Abstract
In-silico approaches are routinely adopted to predict the effects of genetic variants and their relation to diseases. The Critical Assessment of Genome Interpretation (CAGI) has established a common framework for the assessment of available predictors of variant effects on specific problems and our group has been an active participant of CAGI since its first edition.
In this paper, we summarize our experience and lessons learned from the last edition of the experiment (CAGI-5). In particular, we analyse prediction performances of our tools on five CAGI-5 selected challenges grouped into three different categories: prediction of variant effects on protein stability, prediction of variant pathogenicity and prediction of complex functional effects. For each challenge, we analyse in detail the performance of our tools, highlighting their potentialities and drawbacks. The aim is to better define the application boundaries of each tool.
Keywords: CAGI, genetic variants, prediction of variant effects, prediction of protein stability change upon variations, variant pathogenicity prediction, machine learning
1. Introduction
Computational tools for predicting the effects of genetic variants are of invaluable importance for complementing experimental approaches in the dissection of the complexity underlying many human diseases. The development of tools for the prediction of variant effects is nowadays a major line of research in Bioinformatics and, therefore, many different methods have been described in the past years (Niroula & Vihinen, 2016). One major issue concerns the ability to effectively assess the prediction performances of available tools in order to highlight potentialities and drawbacks of each method with respect to different variant-effect-related prediction tasks.
The Critical Assessment of Genome Interpretation (CAGI) is a community-wide, international experiment aiming at assessing different methods and approaches for predicting and interpreting the effects of genetic variants. The CAGI is a periodic experiment (typically ran every two years), which has reached its fifth edition. The first one has been carried-out in 2010 and the last one, the CAGI-5, took place in 2018 and consisted of 14 different prediction challenges covering a wide spectrum of biological problems related to variant effect prediction. Over the years, the CAGI experiment has been significantly contributing to the field, acting as a major driver for testing novel methods and stirring new ideas for variant effect prediction and interpretation.
We have been active participants of the CAGI experiment since its first edition. Indeed, our research activity focuses on the development of tools for genetic variant interpretation, for relating variants and diseases, (Capriotti et al., 2006; Calabrese et al., 2009; Casadio et al., 2011), and for evaluating the impact of variations on protein stability, (Capriotti et al., 2008; Fariselli et al., 2015; Savojardo et al., 2016; Casadio et al., 2011).
In this paper, we analyse the results obtained by our group on a selection of five different CAGI-5 prediction challenges involving the following genes: FXN (frataxin), TPMT-PTEN (thiopurine S-methyltransferase and phosphatase and tensin homolog), CHEK2 (checkpoint kinase 2), PCM1 (pericentriolar material 1) and GAA (glucosidase alpha, acid). We classified these challenges into three different categories on the basis of the nature of the underlying prediction task. In particular, we have challenges requiring assessing the impact of variations on protein stability (FXN and TPMT-PTEN), challenges asking to predict variant pathogenicity (CHEK2) and complex challenges requiring assessing different types of functional effects not directly related to the two above categories (PCM1 and GAA). Here, the aim is to summarize our experience on CAGI-5 in order to highlight pros and cons of our approaches for the different challenge categories, as well as trying to define general guidelines for the proper selection of tools when addressing complex prediction tasks.
2. Methods
2.1. SNPs&GO: a predictor for annotating the pathogenicity of a protein variation
SNPs&GO (Calabrese et al., 2009) is a method based on Support Vector Machines (SVMs) for predicting the probability of a Single Amino acid Variation (SAV) to be pathogenic. The method elaborates information extracted from protein sequence, Multiple Sequence Alignment (MSA) of similar proteins and protein function. Sequence features include the SAV type and the composition of the environment around the variant site. Features derived from MSA include the frequencies of wild type and variant residues, the conservation index and the number of aligned sequences. Functional information is encoded by means of a protein-specific (SAV independent) descriptor derived from the distribution of Gene Ontology (GO) annotations in the UniProtKB database. In the pre-processing phase, for each GO term the frequencies of association to proteins carrying pathogenic and neutral SAVs are estimated and the corresponding log-odd value (LGO) is computed. Then, in the prediction phase, a single descriptor is computed by summing the LGO values of the GO term associated to the input protein (see Calabrese et al. (2009) for details).
SNPs&GO is available as web server at https://snps-and-go.biocomp.unibo.it/snps-and-go/.
