Abstract
Background:
High cathepsin D has been associated with poor prognosis in breast cancer; however, the results of many studies are controversial. Here, we assessed the association between high cathepsin D levels and worse breast cancer prognosis by conducting a meta-analysis.
Methods:
A comprehensive search strategy was used to search relevant literature in PUBMED and EMBASE by September 2018. The meta-analysis was performed in Review Manager 5.3 using hazard ratios (HRs) with 95% confidence intervals (CIs).
Results:
A total of 15,355 breast cancer patients from 26 eligible studies were included in this meta-analysis. Significant associations between elevated high cathepsin D and poor overall survival (OS) (HR = 1.61, 95% CI: 1.35–1.92, p < 0.0001) and disease-free survival (DFS) (HR = 1.52, 95% CI: 1.31–2.18, p < 0.001) were observed. In the subgroup analysis for DFS, high cathepsin D was significantly associated with poor prognosis in node-positive patients (HR = 1.38, 95% CI: 1.25–1.71, p < 0.00001), node-negative patients (HR = 1.78, 95% CI: 1.39–2.27, p < 0.0001), early stage patients (HR = 1.73, 95% CI: 1.34–2.23, p < 0.0001), and treated with chemotherapy patients (HR = 1.60, 95% CI: 1.21–2.12, p < 0.001). Interestingly, patients treated with tamoxifen had a low risk of relapse when their cathepsin D levels were high (HR = 0.71, 95% CI: 0.52–0.98, p = 0.04) and a high risk of relapse when their cathepsin D levels were low (HR = 1.50, 95% CI: 1.22–1.85, p = 0.0001).
Conclusions:
Our meta-analysis suggests that high expression levels of cathepsin D are associated with a poor prognosis in breast cancer. Based on our subgroup analysis, we believe that cathepsin D can act as a marker for poor breast cancer prognosis and also as a therapeutic target for breast cancer.
Keywords: breast cancer, cathepsin D, disease-free survival, meta-analysis, overall survival, prognostic biomarker, systematic review
Background
Breast cancer is the most common cancer among women worldwide. In 2018, 2.1 million new cases were diagnosed and approximately 626,000 deaths were reported due to breast cancer.1 Most breast cancer patients in the United States are diagnosed with early stage disease.2 Although the five-year survival rate for breast cancer is close to 100% when detected at an early stage, more aggressive breast cancer is likely to return if a proper adjuvant therapy is not given after surgery.3,4 For this reason, adjuvant therapy after primary surgery plays an important role in the survival of breast cancer patients. Various factors affect breast cancer adjuvant therapy decision making. The factors currently taken into account for adjuvant therapy decision making include tumor size, lymph node status, and tumor characteristics (hormone receptor status, HER2 status, and KI-67 status). However, these factors are not instructive for all patients concerning the decision to get adequate adjuvant therapy. Therefore, new protein and molecular markers have been proposed as decision-making aids.5,6
Cathepsin D (CTSD) was first described by Westley and Rochefort in 1979. It is also termed aspartic endoproteinase and is proteolytically active at low pH.7 CTSD is over-expressed by human epithelial breast cancer cells and results in over-secretion of 52-kDa pro-CTSD into the extracellular environment.8 CTSD secreted into the extracellular environment is automatically activated under acidic conditions, and activated CTSD affects breast cancer progression by increasing breast cancer cell proliferation, fibroblast growth, tumor angiogenesis, tumor growth and metastasis.9–12 Recent studies have shown that CTSD is involved in estrogen receptor activity and tamoxifen’s drug response,13,14 and has a poor prognosis with extensive induction of angiogenesis in both ovarian and breast cancers.10,15 It has also been reported as a biomarker capable of predicting metastasis and tumor-specific extracellular targets suitable for antibody-based therapies.9,16 As a result, CTSD was expected to act as a potential prognostic factor for breast cancer. Many studies have evaluated the prognostic value of CTSD in breast cancer patients, but contrary to expectations, some studies evaluating the prognostic value of CTSD have shown conflicting results. For this reason, we performed a meta-analysis of relevant literature to better quantify the prognostic impact of CTSD expression.
Methods
Search strategy
In this meta-analysis, we selected studies evaluating the relationship between CTSD protein expression and prognosis in breast cancer. We followed the PRISMA standard guidelines to perform the meta-analysis of observational studies and wrote the manuscript according to the PRISMA checklist17 (see Supplemental Table 1). PubMed and EMBASE databases were searched through September 2018 for relevant articles that reported the association between CTSD levels and the hazard ratio of breast cancer. To fulfil our selection criteria, the studies had to have been published as a full paper in English; reference lists and review articles were included. Articles were identified by an electronic PUBMED and EMBASE database search using the following keywords: ‘CTSD’, ‘CD’, ‘Cathepsin D’, ’breast cancer’, ‘breast cancer’, ‘breast carcinoma’, ‘breast neoplasm’, ‘breast tumor’, ‘breast tumour’, ‘hazard ratios’, ‘hazard ratio’, ‘HR’, ‘HRs’, ‘survival’, and ‘prognosis’ (see Supplemental Table 2).
Study selection
The inclusion criteria for the analysis were as follows: studies published as full articles and in the English language on adult patients (at least 20) with breast cancer that reported either the prognostic impact of CTSD evaluated by immuno-histochemistry (IHC), enzyme-linked immunosorbent assay (ELISA), immunoradiometric assay (ELSA), and radioimmunoassay (RIA). Studies that included the hazard ratios and 95% confidence intervals (CIs) for overall survival (OS), disease-free survival (DFS), and relapse-free survival (RFS). In this meta-analysis, the results of DFS and RFS were integrated into DFS. Duplicate publications were excluded. Two reviewers independently evaluated all the titles and abstracts identified by the search. The results were then pooled, and all potentially relevant publications were retrieved in full. The two reviewers then evaluated the complete articles for eligibility. To avoid the inclusion of duplicated or overlapping data, we compared author names and the institutions where the patients were recruited. The reasons to consider articles as non-evaluable were: (a) no univariate analysis reported; (b) no possibility to calculate HR using one of the methods mentioned above because the distribution of CTSD was not reported in the article or CTSD was analyzed in combination with other prognostic markers rendering analysis impossible; and (c) duplicated data was published in different journals.
