Abstract
The incidence of common, metabolic diseases (e.g. obesity, cardiovascular disease, diabetes) with complex genetic etiology has been steadily increasing nationally and globally. While identification of a genetic model that explains susceptibility and risk for these diseases has been pursued over several decades, no clear paradigm has yet been found to disentangle the genetic basis of polygenic/complex disease development. Since the evolution of the eukaryotic cell involved a symbiotic interaction between the antecedents of the mitochondrion and nucleus (which itself is a genetic hybrid), we suggest that this history provides a rational basis for investigating whether genetic interaction and co-evolution of these genomes still exists. We propose that both mitochondrial and Mendelian, or “mito-Mendelian” genetics play a significant role in cell function, and thus disease risk. This paradigm contemplates the natural variation and co-evolution of both mitochondrial and nuclear DNA backgrounds on multiple mitochondrial functions that are discussed herein, including energy production, cell signaling and immune response, which collectively can influence disease development. At the nexus of these processes is the economy of mitochondrial metabolism, programmed by both mitochondrial and nuclear genomes.
1. Introduction
Metabolic homeostasis is crucial to health and is maintained by a complex interaction of mitochondrial biogenesis, bioenergetics, maintenance, and response to both exogenous and endogenous changes in the environment. Since the 1980s the incidence of metabolic diseases (e.g. obesity, cardiovascular disease, diabetes) has been steadily increasing on both national and global stages [1]. While these diseases can occur at any age, metabolic and lifestyle changes associated with aging, poor diet and lack of exercise are implicated as the major contributors to their rising incidence [[2], [3], [4], [5], [6], [7], [8]]. Identification of a genetic paradigm that explains susceptibility and risk for these diseases linked with metabolism has been pursued over several decades, and while multiple single gene mutations in the nuclear genome have been linked to a variety of metabolic diseases [9], the frequencies of these mutations are generally low and do not account for the increasing incidence of metabolic diseases observed in the developed world [3]. One explanation is that pre-existent natural genetic variation is the basis for differential risk among individuals. While genome-wide association studies (GWAS) have identified numerous gene variants associated with several diseases [10], as has whole genome sequencing [11,12], no clear genetic paradigm has yet been found to disentangle the genetic basis of polygenic/complex disease development. Consequently, while greater granularity in our understanding of the nuclear genome has been established, a genetic paradigm for understanding the polygenic basis for common disease has not yet been clearly defined.
Douglas Wallace formally proposed a “mitochondrial paradigm” for human disease in 2005 [13]. In this treatise, he proposed that mitochondrial DNA (mtDNA) mutations in our prehistoric ancestors altered mitochondrial bioenergetics, and along with changes in diet, enabled ancient humans to adapt to climatic changes as they radiated out of Africa. Further, it was hypothesized that many of these prehistoric adaptive mtDNA mutations in contemporary Western societies contribute to risk for metabolic and degenerative disease. A detailed discussion of the specific features of mitochondrial genetics is not the focus of this review – several reviews are already available on that topic [[14], [15], [16], [17], [18], [19]]. Herein, certain characteristics of the mitochondrion in terms of its origins, genetics and functions are discussed which relate to the multifunctional role of the organelle, yet also place something we refer to as “mitochondrial economy” central to these functions. Current thoughts on the origins of the nucleus and how input from both the mtDNA and nuclear genome influence cell metabolism via “mito-Mendelian” processes are also considered. While challenging nuclear-centric theories derived from Mendelian genetics, we propose that the mitochondrial paradigm, when combined with Mendelian concepts, creates a mito-Mendelian genetic paradigm which contemplates the natural variation and co-evolution of both mtDNA and nuclear backgrounds on organelle function that can influence cell function and disease risk [[20], [21], [22], [23], [24]].
1.1. Mitochondrial Origins
The eukaryotic cell typically contains 100's – 100,000's of mitochondria, depending upon cell type. Multiple taxonomic studies suggest that the mitochondrion shares a remote common ancestor with the Rickettsiales lineage, an extant order of aerobic, α-proteobacteria [25,26]; genomic analyses from numerous α-proteobacteria-related clades (a group of biological taxa that can be traced to one common ancestor) suggest that the origins of the mitochondrion predate the diversification of known α-proteobacterial lineages, meaning that mitochondrial endosymbiotic origins are likely more ancient than originally thought [25]. However, it is currently thought that an α-proteobacterial bacteria entered into an endosymbiotic relationship with an archaeon-like host cell (an archaeon is a single celled prokaryote characterized by extremely large genomes – see next section, Nuclear origins) ~1.5 billion years ago [[27], [28], [29], [30], [31], [32], [33]]. While a certain level of controversy also exists regarding the nature of this endosymbiotic relationship in terms of when phagocytosis evolved relative to endosymbiosis [[28], [29], [30], [31], [32], [33], [34]], both phylogenetic and phylogenomic analyses are consistent with a monophyletic origin of mitochondria [25]. In terms of the endosymbiotic theory and when phagocytosis became established, the “mitochondrion-late” formulation posits that phagocytosis evolved in the host prior to endosymbiosis, whereas the “mitochondrion early” model suggests that phagocytosis evolved after establishment of metabolic interactions between the archaeon-like host and its α-proteobacterial resident [34,35]. The latter model, referred to as “anaerobic syntrophy”, is based upon a process common in the recently discovered deep-sea archaea, which are close archaeal relatives to eukaryotes [35]. Regardless of whether endosymbiosis had phagotrophic or syntrophic origins however, our mitochondrial ancestor appears to have been a small, free-living autotrophic, aerobic, α-proteobacterium that established a symbiotic relationship with its host cell to convert nutrients (provided by the host) into large amounts of ATP through oxidative phosphorylation, establishing a mutually beneficial relationship. As the mitochondrial endosymbiont and its host coevolved, the former lost or transferred the majority of its genetic material to the “nucleus” of its host, and while losing the ability to survive autonomously, it retained key catalytic subunits required for electron transport.
The endosymbiotic theory was first published in 1905 by Konstantin Mereschkowsky, a Russian botanist, who postulated that ancestors of plant chloroplasts were free-living cyanobacteria that became symbionts [28,29]. While mitochondria were not part of Mereschkowsky's theory, approximately a decade later (1918) Paul Portier proposed that mitochondria were consequent of a symbiotic process [36], a concept subsequently promoted by Ivan Wallin [31]. However, none of these theories were seriously considered, and were in fact ridiculed by the mainstream scientific community at the time; therefore endosymbiotic origins as an explanation for the chloroplast, the mitochondrion, or eukaryotic cell were abandoned until 1967, when the theory was resurrected by Lynn Sagan (Margulis) in her seminal paper “On the Origin of Mitosing Cells” [30]. However, much like the response 50 years earlier, her theories regarding the origins of eukaryotic cells and mitochondria were met with much skepticism and criticism [32], until protein and DNA sequencing data by Robert Schwartz and Margaret Dayhoff revealed the bacterial origins of mitochondria [33].
1.2. Nuclear origins
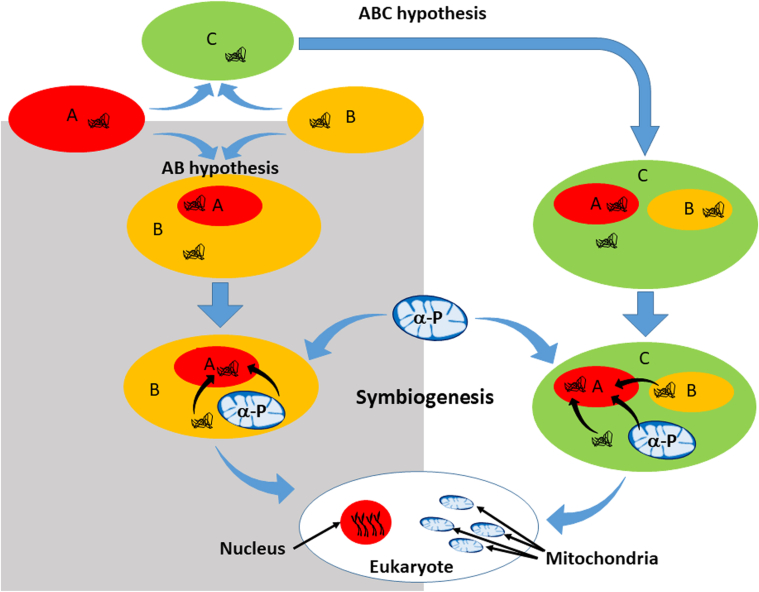
There are currently multiple hypotheses regarding the origins of the nucleus [[37], [38], [39], [40], [41], [42], [43], [44]]. Lopez – Garcia and Moreira have proposed a refined syntrophy (meaning that an interdependency exists) model, that invokes key evolutionary events involving an archaeal - myxobacterial endosymbiosis as the origin of the eukaryotic nucleus [42]. An archaeon is single celled prokaryote that while being morphologically similar to bacteria, is genetically more closely related to eukaryotes; myxobacteria, or “slime bacteria” represent a group of organisms predominantly found in soils and typically feed on insoluble organics; another characteristic of these prokaryotes is extremely large genomes [45]. While the syntropic model (also referred to as the “AB” hypothesis) promotes an archaeal – myxobacterial endosymbiosis for nuclear origins, it also states selective forces of metabolic compartmentation also occurred/followed with the acquisition of the α-proteobacterium (ancestral mitochondrion) and genetic transfer of α-proteobacterial genes to the hybrid archaeal-myxobacterial nuclear material; this was followed by the origin of the nuclear envelope and the endoplasmic reticulum. An alternative hypothesis (called the “ABC” hypothesis), based upon examination of eukaryotic proteins with no significant homology to archaeal or bacterial proteins, includes an additional cell, the “chronocyte” [38] that engulfed both archaea and bacteria, forming the nucleus, and proposes that the host cell was a chronocyte, which already had a cytoskeleton and extensive membrane system, instead of a prokaryotic cell (e.g. bacteria). Another theory proposes that the eukaryotic nucleus evolved from a complex DNA virus, consequent of a “persistent presence” of the virus in an archaeal host that resulted in viral acquisition of host genes that created a merged nuclear genome [37]. Collectively however, while each of these hypotheses propose different processes for the origins of the nucleus, they all: i) involve endosymbiotic events, and ii) note that the nuclear genome represents a genetic hybrid, consequent of symbiogenic events. Hence, the evolution of the eukaryotic cell involved a symbiotic interaction between the antecedents of the mitochondrion and nucleus that were required for survival (Fig. 1). Consequently, it is logical that some form of genetic interaction or co-evolution remains today – in fact, recent studies are now showing that nuclear gene expression, regulation, and metabolism are significantly influenced by different combinations of mtDNA and nuclear genetic backgrounds [20,21,24,[46], [47], [48]].
Fig. 1.
Origins of the eukaryote nucleus based upon the AB/ABC hypothesis. The schematic presents a synthesis of the chronocyte (ABC hypothesis, white shaded area) and syntropic (AB hypothesis, grey shaded area) models. Both concur that the nucleus is an endosymbiont consequent of archaeal (A, red oval) and myxobacterial (B, orange oval) donors, however they differ in that the AB hypothesis proposes the “host” cell came from bacteria (B), whereas the ABC hypothesis concludes that the host cell was a chronocyte (C, green oval), wherein the archaea and myxobacteria were engulfed by the chronocyte. Symbiogenesis occurred (scribbled lines indicate genetic material) either prior and/or subsequent to the acquisition of the α-proteobacterium (α-P, ancestral mitochondrion) generating a hybrid archaeal-myxobacterial-α-proteobacterial nuclear material. While both hypotheses suggest different processes for the origins of the eukaryotic nucleus, they both involve endosymbiotic events, and conclude the nuclear genome is a genetic hybrid, consequent of symbiogenic events.
1.3. Mitochondrial structure
The mitochondrion consists of outer and inner membranes that create an intermembrane space (located between the outer and inner membranes) and a matrix (surrounded by the inner membrane). Collectively it has been estimated that the mitochondrion contains ~1500 proteins in vertebrates, and ~1000 in yeast [49]. The outer mitochondrial membrane is structurally similar to the eukaryotic cell membrane being composed of a 50/50 protein - phospholipid bilayer that acts as a semi permeable barrier (<5000–10,000 MW) to the cytosol [[50], [51], [52]]. The outer mitochondrial membrane contains an abundance of voltage-dependent anion channels (VDACs) or porins which regulate the passage of smaller metabolites and ions into and out of the mitochondrion [51], while larger molecules are imported via the translocation of the outer membrane (TOM) complex, which serves as an entry point for virtually all mitochondrial protein precursors [53]. The outer membrane can also interact with the endoplasmic reticulum, ribosomes, other mitochondria, and the nucleus [50,54], with the mitochondrial distribution and morphology protein (Mdm10) and mitofusin (Mfn) 2, a dynamin related GTPase (also an initiator of fusion) identified as the membrane anchor of the ER-mitochondrion tether structure [53,[55], [56], [57], [58]]. The intermembrane space has a relatively low protein content compared to the matrix, with cytochrome c being a prominent component in addition to other proteins involved in apoptosis, inner membrane remodeling (e.g. Opa1), and additional protein import carrier proteins [50]. The protein rich mitochondrial inner membrane has lipid content consistent with its bacterial origins (e.g. high cardiolipin content) and contains the respiratory complexes (comprised of subunits encoded by both the nuclear and mitochondrial DNA) required for electron transport and ATP production. Due to its low permeability, formation of an electrochemical potential across the inner membrane is possible, which helps drive oxidative phosphorylation. The mitochondrial inner membrane is characterized by the “inner boundary membrane” and the “cristae”. The inner boundary membrane is the part that comes into close proximity with the outer mitochondrial membrane, forming contact sites bridged by outer and inner membrane proteins, including TOM (outer membrane) and inner membrane translocase complexes (translocases of the inner membrane, TIM22 and TIM23), that can form super-complexes for transport of proteins into the inner membrane or matrix [55]. The characteristic wrinkled appearance of the mitochondrial inner membrane which is the consequence of folds within the membrane are known as cristae, which although often described as invaginations of the inner membrane, are actually specialized tubular structures that regulate diffusion of molecules important for oxidative phosphorylation. Cristae that jut out into the matrix and form narrow openings at their base are known as crista junctions [59] – the mitochondrial contact site and cristae organizing system (or MICOS complex) forms the structural basis for crista junctions, which allow for the formation of micro-compartments called intracristal spaces that limit the diffusion of small molecules, ATP and cytochrome c [59]. Cristae are also dynamic – they undergo remodeling that can modulate reaction kinetics of the citric acid cycle and oxidative phosphorylation with changes in metabolism, and during apoptosis alter their structure to facilitate the release of cytochrome c [[60], [61], [62]]. The mitochondrial matrix contains almost 70% of the organelle's proteins, representing hundreds of metabolic enzymes including the citric acid cycle, fatty acid oxidation, heme synthesis and Fe–S biogenesis. Also located within the matrix, anchored to the inner membrane is the mitochondrial DNA (mtDNA). This small, circular DNA encodes key catalytic subunits for electron transport and oxidative phosphorylation, the translational machinery for proper generation of these subunits, and a growing number of mitochondrial derived peptides (MDPs) that function to regulate cellular metabolism [[63], [64], [65]].
