Abstract
Gastric cancer is the most commonly occurring cancer with a rapidly increasing incidence rate worldwide. The underlying molecular mechanisms of gastric cancer require further investigation. MicroRNAs exhibit tissue sensitivity as tumor biomarkers that play a role by promoting tumor growth as oncogenes or tumor suppressor genes. We evaluated the effects of microRNA-12129 on gastric cancer and identified the underlying mechanisms of microRNA-12129. Quantitative real-time polymerase chain reaction was conducted to determine the expression levels of microRNA-12129 and sirtuin 1 in vivo and in vitro, and Western blot analysis was performed to detect sirtuin 1 at the protein level in gastric cancer cell lines. Cell proliferation and cell cycle progression were detected by Cell Counting Kit-8 assay and flow cytometry analysis, respectively. The potential targets of microRNA-12129 were predicted by bioinformatics analysis. The targets of microRNA-12129 were confirmed by luciferase reporter assay and rescue assay. We found that microRNA-12129 was downregulated in gastric cancer tissues and gastric cancer cell lines and was significantly associated with the prognosis of patients with gastric cancer. In addition, microRNA-12129 overexpression suppressed tumor cell proliferation and blocked cell cycle progression. Bioinformatics analysis and luciferase reporter assay suggested that sirtuin 1 was a target of microRNA-12129, and sirtuin 1 expression was negatively related to microRNA-12129. Restoration of sirtuin 1 partly reduced the inhibition of cell proliferation and cell cycle progression induced by microRNA-144. Our results collectively suggested that microRNA-12129 suppressed cell proliferation and cell cycle progression in gastric cancer by targeting sirtuin 1. These findings indicated that manipulation of microRNA-12129 expression could help develop a novel therapeutic strategy for gastric cancer.
Keywords: MiRNA-12129, SIRT1, cell proliferation, cell cycle progression
Introduction
Gastric cancer (GC) ranks fourth as the most prevalent cancer with the third-highest cancer-related mortality among all malignant tumors globally.1,2 More than 1 000 000 new cases are diagnosed as GC, and approximately 700 000 cases die of GC annually.3 With the development of comprehensive therapy and gastroscopy techniques, the prognosis of patients with GC has been significantly improved.4 However, accurate confirmatory diagnosis of patients with early-stage GC presents a challenge, and the majority of patients are diagnosed only when GC is in its progressive stage.5 Moreover, the 5-year relative survival rate of GC in the advanced stage remains limited to barely 20%.6 Therefore, tumor-specific biomarkers have to be urgently identified, and novel strategies for the diagnosis and treatment of GC need to be developed.
MicroRNAs (miRNAs) are a type of small noncoding RNAs and act by binding to the 3′ untranslated region (UTR) of target genes.7,8 Emerging studies have recently shown that miRNAs are abundantly expressed in diverse malignant tumors and function as pivotal regulators of various biological processes in tumors, such as cell proliferation, differentiation, invasion, and apoptosis.9,10 Wang et al identified that miR-216b was downregulated in non-small cell lung cancer tissues and inhibited proliferation, migration, and invasion by binding to the 3′-UTR of FOXM1.11 Wei et al observed that miR-1284 was reduced in GC tissues and cell lines and suppressed the progression of GC by regulating EIF4A1 expression.12
MicroRNA, a type of cancer biomarker, was reported frequently in GC. We also screened some new potential microRNA biomarkers with microarray, data mining, and so on. We sought to identify the gene or microRNA, which causes significant changes between the GC tissue and the normal tissue. MicroRNA-12129 is a novel microRNA we found in biomarkers for screening GC. It was lower in GC tissues than in normal tissues and could act as a biomarker for the diagnosis and prognosis of GC. However, no research has been reported on its function and mechanism in any cancer field. In the current study, we showed miR-12129 expression both in vivo and in vitro and explored the functions and underlying mechanism of miR-12129 in GC. The results revealed that miR-12129 repressed cell proliferation and cell cycle progression by targeting sirtuin 1 (SIRT1), which may provide novel insights into the treatment of GC.
Methods and Materials
Patients and Specimens
Thirty pairs of newly diagnosed GC tissues and adjacent nontumor tissues were provided by The First Affiliated Hospital of Nanchang University and immediately immersed into liquid nitrogen after resection and stored at −80 °C for future use. This study was approved by Medical Ethics Committee of Nanchang University (No.NUF12020127). All patients provided written informed consent prior to enrollment in the study.
