Abstract
Hepatocellular carcinoma (HCC) is the most common type of malignant neoplasm of the liver with high morbidity and mortality. Extensive research into the pathology of HCC has been performed; however, the molecular mechanisms underlying the development of hepatitis B virus-associated HCC have remained elusive. Thus, the present study aimed to identify critical genes and pathways associated with the development and progression of HCC. The expression profiles of the GSE121248 dataset were downloaded from the Gene Expression Omnibus database and the differentially expressed genes (DEGs) were identified. Gene Ontology (GO) and Kyoto Encyclopedia of Gene and Genome (KEGG) analyses were performed by using the Database for Annotation, Visualization and Integrated Discovery. Subsequently, protein-protein interaction (PPI) networks were constructed for detecting hub genes. In the present study, 1,153 DEGs (777 upregulated and 376 downregulated genes) were identified and the PPI network yielded 15 hub genes. GO analysis revealed that the DEGs were primarily enriched in ‘protein binding’, ‘cytoplasm’ and ‘extracellular exosome’. KEGG analysis indicated that DEGs were accumulated in ‘metabolic pathways’, ‘chemical carcinogenesis’ and ‘fatty acid degradation’. After constructing the PPI network, cyclin-dependent kinase 1, cyclin B1, cyclin A2, mitotic arrest deficient 2 like 1, cyclin B2, DNA topoisomerase IIα, budding uninhibited by benzimidazoles (BUB)1, TTK protein kinase, non-SMC condensin I complex subunit G, NDC80 kinetochore complex component, aurora kinase A, kinesin family member 11, cell division cycle 20, BUB1B and abnormal spindle microtubule assembly were identified as hub genes based on the high degree of connectivity by using Cytoscape software. In addition, overall survival (OS) and disease-free survival (DFS) analyses were performed using the Gene Expression Profiling Interactive Analysis online database, which revealed that the increased expression of all hub genes were associated with poorer OS and DFS outcomes. Receiver operating characteristic curves were constructed using GraphPad prism 7.0 software. The results confirmed that 15 hub genes were able to distinguish HCC form normal tissues. Furthermore, the expression levels of three key genes were analyzed in tumor and normal samples of the Human Protein Atlas database. The present results may provide further insight into the underlying mechanisms of HCC and potential therapeutic targets for the treatment of this disease.
Keywords: bioinformatics analysis, hepatocellular carcinoma, biomarker, differentially expressed genes, hepatitis B virus
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-associated mortality worldwide and is highly associated with hepatitis C and B infection, as well as alcohol abuse (1). In East Asia and sub-Saharan Africa, chronic hepatitis B infection is a vital factor in the pathogenesis of liver cancer (2). To date, numerous studies have identified hepatitis B virus (HBV)-associated factors, including hepatitis B e antigen and the hepatitis B surface antigen, as key predictors of HBV-associated HCC development in patients with hepatitis B infection (3). In addition, the viral load of HBV, duration of infection and severity of liver disease have been linked to the risk of developing cirrhosis (2,4); however, the mechanisms of HBV-associated HCC development remain to be fully elucidated, which poses challenges for the clinical treatment of this disease. Post-operative recurrence is the most imperative issue to overcome (5). Bioinformatics analyses may be applied to determine the potential mechanisms underlying the development of HBV-associated HCC. Furthermore, predictive biomarkers may improve the prediction of patient prognosis and the understanding of the molecular mechanisms underlying this disease (6).
α-Fetoprotein, associated with gene duplications (7), was the sole biomarker reported in randomized controlled trials (8). As only a small number of biomarkers have been identified, developments in the treatment and diagnosis of HCC, as well as prognostic evaluation, are restricted. With recent advances in high-throughput technologies, an increasing number of gene chips are available for the detection of differentially expressed genes (DEGs); thus, bioinformatics analyses of associated data are frequently being performed (9). Based on these novel techniques, databases of vast core gene data have been produced, including the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) and the Cancer Genome Atlas (10). Analysis of these freely available gene expression data may lead to identification of DEGs in tumor vs. normal tissue, which may provide novel insight into the development of cancers. Jiao et al (11) reported that upregulated eukaryotic translation initiation factor 2B subunit ε (EIF2B5) expression was associated with HCC development based on GSE54236 and GSE76427 microarray data, indicating that EIF2B5 may be employed as a novel biomarker for patients with liver cancer.
In the present study, the gene expression dataset GSE121248, including hepatitis B-induced HCC and adjacent normal samples, was subjected to bioinformatics analysis. The DEGs were screened and the functions and associated pathways in HBV-associated HCC were further evaluated. The core genes identified in the present study may be considered as potential novel targets for the treatment of HBV-associated HCC. The present results may also improve the understanding of the development and recurrence of this disease, and provide a basis for developments regarding its clinical treatment.
Materials and methods
Data sourcing and identification of DEGs
The GSE121248 microarray dataset was acquired and downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/), on the basis of the GPL570 platform analyzed by the Affymetrix Human Genome U133 Plus 2.0 Array, and included the data of 70 HBV-associated HCC tumor samples and 37 adjacent normal tissue samples. Simultaneously, the Series Matrix File of GSE121248 was downloaded. In order to obtain more meaningful targets for the clinical application, |log fold change (FC)|>1(12) was set as the limit and GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used. Genes were identified as significant DEGs based on adjusted P<0.05(13), which was applied to reduce the false-positive rate.
Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs
GO analysis in the categories cellular component, molecular function and biological process (14) was performed for the functional annotation of genes. KEGG was used to determine the biological pathways associated with the DEGs (15). The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov) (16), a free online biological database, was employed to perform GO and KEGG analyses with the Bingo plug-in to obtain systematic and comprehensive information for all DEGs. P<0.05 was considered to indicate a statistically significant difference.
Construction of the protein-protein interaction (PPI) network
To investigate the mechanism underlying the development of HCC, the Search Tool for the Retrieval of Interacting Genes (https://string-db.org/), a free online tool that may be employed to evaluate and construct a PPI network of target genes and DEGs, including downregulated and upregulated genes, was used. Subsequently, Cytoscape software (17) was utilized to select hub genes in the PPI network. In addition, DAVID was used to perform GO and KEGG analyses to obtain additional information on the hub genes.