1.4. Genome structure and organization
Within each mitochondrion are several copies of closed circular mtDNA. As discussed in the “Mitochondrial Origins” section, the mitochondrion has a monophyletic origin in that it arose from a α-proteobacterial ancestor; the original genome contained hundreds of genes, which over millions of years were lost or transferred to the host (nuclear) genome [25,66]. Consequently, the “original” mitochondrial genome now resides in two compartments, the mitochondrion and the nucleus. In contemporary vertebrates, the mtDNA is a double stranded, circular DNA that ranges from 16 to 18 kb (16.5 kb in humans) and consists of a guanine rich “heavy strand” and a cytosine rich “light strand”. There are 2–15 copies of mtDNA in each mitochondrion, resulting in thousands of copies of mtDNA per cell [[67], [68], [69], [70], [71]]. The mtDNA encodes 13 proteins that are key catalytic subunits of the electron transport chain and ATP synthase. By virtue of having a slightly different genetic code from the nucleus, 22 transfer RNAs (tRNAs) and 2 ribosomal RNAs (rRNAs) also reside in the mtDNA that are required for translation of mtDNA transcripts. Except for the succinate dehydrogenase complex (complex II), each electron transport chain complex (complexes I, III, IV and V) contains mtDNA encoded subunits – all complex II subunits are encoded by the nucleus with exception of the mtDNAs from a photosynthetic red alga, Porphyra purpurea and a heterotrophic zooflagellate, Reclinomonas americana [72]. The heavy strand encodes both rRNAs (12S and 16S), 14 tRNAs, and 12 polypeptide subunits for electron transport chain complexes I, III and IV, and ATP synthase (complex V), while the light strand encodes a complex I subunit (ND6) and 8 tRNAs. In addition to the 13 ETC subunit proteins encoded by the mtDNA, 8 distinct short open reading frames have been identified in the mtDNA which encode biologically active peptides, known as mitochondrial derived peptides (MDPs) [[73], [74], [75], [76], [77], [78], [79], [80]]. The first of these MDPs to be identified, humanin, is encoded within the 16S rRNA, which is exported from the organelle into the cytosol where it has been shown to have cytoprotective effects [74,77,[80], [81], [82], [83]]. The 16S rRNA also encodes 6 small humanin-like peptides (SHLPs 1–6) which have roles in cell proliferation, mitochondrial metabolism and apoptosis [74,75,80]. Unlike the other MDPs discovered thus far, mitochondrial open reading frame within the twelve S rRNA type-c (MOTS-c) is encoded in the 12S rRNA and is a key regulator of metabolic homeostasis of glucose, fatty acids and protein within the cell, with decreased levels of MOTS-c implicated in several metabolic diseases including diabetes and cardiovascular disease [74,79,80]. Currently, MDPs appear to play roles in regulating metabolism and/or cytoprotection, and importantly, can respond to metabolic stress [64,65,74,79,[84], [85], [86]]. While the majority of the mtDNA codes for proteins or translational machinery, there is a ~1 kb “non-coding region” or NCR, which is also known as the “control region” as it contains sites for the origin of heavy strand DNA replication and transcriptional promoter sites for both heavy and light strands [87,88]. The NCR also encompasses the displacement loop (D-loop, a triple stranded region seen in replicating mtDNAs) region and 3 mtDNA hypervariable sequence regions, sometimes referred to as “mutational hotspots” in the mtDNA [89].
1.5. Genome replication
The mtDNA is maternally inherited, polyploid, replicates and transcribes simultaneously with the accessory subunit of DNA polymerase γ and mitochondrial transcription factor A (TFAM) being necessary for both replication and transcription [90]. In contrast to the nuclear DNA, mtDNA replication is not intrinsically tied to cell cycle; further, it is not clear whether any level of control exists for mtDNA sorting during cytokinesis. For replication, mtDNA aggregate with TFAM, mitochondrial single-strand binding protein (mtSSBP) and the mtDNA helicase Twinkle, to form nucleoids [91] which are anchored to the inner mitochondrial membrane. Each nucleoid can contain 5-7 copies of mtDNA packed into a 70 nm diameter structure, reminiscent of packing densities found in bacteria [69] and the number of nucleoids per mitochondrion vary by tissue [90]. There are two proposed models for mtDNA replication: i) the strand-displacement mechanism (SDM), or ii) RNA incorporated throughout the lagging strand (RITOLS) replication [87,[92], [93], [94]]. In the SDM, mtDNA helicase Twinkle unwinds the double stranded mtDNA at the replication fork, allowing the DNA polymerase γ to initiate synthesis of the heavy strand (leading strand) at the origin (OH) [87,[92], [93], [94]]. This displaces the parental heavy strand which is maintained, stabilized and protected as a single strand by mtSSBP, until the light strand origin (OL) is exposed (after about 2/3 of the new heavy strand has been replicated), and synthesis of the light strand (lagging strand) is initiated [87,[92], [93], [94]]. In the RITOLS model, as the parental heavy strand is displaced, it is used as a template for RNA which begins to form the newly synthesized light strand (lagging strand). When the OL is exposed, DNA replaces the RNA to form the new light strand, a process called RNA maturation [87,[92], [93], [94]]. This model (RITOLS) was developed in response to the observation of apparent double-stranded “intermediates” forming during replication [87,95], which are preserved when subjected to protease and phenol-chloroform extraction but are susceptible to the degrading effects of RNAse H, which only degrades RNA/DNA hybrids.
Under conditions of nutrient excess, mitochondria appear to be more fragmented (undergo fission), and therefore mtDNA replication is more active [96]. Conversely, under conditions of high energy demand, mitochondrial elongation (fusion) is favored due to activation of pathways involving mitofusin 2 (Mfn2, a protein essential for mitochondrial fusion) and peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1α, a transcription coactivator that plays an essential role in the regulation of metabolism). Also, under conditions of caloric restriction or starvation, dynamin-related protein 1 (Drp1, a GTPase protein involved in the regulation of mitochondrial fission), is inhibited, and mitochondria appear elongated due to unchecked mitochondrial fusion [97,98]. A study exposing pancreatic islet β-cells to varying concentrations of glucose showed that mitochondria from cells exposed to 20mM glucose exhibited robust fragmentation while mitochondria from cells exposed to 5mM glucose appeared tubular [99]. Studies have also shown that under hyperglycemic conditions, expression and/or activity of TFAM is decreased [100]. Specifically, it has been shown that in HepG2 cells exposed to 30mM glucose, mitochondrial biogenesis is decreased as compared to cells treated with 5.5mM glucose, and this observation is concordant with a decrease in TFAM transcripts [101]. Because of its crucial role in mtDNA transcription and replication, it is likely that nutrient excess may regulate mitochondrial replication through its effects on TFAM.
1.6. Mitochondrial functions
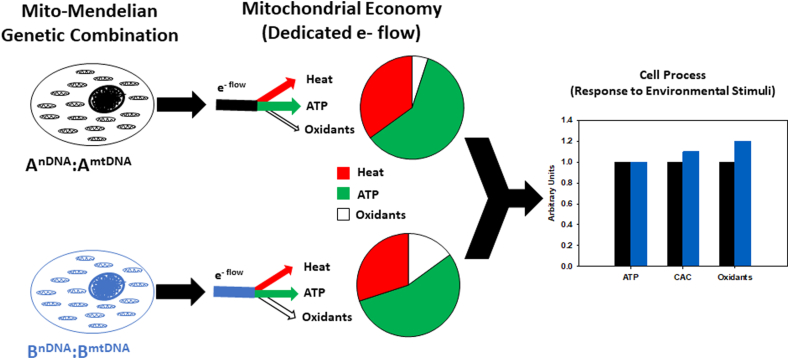
Mitochondria perform a plethora of cell functions beyond the stereotype that they are the “powerplants of the cell”. While indeed important and critical as sources of oxidative energy production, by virtue of their endosymbiotic origins and evolution of the eukaryotic cell, they are multifunctional organelles as well. They are the primary source of heat in endotherms [102,103], enabling us to maintain a constant body temperature and moreover, serve as central supply and signaling stations for the cell, providing energy, oxidants, metabolites and multiple signaling molecules for a variety of biosynthetic, metabolic and immunologic processes required for survival. At the nexus of these processes is mitochondrial metabolism, or, the electron transport chain (ETC) and how efficiently it functions. This mitochondrial efficiency, or “economy” (utilization of electron flow for ATP generation) can affect a variety of cell processes, including ATP generation, citric acid cycle function and oxidant production that impact numerous downstream pathways including cell signaling, calcium regulation, apoptosis, immune response, glucose homeostasis, and nuclear gene expression [[104], [105], [106], [107], [108], [109], [110]]. In this respect, reports have shown that different mtDNA – nDNA combinations associate with different mitochondrial economies [21,24,47,111]. The mito-Mendelian paradigm proposes that the combined natural variation of both the mitochondrial and nuclear genomes serve as the genetic basis for programming mitochondrial metabolism, and thus, different mtDNA – nDNA combinations modulate metabolic homeostatic systems (Fig. 2) that in turn, influence cellular response to stimuli [20,22]. The following sections selectively discuss a few of the many functions performed by the mitochondrion, and conclude with a discussion on mitochondrial – nuclear (mito-Mendelian) genetic interactions.
Fig. 2.
mito-Mendelian genetics influences mitochondrial economy for ATP generation, and thus, can modulate metabolic homeostatic systems that influence cellular response to stimuli. The figure illustrates 2 different nuclear – mtDNA combinations (AnDNA:AmtDNA – black; BnDNA:BmtDNA – blue), that yield different dedicated electron flow (e- flow) levels (arrow width) for the generation of heat (red), ATP (green) and oxidant (no fill) production. In the example, AnDNA:AmtDNA dedicates more e- flow for ATP production relative to BnDNA:BmtDNA (pie chart), yet both generate the same levels of ATP (bar graph), indicating a greater mitochondrial economy for the BnDNA:BmtDNA combination (less e- flow/ATP produced). Because the BnDNA:BmtDNA nuclear-mtDNA combination utilizes less electrons to generate the same amount of ATP (relative to the AnDNA:AmtDNA combination), differences are observed in oxidant production and CAC metabolic intermediates under conditions of energy sufficiency.
