Abstract
Neuronal development is a complex multistep process that shapes neurons by progressing though several typical stages, including axon outgrowth, dendrite formation, and synaptogenesis. Knowledge of the mechanisms of neuronal development is mostly derived from the study of animal models. Advances in stem cell technology now enable us to generate neurons from human induced pluripotent stem cells (iPSCs). Here we provide a mass spectrometry-based quantitative proteomic signature of human iPSC-derived neurons, i.e., iPSC-derived induced glutamatergic neurons and iPSC-derived motor neurons, throughout neuronal differentiation. Tandem mass tag 10-plex labeling was carried out to perform proteomic profiling of cells at different time points. Our analysis reveals significant expression changes (FDR < 0.001) of several key proteins during the differentiation process, e.g., proteins involved in the Wnt and Notch signaling pathways. Overall, our data provide a rich resource of information on protein expression during human iPSC neuron differentiation.
Keywords: iPSC, human development, neuronal differentiation, neuronal development, neuroproteomics, proteome, quantitative proteomics, mass spectrometry, tandem mass tags, TMT 10-plex
Introduction
The human brain is a complex system with different regions and cell types, having billions of cells and trillions of synapses.1,2 This diversity presents a great challenge in understanding the molecular and cellular function of this organ. As recent studies have revealed, perturbation of brain development underlies many neurological disorders, such as autism and schizophrenia; however, much of our current knowledge is derived from the rodent brain.3−5 Human neural development remains difficult to study given the ethical constraints with use of primary human brain tissues, together with the paucity of high-quality post-mortem tissue. Moreover, the degree of cell and tissue heterogeneity, in combination with complex developmental and environmental factors, further complicates (human) neuronal research.6,7 One approach with great promise to study neurological disorders is the use of induced pluripotent stem cells (iPSCs) from, for example, human fibroblasts.8 Ever since the first report on iPSCs, major efforts have been directed toward developing differentiation protocols to induce neurons.9,10 Given the rapid developments in the field of iPSC-derived neurons, a comprehensive understanding of the mechanism underlying iPSC differentiation toward neurons is required. Neuronal development is coordinated by morphogens and neurogenic factors that can be captured in vitro using iPSCs.11,12 Over the last years, major advances in iPSC differentiation improved the generation of a homogeneous population of neurons, which has been used to study various neurological disorders.13 Although many of these regulatory pathways involved in neuronal development have been studied in genomic and transcriptomic studies, their mechanisms at protein levels have not.14 Since proteins are the final molecular effectors of cellular processes and their perturbation is linked to pathological states, their investigation is essential. Multiple protocols exist for generating neurons from iPSCs. Here, to monitor the differentiation process of iPSC-derived neurons by high-resolution proteomics, we adapted two different approaches, often used to model neuronal development and neurological disorders.15−18 Forced expression of a single neurogenic transcription factor (Ngn2) causes rapid differentiation of human iPSCs into functional excitatory cortical neurons (iN cells).19 This approach shows, within 10 days, rapid and reproducible production of a homogeneous population of glutamatergic neurons. In addition, extrinsic-factor-based strategies of different morphogens, such as Wnt, fibroblast growth factor (FGF), retinoic acid (RA), and Sonic Hedgehog (SHH), can be used to generate neuronal subtypes.20 Here, the course of differentiation is a three-step process, with neural crest cell activation by dual SMAD inhibition, caudalization by RA signaling, and ventralization by SHH signaling. We will refer to these neurons as motor neurons (MNs). Both approaches can be used as model systems to study the molecular mechanisms during neuronal development.
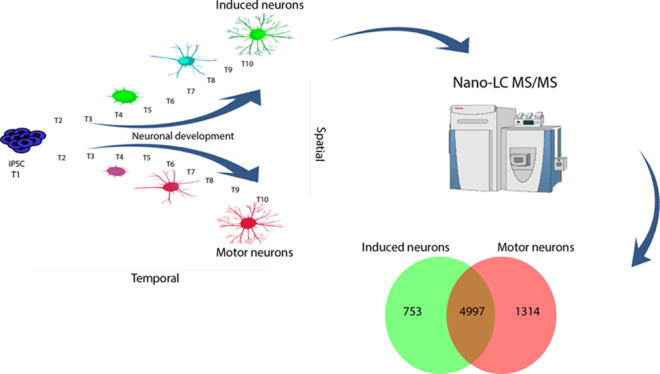
The research presented here quantitatively probes proteome changes during differentiation of iN cells and MNs at 10 different time points (Figure 1). We observe a two-step resetting of the global proteome, showing abundant proteins in iPSCs decreasing and neuronal proteins increasing over time. We highlight both well-established and novel proteins up- and downregulated during differentiation. Additionally, we show the relative fold change of proteins associated with signaling pathways such as Wnt, Notch, and Hedgehog signaling. Finally, we illustrate which proteins are specifically changing during differentiation of iPSCs into either iN cells or MNs.
Figure 1.
Workflow of MS-based quantitative proteomics during neuronal differentiation. Differentiation of iPSCs toward iN cells was performed using doxycycline-induced expression of Ngn2. Differentiation of MNs was performed using the action of small molecules for neural induction and cell fate determination. Proteins extracted at 10 time points from 2 biological replicates were digested and tandem mass tag (TMT) 10-plex labeled. Peptides were combined and fractionated using high-pH fractionation. The resulting fractions were analyzed by high-resolution nano-LC–MS/MS, and quantification was achieved using TMT 10-plex isobaric labeling.
Materials and Methods
Experimental Design and Statistical Rationale
For LC–MS/MS analysis, two biological replicates were used for both iN and MN differentiations. Samples derived from 10 time points were extracted and time point 1 was used for normalization within each biological replicate. In total, 40 samples were collected and processed further with high-pH fractionation. The 10 time point samples were tryptic digested into peptides, TMT-labeled, and mixed at equal ratios. Each mix was processed further with high-pH fractionation and each fraction was run on an Orbitrap Fusion mass spectrometer.
Cell Culture
iPSC Generation
The Medical Ethical Committee of the University Medical Center Utrecht granted approval for iPSC line generation. Generation of iPSCs was performed using a previously established protocol.21 Briefly, skin biopsies from healthy individuals were taken and maintained in mouse embryonic fibroblast (MEF) medium containing DMEM GlutaMAX (Life Technologies), 10% fetal bovine serum (Sigma-Aldrich), and 1% penicillin/streptomycin (Life Technologies). The iPSCs were generated by lentiviral transduction expressing OCT4, KLF4, SOX2, and c-MYC in MEF medium containing 4 mg/mL hexadimethrine bromide (Sigma). After 24 h of incubation, cells were cultured in MEF medium for another 5 days. Subsequently, cells were detached with trypsin–EDTA (Life Technologies) and cultured in a 10 cm dish containing irradiated MEFs in human embryonic stem cell (huES) medium containing DMEM-F12 (Life Technologies), knockout 10% serum replacement (Life Technologies), 1% penicillin/streptomucin (Life Technologies), 2% l-glutamine (Life Technologies), 0.1% β-mercaptoethanol (Merck Millipore), and 20 ng/mL recombinant human fibroblast growth factor-basic (Life Technologies). After 3–6 weeks, colonies were picked manually and maintained in huES medium on irradiated MEFs for another 3–6 weeks. The iPSCs were passaged using Accutase (Innovative Cell Technologies) and cultured feeder-free on Geltrex (Life Technologies) coated dishes in mTeSR1 medium (STEMCELL technologies). Medium change was done every other day.
Virus Generation
Lentiviruses were produced as described previously.22 HEK293T cells plated on a 500 cm2 dish were cotransfected using MAXPEI solution with 35 μg of pMD2.G, 65 μg of psPAX, and 100 μg of pSIN-FUW-TeTO-Ngn2-P2A-EGFP-T2A-Puromycin or pSIN-FUW-M2rtTA in OPTI-MEM (Thermo Fisher Scientific). Six hours after transfection, the medium was replaced with OPTI-MEM supplemented with penicillin/streptomycin 1% (Thermo Fisher Scientific). Lentiviral particles were harvested 48 h after transfection, concentrated by tangential flow filtration using a 100 kDa cutoff spin filter (Millipore), and resuspended in phosphate-buffered saline (PBS).