Association of hub genes with clinical outcomes and diagnostic value
Overall survival (OS) and disease-free survival (DFS) analyses of hub genes were performed using the Gene Expression Profiling Interactive Analysis (GEPIA) web server (http://gepia.cancer-pku.cn/); log-rank P<0.05 was considered to indicate statistical significance. Immunohistochemical data for three hub genes with the highest degrees in patients with or without HCC were then downloaded from the Human Protein Atlas (HPA; https://www.proteinatlas.org/). In addition, GraphPad prism 7 software (GraphPad Software, Inc.) was used to generate receiver operating characteristic (ROC) curves to determine the diagnostic value of the 15 hub genes in HCC.
Results
Identification of DEGs in HBV-associated HCC
The dataset comprised 107 samples, including 70 tumor samples and 37 adjacent normal tissue samples. The GEO2R tool was used to identify the DEGs, with adjusted P<0.05 and |logFC|>1 selected as the cut-off criteria. A total of 1,153 DEGs, comprising 376 downregulated and 777 upregulated genes, were retrieved by analyzing the GSE121248 dataset. All DEGs were included in the subsequent analysis.
GO functional and KEGG pathway enrichment analyses of DEGs
After the DEGs were identified, DAVID was employed to perform GO functional and KEGG pathway enrichment analyses of the DEGs. The data of the DEGs were analyzed with the tool; in the GO analysis, the functional terms in three different categories, namely cellular component, biological process and molecular function enriched by the DEGs were determined. In summary, the DEGs were significantly accumulated in cellular component terms including ‘protein binding’ (GO:0005515), ‘cytoplasm’ (GO:0005737), ‘extracellular exosome’ (GO:0070062) and ‘cytosol’ (GO:005829). Specifically, upregulated genes were mainly enriched in cellular component terms, including ‘extracellular region’ (GO:0005576), ‘extracellular exosome’ (GO:0070062) and ‘extracellular space’ (GO:0005615), while downregulated genes were mainly enriched in biological process terms, including ‘cell division’ (GO:0051301) and ‘mitotic nuclear division’ (GO:0007067). In Table I, the top 10 GO terms of the upregulated and downregulated genes are listed according to their P-value from the lowest to the highest value.
Table I.
GO analysis of differentially expressed genes.
| A, Upregulated genes | ||||
|---|---|---|---|---|
| Category | Term | Count | P-value | FDR |
| GOTERM_CC_DIRECT | GO:0005576-extracellular region | 119 | 3.20x10-20 | 4.34x10-17 |
| GOTERM_CC_DIRECT | GO:0070062-extracellular exosome | 166 | 1.25x10-18 | 1.69x10-15 |
| GOTERM_CC_DIRECT | GO:0005615-extracellular space | 99 | 2.59x10-16 | 3.00x10-13 |
| GOTERM_CC_DIRECT | GO:0072562-blood microparticle | 29 | 1.31x10-14 | 1.78x10-11 |
| GOTERM_BP_DIRECT | GO:0055114-oxidation-reduction process | 58 | 4.38x10-14 | 7.78x10-11 |
| GOTERM_MF_DIRECT | GO:0016705-oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 18 | 5.79x10-13 | 9.05x10-10 |
| GOTERM_BP_DIRECT | GO:0019373-epoxygenase P450 pathway | 12 | 6.99x10-13 | 1.24x10-9 |
| GOTERM_MF_DIRECT | GO:0020037-heme binding | 25 | 3.51x10-12 | 5.48x10-9 |
| GOTERM_MF_DIRECT | GO:0019825-oxygen binding | 16 | 4.49x10-12 | 7.01x10-9 |
| GOTERM_CC_DIRECT | GO:0031090-organelle membrane | 20 | 1.06x10-11 | 1.44x10-8 |
| B, Downregulated genes | ||||
| Category | Term | Count | P-value | FDR |
| GOTERM_BP_DIRECT | GO:0051301-cell division | 38 | 7.08x10-20 | 1.18x10-16 |
| GOTERM_BP_DIRECT | GO:0007067-mitotic nuclear division | 29 | 6.27x10-16 | 1.11x10-12 |
| GOTERM_CC_DIRECT | GO:0000777-condensed chromosome kinetochore | 16 | 4.44x10-12 | 5.98x10-9 |
| GOTERM_BP_DIRECT | GO:0007062-sister chromatid cohesion | 17 | 9.20x10-12 | 1.53x10-8 |
| GOTERM_CC_DIRECT | GO:0030496-midbody | 18 | 1.46x10-11 | 1.97x10-8 |
| GOTERM_BP_DIRECT | GO:0000082-G1/S transition of mitotic cell cycle | 16 | 9.34x10-11 | 1.55x10-7 |
| GOTERM_CC_DIRECT | GO:0000775-chromosome, centromeric region | 12 | 9.48x10-10 | 1.28x10-6 |
| GOTERM_MF_DIRECT | GO:0005515-protein binding | 189 | 5.91x10-9 | 8.35x10-6 |
| GOTERM_BP_DIRECT | GO:0000281-mitotic cytokinesis | 9 | 1.37x10-8 | 2.28x10-5 |
| GOTERM_CC_DIRECT | GO:0000776-kinetochore | 12 | 4.49x10-8 | 6.04x10-5 |
FDR, false discovery rate; GO, gene ontology; CC, cellular component; BP, biological process; MF, molecular function.
Furthermore, KEGG pathway analysis revealed that the DEGs were mainly involved in ‘metabolic pathways’ [Homo sapiens (hsa)01100], ‘chemical carcinogenesis’ (hsa05204) and ‘fatty acid degradation’ (hsa00071). In addition, the upregulated genes were mainly enriched in ‘retinol metabolism’ (hsa00830), which is associated with the development of diabetes (18). Downregulated genes were accumulated in the pathways of ‘cell cycle’ (hsa04110) and ‘extracellular matrix-receptor interaction’ (hsa04512), as well as ‘p53 signaling pathway’ (hsa04115) (Table II).