1.7. Citric acid cycle
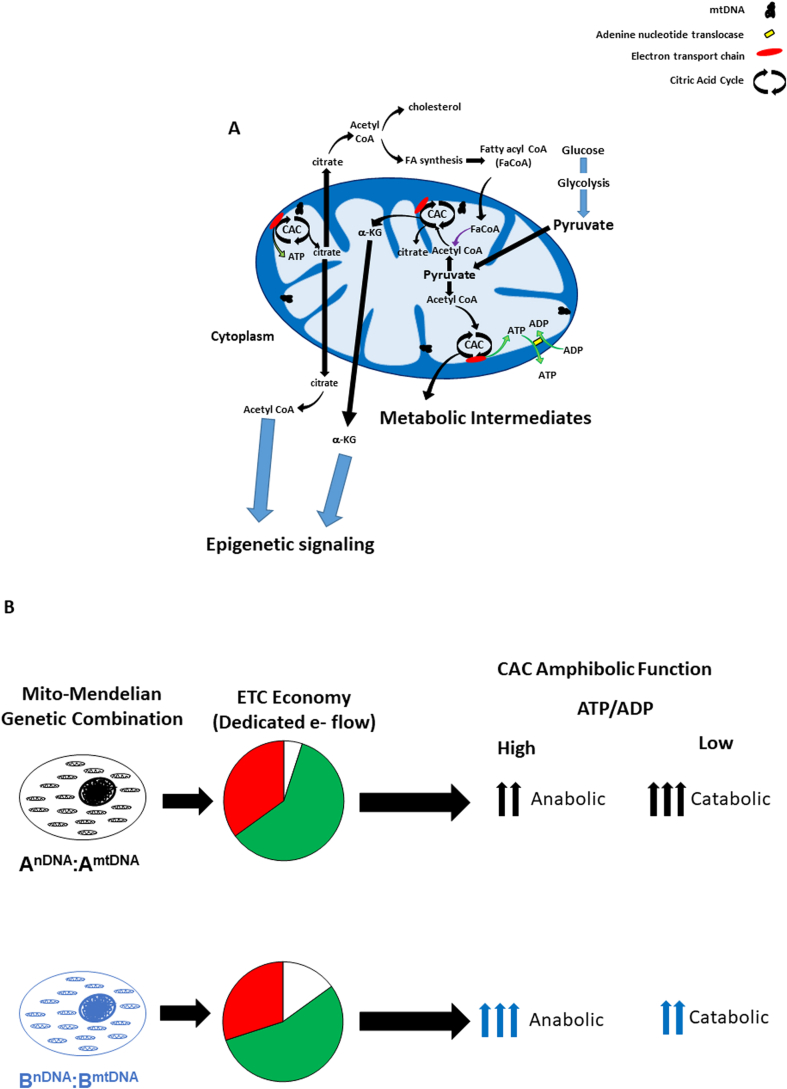
Also known as the tricarboxylic acid or Krebs cycle, the citric acid cycle (CAC) is an amphibolic cycle – it is the hub of eukaryotic cellular metabolism, serving as a source of substrates for both catabolic and anabolic metabolism that are essential for energy generation and multiple biosynthetic processes including the synthesis of fatty acids, cholesterol, amino acids, purines, porphyrins and carbohydrates that participate in lipogenesis, protein biosynthesis, and gluconeogenesis [105] (Fig. 3A). Anaplerotic reactions replace CAC intermediates that are utilized in biosynthesis pathways, while cataplerotic reactions remove excess intermediates [112]. Glucose and glucogenic amino acid carbon skeletons and triglycerides can be metabolized to pyruvate which is oxidatively decarboxylated by pyruvate dehydrogenase to acetyl-CoA (coenzyme A) [105]. Additionally, ketogenic amino acid carbon skeletons and fatty acids can be converted directly to acetyl-CoA [105]. In the initiating reaction of the CAC, citrate synthase catalyzes the reaction of acetyl-CoA with a 4-carbon oxaloacetate to form citrate and CoA [105]. Glucogenic amino acid carbon skeletons can also enter the CAC by being metabolized into intermediates such as α-ketoglutarate, succinyl CoA, fumarate or oxaloacetate [105] which is especially important for anaplerotic maintenance of CAC intermediate levels during gluconeogenesis and lipogenesis [112]. During gluconeogenesis, malate (a CAC intermediate) is shuttled out of the mitochondrion into the cytosol and oxidized (NADH:malate dehydrogenase) to form oxaloacetate (while oxaloacetate is a CAC intermediate, it is not transported across the mitochondrial membrane) that can be decarboxylated by PEP carboxykinase (PEPCK) to form phosphoenolpyruvate (PEP) and converted to glucose [112]. Similarly, during lipogenesis, citrate is shuttled from the mitochondrion into the cytosol and cleaved by ATP-citrate lyase to yield acetyl-CoA and oxaloacetate. Oxaloacetate can be reduced to malate (malate dehydrogenase) and converted to pyruvate by malic enzyme which generates NADPH, which in combination with malonyl-CoA (formed via carboxylation of acetyl-CoA) and acetyl-CoA form the building blocks of fatty acids which combine with glycerol to make lipids [113].
Fig. 3.
Mitochondrial citric acid cycle (CAC) metabolism provides fuel for multiple cellular processes (see text for greater detail). A) Glucose, glucogenic amino acid carbon skeletons and triglycerides can be metabolized to pyruvate which is oxidatively decarboxylated to form acetyl-CoA. Mitochondrial ATP is translocated out of the mitochondrion with exchange of ADP by the adenine nucleotide translocase (ANT). During lipogenesis, citrate is shuttled out of the mitochondrion into the cytosol and cleaved to yield acetyl-CoA that can utilized for fatty acid or cholesterol synthesis. In turn, fatty acid oxidation in the mitochondrion (fatty acyl CoA, FaCoA) generates acetyl CoA from fatty acids. Multiple CAC metabolic intermediates can be used for a variety of cellular biosynthetic pathways, including citrate and α-ketoglutarate, which play important roles in epigenetic processes (see text, Epigenetic Signaling). B) The mito-Mendelian paradigm hypothesizes that genetic mutations which convey changes in mitochondrial economy (e.g. dedicated e- flow for ATP production) impact the amphibolic balance of the CAC. For example, the bioenergetic profile from the AnDNA:AmtDNA combination shows greater dedicated e- flow for ATP production relative to the BnDNA:BmtDNA combination (pie charts). Hence, as ATP/ADP ratios increase, the demand for CAC generated reducing equivalents for energy production (oxidative phosphorylation) declines, and CAC flux shifts towards a more anabolic balance – yet because the AnDNA:AmtDNA combination utilizes more e- flow for ATP production compared to the BnDNA:BmtDNA combination, less metabolic intermediates are available for anabolic processes relative to the BnDNA:BmtDNA combination. Conversely, as ATP/ADP ratios decrease, the AnDNA:AmtDNA combination exhibits higher level of catabolism relative to the BnDNA:BmtDNA combination to meet energetic needs due to its lower mitochondrial economy.
Flux of metabolites through the CAC change with energy demand; as ATP levels increase with a concomitant decrease in ADP, the demand for CAC generated reducing equivalents (NADH and FADH2) for energy production (oxidative phosphorylation) declines, and CAC flux shifts towards a more anabolic balance (both citrate and α-ketoglutarate, two of the major CAC metabolites used for a variety of cellular processes, are generated on the “front end” of the CAC, after production of 1 NADH). Consequently, a decrease in reducing equivalent production will lead to greater availability of metabolites (e.g. citrate and α-ketoglutarate) for other cell processes. Conversely, as energy demand increases and ADP levels increase, multiple fuel sources can be utilized via the CAC for NADH and FADH2 production to provide electrons for ETC and oxidative phosphorylation. The mito-Mendelian paradigm predicts that genetic mutations which convey differences in mitochondrial economy between individuals will therefore impact the amphibolic balance of the CAC, and thus multiple cellular processes and systems (Fig. 3B).
1.8. ATP production
Reducing equivalents (NADH and FADH2) generated via the CAC donate electrons to complex I (NADH-coenzyme Q, or NADH dehydrogenase) and complex II (succinate dehydrogenase, a component of both the CAC and ETC), respectively [105].
These electrons reduce ubiquinone (coenzyme Q) to form ubiquinol, that transfers electrons to complex III (cytochrome bc1 complex or ubiquinol-cytochrome c oxidoreductase), which in a two-step process known as the Q-cycle (involving three subunits of complex III: cytochrome b, cytochrome c1 and the Rieske protein), provides electrons sequentially to reduce cytochrome c [105]. Next, reduced cytochrome c is oxidized at complex IV (cytochrome c oxidase), which donates electrons to oxygen, the final electron acceptor, producing water [105]. As electrons are shuttled through the electron transport, energy is utilized to: i) generate heat, and ii) pump protons from the matrix into the intermembrane space, creating an electrochemical gradient across the inner membrane. The former contributes to our ability to maintain thermostability and the “proton” gradient created by the latter is utilized at ATP synthase (complex V) to convert ADP + Pi to generate ATP through oxidative phosphorylation. ATP synthase consists of a hydrophobic Fo subunit (oligomycin sensitive) that forms a membrane spanning pore for proton movement attached to the hydrophilic F1 subunit located in matrix that hydrolyzes ADP to form ATP. Recent studies have shown that different nuclear – mtDNA combinations can significantly influence individual mitochondrial economy for ATP production, in vivo [21,24], and in vitro [111]. Interestingly, mtDNA SNPs linked to these changes in humans [111] have previously been linked to differences in mitochondrial matrix pH and intra-mitochondrial calcium levels, which can impact ATP synthesis [114,115]. As discussed below (Oxidant production and signaling), as ATP levels rise and ADP decline, electron carriers remain in the reduced state (in coupled mitochondria), generating oxidants.
1.9. Heat production
As electrons shuttle through the ETC, energy is used for work (pumping protons) and lost as heat. In this fashion, mitochondrial respiratory function can be modulated to aid thermogenic homeostasis. Albeit controversial, Chretien and coworkers have reported that a temperature shift exists between the mitochondrion and surrounding cytosol with up to a 10 °C differential, which could be modulated through depletion of the mtDNA or inhibition of respiration [102,103]. In 2004, Ruiz-Pesini and coworkers hypothesized that mutations in the mtDNA enhanced the thermogenic capacity of the organelle and thus, enabled the expansion and survival of prehistoric humans in colder climates as they migrated out of Africa [116]. As part of the mitochondrial paradigm for common diseases, Wallace proposed the bioenergetic changes that accompanied increased/decreased thermogenesis impacted individual risk of contemporary disease development [13]. Several reports using insect models have provided experimental results consistent with the concept that environmental characteristics (e.g. temperature) can impact mitochondrial function and mtDNA haplotype fitness [[117], [118], [119], [120], [121], [122], [123]], as have correlative studies in humans regarding adaptation to cold climates [115,116,124,125]. Interestingly, two mtDNA polymorphisms (np 10,398 – ND3; np 8701 – ATP6), previously shown to have a “robust” association with climate [115] have also been linked to differences in longevity and susceptibility to a variety of neurological diseases, cancer and diabetes [[126], [127], [128], [129], [130], [131], [132], [133]]. These same polymorphisms have also been associated with differences in ATP production, matrix pH and intra-organelle calcium levels [111,114]. Additionally, another study that compared wild-type nematodes (Caenorhabditis elegans) from different climates (England and Hawaii) found that mtDNA haplotype variation directly impacted several parameters of mitochondrial metabolism in response to environmental temperatures – further, those studies also examined the effects of different mtDNAs on the same nuclear background and found effects on lifespan [134]. Consequently, while clearly provocative – accumulating evidence appears consistent with the concept that climatic challenges can influence mitochondrial genome variation, and importantly, can convey changes in organelle function.
Manipulation of mitochondrial metabolism using uncoupling agents such as 2, 4 dinitrophenol (DNP) as a means for weight loss was widespread in the U.S. in the 1930s [135,136], but also had adverse side effects including fatal hyperthermia, resulting in action by the FDA by 1938, declaring DNP toxicity was too great to be used under any circumstance [137]. More recently, low dose DNP or MP201 (prodrug of DNP) administration has been used as a tool for inducing mitochondrial uncoupling in animal disease models with noted reported beneficial effects in neurodegenerative disease (attributed to adaptive responses, including decreased oxidants and stabilization of membrane potentials) and diet induced obesity [[138], [139], [140], [141]]. While perhaps contentious (in the use of DNP), these studies suggest that pharmaceutical targeting of mitochondrial metabolism may be of potential utility for treating or preventing common diseases.
1.10. Oxidant production and signaling
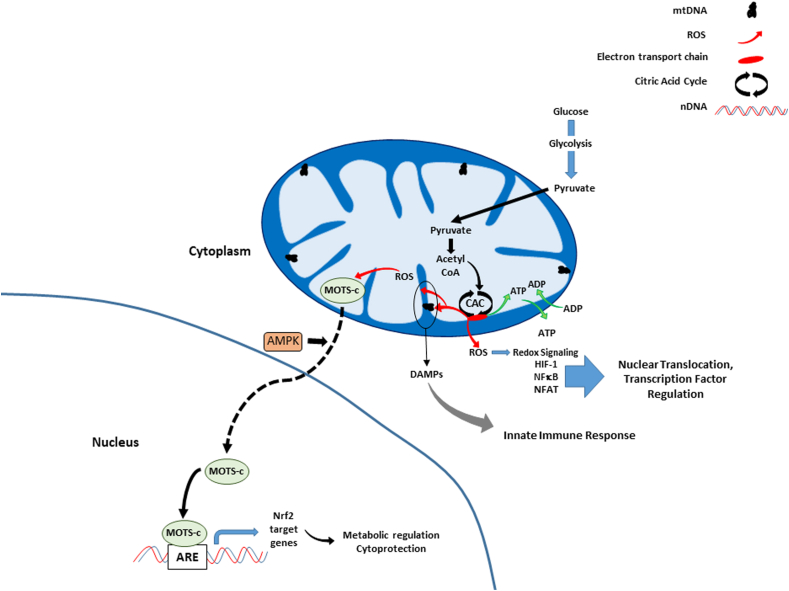
Fig. 4 summarizes the various effects of mitochondrial generated reactive oxygen species (ROS) on cell signaling. ROS are produced by mitochondria as a consequence of electron transport and oxidative phosphorylation [[142], [143], [144], [145], [146], [147]], usually in the form of the free radical superoxide (O2.-), that can be spontaneously converted to the freely diffusible oxidant hydrogen peroxide (H2O2) [[148], [149], [150], [151], [152]] and even more rapidly in the presence of superoxide dismutase of which mitochondria have 2 isoforms: SOD1 and SOD2 found in the intermembrane space and mitochondrial matrix, respectively [[153], [154], [155], [156], [157], [158]]. Thus far 11 sites of ROS production have been identified in mammals including 4 NAD-linked dehydrogenases, 4 ubiquinone-linked dehydrogenases, two sites on complex I and one on complex III of the electron transport chain [149,150]. Although previously thought of as harmful by-products, at low levels mitochondrial ROS participate in cell signaling [106,153,[159], [160], [161], [162]], and acutely at higher levels, are important in cellular defense mechanisms (apoptosis, autophagy, mitophagy and the inflammatory response) [148,153,[163], [164], [165]]. Interestingly, MDPs have also been implicated in ROS signaling – Kim and coworkers showed that one MDP, mitochondrial open reading frame of the 12S rRNA-c (MOTS-c), can translocate from the mitochondrion to the nucleus in response to oxidative stress [85]. This translocation was AMP activated protein kinase (AMPK) dependent, and once inside the nucleus, MOTS-c directly bound at antioxidant response element (ARE)-containing promoter regions of target genes for Nuclear factor erythroid 2-related factor 2 (Nrf2), a transcription factor which responds to ROS. In this manner it was found that MOTS-c was able to regulate gene expression in manner that increased cellular resistance to oxidative stress.
Fig. 4.
