Abstract
Leptospirosis is a re-emerging, worldwide zoonosis, and wild boar (Sus scrofa) are involved in its epidemiology as the reservoir. The aim of this study was to investigate the prevalence of Leptospira with serological, bacteriological, and molecular assays in wild boar hunted in Tuscany (Italy) during two hunting seasons. In total, 287 specimens of sera, kidneys, and liver were collected to perform microscopic agglutination tests (MATs), isolation, and RealTime PCR to detect pathogenic (lipL32 gene), intermediate (16S rRNA gene), and saprophytic (23S rRNA gene) Leptospira. Within sera, 39 (13.59%) were positive to the MAT, and Australis was the most represented serogroup (4.88%), followed by Pomona (4.18%), and Tarassovi (3.14%). Moreover, four Leptospira cultures were positive, and once isolates were identified, one was identified as L. borgpetersenii serovar Tarassovi, and three as L. interrogans serovar Bratislava. Pathogenic Leptospira DNA were detected in 32 wild boar kidneys (11.15%). The characterization through the amplification of the rrs2 gene highlighted their belonging to L. interrogans (23 kidneys), L. borgpetersenii (four), and L. kirschneri (one), while nine kidneys (3.14%) were positive for intermediate Leptospira, all belonging to L. fainei. The results of this study confirmed the importance of wild boar in the epidemiology of leptospirosis among wildlife in Central Italy.
Keywords: leptospirosis, zoonosis, infectious disease, multilocus sequence typing (MLST), wildlife, Leptospira fainei, MAT, intermediate Leptospira
1. Introduction
Wild boar (Sus scrofa) is a large ungulate mammal with worldwide distribution. It can live in several types of habitat, including urban and suburban areas [1,2]. Due to their high adaptability, wild boar populations have rapidly increased in number during recent years, in Europe, and especially in Italy [1,3]. In Italy, wild boar is largely spread in all areas, from the Alps to the southern part of the Italian peninsula, including the islands. There is a high density, particularly in specific regions, such as Tuscany [3,4,5]. The abundant presence of wild boar in the Tuscany region, as well in Central Italy, is suggested by the very high number of animals hunted in this area; every year the hunting of about 42,000 specimens is registered [1,3,4,5]. The massive presence of wild boar in particular areas, other than representing an important source of damage for agriculture [6], can be a severe risk to human and animal health, due to the identification of wild boar as reservoir for many etiological agents; among them typical zoonoses, such as Leptospira [7,8,9].
Leptospirosis is a re-emerging zoonotic disease with worldwide spread. It is caused by Leptospira spp., a Gram-negative spirochetal bacterium [10,11,12]. The genus Leptospira is divided into more than 260 antigenically-different serovars, classified as pathogenic, intermediate, and saprophytic, with different levels of pathogenicity for animals and humans [13,14]. While pathogenic Leptospira cause mild or severe infection, intermediate Leptospira could possibly be pathogenic, causing mild infection, while saprophytic Leptospira are present in the environment and are non-pathogenic [13,14]. Intermediate and saprophytic Leptospira could be important due to the strictly-contact and recombination events with pathogenic serovars [15,16,17]. Leptospirosis occurs in tropical, subtropical, and temperate zones, where it is maintained by a large variety of both wild and domestic mammals which can play the role of Leptospira maintenance host [18,19,20,21]. The reservoir organisms generally do not develop symptoms, except after a long time [11,12].
Leptospira renal-carrying/-colonization/-localization in asymptomatic animals contributes to the maintenance of infection in a particular environment by constantly shedding bacteria through their urine. Accidental contact with Leptospira-infected animal urine causes incidental infection, and produces clinical diseases in so-called “incidental hosts” [11,21].
Swine, including wild boar and pig, are recognized as maintenance hosts for Pomona, Tarassovi, and Bratislava serovars [21], but can be infected by other Leptospira serovars, in relation to both geographic area where the population lives and their behavior [22,23,24,25,26]. The epidemiology of leptospirosis may change over time in domestic and wild animals, and some serovars seems to be prevalent and emerging [26,27]. Moreover, intermediate Leptospira DNA has been detected in the kidneys of wild boar hunted in Liguria region (Italy), suggesting a possible infection [7].
Tuscany, as well as all of Central Italy, is a geographic area that promotes the presence and the persistence of Leptospira in the ecosystem. The features of Leptospira-spreading are the presence of several wild animals involved as reservoir, domestic animals bred in extensive farms in contact with wildlife, high presence of hunting activity, and abundance of wetlands, such as marshes, ponds, and irrigation canals [9,26,28,29,30,31,32,33].
The aim of this investigation was to detect and characterize pathogenic, intermediate, and saprophytic Leptospira in wild boar hunted in Tuscany region during two hunting seasons (2018/ 2019 and 2019/2020), in order to delineate the risk for the transmission and spreading of leptospirosis to domestic animals and humans.
2. Results
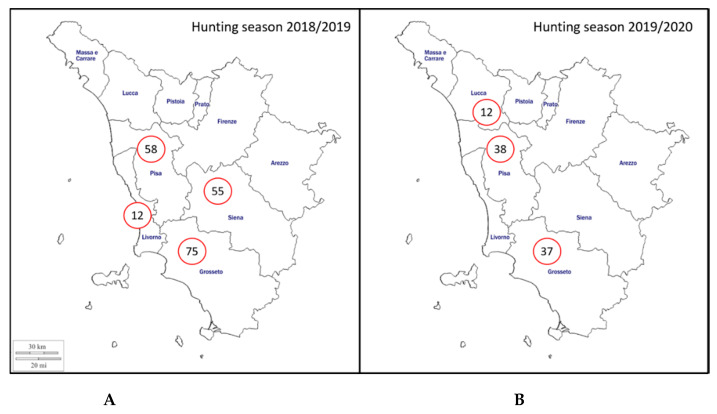
Serum, kidney, and liver samples were collected from a total of 287 hunted wild boar. Two hundred wild boar were sampled during 2018/2019 hunting season— 75 from Grosseto province, 58 from Pisa province, 55 from Siena province, and 12 from Livorno province (Figure 1). In addition, 87 specimens were sampled during 2019/2020 hunting seasons with 38, 37, and 12 from Pisa, Grosseto, and Lucca provinces, respectively (Figure 1).
Figure 1.
Geographical distribution of the sampling area included in the study (Tuscany region, Italy). The number of sampled hunted wild boar per province is indicated in relation to hunting seasons. (A) Hunting season 2018/2019; (B) Hunting season 2019/2020.
Results on distribution of positive sera and kidney for pathogenic Leptospira in relation to hunting season, province, sex, and age class are reported in Table 1.
Table 1.
Distribution of positive sera and kidney for pathogenic Leptospira in relation to hunting season, province, sex, and age class.