2.2. INPS and INPS-3D predictors
INPS (Impact of Non-synonymous mutations on Protein Stability) (Fariselli et al., 2015) is a method for predicting the impact of SAVs on protein stability starting from protein sequence. In particular, it estimates the difference of Gibbs free energy change (ΔG) between wild-type and variant proteins (ΔΔG). INPS adopts a Support Vector Regression (SVR) approach trained on seven features, six of which are extracted from sequence and one from a Hidden Markov Model (HMM), whose parameters are estimated from the MSA of chains, sharing similarity with the input protein. The features derived from sequence include i) the BLOSUM62 score corresponding to the substitution from wild type to variant residues, ii) the Dayhoff mutability index of the wild-type residue, iii, iv) the molecular weights, and v, vi) Kyte-Doolittle hydrophobicity values of wild type and of variant residues, respectively. The seventh feature stems from the difference of the HMM-computed Viterbi scores of wild type and variant sequences.
When necessary, we used INPS-3D (Savojardo et al., 2016), which extends INPS by including, when available, features extracted from the protein 3D structure. These consider the relative solvent accessibility as computed by DSSP (Kabsch & Sander, 1983) and the local energy change upon variation, estimated as the difference between average pairwise residue-contact potential (Bastolla et al., 2001) of wild type and variant proteins.
INPS and INPS-3D are both available through the INPS-MD web server at https://inpsmd.biocomp.unibo.it.
2.3. Pathogenicity and perturbation: Pd and Pp indexes
The disease and perturbation probability indexes (Pd and Pp) (Casadio et al., 2011) associate each SAV type (i.e. wild type and variant residue pair) to the probability of being disease related (Pd) and of perturbing the protein stability (Pp), respectively. The probability indexes were statistically derived from a dataset of 17,170 SAVs in 5,305 proteins retrieved from data available at UniProtKB (release 2010_04), dbSNP (build 132), OMIM and ProTherm (Kumar et al., 2006). The databases include variations related to disease, neutral variants as well as effects of variants on protein thermodynamic stability (Casadio et al., 2011). Only SAVs deriving from single-nucleotide variations (SNPs) are considered. Moreover, SAV types lacking associated thermodynamic data in ProtTherm were filtered out. As a result, Pd and Pp are available for 141 SAV types (Casadio et al., 2011).
2.4. CAGI-5 Challenges
Our research group participated to several challenges of the fifth edition of the Critical Assessment of Genome Interpretation (CAGI-5), which took place in 2018. Our submissions were based (directly or indirectly) on previously developed tools for assessing whether or not protein variations are related to disease, including SNPs&GO and the disease probability indexes (Pd), and tools for the prediction of the impact of protein variants on protein stability like INPS/INPS-3D predictors and the perturbation probability index (Pp).
In this paper, we analyse submitted as well as newly generated predictions for five different CAGI-5 challenges: Frataxin, TPMT-PTEN, CHEK2, PCM1 and GAA, which are classified into three different categories:
challenges related to prediction of the effect of variations on protein stability, measured both directly (Frataxin) or indirectly (TPMT-PTEN) using different experimental methods;
challenges related to the evaluation of the pathogenicity of protein variations, as assessed directly in humans (CHEK2);
challenges that require to address complex problems of different nature, including the evaluation of functional effects of variations as assessed on model organisms different from humans (PCM1) or functional effects not directly related to protein stability and/or disease onset (GAA).
In the following, we will describe our approaches for each of the challenges.
2.4.1. Frataxin challenge
For the CAGI-5 Frataxin challenge, participants were provided with a dataset comprising 8 somatic SAVs of the frataxin protein (FXN), extracted from the COSMIC (Catalog of Somatic Mutations in Cancer) database (Tate et al., 2018) and already known as involved in neoplastic disease and/or cancer. Predictors were asked to submit, for each SAV in the dataset, ΔΔG values in kcal/mol, namely the difference of unfolding free energies (ΔG) between mutant and wild-type proteins, extrapolated at concentration zero of denaturant. Experimental ΔΔGs were obtained at the Sapienza University, Rome, Italy.
We tackled the Frataxin challenge running the INPS-3D predictor directly on the available reference PDB structure (1EKG) and obtaining a ΔΔG value for each SAV. Raw INPS-3D predictions were then submitted without any post-processing.
2.4.2. TPMT-PTEN challenge
In the CAGI-5 TPMT-PTEN challenge, predictors were asked to estimate the impact on protein stability of a large panel of SAVs of human thiopurine S-methyltransferase (TPMT) and phosphatase and tensin homolog (PTEN) proteins. Experimental stability scores were assessed by data providers using a multiplexed Variant Stability Profiling (VSP) assay, which uses a fluorescent reporter system to measure the steady-state abundance of missense protein variants (Yen et al., 2008). Submitted predictions needed to be scaled in the range [0,+∞[, where a value equal to 0 means that the variant is totally unstable, 1 means wild-type stability (neutral) and >1 means stability greater than the wild type.
We predicted the impact on protein stability using both INPS and INPS-3D predictors. In particular, we firstly mapped SAVs on available 3D structures from PDB and we predicted ΔΔGs with INPS-3D. All remaining SAVs that could not be mapped on 3D structures, were predicted using INPS on the PTEN and TPMT sequences available at UniProtKB.
