective toxicity [38], [39] (Supplementary Fig. 4). These observations confirm the utility of our combined in vitro/in vivo method in reliably identifying genes required for cancer cell survival.
Fig. 2.
Identification of genes essential for in vivo tumorigenesis. (A) Comparison of shRNA abundance in the in vitro screen vs the in vivo screen. ShRNAs that were depleted in vivo but not in vitro are in the lower right quadrant. (B) Ranked plot of −log10p-values from a one-sided KS-test for each gene, comparing the distribution of log2 tumor/T4 fold changes in shRNAs against the gene to shRNAs of all other genes. Significance threshold (p ≤ 0.05; −log10 p ≥ 1.3) is indicated by a red dotted line. (C) Comparison of −log10p-values from KS-test for each gene in the in vitro screen (log2 T4/T0) vs the in vivo screen (log2 tumor/T4). Significance thresholds are indicated by red dotted lines (p ≤ 0.05; −log10p ≥ 1.3). Genes which scored as significantly depleted in vivo but not in vitro are represented in the upper left quadrant. (D) Cumulative distribution plots for the 9 screening hits which were significantly depleted in vivo but not in vitro. Left, in vivo plots; right, in vitro plots. The distribution of log2 fold changes of all shRNAs for that gene (red) are compared to the distribution of log2 fold changes of all other shRNAs. KS-test p-values are shown above each plot. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Selection of candidate genes for follow-up
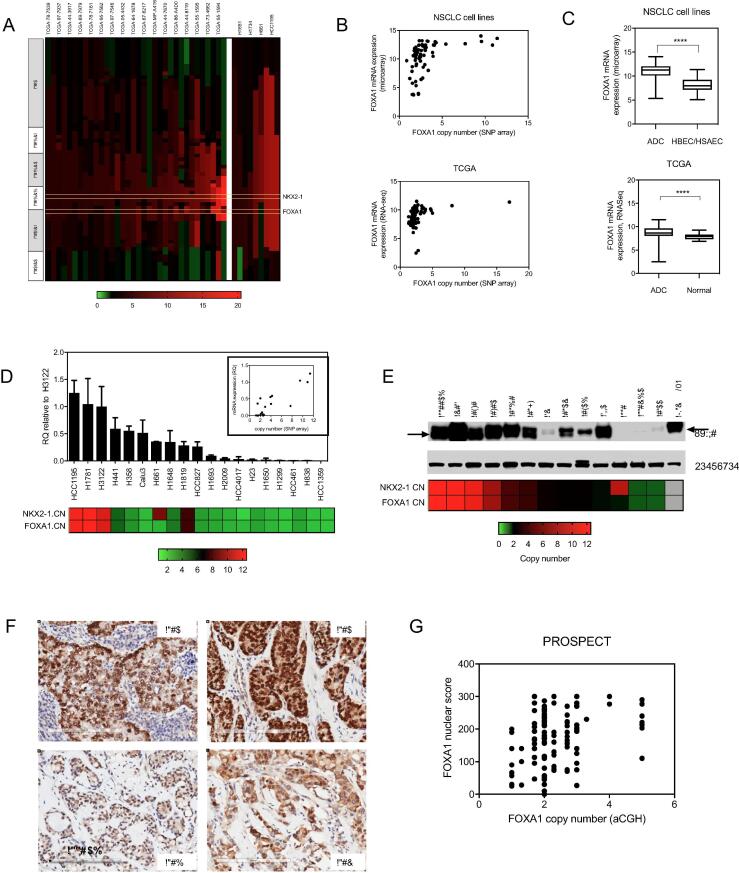
None of the in vitro or in vivo candidate genes have been identified as frequently mutated in lung cancer (3). Analysis of high resolution SNP array profiling across a panel of NSCLC cell lines (N = 63) revealed that for six of the candidate genes, there was at least one cell line with copy number variation (CNV) >6 (Supplementary Fig. 5). Of these genes, only one was a candidate in vivo essential gene: FOXA1. Expanding the SNP analysis to include primary lung adenocarcinomas from TCGA (N = 514) confirmed that amplification is also present in a subset of NSCLC tumors (Fig. 3). Notably, the FOXA1 locus is located on 14q21.1, ∼1 Mb downstream from NKX2-1 on 14q13.3. NKX2-1 is a known lineage-specific oncogene in lung cancer, and focal amplification peaking at the 14q13.3 band is one of the most frequently recurring genomic events observed in lung adenocarcinomas [3], [40], [41]. In cell lines and tumors with amplification of NKX2-1 of 4 or more copies, the amplicon extends to the FOXA1 locus in 89% (8/9 cell lines) and 79% of TCGA cases (27/34 tumor samples), respectively (Fig. 3A). Amplification of NKX2-1 is observed in ∼12% of all lung adenocarcinoma patients [33], [34], [35], [36], thus a NKX2-1:FOXA1 co-amplification frequency of 79–89% in this cohort would represent ∼9–11% of affected lung adenocarcinomas overall. This is consistent with previously reported analyses of TCGA data [42]. SNP array-derived copy number correlated well with FOXA1 mRNA expression levels in both cell lines and tumors (Fig. 3B,D). Comparison of FOXA1 mRNA expression in malignant versus non-malignant tissues also revealed that FOXA1 is more highly expressed in tumor cells compared to normal tissues (Fig. 3C). We also confirmed that FOXA1 protein expression is correlated with gene amplification in cell lines and by immunohistochemical analyses in primary lung adenocarcinomas (Fig. 3E–G). Thus, lung adenocarcinomas harboring 14q amplification exhibit high expression levels of FOXA1, and overall expression of FOXA1 is higher in tumors than in normal tissues. Taken together, these observations are consistent with a possible oncogenic role for FOXA1, thus it was prioritized for follow-up studies.
Fig. 3.
FOXA1 is amplified and overexpressed in NSCLCs. (A) FOXA1 is frequently co-amplified with NKX2-1 in NSCLC tumors. Segmented copy number profiles of the genomic region that includes NKX2-1 and FOXA1, from SNP array datasets on primary lung adenocarcinomas from TCGA (left), and on NSCLC cell lines (right). Samples with ≥4 copies of NKX2-1 are shown in rank order of NKX2-1 amplification from left to right, with corresponding chromosome band regions indicated on the left. (B) Scatter plots comparing FOXA1 copy number with FOXA1 mRNA expression level in cell lines (top), and tumors (bottom). (C) Average FOXA1 expression levels in cell lines compared to HBEC and HSAEC cell lines (top), and in primary tumors compared to normal tissues (bottom). (D) FOXA1 mRNA levels measured by Q-RTPCR in a panel of NSCLC cell lines. RQ = relative quantification, normalized to GAPDH and calculated from three technical replicates. Error bars indicate the upper and lower limits of the RQ value. Copy number for NKX2-1 and FOXA1 are indicated on the bottom. Inset: Scatter plot comparing copy number levels in NSCLC cell lines with mRNA levels. (E) FOXA1 protein expression levels in a panel of cell lines, with copy number indicated on the bottom. (F) Representative images of lung adenocarcinomas from the PROSPECT tumor array harboring 14q-ampflication and high FOXA1 expression (top), or no amplification and low expression (bottom). (G) Scatter plot comparing FOXA1 copy number (aCGH) with quantified IHC staining for FOXA1 on PROSPECT tumor array specimens.