Table II.
KEGG analysis of differentially expressed genes (top 5 according to P-value).
| A, Upregulated genes | ||
|---|---|---|
| Term | Count | P-value |
| hsa01100:Metabolic pathways | 107 | 2.83x10-14 |
| hsa05204:Chemical carcinogenesis | 21 | 9.07x10-11 |
| hsa00830:Retinol metabolism | 17 | 7.29x10-9 |
| hsa04610:Complement and coagulation cascades | 17 | 2.35x10-8 |
| hsa00980:Metabolism of xenobiotics by cytochrome P450 | 17 | 6.78x10-8 |
| B, Downregulated genes | ||
| Term | Count | P-value |
| hsa04110:Cell cycle | 21 | 6.49x10-15 |
| hsa04512:ECM-receptor interaction | 11 | 1.44x10-6 |
| hsa04115:p53 signaling pathway | 9 | 1.28x10-5 |
| hsa05200:Pathways in cancer | 20 | 2.06x10-5 |
| hsa03030:DNA replication | 7 | 2.36x10-5 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens; ECM, extracellular matrix.
PPI network analysis of DEGs
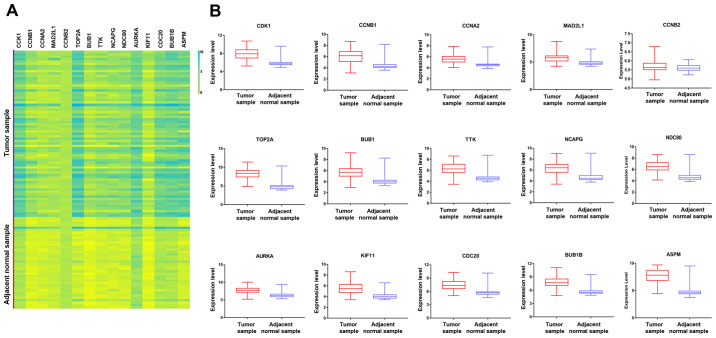
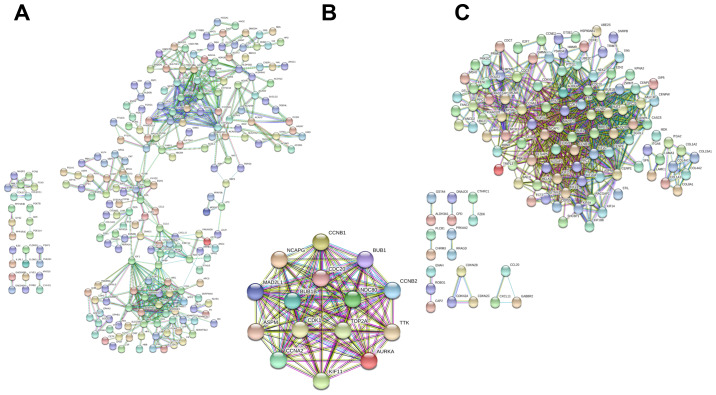
As the minimum interaction score required, 0.900 was applied. The PPI network of all the DEGs contained 677 nodes and 1,118 edges (enrichment P<1.0x10-16). Cytoscape software was then employed to identify the top 15 hub genes that may be associated with HBV-associated HCC according to their high degree of connectivity from high to low based on the results of PPI network. These genes included abnormal spindle-like microcephaly-associated protein (ASPM), aurora kinase A (AURKA), budding uninhibited by benzimidazoles (BUB)1, BUB1B, cyclin A2 (CCNA2), CCNB1, CCNB2, cell division cycle 20 (CDC20), cyclin-dependent kinase 1 (CDK1), kinesin family member 11 (KIF11), mitotic arrest deficient 2 like 1 (MAD2L1), non-SMC condensin I complex subunit G (NCAPG), NDC80 kinetochore complex component (NDC80), DNA topoisomerase II α (TOP2A) and TTK protein kinase (TTK) (Table III). To further investigate the hub genes, a heatmap and box plot of the expression of the top 15 hub genes in HBV-associated HCC tumor samples and adjacent normal tissue samples were generated using GraphPad prism 7 software (Fig. 1). The PPI networks of upregulated and downregulated genes, as well as hub genes, are presented in Fig. 2.
Table III.
Top 15 hub genes in the protein-protein interaction network ranked by degree.
| Gene symbol | Degree |
|---|---|
| CDK1 | 90 |
| CCNB1 | 79 |
| CCNA2 | 75 |
| MAD2L1 | 73 |
| CCNB2 | 70 |
| TOP2A | 70 |
| BUB1 | 69 |
| TTK | 67 |
| NCAPG | 67 |
| NDC80 | 66 |
| AURKA | 66 |
| KIF11 | 66 |
| CDC20 | 65 |
| BUB1B | 63 |
| ASPM | 63 |
CDK1, cyclin-dependent kinase 1; CCNA2, cyclin A2; MAD2L1, mitotic arrest deficient 2 like 1; TOP2A, DNA topoisomerase IIα; BUB1, budding uninhibited by benzimidazoles 1; TTK, TTK protein kinase; NCAPG, non-SMC condensin I complex subunit G; NDC80, NDC80 kinetochore complex component; AURKA, aurora kinase A; KIF11, kinesin family member 11; CDC20, cell division cycle 20; ASPM, abnormal spindle microtubule assembly.
Figure 1.
(A) Volcano map of 15 hub genes in the protein-protein interaction network according to the highest degree. (B) Expression of various genes in the tumor samples and adjacent normal samples, including ASPM (P<0.0001), AURKA (P<0.0001), BUB1 (P<0.0001), BUB1B (P<0.0001), CCNA2 (P<0.0001), CCNB1 (P<0.0001), CCNB2 (P=0.1067), CDC20 (P<0.0001), CDK1 (P<0.0001), KIF11 (P<0.0001), MAD2L1 (P<0.0001), NCAPG (P<0.0001), NDC80 (P<0.0001), TOP2A (P<0.0001) and TTK (P<0.0001). CDK1, cyclin-dependent kinase 1; CCNA2, cyclin A2; MAD2L1, mitotic arrest deficient 2 like 1; TOP2A, DNA topoisomerase IIα; BUB1, budding uninhibited by benzimidazoles 1; TTK, TTK protein kinase; NCAPG, non-SMC condensin I complex subunit G; NDC80, NDC80 kinetochore complex component; AURKA, aurora kinase A; KIF11, kinesin family member 11; CDC2, cell division cycle 20; ASPM, abnormal spindle microtubule assembly; Adj, adjacent.