Mitochondrial oxidant production and cell signaling. Mitochondrial ROS generated during electron transport diffuse out of the organelle, affecting expression of several transcription factors, including HIF-1, NFκB, and NFAT. At high or chronic levels, ROS causes damage to the mtDNA and organelle proteins, which can initiate innate immune responses via damage associated molecular patterns (DAMPs) release (see text, Immune functions and cell signaling). Similarly, in response to oxidative stress, a mitochondrial derived peptide MOTS-c, translocates from the mitochondrion to the nucleus in an AMP-kinase (AMPK) dependent manner where it binds several genes, including those with antioxidant response element (ARE)-containing promoter regions, and interacts with associated transcription factors such as Nrf2 to protect the cell against oxidative damage.
However under chronic conditions, sustained oxidant production leads to mitochondrial and cellular damage, can trigger innate immune response (covered in next section), and be pathogenic [159,161,166,167]. Indeed, increased mtDNA damage has been linked to several common diseases known to have an oxidative stress component, including diabetes, cardiovascular disease, neurodegenerative disease, and aging [[168], [169], [170], [171]]. Similarly, sustained increases in mitochondrial oxidant stress have been shown to increase disease susceptibility [169,172]. Mitochondrial oxidant production is linked to organelle function and mitochondrial economy under coupled conditions [24,111]. For example, under conditions of low ATP/ADP and high energy demand, electron carriers remain in a more oxidized state, with low electron flow resistance that results a relative decrease in oxidant production; as energy demands are met and ATP/ADP levels increase, electron carriers remain in a more reduced state which increases electron flow resistance. With increased resistance, more electron “leak” from the ETC occurs, and reaction with oxygen forms, superoxide (O2.-), which is converted to hydrogen peroxide (H2O2), a freely diffusible oxidant that can act as cell signaling molecule [70,173]. Thus, mitochondrial oxidant production serves as a metabolic signal to the cell, regulating energy production, storage and other cellular functions [108,[174], [175], [176], [177]]. In this respect, different nuclear – mtDNA combinations that convey differences in mitochondrial economy will also have effects upon organelle oxidant production (higher economy will also be linked to increased levels reduced electron carriers) [21,24,111]. This may help to explain the disparity in susceptibility to metabolic diseases reported in the literature where African Americans have been shown to have increased susceptibility to metabolic diseases including cardiovascular disease, type 2 diabetes and obesity compared to European Americans [[178], [179], [180], [181], [182]]. Human umbilical vein endothelial cells generated from African Americans have more tightly coupled (or more economical) mitochondrial electron transport, having a lower oxygen consumption rate to produce the same level of ATP, as well as increased mitochondrial DNA damage compared to European American derived cells [183]. Work in mitochondrial nuclear exchange mice [184] have shown that switching mitochondrial DNA background can alter mitochondrial economy and/or disease progression in mouse models of non-alcoholic fatty liver disease, cardiovascular disease, obesity, breast cancer and premature infant lung development [20,24,46,47,185].
1.11. Immune functions and cell signaling
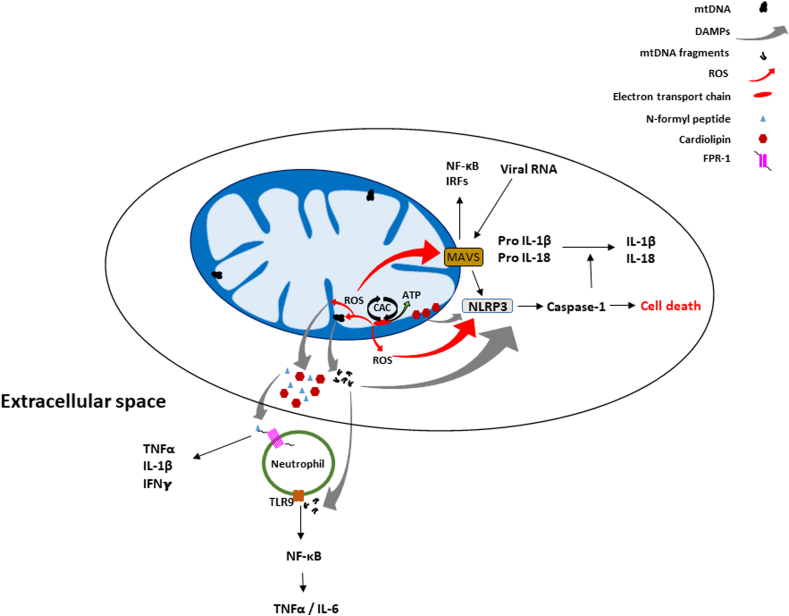
It has been shown that components of the mitochondrion can initiate innate immune response and related signaling cascades (Fig. 5) [109,186]. The α-proteobacterial origin of the mitochondrion is likely the basis of these responses; mtDNA and other mitochondrial constituents such as N-formyl peptides or cardiolipin, upon release from the organelle, can elicit the innate immune response, acting as damage-associated molecular patterns (DAMPs) [109,186]. Outside the mitochondrion, the mtDNA interacts with TLR9 (toll-like receptor 9) which activates NF-κB (nuclear factor kappa B) signaling and transcription of proinflammatory cytokines such as TNFα (tumor necrosis factor alpha) and IL-6 (interleukin 6) [[187], [188], [189]]. Both mtDNA and mitochondrial reactive oxygen species (mROS) can activate the nucleotide-binding oligomerization domain (NOD), leucine-rich repeat (LRR)- containing protein (NLR)-like receptor family pyrin domain containing 3 (NLRP3) inflammasome triggering inflammation (production of IL-1β and IL-18) and cell death [[190], [191], [192]]. N-formyl peptides stimulate the secretion of cytokines by activating the formyl peptide receptor 1 (FPR-1) receptor [188]. Mitochondria also have mitochondrial anti-viral signaling (MAVS) proteins on their outer mitochondrial membrane which are activated by proteins that detect viral RNA [193]. MAVS proteins can induce immune and inflammatory gene expression through the regulation of NF-κB and interferon regulatory factor [194,195]. As with the NLRP3 inflammasome, mROS can activate MAVS proteins, independently of the cytosolic viral sensors [186]. MAVS also promotes the oligomerization and activation of the NLRP3 inflammasome, while cardiolipin recruits NLRP3 to the outer membrane during mitochondrial membrane depolarization when it translocates from the inner to the outer membrane [196].
Fig. 5.
Mitochondrial mediated immune functions and cell signaling. Mitochondria can release molecules such as N-formyl peptides, mtDNA, ROS, and cardiolipin that initiate an immune response and related signaling pathways. N-formyl peptides are released and act as chemoattractants for neutrophils, binding to the FPR-1 receptor and promoting their activation and release of pro-inflammatory cytokines including TNFα, IL-1β, and IFNγ. MtDNA fragments (e.g. when the mtDNA is damaged by ROS) released from the organelle can also bind TLR9 receptors on neutrophils and likewise initiate the NF-κB pathway and release of TNFα and additional cytokines. Mitochondrial anti-viral signaling (MAVS) located on the outer membrane can be activated by viral RNA and/or. ROS, also triggering activation of the NF-κB pathway and the release of interferon regulating factors (IRFs). Additionally, MAVS can promote the oligomerization and activation of the NLRP3 inflammasome which triggers Caspase-1 cell death pathways and the activation of the pro-inflammatory forms of IL-1β and IL-18. In addition to being activated by MAVS, NLRP3 can also be activated by ROS, mtDNA, and cardiolipin.
1.12. Calcium regulation
Ca2+ uptake and storage by the mitochondrion regulates cell survival, metabolism, secretion and signaling. It was in the 1960's when the initial papers describing the capacity of mitochondria to uptake Ca2+ [197,198]. Interestingly, when the uptake system was better characterized, it was found that it had a relatively low affinity for Ca2+, and it was not until the advent of targeted Ca2+ probes to the mitochondrion in living cells [199] that it was discovered this low affinity for Ca2+ was overcome via direct organelle interactions with the endoplasmic reticulum (ER), creating microdomains of high [Ca2+] [[200], [201], [202]]. Mitochondria can form junctions with the endoplasmic reticulum (ER), sarcoplasmic reticulum or plasma membrane to regulate Ca2+ levels in these microenvironments [110]. The formation of junctions in the ER involves the association of mitochondrial fusion proteins mitofusin 1 and 2 (MFN1 and MFN2) on both membranes [203,204]. It is also well established that regulation of mitochondrial Ca2+ levels impacts cell survival. Whereas the processes and proteins responsible for the control and regulation of Ca2+ flux in the mitochondrion have been sought for over 4 decades, a series of reports, initially describing the mitochondrial Na+/Ca2+ exchanger (Na+/Ca2+ Li+ -permeable Exchanger – NCLX) [205], which regulates mitochondrial Ca2+ efflux, the mitochondrial calcium uptake 1 protein (MICU1, also known as CBARA1 or EFHA3), involved in organelle Ca2+ uptake [206], and the identification of the Ruthenium Red sensitive Ca2+ channel that resides in the mitochondrial inner membrane, called the mitochondrial calcium uniporter (MCU) [207,208] which is conserved in metazoans (e.g. multicellular animals with differentiated tissues) [209], were published, followed by several papers [[210], [211], [212], [213], [214]] identifying additional regulators of mitochondrial Ca2+ influx. Overall, there is agreement that the MCU complex is a multi-molecular complex that functions as a regulator of Ca2+ uptake in vivo, and perhaps not surprisingly, its components may be tissue/cell specific. While beyond the scope of this review, several reviews are available regarding Ca2+ regulation in the mitochondrion and the MCU complex [[215], [216], [217], [218], [219]].
Ca2+ overload with increased oxidant production influences opening of the permeability transition pore (PTP) that causes collapse of membrane potential and mitochondrial swelling, loss of pyridine nucleotides, and cytochrome c which result in bioenergetic failure and necrotic death [220,221]. Sustained Ca2+ level elevation in the mitochondrion can also result in apoptosis, both by triggering PTP opening and by causing fission of mitochondria which results in cytochrome c release [222,223]. Increased expression of anti-apoptotic proteins such as Bcl-2 (B cell lymphoma 2) which reside in both the mitochondrion (where it interacts with VDAC2) and the ER (where it interacts with ER Ca2+ transporters to inhibit ER Ca2+ release), to inhibit apoptosis can provide a degree of protection [110,224] – aspects of mitochondrial mediated apoptosis are discussed below.
1.13. Apoptosis
Mitochondria are involved in both intrinsic and extrinsic programmed cell death pathways [162]. In the intrinsic pathway, pro-apoptotic stimuli activate Bcl-2-like protein 4 (BAX) and Bcl-2 homologous antagonist/killer (BAK) proteins, which inhibit the protective effects of Bcl-2, causing an influx of calcium into the mitochondrion while also promoting outer mitochondrial membrane permeabilization, inner mitochondrial membrane cristae remodeling and permeability transition pore opening, which releases cytochrome c and second mitochondria-derived activator of caspases (SMAC) [110,162]. Cytochrome c is a caspase cofactor that is sequestered in inner mitochondrial membrane cristae, however, when released into the cytosol it interacts with apoptotic protease-activating factor 1 (APAF1) to form an apoptosome which recruits pro-caspase 9 and results in a proteolytic cascade and apoptosis. SMAC binds to the inhibitor of apoptosis proteins (IAP), resulting in the activation of caspases and initiating apoptosis [162]. In extrinsic apoptosis, the death receptor, located in the cell membrane, binds to the ligand which activates caspase-8 in the cytosol at the intracellular tail of the receptor [162]. Some cells require mitochondrial outer membrane permeabilization (MOMP) for extrinsic apoptosis (type II cells) while other cells do not require MOMP and can activate executioner caspases directly causing more rapid apoptosis (type I cells) [162]. In type II cells, cardiolipin in the outer mitochondrial membrane recruits and activates caspase-8 [162]. With caspase-8 in close proximity to the mitochondrion, it cleaves and activates the BH3 (Bcl-2 homology domain 3) interacting-domain death agonist (BID), leading to BAX and BAK activation, mitochondrial outer membrane permeabilization and apoptosis [225].
1.14. Glucose homeostasis
Mitochondria play an integral part in normal glucose metabolism and homeostasis. When blood glucose levels increase, glucose is converted to reducing equivalents which the mitochondrion uses to make ATP. This ATP binds to ATP-sensitive potassium (KATP) channels in the insulin secreting pancreatic beta cells, closing them and causing an influx of calcium through voltage dependent calcium channels and the exocytosis of insulin containing vesicles into the bloodstream, whereby insulin can bind insulin receptors in the muscle, liver and adipose tissues leading to increased expression of the glucose transporter, GLUT4 in muscle and adipose, decreased gluconeogenesis and glycogenolysis in the liver, increased glycogen synthesis in liver and muscle, increased fat storage (increased lipid synthesis, decreased lipolysis, increased esterification of fatty acids), decreased proteolysis and increased amino acid uptake via the CAC to provide intermediates for biosynthetic pathways including those stimulated by insulin such as gluconeogenesis and lipogenesis [112].
1.15. Epigenetic signaling
Mitochondria can regulate, and be regulated by the epigenome [226]. While histone acetylation influences chromatin structure and thus, can regulate gene expression [227,228], mitochondrial metabolism regulates the level of acetyl CoA, which is utilized by histone acetyltransferases; further, levels of α-ketoglutarate and succinate impact both histone demethylases (Jumonji-C domain, JmjC) and the ten-eleven translocation (TET) family of dioxygenase activities, which play roles in histone and DNA demethylation, respectively (Fig. 6) [108,229,230]. In addition to α-ketoglutarate and succinate, it has been reported that other intermediates and metabolites (i.e. fumarate, 2-hydroxyglutarate, and NAD+) also have roles in epigenetic signaling [[231], [232], [233]]. For example, cancer studies have shown that when fumarate hydratase is mutated there is an increase in fumarate (a CAC cycle intermediate) which is a competitive inhibitor of several α-ketoglutarate dependent dioxygenases including histone demethylases [231,232]. D-2-hydroxyglutarate, formed from α-ketoglutarate in response to mutations in IDH and IDH2 [234,235], is able to competitively inhibit dioxygenases due to its structural similarity to α-ketoglutarate [233,236]. NAD+ regulates epigenetic signaling through its interaction with sirtuins [[237], [238], [239]]. Sirtuins are a class of histone deacetylases which remove acetyl groups from lysine residues on proteins and transfers them to the ADP-ribose component of NAD+ [237]. Therefore, these proteins are classified as NAD+ dependent deacetylases [238]. The activity of sirtuins has been shown to be dependent on several factors including nutrient availability. For example, caloric restriction results in an increased NAD+:NADH ratio and therefore high sirtuin activity and increased deacetylation of histones; whereas caloric excess results in a decreased NAD+:NADH ratio, low sirtuin activity and decreased deacetylation [239].