Differentiation
Generation of iN cells was performed using a slightly modified version of the protocol described by Zhang et al., in 2013.19 In short, on day 0, iPSCs were treated with Accutase and plated at a density of 50 × 103 cells/well in a 24-well plate on top of a Matrigel (BD Biosciences) coated coverslip in mTeSR containing 10 μM Y27632 (Miltenyibiotec). On day 1, the medium was changed to mTeSR supplemented with 2.5 μL of lentivirus and 8 μg/μL Polybrene (Sigma-Aldrich). On day 2, the medium was replaced with N2/DMEM/F12/NEAA (Invitrogen) supplemented with 10 μg/L BDNF (R&D Systems), 10 μg/L human NT-3 (ReproTech), 0.2 mg/L mouse laminin (Invitrogen), and 2 g/L doxycycline (Clontech). On day 3, 1 mg/L puromycin (Sigma-Aldrich) was added to the cells. On day 4, coverslips with neurons were transferred on top of a 12-well plate cultured with glial cells in neurobasal medium containing B27 (ThermoFisher), Glutamax (Invitrogen), BDNF, NT-3, doxycycline, and 2 g/L Ara-C (Sigma-Aldrich). Every other day thereafter, 50% of the medium was changed. At predetermined time points, cells were fixed for immunohistochemistry or for proteomics approaches.
MN differentiation was performed using a slightly modified version of a previously described protocol.20 Briefly, on day 0, iPSCs were dissociated with Accutase and resuspended in differentiation medium containing DMEM F-12, Neurobasal (1:1; v/v), N2 supplement (Life Technologies), B27 without vitamin A (Life Technologies), 1% penicillin/streptomucin, ascorbic acid (0.5 μM, Sigma-Aldrich), and 5 μM Y27632 (STemGent). Embryoid body (EB) formation was accompanied by a standardized microwell assay.23 IPSCs were seeded at a density of 150 cells/microwell in differentiation medium. Chir-99021 (Tocris), LDN193189, SB-431542, smoothened agonist (SAG, Calbiochem), retinoic acid (RA, Sigma-Aldrich), DAPT (Tocris), BDNF (Peprotech), and GDNF (Peprotech) were added at indicated time points and concentrations. Medium was changed every other day. After 2–3 days, EBs were flushed out of the microwell and transferred to a nonadherent 10 cm Petri dish (Greiner Bio-one). On day 15, EBs were dissociated into single cells using papain (Worthington Biochemical Corp.) and DNase (Worthington Biochemical Corp.). Cells were plated on PDL (20 μg/mL, Sigma-Aldrich) and laminin (5 μg/mL, Invitrogen) coated coverslips at 60–70% confluency.
Glial Cells
Newborn mouse forebrain homogenates were incubated with trypsin for 5 min at 37 °C. Following harsh trituration, cells were plated in MEM Alpha medium containing 20% d-glucose, 10% FBS, and 1% penicillin/streptomucin. After three passages, glial cells were used for coculture purposes.
Immunofluorescence
At the time points of interest, cells were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature (RT) and washed with PBS. Cells were blocked for 45 min at RT in blocking solution [0.1% Tween in PBS containing 2% bovine serum albumin (BSA) and 20% goat serum]. Cells were washed three times in PBS, and primary antibody was incubated for 1 h at RT. After washing the cells three times with PBS, secondary antibody was applied with Hoechst or DAPI (Invitrogen) and incubated for 1 h at RT in darkness. Primary and secondary antibodies were mixed in staining solution (0.1% Tween in PBS containing 5% goat serum). Cells were washed again with PBS, fixed with ProLong Gold, and mounted on glass slides. The following commercial antibodies were used: rabbit anti-tubulin-β3 (1:2000) (Sigma), mouse anti-Isl-1 (1:1000) (DSHB), rabbit anti-FOXG1 (1:1000) (Abcam), and mouse anti-TuJ1 (1:2000) (Covance). Alexa 488 and Alexa 546 conjugated secondary antibodies were obtained from Invitrogen.
Western Blot Analysis
Cells were washed in cold PBS and scraped into cold lysis buffer [20 mM Tris (pH 8), 150 mM KCl, 10% glycerol, 1% Triton X-100, 1% NP-40, phosphatase inhibitor cocktail (Sigma), complete protease inhibitor cocktail (Roche)]. After incubation on ice for 15 min, lysates were cleared by centrifugation. Protein concentration in the supernatant was measured using a BCA protein assay (Pierce). The proteins were heated in NuPAGE LDS sample buffer containing 10% 2-mercaptoethanol for 10 min at 70 °C. Proteins were separated using SDS–PAGE (Novex NuPAGE 4–12% Bis-Tris precast gel, Invitrogen) and transferred onto a nitrocellulose membrane (Amersham). For normalization, lanes were quantified using REVERT Total Protein Stain (LI-COR). Membranes were incubated in blocking buffer (TBS, 0.05% Tween, and 5% milk powder) for 30 min at RT, followed by incubation with primary antibodies [mouse anti-NCAM1 (1:2000; Millipore, clone 11G7.1) and mouse anti-α-tubulin (1:2000; Sigma, clone B-5-1-2) in blocking buffer] overnight at 4 °C. After washes with TBS-T, membranes were incubated with appropriate peroxidase-conjugated secondary antibodies in TBS for 1 h at RT. Blots were washed in TBS and developed using the Supersignal West Dura Extended Duration Substrate detection system (Pierce) before being visualized using a FluorChem E Imager (ProteinSimple). Membranes were stripped and reprobed with mouse anti-TUBB3 (1:1000; BioLegend) for TUBB3 measurements. For quantification, intensity measurements of protein bands were performed with ImageJ.
Sample Preparation for MS Analysis
Samples (n = 2) for each approach were collected at the indicated time points (10 time points) for MS analysis as follows: cells were lysed in lysis buffer containing 8 M urea, 50 mM ammonium bicarbonate, and 1× cOmplete Mini Protease Inhibitor (Roche). The lysate was sonicated on ice using a Bioruptor (Diagenode) and centrifuged at 2500g for 10 min at 4 °C. Supernatant with the proteins was reduced with 4 mM dithiothreitol at 56 °C for 30 min and alkylated with 8 mM iodoacetamide at RT for 30 min in darkness. The lysate was enzymatically predigested with Lys-C (1:75; Wako) incubation for 4 h at 37 °C. The mixture was 4-fold diluted with ammonium bicarbonate and digested with trypsin (1:100; Promega) at 37 °C. The sample was quenched by acidification with formic acid (FA, final concentration 10%), and peptides were desalted using a Sep-Pak C18 column (Waters). Peptides were dried in vacuum and resuspended in 50 mM triethylammonium bicarbonate at a final concentration of 5 mg/mL.
Tandem Mass Tag (TMT) 10-Plex Labeling
Aliquots of ∼100 μg of each sample were chemically labeled with TMT reagents (Thermo Fisher) according to Table 1. Peptides were resuspended in 80 μL of resuspension buffer containing 50 mM HEPES buffer and 12.5% acetonitrile (ACN, pH 8.5). TMT reagents (0.8 mg) were dissolved in 80 μL of anhydrous ACN of which 20 μL was added to the peptides. Following incubation at RT for 1 h, the reaction was quenched using 5% hydroxylamine in HEPES buffer for 15 min at RT. The TMT-labeled samples were pooled at equal protein ratios followed by vacuum centrifuge to near dryness and desalting using Sep-Pak C18 cartridges.
Table 1. TMT 10-Plex Label Reagents Each Corresponding to a Time Point of Differentiation.
| sample | label reagent | sample | label reagent |
|---|---|---|---|
| T1 | TMT10-126 | T6 | TMT10-129N |
| T2 | TMT10-127N | T7 | TMT10-129C |
| T3 | TMT10-127C | T8 | TMT10-130N |
| T4 | TMT10-128N | T9 | TMT10-130C |
| T5 | TMT10-128C | T10 | TMT10-131 |
Off-Line Basic pH Fractionation
We fractionated and pooled samples using basic pH reverse-phase HPLC. Samples were solubilized in buffer A (5% ACN, 10 mM ammonium bicarbonate, pH 8.0) and subjected to a 50 min linear gradient from 18% to 45% ACN in 10 mM ammonium bicarbonate pH 8 at flow rate of 0.8 mL/min. We used an Agilent 1100 pump equipped with a degasser, a photodiode array (PDA) detector, and an Agilent 300 Extend C18 column (5 μm particles, 4.6 mm i.d., and 20 cm in length). The peptide mixture was fractionated into 96 fractions and consolidated into 24 fractions. Samples were acidified with 10% formic acid and vacuum-dried, followed by redissolving with 5% formic acid/5% ACN for LC–MS/MS processing.