Cell Culture and Transfection
Human GC cell lines (SNU-5, SNU-16, and NCI-N87) and a normal gastric mucosa cell line (HS 738 ST/Int) were purchased from American Type Culture Collection. Human GC cell lines (MGC80-3 and SGC-7901) were provided by the Chinese Academy of Sciences. HS 738 ST/Int was maintained in Dulbecco’s Modified Eagle Medium (Invitrogen; Thermo Fisher Scientific, Inc). SNU-5 was cultured in Iscove’s Modified Dulbecco’s Media (Invitrogen; Thermo Fisher Scientific, Inc), and others were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc), 100 U/mL penicillin, and 100 μg/mL streptomycin (Solarbio). All cell lines were placed in a humidified incubator with 5% CO2 at 37 °C.
MicroRNA-12129 mimics, small interfering RNA-SIRT1, and negative control (NC) mimics were purchased from GenePharma. The detailed sequences are described in Table 1. Cell lines (SNU-5, NCI-N87) were transfected in 6-well plates with the Lipofectamine 2000TM reagent (Invitrogen; Thermo Fisher Scientific, Inc) in accordance with the manufacturer protocol.
Table 1.
Correlation Between miR-12129 Expression and Clinical Factors of Patients With GC.
| Clinical factors | Total | MiR-12129 expression | P value | |
|---|---|---|---|---|
| Low (n = 15) | High (n = 15) | |||
| Gender | .27 | |||
| Male | 14 | 6 | 8 | |
| Female | 16 | 8 | 8 | |
| Age | .25 | |||
| ≥60 | 21 | 12 | 9 | |
| <60 | 9 | 4 | 5 | |
| Tumor size (cm) | .21 | |||
| ≥2 | 22 | 10 | 12 | |
| <2 | 8 | 2 | 6 | |
| TNM stage | .04 | |||
| I-II | 7 | 2 | 5 | |
| III-IV | 23 | 17 | 6 | |
| Lymph node metastasis | .02 | |||
| Positive | 18 | 15 | 3 | |
| Negative | 12 | 5 | 7 | |
Abbreviations: GC, gastric cancer; miR, microRNA.
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc) was used to analyze cell proliferation as instructed by the manufacturer. Cells were cultured on a 96-well plate. At the density of 4 × 103 cells per well, 10% CCK-8 was added and incubated for 2 to 4 hours at 37 °C. Proliferation rates were determined from day 1 to day 4 after transfection, with the microplate reader set at 450 nm. The experiment was repeated 3 times.
RNA Isolation and Quantitative Real-Time PCR
Total RNA was extracted from fresh tissues and cell lines by using TRIzol Reagent (Thermo Fisher Scientific, Inc). Reverse transcription was performed using the RNA PCR Kit (AMV; Takara Biotechnology Co, Ltd). In accordance with the instructions provided by the manufacturer, miRNA and mRNA were quantified using quantitative real-time polymerase chain reaction (qRT-PCR) with SYBR Premix Ex Taq (Takara Biotechnology Co), with U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the internal control. All reactions were performed in triplicate.
Western Blot Analysis
Antihuman antibodies of GAPDH and SIRT1 were purchased from Abcam. After transfection for 48 to 72 hours, proteins were collected from the NCI-N87 and SNU-5 cell lines with RIPA buffer (Beyotime Biotechnology) and then measured using the BCA Protein Assay Kit (Beyotime Biotechnology) in accordance with the instructions provided by the manufacturer, with GAPDH as the internal control. All experiments were repeated at least 3 times.
Flow Cytometry Detection of Cell Cycle Distribution
For cell cycle analysis, the NCI-N87 and SNU-5 cell lines were seeded into 6-well plates at a density of 1 × 104 cells per well and then harvested. After fixation in 70% ethanol overnight at 4 °C, the cell lines were treated with 0.5 mg/mL RNase A (Keygen Biotech) and stained with 1 mg/mL propidium iodide (Biosciences) for 0.5 hours at 37 °C. The distribution of cell cycle phases was determined using a FACSCalibur Flow Cytometer (Becton, Dickinson and Company). The percentages of cells in the G0/G1, S, and G2/M phases were calculated.