ΔΔG values predicted by either INPS or INPS-3D were calibrated and rescaled in the required range using data from a functional characterization study of Salavaggione et al. (2005). In this study, functional effects were experimentally evaluated for 11 TPMT SAVs (not included in the challenge dataset). We used this experimental evidence for estimating a linear model to map INPS and INPS-3D ΔΔGs onto the requested range (1=wild type, 0=totally destabilizing, >1 stabilizing). The same calibration procedure was applied to both proteins. For sake of comparison, we complemented our predictions including the protein stability perturbation probability index (Pp).
2.4.3. CHEK2 challenge
The CAGI-5 CHEK2 challenge focus on variants of the human checkpoint kinase 2 (CHEK2), which is involved in breast cancer. Data provided include a panel of 34 SAVs obtained from targeted resequencing study on 1000 Latina breast cancer cases and 1000 ancestry matched controls. Predictors were asked to provide the probability pcase for a variant to occur in a case. A pcase > 0.5 means that the variation is pathogenic, a pcase = 0.5 means that the variation is neutral (occurring with the same frequency in both populations) while a value below 0.5 indicate that the variation is protective.
For this challenge, we used both the disease probability index (Pd) and SNPs&GO to assess pathogenicity of each variant. Furthermore, we complemented the above approaches with methods for assessing protein perturbation, including INPS and the perturbation probability index (Pp).
All the methods did not provide information about protective variants, hence predictions are limited to pcase ≥ 0.5. When Pd or Pp are used, we predicted a pcase = 1 for all variations having Pd (Pp) ≥ 0.8 and a pcase = 0.5 for all variations with Pd (Pp) ≤ 0.4, while values 0.4 < Pd (Pp) < 0.8 were linearly rescaled in the range ]0.5,1[.
From SNPs&GO output, we derived a pcase in the range [0.5,1] using class predictions (C, neutral or disease) and reliability indexes (RI, from 0 to 10). In particular, we linearly mapped SNPs&GO output to [0.5,1] such that a prediction (C=Neutral, RI=8) corresponds to pcase = 0.5 and a prediction (C=Disease, RI=8) corresponds to pcase = 1. We set the maximum RI to 8 because this is the maximum value found in this particular set of predictions.
INPS ΔΔG output was rescaled in the range [0.5,1] such that pcase = 1 if |ΔΔG| ≥ 1 kcal/mol, pcase = 0.5 if 0.0 kcal/mol ≤ |ΔΔG| < 0.5 kcal/mol while any 0.5 kcal/mol < |ΔΔG| < 1.0 kcal/mol was identically mapped in the range ]0.5,1[.
2.4.4. PCM1 challenge
The CAGI-5 PCM1 challenge required to predict the effect of a set of SAVs on zebrafish brain development. In particular, a panel of 38 variants within the pericentriolar material 1 (PCM1) gene were assayed on a zebrafish model in order to determine their impact on the volume of the posterior ventricle area. SAVs were then classified in three different categories: benign (having no impact on zebrafish brain formation), pathogenic (completely disrupting brain formation), and hypomorphic, characterized by a partial loss of function.
Our submission was based on predictions obtained using the disease probability index (Pd), which assigns to each SAV the probability of being associated to disease. According to the Pd values, variants were assigned with a functional effect, considering a SAV as pathogenic when Pd ≥ 0.6, hypomorphic when 0.4 < Pd < 0.6 and benign if Pd ≤ 0.4.
We complemented the above approach with predictions based on the assessment of the impact of SAVs on protein stability. In particular, we adopted the perturbation probability index (Pp) and the INPS predictor (using the entry UniProtKB Q15154). Similarly to Pd, thresholds on values of Pp were defined for assigning variant function effects (the same thresholds, 0.6 and 0.4, were adopted). INPS ΔΔG predictions were mapped to three classes according to the following scheme: pathogenic if |ΔΔG| ≥ 1 kcal/mol, hypomorphic if 0.5 kcal/mol ≤ |ΔΔG| < 1 kcal/mol and benign if |ΔΔG| < 0.5 kcal/mol.
2.4.5. GAA challenge
The CAGI-5 GAA challenge focused on predicting the effect of naturally occurring variations on enzymatic activity of the human glucosidase alpha, acid (GAA). Experimental enzymatic activity was assessed by data providers (BioMarin Pharmaceutical) for 356 novel missense mutations extracted from the ExAC dataset. Plasmids containing cDNAs encoding each of the mutant proteins were transfected into an immortalized Pompe patient fibroblast cell line, with no GAA activity. After 72 hours, cells were lysed, and GAA activity in the lysate was assessed with a fluorogenic substrate. Participants were asked to submit, for each variant in the panel, a numeric value v ≥ 0 representing relative enzymatic activity with respect to wild type: v=0 indicates no activity, v=1 wild-type activity, v>1 increased activity with respect to wild type.