Mass Spectrometry Analysis
Mass spectrometry was performed on Orbitrap Fusion (for iN cells) and Orbitrap Fusion Lumos (for MNs) mass spectrometers (Thermo Fisher Scientific) coupled to an Agilent 1290 HPLC system (Agilent Technologies). Peptides were separated on a double-frit trap column of 20 mm × 100 μm inner diameter (ReproSil C18, Dr Maisch GmbH, Ammerbuch, Germany). This was followed by separation on a 40 cm × 50 μm inner diameter analytical column (ReproSil Pur C18-AQ, Dr Maisch GmbH, Ammerbuch, Germany). Both columns were packed in-house. Trapping was done at 5 μL/min in 0.1 M acetic acid in H2O for 10 min, and the analytical separation was done at 100 nL/min for 2 h by increasing the concentration of 0.1 M acetic acid in 80% acetonitrile (v/v). The instrument was operated in a data-dependent mode to automatically switch between MS and MS2 for MNs or MS and MS3 for iN cells. Full-scan MS spectra were acquired in the Orbitrap from m/z 350 to 1500 with a resolution of 60 000 full half-maximum width (fhmw), automatic gain control (AGC) target of 200 000, and maximum injection time of 50 ms. For the MS2 analysis, the 10 most intense precursors at a threshold above 5000 were selected with an isolation window of 1.2 Th after accumulation to a target value of 30 000 (maximum injection time was 115 ms). Fragmentation was carried out using higher-energy collisional dissociation (HCD) with a collision energy of 38% and activation time of 0.1 ms. Fragment ion analysis was performed on the Orbitrap with a resolution of 60 000 fhmw and a low-mass cutoff setting of 120 Th. For the MS3 analysis, MS2 was first performed with CID fragmentation on the top 10 most intense ions with an AGC target of 10 000 and isolation window of 0.7 Th, followed by a MS3 scan for each MS2 scan with HCD fragmentation with 35% collision energy. The MS isolation window was set to 2 m/z and the AGC target to 100 000, and the maximum injection time was 120 ms. When using TMT-labeling, there is a known common drawback, the so-called coisolation issue. Here, when a peptide is selected for fragmentation, coeluting peptides are present in the isolation window. This leads to a mixed population of peptides that disturbs the ratios of the original reporter ions. Normally this is skewed toward unity, since the majority of the proteins do not change between samples.24 The ratios acquired by the MS2 method are more compressed compared to the MS3 method. Therefore, data analysis and statistics were performed separately for MS2 and MS3 to identify differentially expressed proteins.
Data Processing
Mass spectra were processed using Proteome Discoverer software (version 2.1, Thermo Scientific). the peak list was searched using the Swissprot database (version 2014_08, with 20 156 entities) with the search engine Mascot (version 2.3, Matrix Science). The following parameters were used: the enzyme was specified as trypsin and allowed up to two missed cleavages. Taxonomy was chosen for Homo sapiens and precursor mass tolerance was set to 50 ppm with 0.05 Da fragment mass tolerance for MS2 analysis or 0.6 Da for MS3 analysis. TMT tags on lysine residues and peptide N termini and oxidation of methionine residues were set as dynamic modifications, while carbamidomethylation on cysteine residues was set as a static modification. For the reporter ion quantification, integration tolerance was set to 20 ppm with the most confident centroid method. Results were filtered with a Mascot score of at least 20, and Percolator was used to adjust the peptide-spectrum matches (PSMs) to a false discovery rate (FDR) below 1%. A FDR of 1% was applied at the level of proteins and peptides. Finally, peptides with less than six amino acid residues were discarded.
Data Visualization
Perseus software was used to generate the plots and heatmaps and to calculate Pearson correlations. Normalized protein and peptide abundances were extracted from PD2.2 and further analyzed using Perseus software. For quantitative analysis, the TMT reporter intensity values of proteins at each time point were normalized to the reference intensity value of T1 (reference), within each replicate. The ratios of each replicate were then log2 transformed. All the peptide ratios were normalized against the median. At least one peptide was required to identify a protein. A volcano plot for each time point was generated, and up- or downregulated proteins were considered significant with FDR < 0.05. Z scores were used to generate heatmaps. Functional analysis to enrich to GO, MF, and CO terms was done using the GProX open-source software package.25 Clustering parameters such as fuzzification, regulation threshold, and number of clusters were set to 2.00, upper limit = −0.58/lower limit = 0.58, and 3, respectively. For the identification of time-specific proteins between iN and MNs, volcano plots were generated comparing the two groups for each time point, and proteins were considered to be significant with a fold change cutoff ≥1.5 and a p-value ≤0.1. Furthermore, protein classification was performed using the PANTHER26 classification system and GO analysis, and classification of transcription factors and cytoskeletal proteins was done using the DAVID database. The p-values are corrected for multiple hypothesis testing using the Benjamin–Hochberg method. In addition, Reactome pathway database analysis was used to identify pathways enriched in several clusters.
Data Availability
All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD013399
Results
Differentiation toward iN Cells and MNs
Differentiation into iN cells was performed by doxycycline-induced expression of neurogenin-2 (Ngn2).19 At day 7, the iNs showed mature neuronal morphology and were positive for the neuronal marker TuJ1 and the cortical marker FOXG1 (Figure 2A). In addition, these cells were negative for the MN marker ISLET1. MNs were generated by the combined actions of small molecules for neural induction and cell fate determination20 (Figure 2B). At day 20, cells were positive for βIII-tubulin (>80%) and ISL1 (∼40%) markers. For mass spectrometry-based quantitative proteomics, we performed TMT 10-plex labeling of the tryptic peptides originating from 10 distinct time points within the differentiation process of either iN cells or MNs, resulting in a total of two biological replicates (Figure 1). After separation by high-pH fractionation, the labeled peptides were analyzed with high-resolution tandem mass spectrometry (LC–MS/MS).
Figure 2.

Bright-field images and immunocytochemistry of iN cells and MNs at indicated time points. (A). Bright-field images of iPSCs at day 1 and iN cells at day 7 and immunostainings for neuronal marker (TuJ1), cortical marker (FOXG1), and motor neuron marker (Islet1). Scale bars: day 1, 100 μm; day 7, 40 μm. (B). Bright-field images of iPSCs at day 1, embryonic bodies at day 9, and MNs at day 25. Immunocytochemistry staining for neuronal marker (Tubulin beta 3), motor neuron marker (Islet1), and nuclear marker (Dapi). Scale bars: day 1, 100 μm; day 9, 200 μm; day 25, 40 μm.
MS-Based Quantitative Proteomics
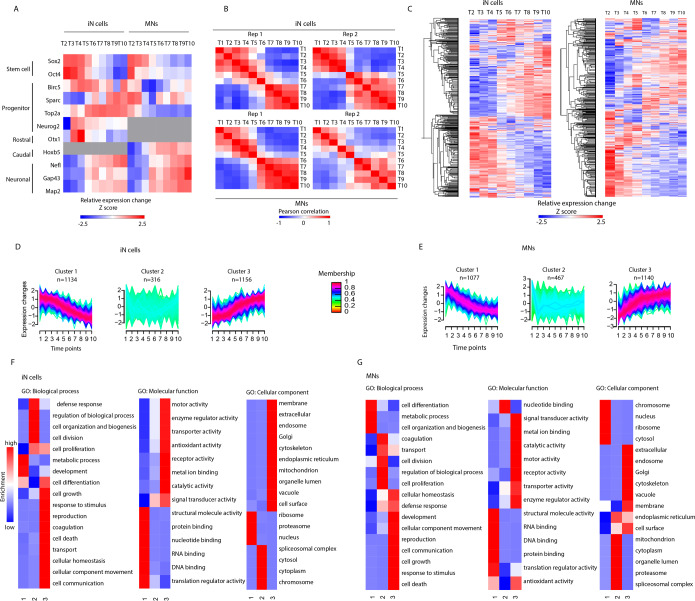
We identified 5750 and 6311 protein groups with a false discovery rate of <1%, in iN cells and MNs, respectively [Figure S1A,B, Supporting Information (SI)]. The proteins that were identified in just one of the replicates within each approach were discarded. The relative abundances of the quantified proteins span more than 4 orders of magnitude, indicating a broad dynamic range in our quantitative measurement. Tissue enrichment analysis of the 25% most abundant proteins against the whole human proteome background revealed enrichment for “Cajal–Retzius cell”, “fetal brain cortex”, and “epithelium” (Figure S1C, SI). The expression of Cajal–Retzius cells is not surprising, since these cells are involved in the organization of brain development.27 The enrichment associated with epithelium may be caused by the origin of the fibroblast. Finally, the annotation of protein class for all identified proteins revealed a wide coverage, including typically lowly abundant protein classes such as transcription factors and storage proteins (Figure S1D, SI). Furthermore, the ratio distribution after normalization was observed between biological replicates both for iN cells and MN differentiation (Figure S1E, SI). The TMT reporter intensity values of proteins identified in each time point were normalized to the reference intensity value of these proteins before initiation of differentiation (T1). As a consequence, all data throughout the study reflects a ratio change relative to T1. After global analysis of our proteomics data, we set out to confirm the quality of the neuronal differentiation. We investigated known stem cell markers (e.g., SOX2 and OCT4) that indeed decreased during differentiation while, as expected, several neuronal markers (e.g., NEFL, GAP43, and MAP2) increased in both neuron types (Figure 3A).28−34 Progenitor markers (e.g., BIRC5, SPARC, and TOP2A) that are required for fetal development and regulation of cortex development show, in our data, higher expression in iN cells compared to MNs.35−38 Furthermore, the rostral marker OTX1 was only identified in iN cells and the caudal marker HOXB5 only in MNs. Finally, NEUROG2 expression was only observed in iN cells, where it is overexpressed, and previous studies showed its involvement in cortical specification.39 Taken together, these data confirm the validity of our approach, showing neuronal subtype-specific development.