| Hunting Season | Province | Sex | Age Class | Examined Wild Boar | MAT-Positive Sera (%) | PCR-Positive Kidneys (%) |
|---|---|---|---|---|---|---|
| 2018/2019 | Pisa | Male | Adult | 9 | 2 (22.2) | 1 (11.1) |
| (n = 200) | (n = 58) | (n = 30) | Subadult | 10 | 2 (20.0) | 4 (40.0) |
| Young | 11 | 3 (27.3) | 0 | |||
| Female | Adult | 14 | 2 (14.3) | 2 (14.3) | ||
| (n = 28) | Subadult | 5 | 1 (20.0) | 2 (40.0) | ||
| Young | 9 | 1 (11.1) | 2 (22.2) | |||
| Grosseto | Male | Adult | 10 | 2 (20.0) | 2 (20.0) | |
| (n = 75) | (n = 29) | Subadult | 5 | 1 (20.0) | 0 | |
| Young | 14 | 1 (7.1) | 3 (21.4) | |||
| Female | Adult | 22 | 2 (9.09) | 1 (4.6) | ||
| (n = 46) | Subadult | 5 | 0 | 0 | ||
| Young | 19 | 2 (10.5) | 4 (21.1) | |||
| Siena | Male | Adult | 10 | 2 (20.0) | 0 | |
| (n = 55) | (n = 22) | Subadult | 4 | 1 (25.0) | 0 | |
| Young | 8 | 0 | 1 (12.5) | |||
| Female | Adult | 21 | 5 (23.8) | 3 (14.3) | ||
| (n = 33) | Subadult | 2 | 0 | 0 | ||
| Young | 10 | 1 (10.0) | 3 (30.0) | |||
| Livorno | Male | Adult | 2 | 0 | 0 | |
| (n = 12) | (n = 4) | Subadult | 0 | 0 | 0 | |
| Young | 2 | 1 (50.0) | 1 (50.0) | |||
| Female | Adult | 4 | 1 (25.0) | 1 (25.0) | ||
| (n=8) | Subadult | 0 | 0 | 0 | ||
| Young | 4 | 2 (50.0) | 1 (25.0) | |||
| 2019/2020 | Pisa | Male | Adult | 6 | 0 | 0 |
| (n = 87) | (n = 38) | (n = 13) | Subadult | 4 | 0 | 0 |
| Young | 3 | 0 | 0 | |||
| Female | Adult | 21 | 2 (9.52) | 0 | ||
| (n = 25) | Subadult | 1 | 1 (100) | 0 | ||
| Young | 3 | 0 | 1 (33.3) | |||
| Grosseto | Male | Adult | 11 | 1 (9.09) | 0 | |
| (n = 37) | (n = 16) | Subadult | 1 | 0 | 0 | |
| Young | 4 | 0 | 0 | |||
| Female | Adult | 10 | 1 (10.0) | 0 | ||
| (n = 21) | Subadult | 5 | 1 (20.0) | 0 | ||
| Young | 6 | 1 (16.7) | 0 | |||
| Lucca | Male | Adult | 1 | 0 | 0 | |
| (n=12) | (n = 4) | Subadult | 0 | 0 | 0 | |
| Young | 3 | 0 | 0 | |||
| Female | Adult | 4 | 0 | 0 | ||
| (n = 8) | Subadult | 0 | 0 | 0 | ||
| Young | 4 | 0 | 0 |
2.1. Microscopic Agglutination Test (MAT)
Overall, 39 out of 287 sera (13.59%) were positive in the MAT (Table 2). Considering each hunting season, seropositivity of 16% (32 out of 200 sera) was recorded during 2018/2019, while 8.05% (7 out of 87) was recorded during 2019/2020. Considering wild boar sex, 16 out of 118 male sera (13.55%) and 23 out of 169 (13.61%) were positive in the MAT. Moreover, in relation to age class, 20 out of 142 adult specimens’ sera (14.08%), 7 out of 42 subadult specimens’ sera (16.67%) and 12 out of 100 young specimens’ sera (12.00%) were positive in serological analysis.
Table 2.
Numbers of positive serological reactions detected for wild boar sera in relation to different Leptospira serogroups at low (1:100) and high titers (1:12,800).
| Leptospira Serogroup | Titer | Total (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 100 | 200 | 400 | 800 | 1600 | 3200 | 6400 | 12800 | ||
| Icterohaemorrhagiae | 1 | 1 (2.56%) | |||||||
| Canicola | 1 | 1 | 2 (5.13%) | ||||||
| Pomona | 8 | 1 | 3 | 12 (30.8%) | |||||
| Grippotyphosa | |||||||||
| Tarassovi | 4 | 1 | 1 | 1 | 1 | 1 | 9 (23.1%) | ||
| Australis | 5 | 5 | 2 | 1 | 1 | 14 (35.9%) | |||
| Sejroe | |||||||||
| Ballum | 1 | 1 (2.56%) | |||||||
| Total | 17 | 3 | 10 | 2 | 2 | 2 | 2 | 1 | 39 (100%) |
Australis resulted the most-recorded serogroup (4.88%), followed by Pomona (4.18%), and Tarassovi (3.14%). Other antibody titers were reported for serogroup Canicola (0.70%) and for serogroups Icterohaemorrhagiae and Ballum (0.45%). The highest titer detected was 1:12,800 for serogroup Tarassovi, followed by titer of 1:6400, which was reported for serogroups Tarassovi and Ballum.
Results on distribution of positive sera detected by MAT in relation to hunting season, province, sex, and age class are reported in Table 1. No statistical differences (p > 0.05) were reported for the serological positivity considering hunting seasons, provinces, and wild boar sex and age class. Moreover, comparing all parameters, no statistical differences (p > 0.05) were showed in Pisa and Grosseto during the two different hunting seasons.
2.2. Molecular Analysis
Concerning pathogenic Leptospira, DNA was detected in 11.15% (32 out of 297) of wild boar kidneys. Table 1 shows PCR-positive kidneys in relation to hunting seasons, province and wild boar sex and age class. During the 2018/2019 and 2019/2020 hunting seasons, 15.5% (31 out of 200) and 1.15% (1 out of 87) of PCR positivity was reported among kidneys samples, respectively. Considering wild boar sex, 12 out of 118 male sera (10.16%) and 20 out of 169 (11.83%) scored positive. Moreover, in relation to age class, 10 out of 142 adult specimens’ kidneys (7.04%), 6 out of 42 subadult specimens’ kidneys (14.28%), and 16 out of 100 young specimens’ kidneys (16.00%) gave positive results in serological analysis.
No statistical differences (p > 0.05) were highlighted comparing province, wild boar sex, or age class. Conversely, the incidence of pathogenic Leptospira-positive kidneys was statically higher (p ≤ 0.01) during 2018/2019 hunting season compared to the 2019/2020 ones.
The detection of pathogenic Leptospira DNA was higher (p ≤ 0.01) during 2018/2019 hunting season in both Pisa and Grosseto provinces compared to the second hunting season. On the contrary, there were no statistical differences (p > 0.05) in the Pisa and Grosseto provinces during the two different hunting seasons, comparing sex and age class of wild boar.
The 3.14% (9 out of 287) of kidneys were positive for intermediate Leptospira. The positivity in relation to hunting seasons, province, wild boar sex, and age class are showed in Table 1. All the intermediate Leptospira-positive kidneys (4.5%; 9 out of 200) were collected in 2018/2019, highlighting a statistical difference (p ≤ 0.01) in relation to 2019/2020 hunting season. Also, the results showed a statistically-higher infection rate (p ≤ 0.01) in male compared to female, and in Pisa province compared to other provinces. No statistical difference (p > 0.05) were noted among age classes.
No saprophytic Leptospira DNA was detected in kidney samples. No positive reactions were recorded in wild boar livers across all specimens during the two year of investigation.
2.3. Leptospira spp. Isolation, Characterization and Genotyping
Four Leptospira cultures were positive after 30 days of incubation. The results, reported in Table 3, show that three isolates were obtained from subadult males hunted in Pisa province, while the other one was from an adult female hunted in Livorno. Through multilocus sequence typing (MLST) analysis, one isolate was identified as Leptospira borgpetersenii serogroup Tarassovi serovar Tarassovi (Sequence Type 153), while the other three were identified as L. interrogans serogroup Bratislava serovar Bratislava (ST 24), as reported in Table 3. Moreover, the amplification of the rrs2 gene from kidney tissue highlights that the species belonged to L. borgpetersenii and L. interrogans, respectively.
Table 3.