Data extraction and quality assessment
Information was extracted from all publications. The meta-analysis was initially conducted for all the included studies for each of the endpoints of interest. DFS was the primary outcome of interest and OS was the secondary outcome of interest. The following data were collected from each study: author names, publication date, follow-up, detection method, staining location, and the CTSD cut-off value used for analysis. High CTSD was defined according to the cut-off chosen by each author. Subgroup analyses were conducted for node-positive, node-negative, early stage, treated with adjuvant chemotherapy, and treated with tamoxifen subgroups and if there were at least two papers for each subgroup. The quality of each nonrandomized study was evaluated using the validated Newcastle–Ottawa Scale (NOS) in this meta-analysis18 (see Supplemental Table 3). This scale awards a maximum of nine points to each cohort study (four for quality of selection, two for comparability, and three for quality of outcome and adequacy of follow-up). Studies with an NOS score of 6 were classified as high quality and only such studies were included in our meta-analysis.
Statistical analyses
In this meta-analysis, we included articles that have information including HR and its 95% CI or Kaplan–Meier curve. HRs were calculated based on the high expression of CTSD protein (HR > 1). A HR > 1 implied poor prognosis for patients with breast cancer. The heterogeneity of the studies was evaluated using the I2 value, as described before.19 We pooled the information with a random or fixed-effect model according to the I2 value. The fixed-effects model method was used when I2 < 50%, indicating a lack of heterogeneity among studies. When heterogeneity was observed, the random-effects model was applied.20 Publication bias was visually estimated by assessing funnel plots.21,22 The extracted data were aggregated for a meta-analysis using the RevMan5.3 software (Cochrane Collaboration, Copenhagen, Denmark).23 The prognosis was plotted as a Kaplan–Meier curve and the digitizer Engauge 4.0 software (http://engauge-digitizer.software.informer.com/) was used to digitize and extract the data.
Results
Study characteristics
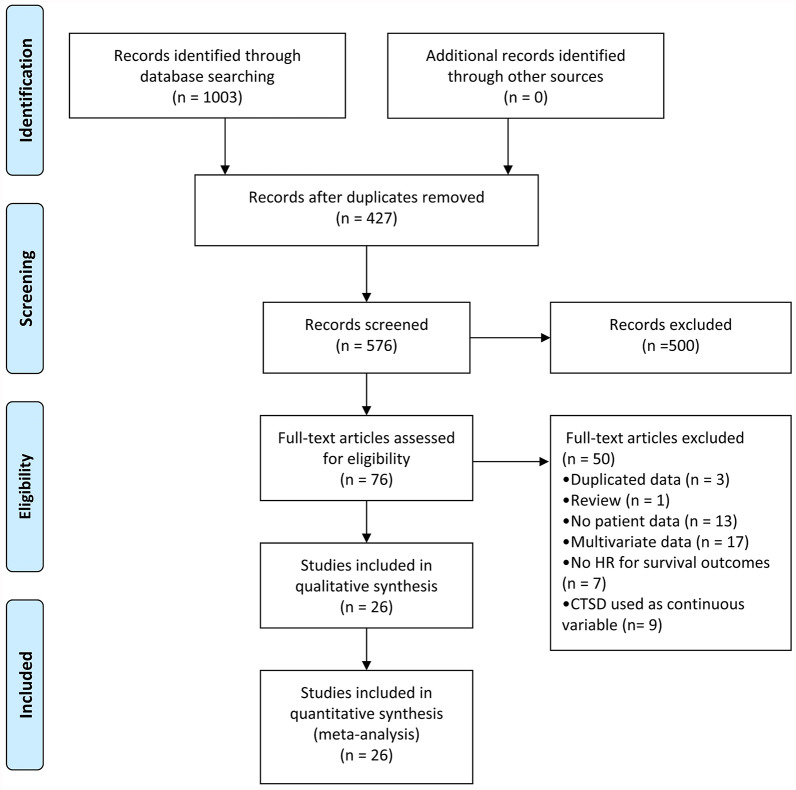
A flowchart of the studies included in the meta-analysis is presented in Figure 1. A computer-based literature search using the PUBMED and EMBASE databases identified a total of 1003 studies. Among these, 427 studies were eliminated as they were non-human studies, conference abstracts, or articles written in a language other than English. Of the 76 full-text articles evaluated, 50 were eliminated because they contained duplicate data, were review articles, or lacked data necessary for estimating the HR at 95% CI. Finally, 26 studies were included in this meta-analysis. In Tables 1 and 2, the characteristics of included studies are described. The different cut-off values used were those of the authors. Threshold definitions were mean or median values, the best cut-off value, or an established arbitrary value.
Figure 1.
Flow chart of the study selection process.
CTSD, cathepsin D; HR, hazard ratio.
Table 1.
Characteristics of the studies on overall survival outcomes of breast cancer patients according to cathepsin D status.