Figure 2.
(A) PPI network of upregulated genes, including 386 nodes and 1,105 edges with a PPI enrichment P<1.0x10-16. (B) PPI network of 15 hub genes. (C) PI network of downregulated genes, including 297 nodes and 3,910 edges with a PPI enrichment P<1.0x10-16. PPI, protein-protein interaction.
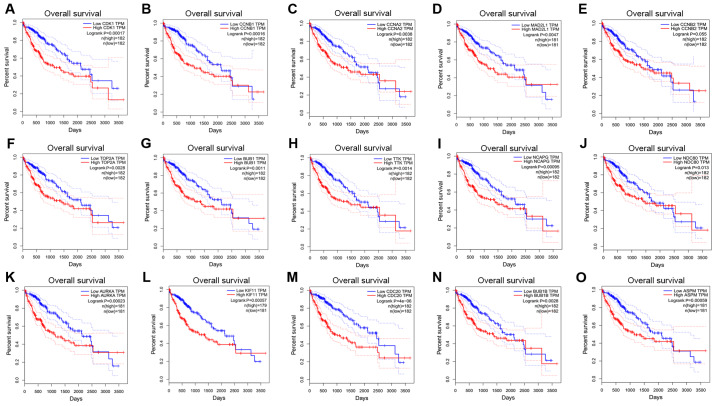
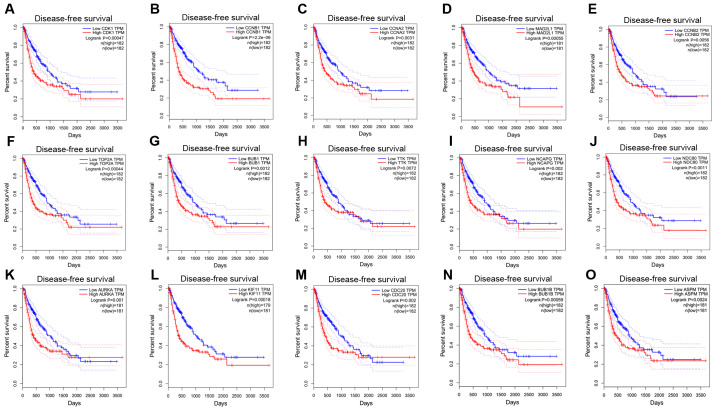
Survival analyses of hub genes
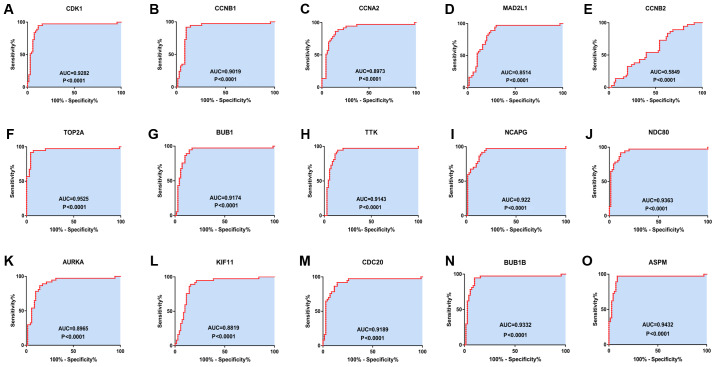
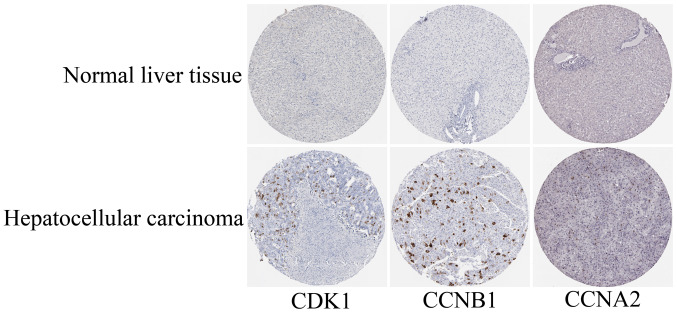
The GEPIA online database, a web-based tool, was designed to rapidly obtain customizable functionalities (19). Utilizing this tool, OS (Fig. 3) and the DFS (Fig. 4) curves were obtained and the log-rank P-values of the 15 hub genes were determined. The results indicated that all of the hub genes except CCNB2 were significantly associated with OS. Subsequently, the series matrix data downloaded from the GEO database were analyzed and ROC curves were generated to gain a comprehensive understanding of the predictive value of the hub genes. The results demonstrated that all the hub genes were able to efficiently distinguish HCC tissues from normal tissues (Fig. 5). In addition, the top three genes were selected based on the degree of connectivity in the PPI network for analysis of the immunohistochemical data in the HPA. Additionally, representative images indicated that the expression of all three hub genes were upregulated in HCC tissues (Fig. 6).
Figure 3.
Overall survival curves for 15 hub genes. (A) CDK1, (B) CCNB1, (C) CCNA2, (D) MAD2L1, (E) CCNB2, (F) TOP2A, (G) BUB1, (H) TTK, (I) NCAPG, (J) NDC80, (K) AURKA, (L) KIF11, (M) CDC20, (N) BUB1B and (O) ASPM. CDK1, cyclin-dependent kinase 1; CCNA2, cyclin A2; MAD2L1, mitotic arrest deficient 2 like 1; TOP2A, DNA topoisomerase IIα; BUB1, budding uninhibited by benzimidazoles 1; TTK, TTK protein kinase; NCAPG, non-SMC condensin I complex subunit G; NDC80, NDC80 kinetochore complex component; AURKA, aurora kinase A; KIF11, kinesin family member 11; CDC20, cell division cycle 20; ASPM, abnormal spindle microtubule assembly; TPM, transcripts per million.