Our submission for this challenge was based on SNPs&GO whose output was linearly rescaled in the range [0,1]. Here we included predictions obtained with the disease and perturbation probability indexes (Pd and Pp), and INPS. Pd and Pp were directly used without any preprocessing. INPS ΔΔG outputs were linearly remapped in the range [0,1].
2.5. Scoring the predictions
In order to score the prediction, we divided the 5 CAGI challenges into two categories: Frataxin, TPMT-PTEN and GAA are regression tasks, whereas CHEK2 and PCM1 are binary classification problems.
Regression challenges were scored using the following scoring measures:
Pearson Correlation Coefficient (ρ)
Spearman Rank Correlation Coefficient (rs)
Kendall Tau Rank Correlation Coefficient (τ)
Root Mean Square Error (RMSE)
Mean Absolute Error (MAE)
Binary classification tasks were evaluated using standard scoring indexes (Vihinen, 2012):
Sensitivity (SEN)
Specificity (SPE)
Positive Predictive Value (PPV)
Negative Predictive Values (NPV)
Accuracy (ACC)
Matthews Correlation Coefficient (MCC)
F1 measure (F1)
3. Results
3.1. Prediction of SAV effect on protein stability: Frataxin and TPMT-PTEN challenges
3.1.1. Frataxin challenge results
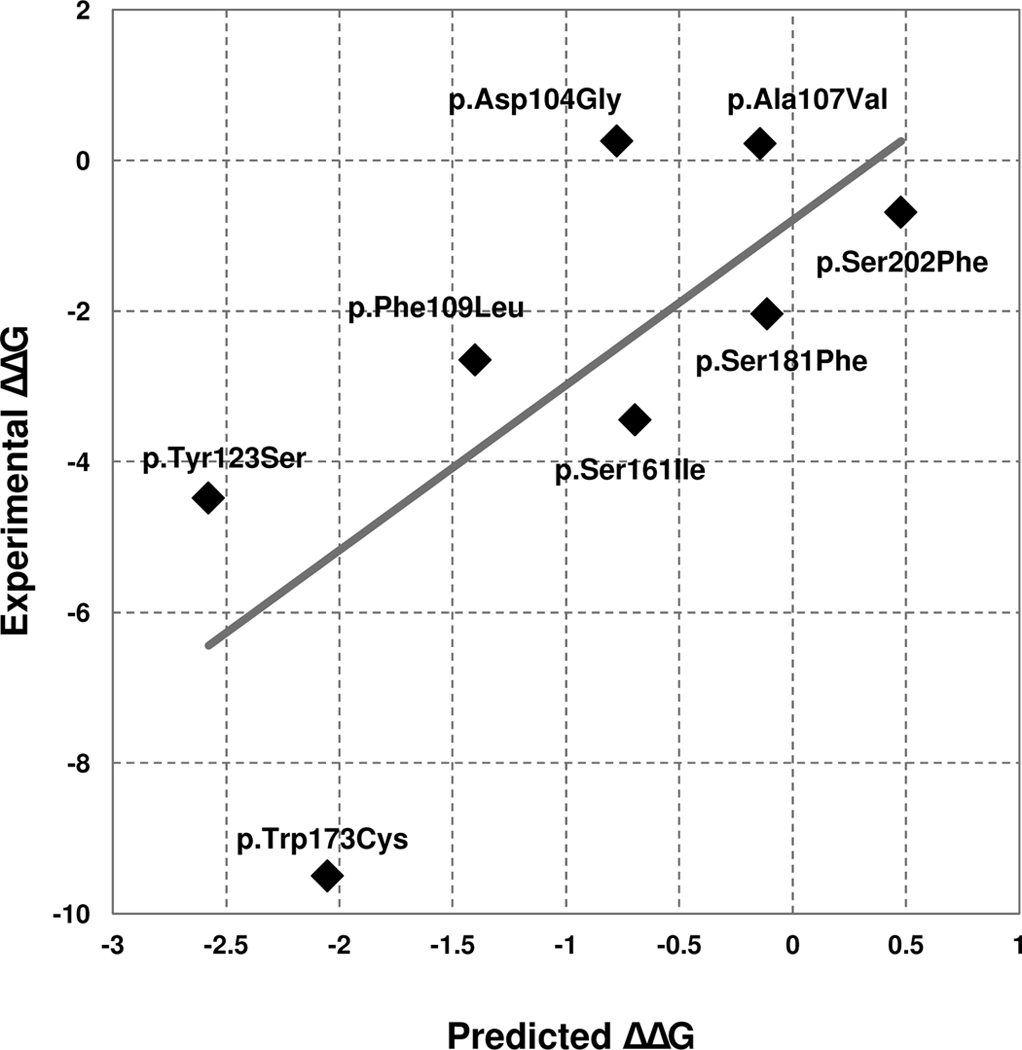
As detailed in Methods, the Frataxin challenge required to submit ΔΔG predictions for 8 SAV of the human frataxin protein (FXN). We evaluated the performance of our INPS-3D predictor with the regression analysis (Table 1). A comparison of experimental and predicted ΔΔG is shown (Figure 1). When INPS-3D is scored as binary classifier, the 8 variants are split into in two subsets, corresponding to destabilizing and non-destabilizing SAVs. In particular, adopting a threshold of −1 kcal/mol on experimental ΔΔG, 5 out of 8 variants are destabilizing. The same threshold is applied to INPS-3D predictions. Table 1 lists classification scoring indexes.