| Author | No. of patients High CTSD/low CTSD (Total patients) |
Median follow-up (months) | Survival analysis test | Detection method | Staining location | Cut-off value (low/high level) |
|---|---|---|---|---|---|---|
| Namer et al.24 | 209/204 (413) | 68 | KM plot | ELSA | cytosol | High (>35 pmol/mg) |
| Granata et al.25 | 67/68 (135) | 87 | KM plot | ELSA | cytosol | High (>40 pmol/mg) |
| Duffy et al.26 | 330 (Total patients) | 47 | KM plot | ELSA | cytosol | High (>40 pmol/mg) |
| Domagala et al.27 | 81/55 (136) | 84 (Mean) | KM plot | IHC | cytosol | High (stained 10%) |
| Pujol et al.28 | 64/59 (123) | 60 (Mean) | KM plot | ELSA | cytosol | High (>20 pmol/mg) |
| Winstanley et al.29 | 265/94 (359) | 132 (Mean) | KM plot | IHC | cytosol | NA |
| Isola et al.30 | 95/167 (262) | 98 (Mean) | KM plot | ELISA | cytosol | High (stained 10%) |
| Donoghue et al.31 | 75/28 (103) | 60 | KM plot | IHC | stromal cell | High (stained 25%) |
| Joensuu et al.32 | 161/52 (213) | 372 | KM plot | ELSA | stromal cell | NA |
| Foekens et al.33 | 1405/1405 (2810) | 88 | KM plot | ELSA | cytosol | High (>45.2 pmol/mg) |
| Harbeck et al.34 | 61/60 (121) | 72 | KM plot | ELSA | cytosol | High (>41 pmol/mg) |
| Kute et al.35 | 552 (Total patients) | 94 | Univariate | RIA | cytosol | High (>10 pmol/mg) |
| Rodriguez et al.36 | 307/696 (1003) | 54 | KM plot | ELSA | cytosol | High (>59 pmol/mg) |
| Mazouni et al.37 | 316 (Total patients) | 75 | KM plot | ELSA | cytosol | High (>41 ng/mg) |
| Mazouni et al.38 | 94/85 (179) | 78 | Univariate | ELSA | cytosol | High (>39 pmol/mg) |
| Jacobson et al.39 | 252/18 (270) | 126 | Univariate | IHC | cytosol | High (> third quartile) |
| Chen et al.40 | 155/44 (199) | 60 | Univariate | IHC | cytosol | High (stained 10%) |
| Huang et al.41 | 140/45 (185) | 66 | Univariate | IHC | cytosol | High (stained 20%) |
| Giatromanolaki et al.42 | 28/72 (100) | 89 | KM plot | IHC | cytosol | High (stained 50%) |
CTSD, cathepsin D; ELSA, immunoradiometric assay; ELISA, enzyme-linked immunosorbent assay; IHC, immunohistochemistry; KM plot, Kaplan–Meier plot; RIA; radioimmunoassay.
Table 2.
Characteristics of the studies on disease-free survival outcomes of breast cancer patients according to cathepsin D status.
| Author | No. of patients High CTSD/low CTSD (Total patients) |
Median follow-up (months) | Survival analysis test | Detection method | Staining location | Cut-off value (low/high level) |
|---|---|---|---|---|---|---|
| Granata et al.43 | 67/68 (135) | 87 | KM plot | ELSA | cytosol | High (>40 pmol/mg) |
| Pujol et al.28 | 64/59 (123) | 59 | KM plot | ELSA | cytosol | High (>20 pmol/mg) |
| Tetu et al.44 | 262/376 (638) | 58 | KM plot | IHC | cytosol | High (stained 10%) |
| Ferno et al.45 | 184/70 (153) | 37 | KM plot | ELSA | cytosol | High (>45 pmol/mg) |
| Donoghue et al.31 | 75/28 (103) | 60 | KM plot | IHC | stromal cell | High (stained 25%) |
| Foekens et al.33 | 1405/1405 (2810) | 88 | KM plot | ELSA | cytosol | High (>45.2 pmol/mg) |
| Jahkola et al.46 | 54/65 (119) | 94 | Univariate | IHC | stromal cell | High (stained 10%) |
| Billgren et al.47 | 707/214 (921) | 59 | Univariate | ELSA | cytosol | High (>10 pmol/mg) |
| Rodriguez et al.36 | 307/696 (1003) | 54 | KM plot | ELSA | cytosol | High (>59 pmol/mg) |
| Jacobson et al.39 | 252/18 (270) | 126 | Univariate | IHC | cytosol | High (> third quartile) |
| Chen et al.40 | 155/44 (199) | 60 | Univariate | IHC | cytosol | High (stained 10%) |
| Markićević et al.48 | 39/19 (58) | 18 | KM plot | ELSA | cytosol | High (>39 pmol/mg) |
| Tazhibi et al.49 | 637/38 (675) | 59 | KM plot | IHC | cytosol | High (stained 20%) |
| Huang et al.41 | 140/45 (185) | 66 | Univariate | IHC | cytosol | High (stained 20%) |
| Sun et al.50 | 91/64 (155) | NA | KM plot | IHC | cytosol | High (stained 26%) |
CTSD, cathepsin D; ELSA, immunoradiometric assay; IHC, immunohistochemistry; KM plot, Kaplan–Meier plot.
In total, 19 evaluable studies24–42 for OS (7809 patients) and 15 evaluable studies28,31,33,35,39–41,43–50 for DFS (7546 patients) were included. Subgroup analysis for OS was possible using five studies with 784 node-positive patients,27,28,36,43,51 five studies with 1193 node-negative patients,28,30,34,35,43 and four studies with 575 adjuvant chemotherapy-treated patients.34,36,40,51 Subgroup analysis for DFS was possible for six studies with 2633 node-positive patients,33,36,44,45,48,51 six studies with 2775 node-negative patients,24,25,30,35,36,52 four studies with 657 early stage patients,42,46,48,52 three studies with 459 adjuvant chemotherapy-treated patients,36,44,46 and two studies with 1747 tamoxifen-treated patients.45,47
Analysis of OS or DFS for all patients
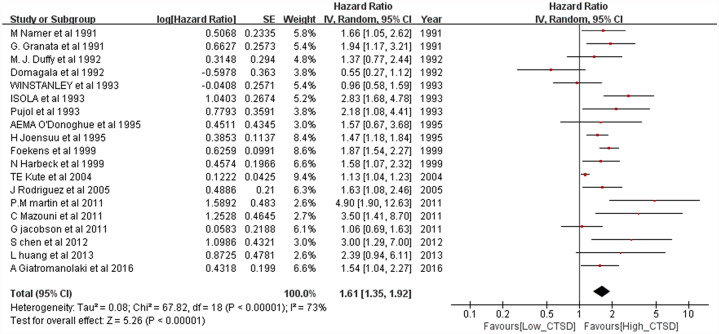
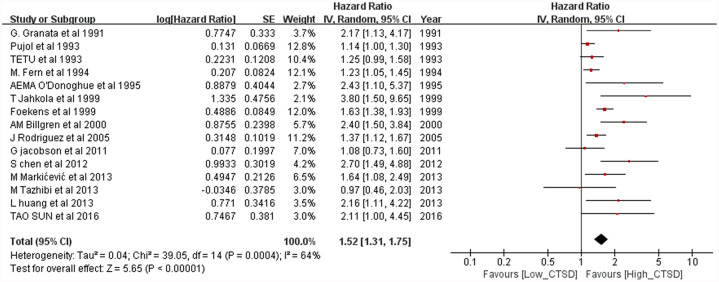
The meta-analysis results of the overall population for OS are shown in Figure 2. For the overall population, worse OS (HR = 1.61, 95% CI: 1.35–1.92; p < 0.00001) was observed among patients considered as CTSD positive. Heterogeneity was high (p < 0.00001, I2 = 73%) for these patients; thus, a random-effects model was used. The meta-analysis results of the overall population for DFS is shown in Figure 3. Worse DFS (HR = 1.52, 95% CI: 1.31–1.75; p < 0.00001) was observed among patients considered as CTSD positive. Heterogeneity was high (p = 0.0004, I2 = 64%) for these patients; thus, a random-effects model was used.