Validation of FOXA1 as an essential gene for in vivo growth
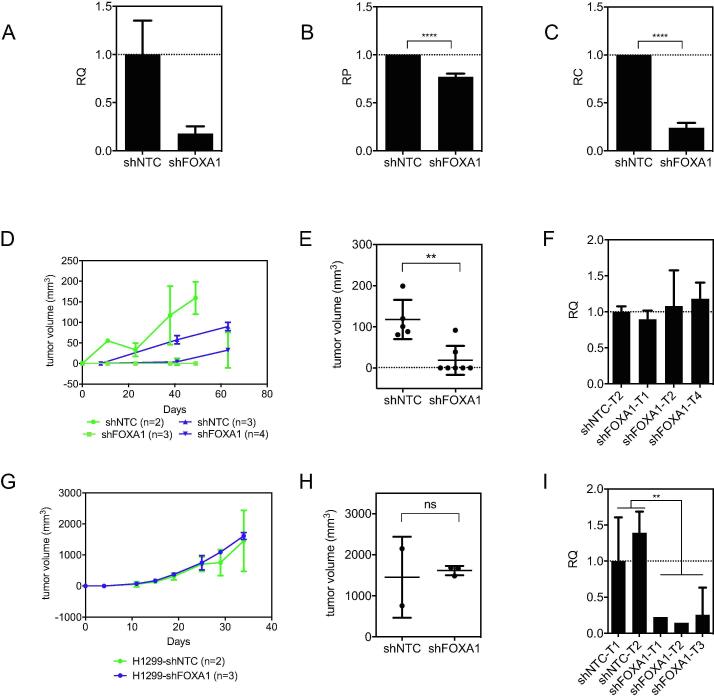
In order to validate FOXA1 as required specifically in vivo, we tested three different shRNAs from the library for their ability to knockdown FOXA1 in H1819. All three shRNAs reduced FOXA1 mRNA levels, and fold mRNA reduction correlated with fold depletion observed in the screen (Supplementary Fig. 6A,B). This direct functional link between expression of the shRNA target and phenotype suggests that the shRNA effects are "on target". Reduction in FOXA1 protein levels was confirmed using the shRNA which produced the highest levels of knockdown (Supplementary Fig. 6C), and this shRNA was used for all subsequent experiments. We also used two other dox-inducible shRNAs against FOXA1 based in the TRIPZ vector to confirm the identity of the FOXA1 band (Supplementary Fig. 6D). Knockdown of FOXA1 had a moderate effect (∼20–25% decrease) on in vitro proliferation as measured by the 5 day mass culture 96 well MTS assay (Fig. 4A,B). However, a much more striking reduction (∼4–5 fold decrease) in tumor cell clonogenicity was observed when cells were plated at low density (Fig. 4C). Of greatest biologic importance, depletion of FOXA1 essentially abolished the ability of H1819 cells to form tumors in mice (Fig. 4D,E). In fact, tumors which did grow out from injection of H1819-shFOXA1 cells arose from tumor cells that escaped FOXA1 inhibition and re-established FOXA1 expression levels comparable to those in H1819 tumors with non-targeting control (shNTC) (Fig. 4F). As a biologic control, knockdown of FOXA1 in H1299, a NSCLC line with detectable, but very low levels of FOXA1 protein expression (Fig. 3E), did not exhibit a change in vivo growth rate, despite maintenance of suppressed FOXA1 levels in vivo (Fig. 4G–I). These results show that only some NSCLCs are dependent on FOXA1 for survival, and also supports an “on target” effect for growth inhibition by the shRNA.
Fig. 4.
Knockdown of FOXA1 reduces clonogenicity and xenograft growth in H1819 but not in H1299. (A) QPCR for FOXA1 mRNA with shFOXA1. RQ = relative quantification, normalized to GAPDH and calculated from three technical replicates. Error bars indicate upper and lower limits of the RQ value. (B) Proliferation over 5 days as measured by MTS. RP = relative proliferation, calculated from at least three technical replicates. Error bars indicate standard deviations. Statistical significance was determined by unpaired t-test (p < 0.001). (C) 2D colony formation assay. RC = relative clonogenicity, calculated from three technical replicates. Error bars indicate standard deviations. Statistical significance was determined by unpaired t-test (p < 0.001). (D) Growth curves for H1819 xenografts in two separate experiments (blue and green, respectively). 10 million cells were injected subcutaneously into female NOD-SCID mice. Number of replicates are indicated in the legend. (E) Tumor weights after a minimum of 50 days growth in vivo. Error bars indicate standard deviations. Statistical significance was determined by unpaired t-test (p = 0.002). (F) QPCR results for FOXA1 in H1819 xenograft tumors which grew out (harvested at 50 days or greater). (G) Growth rate of H1299 xenograft tumors from subcutaneous injections of 5 × 106 cells with either non-targeting shRNA (shNTC) or shFOXA1. Number of replicates are indicated in the legend. Error bars indicate standard deviations. (H) Tumor weights after a minimum of 30 days growth in vivo. Error bars indicate standard deviations. Statistical significance was determined by unpaired t-test; ns = not significant. (I) QPCR for FOXA1 mRNA in H1299 xenograft tumors. Statistical significance was determined by unpaired t-test (p < 0.0074). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to further confirm an on-target effect for the pLKO-based shRNAs we used in the screen, we tested three additional shRNAs against FOXA1 from the TRIPZ dox-inducible vector system in three FOXA1-amplified cell lines: H3122, H1781, and H1819. These shRNA clones exhibited varying levels of knockdown efficiency across this panel of cell lines, however we confirmed that each shRNA was able to reduce FOXA1 protein levels in at least one cell line (Supplementary Fig. 7). Consistent with our prior observations using the pLKO vector system, induction of the shFOXA1 constructs resulted in decreased expression of the middle band on western blots, confirming our assignment of this band to FOXA1.
We then took two of the TRIPZ-shFOXA1 constructs and performed colony formation assays in three FOXA1-amplified lines (H3122, H1781, and H1819) and one NSCLC line expressing trace amounts of FOXA1 (H1299). Clonogenicity of the FOXA1-amplified lines was reduced after treatment with dox, while clonogenicity of H1299 was not affected (Supplementary Figs. 7, 8). These results confirm that the same FOXA1-dependencies are observed with multiple different shRNAs. When taken together with the observation that only some NSCLCs are dependent on FOXA1 for survival, these data support an “on target” effect for growth inhibition by the shRNAs against FOXA1.