Bioinformatics Target Prediction
The targets of miR-12129 were predicted by searching TargetScan (http://www.targetscan.org/vert_72/) and miRDB (http://mirdb.org).
Luciferase Reporter Assay
NCI-N87 and SNU-5 cell lines were cultured on 24-well plates and co-transfected with the miR-12129 mimic or miR-NC, together with the wild-type or mutant 3′-UTR of SIRT1 with the Lipofectamine 2000TM reagent (Invitrogen; Thermo Fisher Scientific, Inc). After 48 hours, luciferase activity was detected using the Dual-Luciferase Reporter Assay System (Promega) and normalized to that of Renilla luciferase activity.
Statistical Analysis
All data were presented as means ± SD and analyzed using SPSS version 19.0. Differences between different groups were calculated using the t test. The relationship between miR-12129 expression and SIRT1 expression was assessed by Spearman correlation analysis. Kaplan–Meier analysis and the log-rank test were conducted to determine the association between overall survival and miR-12129 expression. P < .05 was considered statistically significant.
Results
MicroRNA-12129 Expression Was Downregulated and Related to Outcome in GC
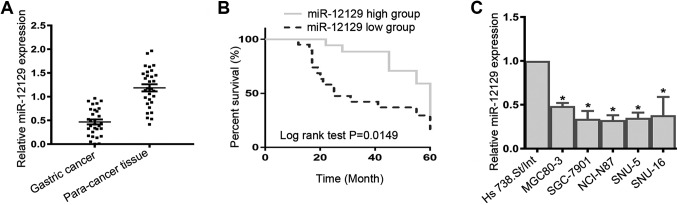
To confirm whether miR-12129 was abnormally expressed in GC, tissues and cell lines were used to examine the relative miR-12129 expression by qRT-PCR. We found that miR-12129 expression was reduced both in the GC tissues (Figure 1A) and cell lines (Figure 1C). The result of survival analysis indicated that the prognosis of patients with GC with high miR-12129 expression was better than that with low miR-12129 expression (P < .001, Figure 1B).
Figure 1.
Expression of miR-12129 in Gastric cancer tissues and cell lines. A, Quantitative real-time polymerase chain reaction analysis of miR-12129 expression in gastric cancer tissues (N = 30). B, Kaplan–Meier curve and log-rank test were conducted to assess the effects of miR-12129 expression on the overall survival of patients with gastric cancer. C, Quantitative real-time polymerase chain reaction analysis of miR-12129 expression in gastric cancer cell lines. *P < .05 versus normal group.
MicroRNA-12129 Overexpression Inhibited Proliferation and Induced G0/G1 Arrest in GC Cells
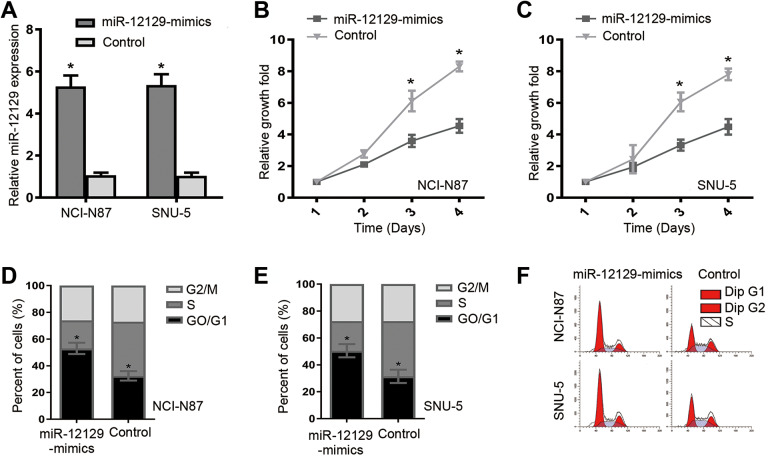
NCI-N87 and SNU-5 cell lines with reduced miR-12129 expression were used for further study. After transfection, significantly increased miR-12129 expression was confirmed in NCI-N87 and SNU-5 cell lines (Figure 2A). Cell Counting Kit-8 assay and flow cytometry were performed to evaluate the effects of miR-12129 on cell proliferation and cell cycle, respectively. Cell Counting Kit-8 assay results showed that after transfection, exogenous miR-12129 expression repressed cell proliferation in both NCI-N87 and SNU-5 (Figure 2B and C). Meanwhile, flow cytometry showed that miR-12129 overexpression increased the G0/G1 phase fraction (Figure. 2D-F). These data suggested that miR-12129 overexpression inhibited proliferation and induced G0/G1 arrest in GC cells.