Fig. 6.
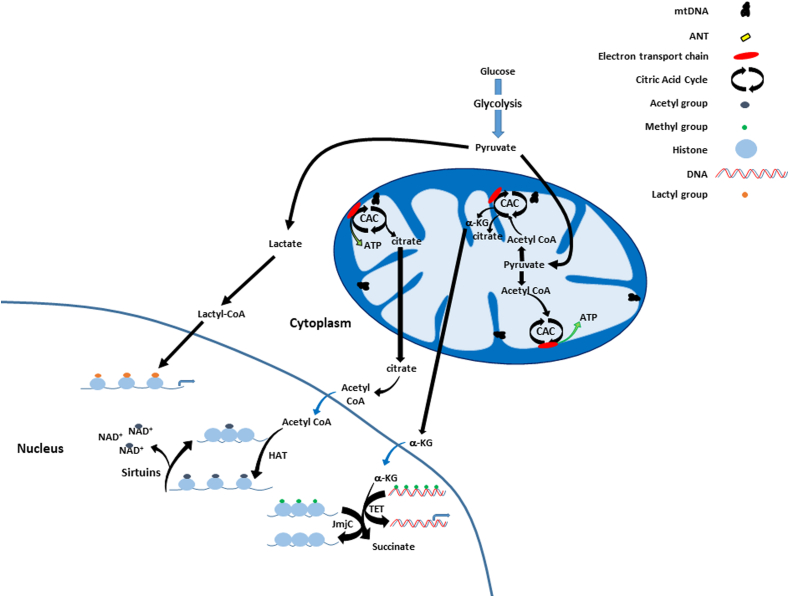
Mitochondrial regulation of the epigenome. Mitochondria produce citrate which when converted to acetyl CoA is utilized by histone acetyltransferases (HAT) for histone acetylation. Levels of α-ketoglutarate (α-KG), succinate, fumarate and D-2-hydroxyglutarate (see text) also impact histone demethylase activities (Jumonji-C domain, JmjC) and the ten-eleven translocation (TET) family of dioxygenases, which regulate histone and DNA demethylation, respectively. Sirtuins act as deacetylases by transferring acetyl groups from histones to NAD+. Through histone lactylation, lactate-derived (Lactyl-CoA) lactyl groups are transferred to histones promoting transcription of homeostatic genes.
Another form of epigenetic modification has recently been discovered – histone lactylation, which involves the transfer of lactyl groups to lysine residues in histones [240,241]. When mitochondrial metabolism is increased, pyruvate from glycolysis is transported into the mitochondrion where it enters the CAC. However, under hypoxic conditions or when mitochondrial respiration is inhibited (i.e. by rotenone), pyruvate is converted to lactate which after conversion to lactyl-CoA, transfers a lactyl group to histones [240]. Histone lactylation promotes transcription of homeostatic genes including Tnf. It has been suggested that histone acetyltransferase p300 may be the enzyme which facilitates lactylation [240].
Mitochondrial function also regulates histone/DNA methylation via S-adenosyl methionine (SAM, is required for both histone and DNA methylation) levels which are dependent upon both the folate cycle (required for the conversion of homocysteine to methionine) and ATP (required for phosphorylation of methionine to SAM). Interestingly, it has been reported that enzymes important for demethylation have been found in the mitochondrion – consistent with reports that the mtDNA has no or very low levels of methylation compared to the nuclear DNA [[242], [243], [244]]. The physiologic significance of these differences in methylation between the mtDNA and nuclear genome currently remain unclear. Recently, changes in heteroplasmy associated with a known pathogenic mtDNA mutation [tRNALeu(UUR) 3243A > G] have been shown to not only affect mitochondrial metabolic function (as expected), but also alter nuclear gene expression through histone modifications that are regulated by acetyl CoA and a-ketoglutarate levels [245]. Because mitochondrial function has been shown to decline with age (which is also accompanied by increased mtDNA damage and somatic mutagenesis), and since differences in metabolism and gene expression have been linked with mtDNA–nDNA background combinations [20,21,24,246], it is reasonable to consider they are integrated in a manner that also impacts the epigenome. Consequently, different nuclear – mtDNA genetic combinations capable of altering organelle economy should also significantly impact the availability of CAC intermediates such as citrate and α-ketoglutarate, and thus, impact individual levels of epigenetic modifications.
1.16. Mito-Mendelian genetics
As the mitochondrion and “host” co-evolved, the former lost the majority of its genetic material to the nucleus, but retained key catalytic subunits for ETC and essential components for the translation (tRNAs, rRNAs) of the mtDNA encoded subunits. Many studies have examined the impact of pathogenic mtDNA mutations upon cell function and disease development [171,[247], [248], [249], [250], [251], [252], [253], [254]]. While studies coming from the viewpoint of “normal” mtDNA polymorphisms or heterogeneity on cell function are less common, reports of mitochondrial – nuclear interactions as a normal process for maintaining metabolic homeostasis beyond anterograde or retrograde signaling are perhaps even less frequent, and studies that consider genetic interaction between the mitochondrion and nucleus as the basis for common disease risk even more rare. This is likely due to the taught viewpoint that the genetic etiology for normal cell physiology is based upon Mendelian paradigms originally defined and developed a century before the discovery of the mtDNA [255]. Studies in the cancer literature were perhaps among the first to reveal a direct role for nuclear-mtDNA heterogeneity in disease progression using in vitro and orthotopic approaches [256,257], which were later followed using “Mitochondrial – Nuclear eXchanged” (MNX) in vivo models [21,258]. Use of congenic breeding strategies has also provided evidence consistent with the concept that different nuclear – mitochondrial genetic background combinations can impact longevity and metabolism [48,[259], [260], [261]]. More direct approaches utilizing MNX models have now shown that different nuclear – mitochondrial combinations impact cancer, heart failure, whole body metabolism, body composition, gene expression, and susceptibility to hyperoxic injury [20,21,24,47,258]. A recent study has shown that nuclear – mitochondrial background combination collectively forms a genetic complement that regulates response to stimuli which is consistent with the notion of nuclear and mitochondrial co-evolution. Dunham-Snary and coworkers have shown that the presence of either the nucleus or mtDNA from particular mouse strains directionally altered differential expression of genes in response to diet, and importantly, while ~40% of gene expression could be linked with either a particular nuclear or mtDNA background individually, the majority (~60%) of the genes found to be differentially expressed were linked with specific nuclear-mtDNA genetic combinations – further, differential expression of genes associated with metabolic processes and biological regulation were those most impacted by different nuclear-mtDNA genetic combinations. Interestingly, these differences were also linked to changes in adiposity and whole body metabolism [20]. Another recent report by Kopinski and coworkers revealed that changes in mitochondrial bioenergetics, mediated by a pathogenic mtDNA mutations impacted epigenetic signaling and thus gene expression [245]. Overall, the notion that mito-Mendelian genetic processes define cellular response to environmental stimuli is consistent with these findings, and importantly, the symbiogenic origins of the eukaryote [20,22].
Of the 89 known protein subunits involved in mammalian ETC and ATP synthase, 13 and 76 are encoded by the mtDNA and nucleus, respectively. Studies that have compared the mutation rates between the two genomes have consistently shown a higher mutation/fixation rate in the mtDNA compared to its nuclear counterpart [[262], [263], [264], [265], [266]]. This poses an interesting dynamic in terms of their co-evolution which suggests the higher rate of mutation in the mtDNA may drive selection and fixation, and thus, adaptation; several studies have shown a strong correlation between nuclear encoded proteins that interact with mtDNA encoded ETC subunits [[263], [264], [265],[267], [268], [269]]. In this respect, studies have shown that co-evolution or co-adaptation occurs in a manner that alters metabolic function in a fashion favorable to environmental adaptation. Studies in several vertebrate species have supported these concepts [[270], [271], [272], [273], [274]], consistent with the hypothesis that genes associated with metabolism (mitochondrial and nuclear encoded), play a role in adaptive metabolism and natural selection in vertebrates. Because the mutational load of mtDNA exceeds that of nDNA by an order of magnitude, the higher mutational load of mtDNA may be compensated by transcriptional flexibility that involves coadaptation of pleiotropic genes. Consequently, the mitochondrial genome appears to have evolved to modulate key aspects of organelle function, which based upon environmental stimuli, can activate processes that regulate many aspects of cell function, including gene expression and adaptive response. Interestingly, while these concepts have been considered for many years in the field of evolutionary biology and genetics, a lag exists regarding their embracement concerning the basis of normal physiologic response or development of disease. A cognitive gauntlet is the lack of appreciation of how a single, non-pathogenic mtDNA mutation can change mitochondrial metabolism. As discussed herein, we propose that even subtle changes in mitochondrial economy (utilization of electron flow for ATP generation) can manifest in differences in oxidant signaling, metabolites and immune response, which over time, collectively impact a multitude of cellular pathways/functions (Fig. 7). From a genetic perspective, there are mutations (e.g. pathogenic) in both genomes that are highly penetrant, and therefore exhibit either Mendelian or mitochondrial genetic transmission [248,[275], [276], [277], [278]]; however, we predict that the bulk of heretofore “complex polygenic” diseases will have a mito-Mendelian component in which penetrance of potentially pathogenic mutations will be modulated by mitochondrial metabolism that is guided by mtDNA – nDNA background combination and environmental stressors (Fig. 7). While features of anterograde and retrograde signaling have been known for some time [279], and have been investigated and discussed as a means of influencing cell function in response to significant or acute cell stress, the concept of nuclear - mitochondrial genetic interaction as a means of central control for cellular function has not been generally considered outside of evolutionary biology. However, because the genetics of the eukaryote are the result of endosymbiotic events [280,281], it is logical to consider endosymbiotic or mito-Mendelian genetics as a means for explaining complex disease development and susceptibility.
Fig. 7.
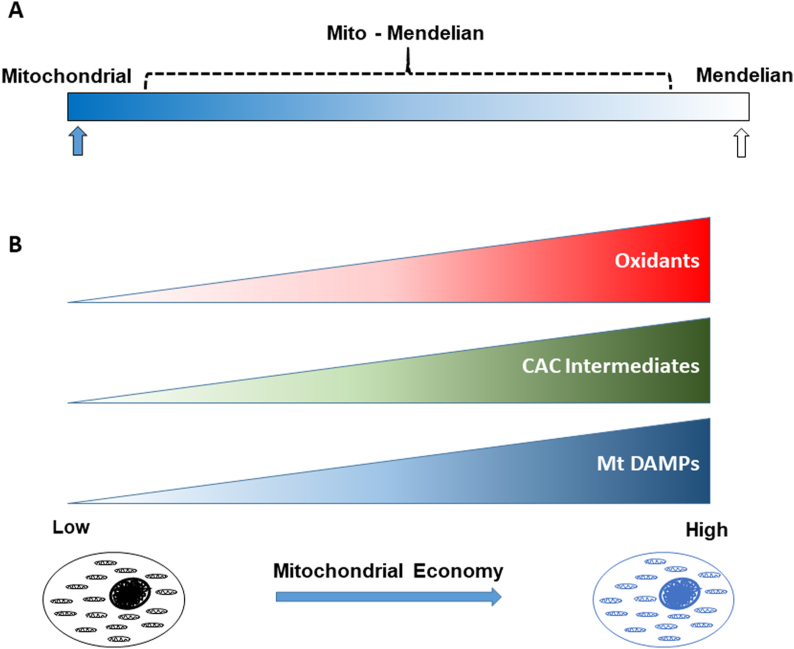
mito-Mendelian genetics and mitochondrial economy in penetrance and metabolism. A) Genetic mutations exist in both genomes (Mitochondrial and Nuclear) that irrespective of their genomic combination, will be highly penetrant and therefore exhibit either Mendelian or mitochondrial genetic transmission (indicated by blue and open arrows, respectively). By contrast, we predict that many common metabolic diseases will follow a mito-Mendelian form (indicated by bracket) of inheritance in which penetrance will be modulated by mitochondrial metabolism that is guided by mtDNA – nDNA background combination (in response to stress factors). B) After the energetic requirements of the cell are met, mtDNA-nDNA combinations that are linked with low (more utilization of electrons for ATP generation) or high (less utilization of electrons for ATP generation) levels of mitochondrial economy will be also linked to lower or higher levels of oxidant production, CAC intermediates, and mitochondrial (Mt) DAMPs with stress (e.g. positive energy balance), respectively.