FOXA1 is required for clonogenicity in cell lines with amplification on 14q
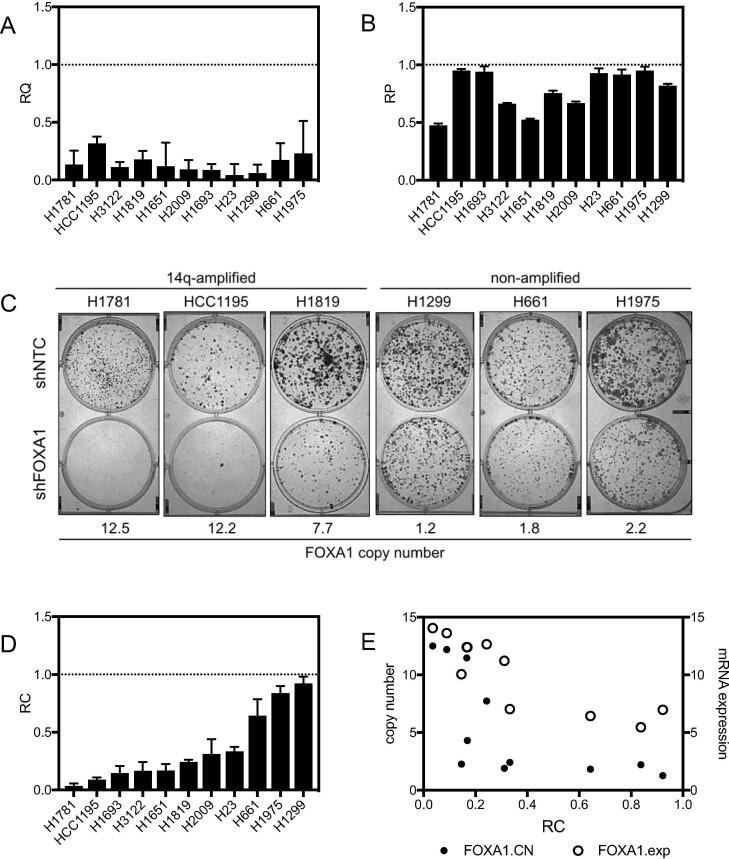
To determine whether amplification and/or expression of FOXA1 could serve as biomarkers for dependence on FOXA1, we tested a panel of NSCLC cell lines representing a range of FOXA1 copy number and expression levels for clonal growth dependency on FOXA1. FOXA1 expression was reduced using the pLKO1-based shRNA construct with the highest level of knockdown, as described above. When tested against a panel of NSCLCs, shRNA-mediated knockdown of FOXA1 in some cases inhibited bulk cell proliferation, however, this knockdown significantly reduced clonogenicity (∼5–10 fold reduction) in all FOXA1 expressing NSCLC lines (Fig. 5A–D). There was a strong correlation between the effect of FOXA1 knockdown on clonogenicity and initial FOXA1 expression and copy number (Fig. 5C–E). We observed that 3 NSCLC lines (H23, H2009, H1693) in which we did not detect FoxA1 protein expression in the population as a whole (Fig. 3E), still exhibited inhibition of colony formation by shRNA-mediated FOXA1 knockdown (Fig. 5C). It is possible that these cell lines are dependent for clonal growth on very low levels of FOXA1 protein, or that a subpopulation of highly clonogenic tumor cells, such as the cancer initiating cell subpopulation, express and are dependent on FOXA1 protein. Several NSCLC lines also provided important controls for the biologic relevance of FOXA1 knockdown. In the first, clonogenicity was only slightly affected (∼20–25%) by knockdown of FOXA1 in the NSCLC line NCI-H661, which harbors focal amplification of NKX2-1 but not of FOXA1. The second was seen in the knockdown of FOXA1 in H1299, a NSCLC line with low levels of FOXA1, which did not exhibit a change in in vivo growth rate. Taken together, these data suggest that dependency on FOXA1 follows an mRNA and protein expression level and gene dosage-dependent model of oncogene addiction, where cells with higher FOXA1 copy number and or higher FOXA1 expression exhibit dependency on the gene, while tumor cells expressing lower levels of FOXA1 do not. Overall, high FOXA1 expression or amplification of the 14q13.3–21.1 locus serve as correlative biomarkers for FOXA1 dependency in NSCLC tumors, and this subset of NSCLCs appears therapeutically vulnerable to inhibition of FOXA1.
Fig. 5.
A panel of NSCLC cell lines exhibits differential dependencies on FOXA1. (A) Knockdown of FOXA1 in a panel of cell lines was verified by Q-RTPCR in all cell lines. RQ = relative quantification, internally normalized to GAPDH, calculated from three technical replicates. (B) Proliferation rates after knockdown of FOXA1 as measured by MTS assay, calculated from at least three technical replicates. RP = relative proliferation, normalized to non-targeting control shRNA (shNTC). (C) Representative colony formation assays in a panel of NSCLC cell lines with knockdown of FOXA1. (D) Quantitation of clonogenicity after knockdown of FOXA1. RC = relative clonogenicity, normalized to shNTC, calculated from at least three technical replicates. (E) Relative clonogenicity compared to FOXA1 copy number and FOXA1 Illumina array-based mRNA expression levels.
Characterization of the FOXA1/NKX2-1 cistrome in 14q-amplified lung cancers identifies 223 genes co-regulated by FOXA1 and NKX2-1 in NSCLCs
To understand the mechanism of FOXA1 vulnerability and explore potential clinical translation applications, we next sought to investigate whether specific signaling pathways may be regulated by FOXA1 in the 14q-amplified context. First, we identified FOXA1 binding sites within the genome using chromatin immunoprecipitation with a FOXA1 antibody followed by massively parallel sequencing (ChIP-Seq) in two NSCLC lines with high FOXA1 copy number: H1819 and H1781. To gain a comprehensive overview of the FOXA1 cistrome in 14q-amplified NSCLCs, we also analyzed a previously published Chip-Seq dataset for FOXA1 in H3122, another NSCLC line with 14q-amplificiation [43]. Intersection of FOXA1-occupied loci in H1819, H1781, and H3122 identified genomic regions in common among all three cell lines, representing ∼42%, ∼13%, and ∼13% of total peaks in each cell line, respectively (Supplementary Fig. 9A; Supplementary Table 5). De novo motif analysis within these regions identified the Forkhead motif as the most significantly enriched motif (p < 1e−7, Supplementary Fig. 9B). Other motifs were also identified as significantly enriched at FOXA1 bound sites, including AP-1 and TEA binding sequences. The NKX2-2 binding motif was also found to be significantly enriched at FOXA1 bound sites, which shares a high degree of similarity with the NKX2-1 consensus motif (p < 1e−7, Supplementary Fig. 9C).
FOXA1 has previously been shown to cooperate with NKX2-1 to regulate expression of NKX2-1 transcriptional targets [44], [45], [46]. To explore the FOXA1/NKX2-1 cistrome in 14q-amplified cells, we analyzed the intersection between genomic regions found to be commonly occupied by FOXA1 in H1819, H1781, and H3122, with NKX2-1 occupied regions from previously published NKX2-1 ChIPs performed in NSCLC lines that harbor 14q amplification: H1819, H3122, H2087 [43], HCC1195, and H661 [47]. The FOXA1 ChIP-Seq datasets were collected using the same antibody, while the studies reporting the NKX2-1 datasets utilized two different antibodies. Importantly, H1819, H1781, H3122, H2087, and HCC1195 all harbor co-amplification of FOXA1 and NKX2-1, while H661 harbors only focal amplification of NKX2-1. Intersection of all NKX2-1 peaks from cell lines harboring co-amplification of FOXA1 and NKX2-1 (H1819, H3122, H2087, and HCC1195) identified 786 loci bound by NKX2-1 in this context (Supplementary Fig. 10A; Supplementary Table 6). We then intersected this set with the list of FOXA1-occupied loci and identified 252 genomic regions that are co-occupied by FOXA1 and NKX2-1 in 14q-amplified cells (Supplementary Fig. 10B). Searching for the nearest genes with a transcriptional start site (TSS) within 10 kb of these peaks identified 223 genes which are potentially co-regulated by both FOXA1 and NKX2-1 (Supplementary Table 7). Notably this list included LMO3, which was previously identified as a NKX2-1/FOXA1 target [43]. Gene set enrichment analysis of Hallmark sets in MSigDB revealed enrichment of genes regulating the NF-kB pathway, UV response, and mTOR signaling, among others (Supplementary Fig. 10).