Figure 4.
Disease-free survival curves of 15 hub genes. (A), CDK1, (B) CCNB1, (C) CCNA2, (D) MAD2L1, (E) CCNB2, (F) TOP2A, (G) BUB1, (H) TTK, (I) NCAPG, (J) NDC80, (K) AURKA, (L) KIF11, (M) CDC20, (N) BUB1B and (O) ASPM. CDK1, cyclin-dependent kinase 1; CCNA2, cyclin A2; MAD2L1, mitotic arrest deficient 2 like 1; TOP2A, DNA topoisomerase IIα; BUB1, budding uninhibited by benzimidazoles 1; TTK, TTK protein kinase; NCAPG, non-SMC condensin I complex subunit G; NDC80, NDC80 kinetochore complex component; AURKA, aurora kinase A; KIF11, kinesin family member 11; CDC20, cell division cycle 20; ASPM, abnormal spindle microtubule assembly; TPM, transcripts per million.
Figure 5.
Receiver operating characteristics curves for the 15 hub genes to identify HCC tissue from normal tissue. (A) CDK1 (AUC=0.9282, P<0.0001), (B) CCNB1 (AUC=0.0.9019, P<0.0001), (C) CCNA2 (AUC=0.8973, P<0.0001), (D) MAD2L1 (AUC=0.8514, P<0.0001), (E) CCNB2 (AUC=0.5849, P=0.1496), (F) TOP2A (AUC=0.9525, P<0.0001), (G) BUB1 (AUC=0.9174, P<0.0001), (H) TTK (AUC=0.9143, P<0.0001), (I) NCAPG (AUC=0.922, P<0.0001), (J) NDC80 (AUC=0.9363, P<0.0001), (K) AURKA (AUC=0.8965, P<0.0001), (L) KIF11 (AUC=0.8819, P<0.0001), (M) CDC20 (AUC=0.9189, P<0.0001), (N) BUB1B (AUC=0.9332, P<0.0001) and (O) ASPM (AUC=0.9432, P<0.0001). CDK1, cyclin-dependent kinase 1; CCNA2, cyclin A2; MAD2L1, mitotic arrest deficient 2 like 1; TOP2A, DNA topoisomerase IIα; BUB1, budding uninhibited by benzimidazoles 1; TTK, TTK protein kinase; NCAPG, non-SMC condensin I complex subunit G; NDC80, NDC80 kinetochore complex component; AURKA, aurora kinase A; KIF11, kinesin family member 11; CDC20, cell division cycle 20; ASPM, abnormal spindle microtubule assembly; AUC, area under the curve.
Figure 6.
Representative histological images from the Human Protein Atlas database (https://www.proteinatlas.org/). The normal liver tissue with staining for CDK1 was from a female subject aged 29 years (patient ID: 1899; staining: Not detected; intensity: Negative; quantity: Negative; location: None; magnification, not available) and the HCC tissue was from a female patient aged 52 years (patient ID: 2399; staining: Medium; intensity: Strong; quantity: <25%; location: Cytoplasmic/membranous nuclear; magnification, not available). The normal liver tissue with staining for CCNB1 was from a female subject aged 32 years (patient ID: 1846; staining: Not detected; intensity: Negative; quantity: Negative; location: None; magnification, not available) and the HCC tissue was from a male patient aged 49 years (patient ID: 929; staining: Medium; intensity: Strong; quantity: <25%; location: Cytoplasmic/membranous; magnification, not available). The normal liver tissue with staining for CCNA2 was from a female subject aged 50 years (patient ID: 2251; staining: Not detected; intensity: Negative; quantity: Negative; location: None; magnification, not available) and the HCC tissue was from a female patient aged 73 years (patient ID: 5192; staining: Low; intensity: Moderate; quantity: <25%; location: Nuclear; magnification, not available). CDK1, cyclin-dependent kinase 1; CCNA2, cyclin A2; HCC, hepatocellular carcinoma.
Discussion
Previous studies have investigated biomarkers and therapeutic targets for HBV-associated HCC (2); however, the markers currently available are unsatisfactory. Thus, it is of importance to identify additional targets for improving the therapeutic efficacy of treatments for this disease. In the present study, the gene expression data of the GSE121248 dataset including the gene expression data of 107 samples comprising 70 HBV-associated HCC tumor samples and 37 adjacent normal tissue samples were analyzed. A total of 15 core genes were identified, namely CDK1, CCNB1, CCNA2, MAD2L1, CCNB2, TOP2A, BUB1, TTK, NCAPG, NDC80, AURK1, KIF11, CDC20, BUB1B and ASPM. The majority of these hub genes were significantly associated with the development and progression of HBV-associated HCC, among which CDK1, CCNB1 and CCNA2 were the top three hub genes based on their degree in the PPI network.