Characterization of wild boar Leptospira isolates tested with anti-sera and multilocus sequence typing (MLST).
| Sample | Wild Boar | Isolates Characterization | |||
|---|---|---|---|---|---|
| Sex | Age Class | Province | Anti-Serum MAT Serogroup | MLST (Sequence Type) | |
| Kidney 5 | Male | Subadult | Pisa | Tarassovi | Tarassovi (ST 153) |
| Kidney 14 | Male | Subadult | Pisa | Australis | Bratislava (ST 24) |
| Kidney 15 | Male | Subadult | Pisa | Australis | Bratislava (ST 24) |
| Kidney 22 | Female | Adult | Livorno | Australis | Bratislava (ST 24) |
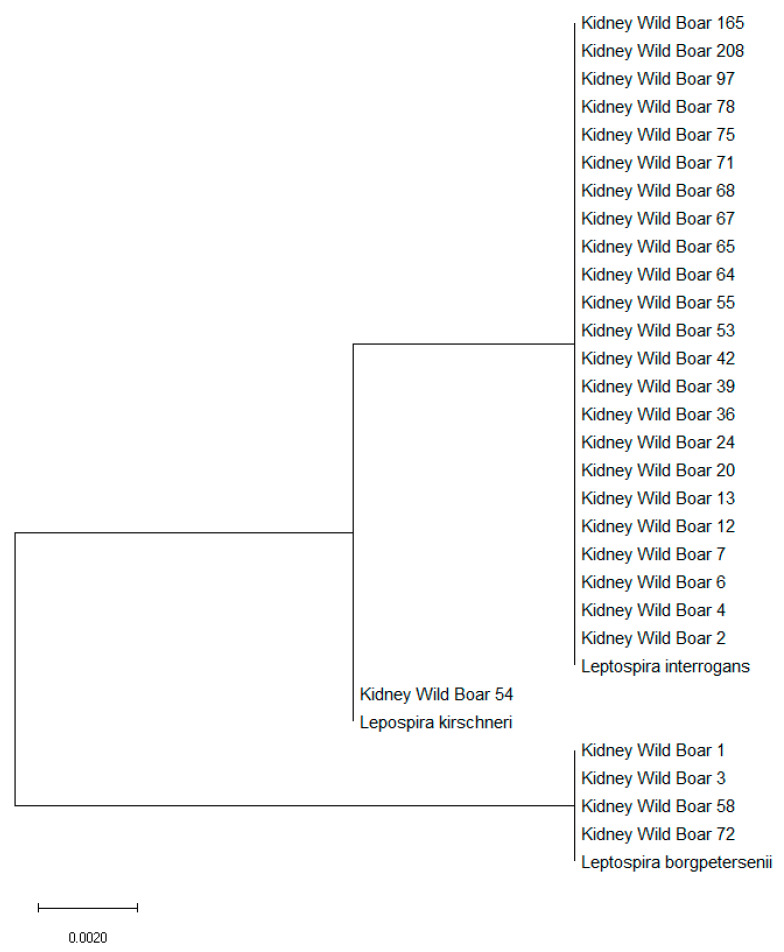
With regard to characterization of PCR-positive samples, amplification of the rr2 gene highlighted that pathogenic Leptospira belonged to L. interrogans (23 kidneys), L. borgpetersenii (four) and L. kirschneri (one) (Table 4). Moreover, phylogenetic analysis identified the close relationship to their respective Leptospira species. (Figure 2).
Table 4.
Characterization of Leptospira species in wild boar pathogenic Leptospira-positive PCR-amplifying rrs2 gene.
| Sample | Wild Boar | Isolate Characterization | ||
|---|---|---|---|---|
| Sex | Age Class | Province | Leptospira Species | |
| Kidney 1 | Female | Young | Pisa | L. borgpetersenii |
| Kidney 2 | Female | Subadult | Pisa | L. interrogans |
| Kidney 3 | Male | Adult | Pisa | L. borgpetersenii |
| Kidney 4 | Female | Young | Pisa | L. interrogans |
| Kidney 6 | Male | Young | Siena | L. interrogans |
| Kidney 7 | Female | Young | Siena | L. interrogans |
| Kidney 12 | Female | Young | Siena | L. interrogans |
| Kidney13 | Female | Adult | Siena | L. interrogans |
| Kidney 20 | Male | Young | Grosseto | L. interrogans |
| Kidney 24 | Female | Young | Livorno | L. interrogans |
| Kidney 36 | Female | Adult | Grosseto | L. interrogans |
| Kidney39 | Female | Young | Grosseto | L. interrogans |
| Kidney 42 | Female | Young | Siena | L. interrogans |
| Kidney 53 | Female | Young | Grosseto | L. interrogans |
| Kidney 54 | Male | Young | Grosseto | L. kirschneri |
| Kidney 55 | Male | Young | Grosseto | L. interrogans |
| Kidney 58 | Female | Adult | Pisa | L. borgpetersenii |
| Kidney 64 | Female | Adult | Siena | L. interrogans |
| Kidney 65 | Female | Adult | Siena | L. interrogans |
| Kidney 67 | Female | Adult | Pisa | L. interrogans |
| Kidney 68 | Female | Subadult | Pisa | L. interrogans |
| Kidney 71 | Male | Adult | Grosseto | L. interrogans |
| Kidney 72 | Female | Young | Grosseto | L. borgpetersenii |
| Kidney 75 | Male | Subadult | Pisa | L. interrogans |
| Kidney 78 | Male | Young | Livorno | L. interrogans |
| Kidney 97 | Male | Adult | Grosseto | L. interrogans |
| Kidney 165 | Female | Young | Grosseto | L. interrogans |
| Kidney 208 | Female | Young | Pisa | L. interrogans |
Figure 2.
Molecular phylogenetic analysis for the rrs2 gene of Leptospira interrogans, Leptospira borgpetersenii, and Leptospira kirschneri by the maximum likelihood method, based on the Tamura–Nei model. The branch lengths of the tree measured the number of substitutions per site. The analysis involved 31 nucleotide sequences. There was a total of 452 positions in the final dataset.
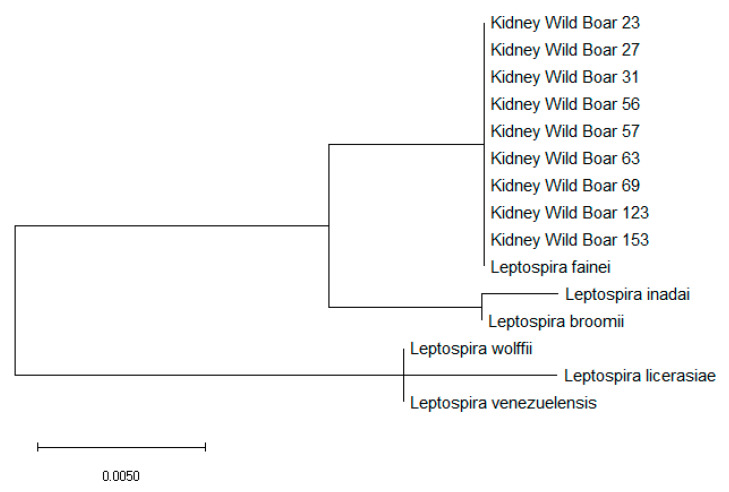
Moreover, the amplification of intermediate Leptospira 16s rRNA DNA of PCR-positive specimens showed L. fainei in all nine kidneys (Table 5). Furthermore, the phylogenetic analysis identified the close relationship to L. fainei specie. (Figure 3).
Table 5.
Characterization of Leptospira species in wild boar intermediate Leptospira-positive PCR-amplifying 16s rRNA gene.
| Sample | Wild Boar | Isolate Characterization | ||
|---|---|---|---|---|
| Sex | Age Class | Province | Leptospira Species | |
| Kidney 23 | Male | Young | Livorno | L. fainei |
| Kidney 27 | Male | Adult | Pisa | L. fainei |
| Kidney 31 | Female | Adult | Pisa | L. fainei |
| Kidney 56 | Male | Young | Grosseto | L. fainei |
| Kidney 57 | Male | Adult | Pisa | L. fainei |
| Kidney 63 | Male | Adult | Siena | L. fainei |
| Kidney 69 | Female | Subadult | Pisa | L. fainei |
| Kidney123 | Male | Adult | Livorno | L. fainei |
| Kidney 153 | Male | Adult | Siena | L. fainei |
Figure 3.
Molecular phylogenetic analysis for 16s rRNA gene of Leptospira fainei, Leptospira inadai, Leptospira broomii, Leptospira wolffii, Leptospira licerasiae, and Leptospira venezuelensis by the maximum likelihood method based on the Tamura–Nei model. The branch lengths of the tree measured the number of substitutions per site. The analysis involved 22 nucleotide sequences. There was a total of 438 positions in the final dataset.
3. Discussion
Leptospirosis is a re-emerging worldwide public health risk, but is underestimated and characterized by a downward trend [34]. Climatic changes, rainfall, modifications of ecological niches, and new potential maintenance hosts all represent important features involved in Leptospira epidemiology.
Wild boar, among wildlife, is an important Leptospira reservoir and, for several areas, represents an appropriate indicator for this zoonotic infectious disease.
In this investigation we reported the results of serological analysis, isolation and molecular investigations performed on 287 hunted wild boar during two hunting seasons (2018/2019 and 2019/2020).