In order to investigate whether co-amplification of FOXA1 with NKX2-1 may affect the NKX2-1 cistrome, we then compared NKX2-1 ChIP-Seq data in co-amplified cell lines H1819, H3122, H2087 [43], and HCC1195 [47], to NKX2-1 ChIP data in the cell line H661, which harbors focal amplification of NKX2-1 but not FOXA1 [47]. Intersection of NKX2-1-occupied loci from the focally amplified cell line H661 with the common FOXA1 loci identified in the four co-amplified cell lines revealed only 115 regions in common among all five cell lines, representing only ∼15% of sites that are co-occupied by FOXA1 and NKX2-1 in (Supplementary Fig. 10C). The overlap of NKX2-1 occupied loci in H661 with NKX2-1-occupied loci in other cell lines ranged from 5-15% of total peaks (Supplementary Table 8), and the vast majority of peaks identified in H661 (25,101/21,658, or 86%) were unique to that cell line. The only other cell line with a comparably sized unique space was H3122, and this may be due to the unusually large number of peaks called in that cell line. These data suggest that in NSCLCs harboring co-amplification of FOXA1 and NKX2-1, the NKX2-1 cistrome may be markedly divergent from NSCLCs in which only NKX2-1 is amplified.
Identification of a FOXA1 copy number-dependent transcriptional program
Next, we investigated whether FOXA1 copy number gain can be correlated with specific transcriptional changes in NSCLCs. Unsupervised clustering analyses of expression data in the NSCLC cell line panel reveal that 14q-amplified cell lines tend to cluster together (Supplementary Fig. 12A). A comparison of expression profiles in NSCLC lines with high FOXA1 copy number (copy number increases ≥4) to NSCLC lines with normal FOXA1 copy number (copy number increases ≥1.5 and ≤2.5) identified 3025 differentially expressed genes, of which there were 1001 genes with at least 2-fold expression differences (p < 0.05; Supplementary Tables 9 and 10). Among the upregulated differentially expressed genes, we observed significant enrichment of genes located on chr14q13 (hypergeometric test for positional gene sets in the MSigDB, p = 3.9 e−19). A significant portion of the differentially expressed genes also share directional expression with a previously reported expression signature associated with NKX2-1 amplification (89/185 genes; Supplementary Fig. 12B) [43]. This is expected, given frequent co-amplification of NKX2-1 and FOXA1 and correlation of FOXA1 amplification with expression. We also observed overlap with an epithelial-to-mesenchymal (EMT) gene signature by Byers and colleagues (17/76 genes among upregulated genes, and 3/76 genes among down regulated genes; Supplementary Tables 9 and 10) [48]. Indeed, nearly all NSCLC lines with high FOXA1 expression are classified as epithelial as assessed by e-cadherin and vimentin protein expression (Supplementary Fig. 12A). These observations are consistent with prior studies which implicate NKX2-1 and FOXA1 in positively regulating epithelial-specific markers [46], and which correlate NKX2-1 expression in lung tumors with differentiation state [45], [49]. FOXA1 and its related family member FOXA2 have also been previously implicated in epithelial-to-mesenchymal transition in various contexts [50], [51], [52], [53], [54]. However, here we found no evidence of EMT induction after knockdown of FOXA1 in NSCLCs with 14q-amplification, as assessed by changes in E-cadherin and vimentin expression (Supplementary Fig. 12C). Likewise, comparing gene expression profiles from 14q-amplified tumors in the SPORE lung adenocarcinoma panel (aCGH-based copy number ≥4) to tumors with normal copy number in this region (aCGH-based copy number ≤2.5 and ≥1.5) revealed a gene expression profile with notable overlap to the profile identified in 14q-amplified NSCLC cell lines (Supplementary Tables 11 and 12). These observations suggest that NSCLCs which harbor 14q (FOXA1 and NKX2-1) amplification may be defined by a transcriptional program that is distinct from tumors without 14q amplification or with only focal amplification of NKX2-1.
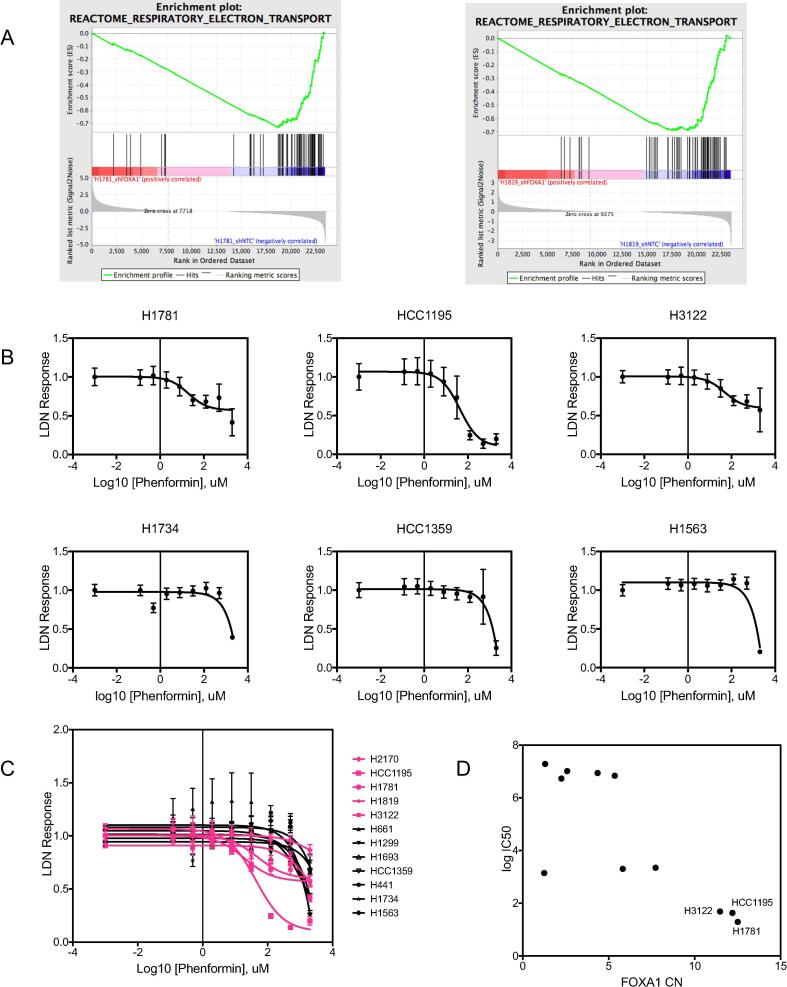
We then identified genes which are differentially expressed (DEGs) by RNAseq analyses after FOXA1 knockdown in the cell lines H1781 and H1819, (with FOXA1 copy numbers of 12.5 and 7.5, respectively), and both of which also exhibit dramatic clonal growth inhibition by shRNA-mediated FOXA1 knockdown (Supplementary Tables 13 and 14). Gene set enrichment analysis (GSEA) on differentially expressed genes in each cell line after knockdown of FOXA1 identified genes involved in electron transport as the top scoring set (NES ≤ −2) enriched in both control samples compared to the FOXA1 knockdown samples (Fig. 6A). This suggests that reducing FOXA1 expression results in a decrease in cellular respiration and led us to hypothesize that FOXA1-amplified cell lines may be preferentially sensitive to mitochondrial respiratory complex I (NADH:ubiquinone oxidoreductase) inhibition. In agreement with this, we found a positive correlation between high FOXA1 copy number level and sensitivity to phenformin (lower IC50) in a panel of NSCLC cell lines (Pearson r = −0.76; Fig. 6B-C; individual dose response curves are shown in Supplementary Fig. 13, and IC50 values are reported in Supplementary Table 15).