Table 1.
Regression and classification performances of INPS-3D on the Frataxin challenge.
| Method | Regression † | Classification † | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ | rs | τ | RMSE | MAE | SEN | SPE | PPV | NPV | ACC | MCC | F1 | |
| INPS-3D | 0.71 | 0.62 | 0.43 | 3.05 | 2.24 | 0.6 | 1.0 | 1.0 | 0.6 | 0.75 | 0.6 | 0.75 |
For scoring indexes definition see the Scoring the predictions paragraph in the Method Section
Figure 1.
Predicted vs experimental ΔΔG values for the 8 variants of the human frataxin protein (FXN). Predictions were obtained using INPS-3D.
Results indicate that INPS-3D performs very well on the task, achieving very high performances in both regression and classification schemes. According to the official CAGI-5 assessment, INPS-3D is among the top-performing methods participating to this challenge.
Our method fails on predicting the single SAV p.Trp173Cys (Figure 1). This variant is associated to a very low experimental ΔΔG value of −9.5 kcal/mol. As stated during the official assessment, the protein variant p.Trp173Cys corresponds to a clear unfolded state of the protein as experimentally determined (Petrosino et al., 2019; Savojardo et al., 2019). For this reason, the data providers assigned to the variant an arbitrary ΔG of 0 kcal/mol and, as a consequence, a very low ΔΔG value. Removing the outlier from the evaluation, RMSE and MAE decrease down to 1.64 and 1.48 kcal/mol, respectively. These values are much lower than the ones reported in Table 1 and closer to performances reported by INPS-3D in other benchmarks (Savojardo et al., 2016).
Overall, we can conclude that the performance of INPS-3D in the challenge is similar to the one already described in previous papers when predicting experimentally determined ΔΔG values, as expected, given that our method was trained on the same type of data (Savojardo et al., 2016).
INPS-3D predictions for FXN SAVs are reported in Supplementary Table S1.
3.1.2. TPMT-PTEN challenge results
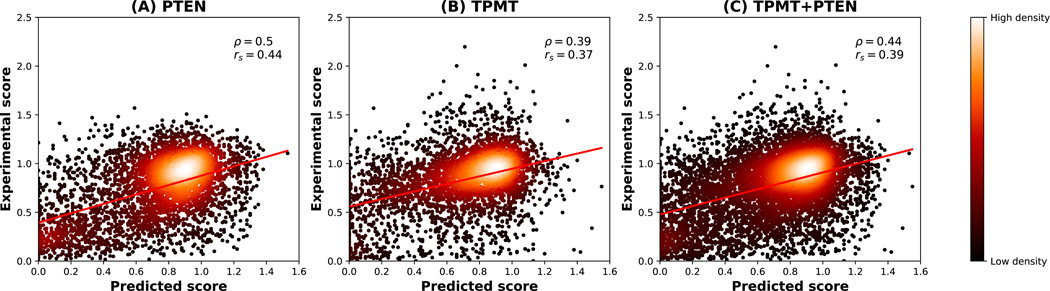
For the TPMT-PTEN challenge, a set of SAVs of human thiopurine S-methyltransferase (TPMT) and phosphatase and tensin homolog (PTEN) were provided to participants. The task was to compute the effect of each SAV on protein stability (i.e. a numeric score >0), representing relative stability with respect to wild type (see Methods for details). During challenge evaluation, after excluding SAVs with negative experimental scores as well as stop-gain variants, assessors retained 7473 SAVs, 3860 and 3613 on PTEN and TPMT, respectively. This dataset was predicted, adopting a combined approach based on INPS and INPS-3D (the method used for our official submission) as well as on the perturbation probability index (Pp). Table 2 lists results of the regression analysis. Since Pp is only defined for 141 SAV types, we were able to provide predictions for a subset of 2982 out of 7473 SAVs in the dataset (1556 from PTEN and 1426 from TPMT). In Figure 2 results obtained with INPS+INPS-3D are shown adopting a scatter plot between experimental and predicted stability scores.
Table 2.