Figure 2.
Overexpression of miR-12129 inhibited proliferation and induced G0/G1 arrest in gastric cancer cells. A, Quantitative real-time polymerase chain reaction analysis of miR-12129 expression. B, Proliferation ability was tested using the CCK-8 in NCI-N87 cell lines, (C) SNU-5 cell lines. D, The percentages of cells in G0/G1, S and G2/M phase in NCI-N87 cell lines, (E) SNU-5 cell lines. F, Images of cells in a cell cycle after transfection with miR-12129 mimics was assessed by Flow cytometry in NCI-N87 and SNU-5 cell lines. *P < .05 versus control. CCK-8 indicates Cell Counting Kit-8; miR, microRNA.
Sirtuin 1 Was a Direct Target of miR-12129 in GC
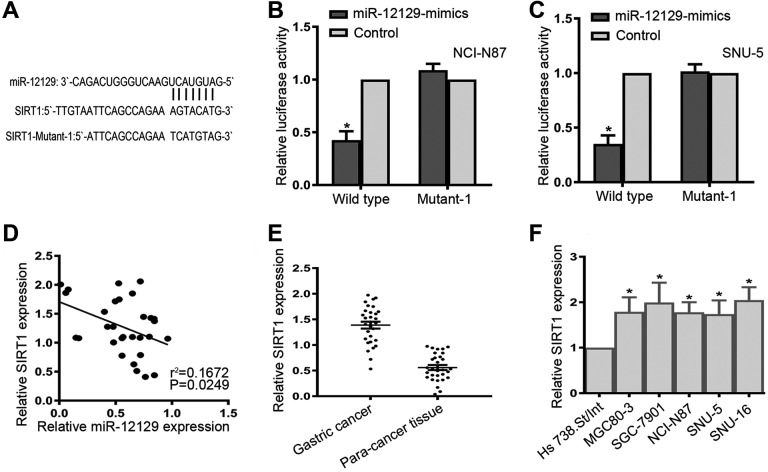
To elucidate the molecular mechanisms of miR-12129, we predicted the potential targets of miR-12129 by bioinformatics analysis. As shown in Figure 3A, the sequence of SIRT1 was a superior match for miR-12129 at the 3′-UTR. We also performed luciferase reporter assay to verify whether SIRT1 was a target of miR-12129. The result revealed that miR-12129 inhibited the activity of the reporter gene SIRT1 in both NCI-N87 and SNU-5 cell lines (Figure 3B and C).
Figure 3.
Sirtuin 1 was a Direct Target of miR-12129. A, MicroRNA-12129 binding site within the 3’-UTR of SIRT1 was predicted. B, Relative activities of luciferase reporters in NCI-N87 cell lines, C, SNU-5 cell lines. D, Pearson’s correlation analysis of the relative expression levels of miR-12129 and the relative SIRT1 mRNA levels in gastric cancer tissues (e) qRT-PCR analysis of SIRT1 expression in gastric cancer tissues (N = 30). F, Quantitative real-time polymerase chain reaction analysis of SIRT1 expression in gastric cancer cell lines. *P < .05 vs Control. miR indicates microRNA; qRT-PCR, quantitative real-time polymerase chain reaction; SIRT1, sirtuin 1; 3’-UTR, 3′ untranslated region.
Moreover, we evaluated SIRT1 expression at the mRNA level in tissue samples and cell lines. The result indicated that SIRT1 was significantly upregulated in both the GC tissues and GC cell lines (Figure 3E and F). We further found that SIRT1 expression was negatively related to miR-12129 expression in GC tissues (r2 = .1672, P = .0249, Figure 3D).