To date, the association between the expression of CDK1, CCNB1, CCNA2, KIF11 and CCNB2, and the development of HCC deserves further investigation. Prevo and Pirovano (20) reported that CDK1 is able to mediate the transition from G2 phase to mitosis via the CDK1/PI3K/β-catenin signaling pathway. Haider et al (21) assessed the inhibition of CDK1 by the kinase inhibitor compounds BA-12 and B1-14, which led to necrosis and apoptosis of tumor cells without toxicity to adjacent normal cells. In addition, the current study reported that CDK1 was involved in the p53 signaling pathway. However, the true mechanism underlying the correlation between CDK1 and HCC remains unclear (22). Previous studies have indicated that the CCNB1-Cdk1 complex is a key regulator of mitotic entry (23,24). Chai et al (24) revealed that CCNB1 is highly expressed in HCC and is closely associated with the poor prognosis of patients with HCC, which was consistent with the present results. This indicated the plausibility of CCNB1 as a potential therapeutic target for HCC. However, Weng et al (25) demonstrated that there was no significant difference in CCNB1 expression between patients with non-recurrent HCC and healthy subjects. The study concluded that the role of CCNB1 overexpression in oncogenesis and the progression of HCC remains unclear. Thus, further investigation is required to determine the association between the expression of CCNB1 and the development of HCC. Of note, CCNA2 was reported to be associated with the formation of the HBV-CCNA2 chimeric transcript, which may accelerate the cell cycle and result in tumor development (26). In the present study, after constructing the CCNA2-associated Kaplan-Meier curves for OS and DFS, which indicated a significant association between CCNA2 expression and the prognosis of HCC, it was concluded that high expression levels of CCNA2 are associated with poor prognosis of patients with HCC. Furthermore, the results of ROC curve analysis demonstrated the ability of CCNA2 to distinguish HCC samples from normal samples. KIF11 serves an essential role in centrosome and chromosome dynamics in mitosis (27). To the best of our knowledge, the association between the expression levels of KIF11 and the development of HBV-associated HCC has remained elusive. In the GO analysis of the present study, KIF11 was mainly enriched in the terms ‘cytoplasm’, ‘cytosol’ and ‘cell division’, as well as ‘spindle formation’ and ‘carcinogenesis’; however, KEGG pathway enrichment analysis did not suggest any association between a particular pathway and KIF11. Furthermore, the Kaplan-Meier curves for OS and DFS suggested that upregulated KIF11 expression was linked to poor prognosis in HBV-associated HCC. Chen et al (28) identified that the overexpression of KIF11 was significantly associated with shorter relapse-free survival times; however, the function of KIF11 and the mechanism involved in HCC remains unknown. In addition, Li et al (29) reported that downregulated CCNB2 expression led to inhibition of the progression of malignant neoplasms. Conversely, to the best of our knowledge, the biological function of CCNB2 in HCC is largely unknown. In the present study, low expression levels of CCNB2 were associated with improved clinical outcomes of patients with HBV-associated HCC, including DFS.
The roles of MAD2L1, TOP2A, BUB1, TTK, NCAPG, NDC80, AURKA, CDC20, BUB1B and ASPM in HCC have been well reported in previous studies. As for MAD2L1, Li et al (30) identified this gene as a vital mediator of pathways underlying chromosomal regulation in HCC. Yun et al (31) demonstrated that inhibition of MAD2L1 expression led to the suppression of HCC cell proliferation, migration and invasion, suggesting that MAD2L1 may serve as a potential target for the clinical treatment and prognostic evaluation of patients with HCC. In agreement with a previous study (32), TOP2A may be a valuable prognostic marker and predictor of poor survival in patients with HCC. Based on previous studies and the present results, it was concluded that TOP2A may be a potential biomarker linked to the development of HBV-associated HCC. BUB1 is a serine/threonine kinase that binds centromeres during mitosis (33), and has been associated with the cell cycle and apoptosis (34), as well as reduced OS in patients with HCC (35). The expression levels of protein kinase human monopolar, also known as TTK, were previously reported to be markedly increased in HBV-associated HCC (36); TTK was reported to predict poorer outcomes for patients with HBV-associated HCC (37). The first TTK inhibitors to be developed in combination with taxane chemotherapy have entered phase 1 dose-escalation studies (38). NCAPG, which mediates the coiling topology of individual chromatids (39), was proposed to be associated with cell growth, proliferation and migration in HCC (40). NDC80 was reported to be associated with the progression of HCC (41). Of note, downregulation of NDC80 was able to suppress the replication of HBV-associated HCC cells (42). Bound to MYC, AURKA, which has two functional non-synonymous polymorphisms (lle31Phe and Val57lle), was able to regulate tumor cell growth at the genetic level (43). It has been reported that AURKA lle31Phe enhances the proneness of HBV-infected individuals to develop liver cancer (44). The expression levels of CDC20 have been indicated to be linked to the development of liver cancer and the prognosis of affected patients (45). Thus, it may be a candidate prognostic factor in patients with HBV-associated HCC (46). BUB1B was reported to be involved in the regulation of the cell cycle of tumor cells (47). As for ASPM, its upregulation has been suggested to serve as a novel marker for predicting the progression of HCC, early tumor recurrence and poor prognosis (48).
Of note, the present study had certain limitations. Data sourcing was performed using only one dataset, which may not be sufficient to provide a convincing hypothesis. Analyses with multiple datasets should be considered in the future. Hepatitis B infection is considered an independent factor in the development of HCC in the Cancer Genome Atlas database (TCGA) and therefore, TCGA-based research should be conducted in the future. Furthermore, the sample size was relatively small, which may result in unavoidable bias. Finally, the gene chip used in the present study did not contain any information on the pathological sections of HBV-associated HCC, and therefore, it was not possible to determine the association between HBV infection and HCC pathological grading sample by sample. It is indicated that HBV infection, even if temporary, may have a role in promoting the development of liver cancer; at the same time, a longer duration of HBV infection may accelerate the process of HCC (49). However, to the best of our knowledge, no biomarker can predict the association between HBV infection and the extent of HCC pathological severity.
In conclusion, the present bioinformatics analysis of a microarray dataset identified 15 core genes involved in the development and progression of HBV-associated HCC. Assessment of the association of these key genes with clinical outcomes indicated that these genes may be potential therapeutic targets for HCC, which may contribute to the development of treatments for HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81471765 and 81771950).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on request.