Figure 3.
Proteome dynamics during neuronal differentiation. (A). Differential expression of general iPSC, progenitor, and neuronal markers represented in a heatmap. Each row represents average protein log2 ratios relative to iPSCs (T1) in both biological replicates for each approach. (B). Pearson correlation within each replicate. Note for each replicate the high correlation between early time points (T1–T5) and late ones (T6–T10). (C). Heatmap of all proteins identified in iN cells and MNs showing the expression changes along the course of differentiation for each approach relative to T1. Clusters of the protein dynamics during differentiation toward iN cells (D) and MNs (E). Three clusters with increasing, decreasing, and unchanging profiles were revealed in both differentiation approaches. An upper and lower ratio limit of log2(0.5) and log2(−0.5) was selected for inclusion into a cluster. The number of proteins is represented by n, while the membership value indicates how well each protein fits to the average cluster profile. (F, G) Gene ontology enrichment analysis of each cluster tested for over-represented biological processes (BP), molecular function (MF), and cellular component (CC) compared to unregulated proteins.
To obtain a global proteome view, changes of proteins along the course of differentiation for the two approaches were evaluated (Figure 3B,C). We observed that proteins highly expressed in iPSCs show downregulation over time, and vice versa, proteins with low expression in iPSCs increase over time. From these observations it appears that neurons are differentiated around T5 and maintain their neuronal identity onward. This distinct separation of the proteome, which happens around the intermediate stage (T5), independent of the differentiation approach, can be further emphasized by performing Pearson correlation within each replicate (Figure 3B). Here, an anticorrelation (R between −0.58 and −0.83) was found between T1 and T10, which was similarly observed during reprogramming of somatic cells to pluripotency.40 Next, we performed an unsupervised clustering of the protein expression across the time points for both cell types iN cells and MNs. (Figure 3D,E). Gene ontology (GO) enrichment analysis of these clusters (Figure 3F,G) for both iN cells and MNs respectively revealed biological processes such as cell communication, growth, and movement as being upregulated (cluster 3), while biogenesis is slowly downregulated (cluster 1). Proteins associated with cell differentiation in MNs are downregulated, whereas in iN cells, most of these proteins belong to cluster 3 and few to cluster 1. For both iN cells and MNs, closer inspection of the corresponding proteins in cluster 3 revealed pan-neuronal proteins, such as tubulin-beta-3, neurofilament, and MAP2. For iN cells and MNs, cluster 3 also reveals increased expression of proteins localized in the membrane, cytoskeleton, and several organelles, as well as molecular functions related to transport, enzymatic activity, and signaling. Cluster 1 contains nuclear, chromosomal, and ribosomal proteins that gradually decrease over time, having a function in RNA and DNA binding. Downregulation of RNA processing during differentiation was previously shown in mouse by proteomics analysis and in human by transcriptomic analysis.14,41 Cluster 2 includes a group of proteins with general terms such as regulation of biological processes or cell division.
Proteomic Changes across Time Points during Neuronal Differentiation
To identify differentially expressed proteins during neuronal differentiation in the two approaches, we compared T10 relative to T1 and performed a t test for both neuronal subtypes separately. Figure 4A,C shows the volcano plots with the most extreme comparison of T10 (mature neurons) against T1 (iPSC) for both approaches, depicting proteins with a FDR below 0.1%. This resulted in 696 significantly downregulated proteins and 686 upregulated proteins in iN cells and 706 significantly downregulated proteins and 620 upregulated proteins in MNs (Tables S1 and S2, SI). Close inspection of the proteins significantly enriched in T10 reveals general microtubule-associated proteins necessary for neuronal stabilization, outgrowth, and migration, whereas specific iPSC-enriched proteins consisted of transcription factors and proteins having a role in embryonic development. Furthermore, we highlight the top 15 proteins showing the largest fold change over time in both neuronal differentiation approaches (Figure 4B,D). As expected, proteins such as SOX2, POU5F1 (OCT4) (iPSC markers) and MAP2, DCX (neuronal markers) were enriched. Moreover, several proteins (e.g., INA, DPYSL4, and RPS19) were identified that have less established roles in the context of pluripotency and differentiation.42−44 Furthermore, a protein interaction network was visualized using Cytoscape, Genenmania plugin. Protein network analysis of these regulated proteins shows an interaction network around the proteins INA, NCAM, NEFM, and NEFL, being upregulated, and POU5F1, SOX2, DNMT3B, and DPPA4, being downregulated (Figure 4E). To further validate the utility of this data set, we compared two highly expressed proteins during neuronal development via Western blots. We validated the expression changes for NCAM and TUBB3 on Western blots for human MN and mouse Neuro2a cell line, which have many properties of neurons and have been used to study, for example, neurite outgrowth (Figure 4F). Both NCAM1 and TUBB3 expression was higher in human iPSC-derived MNs compared to iPSCs and to a lesser extent in mouse Neuro2a cells. Several of our highest top 15 upregulated proteins are increased in expression for rat hippocampal neurons as well as during human brain development, indicating that these proteins might serve as general neuronal markers (e.g., NEFL, NEFM, CNRIP1, and NAPB).
Figure 4.

Significance analysis of protein expression changes during neuronal differentiation. (A) Volcano plot illustrating differentially expressed proteins during iN cell differentiation. The −log10P value is plotted against the log2 fold change of T1 (iPSCs) and T10 (mature iN cells) with a significant threshold t test with FDR < 0.001 and S = 0.5. Red represents proteins upregulated in T10 and blue represents proteins downregulated in T10. Black represents the top 15 proteins with the highest fold change in both differentiation approaches. (B) Profile plot highlighting the top 15 significant up- and downregulated proteins in iN cells. (C) Volcano plot illustrating differentially expressed proteins during MN differentiation. The −log10P value is plotted against the log2 fold change of T1 and T10 with a significant threshold t test with FDR < 0.001 and S = 0.5. Red represents proteins upregulated in T10 and blue represents proteins downregulated in T10. Black represents the top 15 proteins with the highest fold change in both differentiation approaches. (D) Profile plot highlighting the top 15 significant up- and downregulated proteins in MN differentiation. (E) Protein network analysis showing the top 15 upregulated or downregulated proteins that interact with each other. (F) Western blot analysis of iPSCs and iPSC-derived MN (DIV14) protein lysates reveals changes in protein expression during neuronal differentiation. iPSC-MN samples were diluted to avoid smears. Graphs showing the quantifications of relative intensities of the Western blot analysis in n = 3 experiments per condition. Error bars indicate the mean ± SEM.