Funding
This work is supported by NIH grants RO1 HL103859 (SWB), NIH Predoctoral Training Program in Cardiovascular Pathophysiology T32 HL007918 (MJS), A.G. and Minnie L. Gaston Fellowship (JAB) and a BCRFA award (SWB).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.redox.2020.101568.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Saklayen M.G. The global epidemic of the metabolic syndrome. Curr. Hypertens. Rep. 2018;20(2):12. doi: 10.1007/s11906-018-0812-z. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC . 2012. Overweight and Obesity. 2013. [Google Scholar]
- 3.CDC . 2012. Behavior, Environment, and Genetic Factors All Have a Role in Causing People to Be Overweight and Obese. [Google Scholar]
- 4.Schonfeld-Warden N., Warden C.H. Pediatric obesity. An overview of etiology and treatment. Pediatr. Clin. 1997:339–361. doi: 10.1016/s0031-3955(05)70480-6. 1997/04/01. [DOI] [PubMed] [Google Scholar]
- 5.Hill J.O., Peters J.C. 1998. Environmental Contributions to the Obesity Epidemic; pp. 1371–1374. 1998/06/20. [DOI] [PubMed] [Google Scholar]
- 6.Hu F.B. Plant-based foods and prevention of cardiovascular disease: an overview. Am. J. Clin. Nutr. 2003;78(3):544S–551S. doi: 10.1093/ajcn/78.3.544S. [DOI] [PubMed] [Google Scholar]
- 7.Hu F.B. Sedentary lifestyle and risk of obesity and type 2 diabetes. Lipids. 2003:103–108. doi: 10.1007/s11745-003-1038-4. 2003/05/08. [DOI] [PubMed] [Google Scholar]
- 8.Hu F.B. Television watching and other sedentary behaviors in relation to risk of obesity and type 2 diabetes mellitus in women. J. Am. Med. Assoc. : J. Am. Med. Assoc. 2003:1785–1791. doi: 10.1001/jama.289.14.1785. 2003/04/10. [DOI] [PubMed] [Google Scholar]
- 9.Farooqi I.S., O'Rahilly S. 2004. Monogenic Human Obesity Syndromes. Recent Progress in Hormone Research; pp. 409–424. 2004/01/30. [DOI] [PubMed] [Google Scholar]
- 10.Visscher P.M. 10 Years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 2017;101(1):5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins B.A. Precision medicine screening using whole-genome sequencing and advanced imaging to identify disease risk in adults. Proc. Natl. Acad. Sci. Unit. States Am. 2018;115(14):3686. doi: 10.1073/pnas.1706096114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzaga-Jauregui C., Lupski J.R., Gibbs R.A. Human genome sequencing in health and disease. Annu. Rev. Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005:359–407. doi: 10.1146/annurev.genet.39.110304.095751. 2005/11/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart J.B., Chinnery P.F. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 2015;16(9):530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 15.Bray A.W., Ballinger S.W. Mitochondrial DNA mutations and cardiovascular disease. Curr. Opin. Cardiol. 2017;32(3):267–274. doi: 10.1097/HCO.0000000000000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fetterman J.L., Sammy M.J., Ballinger S.W. Mitochondrial toxicity of tobacco smoke and air pollution. Toxicology. 2017;391:18–33. doi: 10.1016/j.tox.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aryaman J., Johnston I.G., Jones N.S. Mitochondrial heterogeneity. Front. Genet. 2019;9:718. doi: 10.3389/fgene.2018.00718. 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace D.C. Mitochondrial DNA variation in human radiation and disease. Cell. 2015;163(1):33–38. doi: 10.1016/j.cell.2015.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace D.C., Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harbor Perspec. Biol. 2013;5(11):a021220. doi: 10.1101/cshperspect.a021220. a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunham-Snary K.J. Mitochondrial - nuclear genetic interaction modulates whole body metabolism, adiposity and gene expression in vivo. EBioMedicine. 2018;36:316–328. doi: 10.1016/j.ebiom.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feeley K.P. Mitochondrial genetics regulate breast cancer tumorigenicity and metastatic potential. Canc. Res. 2015;75(20):4429–4436. doi: 10.1158/0008-5472.CAN-15-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunham-Snary K.J., Ballinger S.W. GENETICS. Mitochondrial-nuclear DNA mismatch matters. Science. 2015;349(6255):1449–1450. doi: 10.1126/science.aac5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunham-Snary K.J., Ballinger S.W. Mitochondrial genetics and obesity: evolutionary adaptation and contemporary disease susceptibility. Free Radic. Biol. Med. 2013;65:1229–1237. doi: 10.1016/j.freeradbiomed.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fetterman J.L. Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem. J. 2013;455(2):157–167. doi: 10.1042/BJ20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray M.W. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4(9):a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray M.W., Burger G., Lang B.F. The origin and early evolution of mitochondria. Genome Biol. 2001;2(6) doi: 10.1186/gb-2001-2-6-reviews1018. REVIEW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Wu M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 2015;5:7949. doi: 10.1038/srep07949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin W., Kowallik K.V. Annotated English translation of Mereschkowsky's 1905 paper ‘Über Natur und Ursprung der Chromatophoren im Pflanzenreiche’. Eur. J. Phycol. 1999;34(3):287–295. [Google Scholar]
- 29.Mereschkowsky, C., Über Natur und Ursprung der Chromatophoren imPflanzenreiche. Eur. J. Phycol.. 34(3): p. 287-295.
- 30.Sagan L. On the origin of mitosing cells. J. Theor. Biol. 1967;14(3):255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 31.Wallin I.E. The Williams & Wilkins Company; Baltimore, MD., USA: 1927. Symbionticism and the Origin of Species; p. 171. [Google Scholar]
- 32.Lake J.A. Lynn margulis (1938-2011) Nature. 2011;480(7378):458. doi: 10.1038/480458a. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz R.M., Dayhoff M.O. Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science. 1978;199(4327):395–403. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- 34.Martin W.F. The physiology of phagocytosis in the context of mitochondrial origin. Microbiol. Mol. Biol. Rev. 2017;81(3) doi: 10.1128/MMBR.00008-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin W.F., Garg S., Zimorski V. Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370(1678):20140330. doi: 10.1098/rstb.2014.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portier P. Masson; Paris: 1918. Les Symbiotes; p. 315. [Google Scholar]
- 37.Bell P.J. Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? J. Mol. Evol. 2001;53(3):251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- 38.Hartman H., Fedorov A. The origin of the eukaryotic cell: a genomic investigation. Proc. Natl. Acad. Sci. U. S. A. 2002;99(3):1420–1425. doi: 10.1073/pnas.032658599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McInerney J.O., Pisani D. Genetics. Paradigm for life. Science. 2007;318(5855):1390–1391. doi: 10.1126/science.1151657. [DOI] [PubMed] [Google Scholar]
- 40.Pisani D., Cotton J.A., McInerney J.O. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol. Biol. Evol. 2007;24(8):1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- 41.Ku C. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524(7566):427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Garcia P., Moreira D. Selective forces for the origin of the eukaryotic nucleus. Bioessays. 2006;28(5):525–533. doi: 10.1002/bies.20413. [DOI] [PubMed] [Google Scholar]
- 43.Takemura M. Poxviruses and the origin of the eukaryotic nucleus. J. Mol. Evol. 2001;52(5):419–425. doi: 10.1007/s002390010171. [DOI] [PubMed] [Google Scholar]
- 44.Devos D.P., Graf R., Field M.C. Evolution of the nucleus. Curr. Opin. Cell Biol. 2014;28:8–15. doi: 10.1016/j.ceb.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reichenbach H. Myxobacteria, producers of novel bioactive substances. J. Ind. Microbiol. Biotechnol. 2001;27(3):149–156. doi: 10.1038/sj.jim.7000025. [DOI] [PubMed] [Google Scholar]
- 46.Betancourt A.M. Mitochondrial-nuclear genome interactions in nonalcoholic fatty liver disease in mice. Biochem. J. 2014;46(2):223–232. doi: 10.1042/BJ20131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kandasamy J. Mitochondrial DNA variation modulates alveolar development in newborn mice exposed to hyperoxia. Am. J. Physiol. Lung Cell Mol. Physiol. 2019;317(6):L740–L747. doi: 10.1152/ajplung.00220.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Latorre-Pellicer A. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016;535:561–565. doi: 10.1038/nature18618. [DOI] [PubMed] [Google Scholar]
- 49.Zhao J., Lendahl U., Nistér M. Regulation of mitochondrial dynamics: convergences and divergences between yeast and vertebrates. Cell. Mol. Life Sci. 2013;70(6):951–976. doi: 10.1007/s00018-012-1066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cogliati S., Enriquez J.A., Scorrano L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci. 2016;41(3):261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Kuhlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015;13:89. doi: 10.1186/s12915-015-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fontanesi F. eLS; 2015. Mitochondria: Structure and Role in Respiration; pp. 1–13. [Google Scholar]
- 53.Krüger V. Identification of new channels by systematic analysis of the mitochondrial outer membrane. J. Cell Biol. 2017;216(11):3485–3495. doi: 10.1083/jcb.201706043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuznetsov A.V., Margreiter R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int. J. Mol. Sci. 2009;10(4):1911–1929. doi: 10.3390/ijms10041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wenz L.-S. Cooperation of protein machineries in mitochondrial protein sorting. Biochim. Biophys. Acta Mol. Cell Res. 2015;1853(5):1119–1129. doi: 10.1016/j.bbamcr.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 56.Bach D. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J. Biol. Chem. 2003:17190–17197. doi: 10.1074/jbc.M212754200. 2003/02/25. [DOI] [PubMed] [Google Scholar]
- 57.Chen H. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160(2):189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filadi R. Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proc. Natl. Acad. Sci. U. S. A. 2015;112(17):E2174–E2181. doi: 10.1073/pnas.1504880112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Laan M., Horvath S.E., Pfanner N. Mitochondrial contact site and cristae organizing system. Curr. Opin. Cell Biol. 2016;41:33–42. doi: 10.1016/j.ceb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 60.Scorrano L. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell. 2002;2(1):55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 61.Cogliati S. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155(1):160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lizana L., Bauer B., Orwar O. Controlling the rates of biochemical reactions and signaling networks by shape and volume changes. Proc. Natl. Acad. Sci. U. S. A. 2008;105(11):4099–4104. doi: 10.1073/pnas.0709932105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cobb L.J. Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging. 2016;8(4):796–809. doi: 10.18632/aging.100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S.J. Mitochondrially derived peptides as novel regulators of metabolism. J. Physiol. 2017;595(21):6613–6621. doi: 10.1113/JP274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S.J. Mitochondrial peptides modulate mitochondrial function during cellular senescence. Aging. 2018;10(6):1239–1256. doi: 10.18632/aging.101463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ku C. Endosymbiotic gene transfer from prokaryotic pangenomes: inherited chimerism in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 2015;112(33):10139–10146. doi: 10.1073/pnas.1421385112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robin E.D., Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell. Physiol. 1988;136(3):507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- 68.Satoh M., Kuroiwa T. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res. 1991;196(1):137–140. doi: 10.1016/0014-4827(91)90467-9. [DOI] [PubMed] [Google Scholar]
- 69.Iborra F.J., Kimura H., Cook P.R. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krzywanski D.M. The mitochondrial paradigm for cardiovascular disease susceptibility and cellular function: a complementary concept to Mendelian genetics. Lab. Invest. 2011;91(8):1122–1135. doi: 10.1038/labinvest.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burger G. Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 1996;93(6):2328–2332. doi: 10.1073/pnas.93.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Y. The role of mitochondria-derived peptides in cardiovascular disease: recent updates. Biomed. Pharmacother. 2019;117:109075. doi: 10.1016/j.biopha.2019.109075. [DOI] [PubMed] [Google Scholar]
- 74.Reynolds J.C., Bwiza C.P., Lee C. Mitonuclear genomics and aging. Hum. Genet. 2020;139(3):381–399. doi: 10.1007/s00439-020-02119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cobb L.J. Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging. 2016;8(4):796–809. doi: 10.18632/aging.100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hashimoto Y. Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein. Biochem. Biophys. Res. Commun. 2001;283(2):460–468. doi: 10.1006/bbrc.2001.4765. [DOI] [PubMed] [Google Scholar]
- 77.Hashimoto Y. Proceedings of the National Academy of Sciences. vol. 98. 2001. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Aβ; p. 6336. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee C., Yen K., Cohen P. Humanin: a harbinger of mitochondrial-derived peptides? Trends Endocrinol. Metabol. 2013;24(5):222–228. doi: 10.1016/j.tem.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee C. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metabol. 2015;21(3):443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hill S., Sataranatarajan K., Van Remmen H. Role of signaling molecules in mitochondrial stress response. Front. Genet. 2018;9(225) doi: 10.3389/fgene.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo B. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423(6938):456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 82.Guo B. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423(6938):456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 83.Ikonen M. Interaction between the Alzheimer's survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100(22):13042–13047. doi: 10.1073/pnas.2135111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim S.J. The mitochondrial-derived peptide MOTS-c is a regulator of plasma metabolites and enhances insulin sensitivity. Phys. Rep. 2019;7(13) doi: 10.14814/phy2.14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim K.H. The mitochondrial-encoded peptide MOTS-c translocates to the nucleus to regulate nuclear gene expression in response to metabolic stress. Cell Metabol. 2018;28(3):516–524 e7. doi: 10.1016/j.cmet.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim S.-J. Mitochondrially derived peptides as novel regulators of metabolism. J. Physiol. 2017;595(21):6613–6621. doi: 10.1113/JP274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holt I.J., Reyes A. Human mitochondrial DNA replication. Cold Spring Harbor Perspect. Biol. 2012;4(12):a012971. doi: 10.1101/cshperspect.a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Falkenberg M. Mitochondrial DNA replication in mammalian cells: overview of the pathway. Essays Biochem. 2018;62(3):287–296. doi: 10.1042/EBC20170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am. J. Hum. Genet. 2000;67(4):1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clay Montier L.L., Deng J., Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J. Gene. Genomics. 2009;36(3):125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y., Bogenhagen D.F. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281(35):25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 92.Fonseca M.M., Harris D.J., Posada D. The inversion of the Control Region in three mitogenomes provides further evidence for an asymmetric model of vertebrate mtDNA replication. PloS One. 2014;9(9) doi: 10.1371/journal.pone.0106654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ciesielski G.L., Oliveira M.T., Kaguni L.S. Animal mitochondrial DNA replication. Enzymes. 2016;39:255–292. doi: 10.1016/bs.enz.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McKinney E.A., Oliveira M.T. Replicating animal mitochondrial DNA. Genet. Mol. Biol. 2013;36(3):308–315. doi: 10.1590/S1415-47572013000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yasukawa T. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25(22):5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Molina A.J.A. Mitochondrial networking protects β-cells from nutrient-induced apoptosis. Diabetes. 2009;58(10):2303–2315. doi: 10.2337/db07-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gomes L.C., Benedetto G.D., Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rambold A.S. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108(25):10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liesa M., Shirihai Orian S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metabol. 2013;17(4):491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Suarez J. Alterations in mitochondrial function and cytosolic calcium induced by hyperglycemia are restored by mitochondrial transcription factor A in cardiomyocytes. Am. J. Physiol. Cell Physiol. 2008;295(6):C1561–C1568. doi: 10.1152/ajpcell.00076.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Palmeira C.M. Hyperglycemia decreases mitochondrial function: the regulatory role of mitochondrial biogenesis. Toxicol. Appl. Pharmacol. 2007;225(2):214–220. doi: 10.1016/j.taap.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 102.Lane N. Hot mitochondria? PLoS Biol. 2018;16(1) doi: 10.1371/journal.pbio.2005113. e2005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chretien D. Mitochondria are physiologically maintained at close to 50 degrees C. PLoS Biol. 2018;16(1) doi: 10.1371/journal.pbio.2003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lenaz G., Genova M.L. Structure and organization of mitochondrial respiratory complexes: a new understanding of an old subject. Antioxidants Redox Signal. 2010;12(8):961–1008. doi: 10.1089/ars.2009.2704. [DOI] [PubMed] [Google Scholar]