Fig. 6.
Knockdown of FOXA1 in 14q-amplified cell lines reveals a correlation between FOXA1 function and cellular respiration. (A) Top GSEA result for differentially expressed genes after FOXA1 knockdown in both H1819 and H1781, showing enrichment of electron transport gene expression in shNTC control samples as compared to shFOXA1 samples. (B) Representative dose response curves to phenformin treatment, in three 14q-amplified NSCLC cell lines (top panel), and three NSCLC cell lines without amplification (bottom panel). Profileration was measured by MTS, normalized to the lowest dose. Mean values are calculated from 8 technical replicates. Error bars indicate standard deviation. Data were fit to non-linear regression models using three parameters. (C) Overlay of all drug response curves in a panel of 11 NSCLC cell lines (pink = 14q amplified, black = non-amplified). (D) Correlation between FOXA1 copy number and phenformin IC50s, with select cell lines highlighted.
Identification of 29 genes that are direct transcriptional targets of FOXA1 and differentially regulated by FOXA1 knockdown
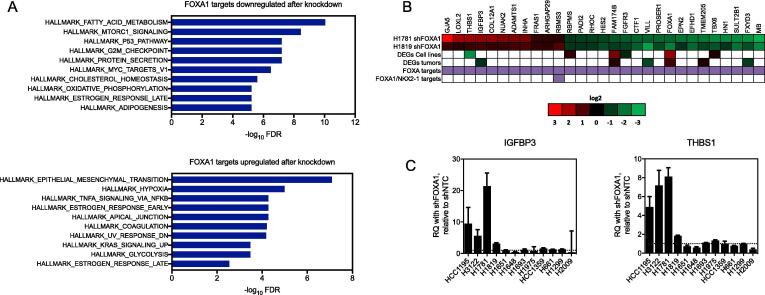
Finally, we integrated these transcriptomic and cistromic analyses together by intersecting the list of genes found near FOXA1 peaks in H1819, H1781 and H3122 with genes sharing directional differential expression after FOXA1 knockdown in H1819 and H1781, and compared the overlap to DEGs in 14q-amplified NSCLCs vs. non-amplified and the list of FOXA1/NKX2-1 co-regulated genes. This analysis identified 29 direct FOXA1 targets which share directional expression changes after FOXA1 knockdown in both cell lines, 1 of which was also an NKX2-1 target (Fig. 7B, Supplementary Table 16). Thus, these 29 genes represent direct transcriptional targets which may mediate downstream function (MSigDB Hallmark gene sets, Fig. 7A) in lung cancer cell lines with high FOXA1 expression, for the most part independently of NKX2-1. These 29 genes were submitted to the Drug Gene Interaction database (DGIdb, http://www.dgidb.org/) and 13 had drug-related hits that are the subject of future work (Supplementary Table 17).
Fig. 7.
Intersection of transcriptomic and cistromic characterizations of FOXA1 function identifies transcriptional targets in 14q-amplified NSCLCs. (A) Overlap of MSigDB Hallmark gene sets with genes that are positively regulated by FOXA1 (downregulated after FOXA1 knockdown in H1781; top panel) and negatively regulated by FOXA1 (upregulated after FOXA1 knockdown in H1781; bottom panel). (B) Intersection between differentially expressed genes after knockdown of FOXA1 and FOXA1 targets as identified by Chip-Seq in H1819 and H1781 identifies 29 direct FOXA1 targets. Heatmap values in the first two columns are log2 fold changes in genes with matched directional expression changes in H1819-shFOXA1 and H1781-shFOXA1 that were also identified as FOXA1 targets. Genes identified as FOXA1 targets or both FOXA1/NKX2-1 by Chip-seq analysis are indicated with a purple box. (C) QPCR results showing differential expression of IGFBP3 and THBS1 in response to knockdown of FOXA1. RQ = relative quantification, normalized to GAPDH, calculated from three technical replicates. Error bars indicate the upper and lower limits of the RQ value.
In order to test whether this FOXA1 transcriptional program is maintained in other 14q-amplified lines, we selected two of the 29 putative FOXA1 targets, IGFBP3 and THBS1, and tested for mRNA expression in a panel of NSCLC lines with and without FOXA1 knockdown. We found that expression of IGFBP3 and THBS1 mRNA increased in 14q-amplified NSCLC lines after FOXA1 inhibition, but not in non-amplified NSCLC lines, including NSCLCs where FOXA1 is expressed (Fig. 7B, C). Providing further specificity, we note that NSCLC lines H661 and H1693, which were not growth inhibited by FOXA1 knockdown, are among those which did not exhibit increased expression of IGFBP3 and THBS1 after FOXA1 knockdown. Again, we note that H661 harbors a focal amplification of NKX2-1 (copy number ∼9), but normal copy number levels of FOXA1 (copy number ∼1.9). Even more interesting, we point out as an “experiment of nature” that NCI-H1693 is a NSCLC line derived from the same patient and nearly isogenic with NSCLC NCI-H1819, but derived prior to neoadjuvant chemotherapy treatment, while NCI-H1819 was derived from a tumor sample after neoadjuvant treatment. H1693 does not harbor amplification on 14q while H1819 derived after chemotherapy does. Taken together, these data suggest that FOXA1 acquires a neomorphic function in lung adenocarcinomas when amplified at the gene copy number level.
Discussion
This study sought to identify acquired vulnerabilities to nuclear hormone receptors, their co-regulators, and associated transcription factors in non-small cell lung cancer through the use of parallel in vitro and in vivo RNAi screening. We reasoned that important cancer vulnerabilities might be identified in the in vivo tumor physiological context that might otherwise not be discoverable in tumor cells cultured in vitro at high density, on plastic and in nutrient-rich media. We found in a parallel in vitro and in vivo functional genomics screen that FOXA1 is a key regulator of tumorigenesis in a subset of lung adenocarcinomas. Importantly, FOXA1 may not have been identified in a loss of function screen using standard in vitro methods in lung cancer cells, as a FOXA1-dependent growth defect was most pronounced in clonogenicity and xenograft growth assays.