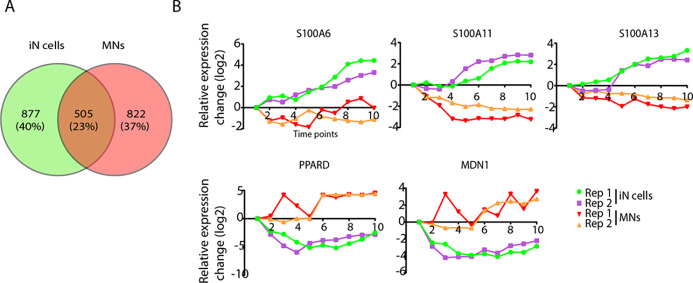
Next, we compared the significantly altered proteins during differentiation toward iN cells or MNs and observed only moderate overlap (23%) (Figure 5A). Of the significant differentially expressed proteins, 38 proteins were shown to be expressed in the opposite direction. Twenty-three proteins were significantly downregulated in iN cells and upregulated in MNs, whereas 15 proteins were upregulated in iN cells and downregulated in MNs (Table S3, SI). The differences in protein expression might suggest specificity toward a neuronal subtype or differentiation approach. Proteins that increase most in iN cells (compared to MNs), are S100A11, S100A13, and S100A6 (Figure 5B). The S100 family of calcium binding proteins was first identified in the brain, regulating processes such as cell cycle progression and differentiation.45 They have been found in subcortical structures and in a subpopulation of astrocytes.46−49 Proteins highly enriched in MNs are PPARD and MDN1. They act downstream of the Wnt-β-catenin and Notch signaling pathway.50,51 We then selected proteins exclusively identified either in iN cells or MNs that show a minimum 2-fold change in at least one time point during differentiation. We selected proteins that were identified in one or two replicates from one approach and were not identified in both replicates from the other approach (Figure S2, SI). The highly expressed protein SULF2 in iN cells is involved in brain development and neurite outgrowth in mice,52,53 but its involvement in human neuronal development is unclear. Furthermore, highly expressed proteins in MNs (e.g., PBX3 and HOXB5) regulate dorsal spinal cord development, which is in line with the neuronal origin.54
Figure 5.
Differentially expressed proteins in iN cells and MNs. (A) Venn diagram indicating the overlap between the differentially expressed proteins in iN cells or MNs (FDR 0.1%). (B) Example of proteins showing the largest differences between iN cells and MNs during differentiation.
Transcription Factors and Cytoskeletal Proteins
During neuronal differentiation, we showed that most of the up- and downregulated proteins are cytoskeletal and transcription factors (TFs), respectively. To better categorize the involvement of these proteins in neuronal development, we aimed to capture all TFs and cytoskeletal proteins identified in our proteomic data (Figure S3, SI). TFs play essential roles in both reprogramming and neuronal development and have been used to induce neuronal differentiation from multiple cell lineages, such as human fibroblasts, neuronal progenitors, and stem cells.55−57 We identified 259 regulated TFs (with a fold change cutoff ≥2 in at least in one time point for both approaches separately), of which PHC2, CRTC1, NCOA3, and SMAD3 are the most upregulated in both neuronal differentiation approaches (Figure S3A, SI). Interestingly, PHC2 has been reported to have an early developmental role in Drosophila, while its specific function in human cells has not been determined. CRTC1 is ubiquitously expressed in fetal brain and acts as a coactivator of the CREM (cAMP-responsive element binding)-dependent gene transcription pathway.58 Furthermore, CRTC1 has been associated with different neuronal functions, such as synaptic plasticity and dendritic growth in cortical neurons, and is also downregulated in Alzheimer’s disease.59,60 NCOA3 has been identified as a novel microRNA regulator of dendritogenesis in mouse cortical neurons.61 In line with this, we show that NCOA3 increases over the course of human neuronal differentiation. SMAD3, which was shown to promote neuronal differentiation in the spinal cord of zebrafish,62 was found to be more highly expressed in MNs compared to iN cells. In addition, several TFs are upregulated at early time points of neuronal differentiation. NFXL1 was upregulated at the early time points of both neuronal subtypes, while CCNT1 and LITAF were upregulated for iN cells and ZIC5, NFATC4, and ZBTB40 were upregulated for MNs. Both ZIC5 and NFATC4 are essential in neural development and survival; however, their association with early stages of caudalization, as seen here, has not been studied yet.63,64
In addition, we captured 256 cytoskeletal proteins, as shown in Figure S3B (SI), with TUBB3, TAGLN3, and MAP1LC3A being the most upregulated in both differentiation approaches. TUBB3 and MAP1LC3A are highly expressed in neurons and function in neurite formation and stabilization.65,66 TAGLN3 is an actin-binding protein involved in cytoskeletal organization.67 Interestingly, the transgelin family members TAGLN, TAGLN2, and TAGLN3 show differential expression between iN cells and MN differentiation. (Figure S3C, SI). TAGLN decreases during MN differentiation, while in iN cells, there is an increase only in the early time points. TAGLN2 has a moderate increase in iN cells and decreases in the early time points of MN differentiation. Only the third member of the transgelin family (TAGLN3) shows strong upregulation during neuronal differentiation in both approaches.
Signaling Pathways
In the last 30 years, several studies have demonstrated the importance of signaling pathways for regulating the developmental program, specifying cell fate and patterning.68,69 Some of these signaling pathways will be discussed here, displaying the expression of their associated proteins in heatmaps for both iN cells and MNs (Figure S4, SI). The TGFβ/BMP signaling is known to inhibit neuronal development by blocking the proliferation of precursor cells in the adult brain. The dual-SMAD inhibition with LDN-193189 and SB-431542 (SMADi) induces the specification of cells with neural plate identity by selectively inhibiting the TGFβ/BMP signaling pathway.70 We applied this to our MN differentiation protocol (at T2) and identified expression changes for all SMAD members, as well as SMAD interacting protein 1 (SNIP1) over time (Figure S4A, SI). SMAD3 rapidly increases at the late time points of MN differentiation, whereas the other members remain more constant. Interestingly, we observed in our iN cell differentiation protocol, which did not use SMADi, a similar expression pattern over time for most of the SMAD family members. This might indicate that NgN2 protein is an upstream regulator of TGFβ signaling. Furthermore, Wnt signaling regulates cell migration, cell polarity, and neuronal development during embryonic development and has an essential role in body axis formation, particularly in the formation of anteroposterior and dorsoventral axes.71−73 Furthermore, the combinatorial effects of Wnt, retinoic acid (RA), and Hedgehog (HH) signaling specify neuronal subtypes.74,75 Upon activation of Wnt signaling in MNs (by Chir-99021 at T2 for 4 days), elevated levels of WNT5A, WLS, ZBTB16, and NFATC4 are observed in the early time points of MNs, while not identified or very lowly expressed in iN cells (Figure S4B, SI). These proteins are involved in cell fate pattering and development.76−79 PLCB1 and PLCB4 are highly expressed in iN cells, compared to MNs. A previous study showed their high expression and differential distribution in several regions of the brain, including cortex, suggesting a specific role in different regions of the brain.80 Proteins associated with HH and RA signaling act as the posteriorizing agent for spinal cord development.81,82 Both HH and RA are induced at T3 in the MN differentiation protocol and remained until the end of the experiment. We looked for proteins associated with HH and RA and observed that the majority of these proteins are slightly elevated in MNs compared to iN cells (Figure S4C,D, SI). CSNK1E and GSK3B together with CCND2 protein are higher in expression in the MNs compared to iN cells. GSK3B has a critical role in axonal regulation and CCND2 has a function in neuronal development.83,84
The expression of cellular retinoic acid-binding protein 1 and 2 (CRABP1 and CRABP2) is increased in the MNs after addition of RA. Interestingly, CRABP1 expression was restricted to certain neuronal subtypes in the hypothalamus, suggesting a cell-type-specific function in the brain.85 Homeobox proteins, which promote the expression of posterior neural genes, are all identified in the MNs but not in the iN cells (Figure S4E, SI).86
Inhibition of Notch signaling is induced (at T4) for the MN differentiation, which is necessary to induce proneural genes.87,88 We identified 81 proteins involved in Notch signaling, of which THBS1 showed a direct response after induction in the MNs (Figure S4F, SI). THBS1 has been shown to play roles in neuronal development, such as in neurite outgrowth and cell migration.89,90 We further explored our data for enrichment of axon guidance proteins. They are important in regeneration along the anterior–posterior axis.91 We identified 36 proteins associated with axon guidance, of which the majority increases along the course of MN differentiation (Figure S4G, SI). As described previously, axonal guidance cues are often categorized as “attractive” or “repulsive”.92 NTN1 and its receptor (DCC), having attractive roles, are both increased in expression only in MNs, whereas ENAH and VASP, which function downstream of the repulsive axon guidance receptor Robo, are downregulated along the course of differentiation for both approaches.93
Neurogenesis Associated Proteins
To further investigate proteins involved in specific processes related to neuronal differentiation we next looked for enrichment of proteins associated with neurogenesis using the DAVID database.94 This analysis revealed 129 proteins for which we had a closer look at their differentiation profile, plotting their expression over time in a heatmap (Figure S5, SI). These neurogenesis-associated proteins for the most part increase their expression over time, such as DCX, CPLX2, and ELAVL3. Upregulated microtubule-associated protein DCX has been increasingly used as a marker for neurogenesis, and ELAVL3 is a neuron-specific protein.95,96 Several proteins also show downregulation, such as SLC7A5 and the transcription factor ZIC2, known to play a role in neuronal cell proliferation (neurogenesis) and early stages of central nervous system development.97,98 Additionally, ARF6 is known to regulate neuronal development in the brain via regulation of actin dynamics and synaptic plasticity. Its relatively lower expression here, however, might indicate that additional factors are needed to fully recapitulate the neuronal development.