With regard to serological assay, the prevalence of Leptospira infection, recorded in both hunting seasons, was very similar to other studies carried out on wild boar in Tuscany [9,22,26,31,33]. Moreover, the seroprevalence reported in this investigation was very close to other data obtained in different Italian regions [27,35,36,37,38]. Unfortunately, serological data about leptospirosis in wildlife, especially regarding wild boar, are available just in some regions. It also seems that Leptospira serovars/serogroups have a different geographical distribution, suggesting a distinct circulation/epidemiology in other environments/ecosystems. Examining each region, Australis, Pomona, and Tarassovi, are the most-detected serogroups in the Tuscany region [22,26,31,33], In the Lombardy and Emilia Romagna regions it is Bratislava [37,38,39], in the Campania region it is Tarassovi [35], whereas in the Sardinia region it is Pomona and Grippotyphosa [36].
The distribution of Leptospira serovars in wild boar in Europe is also not homogeneous; high levels of Pomona infection was recorded in Germany, Croatia, Poland, and Spain [23,40,41,42]. Bratislava was the most-detected serovar in Sweden [25], Tarassovi in Portugal and Slovenia [24,43], Grippotyphosa in Czech Republic [44], and Hardjo in Poland [23].
Little information is available on Leptospira isolation in wild boar, especially in Italy [38]. The obtained Leptospira isolates, identified by MLST, confirm the circulation of Tarassovi and Bratislava serogroups within wild boar in Tuscany. Bratislava isolation is commonly performed in wildlife due to the high spectrum of maintenance hosts [45,46,47], while Tarassovi is rarely isolated and detected through serology. Indeed, Tarassovi is strictly a swine-specific serovar; its isolation, reported in this investigation, seems to confirm the hypothesis that wild boar could serve as reservoir of Tarassovi [26,43,48]. Only two of them (Bratislava, isolated from subadult from Pisa province; Table 3) reported correlated serological positivity for serogroup Australis at titer 1:100, while the other two gave negative results in the MAT. No correlation was found between the MAT and PCR-positive results. The seronegativity of Leptospira-positive kidneys has been previously reported for other animal species [49,50,51,52], including swine [47], suggesting an early or chronic infection.
Conversely to serological results, very few studies were performed on pathogenic Leptospira DNA in wild boar kidneys. In spite of this, the prevalence of pathogenic Leptospira infection reported during the years of this investigation was very close to the results obtained in Northern Italy (11.02%) [38] and in the Liguria region (12.13%) [7]. Moreover, prevalences of 10.30% and 15.3% were found in two different investigations performed in Japan [53,54], while 3.40% was reported in the USA [55]. Based on phylogenetic analysis, pathogenic Leptospira DNA in wild boar kidney belong to L. interrogans, L. borgpetersenii, and L. kirschneri. With respect to the serovars in Italy that are more often detected through isolation or serology, [27,38,56,57,58,59] and the other serovars that are rarely seropositive [28,60], it might be hypothesized that L. kirschneri species found in wild boar kidneys could be related to serogroup Grippotyphosa, while L. borgpetersenii species could be related to the serogroups Tarassovi or Ballum. On the other hand, it is very difficult to infer the serogroup related to L. interrogans positivity, due to the inclusion of Icterohaemorrhagiae, Canicola, Pomona, Australis, and Sejroe serogroups in this species. Probably, in relation to serological and bacteriological results obtained in this study, the identified L. interrogans could belong to serogroup Australis.
The data reported in this investigation suggest that the liver does not seem be a Leptospira target organ in wild boar. Furthermore, it could exclude an early stage of infection (leptospiremia) and confirm that positive animals are only chronic renal carriers, as also suggested by isolation from kidneys.
If very little information is available on pathogenic Leptospira DNA in wild boar, there is even less data on intermediate Leptospira. To the best of these authors’ knowledge, it was only in the Liguria region of Italy that 0.49% of wild boar kidneys were positive for intermediate Leptospira DNA in the same year of this investigation [7]. As Liguria and Tuscany are two adjoining Italian regions, a large wild boar movement could be a feature of these regions [61,62,63]. Even though the species of intermediate Leptospira from Liguria were not identified, those found in this investigation belong to L. fainei species. L. fainei was isolated for the first time from fig and was detected in human sera in Australia [64,65] and a human infection with febrile status was reported in France (from a Portuguese citizen) and in two patients in Denmark [66,67]. Considering wild boar behavior and its ability to live in anthropomorphic environment, a transmission between human and wildlife could be possible. As these are the first determination in European wildlife, more studies are needed to understand the epidemiology of this intermediate Leptospira that could causes severe infection in humans [65,66,67].
The statistical difference presented during the hunting seasons between pathogenic and intermediate Leptospira incidence in wild boar could be related to the temperature and the amount of rainfall recorded in Tuscany during these periods. As reported in literature, rainfall and temperature influence the incidence of leptospirosis in humans and animals [12,68,69,70,71,72,73,74,75]. Indeed, from 2018 to 2019, the temperatures and the rainfall were both higher than those from 2019 to 2020 [76,77,78,79,80,81], suggesting that these atmospheric phenomena could be involved in these seasonality incidence differences.
4. Materials and Methods
4.1. Sample Collection
During two hunting seasons (the first from November 2018 to January 2019 and the second from November 2019 to January 2020) hunted wild boar blood, kidney, and liver were sampled. Blood samples were collected by ocular puncture [82]. The boar’s age class was determined after assessing the degree of tooth eruption and the wear and tear of teeth of the lower jaw, considering three age classes: young (under 12 months old), sub-adult (between 12 and 24 months), and adult (over 24 months old). The animal’ sex was also recorded [83].
All animals were hunted in the Tuscany region during authorized hunting seasons (November–January), following the regional hunting law (Regolamento di attuazione della legge regionale 12 gennaio 1994, n. 3 D.P.G.R. 48/R/2017). No animals were specifically sacrificed for this study purpose. Animals did not present gross lesions related to infectious disease at postmortem examination, performed during sampling operations.
4.2. Microscopic Agglutination Test (MAT)
Blood samples were centrifugated at 10,000 rpm for 10 minutes to obtain the serum. In order to detect Leptospira antibodies, sera were tested through microscopic agglutination test (MAT) [84]. Titer of 1:100 was considered as positive. For the MAT, live Leptospira antigens used were: Leptospira interrogans serovar Icterohaemorrhagiae (serogroup Icterohaemorrhagiae, strain RGA), L. interrogans serovar Canicola (serogroup Canicola, strain Alarik), L. interrogans serovar Pomona (serogroup Pomona, strain Mezzano), L. kirschneri serovar Grippotyphosa (serogroup Grippotyphosa, strain Moskva V), L. borgpetersenii serovar Tarassovi (serogroup Tarassovi, strain Mitis Johnson), L. interrogans serovar Bratislava (serogroup Australis, strain Riccio 2), L. interrogans serovar Hardjo (serogroup Sejroe, serovar Hardjoprajitno), and L. borgpetersenii serovar Ballum (serogroup Ballum, strain Mus 127).
4.3. Leptospira spp. Isolation
Each wild boar organ was cultured in Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (Difco, Detroit, MI, USA). Approximately 10 cm³ from each organ was homogenized with 5 mL of sterile water and 1 mL of homogenate was cultured in 5 mL of EMJH. Cultures were incubated at 30 °C ± 1 °C for 120 days and observed every 10 days under dark-field microscopy to evaluate possible bacterial growth.
4.4. Molecular Analysis
From each kidney and liver, DNA was extracted using Quick-DNA Plus Kits (Zymo Research, Irvine, CA, USA) following the manufacturer’s instructions.