Prediction performances of INPS+INPS-3D and Pp on the TPMT-PTEN challenge.
| Method | Dataset | ρ | rs | τ |
|---|---|---|---|---|
| INPS+INPS-3D | PTEN | 0.50 | 0.44 | 0.30 |
| INPS+INPS-3D | TPMT | 0.39 | 0.37 | 0.25 |
| INPS+INPS-3D | TPMT+PTEN | 0.44 | 0.39 | 0.27 |
| Pp | PTEN† | 0.18 | 0.16 | 0.11 |
| Pp | TPMT‡ | 0.17 | 0.16 | 0.11 |
| Pp | TPMT+PTEN§ | 0.18 | 0.16 | 0.11 |
Predictions obtained on the subset of 1556 PTEN SAVs for which Pp is defined.
Predictions obtained on the subset of 1426 TPMT SAVs for which Pp is defined.
Predictions obtained on the subset of 2982 PTEN+TPMT SAVs for which Pp is defined.
Figure 2.
Predicted vs experimental stability scores for the 7473 variants of human human thiopurine S-methyltransferase (TPMT, 3613 variants) and phosphatase and tensin homolog (PTEN, 3860 variants). Predictions obtained with INPS+INPS-3D are shown individually for PTEN (A) and TPMT (B) variants and for the whole dataset (C).
When comparing the two approaches, it appears that INPS+INPS-3D (based on machine learning) outperforms Pp, which is a simple statistical approach. ΔΔG predictions obtained with INPS+INPS-3D (which were rescaled linearly in the required range) significantly correlate with experimental values, achieving, on the whole dataset of SAVs, Pearson’s and Spearman’s correlation coefficients of 0.44 and 0.39, respectively.
Our strategies show performances different on the two proteins, with correlations that are lower for TPTM and higher for PTEN. Overall, our INPS+INPS-3D submissions are in the top 50% among challenge participants as highlighted in the assessment.
Comparing results of Frataxin and TPMT-PTEN challenges, it is worth noting that, using the same prediction approach, we achieved very different levels of performance (compare correlation coefficients in Tables 1 and 2). Moreover, prediction performance of INPS and INPS-3D in TPMT-PTEN are far below those previously achieved with the same methods in several benchmark datasets (Fariselli et al., 2015; Savojardo et al., 2016). A possible interpretation of the results is that when the methods are adopted to predict experimental thermodynamic stability data (ΔΔG), they perform better since they have been trained on the same type of data. As soon as the data type differs (for TPMT-PTEN, the impact of SAVs on protein stability was measured using a large-scale multiplexed Variant Stability Profiling (VSP) assay, the performance decreases.
Predictions for TPMT-PTEN SAVs are reported in Supplementary Table S2.
3.2. Prediction of SAV pathogenicity: CHEK2 challenge
For the CHEK2 challenge participants were asked to provide predictions of pathogenicity for 34 SAVs of the human checkpoint kinase 2 (CHEK2). Here, we evaluated performances of methods devised to predict the relation of SAVs with diseases, such as Pd and SNPs&GO, as well as methods devised to predict impact of SAVs on protein stability like Pp and INPS. Outputs of all methods were rescaled, so as to provide a numerical value, referred to as pcase, which represents the probability of each SAV to be pathogenic (see Methods for details). Binary classification of SAVs in the dataset was obtained by applying a threshold on the pcase value. Table 3 lists the results obtained with pcase -threshold set to 0.75.
Table 3.
Comparison of Pd, SNPs&GO, Pp and INPS on the CHEK2 challenge. Classification scoring indexes were computed setting pcase threshold to 0.75 for identifying pathogenic variants.
| Method | SEN | SPE | PPV | NPV | ACC | MCC | F1 |
|---|---|---|---|---|---|---|---|
| Pd | 0.52 | 0.69 | 0.73 | 0.47 | 0.59 | 0.21 | 0.61 |
| SNPs&GO | 0.71 | 0.85 | 0.88 | 0.65 | 0.76 | 0.54 | 0.79 |
| Pp | 0.19 | 0.77 | 0.57 | 0.37 | 0.41 | −0.05 | 0.29 |
| INPS | 0.52 | 0.69 | 0.73 | 0.47 | 0.59 | 0.21 | 0.61 |
Among the different approaches evaluated, SNPs&GO is the best performing one, reporting an MCC value of 0.54. The pattern of mis-predictions of SNPs&GO in this challenge is very similar to what already assessed for the predictor in much larger datasets (Calabrese et al., 2008). As a rule of thumb, SNPs&GO tends to more precise than sensitive (i.e. it is characterized by a high positive predictive value and a lower sensitivity). The same behavior can be observed in the CHEK2 challenge, where sensitivity and positive predictive values are 0.71 and 0.88, respectively. During the official challenge assessment, SNPs&GO was scored as the top-performing method.
