Abstract
Purpose
Radiation dose to cardiac substructures is related to radiation-induced heart disease. However, substructures are not considered in radiation therapy planning (RTP) due to poor visualization on CT. Therefore, we developed a novel deep learning (DL) pipeline leveraging MRI’s soft tissue contrast coupled with CT for state-of-the-art cardiac substructure segmentation requiring a single, non-contrast CT input.
Materials/methods
Thirty-two left-sided whole-breast cancer patients underwent cardiac T2 MRI and CT-simulation. A rigid cardiac-confined MR/CT registration enabled ground truth delineations of 12 substructures (chambers, great vessels (GVs), coronary arteries (CAs), etc.). Paired MRI/CT data (25 patients) were placed into separate image channels to train a three-dimensional (3D) neural network using the entire 3D image. Deep supervision and a Dice-weighted multi-class loss function were applied. Results were assessed pre/post augmentation and post-processing (3D conditional random field (CRF)). Results for 11 test CTs (seven unique patients) were compared to ground truth and a multi-atlas method (MA) via Dice similarity coefficient (DSC), mean distance to agreement (MDA), and Wilcoxon signed-ranks tests. Three physicians evaluated clinical acceptance via consensus scoring (5-point scale).
Results
The model stabilized in ~19 h (200 epochs, training error <0.001). Augmentation and CRF increased DSC 5.0 ± 7.9% and 1.2 ± 2.5%, across substructures, respectively. DL provided accurate segmentations for chambers (DSC = 0.88 ± 0.03), GVs (DSC = 0.85 ± 0.03), and pulmonary veins (DSC = 0.77 ± 0.04). Combined DSC for CAs was 0.50 ± 0.14. MDA across substructures was <2.0 mm (GV MDA = 1.24 ± 0.31 mm). No substructures had statistical volume differences (P > 0.05) to ground truth. In four cases, DL yielded left main CA contours, whereas MA segmentation failed, and provided improved consensus scores in 44/60 comparisons to MA. DL provided clinically acceptable segmentations for all graded patients for 3/4 chambers. DL contour generation took ~14 s per patient.
Conclusions
These promising results suggest DL poses major efficiency and accuracy gains for cardiac substructure segmentation offering high potential for rapid implementation into RTP for improved cardiac sparing.
Keywords: cardiotoxicity, deep learning, magnetic resonance imaging, radiotherapy, segmentation
1. INTRODUCTION
Increased risks of radiation-induced heart disease (RIHD) including acute (pericarditis) and late (congestive heart failure, coronary artery disease (CAD), and myocardial infarction) cardiotoxicities have been linked to dose from thoracic radiation therapy (RT) for lymphoma, lung, breast, and esophageal cancers.1–4 RIHD presents earlier than previously expected, beginning only a few years after RT and with elevated risk persisting for ~20 yr.4 Importantly, dose escalation evaluation for locally advanced non-small cell lung cancer in RTOG 06175 revealed that the volumes of the heart receiving ≥5 and ≥30 Gy were independent predictors of survival.5 Furthermore, heart dose/volume metrics are significantly associated with quality of life.6
At present, dosimetric evaluation is currently limited to simplified heart volume/dose relationships, such as those recommended by QUANTEC, where the heart is considered a single organ. It is currently recommended that <10% of the heart receive >25 Gy with the clinical endpoint of long-term cardiac mortality.7 Despite having whole-heart dose limits, evidence suggests that dose to sensitive cardiac substructures may lead to cardiac toxicities4,8,9 including cardiomyopathy, CAD, as well as pericardial, and conduction system diseases.10 Specifically, an increased rate of cardiac events and ischemic diseases have been associated with increased radiation dose to the left ventricle,8 left atrium,9 and left anterior descending artery (LADA).11 Patel et al. found that a maximum dose >10 Gy to the LADA was a significant threshold for increased odds of developing coronary artery calcification (CAC). When compared to mean heart dose, maximum dose to the LADA had a stronger association with CAC onset.12 However, following these dosimetric thresholds is currently limited by the poor visualization and ability to delineate these sensitive cardiac substructures on non-contrast CT simulation (CT-SIM) scans.