Authors' contributions
CZ and BL designed the current study. YL, XG, LZ and DZ acquired and analyzed the data. LZ and JM wrote and revised the manuscript and analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16(1) doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 3.Geier A, Gartung C, Dietrich CG. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347:1721–1722. doi: 10.1056/NEJMoa013215. [DOI] [PubMed] [Google Scholar]
- 4.EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu; European Association for the Study of the Liver. [DOI] [PubMed] [Google Scholar]
- 5.Fu PY, Hu B, Ma XL, Yang ZF, Yu MC, Sun HX, Huang A, Zhang X, Wang J, Hu ZQ, et al. New insight into BIRC3: A novel prognostic indicator and a potential therapeutic target for liver cancer. J Cell Biochem. 2019;120:6035–6045. doi: 10.1002/jcb.27890. [DOI] [PubMed] [Google Scholar]
- 6.Long J, Zhang L, Wan X, Lin J, Bai Y, Xu W, Xiong J, Zhao H. A four-gene-based prognostic model predicts overall survival in patients with hepatocellular carcinoma. J Cell Mol Med. 2018;22:5928–5938. doi: 10.1111/jcmm.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauzay C, Petit A, Bourgeois AM, Barbare JC, Chauffert B, Galmiche A, Houessinon A. Alpha-foetoprotein (AFP): A multi-purpose marker in hepatocellular carcinoma. Clin Chim Acta. 2016;463:39–44. doi: 10.1016/j.cca.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10573–10583. doi: 10.3748/wjg.v21.i37.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan P, He Y, Xie K, Kong S, Zhao W. In silico analyses for potential key genes associated with gastric cancer. PeerJ. 2018;6(e6092) doi: 10.7717/peerj.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teufel A. Bioinformatics and database resources in hepatology. J Hepatol. 2015;62:712–719. doi: 10.1016/j.jhep.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 11.Jiao Y, Fu Z, Li Y, Meng L, Liu Y. High EIF2B5 mRNA expression and its prognostic significance in liver cancer: A study based on the TCGA and GEO database. Cancer Manag Res. 2018;10:6003–6014. doi: 10.2147/CMAR.S185459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen S, Kong J, Qiu Y, Yang X, Wang W, Yan L. Identification of core genes and outcomes in hepatocellular carcinoma by bioinformatics analysis. J Cell Biochem. 2019;120:10069–10081. doi: 10.1002/jcb.28290. [DOI] [PubMed] [Google Scholar]
- 13.Zhou SL, Hu ZQ, Zhou ZJ, Dai Z, Wang Z, Cao Y, Fan J, Huang XW, Zhou J. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63:1560–1575. doi: 10.1002/hep.28445. [DOI] [PubMed] [Google Scholar]
- 14.Gaudet P, Skunca N, Hu JC, Dessimoz C. Primer on the gene ontology. Methods Mol Biol. 2017;1446:25–37. doi: 10.1007/978-1-4939-3743-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wixon J, Kell D. The Kyoto encyclopedia of genes and genomes-KEGG. Yeast. 2000;17:48–55. doi: 10.1002/(SICI)1097-0061(200004)17:1<48::AID-YEA2>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Baryshnikova A. doi: 10.1101/pdb.prot077644. Exploratory analysis of biological networks through visualization, clustering, and functional annotation in cytoscape. Cold Spring Harb Protoc 2016, 2016. [DOI] [PubMed] [Google Scholar]
- 18.McClinton KJ, Aliani M, Kuny S, Sauvé Y, Suh M. Differential effect of a carotenoid-rich diet on retina function in non-diabetic and diabetic rats. Nutr Neurosci. 2019;11:1–11. doi: 10.1080/1028415X.2018.1563664. [DOI] [PubMed] [Google Scholar]
- 19.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prevo R, Pirovano G, Puliyadi R, Herbert KJ, Rodriguez-Berriguete G, O'Docherty A, Greaves W, McKenna WG, Higgins GS. CDK1 inhibition sensitizes normal cells to DNA damage in a cell cycle dependent manner. Cell Cycle. 2018;17:1513–1523. doi: 10.1080/15384101.2018.1491236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haider C, Grubinger M, Řezníčková E, Weiss TS, Rotheneder H, Miklos W, Berger W, Jorda R, Zatloukal M, Gucky T, et al. Novel inhibitors of cyclin-dependent kinases combat hepatocellular carcinoma without inducing chemoresistance. Mol Cancer Ther. 2013;12:1947–1957. doi: 10.1158/1535-7163.MCT-13-0263. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Li Y, Hao H, Wang Y, Zhou Z, Wang Z, Chu X. Screening Hub genes as prognostic biomarkers of hepatocellular carcinoma by bioinformatics analysis. Cell Transplant. 2019;11(963689719893950) doi: 10.1177/0963689719893950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama Y, Yamaguchi N. Role of cyclin B1 levels in DNA damage and DNA damage-induced senescence. Int Rev Cell Mol Biol. 2013;305:303–337. doi: 10.1016/B978-0-12-407695-2.00007-X. [DOI] [PubMed] [Google Scholar]
- 24.Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang M, Liu J, Zhang XF, Zhang X, Yin H, et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun. 2018;500:924–929. doi: 10.1016/j.bbrc.2018.04.201. [DOI] [PubMed] [Google Scholar]
- 25.Weng L, Du J, Zhou Q, Cheng B, Li J, Zhang D, Ling C. Identification of cyclin B1 and Sec62 as biomarkers for recurrence in patients with HBV-related hepatocellular carcinoma after surgical resection. Mol Cancer. 2012;11(39) doi: 10.1186/1476-4598-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu YT, Wong JK, Choi SW, Sze KM, Ho DW, Chan LK, Lee JM, Man K, Cherny S, Yang W, et al. Novel pre-mRNA splicing of intronically integrated HBV generates oncogenic chimera in hepatocellular carcinoma. J Hepatol. 2016;64:1256–1264. doi: 10.1016/j.jhep.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Jin Q, Dai Y, Wang Y, Zhang S, Liu G. High kinesin family member 11 expression predicts poor prognosis in patients with clear cell renal cell carcinoma. J Clin Pathol. 2019;72:354–362. doi: 10.1136/jclinpath-2018-205390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Li S, Zhou S, Cao S, Lou Y, Shen H, Yin J, Li G. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther. 2017;13:651–659. doi: 10.4103/jcrt.JCRT_491_17. [DOI] [PubMed] [Google Scholar]