Discussion
Our study provides insight into the remodeling of the proteome during human neuronal development. To our knowledge, this is the first comprehensive proteomic profiling of human iPSC differentiation toward neurons. The rapid neurogenesis through transcriptional activation toward iN cells and the introduction of small molecules toward MNs allowed us to identify regulatory proteins involved in differentiation. Quantitative proteomics was applied to profile the dynamic changes of proteins during neuronal differentiation by using TMT 10-plex labeling coupled with high-resolution LC–MS/MS, which resulted in the identification of 7230 proteins. There are several challenges in the study of cell differentiation with MS-based proteomics. In this study, we collected different time points (temporal resolution) during neuronal development to visualize proteins having specific expression profiles. However, cells are not completely synchronized at each time point, leading to potential dilution of differentiation stage in the same culture dish. Here, the protein expression profiles are therefore an average of multiple cells. Despite this, we do see time-point-specific clusters and progenitor markers appearing at early time points, as expected. Ideally, to achieve a high temporal resolution, single-cell proteomic approaches are needed in the future.
Here, we found a proteome-wide change in expression, which occurs in a two-step fashion, revealing a clear switch in protein expression levels, halfway through our time window, leading to an anticorrelation between iPSCs (T1) and neurons (T10). A similar but opposite trend was observed previously in a study monitoring proteome changes during the course of fibroblast reprogramming to iPSCs.40 According to GO classification, most of the proteins that downregulate during differentiation are located in the nucleus and are involved in RNA and DNA binding. Proteins that upregulate are mainly cytoskeletal proteins involved in cell communication and transport that could play a role in dendritic/axon outgrowth and branching. These findings are in line with our previous study, characterizing the proteome changes of rat hippocampal neuron development.41 The majority of the proteins are steadily upregulated or downregulated along the course of differentiation. We highlighted the most extreme comparison of iPSCs (T1) against mature neurons (T10) and showed the top 15 most up- or downregulated proteins during neuronal differentiation, consisting of proteins such as MAP2, a neuron-specific cytoskeletal protein having an established role in neurodevelopment.99 Moreover, we detect several proteins (e.g., INA, DPYSL4, and RPS19) being highly upregulated with less established association with neuronal development, suggesting that they may be considered as novel pan-neuronal markers. Previously, Djuric et al. used proteomics based on label-free quantification MS to profile neuronal development in IPSCs, neural precursor cells, and cortical neurons.100 In line with this study, extracellular membrane proteins were enriched in neurons, and nuclear proteins were enriched in early time points. Despite the different protocols, among the most differentially upregulated proteins were TUBB3, DCX, MAP2, and NCAM1. In conjunction, we captured all TFs and cytoskeletal proteins in our data and illustrated their expression changes during neuronal differentiation toward the two subtypes. Several of these TFs (PHC2, CRTC1, NCOA3, and SMAD3) have been reported to promote neuronal differentiation in zebrafish and mice; however, their association with human neuronal differentiation was not detected. In addition, we illustrated several subunits of a protein family having differential expression profiles in both cell types, such as strong upregulation of TAGLN3 in both cell types and the upregulation of TAGLN and TAGLN2 only in iN cells. TAGLN was previously shown to regulate cytoskeleton organization101 and was differentially expressed in neuronal subpopulations of the rat central nervous system,102 highlighting this protein as an interesting target for further investigation. Furthermore, we provide a rich source of information on proteins associated with several signaling pathways, such as Wnt and Notch, involved in neuronal development. We identified several proteins associated with these signaling pathways as being upregulated toward one of the two neuronal subtypes and being involved in cell fate pattering and development, further emphasizing their critical role in neuronal (subtype) differentiation.
NgN2 is a transcription factor essential for neurogenesis and its expression is regulated by Notch signaling. Upregulation of Notch represses NgN2 expression in early time points (iPSCs or neural precursor stages) and keeps cells in a proliferative state. During neuronal development, inhibition of Notch leads to upregulation of NgN2.103 Proteins associated with the Notch signaling pathway are lower in expression in iN cells compared to MNs. At T2, NgN2 is induced in iN cells, and THBS1 is downregulated as a consequence within this pathway, which was shown to play different roles in neuronal development, such as in neurite outgrowth and cell migration.89,90 It might be interesting to inhibit THBS1 and further evaluate whether neuronal differentiation is possible. Inhibition of Notch was previously shown to upregulate NgN2 expression. However, upregulation of NgN2 (and as a consequence downregulation of Notch) seems to be more effective and efficient (neuronal differentiation within 7 days). NgN2 is a key transcription factor regulating other neuronal transcription factors and repressing inhibitors of neurogenesis. Combining the overexpression of two transcription factors (NgN1 and NgN2) resulted in rapid neuronal differentiation within 4 days.14 This may regulate neuronal genes downstream of NgN, resulting in concerted activation of neuronal development. Several transcription factors, such as PHC2 and CCNT1, are shown to be elevated after NgN2 induction, which might be of importance in neurogenesis. In summary, we provide a quantitative overview of key proteins that promote the loss of pluripotency and rapid neuronal development along the course of differentiation.
Overall, we show that neuronal development is a complex process involving many protein expression changes. During differentiation, especially cytoskeletal proteins associated with cell communication and movement are upregulated, whereas nuclear proteins involved in embryogenesis were downregulated. Several proteins are known pan-neuronal markers, but we also identified multiple candidate proteins that could be involved in the regulation of differentiation. Many of the upregulated proteins were localized in the extracellular membrane, which could be useful for future extracellular cell surface protein-based sorting strategies. This study provides a valuable discovery platform for novel markers of neuronal development. Especially, transcription factors, identified in this study to show upregulation during differentiation, can be interesting candidates for further studies to investigate their ability to influence neuronal differentiation. Our data further supports the important role of signaling pathways such as Notch, Wnt, and Hedgehog in neuronal differentiation and in neuronal subtype specification. Although the two approaches (iN cells and MNs) are different in their differentiation protocol, we show many proteins that are similarly regulated. This indicates that these proteins have a general neuronal function rather than being subtype-specific. We further highlighted proteins showing opposite trends in expression between the two subtypes, which could be interesting candidates for future studies for their subtype-specific roles. We also highlighted several proteins with time-point-specific expression patterns, which might be important in precise tuning of developmental stages. These data constitute a rich resource that can be used to understand specific molecular mechanisms involved in neurodevelopment.
Acknowledgments
This work was supported by The Netherlands Organization for Scientific Research (NWO) through a VIDI grant for M.A. (723.012.102) and Proteins@Work, a program of the National Roadmap Large-Scale Research Facilities of The Netherlands (project number 184.032.201). We thank the MIND facility at the UMC Utrecht for the access to and help with the cell culture.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00070.
Figures showing the global proteome analysis, proteins only identified in iN cells or MNs, transcription factors and cytoskeletal proteins, proteins related to signaling pathways, neurogenesis-associated proteins, and uncropped blots (Figures S1–S6) and titles for Tables S1–S4 (PDF)
Table S1. Expression changes of proteins during iN cell differentiation (XLSX)
Table S2. Expression changes of proteins during MN differentiation (XLSX)
Table S3. Significant differentially expressed proteins in the opposite direction (XLSX)
Table S4. Proteins significantly downregulated in iN cells and MNs (XLSX)
The authors declare no competing financial interest.
Notes
All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD013399.