Figure 2.
Forest plot for overall survival according to cathepsin D (CTSD) expression.
CI, confidence interval.
Figure 3.
Forest plot for disease-free survival according to cathepsin D (CTSD) expression.
CI, confidence interval.
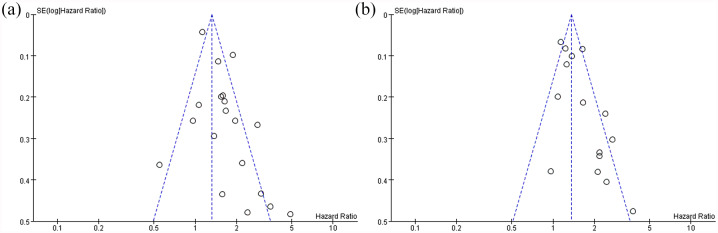
Publication bias
Publication bias was reported via funnel plots; the asymmetry of the funnel plots may have arisen through heterogeneity. The funnel plots of the overall population for OS and DFS are shown in Figure 4. The funnel plots showed an asymmetrical distribution for CTSD among the studies, revealing that publication bias might exist. The funnel plots of subgroup analyses are shown in Supplement Figures 3–5. In the subgroup analyses funnel plots, only the node-negative patients showed an asymmetrical distribution for OS; the remaining groups showed a symmetrical distribution.
Figure 4.
Funnel plots of the 27 studies included in the meta-analysis. (a) overall survival and (b) disease-free survival.
Subgroup analyses of OS
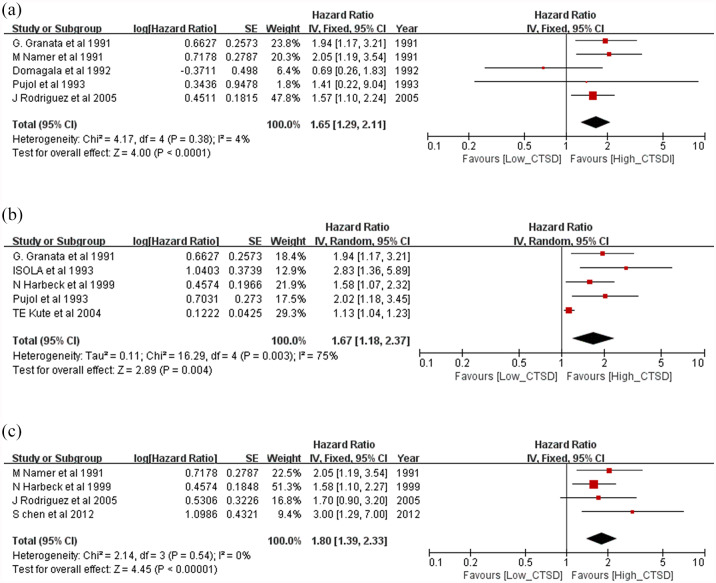
In the subgroup analyses for OS, a worse prognosis was observed independently for node-positive patients (HR = 1.65, 95% CI: 1.29–2.11, p < 0.0001; Figure 5a) and node-negative patients (HR = 1.67, 95% CI: 1.18–2.37; p < 0.00001; Figure 5b). Moreover, adjuvant chemotherapy-treated patients showed a worse prognosis (HR = 1.8, 95% CI: 1.39–2.33; p < 0.00001; Figure 5c). Characteristics of the studies included in the subgroup analyses are shown in Table 3.
Figure 5.
Forest plots of subgroup analysis for overall survival. (a) node-positive patients, (b) node-negative patients and (c) adjuvant chemotherapy-treated patients.
CI, confidence interval; CTSD, cathepsin D.
Table 3.
Summarized hazard ratios of overall and subgroup analyses for overall survival and disease-free survival.
| Group | No. of studies | No. of patients | HR (95% CI) | p | Heterogeneity x2 | I2 (%) | p Heterogeneity |
|---|---|---|---|---|---|---|---|
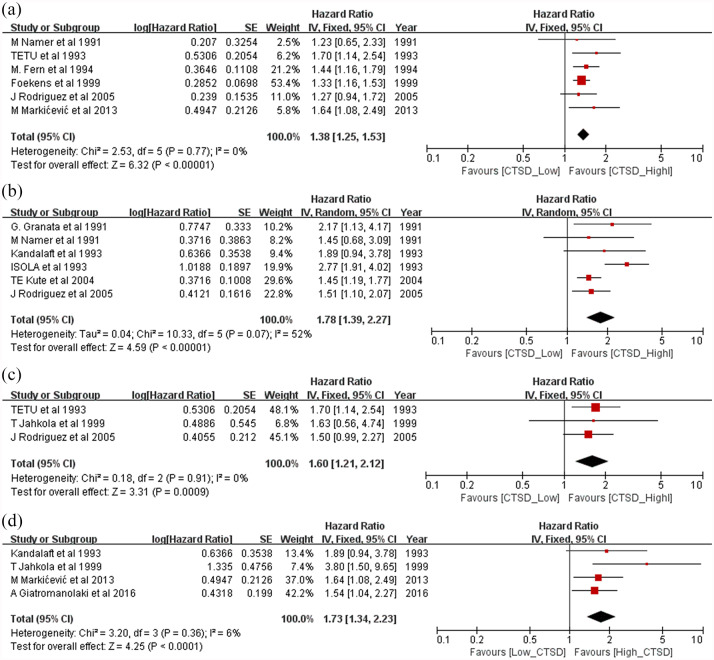
| Disease-free survival | |||||||
| N– patients | 6 | 2775 | 1.78 (1.39–2.27) | <0.00001 | 10.33 | 52 | 0.07 |
| N+ patients | 6 | 2633 | 1.38 (1.25–1.53) | <0.00001 | 2.53 | 0 | 0.77 |
| Early stage patients | 5 | 657 | 1.73 (1.34–2.23) | <0.0001 | 3.2 | 6 | 0.36 |
| Chemotherapy patients | 3 | 459 | 1.60 (1.21–2.12) | 0.0009 | 0.18 | 0 | 0.91 |
| Overall survival | |||||||
| N– patients | 5 | 1193 | 1.67 (1.18–2.37) | 0.004 | 16.29 | 75 | 0.003 |
| N+ patients | 5 | 784 | 1.65 (1.29–2.11) | <0.0001 | 4.17 | 4 | 0.38 |
| Chemotherapy patients | 4 | 575 | 1.80 (1.39–2.33) | <0.00001 | 2.14 | 0 | 0.54 |
CI, confidence interval; HR, hazard ratio; N–, node-negative; N+, node-positive.