Several studies have assessed atlas-based segmentation of cardiac substructures in rt13–15 to avoid the time consuming (6–10 h/patient) and tedious task of manual delineation.16 However, most atlases fail in segmenting the coronary arteries, with Dice similarity coefficients (DSCs) between ground truth and auto-segmentation of the LADA ranging from 0.09 to 0.27.13–15,17 Incorporating multiple imaging modalities (i.e., contrast enhanced CT and magnetic resonance imaging (MRI)) has improved visualization and yielded successful chamber and great vessel segmentation, yet coronary artery segmentation remains an unmet need.13,14
Recently, deep neural networks (DNNs), such as U-Net,18 have shown great promise for generating accurate and rapid delineations for RT.19 Here, a DNN learns a mapping function between an image and a corresponding feature map (i.e., segmented ground truth). Payer et al. implemented a U-Net for substructure segmentation and obtained a DSC of 94% in the aorta as compared to ground truth.20 Various DNNs have been applied to medical image segmentation,19 specifically for cardiac substructure segmentation. These include deep convolutional neural networks (CNNs) with adaptive fusion21 or multi-stage20 strategies, as well as generative adversarial networks (GANs)22. Adaptation of these segmentation strategies has greatly improved cardiac chamber20 and pulmonary artery21 segmentations on contrast-enhanced CTs (DSCs > 85%). Additionally, deep residual learning techniques are currently being used to generate cardiac substructure segmentation models that are robust against the presence or absence of image contrast.23 However, most of these models have not been applied to conventional CT-SIM images and have yet to implement segmentations of the pulmonary veins (PVs) and coronary arteries.
The current work builds upon recent DNN results to develop an efficient and accurate cardiac substructure deep learning (DL) segmentation pipeline that can be implemented into routine practice on standard, non-contrast CT-SIMs, thus requiring no additional image acquisitions. Here, training is performed via labeled MRI/CT pairs inputted into a three-dimensional (3D) U-Net coupled to predict cardiac substructure segmentations using a single non-contrast CT-SIM input. We further improve agreement to ground truth delineations by introducing a 3D dense conditional random field (CRF) as a post-processing step, which has been recently merged with DNNs for state-of-the-art results in medical image segmentation.24 Overall, the overarching goal is to enable widespread implementation of DL to improve cardiac sparing in RT planning (RTP) accomplished via cardiac sparing trials and improved risk assessment evaluation.
2. METHODS
2.A. Imaging and ground truth contour delineation
Thirty-two left-sided whole-breast cancer patients, with 36 unique datasets, were consented to an Institutional Review Board approved study and underwent cardiac MRI scans (two-dimensional T2 single-shot turbo spin echo sequence, TR = 927.9 ms, TE = 81 ms, voxel size = 0.7 × 0.7 × 8.0 mm3) at end-expiration (EE) on a 3T Philips Ingenia (Philips Medical Systems, Cleveland, OH). Imaging was completed in a single breath hold (acquisition time = 22.1 ± 4.4 s). Non-contrast CT-SIM images were acquired on a Brilliance Big Bore CT simulator (Philips Medical Systems, Cleveland, OH) (voxel size = 1.1 × 1.1 × 3. mm3−1.4 × 1.4 × 3.0 mm3, 120–140 kVp, and 275–434 mAs) with patients immobilized in the supine position on a Posiboard (Civco, The Netherlands). Twenty-four patients were imaged under free breathing conditions, while the other eight underwent four-dimensional CT (4DCT).
To develop a mutual coordinate system between datasets, an automatic global rigid registration between the T2 MR (moving image) and CT-SIM (target image) images was performed in MIM (version 6.9.1, MIM Software Inc., Cleveland, OH). An automated local rigid registration was then applied via a manually drawn cardiac-confined bounding box. For both rigid registrations, normalized mutual information was used as the similarity metric as it has been shown to perform well with multi-modality image registration tasks.25 For patients who underwent 4DCT, the 50% phase was used as it most closely matched the EE MRI.
The evaluation and approval of the co-registration of the T2 MRI to the non-contrast CT were performed through visual verification by a radiation oncologist. To generate the contours, a consensus atlas was followed26 as implemented in our previous work.13 In brief, 12 cardiac substructures (left/right ventricles (LV, RV) and atria (LA, RA), superior/inferior venae cavae (SVC, IVC), pulmonary artery/veins (PA, PV), ascending aorta (AA), right coronary artery (RCA), left main coronary artery (LMCA), and LADA) were manually delineated by a radiation oncologist and verified by a radiologist with a cardiac subspecialty. Due to the enhanced soft tissue contrast that MRI provides, preference was given to anatomical information from the MRI.
2.B. Data preparation
All work was performed using an NVIDIA Quadro M4000 graphical processing unit (NVIDIA, Santa Clara, CA). To improve generalizability, zero-mean normalization27 (i.e., subtracting the mean intensity from the image and dividing by the standard deviation of the image) was performed to maintain intensity consistency across MRI/CT datasets and patients. To generate ground truth images, substructure masks were combined into a single image volume (ground truth (GT) image in Fig. 1) with intensity values for the 12 substructures indexed every 20 grayscale values from 35 to 255 with no overlap among substructures. MR and CT images were all resampled to a 650 × 650 mm in-plane resolution using bilinear interpolation. Bilinear interpolation was also used to interpolate MR images in the z-direction to match the 3 mm CT slice thickness (final voxel size of 1.27 × 1.27 × 3 mm3). Registered MR and CT image volumes were cropped to 64 slices (in-plane dimension of 128 × 128 pixels), centered on the centroid of the whole heart, and padded with 32 blank slices both superiorly and inferiorly for a final size of 128 × 128 × 128 pixels.