- 29.Li R, Jiang X, Zhang Y, Wang S, Chen X, Yu X, Ma J, Huang X. Cyclin B2 overexpression in human hepatocellular carcinoma is associated with poorprognosis. Arch Med Res. 2019;50:10–17. doi: 10.1016/j.arcmed.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Bai W, Zhang J. MiR-200c-5p suppresses proliferation and metastasis of human hepatocellular carcinoma (HCC) via suppressing MAD2L1. Biomed Pharmacother. 2017;92:1038–1044. doi: 10.1016/j.biopha.2017.05.092. [DOI] [PubMed] [Google Scholar]
- 31.Yun MY, Kim SB, Park S, Han CJ, Han YH, Yoon SH, Kim SH, Kim CM, Choi DW, Cho MH, et al. Mutation analysis of p31comet gene, a negative regulator of Mad2, in human hepatocellular carcinoma. ExperimeExp Mol Med. 2007;39:508–513. doi: 10.1038/emm.2007.56. [DOI] [PubMed] [Google Scholar]
- 32.Wong N, Yeo W, Wong WL, Wong NL, Chan KY, Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124:644–652. doi: 10.1002/ijc.23968. [DOI] [PubMed] [Google Scholar]
- 33.Xu B, Xu T, Liu H, Min Q, Wang S, Song Q. MiR-490-5p suppresses cell proliferation and invasion by targeting BUB1 in hepatocellular carcinoma cells. Pharmacology. 2017;100:269–282. doi: 10.1159/000477667. [DOI] [PubMed] [Google Scholar]
- 34.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–1064. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen QF, Xia JG, Li W, Shen LJ, Huang T, Wu P. Examining the key genes and pathways in hepatocellular carcinoma development from hepatitis B virus-positive cirrhosis. Mol Med Rep. 2018;18:4940–4950. doi: 10.3892/mmr.2018.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Liao W, Yuan Q, Ou Y, Huang J. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget. 2015;6:34309–34320. doi: 10.18632/oncotarget.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baffy G. Decoding multifocal hepatocellular carcinoma: An opportune pursuit. Hepatobiliary Surg Nutr. 2015;4:206–210. doi: 10.3978/j.issn.2304-3881.2014.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaman GJR, de Roos JADM, Libouban MAA, Prinsen MBW, de Man J, Buijsman RC, Uitdehaag JCM. TTK inhibitors as a targeted therapy for CTNNB1 (β-catenin) mutant cancers. Mol Cancer Ther. 2017;16:2609–2617. doi: 10.1158/1535-7163.MCT-17-0342. [DOI] [PubMed] [Google Scholar]
- 39.Liu W, Liang B, Liu H, Huang Y, Yin X, Zhou F, Yu X, Feng Q, Li E, Zou Z, Wu L. Overexpression of non-SMC condensin I complex subunit G serves as a promising prognostic marker and therapeutic target for hepatocellular carcinoma. Int J Mol Med. 2017;40:731–738. doi: 10.3892/ijmm.2017.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu K, Li Y, Yu B, Wang F, Mi T, Zhao Y. Silencing non-SMC chromosome-associated polypeptide G inhibits proliferation and induces apoptosis in hepatocellular carcinoma cells. Can J Physiol Pharmacol. 2018;96:1246–1254. doi: 10.1139/cjpp-2018-0195. [DOI] [PubMed] [Google Scholar]
- 41.Ju LL, Chen L, Li JH, Wang YF, Lu RJ, Bian ZL, Shao JG. Effect of NDC80 in human hepatocellular carcinoma. World J Gastroenterol. 2017;23:3675–3683. doi: 10.3748/wjg.v23.i20.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B, Yao Z, Hu K, Huang H, Xu S, Wang Q, Yang Y, Ren J. ShRNA-mediated silencing of the Ndc80 gene suppress cell proliferation and affected hepatitis B virus-related hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2016;40:297–303. doi: 10.1016/j.clinre.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Huang Y, Ling J, Zhuo W, Yu Z, Shao M, Luo Y, Zhu Y. Screening and function analysis of hub genes and pathways in hepatocellular carcinoma via bioinformatics approaches. Cancer Biomark. 2018;22:511–521. doi: 10.3233/CBM-171160. [DOI] [PubMed] [Google Scholar]
- 44.Bao Z, Lu L, Liu X, Guo B, Zhai Y, Li Y, Wang Y, Xie B, Ren Q, Cao P, et al. Association between the functional polymorphism Ile31Phe in the AURKA gene and susceptibility of hepatocellular carcinoma in chronic hepatitis B virus carriers. Oncotarget. 2017;8:54904–54912. doi: 10.18632/oncotarget.18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Gao JZ, Du JL, Huang ZX, Wei LX. Increased CDC20 expression is associated with development and progression of hepatocellular carcinoma. Int J Oncol. 2014;45:1547–1555. doi: 10.3892/ijo.2014.2559. [DOI] [PubMed] [Google Scholar]
- 46.Chen PF, Li QH, Zeng LR, Yang XY, Peng PL, He JH, Fan B. A 4-gene prognostic signature predicting survival in hepatocellular carcinoma. J Cell Biochem. 2019;120:9117–9124. doi: 10.1002/jcb.28187. [DOI] [PubMed] [Google Scholar]
- 47.Wen DY, Lin P, Pang YY, Chen G, He Y, Dang YW, Yang H. Expression of the long intergenic non-protein coding RNA 665 (LINC00665) gene and the cell cycle in hepatocellular carcinoma using the cancer genome atlas, the gene expression omnibus, and quantitative real-time polymerase chain reaction. Med Sci Monit. 2018;24:2786–2808. doi: 10.12659/MSM.907389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin SY, Pan HW, Liu SH, Jeng YM, Hu FC, Peng SY, Lai PL, Hsu HC. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res. 2008;14:4814–4820. doi: 10.1158/1078-0432.CCR-07-5262. [DOI] [PubMed] [Google Scholar]
- 49.Okuda K, Nakashima T, Sakamoto K, Ikari T, Hidaka H, Kubo Y, Sakuma K, Motoike Y, Okuda H, Obata H. Hepatocellular carcinoma arising in noncirrhotic and highly cirrhotic livers: A comparative study of histopathology and frequency of hepatitis B markers. Cancer. 1982;49:450–455. doi: 10.1002/1097-0142(19820201)49:3<450::aid-cncr2820490310>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on request.