- 105.Berg J.M.T., Tymoczko J.L., Stryer L. W H Freeman; New York: 2002. Biochemistry. [Google Scholar]
- 106.Chandel N.S. Mitochondria as signaling organelles. BMC Biol. 2014;12:34. doi: 10.1186/1741-7007-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cloonan S.M., Choi A.M. Mitochondria: commanders of innate immunity and disease? Curr. Opin. Immunol. 2012;24(1):32–40. doi: 10.1016/j.coi.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 108.Mehta M.M., Weinberg S.E., Chandel N.S. Mitochondrial control of immunity: beyond ATP. Nat. Rev. Immunol. 2017;17(10):608–620. doi: 10.1038/nri.2017.66. [DOI] [PubMed] [Google Scholar]
- 109.Weinberg S.E., Sena L.A., Chandel N.S. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rizzuto R. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 2012;13(9):566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 111.Krzywanski D.M. Endothelial cell bioenergetics and mitochondrial DNA damage differ in humans having african or west eurasian maternal ancestry. Circ. Cardiovasc. Genet. 2016;9(1):26–36. doi: 10.1161/CIRCGENETICS.115.001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Owen O.E., Kalhan S.C., Hanson R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 113.Rui L. Energy metabolism in the liver. Comprehen. Physiol. 2014;4(1):177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kazuno A.A. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2(8):e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Balloux F. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc. Biol. Sci. 2009;276(1672):3447–3455. doi: 10.1098/rspb.2009.0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ruiz-Pesini E. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 117.Doi A., Suzuki H., Matsuura E.T. Genetic analysis of temperature-dependent transmission of mitochondrial DNA in Drosophila. Heredity. 1999;82(Pt 5):555–560. doi: 10.1038/sj.hdy.6885080. [DOI] [PubMed] [Google Scholar]
- 118.Matsuura E.T., Niki Y., Chigusa S.I. Temperature-dependent selection in the transmission of mitochondrial DNA in Drosophila. Jpn. J. Genet. 1993;68(2):127–135. doi: 10.1266/jjg.68.127. [DOI] [PubMed] [Google Scholar]
- 119.Arnqvist G. Genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution. 2010;64(12):3354–3363. doi: 10.1111/j.1558-5646.2010.01135.x. [DOI] [PubMed] [Google Scholar]
- 120.Dowling D.K., Abiega K.C., Arnqvist G. Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution. 2007;61(1):194–201. doi: 10.1111/j.1558-5646.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- 121.Wolff J.N. Mitonuclear interactions, mtDNA-mediated thermal plasticity, and implications for the Trojan Female Technique for pest control. Sci. Rep. 2016;6:30016. doi: 10.1038/srep30016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lajbner Z. Experimental evidence that thermal selection shapes mitochondrial genome evolution. Sci. Rep. 2018;8(1):9500. doi: 10.1038/s41598-018-27805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Camus M.F. Experimental support that natural selection has shaped the latitudinal distribution of mitochondrial haplotypes in Australian Drosophila melanogaster. Mol. Biol. Evol. 2017;34(10):2600–2612. doi: 10.1093/molbev/msx184. [DOI] [PubMed] [Google Scholar]
- 124.Mishmar D. Proceedings of the National Academy of Sciences of the United States Of America. 2003. Natural selection shaped regional mtDNA variation in humans; pp. 171–176. 2003/01/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo Y. Mitochondrial nt3010G-nt3970C haplotype is implicated in high-altitude adaptation of Tibetans. Mitochondrial DNA. 2011;22(5–6):181–190. doi: 10.3109/19401736.2011.632771. [DOI] [PubMed] [Google Scholar]
- 126.Niemi A.K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum. Genet. 2003;112(1):29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 127.van der Walt J.M. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci. Lett. 2004;365(1):28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 128.van der Walt J.M. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am. J. Hum. Genet. 2003;72(4):804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Canter J.A. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Canc. Res. 2005;65(17):8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 130.Darvishi K. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Canc. Lett. 2007;249(2):249–255. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 131.Niemi A.K. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur. J. Hum. Genet. 2005;13(2):166–170. doi: 10.1038/sj.ejhg.5201308. [DOI] [PubMed] [Google Scholar]
- 132.Mims M.P. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Canc. Res. 2006;66(3):1880. doi: 10.1158/0008-5472.CAN-05-3774. author reply 1880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rai E. Interaction between the UCP2-866G/A, mtDNA 10398G/A and PGC1alpha p.Thr394Thr and p.Gly482Ser polymorphisms in type 2 diabetes susceptibility in North Indian population. Hum. Genet. 2007;122(5):535–540. doi: 10.1007/s00439-007-0421-4. [DOI] [PubMed] [Google Scholar]
- 134.Dingley S.D. Mitochondrial DNA variant in COX1 subunit significantly alters energy metabolism of geographically divergent wild isolates in Caenorhabditis elegans. J. Mol. Biol. 2014;426(11):2199–2216. doi: 10.1016/j.jmb.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cutting W.C., Mehrtens H.G., Tainter M.L. Actions and uses OF dinitrophenol: promising metabolic applications. J. Am. Med. Assoc. 1933;101(3):193–195. [Google Scholar]
- 136.Tainter M.L., Cutting W.C., Stockton A.B. Use of dinitrophenol in nutritional disorders : a critical survey of clinical results. Am. J. Public Health Nation's Health. 1934;24(10):1045–1053. doi: 10.2105/ajph.24.10.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Colman E. Dinitrophenol and obesity: an early twentieth-century regulatory dilemma. Regul. Toxicol. Pharmacol. 2007;48(2):115–117. doi: 10.1016/j.yrtph.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 138.Khan R.S. Mitochondrial uncoupler prodrug of 2,4-dinitrophenol, MP201, prevents neuronal damage and preserves vision in experimental optic neuritis. Oxid. Med. Cell Longev. 2017 doi: 10.1155/2017/7180632. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu B. 2,4 DNP improves motor function, preserves medium spiny neuronal identity, and reduces oxidative stress in a mouse model of Huntington's disease. Exp. Neurol. 2017;293:83–90. doi: 10.1016/j.expneurol.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee Y. Neuroprotective effects of 2,4-dinitrophenol in an acute model of Parkinson's disease. Brain Res. 2017;1663:184–193. doi: 10.1016/j.brainres.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 141.Goldgof M. The chemical uncoupler 2,4-dinitrophenol (DNP) protects against diet-induced obesity and improves energy homeostasis in mice at thermoneutrality. J. Biol. Chem. 2014;289(28):19341–19350. doi: 10.1074/jbc.M114.568204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jensen P.K. Antimycin-insensitive oxidation of succinate and reduced nicotinamide-adenine dinucleotide in electron-transport particles. I. pH dependency and hydrogen peroxide formation. Biochim. Biophys. Acta. 1966;122(2):157–166. doi: 10.1016/0926-6593(66)90057-9. [DOI] [PubMed] [Google Scholar]
- 143.Loschen G., Flohé L., Chance B. Respiratory chain linked H(2)O(2) production in pigeon heart mitochondria. FEBS Lett. 1971;18(2):261–264. doi: 10.1016/0014-5793(71)80459-3. [DOI] [PubMed] [Google Scholar]
- 144.Boveris A., Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem. J. 1973;134(3):707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weisiger R.A., Fridovich I. Superoxide dismutase. Organelle specificity. J. Biol. Chem. 1973;248(10):3582–3592. [PubMed] [Google Scholar]
- 146.Loschen G. Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett. 1974;42(1):68–72. doi: 10.1016/0014-5793(74)80281-4. [DOI] [PubMed] [Google Scholar]
- 147.Forman H.J., Kennedy J.A. Role of superoxide radical in mitochondrial dehydrogenase reactions. Biochem. Biophys. Res. Commun. 1974;60(3):1044–1050. doi: 10.1016/0006-291x(74)90418-5. [DOI] [PubMed] [Google Scholar]
- 148.Finkel T. Signal transduction by mitochondrial oxidants. J. Biol. Chem. 2012;287(7):4434–4440. doi: 10.1074/jbc.R111.271999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Brand M.D. The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 2010;45(7–8):466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brand M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016;100:14–31. doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 151.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244(22):6049–6055. [PubMed] [Google Scholar]
- 152.Miao L., St Clair D.K. Regulation of superoxide dismutase genes: implications in disease. Free Radic. Biol. Med. 2009;47(4):344–356. doi: 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sena L.A., Chandel N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fischer L.R. SOD1 targeted to the mitochondrial intermembrane space prevents motor neuropathy in the Sod1 knockout mouse. Brain. 2011;134(Pt 1):196–209. doi: 10.1093/brain/awq314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weisiger R.A., Fridovich I. Mitochondrial superoxide simutase. Site of synthesis and intramitochondrial localization. J. Biol. Chem. 1973;248(13):4793–4796. [PubMed] [Google Scholar]
- 156.Sturtz L.A. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 2001;276(41):38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 157.Iñarrea P. Purification and determination of activity of mitochondrial cyanide-sensitive superoxide dismutase in rat tissue extract. Methods Enzymol. 2002;349:106–114. doi: 10.1016/s0076-6879(02)49326-3. [DOI] [PubMed] [Google Scholar]
- 158.Vijayvergiya C. Mutant superoxide dismutase 1 forms aggregates in the brain mitochondrial matrix of amyotrophic lateral sclerosis mice. J. Neurosci. 2005;25(10):2463–2470. doi: 10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pi J. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes. 2007;56(7):1783–1791. doi: 10.2337/db06-1601. [DOI] [PubMed] [Google Scholar]
- 160.Cadenas E. Mitochondrial free radical production and cell signaling. Mol. Aspect. Med. 2004;25(1–2):17–26. doi: 10.1016/j.mam.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 161.Leloup C. Mitochondrial reactive oxygen species are obligatory signals for glucose-induced insulin secretion. Diabetes. 2009;58(3):673–681. doi: 10.2337/db07-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tait S.W., Green D.R. Mitochondria and cell signalling. J. Cell Sci. 2012;125(Pt 4):807–815. doi: 10.1242/jcs.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fleury C., Mignotte B., Vayssiere J.L. Mitochondrial reactive oxygen species in cell death signaling. Biochimie. 2002;84(2–3):131–141. doi: 10.1016/s0300-9084(02)01369-x. [DOI] [PubMed] [Google Scholar]
- 165.Lee J., Giordano S., Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 2012;441(2):523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82(1):47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 167.Rhee S.G. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312(5782):1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 168.Fetterman J.L. Mitochondrial DNA damage and vascular function in patients with diabetes mellitus and atherosclerotic cardiovascular disease. Cardiovasc. Diabetol. 2016;15:53. doi: 10.1186/s12933-016-0372-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ballinger S.W. Mitochondrial integrity and function in atherogenesis. Circulation. 2002:544–549. doi: 10.1161/01.cir.0000023921.93743.89. [DOI] [PubMed] [Google Scholar]
- 170.Corral-Debrinski M. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat. Res. 1992;275:169–180. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 171.Corral-Debrinski M. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 1992;2(4):324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 172.Harrison C.M. Mitochondrial oxidative stress significantly influences atherogenic risk and cytokine-induced oxidant production. Environ. Health Perspect. 2011;119(5):676–681. doi: 10.1289/ehp.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen Q. Production of reactive oxygen species by mitochondria: central role of complex III. J. Biol. Chem. 2003;278(38):36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 174.Sena L.A., Chandel N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sena L.A. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38(2):225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chandel N.S. Mitochondria as signaling organelles. BMC Biol. 2014;12:34. doi: 10.1186/1741-7007-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Weinberg S.E., Sena L.A., Chandel N.S. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Prevention C.f.D.C.a. U.S. Dept of Health and Human Services; 2020. National Diabetes Statistics Report; p. 30. 2020. [Google Scholar]
- 179.An R. ISRN Obes; 2014. Prevalence and Trends of Adult Obesity in the US, 1999-2012; p. 185132. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li C. Estimates of body composition with dual-energy X-ray absorptiometry in adults. Am. J. Clin. Nutr. 2009;90(6):1457–1465. doi: 10.3945/ajcn.2009.28141. [DOI] [PubMed] [Google Scholar]