Fig. 1.
3D U-Net architecture with CT and MR inputs in different image channels, along with the ground truth (GT) labels. Prediction maps are outputted for each substructure. [Color figure can be viewed at wileyonlinelibrary.com]
2.C. Neural network architecture and training
The proposed 3D U-Net shown in Fig. 1 was based on an existing architecture designed for brain tumor auto-segmentation28 with several customizations as follows: (a) including deep supervision, (b) training using the entire 3D image volume simultaneously with multi-channel data inputs (i.e., MRI, CT, and cardiac substructure ground truth masks), (c) optimizing hyperparameters of a Dice-weighted multi-class loss function,28 (d) utilizing deconvolution in the upsampling process, and (e) optimizing the number of feature maps used in the first layer.
This 3D U-Net is composed of a contraction pathway (Fig. 1, left) to aggregate high level information using context modules and an expansion pathway (Fig. 1, right) to combine feature and spatial information for localization.28 Context modules (Fig. 1, left) were composed of a dropout layer with 30% probability between two 3 × 3 × 3 convolutional layers. Deep supervision was implemented by adding segmentation layers at each step of the localization pathway (Fig. 1, right). Deep supervision allows for the injection of gradient signals deep into the network,29 as it speeds up convergence and enhances training efficiency when there is a small amount of available labeled training data.28,30 An elementwise summation with upsample was then applied across all added segmentation layers to generate the final segmentation. As coarse segmentation results may yield unrealistic results, skip connections were applied (i.e., concatenation) by fusing earlier layers in the network where the downsampling factor is smaller to recover the original spatial resolution.31 To rebuild high-resolution feature maps, deconvolution was used in the localization pathway in order to learn the upsampling.32
To prevent model overfitting (i.e., ensuring the model remains generalizable to the hold-out dataset after being tuned to a training set), data augmentation18 including flipping, rotating (0–30°, 1° increments), scaling (±25%, 1% increments), and translating (10 pixels in the left-right, anterior-posterior, and superior-inferior directions) was applied. Originally proposed as a novel objective function based on DSC,33 a Dice-weighted multi-class loss function was used28,34 to manage the different image features among substructures, as shown in Eq. 1:
| (1) |
As label encoding is not sufficient for model training, y represents the ground truth segmentation map converted from categorical to binary variables (i.e., one-hot encoding). For training voxel a in class b, xa,b and ya,b represent the prediction and ground truth, respectively. As a larger DSC represents better overlap between ground truth and the prediction, the loss function is negative due to it being minimized during the training. Each value of correspondence between both the training and validation datasets to ground truth is represented by an average across all 12 substructures.
An adaptive momentum estimation (Adam) optimizer35 was used along with randomly initialized weights. Patience (i.e., number of epochs to wait without validation loss improvement before reducing the learning) was also implemented during training. An epoch is defined as one forward pass and one backward pass (i.e., backpropagation process) of all the training samples.36 Optimized hyperparameters included an initial learning rate of 5 × 10−4, 50% learning rate reduction, a batch size of 1, patience of 10 epochs, and 16 base filters in the first layer of the localization pathway.
Patient data were split into 25 patients for training data and 7 (11 unique datasets) patients for a hold-out dataset for network testing. No hold-out datasets used for testing were implemented in the network training. Training data were split via random assignment into 80% training and 20% validation data. Paired MRI and CT data were placed into separate image channels along with indexed ground truth labels for 25 patients to train the 3D U-Net using the entire 3D MR and CT images and all substructures simultaneously. Training was considered to be sufficiently converged when the training error between two adjacent epochs (i.e., one forward and backward pass of all the training samples) was <0.001.36
All work was performed using Windows 10 operating system in Python version 3.6. The 64-bit Microsoft Windows system is equipped with a quad-core Intel® Xeon® CPU-E5–1630 v4 at 3.70 GHz and 16 GB of memory. The employed graphics processing unit was an NVIDIA Quadro M4000 with 8 GB of RAM and 1664 CUDA cores where Keras 2.0 was implemented with a Tensorflow backend.
2.D. Contour post-processing and optimization
Coarse output maps from the DL network containing holes and spurious predictions from neural networks are common.37 Thus, contour post-processing was performed on the 3D U-Net output using a fully connected CRF38 that imposes regularization constraints through minimizing an energy function.39 A 3D-CRF model was developed based on an initial 2D implementation39 and optimized to refine segmentations by smoothing, filling holes, and removing false positives, such as small remote regions.