Two different multiplex Realtime-PCR were employed; the first, targeting Leptospira spp. (16S rRNA gene) and pathogenic Leptospira (lipL32 gene), was performed on all samples [85,86]. The second protocol was only performed on positive Leptospira spp. and negative lipL32 samples, targeting intermediate Leptospira (16S rRNA gene) and saprophytic Leptospira (23S rRNA gene) [16,86]. As a positive control for the lipL32 gene, DNA extracted from a pure culture of Leptospira interrogans serogroup Pomona strain Mezzano was used. As a positive control for the 23S rRNA gene for saprophytic Leptospira, DNA extracted from a pure culture of Leptospira biflexa serogroup Patoc strain Patoc I was used. As a negative control, sterilized saline water was used. A total reaction volume of 15 μL was prepared by using 2x QuantiTect Probe PCR Master Mix (Qiagen, Hilden, Germany), 2 μM of each primer, 500 nM of each probe, and 3 μL of DNA, as previously reported [7]. The RealTime-PCR assay was performed on a Rotorgene Corbett 6000 (Corbett Research, Sydney, Australia) with the following thermal conditions: a holding stage of 95 °C for 5 min and 45 cycles of 95 °C for 15 s and 60 °C for 30 s. Samples with Ct lipL32 < 35 were considered positive and those samples with 35 < Ct lipL32 ≥ 40 were repeated.
4.5. Leptospira spp. Characterization and Genotyping
First, serogroups of the isolates were determined through the MAT using a panel of eight polyclonal anti-sera against the eight serovars reported in Section 4.2. The agglutination with specific antiserum was used to identify the presumptive strain’s serogroup [84].
Isolated Leptospira were genotyped using a multilocus sequence typing (MLST) scheme based on housekeeping genes [87,88,89].
Moreover, the Leptospira species were identified from positive pathogenic and intermediate Leptospira PCR reactions, using primer for rrs2 gene and 16S rRNA gene, respectively [86,88].
The amplification of each target gene was realized with HotStarTaq Master Mix Kit (Qiagen, Hilden, Germany), and further sequenced (BMR Genomics, Padova, Italy) using the same amplification primer sets and analyzed using BioEdit Software [90]. Phylogenetic analysis was performed by the maximum likelihood method based on the Tamura–Nei model using MEGA 10 software [91].
4.6. Statistical Analysis
Data were analyzed with chi-square (X2) test. The statistical test was used to evaluate the Leptospira infection ratio in relationship to sex (male or female), age class (young, sub-adult, or adult), province (Pisa, Lucca, Livorno, Grosseto, or Siena) and hunting season (2018/2019 or 2019/2020). Statistical significance threshold was set at a p value ≤ 0.05 [92].
5. Conclusions
In conclusion, this investigation confirms through the MAT, isolation, and molecular assays, the role of wild boar in the epidemiology of leptospirosis in Central Italy. Wild boar represents a good indicator of Leptospira circulating in a specific area where many different animal species share the same environment. Furthermore, wild boar populations are able to live in a wide spectrum of habitat types, and, have recently reached sub-urban and urban areas. In Italy, little recent data on human leptospirosis are available; however, some studies investigated the prevalence of infection in risk categories (hunters, farmers, and forestry workers) showing serological positivity to Leptospira [93,94]. Moreover, on the basis of the most recent report on human leptospirosis in Italy [95], a high infection rate was recorded in adult males, and this could indicate that leptospirosis is strictly related to worker activity. Hunters, for example, are usually all male and over 30 years old. In particular, these peoples are exposed to an high risk of infection due to management and slaughtering of dead animals being performed with little health care [96].
Tarassovi and Bratislava are the two main serogroups that circulate within wild boar in Tuscany. Although Bratislava has been more detected, the isolation of Tarassovi suggests that wild boar could be the main reservoir. In addition, as for pathogenic Leptospira, the presence of intermediate species in wild boar kidney underlines the need to perform other studies aimed at understanding the newly- emerging species, L. fainei, in animals and in humans.
Acknowledgments
We thank Mario D’Incau and his team of National Reference Centre for Animal Leptospirosis (Istituto Zooprofilattico Sperimentale della Lombardia e dell’ Emilia Romagna “Bruno Ubertini”, via Bianchi 7/9, 25121 Brescia, Italy) for provision of the Leptospira live strains employed in this investigation.
Author Contributions
Conceptualization, G.C., F.B., and F.F.; investigation, G.C., F.B., and M.A.; data curation, G.C., F.B., M.A., D.C., and F.F.; writing—original draft preparation, G.C., F.B., and F.F.; writing—review and editing, G.C., F.B., M.A., D.C., and F.F.; supervision, D.C. and F.F.; funding acquisition, F.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by University of Pisa, grant number PRA_2018_56.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Massei G., Kindberg J., Licoppe A., Gačić D., Šprem N., Kamler J., Baubet E., Hohmann U., Monaco A., Ozoliņš J., et al. Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Manag. Sci. 2015;71:492–500. doi: 10.1002/ps.3965. [DOI] [PubMed] [Google Scholar]
- 2.Castillo-Contreras R., Carvalho J., Serrano E., Mentaberre G., Fernández-Aguilar X., Colom A., González-Crespo C., Lavín S., López-Olvera J.R. Urban wild boars prefer fragmented areas with food resources near natural corridors. Sci. Total Environ. 2018;615:282–288. doi: 10.1016/j.scitotenv.2017.09.277. [DOI] [PubMed] [Google Scholar]
- 3.Santilli F., Varuzza P. Factors affecting wild boar (Sus scrofa) abundance in southern Tuscany. Hystrix, Ital. J. Mammal. 2013;24:169–173. [Google Scholar]
- 4.Pittiglio C., Khomenko S., Beltran-Alcrudo D. Wild boar mapping using population-density statistics: From polygons to high resolution raster maps. PLoS ONE. 2018;13:e0193295. doi: 10.1371/journal.pone.0193295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carnevali L., Pedrotti L., Riga F., Toso S. Banca Dati Ungulati: Status, distribuzione, consistenza, gestione e prelievo venatorio delle popolazioni di Ungulati in Italia. Rapporto 2001–2005. Biol. e Conserv. della Fauna. 2009;117:1–168. [Google Scholar]
- 6.Lombardini M., Meriggi A., Fozzi A. Factors influencing wild boar damage to agricultural crops in Sardinia (Italy) Curr. Zool. 2017;63:507–514. doi: 10.1093/cz/zow099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cilia G., Bertelloni F., Mignone W., Spina S., Berio E., Razzuoli E., Vencia W., Franco V., Cecchi F., Bogi S., et al. Molecular detection of Leptospira spp. in wild boar (Sus scrofa) hunted in Liguria region (Italy) Comp. Immunol. Microbiol. Infect. Dis. 2020;68:101410. doi: 10.1016/j.cimid.2019.101410. [DOI] [PubMed] [Google Scholar]
- 8.Meng X.J., Lindsay D.S., Sriranganathan N. Wild boars as sources for infectious diseases in livestock and humans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2009;364:2697–2707. doi: 10.1098/rstb.2009.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertelloni F., Mazzei M., Cilia G., Forzan M., Felicioli A., Sagona S., Bandecchi P., Turchi B., Cerri D., Fratini F. Serological Survey on Bacterial and Viral Pathogens in Wild Boars Hunted in Tuscany. Ecohealth. 2020;17:85–93. doi: 10.1007/s10393-020-01475-y. [DOI] [PubMed] [Google Scholar]