- 181.Ogden C.L. Prevalence of childhood and adult obesity in the United States, 2011-2012. Jama. 2014;311(8):806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bhupathiraju S.N., Hu F.B. Epidemiology of obesity and diabetes and their cardiovascular complications. Circ. Res. 2016;118(11):1723–1735. doi: 10.1161/CIRCRESAHA.115.306825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Krzywanski D.M. Endothelial cell bioenergetics and mitochondrial DNA damage differ in humans having african or west eurasian maternal ancestry. Circ Cardiovasc Genet. 2016;9(1):26–36. doi: 10.1161/CIRCGENETICS.115.001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kesterson R.A. Generation of mitochondrial-nuclear eXchange mice via pronuclear transfer. Bio Protoc. 2016;6(20) doi: 10.21769/BioProtoc.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Feeley K.P. Mitochondrial genetics regulate breast cancer tumorigenicity and metastatic potential. Canc. Res. 2015;75(20):4429–4436. doi: 10.1158/0008-5472.CAN-15-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mills E.L., Kelly B., O'Neill L.A.J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 2017;18(5):488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 187.Wei X. Cationic nanocarriers induce cell necrosis through impairment of Na(+)/K(+)-ATPase and cause subsequent inflammatory response. Cell Res. 2015;25(2):237–253. doi: 10.1038/cr.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Q. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang J.Z. Mitochondrial DNA induces inflammation and increases TLR9/NF-kappaB expression in lung tissue. Int. J. Mol. Med. 2014;33(4):817–824. doi: 10.3892/ijmm.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 190.Nakahira K. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhou R. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 192.Shimada K. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Seth R.B. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappa B and IRF3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 195.Buskiewicz I.A. Reactive oxygen species induce virus-independent MAVS oligomerization in systemic lupus erythematosus. Sci. Signal. 2016;9(456) doi: 10.1126/scisignal.aaf1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Iyer S.S. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39(2):311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.DeLuca H.F., Engstrom G.W. Calcium uptake BY rat kidney mitochondria. Proc. Natl. Acad. Sci. Unit. States Am. 1961;47(11):1744–1750. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vasington F.D., Murphy J.V. Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J. Biol. Chem. 1962;237:2670–2677. [PubMed] [Google Scholar]
- 199.Rizzuto R. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358(6384):325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 200.Rizzuto R. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993;262(5134):744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- 201.Giacomello M. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol. Cell. 2010;38(2):280–290. doi: 10.1016/j.molcel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 202.Csordas G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 2010;39(1):121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.de Brito O.M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 204.Szabadkai G. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol. Cell. 2004;16(1):59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 205.Palty R. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc. Natl. Acad. Sci. U. S. A. 2010;107(1):436–441. doi: 10.1073/pnas.0908099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Perocchi F. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467(7313):291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Baughman J.M. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476(7360):341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.De Stefani D. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476(7360):336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bick A.G., Calvo S.E., Mootha V.K. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336(6083):886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Raffaello A. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013;32(17):2362–2376. doi: 10.1038/emboj.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Plovanich M. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PloS One. 2013;8(2) doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mallilankaraman K. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat. Cell Biol. 2012;14(12):1336–1343. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sancak Y. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342(6164):1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hoffman N.E. SLC25A23 augments mitochondrial Ca(2)(+) uptake, interacts with MCU, and induces oxidative stress-mediated cell death. Mol. Biol. Cell. 2014;25(6):936–947. doi: 10.1091/mbc.E13-08-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mammucari C., Gherardi G., Rizzuto R. Structure, activity regulation, and role of the mitochondrial calcium uniporter in health and disease. Front. Oncol. 2017;7(139) doi: 10.3389/fonc.2017.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Stefani D.D., Rizzuto R., Pozzan T. Enjoy the trip: calcium in mitochondria back and forth. Annu. Rev. Biochem. 2016;85(1):161–192. doi: 10.1146/annurev-biochem-060614-034216. [DOI] [PubMed] [Google Scholar]
- 217.Tarasova N.V. Mitochondrial calcium uniporter structure and function in different types of muscle tissues in health and disease. Int. J. Mol. Sci. 2019;20(19) doi: 10.3390/ijms20194823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pathak T., Trebak M. Mitochondrial Ca(2+) signaling. Pharmacol. Ther. 2018;192:112–123. doi: 10.1016/j.pharmthera.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kamer K.J., Mootha V.K. The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 2015;16(9):545–553. doi: 10.1038/nrm4039. [DOI] [PubMed] [Google Scholar]
- 220.O'Rourke B. Pathophysiological and protective roles of mitochondrial ion channels. J. Physiol. 2000;529(1):23–36. doi: 10.1111/j.1469-7793.2000.00023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Di Lisa F., Bernardi P. A CaPful of mechanisms regulating the mitochondrial permeability transition. J. Mol. Cell. Cardiol. 2009;46(6):775–780. doi: 10.1016/j.yjmcc.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 222.Frank S. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1(4):515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 223.Martinou J.C., Youle R.J. Which came first, the cytochrome c release or the mitochondrial fission? Cell Death Differ. 2006;13(8):1291–1295. doi: 10.1038/sj.cdd.4401985. [DOI] [PubMed] [Google Scholar]
- 224.Popgeorgiev N., Jabbour L., Gillet G. Subcellular localization and dynamics of the bcl-2 family of proteins. Front Cell Dev Biol. 2018;6:13. doi: 10.3389/fcell.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pena-Blanco A., Garcia-Saez A.J. Bax, Bak and beyond - mitochondrial performance in apoptosis. FEBS J. 2018;285(3):416–431. doi: 10.1111/febs.14186. [DOI] [PubMed] [Google Scholar]
- 226.Matilainen O., Quiros P.M., Auwerx J. Mitochondria and epigenetics - crosstalk in homeostasis and stress. Trends Cell Biol. 2017;27(6):453–463. doi: 10.1016/j.tcb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 227.Zentner G.E., Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013;20(3):259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 228.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17(8):487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 229.Wu X., Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 2017;18(9):517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 230.Klose R.J., Kallin E.M., Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7(9):715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 231.Laukka T. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J. Biol. Chem. 2016;291(8):4256–4265. doi: 10.1074/jbc.M115.688762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Xiao M. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26(12):1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Shelar S. Biochemical and epigenetic insights into L-2-hydroxyglutarate, a potential therapeutic target in renal cancer. Clin. Canc. Res. : offc. J. Am. Assoc. Canc. Res. 2018;24(24):6433–6446. doi: 10.1158/1078-0432.CCR-18-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ward P.S. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Canc. Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wang Y. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol. Cell. 2015;57(4):662–673. doi: 10.1016/j.molcel.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dang L. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465(7300):966. doi: 10.1038/nature09132. 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fang Y., Tang S., Li X. Sirtuins in metabolic and epigenetic regulation of stem cells. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2019;30(3):177–188. doi: 10.1016/j.tem.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Janke R., Dodson A.E., Rine J. Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol. 2015;31:473–496. doi: 10.1146/annurev-cellbio-100814-125544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Phan A.T., Goldrath A.W., Glass C.K. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity. 2017;46(5):714–729. doi: 10.1016/j.immuni.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang D. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ivashkiv L.B. The hypoxia–lactate axis tempers inflammation. Nat. Rev. Immunol. 2020;20(2):85–86. doi: 10.1038/s41577-019-0259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bellizzi D. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 2013;20(6):537–547. doi: 10.1093/dnares/dst029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hong E.E. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol. Cell Biol. 2013;33(14):2683–2690. doi: 10.1128/MCB.00220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shock L.S. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2011;108(9):3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kopinski P., Janssen K., Schaefer P., Trefely S., Perry C., Potluri P., Tintos-Hernandez J., Singh L., Karch K., Campbell S., Doan M., Jiang H., Nissim I., Nakamaru-Ogiso E., Wellen K., Snyder N., Garcia B., Wallace D.C. Proc Natl Acad Sci U S A. 2019. Regulation of nuclear epigenome by mitochondrial DNA heteroplasmy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Picard M. Mitochondrial functions modulate neuroendocrine, metabolic, inflammatory, and transcriptional responses to acute psychological stress. Proc. Natl. Acad. Sci. U. S. A. 2015;112(48):E6614–E6623. doi: 10.1073/pnas.1515733112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wallace D.C. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242(4884):1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 248.Shoffner J.M. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61(6):931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 249.Ballinger S.W. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat. Genet. 1992;1(1):11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- 250.Van Hove J.L. Mitochondrial myopathy with anemia, cardiomyopathy, and lactic acidosis: a distinct late onset mitochondrial disorder. Am. J. Med. Genet. 1994;51(2):114–120. doi: 10.1002/ajmg.1320510207. [DOI] [PubMed] [Google Scholar]
- 251.Cortopassi G.A., Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cortopassi G.A. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl. Acad. Sci. U. S. A. 1992;89(16):7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Prezant T.R. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 1993;4(3):289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 254.Goto Y., Nonaka I., Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348(6302):651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 255.Nass M.M., Nass S. Intramitochondrial fibers with DNA characteristics. J. Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ishikawa K. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320(5876):661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 257.Ishikawa K. The innate immune system in host mice targets cells with allogenic mitochondrial DNA. J. Exp. Med. 2010;207(11):2297–2305. doi: 10.1084/jem.20092296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Brinker A.E. Mitochondrial haplotype Alters mammary cancer tumorigenicity and metastasis in an oncogenic driver-dependent manner. Canc. Res. 2017;77(24):6941–6949. doi: 10.1158/0008-5472.CAN-17-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Pravenec M. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic strains. Genome Res. 2007:1319–1326. doi: 10.1101/gr.6548207. 2007/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Houstek J. Nonsynonymous variants in mt-Nd2, mt-Nd4, and mt-Nd5 are linked to effects on oxidative phosphorylation and insulin sensitivity in rat conplastic strains. Physiol. Genom. 2012:487–494. doi: 10.1152/physiolgenomics.00156.2011. 2012/03/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Houstek J. Effects of mtDNA in SHR-mtF344 versus SHR conplastic strains on reduced OXPHOS enzyme levels, insulin resistance, cardiac hypertrophy, and systolic dysfunction. Physiol. Genom. 2014;46(18):671–678. doi: 10.1152/physiolgenomics.00069.2014. [DOI] [PubMed] [Google Scholar]
- 262.Neckelmann N. Proceedings of the National Academy of Sciences of the United States Of America. 1987. cDNA sequence of a human skeletal muscle ADP/ATP translocator: lack of a leader peptide, divergence from a fibroblast translocator cDNA, and coevolution with mitochondrial DNA genes; pp. 7580–7584. 1987/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Schmidt T.R. Evolution of nuclear- and mitochondrial-encoded subunit interaction in cytochrome c oxidase. Mol. Biol. Evol. 2001;18(4):563–569. doi: 10.1093/oxfordjournals.molbev.a003836. [DOI] [PubMed] [Google Scholar]
- 264.Pierron D. Evolution of the couple cytochrome c and cytochrome c oxidase in primates. Adv. Exp. Med. Biol. 2012;748:185–213. doi: 10.1007/978-1-4614-3573-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pierron D. Cytochrome c oxidase: evolution of control via nuclear subunit addition. Biochim. Biophys. Acta. 2012;1817(4):590–597. doi: 10.1016/j.bbabio.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Willett C.S., Burton R.S. Evolution of interacting proteins in the mitochondrial electron transport system in a marine copepod. Mol. Biol. Evol. 2004;21(3):443–453. doi: 10.1093/molbev/msh031. [DOI] [PubMed] [Google Scholar]
- 267.Telschow A. Genetic incompatibilities between mitochondria and nuclear genes: effect on gene flow and speciation. Front. Genet. 2019;10:62. doi: 10.3389/fgene.2019.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Uddin M. Molecular evolution of the cytochrome c oxidase subunit 5A gene in primates. BMC Evol. Biol. 2008;8:8. doi: 10.1186/1471-2148-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yan Z., Ye G., Werren J.H. Evolutionary rate correlation between mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins in insects. Mol. Biol. Evol. 2019;36(5):1022–1036. doi: 10.1093/molbev/msz036. [DOI] [PubMed] [Google Scholar]
- 270.Scott G.R., Richards J.G., Milsom W.K. Control of respiration in flight muscle from the high-altitude bar-headed goose and low-altitude birds. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009:R1066–R1074. doi: 10.1152/ajpregu.00241.2009. 2009/08/07. [DOI] [PubMed] [Google Scholar]
- 271.Scott G.R. Molecular evolution of cytochrome C oxidase underlies high-altitude adaptation in the bar-headed goose. Mol. Biol. Evol. 2011;28(1):351–363. doi: 10.1093/molbev/msq205. [DOI] [PubMed] [Google Scholar]
- 272.Peng Q. Mitogenomic analysis of the genus Pseudois: evidence of adaptive evolution of morphological variation in the ATP synthase genes. Mitochondrion. 2012:500–505. doi: 10.1016/j.mito.2012.07.107. 2012/07/24. [DOI] [PubMed] [Google Scholar]
- 273.Shen Y.-Y. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. U. S. A. 2010;107(19):8666–8671. doi: 10.1073/pnas.0912613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Consuegra S. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. : GSE. 2015;47(1):58. doi: 10.1186/s12711-015-0138-0. 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Goto Y., Nonaka I., Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348(6302):651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 276.Shoffner J.M. Subacute necrotizing encephalopathy: oxidative phosphorylation defects and the ATPase 6 point mutation. Neurology. 1992;42(11):2168–2174. doi: 10.1212/wnl.42.11.2168. [DOI] [PubMed] [Google Scholar]
- 277.Nagy R., Sweet K., Eng C. Highly penetrant hereditary cancer syndromes. Oncogene. 2004;23(38):6445–6470. doi: 10.1038/sj.onc.1207714. [DOI] [PubMed] [Google Scholar]
- 278.Mercuri E., Bonnemann C.G., Muntoni F. Muscular dystrophies. Lancet. 2019;394(10213):2025–2038. doi: 10.1016/S0140-6736(19)32910-1. [DOI] [PubMed] [Google Scholar]
- 279.Quiros P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17(4):213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 280.Margulis L. The origin of plant and animal cells. Am. Sci. 1971;59(2):230–235. [PubMed] [Google Scholar]
- 281.Margulis L., Chapman M.J. Endosymbioses: cyclical and permanent in evolution. Trends Microbiol. 1998;6(9):342–345. doi: 10.1016/s0966-842x(98)01325-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.