3D-CRF was implemented on a GPU for improved computation and inference time. Inference here is with regard to the number of iterations applied to minimize the Kullback-Leibler (KL) divergence.39 Both bilateral40 (i.e., appearance kernel) and Gaussian39 (i.e., smoothness kernel) pairwise energies were used to account for the grayscale intensity similarity as well as the spatial proximity of pixels. The applied kernel involves the sum of a smoothness and appearance kernel which are shown in the following equations39:
where P and Q represent intensity and position vectors at pixel (α, β). The smoothness kernel works to remove small remote regions41 and is controlled by scaling factor θx. Parameter θy is an additional scaling factor in the appearance kernel, which controls the degree of similarity in predicted pixels. The appearance and smoothness kernel are equally weighted with weights w1 = w2 = 1.
CRF hyperparameters θx and θy were optimized automatically42 for each substructure by stepping through different parameter values and then comparing the prediction result back to ground truth through DSC. Based on the range of utilized values found in the current literature,39,43,44 full integer values (1–80) were stepped through for θx and in steps of 0.05 from 0–1 for θy, yielding 100 individual tests. Ten inference steps per substructure prediction were used.39,42 Finally, predictions both with and without CRF post-processing were assessed by comparing agreement with ground truth and calculating the Hausdorff distance (HD, maximum nearest neighbor Euclidean distance45).
Network testing on the hold-out dataset was conducted using the remaining 11 test patient CTs containing the heart and thorax from seven unique patients. Binary mask segmentations were converted to contours in Digital Imaging and Communications in Medicine (DICOM) format and imported into MIM.
2.E. Evaluations and statistical assessment
Quantitative evaluations between DL and ground truth segmentations were performed via DSC,46 mean distance to agreement (MDA, average of the shortest distance between all voxels of the predicted and ground truth segmentations47), and centroid displacement in three cardinal axes. DL segmentations were also compared to our previously published multi-atlas (MA) results, which implemented simultaneous truth and performance level estimation (STAPLE) with ten atlas matches13 using a shared cohort of 11 test subjects. Lastly, qualitative consensus scoring of DL segmentations was conducted to evaluate clinical utility. Before the qualitative grading was performed, three physicians (two radiation oncologists and a radiologist with a cardiac subspecialty) reviewed DL segmentations from a patient who was excluded from the grading. The physicians were instructed on the image grading system and a grading consensus scale was established for each substructure and then applied for five unique patients. Qualitative consensus scoring was completed on five of the test subjects as evaluated in our previously published atlas study.13 Scoring was completed using a 5-point scale13,48 as follows: (a) not clinically acceptable, (b) clinically acceptable with major changes, (c) clinically acceptable with moderate changes, (d) clinically acceptable with minor changes, (e) clinically acceptable. Contours were converted to a 0.25 mm high resolution display for final evaluation in MIM.
For volume size similarity assessment, two-tailed Wilcoxon signed-ranks tests were performed between ground truth and auto-segmented DL segmentations, with P < 0.05 considered significantly different. Statistical assessments using two-tailed Wilcoxon signed-ranks tests were also used to compare DL segmentations to our previous MA method via DSC, MDA, and qualitative consensus scores.
3. RESULTS
3.A. Segmentation and post-processing time
The initial manual ground truth delineations of the 12 cardiac substructures required ~3 h per patient. The DL network stabilized in ~19.4 h after training the network for 200 epochs, including ~2 h after implementing data augmentation. Figure 2 shows the results for the training and validation datasets over the 200 epochs. The final training and validation DSC values were 83.1% and 81.5%, respectively (difference <2%), which represents an average over all 12 substructures.
Fig. 2.
3D U-Net training and validation results over 200 epochs. Values for mean Dice similarity coefficient (DSC) represent an average over all 12 substructures. [Color figure can be viewed at wileyonlinelibrary.com]
Augmentation led to an overall DSC increase of 5.0 ± 7.9% across all substructures, with greatest improvements in the coronary arteries (LMCA = 18.6 ± 15.5%, RCA = 8.7 ± 9.1%, LADA = 7.8 ± 7.1%). Substructure contour generation (12 substructures) for a new patient using a single non-contrast CT-SIM dataset input took 5.0 ± 0.6 s. CRF post-processing time from a single test patient using ten inference steps for 12 substructures was 9.3 ± 0.3 s, for a total DL generation time of 14.3 s (range: 13.5–15.6 s).