Restoration of SIRT1 Reverses the Inhibitory Effects of miR-12129
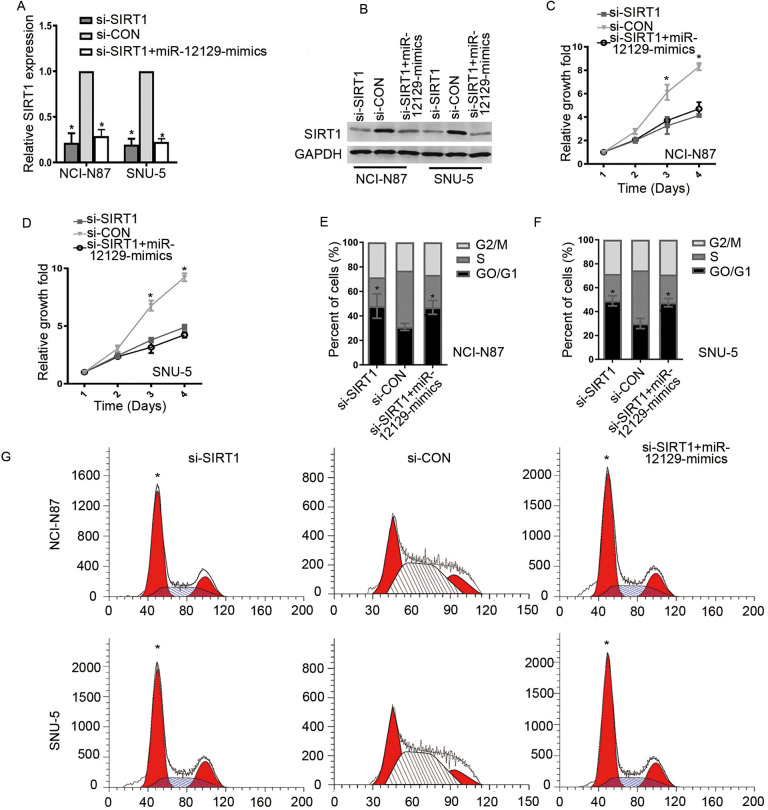
Considering the aforementioned results, we hypothesized that miR-12129 regulated cell proliferation and cell cycle via SIRT1. To verify this hypothesis, we performed several rescue experiments. First, we silenced the SIRT1 expression with si-SIRT1 in NCI-N87 and SNU-5 cell lines. Western blot analysis and qRT-PCR results showed that SIRT1 expression was significantly decreased after SIRT1 silencing (Figures 4A and B). However, co-transfecting si-SIRT1 and miR-12129 mimic did not further minimize SIRT1 expression, compared with transfecting si-SIRT1 alone. The CCK-8 results revealed that cell proliferation in NCI-N87 and SNU-5 cell lines was suppressed after co-transfecting si-SIRT1 with or without miR-12129 mimics (Figure 4C). Moreover, flow cytometry indicated that SIRT1 silencing inhibited the transition of cells in the G1 phase, which was similar to the effect induced by co-transfecting si-SIRT1 and miR-12129 mimics (Figure 4D-F).
Figure 4.
Knockdown of SIRT1 inhibited proliferation and cell cycle in Gastric cancer cells. A, Quantitative real-time polymerase chain reaction analysis of SIRT1 expression in gastric cancer cell lines. B, Western blot analysis of SIRT1 expression in gastric cancer cell lines. C, Proliferation ability was tested using the CCK-8 in NCI-N87 cell lines, (D) SNU-5 cell lines. The percentages of cells in G0/G1, S and G2/M phase in NCI-N87 cell lines, (E) SNU-5 cell lines. F, Images of cells in a cell cycle after transfection with miR-12129 mimics was assessed by Flow cytometry in NCI-N87 and SNU-5 cell lines. *P < .05 versus si-con. CCK-8 indicates Cell Counting Kit-8; con, control; miR, microRNA.