Subgroup analyses of DFS
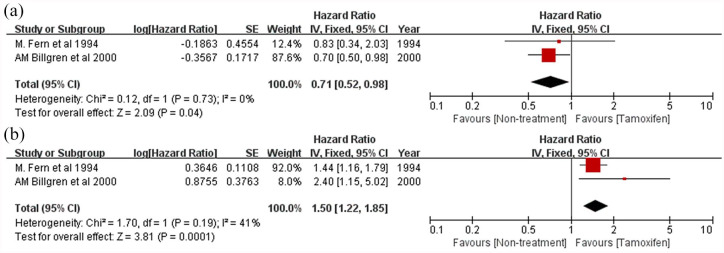
In the subgroup analyses for DFS, a worse prognosis was observed independently for node-positive patients (HR = 1.38, 95% CI: 1.25–1.71, p < 0.00001; Figure 6a) and node-negative patients (HR = 1.66, 95% CI: 1.44–1.91; p < 0.00001; Figure 6b). Worse prognosis was observed independently for early stage patients (HR = 1.41, 95% CI: 1.16–1.70; p = 0.0004; Figure 6c) and adjuvant chemotherapy-treated patients (HR = 1.6, 95% CI: 1.21–2.12; p = 0.0009; Figure 6d). Patients with high CTSD expression levels showed good prognosis when treated with tamoxifen (HR = 0.71, 95% CI: 0.52–0.98, p = 0.04; Figure 7a). However, patients with low CTSD expression levels showed a worse prognosis when treated with tamoxifen (HR = 1.50, 95% CI: 1.22–1.85, p = 0.0001; Figure 7b).
Figure 6.
Forest plot of subgroup analysis for disease-free survival. (a) node-positive patients, (b) node-negative patients, (c) early stage patients and (d) adjuvant chemotherapy-treated patients.
CI, confidence interval; CTSD, cathepsin D.
Figure 7.
Forest plot of subgroup analyses for patients treated with tamoxifen versus non-treated patients. (a) patients with high cathepsin D expression and (b) patients with low cathepsin D (CTSD) expression.
CI, confidence interval.
Discussion
Our meta-analysis confirms that breast cancer patients with high CTSD expression have a worse prognosis in the overall population. The prognostic impact of CTSD was verified through a univariate analysis. Furthermore, our subgroup analysis suggests that CTSD may be helpful to decide the most appropriate adjuvant therapy. To our knowledge, this is the first meta-analysis of published studies to evaluate the association between CTSD expression and prognosis in breast cancer patients.
We found that high CTSD expression in breast cancer was statistically significantly associated with worse prognosis in terms of both OS and DFS. This finding was consistent with most, but not all, of the results of individual studies included this meta-analysis. Prognostic markers are very important for the treatment and prognosis prediction of breast cancer, and we believe that CTSD can be used as a prognostic marker for all breast cancer patients and especially for early stage or node-negative patients. In addition, our subgroup analysis results suggest that CTSD will play an important role in making adjuvant therapy decisions for breast cancer patients.
Adjuvant therapy is currently recommended for all node-positive patients with breast cancer because the 10-year recurrence rate in these patients approaches 70%. In contrast, for node-negative patients with a relatively good prognosis, adjuvant therapy is not recommended. However, even node-negative HER2-positive patients can experience increased recurrence and decreased survival. The prognostic markers considered for adjuvant therapy decision-making for node-negative patients are only HER2 status and tumor size.53,54 More prognostic markers are needed to select the appropriate patients to receive adjuvant therapy. Our study indicates that high CTSD is significantly associated with worse OS and DFS in node-negative patients. These results support previous findings55 and indicate that CTSD has great potential as a potential prognostic marker for the survival and relapse of node-negative patients. We, thus, believe that CTSD should be considered as a prognostic marker for the survival and relapse of node-negative patients.
In our study, patients with high CTSD seemed to be less affected by adjuvant chemotherapy and had higher rates of relapse at an early stage. Chemotherapy reduces the risk of recurrence in women with early stage breast cancer. However, its absolute benefits may be small and not worth the added risk of toxicity among women with a baseline risk of recurrence.56,57 For this reason, the discovery of accurate prognostic markers that can predict early stage relapse and chemotherapy response is important. Our subgroup analysis indicates that high CTSD can act as a prognostic marker for predicting early stage recurrence and chemotherapy response in breast cancer.
One of the interesting results of our subgroup analysis was the tamoxifen drug response. Hormone-positive breast cancer accounts for about 70% of all breast cancers, and these patients are often treated with anti-hormonal drugs. However, approximately 20–30% of breast cancer patients are resistant to this treatment and have a high risk of relapse.58,59 Although there were only two studies included, these showed that patients with high CTSD who were treated with tamoxifen have a low risk of relapse and patients with low CTSD who were treated with tamoxifen have a high risk of relapse. CTSD is a lysosomal protein that helps maintain homeostasis of cell metabolism and is known to be involved in lysosomal membrane permeabilization.60 Previous studies have reported that tamoxifen-resistance cells are less susceptible to lysosomal membrane permeabilization, which is associated with low CTSD. These results indicate that CTSD is potentially associated with tamoxifen-resistance and CTSD, and our results support these studies.61–64 These results suggest that CTSD is one of the potentially important proteins for tamoxifen resistance and that CTSD should be considered as a biomarker for predicting tamoxifen resistance.