3.B. CRF post-processing
CRF hyperparameter optimization revealed that differing values of (θx,θy) provided maximal DSC when three different sets of optimized parameters were employed for (a) coronary arteries and PV (2.0, 0.40), (b) superior/inferior venae cavae (2.0, 0.50), and (c) chambers and great vessels (8.0, 0.55). CRF applications lead to an average improvement in DSC, MDA, and HD over all substructures of 1.2 ± 2.5%, 0.11 ± 0.31 mm, and 5.58 ± 14.25 mm, respectively. The LMCA had the greatest improvement in DSC (6.2 ± 6.6%, range: 1–22%) after CRF application, whereas the RV and RA saw the least improvement (0.3 ± 0.2%, range: 0.0–1.0%). The LV showed the greatest improvement in MDA (0.3 ± 0.5 mm, range: 0.0–1.2 mm) and HD (34.4 ± 23.0 mm, range: 0.1–64.7 mm) after 3D-CRF application. Lastly, after applying CRF, the mean improvement in MDA ranged from 0.04 to 0.21 mm over the 12 substructures.
3.C. Geometric performance of segmentation
DL segmentation results are presented in Table I.
Table I.
Improvement in automatic segmentation performance in Dice similarity coefficient (DSC) after augmentation and in Hausdorff distance (HD) after implementation of conditional random fields (CRF) post-processing.
| Segmentation Improvement |
||||
|---|---|---|---|---|
| Substructures | Augmentation DSC | CRF HD (mm) | Final DSC | Final MDA (mm) |
| Chambers (LA, LV, RA, RV) | 0.03 ± 0.03 | 10.23 ± 19.34 | 0.88 ± 0.03 | 1.53 ± 0.26 |
| Great Vessels (SVC, PA, AA) | 0.03 ± 0.05 | 3.59 ± 10.79 | 0.85 ± 0.03 | 1.24 ± 0.31 |
| Inferior Vena CavaSS | 0.00 ± 0.05 | 6.61 ± 15.23 | 0.78 ± 0.04 | 1.45 ± 0.45 |
| Pulmonary Veins | 0.05 ± 0.04 | 2.82 ± 4.41 | 0.77 ± 0.04 | 1.04 ± 0.21 |
| Coronary Arteries | ||||
| Left Anterior Descending Artery | 0.08 ± 0.07 | 0.16 ± 0.27 | 0.53 ± 0.08 | 1.90 ± 0.90 |
| Right Coronary Artery | 0.09 ± 0.09 | 5.01 ± 15.86 | 0.50 ± 0.09 | 1.97 ± 0.46 |
| Left Main Coronary Artery | 0.19 ± 0.16 | 0.65 ± 0.96 | 0.50 ± 0.18 | 1.27 ± 0.68 |
The table also shows the final agreement to ground truth via DSC and mean distance to agreement (MDA). Additional abbreviations defined in the text.
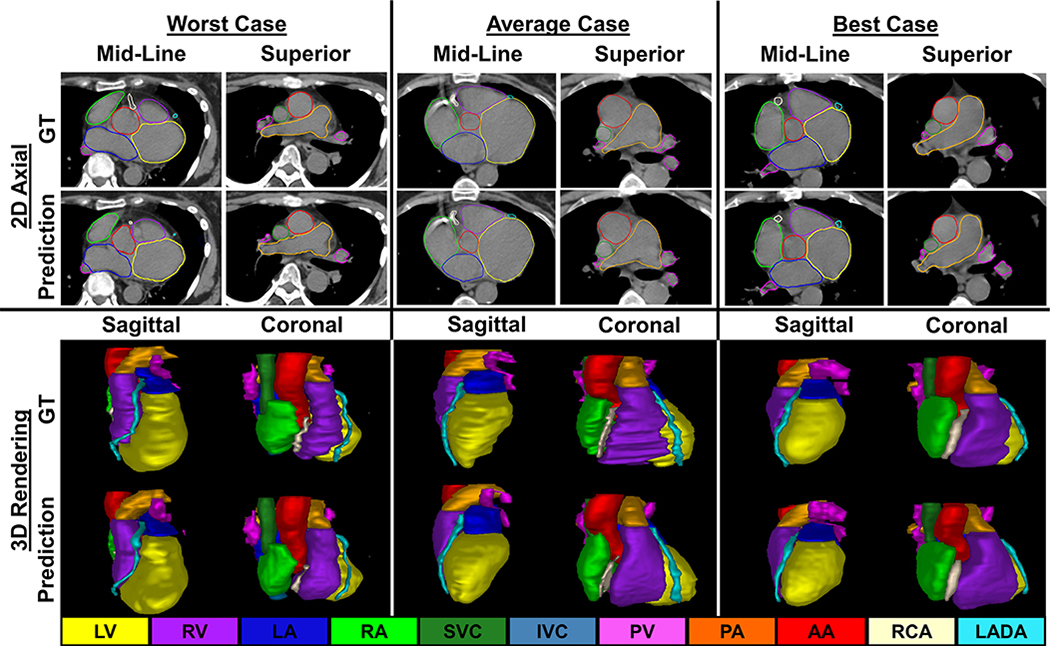
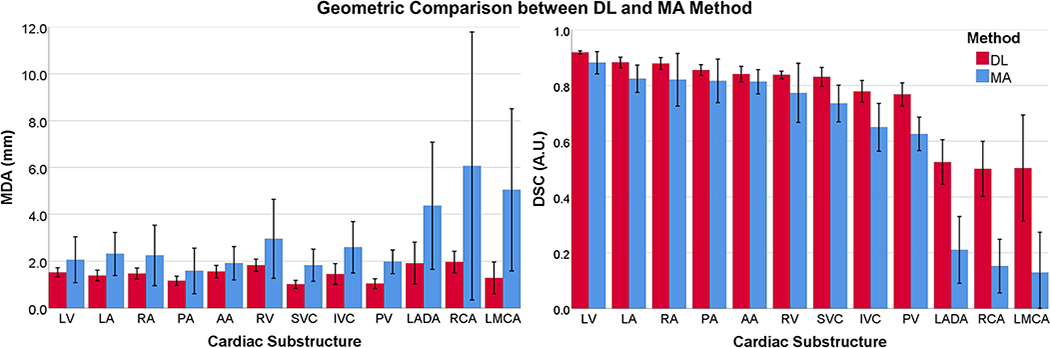