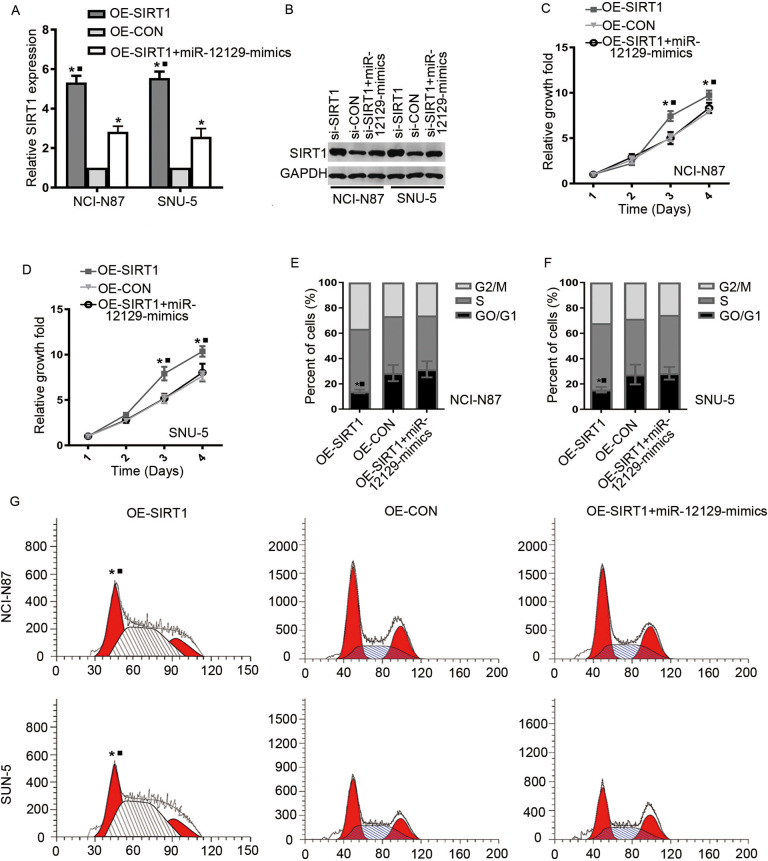
We finally determined whether SIRT1 was involved in miR-12129 regulation of cell proliferation and cell cycle. After SIRT1 overexpression, SIRT1 expression was increased at the mRNA and protein levels both in NCI-N87 and SNU-5 cell lines (Figure 5A and B). However, for cell lines with co-transfecting miR-12129 mimics and SIRT1 overexpression, SIRT1 expression was opposite, indicating that SIRT1 expression was affected by miR-12129. Cell Counting Kit-8 assay then revealed SIRT1 that overexpression could promote cell proliferation and be reversed by miR-12129 overexpression (Figure 5C and D). Moreover, flow cytometry showed that SIRT1 overexpression reversed miR-12129-induced G0/G1 arrest (Figure 5E-G).
Figure 5.
Restoration of SIRT1 reverses the inhibitory effects of miR-12129. A, Quantitative real-time polymerase chain reaction analysis of SIRT1 expression in gastric cancer cell lines. B, Western blot analysis of SIRT1 expression in Gastric cancer cell lines. C, Proliferation ability was tested using the CCK-8 in NCI-N87 cell lines, (D) SNU-5 cell lines. The percentages of cells in G0/G1, S, and G2/M phase in NCI-N87 cell lines, (E) SNU-5 cell lines. F, Images of cells in a cell cycle after transfection with miR-12129 mimics was assessed by flow cytometry in NCI-N87 and SNU-5 cell lines. *P < .05 versus si-con. CCK-8 indicates Cell Counting Kit-8; con indicates control; miR, microRNA.
Discussion
One of the most prevalent digestive malignancies, GC is the third leading cause of cancer-related mortality, particularly in developing countries.1,13 About 1 000 000 new cases and 700 000 deaths are reported annually3 and approximately 50% of patients with GC have undergone metastasis at the time of diagnosis.5 Therefore, finding the molecular pathogenesis of GC bears clinical significance for the diagnosis and treatment of GC.
As a class of small endogenous RNAs of 18 to 25 nucleotides in length, miRNAs play a critical role in transcriptional and posttranscriptional regulation during the initiation and progression of various cancer types.7,14 Several novel treatment strategies based on miRNAs have also been developed and reached the preclinical stage.15 Therapeutic strategies targeting miRNAs have shown potential. We found miR-12129, a new microRNA biomarker, in the GC tissue. No previous studies have been reported regarding its function in cancer. We checked the microRNA microarray data set and found that its expression was lower in most GC tissues than in normal tissues. We found that it was downregulated in GC tissues.