Study limitations
There are some limitations to our study. First, our meta-analysis only evaluated the univariate prognostic value of CTSD. Because the results from multivariate analyses were excluded, our results may have been biased. Second, heterogeneity existed among the selected studies. Although it was impossible to determine all sources of heterogeneity, we excluded some covariates that might contribute to heterogeneity of data due to unavailable data. These covariates included progesterone receptor status, tumor size, age of patients, and others. Third, in the subgroup analysis, some subgroups contained very small studies, which may bias their findings. Fourth, high CTSD is defined according to the cut-off chosen by each author, so there may be a bias towards high-CTSD definitions. Moreover, language bias might exist due to the references being restricted to English publications only.
Conclusion
Despite some limitations, our meta-analysis supports the prognostic role of CTSD in breast cancer by showing a significant association between its expression and the risk of breast cancer recurrence and death in all populations considered and for both DFS and OS. Furthermore, high CTSD expression may be a potential biomarker for DFS of node-negative, early stage patients and may assist clinicians make decisions regarding tamoxifen treatment.
Supplemental Material
Supplemental material, Supplement_Figure_1 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Figure_2 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Figure_3 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Table_1 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Table_2 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Table_3 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Acknowledgments
Not applicable
Footnotes
Author note: Jeong Jun Park is now affiliated with Department of Anesthesiology and Pain Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Author contribution: JK and YY conceived the study and take responsibility for the integrity of the data and accuracy of the data analysis. HL and SJ did the literature research, performed study selection, data extraction, and synthesis. HJH and JJP participated in the analysis and interpretation of the data. HSN wrote the draft review paper. DSK and YHK revised the manuscript critically for important intellectual content and redrafted some of its sections. All the authors read and approved the final version of the manuscript.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by grants from the Basic Science Research Program (NRF-2020R1C1C1003741), Collaborative Genome Program for Fostering New Post-Genome Industry (NRF-2017M3C9A6047610), and Medical Research Center (NRF-2018R1A5A2023879) through the National Research Foundation of Korea grant funded by the Korean government.
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iDs: Junho Kang  https://orcid.org/0000-0003-3430-0960
https://orcid.org/0000-0003-3430-0960
Yun Hak Kim  https://orcid.org/0000-0002-9796-8266
https://orcid.org/0000-0002-9796-8266
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Junho Kang, Interdisciplinary Program of Genomic Data Science, Pusan National University, Yangsan, Republic of Korea.
Yeuni Yu, Interdisciplinary Program of Genomic Data Science, Pusan National University, Yangsan, Republic of Korea.
Seongdo Jeong, Interdisciplinary Program of Genomic Data Science, Pusan National University, Yangsan, Republic of Korea.
Hansong Lee, Interdisciplinary Program of Genomic Data Science, Pusan National University, Yangsan, Republic of Korea.
Hye Jin Heo, Departmment of Anatomy, School of Medicine, Pusan National University, Yangsan, Republic of Korea.
Jeong Jun Park, Departemt of Anesthesiology and Pain Medicine, Korea University College of Medicine, Anam Hospital, Seoul, Republic of Korea.
Hee Sam Na, Department of Oral Microbiology, School of Dentistry, Pusan National University, Yangsan, Republic of Korea.
Dai Sik Ko, Division of Vascular Surgery, Department of Surgery, Gachon University Gil Medical Center, Incheon, Republic of Korea.
Yun Hak Kim, Department of Anatomy and Department of Biomedical Informatics, Pusan National University, 49 Busandaehak-ro, Yangsan 50612, Republic of Korea.
References
- 1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Akram M, Iqbal M, Daniyal M, et al. Awareness and current knowledge of breast cancer. Biol Res 2017; 50: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 4. Geurts YM, Witteveen A, Bretveld R, et al. Patterns and predictors of first and subsequent recurrence in women with early breast cancer. Breast Cancer Res Treat 2017; 165: 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harbeck N, Sotlar K, Wuerstlein R, et al. Molecular and protein markers for clinical decision making in breast cancer: today and tomorrow. Cancer Treat Rev 2014; 40: 434–444. [DOI] [PubMed] [Google Scholar]
- 6. Janes H, Pepe MS, Bossuyt PM, et al. Measuring the performance of markers for guiding treatment decisions. Ann Intern Med 2011; 154: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benes P, Vetvicka V, Fusek M. Cathepsin D–many functions of one aspartic protease. Crit Rev Oncol Hematol 2008; 68: 12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vignon F, Capony F, Chambon M, et al. Autocrine growth stimulation of the MCF 7 breast cancer cells by the estrogen-regulated 52 K protein. Endocrinology 1986; 118: 1537–1545. [DOI] [PubMed] [Google Scholar]
- 9. Ashraf Y, Mansouri H, Laurent-Matha V, et al. Immunotherapy of triple-negative breast cancer with cathepsin D-targeting antibodies. J Immunother Cancer 2019; 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berchem G, Glondu M, Gleizes M, et al. Cathepsin-D affects multiple tumor progression steps in vivo: proliferation, angiogenesis and apoptosis. Oncogene 2002; 21: 5951–5955. [DOI] [PubMed] [Google Scholar]