Comparing performances of the different methods, it is evident that those that directly predict SAV pathogenicity (SNPs&GO and Pd) tend to outperform those that are instead devised to predict impact of SAV on protein stability (INPS and Pp). This suggests that methods implemented for predicting impact of SAVs on thermodynamic stability can be helpful in assessing SAV pathogenicity but, in many cases, they are not sufficient for obtaining accurate predictions.
Predictions for CHEK2 SAVs are reported in Supplementary Table S3.
3.3. Complex prediction challenges: PCM1 and GAA
3.3.1. PCM1 challenge results
The PCM1 challenge required to classify a set of 38 SAVs of the pericentriolar material 1 (PCM1) gene into three different classes (pathogenic, hypomorphic and benign) according to the estimated impact on brain development as measured on a zebrafish model. Following the same approach adopted during the challenge assessment, predictions were scored using a binary classification scheme, which collects into a single class pathogenic and hypomorphic SAVs and evaluates the ability of methods in discriminating them from benign SAVs.
In Table 4, we report classification results obtained in this task with Pd, Pp and INPS.
Table 4.
Prediction performances of Pd, Pp and INPS on the PCM1 challenge.
| Method | SEN | SPE | PPV | NPV | ACC | MCC | F1 |
|---|---|---|---|---|---|---|---|
| Pd | 0.86 | 0.00 | 0.54 | 0.00 | 0.50 | −0.25 | 0.67 |
| Pp | 0.82 | 0.31 | 0.62 | 0.56 | 0.61 | 0.15 | 0.71 |
| INPS | 0.64 | 0.50 | 0.64 | 0.50 | 0.58 | 0.14 | 0.64 |
Results highlight that all methods evaluated are essentially failing in this challenge, reporting MCC scores that are close to randomness and, in some case, even negatives. Interestingly, our official submission for this challenge (the one based on Pd), scoring with an MCC of −0.25, was globally ranked as the third top-performing among all participating methods (global ranks were computed averaging individual ranks computed for each scoring index). Our conclusion would be that our predictors are not suited to capture the complexity of the biological process leading to the different results.
Predictions for PCM1 SAVs are reported in Supplementary Table S4.
3.3.2. GAA challenge results
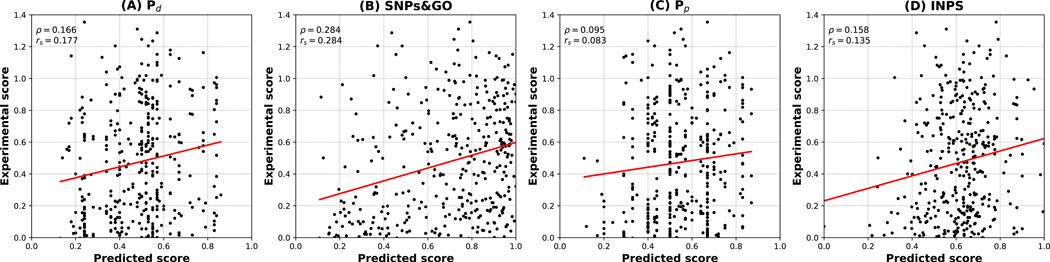
The GAA challenge requires to predict the impact on enzymatic activity of 356 SAVs of the human glucosidase alpha, acid (GAA) protein. In this task, we compared Pd, SNPs&GO, Pp and INPS. Results of regression analyses are reported in Figure 3 and Table 5.
Figure 3.
Predicted vs experimental enzymatic activity scores for 356 variants of human glucosidase alpha, acid (GAA) protein. Predictions were generated with Pd (A), SNPs&GO (B), Pp (C) and INPS (D).
Table 5.
Comparison of prediction performances of Pd, SNPs&GO, Pp and INPS on the GAA challenge.
| Method | ρ | rs | τ | RMSE | MAE |
|---|---|---|---|---|---|
| Pd | 0.17 | 0.18 | 0.12 | 0.36 | 0.30 |
| SNPs&GO | 0.28 | 0.28 | 0.19 | 0.42 | 0.35 |
| Pp | 0.10 | 0.09 | 0.06 | 0.37 | 0.32 |
| INPS | 0.16 | 0.14 | 0.09 | 0.97 | 0.80 |
In our tests, pathogenicity predictors (Pd and SNPs&GO) significantly outperform stability ones (Pp and INPS). However, our submission (based on SNPs&GO), is characterized by a Pearson’s correlation value of 0.28 and interestingly ranked among the top-scoring ones (the fourth in terms of individual submissions and the second in terms of research groups). Our interpretation is that again the complexity of the detection system hampers direct predictions that can be addressed by our tools.
Predictions for GAA SAVs are reported in Supplementary Table S4.