In the current study, we reported its function first in GC and identified its mechanisms in the cancer cell cycle. In this study, we assessed the miR-12129 expression in GC tissues and GC cell lines. The results indicated that miR-12129 was considerably downregulated in GC tissues and GC cell lines. Kaplan–Meier analysis also revealed that miR-12129 was positively correlated with the prognosis of patients with GC. Cell Counting Kit-8 and flow cytometry demonstrated that miR-12129 overexpression inhibited proliferation and induced the G0/G1 phase cell cycle arrest in GC cells. All data suggested that miR-12129 suppressed tumor genes.
To elucidate the molecular mechanisms of miR-12129 in GC, we predicted the target of miR-12129 by bioinformatics analysis and confirmed the results by luciferase reporter assay. The results revealed that SIRT1 was a target of miR-12129. Moreover, SIRT1 expression was negatively related to miR-12129 expression in GC tissues. Sirtuin 1knockout could also suppress cell proliferation and cell cycle. Restored SIRT1 expression exerted an apparent rescue effect on cell proliferation and cell cycle, indicating that the importance of SIRT1 in GC and miR-12129 could inhibit proliferation and induce G0/G1 phase cell cycle arrest via SIRT1.
Sirtuin 1, a member of the sirtuin family, is closely related to histone deacetylases and participates in the coordination of separate cellular functions, such as cell cycle, apoptosis, autophagy, and metabolism.16,17 Sirtuin 1 is also involved in the development of various malignancies, such as ovarian cancer,18 thyroid cancer,19 pancreatic cancer,20 GC,21-23 and hepatocellular carcinoma.24 Noguchi et al found that SIRT1 expression was increased in GC tissues and was closely associated with progression and prognosis, which was consistent with the findings of this study.21 Yang et al observed the contrary result that patients with GC exhibited elevated SIRT1 expression.25 Similar to the current study, several studies reported that SIRT1 could inhibit cell proliferation, migration, invasion, and induced G1 phase arrest in GC cell lines.22,23
In summary, our data revealed that miR-12129 was downregulated in both GC tissues and GC cell lines. MiR-12129 served as a tumor suppressor by inhibiting cell proliferation and inducing G0/G1 arrest via SIRT1 targeting. Therefore, manipulation of miRNA-12129 expression is a potential therapeutic strategy for the treatment of GC.
Abbreviations
- CCK-8
Cell Counting Kit-8
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GC
gastric cancer
- miRNAs
microRNAs
- NC
negative control
- qRT-PCR
quantitative real-time polymerase chain reaction
- UTR
3′ untranslated region
- SIRT1
sirtuin 1
Footnotes
Authors’ Note: Dongning Liu conceived and designed the study. Wei Zhang and Kai Liao performed the experiment. Wei Zhang wrote the paper. Dongning Liu reviewed and edited the manuscript. All authors approved the manuscript and agree to be accountable for all aspects of the research with regard to ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved. The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.Informed consent was obtained from all patients. The present study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Nanchang University and informed consent was obtained from all participants.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Dongning Liu  https://orcid.org/0000-0002-0195-0759
https://orcid.org/0000-0002-0195-0759
References
- 1. Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski WP. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang L, Chang Y, Xu J, Zhang Q. Predictive significance of serum level of vascular endothelial growth factor in gastric cancer patients. Biomed Res Int. 2016;2016:8103019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Park JY, Forman D, Waskito LA, Yamaoka Y, Crabtree JE. Epidemiology of helicobacter pylori and caga-positive infections and global variations in gastric cancer. Toxins. 2018;10(4):E163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu F, Gao H, Liu K, et al. The lncrna ZeB2-as1 is upregulated in gastric cancer and affects cell proliferation and invasion via mir-143-5p/hiF-1α axis. Onco Targets Ther. 2019;12:657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y, Xiao S, Wang B, Li Y, Chen Q. Knockdown of lncRNA TP73-AS1 inhibits gastric cancer cell proliferation and invasion via the WNT/β-catenin signaling pathway. Oncol Lett. 2018;16(3):3248–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wei J, Wang Z, Wang Z, et al. MicroRNA-31 function as a suppressor was regulated by epigenetic mechanisms in gastric cancer. Biomed Res Int. 2017;2017:5348490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu GJ, Dong YQ, Zhang QM, et al. miRNA-221 promotes proliferation, migration and invasion by targeting TIMP2 in renal cell carcinoma. Int J Clin Exp Pathol. 2015;8(5):5224–5229. [PMC free article] [PubMed] [Google Scholar]