Figure 3 presents comparisons between ground truth and DL segmentations across substructures (LMCA not shown). The best case patient (Fig. 3, right) had chamber DSCs > 0.90 and MDAs < 2 mm for all substructures with favorable results for the RCA (DSC = 0.72, MDA = 1.67 mm). MDA (Fig. 5, left) across all 12 substructures was <2.0 mm (MDA = 1.46 ± 0.50 mm).
Fig. 3.
Comparisons between contours generated via deep learning and ground truth (GT) in 2D axial slices (top) and 3D renderings (bottom) for the worst (left), average (center), and best (right) cases. [Color figure can be viewed at wileyonlinelibrary.com]
Fig. 5.
Agreement between manually drawn ground truth and auto-segmentation methods (Blue: Previous multi-atlas method (MA), Red: Novel DL method) over 11 test cases. Left: Mean MDA, Right: Mean DSC. DSC, Dice similarity coefficient; MDA, mean distance to agreement. [Color figure can be viewed at wileyonlinelibrary.com]
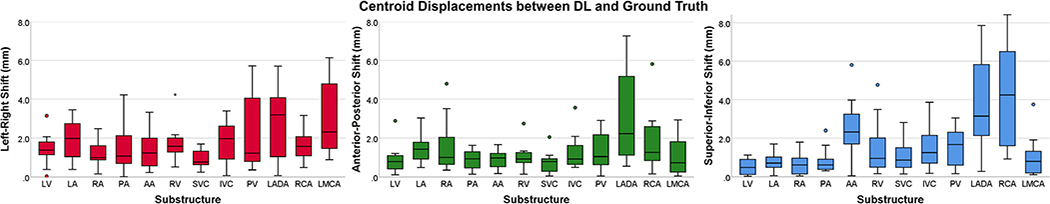
Wilcoxon signed-ranks tests revealed no significant differences in cardiac substructure volumes between DL and ground truth (P > 0.05). Figure 4 summarizes the centroid shifts in all cardinal axes. On average, the smallest displacements (<2 mm) occurred in the anterior-posterior direction for 11 substructures. The largest displacements occurred in the superior-inferior direction.
Fig. 4.
Substructure centroid displacements in the left-right (left), anterior-posterior (center), and superior–inferior (right) directions. Legend: interquartile range = box, median = line, minimum and maximum = whiskers, circles and stars = 1.5 and 3 times the interquartile range, respectively. [Color figure can be viewed at wileyonlinelibrary.com]
3.D. DL vs. MA segmentation
Figure 5 summarizes MDA and DSC results over 11 test cases and compares DL with our previously developed MA method for the same cohort.13 MDA and DSC for all cardiac substructures improved with DL.
Specifically, DSC agreement to ground truth increased 3–7% for chambers, 9–11% for the superior/inferior venae cavae and PV, and reached 23–35% for the coronary arteries. On average, MDA improved by ~1.4 mm with DL, with greatest agreement in the SVC (MDA = 0.99 ± 0.15) and worst agreement in the RCA (MDA = 1.97 ± 0.46). For four test CTs, our DL method yielded LMCA contours, whereas our previous atlas-based model failed to produce any segmentation. Overall, DL provided a significant improvement (P < 0.05) over the previous MA method for every substructure in terms of MDA and DSC.