- 10.Picardeau M. Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 2017;15:297–307. doi: 10.1038/nrmicro.2017.5. [DOI] [PubMed] [Google Scholar]
- 11.Adler B., de la Peña Moctezuma A. Leptospira and leptospirosis. Vet. Microbiol. 2010;140:287–296. doi: 10.1016/j.vetmic.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Levett P.N. Leptospirosis. Clin. Microbiol. Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent A.T., Schiettekatte O., Goarant C., Neela V.K., Bernet E., Thibeaux R., Ismail N., Mohd Khalid M.K.N., Amran F., Masuzawa T., et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019;13:e0007270. doi: 10.1371/journal.pntd.0007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guglielmini J., Bourhy P., Schiettekatte O., Zinini F., Brisse S., Picardeau M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019;13:e0007374. doi: 10.1371/journal.pntd.0007374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balamurugan V., Gangadhar N.L., Mohandoss N., Thirumalesh S.R.A., Dhar M., Shome R., Krishnamoorthy P., Prabhudas K., Rahman H. Characterization of leptospira isolates from animals and humans: Phylogenetic analysis identifies the prevalence of intermediate species in India. Springerplus. 2013;2:362. doi: 10.1186/2193-1801-2-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barragan V., Chiriboga J., Miller E., Olivas S., Birdsell D., Hepp C., Hornstra H., Schupp J.M., Morales M., Gonzalez M., et al. High Leptospira Diversity in Animals and Humans Complicates the Search for Common Reservoirs of Human Disease in Rural Ecuador. PLoS Negl. Trop. Dis. 2016;10:e0004990. doi: 10.1371/journal.pntd.0004990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiriboga J., Barragan V., Arroyo G., Sosa A., Birdsell D.N., España K., Mora A., Espín E., Mejía M.E., Morales M., et al. High Prevalence of Intermediate Leptospira spp. DNA in Febrile Humans from Urban and Rural Ecuador. Emerg. Infect. Dis. 2015;21:2141–2147. doi: 10.3201/eid2112.140659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz-Fons F. A Review of the Current Status of Relevant Zoonotic Pathogens in Wild Swine (Sus scrofa) Populations: Changes Modulating the Risk of Transmission to Humans. Transbound. Emerg. Dis. 2017;64:68–88. doi: 10.1111/tbed.12369. [DOI] [PubMed] [Google Scholar]
- 19.Dupouey J., Faucher B., Edouard S., Richet H., Kodjo A., Drancourt M., Davoust B. Human leptospirosis: An emerging risk in Europe? Comp. Immunol. Microbiol. Infect. Dis. 2014;37:77–83. doi: 10.1016/j.cimid.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Haake D.A., Levett P.N. Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 2015;387:65–97. doi: 10.1007/978-3-662-45059-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis W.A. Animal Leptospirosis. In: Adler B., editor. Leptospira and Leptospirosis. Current Topics in Microbiology and Immunology. Volume 387. Springer; Berlin, Germany: 2015. pp. 99–137. [DOI] [PubMed] [Google Scholar]
- 22.Cerri D., Ebani V.V., Fratini F., Pinzauti P., Andreani E. Epidemiology of leptospirosis: Observations on serological data obtained by a "diagnostic laboratory for leptospirosis" from 1995 to 2001. New Microbiol. 2003;26:383–389. [PubMed] [Google Scholar]
- 23.Żmudzki J., Jabłoński A., Nowak A., Zębek S., Arent Z., Bocian Ł., Pejsak Z. First overall report of Leptospira infections in wild boars in Poland. Acta Vet. Scand. 2015;58:3. doi: 10.1186/s13028-016-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vale-Goncalves H.M., Cabral J.A., Faria M.C., Nunes-Pereira M., Faria A.S., Veloso O., Vieira M.L., Paiva-Cardoso M.N. Prevalence of Leptospira antibodies in wild boars (Sus scrofa) from Northern Portugal: Risk factor analysis. Epidemiol. Infect. 2015;143:2126–2130. doi: 10.1017/S0950268814003331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boqvist S., Bergström K., Magnusson U. Prevalence of Antibody to Six Leptospira Servovars in Swedish Wild Boars. J. Wildl. Dis. 2012;48:492–496. doi: 10.7589/0090-3558-48.2.492. [DOI] [PubMed] [Google Scholar]
- 26.Bertelloni F., Cilia G., Turchi B., Pinzauti P., Cerri D., Fratini F. Epidemiology of leptospirosis in North-Central Italy: Fifteen years of serological data (2002–2016) Comp. Immunol. Microbiol. Infect. Dis. 2019;65:14–22. doi: 10.1016/j.cimid.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Tagliabue S., Figarolli B.M., D’Incau M., Foschi G., Gennero M.S., Giordani R., Giordani R., Natale A., Papa P., Ponti N., et al. Serological surveillance of Leptospirosis in Italy: Two-year national data (2010-2011) Vet. Ital. 2016;52:129–138. doi: 10.12834/VetIt.58.169.2. [DOI] [PubMed] [Google Scholar]
- 28.Coppola F., Cilia G., Bertelloni F., Casini L., D’Addio E., Fratini F., Cerri D., Felicioli A. Crested porcupine (Hystrix cristata L.): A new potential host for pathogenic Leptospira among semi-fossorial mammals. Comp. Immunol. Microbiol. Infect. Dis. 2020;70:101472. doi: 10.1016/j.cimid.2020.101472. [DOI] [PubMed] [Google Scholar]
- 29.Fratini F., Turchi B., Ebani V.V., Bertelloni F., Galiero A., Cerri D. The presence of Leptospira in coypus (Myocastor coypus) and rats (Rattus norvegicus) living in a protected wetland in Tuscany (Italy) Vet. Arh. 2015;85:407–414. [Google Scholar]
- 30.Bertelloni F., Turchi B., Vattiata E., Viola P., Pardini S., Cerri D., Fratini F. Serological survey on Leptospira infection in slaughtered swine in North-Central Italy. Epidemiol. Infect. 2018:1–6. doi: 10.1017/S0950268818001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebani V.V., Cerri D., Poli A., Andreani E. Prevalence of Leptospira and Brucella antibodies in wild boars (Sus scrofa) in Tuscany, Italy. J. Wildl. Dis. 2003;39:718–722. doi: 10.7589/0090-3558-39.3.718. [DOI] [PubMed] [Google Scholar]
- 32.Verin R., Varuzza P., Mazzei M., Poli A. Serologic, molecular, and pathologic survey of pseudorabies virus infection in hunted wild boars (Sus scrofa) in Italy. J. Wildl. Dis. 2014;50:559–565. doi: 10.7589/2013-01-004. [DOI] [PubMed] [Google Scholar]
- 33.Ebani V.V., Bertelloni F., Pinzauti P., Cerri D. Seroprevalence of Leptospira spp. and Borrelia burgdorferi sensu lato in Italian horses. Ann. Agric. Environ. Med. 2012;19:237–240. [PubMed] [Google Scholar]
- 34.Hartskeerl R., Collares-Pereira M., Ellis W.A. Emergence, control and re-emerging leptospirosis: Dynamics of infection in the changing world. Clin. Microbiol. Infect. 2011;17:494–501. doi: 10.1111/j.1469-0691.2011.03474.x. [DOI] [PubMed] [Google Scholar]
- 35.Montagnaro S., Sasso S., De Martino L., Longo M., Iovane V., Ghiurmino G., Pisanelli G., Nava D., Baldi L., Pagnini U. Prevalence of Antibodies to Selected Viral and Bacterial Pathogens in Wild Boar (Sus scrofa) in Campania Region, Italy. J. Wildl. Dis. 2010;46:316–319. doi: 10.7589/0090-3558-46.1.316. [DOI] [PubMed] [Google Scholar]
- 36.Piredda I., Palmas B., Noworol M., Canu M., Fiori E., Picardeau M., Tola A., Pintore A., Ponti N. Indagine sulla prevalenza della leptospirosi nei cinghiali del centro-nord Sardegna; Proceedings of the XIII Congresso Nazionale S.I.Di.L.V.; Trani, Italy. 12–14 October 2011; pp. 63–64. [Google Scholar]
- 37.Figarolli B.M., Gaffuri A., Alborali G.L., D’Incau M., Tagliabue S. Survey on leptospirosis in wild boars (Sus scrofa) in Lombardy, Northern Italy; Proceedings of the 1°ELS meeting; Dubrovnik, Croatian. 30 April –1 May 2012; pp. 63–64. [Google Scholar]