- 8. Wang C, Xu C, Niu R, Hu G, Gu Z, Zhuang Z. MiR-890 inhibits proliferation and invasion and induces apoptosis in triple-negative breast cancer cells by targeting CD147. BMC Cancer. 2019;19(1):577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jia XN, Yin SD, Wei Y, Chen L. MiR-182-5p inhibited proliferation and migration of ovarian cancer cells by targeting BNIP3. Eur Rev Med Pharmacol Sci. 2019;23(8):3270–3276. [DOI] [PubMed] [Google Scholar]
- 10. Zhao J, Li D, Fang L. MiR-128-3p suppresses breast cancer cellular progression via targeting LIMK1. Biomed Pharmacother. 2019;115:108947. [DOI] [PubMed] [Google Scholar]
- 11. Wang L, Wang Y, Du X, Yao Y, Wang L, Jia Y. MiR-216b suppresses cell proliferation, migration, invasion, and epithelial–mesenchymal transition by regulating FOXM1 expression in human non-small cell lung cancer. Onco Targets Ther. 2019;12:2999–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wei W, Cao W, Zhan Z, Yan L, Xie Y, Xiao Q. MiR-1284 suppresses gastric cancer progression. Onco Targets Ther. 2019;12:3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen J, Zhang J, Li X, et al. Downregulation of ADAMTS8 by DNA hypermethylation in gastric cancer and its clinical significance. Biomed Res Int. 2016;2016:5083841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tian X, Zhang J, Yan L, Dong JM, Guo Q. MiRNA-15a inhibits proliferation, migration and invasion by targeting TNFAIP1 in human osteosarcoma cells. Int J Clin Exp Pathol. 2015;8(6):6442–6449. [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y, Liu T, Jin H, Yin L, Yu H, Bi J. MiR-411suppresses the development of bladder cancer by regulating ZnT1. Onco Targets Ther. 2018;11:8695–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Zeng J, Wu W, et al. Nicotinamide N-methyltransferase enhances chemoresistance in breast cancer through SIRT1 protein stabilization. Breast Cancer Res. 2019;21(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frazzi R. SIRT1 in secretory organ cancer. Front Endocrinol (Lausanne). 2018;9:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mvunta DH, Miyamoto T, Asaka R, et al. Overexpression of SIRT1 is associated with poor outcomes in patients with ovarian carcinoma. Appl Immunohistochem Mol Morphol. 2017;25(6):415–421. [DOI] [PubMed] [Google Scholar]
- 19. Li D, Bai L, Wang T, et al. Function of miR-212 as a tumor suppressor in thyroid cancer by targeting SIRT1. Oncol Rep. 2018;39(2):695–702. [DOI] [PubMed] [Google Scholar]
- 20. Tian S, Guo X, Yu C, Sun C, Jiang J. miR-138–5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget. 2017;8(7):11071–11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noguchi A, Kikuchi K, Zheng H, et al. SIRT1 expression is associated with a poor prognosis, whereas DBC1 is associated with favorable outcomes in gastric cancer. Cancer Med. 2014;3(6):1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang Q, Wang B, Zang W, et al. Resveratrol inhibits the growth of gastric cancer by inducing G1 phase arrest and senescence in a sirt1-dependent manner. PLoS One. 2013;8(11):e70627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang S, Yang Y, Huang S, et al. SIRT1 inhibits gastric cancer proliferation and metastasis via STAT3/MMP-13 signaling. J Cell Physiol. 2019;234(9):15395–15406. [DOI] [PubMed] [Google Scholar]
- 24. Zhou X, Chen J, Chen L, et al. Negative regulation of Sirtuin 1 by AMP-activated protein kinase promotes metformin-induced senescence in hepatocellular carcinoma xenografts. Cancer Lett. 2017;411:1–11. [DOI] [PubMed] [Google Scholar]
- 25. Yang Q, Wang B, Gao W, et al. SIRT1 is downregulated in gastric cancer and leads to G1-phase arrest via NF-κB/Cyclin D1 signaling. Mol Cancer Res. 2013;11(12):1497–1507. [DOI] [PubMed] [Google Scholar]