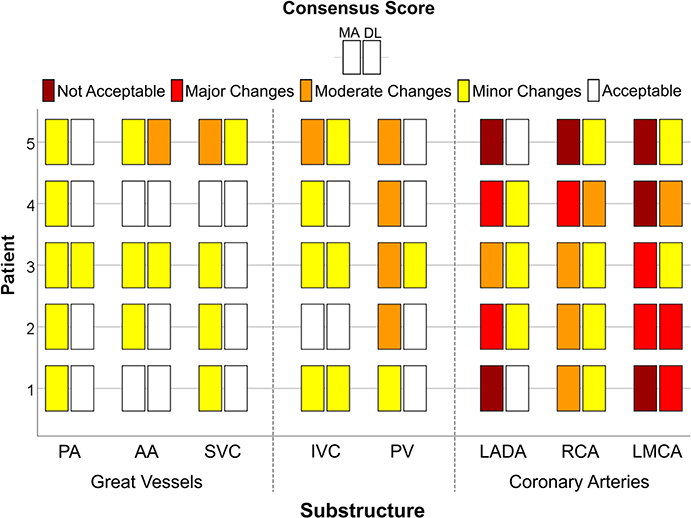
3.E. Qualitative analysis
Physician consensus scores are summarized in Fig. 6. All patients had clinically acceptable contours (score of 5) for the LV, RA, and RV (results not shown), while the SVC, PA, and PV had clinically acceptable contours for 4/5 patients with DL. For the cardiac chambers, 6/20 comparisons between DL and MA methods were equivalent, while all others improved by at least one grade with DL. The LMCA and RCA had the lowest average scores of 3.0 ± 1.0 and 3.8 ± 0.4, respectively, with all other substructures scoring an average of ≥4.4. DL provided significant improvements (P < 0.05) over the MA method for the LADA, RCA, PV, PA, SVC, LA, RA, and RV. Improvements in 44/60 (5 patients, 12 substructures) qualitative scores were observed with DL. For only one instance, DL scored worse than MA (AA: grade 4 to 3). For two LADA segmentations, MA yielded a grade of 1 (clinically unusable) and improved to a 5 (clinically acceptable) with DL (Fig. 6, right).
Fig. 6.
Qualitative consensus scoring (not clinically acceptable, clinically acceptable with major, moderate, and minor changes, and clinically acceptable) of five patients for the multi-atlas (MA) and deep learning (DL) auto-segmentations (chambers not shown). For each substructure, the MA and DL methods are shown on the left and right, respectively. [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
This work presented a novel DL pipeline to segment sensitive cardiac substructures using a 3D U-Net with the principal goal of applying to non-contrast CT-SIM for RT planning. Data augmentation and CRF post-processing improved DL contour agreement with ground truth. Overall, our method provided accurate segmentations of the chambers, great vessels, and PVs, and led to promising results in coronary artery segmentation on non-contrast CT-SIM datasets.
While cardiac substructure segmentation has been explored previously, to our knowledge, none have included paired MR/CT multi-channel data inputs to yield robust segmentations on non-contrast CT inputs. Several atlas segmentation methods have been recently published13–15 and report cardiac chamber DSCs > 0.75. However, these methods have had limited success segmenting coronary arteries as atlas methods rely on image registration quality and are unable to consider large amounts of patient data due to computational demands.49 Our work parallels recent applications of DNNs where CT coronary angiography (CTCA) scans specifically optimized for cardiac imaging were utilized. Here, DSC in the RA (87.8%)20 and PA (85.1%)21 were within 1% of our DL method, while we were within 5% of their chamber segmentation results. Our work adds to the current literature by including additional substructures and allowing for predictions to be made on non-contrast CT-SIM scans.
Data augmentation improved DL segmentation accuracy by ~5% across all substructures. Although no comparison values exist in the literature for cardiac substructure segmentation, this value is consistent with studies performed on liver lesion segmentation.50,51 One extreme outlier (>3 times the interquartile range) observed for the RV in the left-right axis occurred for the worst-case patient (Fig. 4, left), where the heart was rotated clockwise and shifted posteriorly/left. While this patient’s anatomy was an anomaly, this result may be addressed in the future by further augmenting the data (i.e., rotation >30°). Furthermore, both the LADA and RCA had larger centroid shifts in the superior–inferior plane (Fig. 4, right). This can be further visualized in Fig. 3 (left), where the inferior extent of the LADA and RCA were underrepresented with some narrowing of these substructures observed in the midline axial slice. To address this, recent atlas-based methods have standardized the size of the LADA to 4 mm throughout its entire length.17 Nevertheless, our DL pipeline performed well for coronary artery contours on non-contrast CTs (DSC ~ 0.50, MDA < 2.0 mm), particularly as compared to recent atlas results where coronary artery (LADA, RCA, and LMCA) DSCs ranged from 0.09 to 0.2713–15 and had MDAs > 4 mm.14 Coronary artery segmentations may be improved through the use of high resolution (0.78 × 0.78 × 1.6 mm3) CTCA21 that use contrast and yield DSCs ~ 60%.52 Additionally, implementing a Dice loss function weighted on the inverse of the class size may improve the results for smaller substructures such as the coronary arteries. Originally proposed by Crum et al.,53 the generalized Dice loss (GDL) function has been shown to improve hyperparameter robustness for unbalanced tasks (i.e., when each class is not represented equally in the dataset), and improve overall segmentation accuracy for small structures.54
While rare cases involved the removal of spurious remote predictions that resided within the ground truth delineation, 3D-CRF led to an overall improvement in segmentation agreement. The coronary arteries experienced the greatest improvement from CRF post-processing, with the LCMA improving ~6% in DSC. Additionally, there were improvements in MDA up to 1.21 mm and 1.96 mm for the LV and LA, respectively. Aside from removing spurious outlying points, CRFs also improved the smoothed appearance of the segmentations as needed for clinical application.55 CRF tuning required different parameters for cardiac substructures based on size and shape, much like the work completed by Rajchl et al.56 The improvement in segmentation agreement observed, along with the use of a 3D-CRF to remove spurious isolated regions, parallels other emerging uses of 3D-CRF post-processing in medical imaging.57,58 Although this study implemented CRFs as a post-processing step, some current studies have integrated CRFs into the utilized neural network and have seen improved segmentation performance42,43,59 and can be explored in future work for possible coronary artery segmentation improvement.