- 11. Glondu M, Liaudet-Coopman E, Derocq D, et al. Down-regulation of cathepsin-D expression by antisense gene transfer inhibits tumor growth and experimental lung metastasis of human breast cancer cells. Oncogene 2002; 21: 5127–5134. [DOI] [PubMed] [Google Scholar]
- 12. Laurent-Matha V, Maruani-Herrmann S, Prebois C, et al. Catalytically inactive human cathepsin D triggers fibroblast invasive growth. J Cell Biol 2005; 168: 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodman WA, Havran HL, Quereshy HA, et al. Estrogen receptor α loss-of-function protects female mice from DSS-induced experimental colitis. Cell Mol Gastroenterol Hepatol 2018; 5: 630–633e631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bannoud N, Carvelli FL, Troncoso M, et al. Cation-dependent mannose-6-phosphate receptor expression and distribution are influenced by estradiol in MCF-7 breast cancer cells. PLoS One 2018; 13: e0201844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pranjol MZI, Gutowski NJ, Hannemann M, et al. Cathepsin D non-proteolytically induces proliferation and migration in human omental microvascular endothelial cells via activation of the ERK1/2 and PI3K/AKT pathways. Biochim Biophys Acta Mol Cell Res 2018; 1865: 25–33. [DOI] [PubMed] [Google Scholar]
- 16. Tabish TA, Pranjol MZI, Horsell DW, et al. Graphene oxide-based targeting of extracellular cathepsin D and cathepsin L as a novel anti-metastatic enzyme cancer therapy. Cancers (Basel) 2019; 11: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 2009; 339: b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010; 25: 603–605. [DOI] [PubMed] [Google Scholar]
- 19. Kim JS, Pak K, Goh TS, et al. Prognostic value of microRNAs in coronary artery diseases: a meta-analysis. Yonsei Med J 2018; 59: 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sacks HS, Berrier J, Reitman D, et al. Meta-analyses of randomized controlled trials. N Engl J Med 1987; 316: 450–455. [DOI] [PubMed] [Google Scholar]
- 21. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994; 50: 1088–1101. [PubMed] [Google Scholar]
- 22. Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The Cochrane Collaboration. Review manager (version 5.3) [computer software]. Copenhagen, Denmark: The Cochrane Collaboration, 2014. [Google Scholar]
- 24. Namer M, Ramaioli A, Fontana X, et al. Prognostic value of total cathepsin D in breast tumors. A possible role in selection of chemoresistant patients. Breast Cancer Res Treat 1991; 19: 85–93. [DOI] [PubMed] [Google Scholar]
- 25. Granata G, Coradini D, Cappelletti V, et al. Prognostic relevance of cathepsin D versus oestrogen receptors in node negative breast cancers. Eur J Cancer 1991; 27: 970–972. [DOI] [PubMed] [Google Scholar]
- 26. Duffy MJ, Reilly D, Brouillet JP, et al. Cathepsin D concentration in breast-cancer cytosols - correlation with disease-free interval and overall survival. Clin Chem 1992; 38: 2114–2116. [PubMed] [Google Scholar]
- 27. Domagala W, Striker G, Szadowska A, et al. Cathepsin-D in invasive ductal nos breast-carcinoma as defined by immunohistochemistry - no correlation with survival at 5 years. Am J Pathol 1992; 141: 1003–1012. [PMC free article] [PubMed] [Google Scholar]
- 28. Pujol P, Maudelonde T, Daures JP, et al. A prospective-study of the prognostic value of cathepsin-D levels in breast-cancer cytosol. Cancer 1993; 71: 2006–2012. [DOI] [PubMed] [Google Scholar]
- 29. Winstanley JH, Leinster SJ, Cooke TG, et al. Prognostic-significance of cathepsin-D in patients with breast-cancer. Br J Cancer 1993; 67: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Isola J, Weitz S, Visakorpi T, et al. Cathepsin-D expression detected by immunohistochemistry has independent prognostic value in axillary node-negative breast-cancer. J Clin Oncol 1993; 11: 36–43. [DOI] [PubMed] [Google Scholar]
- 31. O’Donoghue AE, Poller DN, Bell JA, et al. Cathepsin-D in primary breast-carcinoma - adverse prognosis is associated with expression of cathepsin-D in stromal cells. Breast Cancer Res Treat 1995; 33: 137–145. [DOI] [PubMed] [Google Scholar]
- 32. Joensuu H, Toikkanen S, Isola J. Stromal cell cathepsin-D expression and long-term survival in breast-cancer. Br J Cancer 1995; 71: 155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Foekens JA, Look MP, Bolt-de Vries J, et al. Cathepsin-D in primary breast cancer: prognostic evaluation involving 2810 patients. Br J Cancer 1999; 79: 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Harbeck N, Dettmar P, Thomssen C, et al. Risk group discrimination in node-negative breast cancer using invasion and proliferation markers: 6-year median follow up. Br J Cancer 1999; 80: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kute TE, Russell GB, Zbieranski N, et al. Prognostic markers in node-negative breast cancer: a prospective study. Cytometry B Clin Cytom 2004; 59: 24–31. [DOI] [PubMed] [Google Scholar]
- 36. Rodriguez J, Vazquez J, Corte MD, et al. Clinical significance of cathepsin D concentration in tumor cytosol of primary breast cancer. Int J Biol Marker 2005; 20: 103–111. [DOI] [PubMed] [Google Scholar]
- 37. Mazouni C, Bonnier P, Romain S, et al. A nomogram predicting the probability of primary breast cancer survival at 2-and 5-years using pathological and biological tumor parameters. J Surg Oncol 2011; 103: 746–750. [DOI] [PubMed] [Google Scholar]
- 38. Mazouni C, Romain S, Bonnier P, et al. Prognostic significance of tumor-related proteases as a function of the estrogen receptor status. Cancer Biol Ther 2011; 11: 277–283. [DOI] [PubMed] [Google Scholar]
- 39. Jacobson-Raber G, Lazarev I, Novack V, et al. The prognostic importance of cathepsin D and E-cadherin in early breast cancer: a single-institution experience. Oncol Lett 2011; 2: 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen S, Chen CM, Yu KD, et al. A prognostic model to predict outcome of patients failing to achieve pathological complete response after anthracycline-containing neoadjuvant chemotherapy for breast cancer. J Surg Oncol 2012; 105: 577–585. [DOI] [PubMed] [Google Scholar]