- 38.Chiari M., Figarolli B.M., Tagliabue S., Alborali G.L., Bertoletti M., Papetti A., D’Incau M., Zanoni M., Boniotti M.B. Seroprevalence and risk factors of leptospirosis in wild boars ( Sus scrofa ) in northern Italy. Hystrix, Ital. J. Mammal. 2016;27 doi: 10.4404/hystrix-27.2-11682. [DOI] [Google Scholar]
- 39.Tagliabue S., Raffo A., Foni E., Candotti R., Barigazzi G. Anticorpi per Leptospira interrogans in sieri di cinghiale selvatico (Sus scrofa) nell’Appennino parmense; Proceedings of the Convegno Nazionale di Ecopatologia della fauna selvatica; Bologna, Italy. 9–11 February 1995. [Google Scholar]
- 40.Espí A., Prieto J.M., Alzaga V. Leptospiral antibodies in Iberian red deer (Cervus elaphus hispanicus), fallow deer (Dama dama) and European wild boar (Sus scrofa) in Asturias, Northern Spain. Vet. J. 2010;183:226–227. doi: 10.1016/j.tvjl.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Jansen A., Nöckler K., Schönberg A., Luge E., Ehlert D., Schneider T. Wild boars as possible source of hemorrhagic leptospirosis in Berlin, Germany. Eur. J. Clin. Microbiol. Infect. Dis. 2006;25:544–546. doi: 10.1007/s10096-006-0174-3. [DOI] [PubMed] [Google Scholar]
- 42.Vicente J., León-Vizcaíno L., Gortázar C., Cubero M.J., González M., Martín-Atance P. Antibodies to selected viral and bacterial pathogens in European wild boars from southcentral Spain. J. Wildl. Dis. 2002;38:649–652. doi: 10.7589/0090-3558-38.3.649. [DOI] [PubMed] [Google Scholar]
- 43.Vengust G., Lindtner-Knific R., Zele D., Bidovec A. Leptospira antibodies in wild boars (Sus scrofa) in Slovenia. Eur. J. Wildl. Res. 2008;54:749–752. doi: 10.1007/s10344-008-0178-7. [DOI] [Google Scholar]
- 44.Treml F., Pikula J., Holešovská Z. Prevalence of antibodies against leptospires in the wild boar (Sus scrofa L., 1758) Vet. Med. (Praha). 2012;48:66–70. doi: 10.17221/5751-VETMED. [DOI] [Google Scholar]
- 45.Arent Z., Gilmore C., Brem S., Ellis W.A. Molecular studies on European equine isolates of Leptospira interrogans serovars Bratislava and Muenchen. Infect. Genet. Evol. 2015;34:26–31. doi: 10.1016/j.meegid.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Rocha T., Ellis W.A., Montgomery J., Gilmore C., Regalla J., Brem S. Microbiological and serological study of leptospirosis in horses at slaughter: First isolations. Res. Vet. Sci. 2004;76:199–202. doi: 10.1016/j.rvsc.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Ellis W.A., McParland P.J., Bryson D.G., Cassells J.A. Boars as carriers of leptospires of the Australis serogroup on farms with an abortion problem. Vet. Rec. 1986;118:563. doi: 10.1136/vr.118.20.563. [DOI] [PubMed] [Google Scholar]
- 48.Slavica A., Cvetnić Z., Konjević D., Janicki Z., Severin K., Dežđek D., Starešina V., Sindičić M., Antić J. Detection of Leptospira spp. serovars in wild boars (Sus scrofa) from continental Croatia. Vet. Arh. 2010;80:247–257. [Google Scholar]
- 49.Agampodi S.B., Matthias M.A., Moreno A.C., Vinetz J.M. Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: Association of level of leptospiremia and clinical manifestations in Sri Lanka. Clin. Infect. Dis. 2012;54:1249–1255. doi: 10.1093/cid/cis035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merien F., Baranton G., Perolat P. Comparison of Polymerase Chain Reaction with Microagglutination Test and Culture for Diagnosis of Leptospirosis. J. Infect. Dis. 1995;172:281–285. doi: 10.1093/infdis/172.1.281. [DOI] [PubMed] [Google Scholar]
- 51.Hall C., Lambourne J. The Challenges of Diagnosing Leptospirosis. J. Travel Med. 2014;21:139–140. doi: 10.1111/jtm.12095_1. [DOI] [PubMed] [Google Scholar]
- 52.Fornazari F., Langoni H., Marson P.M., Nóbrega D.B., Teixeira C.R. Leptospira reservoirs among wildlife in Brazil: Beyond rodents. Acta Trop. 2018;178:205–212. doi: 10.1016/j.actatropica.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 53.Koizumi N., Muto M., Yamada A., Watanabe H. Prevalence of Leptospira spp. in the Kidneys of Wild Boars and Deer in Japan. J. Vet. Med. Sci. 2009;71:797–799. doi: 10.1292/jvms.71.797. [DOI] [PubMed] [Google Scholar]
- 54.Koizumi N., Uchida M., Makino T., Taguri T., Kuroki T., Muto M., Kato Y., Watanabe H. Isolation and characterization of Leptospira spp. from raccoons in Japan. J. Vet. Med. Sci. 2009;71:425–429. doi: 10.1292/jvms.71.425. [DOI] [PubMed] [Google Scholar]
- 55.Pedersen K., Anderson T.D., Bevins S.N., Pabilonia K.L., Whitley P.N., Virchow D.R., Gidlewski T. Evidence of leptospirosis in the kidneys and serum of feral swine (Sus scrofa) in the United States. Epidemiol. Infect. 2017;145:87–94. doi: 10.1017/S0950268816002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scanziani E., Origgi F., Giusti A.M., Iacchia G., Vasino A., Pirovano G., Scarpa P., Tagliabue S. Serological survey of leptospiral infection in kennelled dogs in Italy. J. Small Anim. Pract. 2002;43:154–157. doi: 10.1111/j.1748-5827.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 57.Andreoli E., Radaelli E., Bertoletti I., Bianchi A., Scanziani E., Tagliabue S., Mattiello S. Leptospira spp. infection in wild ruminants: A survey in Central Italian Alps. Vet. Ital. 2014;50:285–291. doi: 10.12834/VetIt.1309.06. [DOI] [PubMed] [Google Scholar]
- 58.Zanzani S.A., Cerbo A.D., Gazzonis A.L., Epis S., Invernizzi A., Tagliabue S., Manfredi M.T. Parasitic and Bacterial Infections of Myocastor coypus in a Metropolitan Area of Northwestern Italy. J. Wildl. Dis. 2016;52:126–130. doi: 10.7589/2015-01-010. [DOI] [PubMed] [Google Scholar]
- 59.Vitale M., Agnello S., Chetta M., Amato B., Vitale G., Bella C.D., Vicari D., Presti V.D.M.L. Human leptospirosis cases in Palermo Italy. The role of rodents and climate. J. Infect. Public Health. 2018;11:209–214. doi: 10.1016/j.jiph.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 60.Ciceroni L., Lombardo D., Pinto A., Ciarrocchi S., Simeoni J. Prevalence of antibodies to Leptospira serovars in sheep and goats in Alto Adige-South Tyrol. J. Vet. Med. B. Infect. Dis. Vet. Public Health. 2000;47:217–223. doi: 10.1046/j.1439-0450.2000.00333.x. [DOI] [PubMed] [Google Scholar]
- 61.Russo L., Massei G., Genov P.V. Daily home range and activity of wild boar in a mediterranean area free from hunting. Ethol. Ecol. Evol. 1997;9:287–294. doi: 10.1080/08927014.1997.9522888. [DOI] [Google Scholar]
- 62.Boitani L., Mattei L., Nonis D., Corsi F. Spatial and Activity Patterns of Wild Boars in Tuscany, Italy. J. Mammal. 1994;75:600–612. doi: 10.2307/1382507. [DOI] [Google Scholar]
- 63.Massolo A., Della Stella R.M. Population structure variations of wild boar Sus scrofa in central Italy. Ital. J. Zool. 2006;73:137–144. doi: 10.1080/11250000600727717. [DOI] [Google Scholar]
- 64.Perolat P., Chappel R.J., Adler B., Baranton G., Bulach D.M., Billinghurst M.L., Letocart M., Merien F., Serrano M.S. Leptospira fainei sp. nov., isolated from pigs in Australia. Int. J. Syst. Bacteriol. 1998;48:851–858. doi: 10.1099/00207713-48-3-851. [DOI] [PubMed] [Google Scholar]
- 65.Chappel R.J., Khalik D.A., Adler B., Bulach D.M., Faine S., Perolat P., Vallance V. Serological titres to Leptospira fainei serovar hurstbridge in human sera in Australia. Epidemiol. Infect. 1998;121:473–475. doi: 10.1017/S095026889800137X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arzouni J.P., Parola P., Scola B.L., Postic D., Brouqui P., Raoult D. Human infection caused by Leptospira fainei. Emerg. Infect. Dis. 2002;8:865–868. doi: 10.3201/eid0808.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petersen A.M., Boye K., Blom J., Schlichting P., Krogfelt K.A. First isolation of Leptospira fainei serovar Hurstbridge from two human patients with Weil’s syndrome. J. Med. Microbiol. 2001;50:96–100. doi: 10.1099/0022-1317-50-1-96. [DOI] [PubMed] [Google Scholar]