The FOXA1 locus is located on chromosome 14q, near NKX2-1, a lung-specific lineage-survival transcription factor that is also the most frequently amplified gene in lung adenocarcinomas [3], [40]. Recent studies have identified FOXA1 amplification as a unique event in NSCLC evolution, though the extent to which this is separate from NKX2-1 amplification is not yet clear [55], [56]. NSCLC lines harboring 14q amplification are known to require NKX2-1 for survival [41], [57], [58], however the specific role of FOXA1 in NKX2-1-amplified lung adenocarcinomas has not yet been systematically investigated. Co-amplification of NKX2-1 and FOXA1 is observed in 9–11% of lung adenocarcinomas (Fig. 3 and reference [43]), and here we report that NSCLC lines harboring this co-amplification are also dependent on FOXA1 for growth in clonal and in vivo xenograft assays. The size of this patient cohort is comparable to patients which harbor EGFR mutation, representing a significant number of patients who may stand to benefit from therapies targeting the biological effects of 14q amplification.
FOXA1/HNF3α is the founding member of the Forkhead box superfamily of transcription factors and belongs to the “pioneer” class of transcription factors. Pioneer factors possesses the unique ability to bind to compacted chromatin and induce an open, permissive state for transcription with cooperating transcription factors [59], [60], [61], [62]. In breast and prostate cancers, FOXA1 is known as a master regulator of steroid response, where it cooperates with ER and AR to globally direct hormone-dependent transcription programs, respectively [63]. FOXA1/2 also interact with ER and AR in the liver, where they are essential for gender-specific occurrences of hepatocellular carcinoma [64]. In our datasets, neither FOXA1 expression nor copy number are highly correlated with ER or AR expression (data not shown). However, several studies have linked FOXA1 function in the lung with NKX2-1, and the two transcription factors are known to be able to physically interact to either positively or negatively regulate NKX2-1 target genes [43], [45], [46].
Previous studies have suggested that NKX2-1 and FOXA1 binding sites share a high degree of overlap in 14q-amplified NSCLC cells [43]. Our integrative analyses of multiple FOXA1 and NKX2-1 ChIP-Seq datasets in 14q-amplified cell lines reveals that the overlap between these two cistromes may be smaller than previously anticipated. Thus, it is possible that a consensus transcription program driven by 14q amplification is defined by a relatively small number of genes. Our analyses across multiple NKX2-1 ChIP-Seq datasets also suggests that the NKX2-1 cistrome in tumors with focal amplification of NKX2-1 may be divergent from the NKX2-1 cistrome in tumors with co-amplification of FOXA1 with NKX2-1. One intriguing interpretation of these results is that the chromatin-opening functions of FOXA1 lead to heterogeneous and divergent effects in different patients when deregulated as a result of gene amplification. It is possible that increased copy number and expression of FOXA1 creates a more permissive transcriptional environment for NKX2-1 and other co-factors, thus inducing a neomorphic, pro-survival transcriptional program that contributes to oncogenesis.
Recruitment of FOXA1 to specific sites may also be dependent on other cis-acting transcription factors besides NKX2-1. In KRAS-driven mouse models of lung cancer, depletion of NKX2-1 results in redirection of FOXA1/2 to other de novo binding sites, and in fact drives gastric differentiation in NKX2-1-negative adenosquamous carcinomas [45], [65]. Our analyses suggest that the neomorphic function of FOXA1 in lung adenocarcinomas with 14q-amplification is at least in part independent of its co-regulatory roles with NKX2-1, as evidenced from the absence of shared binding regions near many FOXA1-regulated genes in 14q-amplified cell lines. De novo motif analyses identified enrichment of other transcription factor motifs near FOXA1 binding sites as well, which warrant further investigation as potential co-regulatory factors by which FOXA1 promotes pro-growth pathways.
The role of FOXA1 in tumorigenesis is known to be complex and highly context-dependent. In ER-positive breast cancer and androgen-dependent prostate cancer, FOXA1 clearly plays a role in driving tumor growth [66], [67]. However, in androgen-independent prostate cancers, FOXA1 appears to be able to play either a pro-growth [68] or anti-metastatic role [54]. Similarly, loss of FOXA1/2 has been shown to be required for the epithelial-to-mesenchymal transition (EMT) in pancreatic cancer cells [50], but more recent studies suggest that FOXA1 may drive metastasis in certain subtypes of pancreatic cancer [69]. In lung cancer, some cell line-based studies have linked loss of FOXA1 expression with EMT [53], [70], while others have implicated FOXA1 as a positive regulator of tumor growth or metastasis [71], [72], [73]. High FOXA1 expression is associated with good prognosis in breast cancer [74], while prognosis in prostate cancer may be context-specific [67], [68], [75], [76]. In lung cancer, one study reported that high FOXA1 expression in lung squamous cell carcinoma is correlated with poor outcome [77]. Intrigingly, high NKX2-1 expression has been shown to correlate with better overall survival in NSCLC [42], [78], however these and other studies have also reported that amplification of NKX2-1 appears to have the opposite effect, correlating with worse prognosis [58], [78], [79]. Given that FOXA1 is frequently co-amplified with NKX2-1, this suggests that in NSCLC, 14q amplification defines a phenotype that is distinctly different from tumors with mere high expression of these transcription factors.
The integrative transcriptomic and cistromic analyses reported here identified 29 genes which are positively or negatively regulated by FOXA1 only in the 14q-amplified context, 13 of which are upregulated by FOXA1 and potentially druggable targets. These findings both identify potential therapeutic targets and suggest that copy number gain at this locus contributes to a neomorphic phenotype in these cells that is distinct from contexts where NKX2-1 and/or FOXA1 are highly expressed through other mechanisms, or where they are expressed at lower levels. Divergence of the NKX2-1 cistrome in cells with co-amplification of FOXA1 with NKX2-1 compared to focal amplification of NKX2-1 suggest that increased copy number of FOXA1 may redistribute the NKX2-1 cistrome towards a distinct neomorphic pro-survival program. While focal amplification of NKX2-1 appears to be sufficient to cause overexpression and dependency on NKX2-1 [40], [41], our data suggest that co-amplification of FOXA1 with NKX2-1 may create a distinct oncogenic program. We also report a correlation between phenformin sensitivity and 14q-amplification in our panel of tested lines, which suggests that amplification of FOXA1 may play a role in regulating cellular respiration in this genetic context. The pharmacological accessibility of these and related pathways warrants further studies into this link to explore potential therapeutic intervention strategies for 14q-amplified NSCLCs.
We focused on a functional analysis of FOXA1 in this report because a clear candidate biomarker for dependency was identified in both the screening cell line and a subset of other lung cancers. However, the other eight candidate in vivo genes (NCOR2, HDAC1, RXRA, RORB, RARB, MTA2, ETV4, and NR1H2) still represent important candidates for follow-up as potential therapeutic targets. Two of these genes, HDAC1 and MTA2, are core components of the nucleosome remodeling and histone deacetylase complex (NuRD) and known to play important roles in tumorigenesis [80]. NCOR2 is also a key regulator of chromatin remodeling through recruitment of histone deacetylase complexes [81]. Additionally, several of the other candidate genes are ligand-associated NHRs and currently under active investigation as drug targets in various cancers [82]. Thus, in future studies it will be important to examine the role of these genes in tumorigenesis and their therapeutic potential in lung cancer.