- 41. Huang L, Liu ZB, Chen S, et al. A prognostic model for triple-negative breast cancer patients based on node status, cathepsin-D and Ki-67 index. PLoS One 2013; 8: e83081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giatromanolaki A, Sivridis E, Kalamida D, et al. Transcription factor EB expression in early breast cancer relates to lysosomal/autophagosomal markers and prognosis. Clin Breast Cancer 2017; 17: e119–e125. [DOI] [PubMed] [Google Scholar]
- 43. Granata G, Coradini D, Cappelletti V, et al. Prognostic relevance of cathepsin D versus estrogen-receptors in node negative breast cancers. Eur J Cancer 1991; 27: 970–972. [DOI] [PubMed] [Google Scholar]
- 44. Tetu B, Brisson J, Cote C, et al. Prognostic-significance of cathepsin-D expression in node-positive breast-carcinoma - an immunohistochemical study. Int J Cancer 1993; 55: 429–435. [DOI] [PubMed] [Google Scholar]
- 45. Ferno M, Baldetorp B, Borg A, et al. Cathepsin-D, both a prognostic factor and a predictive factor for the effect of adjuvant tamoxifen in breast-cancer. Eur J Cancer 1994; 30a: 2042–2048. [DOI] [PubMed] [Google Scholar]
- 46. Jahkola T, Toivonen T, von Smitten K, et al. Cathepsin-D, urokinase plasminogen activator and type-1 plasminogen activator inhibitor in early breast cancer: an immunohistochemical study of prognostic value and relations to tenascin-C and other factors. Br J Cancer 1999; 80: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Billgren AM, Rutqvist LE, Johansson H, et al. The role of cathepsin D and PAI-1 in primary invasive breast cancer as prognosticators and predictors of treatment benefit with adjuvant tamoxifen. Eur J Cancer 2000; 36: 1374–1380. [DOI] [PubMed] [Google Scholar]
- 48. Markicevic M, Kanjer K, Mandusic V, et al. Cathepsin D as an indicator of clinical outcome in early breast carcinoma during the first 3 years of follow-up. Biomark Med 2013; 7: 747–758. [DOI] [PubMed] [Google Scholar]
- 49. Tazhibi M, Fayaz M, Mokarian F. Detection of prognostic factors in metastatic breast cancer. J Res Med Sci 2013; 18: 283–290. [PMC free article] [PubMed] [Google Scholar]
- 50. Sun T, Jiang DQ, Zhang L, et al. Expression profile of cathepsins indicates the potential of cathepsins B and D as prognostic factors in breast cancer patients. Oncol Lett 2016; 11: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Namer M, Ramaioli A, Fontana X, et al. Prognostic value of total cathepsin-D in breast-tumors - a possible role in selection of chemoresistant patients. Breast Cancer Res Treat 1991; 19: 85–93. [DOI] [PubMed] [Google Scholar]
- 52. Kandalaft PL, Chang KL, Ahn CW, et al. Prognostic significance of immunohistochemical analysis of cathepsin D in low-stage breast cancer. Cancer 1993; 71: 2756–2763. [DOI] [PubMed] [Google Scholar]
- 53. Rouanet P, Roger P, Rousseau E, et al. HER2 overexpression a major risk factor for recurrence in pT1a-bN0M0 breast cancer: results from a French regional cohort. Cancer Med 2014; 3: 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Joerger M, Thürlimann B, Huober J. Small HER2-positive, node-negative breast cancer: who should receive systemic adjuvant treatment? Ann Oncol 2010; 22: 17–23. [DOI] [PubMed] [Google Scholar]
- 55. Mirza AN, Mirza NQ, Vlastos G, et al. Prognostic factors in node-negative breast cancer: a review of studies with sample size more than 200 and follow-up more than 5 years. Ann Surg 2002; 235: 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nagaraj G, Ma CX. Adjuvant chemotherapy decisions in clinical practice for early-stage node-negative, estrogen receptor-positive, HER2-negative breast cancer: challenges and considerations. J Natl Compr Canc Netw 2013; 11: 246–251. [DOI] [PubMed] [Google Scholar]
- 57. Denduluri N, Somerfield MR, Eisen A, et al. Selection of optimal adjuvant chemotherapy regimens for human epidermal growth factor receptor 2 (HER2)-negative and adjuvant targeted therapy for HER2-positive breast cancers: an American society of clinical oncology guideline adaptation of the cancer care Ontario clinical practice guideline. J Clin Oncol 2016; 34: 2416–2427. [DOI] [PubMed] [Google Scholar]
- 58. Johnston SR, Saccanijotti G, Smith IE, et al. Changes in estrogen-receptor, progesterone-receptor, and Ps2 expression in tamoxifen-resistant human breast-cancer. Cancer Res 1995; 55: 3331–3338. [PubMed] [Google Scholar]
- 59. Chang M. Tamoxifen resistance in breast cancer. Biomol Ther 2012; 20: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rochefort H, Capony F, Augereau P, et al. The estrogen-regulated 52K-cathepsin-D in breast cancer: from biology to clinical applications. Int J Rad Appl Instrum B 1987; 14: 377–384. [DOI] [PubMed] [Google Scholar]
- 61. Hultsch S, Kankainen M, Paavolainen L, et al. Association of tamoxifen resistance and lipid reprogramming in breast cancer. BMC Cancer 2018; 18: 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Itoh T, Karlsberg K, Kijima I, et al. Letrozole-, anastrozole-, and tamoxifen-responsive genes in MCF-7aro cells: a microarray approach. Mol Cancer Res 2005; 3: 203–218. [DOI] [PubMed] [Google Scholar]
- 63. Liaudet-Coopman E, Beaujouin M, Derocq D, et al. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett 2006; 237: 167–179. [DOI] [PubMed] [Google Scholar]
- 64. Long BJ, van den Berg HW. Reduced levels of cathepsin D associated with tamoxifen resistance and estrogen independence in the ZR-75-1 human breast cancer cell line. Cancer Lett 1996; 99: 233–238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplement_Figure_1 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Figure_2 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Figure_3 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Table_1 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Table_2 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology
Supplemental material, Supplement_Table_3 for Prognostic role of high cathepsin D expression in breast cancer: a systematic review and meta-analysis by Junho Kang, Yeuni Yu, Seongdo Jeong, Hansong Lee, Hye Jin Heo, Jeong Jun Park, Hee Sam Na, Dai Sik Ko and Yun Hak Kim in Therapeutic Advances in Medical Oncology