- 68.Soares T.S.M., do Rosário Dias de Oliveira Latorre M., Laporta G.Z., Buzzar M.R. Spatial and seasonal analysis on leptospirosis in the municipality of São Paulo, Southeastern Brazil, 1998 to 2006. Rev. Saude Publica. 2010;44:283–291. doi: 10.1590/S0034-89102010000200008. [DOI] [PubMed] [Google Scholar]
- 69.Miller D.A., Wilson M.A., Beran G.W. Relationships between prevalence of Leptospira interrogans in cattle, and regional, climatic, and seasonal factors. Am. J. Vet. Res. 1991;52:1766–1768. [PubMed] [Google Scholar]
- 70.Chadsuthi S., Modchang C., Lenbury Y., Iamsirithaworn S., Triampo W. Modeling seasonal leptospirosis transmission and its association with rainfall and temperature in Thailand using time-series and ARIMAX analyses. Asian Pac. J. Trop. Med. 2012;5:539–546. doi: 10.1016/S1995-7645(12)60095-9. [DOI] [PubMed] [Google Scholar]
- 71.Ward M.P. Seasonality of canine leptospirosis in the United States and Canada and its association with rainfall. Prev. Vet. Med. 2002;56:203–213. doi: 10.1016/S0167-5877(02)00183-6. [DOI] [PubMed] [Google Scholar]
- 72.Stoddard R.A., Bui D., Haberling D.L., Wuthiekanun V., Thaipadungpanit J., Hoffmaster A.R. Viability of Leptospira isolates from a human outbreak in Thailand in various water types, pH, and temperature conditions. Am. J. Trop. Med. Hyg. 2014;91:1020–1022. doi: 10.4269/ajtmh.13-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Filho J.G., Nazário N.O., Freitas P.F., Pinto G.A., Schlindwein A.D. Temporal analysis of the relationship between leptospirosis, rainfall levels and seasonality, Santa Catarina, Brazil, 2005-2015. Rev. Inst. Med. Trop. Sao Paulo. 2018;60:e39. doi: 10.1590/S1678-9946201860039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kupek E., de Sousa Santos Faversani M.C., de Souza Philippi J.M. The relationship between rainfall and human leptospirosis in Florianópolis, Brazil, 1991–1996. Braz. J. Infect. Dis. 2000;4:131–134. [PubMed] [Google Scholar]
- 75.Hacker K.P., Sacramento G.A., Cruz J.S., De Oliveira D., Nery N., Lindow J.C., Carvalho M., Hagan J., Diggle P.J., Begon M., et al. Influence of rainfall on leptospira infection and disease in a tropical urban setting, Brazil. Emerg. Infect. Dis. 2020;26:311–314. doi: 10.3201/eid2602.190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Regione Toscana - Settore Idrologico Regionale Analisi dei Dati Termometrici Report Anno 2018. [(accessed on 10 May 2020)];2018 Available online: https://www.sir.toscana.it/supports/download/report/2018_situazione_termometrica.pdf.
- 77.Regione Toscana - Settore Idrologico Regionale Report Pluviometrico Anno 2018. [(accessed on 10 May 2020)];2018 Available online: https://www.sir.toscana.it/supports/download/report/2018_situazione_idrologica.pdf.
- 78.Regione Toscana - Settore Idrologico Regionale Analisi dei Dati Termometrici Report Anno 2019. [(accessed on 10 May 2020)];2019 Available online: https://www.sir.toscana.it/supports/download/report/2019_situazione_termometrica.pdf.
- 79.Regione Toscana - Settore Idrologico Regionale Report Pluviometrico Anno 2019. [(accessed on 10 May 2020)];2019 Available online: https://www.sir.toscana.it/supports/download/report/2019_situazione_idrologica.pdf.
- 80.Regione Toscana - Settore Idrologico Regionale Report Pluviometrico del Mese di Gennaio 2020. [(accessed on 10 May 2020)];2020 Available online: https://www.sir.toscana.it/supports/download/report/2020-01_report_cumulate_mensili.pdf.
- 81.Regione Toscana - Settore Idrologico Regionale Analisi Dati Termometrici Report Gennaio 2020. [(accessed on 10 May 2020)];2020 Available online: https://www.sir.toscana.it/supports/download/report/report_termometria_2020-01.pdf.
- 82.Arenas-Montes A., García-Bocanegra I., Paniagua J., Franco J.J., Miró F., Fernández-Morente M., Carbonero A., Arenas A. Blood sampling by puncture in the cavernous sinus from hunted wild boar. Eur. J. Wildl. Res. 2013;59:299–303. doi: 10.1007/s10344-013-0701-3. [DOI] [Google Scholar]
- 83.Sáez-Royuela C., Gomariz R.P., Luis Tellería J. Age Determination of European Wild Boar. Volume 17 Wiley; Hoboken, NJ, USA: 1989. [Google Scholar]
- 84.OIE Leptospirosis. [(accessed on 10 May 2020)];Man. Diagnostic Tests Vaccines Terr. Anim. 2014 Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.01.12_LEPTO.pdf. [Google Scholar]
- 85.Stoddard R.A., Gee J.E., Wilkins P.P., McCaustland K., Hoffmaster A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009;64:247–255. doi: 10.1016/j.diagmicrobio.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 86.Bedir O., Kilic A., Atabek E., Kuskucu A.M., Turhan V., Basustaoglu A.C. Simultaneous detection and differentiation of pathogenic and nonpathogenic Leptospira spp. by multiplex real-time PCR (TaqMan) assay. Polish J. Microbiol. 2010;59:167–173. doi: 10.33073/pjm-2010-026. [DOI] [PubMed] [Google Scholar]
- 87.Boonsilp S., Thaipadungpanit J., Amornchai P., Wuthiekanun V., Bailey M.S., Holden M.T.G., Zhang C., Jiang X., Koizumi N., Taylor K., et al. A single Multilocus Sequence Typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl. Trop. Dis. 2013;7:e1954. doi: 10.1371/journal.pntd.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahmed N., Devi S.M., de los Á Valverde M., Vijayachari P., Machang’u R.S., Ellis W.A., Hartskeerl R.A. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 2006;5:28. doi: 10.1186/1476-0711-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Varni V., Ruybal P., Lauthier J.J., Tomasini N., Brihuega B., Koval A., Caimi K. Reassessment of MLST schemes for Leptospira spp. typing worldwide. Infect. Genet. Evol. 2014;22:216–222. doi: 10.1016/j.meegid.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 90.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 91.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.R. R Development Core Team A language and environment for statistical computing. R Found. Stat. Comput. Vienna, Austria. 2015 doi: 10.1890/0012-9658(2002)083[3097:CFHIWS]2.0.CO;2. [DOI] [Google Scholar]
- 93.Nuti M., Amaddeo D., Crovatto M., Ghionni A., Polato D., Lillini E., Pitzus E., Santini G.F. Infections in an alpine environment: Antibodies to hantaviruses, leptospira, rickettsiae, and Borrelia burgdorferi in defined Italian populations. Am. J. Trop. Med. Hyg. 1993;48:20–25. doi: 10.4269/ajtmh.1993.48.20. [DOI] [PubMed] [Google Scholar]
- 94.Nuti M., Amaddeo D., Autorino G.L., Crovatto M., Crucil C., Ghionni A., Giommi M., Salvati F., Santini G.F. Seroprevalence of antibodies to hantaviruses and leptospires in selected Italian population groups. Eur. J. Epidemiol. 1992;8:98–102. doi: 10.1007/BF03334979. [DOI] [PubMed] [Google Scholar]
- 95.Istisan R., Graziani C., Duranti A., Morelli A., Busani L., Pezzotti P. Rapoorti ISTISAN Istituto Superiore di Sanità 16/1; Proceedings of the I Simposio internazionale Nuove strategie per gli interventi di prevenzione dello stress da lavoro; Sassari-Alghero, Italy. 8–10 July 2015. [Google Scholar]
- 96.Ngugi J.N., Fèvre E.M., Mgode G.F., Obonyo M., Mhamphi G.G., Otieno C.A., Cook E.A.J. Seroprevalence and associated risk factors of leptospirosis in slaughter pigs; A neglected public health risk, western Kenya. BMC Vet. Res. 2019;15:403. doi: 10.1186/s12917-019-2159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]