Taken together, our findings implicate amplification of FOXA1 as an oncogenic change that may cooperate with other transcription factors to regulate tumorigenesis in lung cancer. Although focal amplification of the NKX2-1 locus (which includes FOXA1 >75% of the time) is the most frequently observed amplification event in lung adenocarcinomas, currently there are no therapies available that specifically target tumors with this abnormality. We found that 14q-amplified NSCLC lines are preferentially dependent on FOXA1 for in vivo xenograft growth, and also in vitro colony formation (but not mass culture growth). Thus, our findings confirm the utility of performing dropout screens in vivo and provide new insight into the functional consequences of 14q amplification in lung adenocarcinomas.
Acknowledgements
The authors gratefully acknowledge members of the Minna laboratory for technical support; Brenda Timmons for assistance with mouse experiments; Ralph Deberadinis for scientific discussion and feedback on the manuscript. This work was supported by the Lung Cancer SPORE P50 CA070907, CPRIT RP120732, Margot Johnson Foundation, and Simmons Cancer Center Support 5P30 CA142543-08, T32 CA124434.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.04.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Pao W., Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw A.T., Yeap B.Y., Solomon B.J., Riely G.J., Gainor J., Engelman J.A. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12(11):1004–1012. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. Nature Publishing Group; 2014;511(7511):543–50. [DOI] [PMC free article] [PubMed]
- 4.Campbell JD, Alexandrov A, Wala J, Pedamallu CS, Shukla SA, Guo G, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nature Genetics. Nature Publishing Group; 2016:1–12. [DOI] [PMC free article] [PubMed]
- 5.Cheung H.W., Cowley G.S., Weir B.A., Boehm J.S., Rusin S., Scott J.A. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci. 2011;108(30):12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcotte R., Brown K.R., Suarez F., Sayad A., Karamboulas K., Krzyzanowski P.M. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discovery. 2012;2(2):172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcotte R., Sayad A., Brown K.R., Sanchez-Garcia F., Reimand J., Haider M. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164(1-2):293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;30(1):140035–140112. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brough R., Frankum J.R., Sims D., Mackay A., Mendes-Pereira A.M., Bajrami I. Functional viability profiles of breast cancer. Cancer Discovery. 2011;1(3):260–273. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald ER III, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell. Elsevier Inc; 20177;170:577–586.e10. [DOI] [PubMed]
- 11.Campbell J., Ryan C.J., Brough R., Bajrami I., Pemberton H.N., Chong I.Y. Large-scale profiling of kinase dependencies in cancer cell lines. Cell Rep. 2016;14(10):2490–2501. doi: 10.1016/j.celrep.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prahallad A., Sun C., Huang S., Di Nicolantonio F., Salazar R., Zecchin D. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman G.R., Rahal R., Buxton F., Xiang K., McAllister G., Frias E. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111(8):3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meacham C.E., Ho E.E., Dubrovsky E., Gertler F.B., Hemann M.T. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat Genet. 2009;41(10):1133–1137. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Possik P.A., Müller J., Gerlach C., Kenski J.C.N., Huang X., Shahrabi A. parallel in vivo and in vitro melanoma RNAi dropout screens reveal synthetic lethality between hypoxia and DNA damage response inhibition. Cell Rep. 2014;9(4):1375–1386. doi: 10.1016/j.celrep.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 17.D'Alesio C., Punzi S., Cicalese A., Fornasari L., Furia L., Riva L. RNAi screens identify CHD4 as an essential gene in breast cancer growth. Oncotarget. 2016;7(49):80901–80915. doi: 10.18632/oncotarget.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans R.M., Mangelsdorf D.J. Nuclear receptors, RXR, the Big Bang. Cell Elsevier Inc. 2014;157(1):255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lonard DM, O'Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol Nature Publishing Group; 2012;8(10):598–604. [DOI] [PMC free article] [PubMed]
- 20.Dasgupta S., Lonard D.M., O'Malley B.W. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2014;65(1):279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. Nature Publishing Group; 2006 Dec; 5(12):993–996. [DOI] [PubMed]
- 22.Jeong Y, Xie Y, Xiao G, Behrens C, Girard L, Wistuba II, et al. Nuclear Receptor Expression Defines a Set of Prognostic Biomarkers for Lung Cancer. Pao W, editor. PLoS Med 2010;7(12):e1000378–13. [DOI] [PMC free article] [PubMed]
- 23.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P. Galaxy: a platform for interactive large-scale genome analysis. Genome Res Cold Spring Harbor Lab. 2005;15(10):1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goecks J., Nekrutenko A., Taylor J. Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol BioMed Central Ltd. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blankenberg D, Kuster Von G, Coraor N, Ananda G, Lazarus R, Mangan M, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 2010;Chapter 19:Unit19.10.1–21. [DOI] [PMC free article] [PubMed]
- 26.Gandhi J, Zhang J, Xie Y, Soh J, Shigematsu H, Zhang W, et al. Alterations in Genes of the EGFR Signaling Pathway and Their Relationship to EGFR Tyrosine Kinase Inhibitor Sensitivity in Lung Cancer Cell Lines. Lewin A, editor. PLoS ONE 2009;4(2):e4576–11. [DOI] [PMC free article] [PubMed]
- 27.Tang H., Xiao G., Behrens C., Schiller J., Allen J., Chow C.W. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19(6):1577–1586. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding L-H, Xie Y, Park S, Xiao G, Story MD. Enhanced identification and biological validation of differential gene expression via Illumina whole-genome expression arrays through the use of the model-based background correction methodology. Nucleic Acids Res Oxford University Press; 2008;36(10):e58–8. [DOI] [PMC free article] [PubMed]
- 29.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aerts S. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31(6):1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pajares M.J., Agorreta J., Larrayoz M., Vesin A., Ezponda T., Zudaire I. Expression of tumor-derived vascular endothelial growth factor and its receptors is associated with outcome in early squamous cell carcinoma of the lung. J Clin Oncol. 2012;30(10):1129–1136. doi: 10.1200/JCO.2011.37.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan J.P., Spinola M., Dodge M., Raso M.G., Behrens C., Gao B. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70(23):9937–9948. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao C., Sullivan J.P., Girard L., Augustyn A., Yenerall P., Rodriguez-Canales J. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Cancer Res. 2014;20(15):4154–4166. doi: 10.1158/1078-0432.CCR-13-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S. Defining a cancer dependency map. Cell. 2017;170(3):564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Rev Drug Discov. 2017;49(12):1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weir B.A., Woo M.S., Getz G., Perner S., Ding L., Beroukhim R. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450(7171):893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwei K.A., Kim Y.H., Girard L., Kao J., Pacyna-Gengelbach M., Salari K. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27(25):3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang X., Kadara H., Behrens C., Liu D.D., Xiao Y., Rice D. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin Cancer Res. 2011;17(8):2434–2443. doi: 10.1158/1078-0432.CCR-10-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe H., Francis J.M., Woo M.S., Etemad B., Lin W., Fries D.F. Integrated cistromic and expression analysis of amplified NKX2-1 in lung adenocarcinoma identifies LMO3 as a functional transcriptional target. Genes Dev. 2013;27(2):197–210. doi: 10.1101/gad.203208.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe H., Meyerson M. Hopping between differentiation states in lung adenocarcinoma. Cancer Cell. 2013;23(6):707–709. doi: 10.1016/j.ccr.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snyder E.L., Watanabe H., Magendantz M., Hoersch S., Chen T.A., Wang D.G. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 2013;50(2):185–199. doi: 10.1016/j.molcel.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minoo P., Hu L., Xing Y., Zhu N.L., Chen H., Li M. Physical and functional interactions between homeodomain NKX2.1 and winged helix/Forkhead FOXA1 in lung epithelial cells. MCB. 2007;27(6):2155–2165. doi: 10.1128/MCB.01133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clarke N, Biscocho J, Kwei KA, Davidson JM, Sridhar S, Gong X, et al. Integrative genomics implicates EGFR as a downstream mediator in NKX2-1 amplified non-small cell lung cancer. Chi J-TA, editor. PLoS ONE 2015;10(11):e0142061–17. [DOI] [PMC free article] [PubMed]
- 48.Byers L.A., Diao L., Wang J., Saintigny P., Girard L., Peyton M. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19(1):279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winslow M.M., Dayton T.L., Verhaak R.G.W., Kim-Kiselak C., Snyder E.L., Feldser D.M. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473(7345):101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song Y., Washington M.K., Crawford H.C. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70(5):2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Y., Shu G., Yuan X., Jing N., Song J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011;21(2):316–326. doi: 10.1038/cr.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C.M.-C., Gocheva V., Oudin M.J., Bhutkar A., Wang S.Y., Date S.R. Foxa2 and Cdx2 cooperate with Nkx2-1 to inhibit lung adenocarcinoma metastasis. Genes Dev. 2015;29(17):1850–1862. doi: 10.1101/gad.267393.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Meyer C.A., Fei T., Wang G., Zhang F., Liu X.S. A systematic approach identifies FOXA1 as a key factor in the loss of epithelial traits during the epithelial-to-mesenchymal transition in lung cancer. BMC Genomics. 2013;14(1):680. doi: 10.1186/1471-2164-14-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin H.J., Zhao J.C., Ogden I., Bergan R.C., Yu J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013;73(12):3725–3736. doi: 10.1158/0008-5472.CAN-12-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B.K., Veeriah S. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 56.Yu H.A., Suzawa K., Jordan E., Zehir A., Ni A., Kim R. Concurrent alterations in EGFR-mutant lung cancers associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res. 2018;24(13):3108–3118. doi: 10.1158/1078-0432.CCR-17-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kendall J., Liu Q., Bakleh A., Krasnitz A., Nguyen K.C.Q., Lakshmi B. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104(42):16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka H., Yanagisawa K., Shinjo K., Taguchi A., Maeno K., Tomida S. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67(13):6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 59.Clark K.L., Halay E.D., Lai E., Burley S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 60.Cirillo L.A. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17(1):244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cirillo L.A., Zaret K.S. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4(6):961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- 62.Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 63.Augello MA, Hickey TE, Knudsen KE. Review FOXA1: master of steroid receptor function in cancer. EMBO J Nature Publishing Group; 2011;30(19):3885–94. [DOI] [PMC free article] [PubMed]
- 64.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell 2012;148(1-2):72–83. [DOI] [PMC free article] [PubMed]
- 65.Camolotto S.A., Pattabiraman S., Mosbruger T.L., Jones A., Belova V.K., Orstad G. FoxA1 and FoxA2 drive gastric differentiation and suppress squamous identity in NKX2-1-negative lung cancer. eLife. 2018;26(7):47. doi: 10.7554/eLife.38579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet Nature Publishing Group 2010;43(1):27–33. [DOI] [PMC free article] [PubMed]
- 67.Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J Nature Publishing Group; 2011;30(19):3962–76. [DOI] [PMC free article] [PubMed]
- 68.Gerhardt J., Montani M., Wild P., Beer M., Huber F., Hermanns T. FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer. Am J Pathol. 2012;180(2):848–861. doi: 10.1016/j.ajpath.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 69.Roe J-S, Hwang C-I, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Elsevier Inc; 2017:1–35. [DOI] [PMC free article] [PubMed]
- 70.Bersaas A., Arnoldussen Y.J., Sjøberg M., Haugen A., Mollerup S. Epithelial-mesenchymal transition and FOXA genes during tobacco smoke carcinogen induced transformation of human bronchial epithelial cells. TIV. 2016;35(C):55–65. doi: 10.1016/j.tiv.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 71.Li J, Zhang S, Zhu L, Ma S. Role of transcription factor FOXA1 in non‑small cell lung cancer. Mol Med Report 2017:1–13. [DOI] [PubMed]
- 72.Ye J.-J., Cheng Y.-L., Deng J.-J., Tao W.-P., Wu L. LncRNA LINC00460 promotes tumor growth of human lung adenocarcinoma by targeting miR-302c-5p/FOXA1 axis. Gene Elsevier. 2019;15(685):76–84. doi: 10.1016/j.gene.2018.10.058. [DOI] [PubMed] [Google Scholar]
- 73.Du H., Chen Y., Hou X., Huang Y., Wei X., Yu X. PLOD2 regulated by transcription factor FOXA1 promotes metastasis in NSCLC. Nat Rev Drug Discov. 2017;8(10) doi: 10.1038/cddis.2017.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shou J., Lai Y., Xu J., Huang J. Prognostic value of FOXA1 in breast cancer: a systematic review and meta-analysis. Breast. 2016;27:35–43. doi: 10.1016/j.breast.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 75.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature Nature Publishing Group; 2011;474(7351):390–4. [DOI] [PMC free article] [PubMed]
- 76.Imamura Y., Sakamoto S., Endo T., Utsumi T., Fuse M., Suyama T. FOXA1 promotes tumor progression in prostate cancer via the insulin-like growth factor binding protein 3 pathway. PLoS ONE. 2012;7(8) doi: 10.1371/journal.pone.0042456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deutsch L., Wrage M., Koops S., Glatzel M., Uzunoglu F.G., Kutup A. Opposite roles of FOXA1 and NKX2-1 in lung cancer progression. Genes Chromosom Cancer. 2012;51(6):618–629. doi: 10.1002/gcc.21950. [DOI] [PubMed] [Google Scholar]
- 78.Barletta J.A., Perner S., Iafrate A.J., Yeap B.Y., Weir B.A., Johnson L.A. Clinical significance of TTF-1 protein expression and TTF-1gene amplification in lung adenocarcinoma. J Cell Mol Med. 2008;13(8b):1977–1986. doi: 10.1111/j.1582-4934.2008.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Inoue Y., Matsuura S., Kurabe N., Kahyo T., Mori H., Kawase A. Clinicopathological and survival analysis of japanese patients with resected non-small-cell lung cancer harboring NKX2-1, SETDB1, MET, HER2, SOX2, FGFR1, or PIK3CA gene amplification. J Thoracic Oncol. 2015;10(11):1590–1600. doi: 10.1097/JTO.0000000000000685. [DOI] [PubMed] [Google Scholar]
- 80.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer Nature Publishing Group; 2011:1–9. [DOI] [PMC free article] [PubMed]
- 81.Perissi V., Jepsen K., Glass C.K., Rosenfeld M.G. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11(2):109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 82.Zhao L., Zhou S., Gustafsson J.-Å. Nuclear receptors: recent drug discovery for cancer therapies. Endocr Rev. 2019;40(5):1207–1249. doi: 10.1210/er.2018-00222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.