4. Conclusion
In this paper we summarized our experience as participants to the fifth edition of CAGI. In particular, we focused on five different challenges which for sake of simplicity, we divided into three different categories: i) prediction of protein stability perturbation upon variation, ii) prediction of variant pathogenicity and iii) prediction of complex functional effects. For each challenge, we analysed prediction performance of our CAGI official submissions as well as performance of other complementing approaches.
Overall, results on the five challenges here considered confirm the superiority of machine-learning based approaches (SNPs&GO and INPS/INPS-3D) over methods based on basic statistical analyses (Pd and Pp). Our methods well perform when the test set contains data homogeneous to those of the training set. As an example, when predicting SAV effects on protein stability, our methods performs better in the case of Frataxin than in the case of TPMT-PTEN. Indeed, these latter ΔΔG values of TPMT-PTEN variations are not directly measured, as in the case of Frataxin, rather indirectly evaluated from a large-scale multiplexed Variant Stability Profiling (VSP) assay (Yen et al., 2008). Again, when predicting variant pathogenicity of CHEK2, our SNPs&GO is satisfactory performing, given the similarity between the training procedure and the required task. However, when predicting on what we call complex prediction challenges (such as the pathogenicity of PCM1 and GAA variants) even machine-learning approaches fail. Our tools are indeed able to capture the binary classification of simple sets of molecular data directly annotated (Calabrese et al., 2009; Casadio et al., 2011). When classification is derived indirectly with in vivo approaches, possibly it implies complex biological processes, which deserve other models for their simulation.
Supplementary Material
Acknowledgements
The CAGI experiment coordination is supported by NIH U41 HG007346 and the CAGI conference by NIH R13 HG006650.
Footnotes
Conflict of Interest
None declared.
References
- Bastolla U, Farwer J, Knapp EW, Vendruscolo M (2001). How to guarantee optimal stability for most representative structures in the Protein Data Bank. Proteins, 44, 79–96. doi: 10.1002/prot.1075 [DOI] [PubMed] [Google Scholar]
- Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R (2009). Functional annotations improve the predictive score of human disease-related mutations in proteins. Human Mutation, 30, 1237–1244. doi: 10.1002/humu.21047 [DOI] [PubMed] [Google Scholar]
- Capriotti E, Calabrese R, Casadio R (2006). Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics, 22, 2729–2734. doi: 10.1093/bioinformatics/btl423 [DOI] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Rossi I, Casadio R (2008). A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics, 9(Suppl 2), S6. doi: 10.1186/1471-2105-9-S2-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio R, Vassura M, Tiwari S, Fariselli P, Martelli PL (2011). Correlating disease-related mutations to their effect on protein stability: a large-scale analysis of the human proteome. Human Mutation, 32, 1161–1170. doi: 10.1002/humu.21555 [DOI] [PubMed] [Google Scholar]
- Fariselli P, Martelli PL, Savojardo C, Casadio R (2015). INPS: predicting the impact of non-synonymous variations on protein stability from sequence, Bioinformatics, 31, 2816–2821. doi: 10.1093/bioinformatics/btv291 [DOI] [PubMed] [Google Scholar]
- Kabsch W & Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. doi: 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- Kumar MD, Bava KA, Gromiha MM, Parabakaran P, Kitajima K, Uedaira H, Sarai A (2006). ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Research, 34, D204–D206. doi: 10.1093/nar/gkj103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niroula A & Vihinen M (2016). Variation Interpretation Predictors: Principles, Types, Performance, and Choice. Human Mutation, 37, 579–597. doi: 10.1002/humu.22987 [DOI] [PubMed] [Google Scholar]
- Petrosino M et al. (2019). Characterization of the human frataxin missense variants in cancer. Human Mutation. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavaggione OE, Wang L, Wiepert M, Yee VC, Weinshilboum RM (2005). Thiopurine S-methyltransferase pharmacogenetics: variant allele functional and comparative genomics. Pharmacogenet Genomics, 15, 801–815. doi: 10.1097/01.fpc.0000174788.69991.6b [DOI] [PubMed] [Google Scholar]
- Savojardo C, Fariselli P, Martelli PL, Casadio R (2016). INPS-MD: a web server to predict stability of protein variants from sequence and structure. Bioinformatics, 32, 2542–2544. doi: 10.1093/bioinformatics/btw192 [DOI] [PubMed] [Google Scholar]
- Savojardo C et al. (2019). Evaluating the predictions of the protein stability change upon single point mutation for the Frataxin CAGI5 challenge. Human Mutation. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ, Forbes SA (2018). COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Research, 47, D941–D947. doi: 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M (2012) How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics, 13, S2. doi: 10.1186/1471-2164-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HCS, Xu Q, Chou DM, Zhao Z, Elledge SJ (2008). Global protein stability profiling in mammalian cells. Science, 322, 918–923. doi: 10.1126/science.1160489 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.