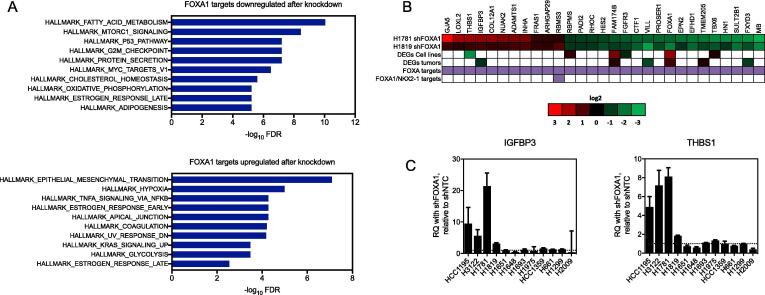
Fig. 7.
Intersection of transcriptomic and cistromic characterizations of FOXA1 function identifies transcriptional targets in 14q-amplified NSCLCs. (A) Overlap of MSigDB Hallmark gene sets with genes that are positively regulated by FOXA1 (downregulated after FOXA1 knockdown in H1781; top panel) and negatively regulated by FOXA1 (upregulated after FOXA1 knockdown in H1781; bottom panel). (B) Intersection between differentially expressed genes after knockdown of FOXA1 and FOXA1 targets as identified by Chip-Seq in H1819 and H1781 identifies 29 direct FOXA1 targets. Heatmap values in the first two columns are log2 fold changes in genes with matched directional expression changes in H1819-shFOXA1 and H1781-shFOXA1 that were also identified as FOXA1 targets. Genes identified as FOXA1 targets or both FOXA1/NKX2-1 by Chip-seq analysis are indicated with a purple box. (C) QPCR results showing differential expression of IGFBP3 and THBS1 in response to knockdown of FOXA1. RQ = relative quantification, normalized to GAPDH, calculated from three technical replicates. Error bars indicate the upper and lower limits of the RQ value.