Supplementary Material
References
- Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Front. Hum. Neurosci. 2009, 3, 31. 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockland K. S. Non-uniformity of extrinsic connections and columnar organization. J. Neurocytol. 2002, 31 (3–5), 247–53. 10.1023/A:1024169925377. [DOI] [PubMed] [Google Scholar]
- Toro R.; et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010, 26 (8), 363–72. 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Hall J.; et al. Associative learning and the genetics of schizophrenia. Trends Neurosci. 2009, 32 (6), 359–65. 10.1016/j.tins.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Harvey M.; Belleau P.; Barden N. Gene interactions in depression: pathways out of darkness. Trends Genet. 2007, 23 (11), 547–56. 10.1016/j.tig.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Willsey A. J.; et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 2013, 155 (5), 997–1007. 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak N. N.; et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 2013, 155 (5), 1008–21. 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K.; et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131 (5), 861–72. 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Han S. S.; Williams L. A.; Eggan K. C. Constructing and deconstructing stem cell models of neurological disease. Neuron 2011, 70 (4), 626–44. 10.1016/j.neuron.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Marchetto M. C.; Gage F. H. Modeling brain disease in a dish: really?. Cell Stem Cell 2012, 10 (6), 642–5. 10.1016/j.stem.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Tao Y.; Zhang S. C. Neural Subtype Specification from Human Pluripotent Stem Cells. Cell Stem Cell 2016, 19 (5), 573–586. 10.1016/j.stem.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N.; Castro D. S.; Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002, 3 (7), 517–30. 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Kelava I.; Lancaster M. A. Stem Cell Models of Human Brain Development. Cell Stem Cell 2016, 18 (6), 736–48. 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Busskamp V.; et al. Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 2014, 10, 760. 10.15252/msb.20145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A.; Ziller M.; Spengler D. Childhood-Onset Schizophrenia: Insights from Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2018, 19 (12), 3829. 10.3390/ijms19123829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoufik E. Synaptic dysfunction in neurodegenerative and neurodevelopmental diseases: an overview of induced pluripotent stem-cell-based disease models. Open Biol. 2018, 8 (9), 180138. 10.1098/rsob.180138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.; et al. Current Advances and Limitations in Modeling ALS/FTD in a Dish Using Induced Pluripotent Stem Cells. Front. Neurosci. 2017, 11, 671. 10.3389/fnins.2017.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sances S.; et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 2016, 19 (4), 542–53. 10.1038/nn.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013, 78 (5), 785–98. 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury Y.; et al. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2015, 33 (1), 89–96. 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- Harschnitz O.; et al. Autoantibody pathogenicity in a multifocal motor neuropathy induced pluripotent stem cell-derived model. Ann. Neurol. 2016, 80 (1), 71–88. 10.1002/ana.24680. [DOI] [PubMed] [Google Scholar]
- Naldini L.; et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272 (5259), 263–7. 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Rivron N. C.; et al. Tissue deformation spatially modulates VEGF signaling and angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (18), 6886–91. 10.1073/pnas.1201626109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altelaar A. F. M.; et al. Benchmarking stable isotope labeling based quantitative proteomics. J. Proteomics 2013, 88, 14–26. 10.1016/j.jprot.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Rigbolt K. T.; Vanselow J. T.; Blagoev B. GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell. Proteomics 2011, 10 (8), O110.007450. 10.1074/mcp.O110.007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H.; et al. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007, 35 (Database), D247. 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielle F.; et al. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat. Neurosci. 2005, 8 (8), 1002–12. 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- Rosskothen-Kuhl N.; Illing R. B. Gap43 transcription modulation in the adult brain depends on sensory activity and synaptic cooperation. PLoS One 2014, 9 (3), e92624 10.1371/journal.pone.0092624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P. N.; Lasek R. J. The slow component of axonal transport. Identification of major structural polypeptides of the axon and their generality among mammalian neurons. J. Cell Biol. 1975, 66 (2), 351–66. 10.1083/jcb.66.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. K.; et al. Dynamic Ca2+-dependent stimulation of vesicle fusion by membrane-anchored synaptotagmin 1. Science 2010, 328 (5979), 760–3. 10.1126/science.1187722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V.; et al. SOX2 functions to maintain neural progenitor identity. Neuron 2003, 39 (5), 749–65. 10.1016/S0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Zeineddine D.; et al. The Oct4 protein: more than a magic stemness marker. Am. J. Stem Cells 2014, 3 (2), 74–82. [PMC free article] [PubMed] [Google Scholar]
- Mazzoni E. O.; et al. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 2013, 16 (9), 1219–27. 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; et al. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 2015, 17 (2), 195–203. 10.1016/j.stem.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Vincent A. J.; Lau P. W.; Roskams A. J. SPARC is expressed by macroglia and microglia in the developing and mature nervous system. Dev. Dyn. 2008, 237 (5), 1449–62. 10.1002/dvdy.21495. [DOI] [PubMed] [Google Scholar]
- Zhou S.; et al. Survivin Improves Reprogramming Efficiency of Human Neural Progenitors by Single Molecule OCT4. Stem Cells Int. 2016, 2016, 4729535. 10.1155/2016/4729535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongidi V.; et al. SPARC-like 1 regulates the terminal phase of radial glia-guided migration in the cerebral cortex. Neuron 2004, 41 (1), 57–69. 10.1016/S0896-6273(03)00818-3. [DOI] [PubMed] [Google Scholar]
- Harkin L. F.; et al. Distinct expression patterns for type II topoisomerases IIA and IIB in the early foetal human telencephalon. J. Anat. 2016, 228 (3), 452–63. 10.1111/joa.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C.; et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004, 23 (14), 2892–902. 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J.; et al. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012, 2 (6), 1579–92. 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese C. K.; et al. Quantitative Map of Proteome Dynamics during Neuronal Differentiation. Cell Rep. 2017, 18 (6), 1527–1542. 10.1016/j.celrep.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M.-L.; et al. Distribution patterns of the zebrafish neuronal intermediate filaments inaa and inab. J. Neurosci. Res. 2019, 97 (2), 202–214. 10.1002/jnr.24347. [DOI] [PubMed] [Google Scholar]
- Quach T. T.; et al. Collapsin response mediator protein 3 increases the dendritic arborization of hippocampal neurons. Mol. Psychiatry 2015, 20 (9), 1027. 10.1038/mp.2015.123. [DOI] [PubMed] [Google Scholar]
- da Rocha Boeira T.; et al. Polymorphism Located in the Upstream Region of the RPS19 Gene (rs2305809) Is Associated With Cervical Cancer: A Case-control Study. J. Cancer Prev 2018, 23 (3), 147–152. 10.15430/JCP.2018.23.3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. W. A soluble protein characteristic of the nervous system. Biochem. Biophys. Res. Commun. 1965, 19 (6), 739–44. 10.1016/0006-291X(65)90320-7. [DOI] [PubMed] [Google Scholar]
- Filipek A.; et al. Calcyclin--Ca(2+)-binding protein homologous to glial S-100 beta is present in neurones. NeuroReport 1993, 4 (4), 383–6. 10.1097/00001756-199304000-00010. [DOI] [PubMed] [Google Scholar]
- Yamashita N.; et al. Distribution of a specific calcium-binding protein of the S100 protein family, S100A6 (calcyclin), in subpopulations of neurons and glial cells of the adult rat nervous system. J. Comp. Neurol. 1999, 404 (2), 235–57. . [DOI] [PubMed] [Google Scholar]
- Chan W. Y.; et al. Differential expression of S100 proteins in the developing human hippocampus and temporal cortex. Microsc. Res. Tech. 2003, 60 (6), 600–13. 10.1002/jemt.10302. [DOI] [PubMed] [Google Scholar]
- Girard F.; et al. The EF-hand Ca(2+)-binding protein super-family: a genome-wide analysis of gene expression patterns in the adult mouse brain. Neuroscience 2015, 294, 116–55. 10.1016/j.neuroscience.2015.02.018. [DOI] [PubMed] [Google Scholar]
- Hupe M.; et al. Gene expression profiles of brain endothelial cells during embryonic development at bulk and single-cell levels. Sci. Signaling 2017, 10 (487), eaag2476. 10.1126/scisignal.aag2476. [DOI] [PubMed] [Google Scholar]
- Chantha S. C.; et al. The MIDASIN and NOTCHLESS genes are essential for female gametophyte development in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2010, 16 (1), 3–18. 10.1007/s12298-010-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalus I.; et al. Sulf1 and Sulf2 Differentially Modulate Heparan Sulfate Proteoglycan Sulfation during Postnatal Cerebellum Development: Evidence for Neuroprotective and Neurite Outgrowth Promoting Functions. PLoS One 2015, 10 (10), e0139853 10.1371/journal.pone.0139853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalus I.; et al. Differential involvement of the extracellular 6-O-endosulfatases Sulf1 and Sulf2 in brain development and neuronal and behavioural plasticity. J. Cell. Mol. Med. 2009, 13 (11–12), 4505–21. 10.1111/j.1582-4934.2008.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J.; Ericson J. Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 2001, 11 (1), 43–49. 10.1016/S0959-4388(00)00172-0. [DOI] [PubMed] [Google Scholar]
- Morrison S. J. Neuronal differentiation: proneural genes inhibit gliogenesis. Curr. Biol. 2001, 11 (9), R349–51. 10.1016/S0960-9822(01)00191-9. [DOI] [PubMed] [Google Scholar]
- Serre A.; et al. Overexpression of basic helix-loop-helix transcription factors enhances neuronal differentiation of fetal human neural progenitor cells in various ways. Stem Cells Dev. 2012, 21 (4), 539–53. 10.1089/scd.2011.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J.; et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 2012, 9 (6), 575–8. 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- Conkright M. D.; et al. TORCs: transducers of regulated CREB activity. Mol. Cell 2003, 12 (2), 413–23. 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Li S.; et al. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J. Neurosci. 2009, 29 (8), 2334–43. 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendioroz M.; et al. CRTC1 gene is differentially methylated in the human hippocampus in Alzheimer’s disease. Alzheimer's Res. Ther. 2016, 8 (1), 15. 10.1186/s13195-016-0183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchel P. H.; et al. A large-scale functional screen identifies Nova1 and Ncoa3 as regulators of neuronal miRNA function. EMBO J. 2015, 34 (17), 2237–54. 10.15252/embj.201490643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casari A.; et al. A Smad3 transgenic reporter reveals TGF-beta control of zebrafish spinal cord development. Dev. Biol. 2014, 396 (1), 81–93. 10.1016/j.ydbio.2014.09.025. [DOI] [PubMed] [Google Scholar]
- Inoue T.; et al. Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev. Biol. 2004, 270 (1), 146–62. 10.1016/j.ydbio.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Quadrato G.; et al. Nuclear factor of activated T cells (NFATc4) is required for BDNF-dependent survival of adult-born neurons and spatial memory formation in the hippocampus. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (23), E1499–E1508. 10.1073/pnas.1202068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S. S.; Hammarback J. A. Gene localization and developmental expression of light chain 3: a common subunit of microtubule-associated protein 1A(MAP1A) and MAP1B. J. Neurosci. Res. 1996, 43 (5), 535–44. . [DOI] [PubMed] [Google Scholar]
- Sullivan K. F.; Cleveland D. W. Identification of conserved isotype-defining variable region sequences for four vertebrate beta tubulin polypeptide classes. Proc. Natl. Acad. Sci. U. S. A. 1986, 83 (12), 4327–31. 10.1073/pnas.83.12.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K.; et al. Neuronal protein NP25 interacts with F-actin. Neurosci. Res. 2004, 48 (4), 439–46. 10.1016/j.neures.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Stiles J.; Jernigan T. L. The basics of brain development. Neuropsychology review 2010, 20 (4), 327–348. 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y.; Zhang S.-C. Neural Subtype Specification from Human Pluripotent Stem Cells. Cell stem cell 2016, 19 (5), 573–586. 10.1016/j.stem.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. M.; et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27 (3), 275–80. 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B. L.; Kimelman D. Wnt signaling and the evolution of embryonic posterior development. Curr. Biol. 2009, 19 (5), R215–9. 10.1016/j.cub.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani L.; Salinas P. C. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 2005, 6 (5), 351–62. 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- Budnik V.; Salinas P. C. Wnt signaling during synaptic development and plasticity. Curr. Opin. Neurobiol. 2011, 21 (1), 151–9. 10.1016/j.conb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y.; et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 2004, 131 (12), 2791–801. 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- Nordstrom U.; et al. An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 2006, 4 (8), e252 10.1371/journal.pbio.0040252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt P. M.; Malgor R. Wnt5a: a player in the pathogenesis of atherosclerosis and other inflammatory disorders. Atherosclerosis 2014, 237 (1), 155–62. 10.1016/j.atherosclerosis.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; et al. Wls-mediated Wnts differentially regulate distal limb patterning and tissue morphogenesis. Dev. Biol. 2012, 365 (2), 328–38. 10.1016/j.ydbio.2012.02.019. [DOI] [PubMed] [Google Scholar]
- Barna M.; Pandolfi P. P.; Niswander L. Gli3 and Plzf cooperate in proximal limb patterning at early stages of limb development. Nature 2005, 436 (7048), 277–81. 10.1038/nature03801. [DOI] [PubMed] [Google Scholar]
- Serfling E.; et al. Trends Immunol. 2006, 27, 461–9. 10.1016/j.it.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Yang Y. R.; et al. Primary phospholipase C and brain disorders. Adv. Biol. Regul 2016, 61, 80–5. 10.1016/j.jbior.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Lara-Ramirez R.; Zieger E.; Schubert M. Retinoic acid signaling in spinal cord development. Int. J. Biochem. Cell Biol. 2013, 45 (7), 1302–13. 10.1016/j.biocel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Cayuso J.; et al. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development 2006, 133 (3), 517–28. 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- Numata-Uematsu Y.; et al. Inhibition of collapsin response mediator protein-2 phosphorylation ameliorates motor phenotype of ALS model mice expressing SOD1G93A. Neurosci. Res. 2019, 139, 63. 10.1016/j.neures.2018.08.016. [DOI] [PubMed] [Google Scholar]
- Kowalczyk A.; et al. The critical role of cyclin D2 in adult neurogenesis. J. Cell Biol. 2004, 167 (2), 209–13. 10.1083/jcb.200404181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.; et al. Single-Cell RNA-Seq Reveals Hypothalamic Cell Diversity. Cell Rep. 2017, 18 (13), 3227–3241. 10.1016/j.celrep.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalopulu N.; Kintner C. A posteriorising factor, retinoic acid, reveals that anteroposterior patterning controls the timing of neuronal differentiation in Xenopus neuroectoderm. Development 1996, 122 (11), 3409–18. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y.; et al. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008, 22 (2), 152–65. 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese L.; et al. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells 2010, 28 (5), 955–64. 10.1002/stem.408. [DOI] [PubMed] [Google Scholar]
- Adams J. C.; Watt F. M. Regulation of development and differentiation by the extracellular matrix. Development 1993, 117 (4), 1183–98. [DOI] [PubMed] [Google Scholar]
- Christopherson K. S.; et al. Thrombospondins Are Astrocyte-Secreted Proteins that Promote CNS Synaptogenesis. Cell 2005, 120 (3), 421–433. 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Rasmussen J. P.; Sagasti A. Learning to swim, again: Axon regeneration in fish. Exp. Neurol. 2017, 287, 318–330. 10.1016/j.expneurol.2016.02.022. [DOI] [PubMed] [Google Scholar]
- Dickson B. J. Molecular Mechanisms of Axon Guidance. Science 2002, 298 (5600), 1959–1964. 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Bashaw G. J.; et al. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell 2000, 101 (7), 703–15. 10.1016/S0092-8674(00)80883-1. [DOI] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4 (1), 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Couillard-Despres S.; et al. Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci 2005, 21 (1), 1–14. 10.1111/j.1460-9568.2004.03813.x. [DOI] [PubMed] [Google Scholar]
- Kim D. H.; et al. Pan-neuronal calcium imaging with cellular resolution in freely swimming zebrafish. Nat. Methods 2017, 14, 1107. 10.1038/nmeth.4429. [DOI] [PubMed] [Google Scholar]
- Abbott N. J.; et al. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010, 37 (1), 13–25. 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Niswander L. Zic2 is required for enteric nervous system development and neurite outgrowth: a mouse model of enteric hyperplasia and dysplasia. Neurogastroenterol. Motil. 2013, 25 (6), 538–541. 10.1111/nmo.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R. L.; et al. Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Mol. Brain Res. 1986, 387 (1), 271–80. 10.1016/0169-328X(86)90033-1. [DOI] [PubMed] [Google Scholar]
- Djuric U.; et al. Spatiotemporal Proteomic Profiling of Human Cerebral Development. Mol. Cell. Proteomics 2017, 16 (9), 1548–1562. 10.1074/mcp.M116.066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsafadi M.; et al. Transgelin is a TGFβ-inducible gene that regulates osteoblastic and adipogenic differentiation of human skeletal stem cells through actin cytoskeleston organization. Cell Death Dis. 2016, 7 (8), e2321 10.1038/cddis.2016.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W.-Z.; et al. The identification of NP25: a novel protein that is differentially expressed by neuronal subpopulations. Mol. Brain Res. 1994, 22 (1), 173–185. 10.1016/0169-328X(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Nelson B. R.; et al. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev. Biol. 2007, 304 (2), 479–498. 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD013399