The overall time to generate DL segmentations on preprocessed CT-SIM data was rapid: 14 s for all 12 substructures. This value can be compared to Mortazi et al. who segmented seven cardiac substructures in ~50 s on high resolution CTCA and 17 s on MRI.21 Moreover, our previous MA method required ~10 min to generate substructure contours per patient without post-processing.13
Although the in-plane resolution was 0.7 × 0.7 mm2, our study may have been limited by the 8 mm slice thickness of the MRI. Despite our data augmentation techniques, increasing the training sample size may further improve segmentation results. However, similar training and testing cohort sizes with augmentation have been used previously.60 While paired cardiac MRI/CT data are commonly limited for cancer patients, the training cohort may be expanded in the future by applying our DL model to generate additional ground truth segmentations. Data quantity may also be increased through utilizing unlabeled images for unsupervised learning via generative models such as a cycle61 or a stacked62 GAN, which implement multiple GANs for data synthesis. Recently, Zhang et al.22 proposed a novel cardiac chamber segmentation method using a GAN integrating cycle and shape consistency. They obtained DSCs comparable to atlas segmentations (DSC ~ 0.75) on CT and MRI by using ~14% real data and augmenting their dataset by incorporating synthetic MRI and CT data into training. Our model may be enhanced similarly by incorporating synthetic images in the network training, while also providing additional substructures, such as pulmonary veins and coronary arteries. Nevertheless, even with the current limited training dataset, our results outperform other currently available approaches. As shown in Fig. 2, training and validation results increased to a point of stability with a difference of <2% after convergence. Moreover, to further limit potential overfitting in this more limited cohort, data augmentation (i.e., flipping, scaling, rotating, and translating) and model regularization (dropout = 0.3) were implemented. Even though the presented dataset was limited in size, overfitting was not an issue in this work.
As both the CT and MR images were acquired in breath hold conditions, respiratory motion is assumed to be negligible during this study. However, due to extended scan times, one limitation of this study is that numerous cardiac cycles are captured during imaging. Thus, the substructures are represented by their average intensity over the course of the scan and cardiac motion is not taken into consideration. Currently, cardiac motion is not managed clinically due to limitations in available treatment technologies. Nevertheless, the magnitude of cardiac motion is on the order of 3–8 mm63 suggesting internal motion may be incorporated into future margin design as has been previously proposed.64,65
As MR-guided RT and MR-only planning become more prevalent, future work will include training an MR-only model. It has been recently recommended that the LADA be included as an avoidance structure in RTP;12 thus, a natural clinical endpoint of this work includes dosimetric analysis and implementing cardiac avoidance strategies via accurate and efficient cardiac substructure segmentation made possible by DL.
5. CONCLUSIONS
These promising results suggest that our novel DL application offers major efficiency and accuracy gains for cardiac substructure segmentation over previously published MA results, using only non-contrast CT inputs. Future work involves further refinement of coronary artery segmentation using conditional random fields as a recurrent neural network and through expanding the patient cohort. Coupled with robust margin design, improved cardiac sparing in treatment planning can be realized.
ACKNOWLEDGMENTS
The authors thank the cardiac and radiation oncology teams at the University of Michigan for their consultation regarding the cardiac imaging protocol, including Dr. Lori Pierce, Dr. Venkatesh Murthy, and Robin Marsh. We also thank Lonni Schultz PhD for assistance in statistical design and analysis.
Footnotes
CONFLICT OF INTEREST
Data acquisition costs were supported by the Breast Cancer Research Foundation. Research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health under award Number R01 CA204189–01A1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additionally, this work was partially supported by US National Science Foundation (NSF) grant CNS-1637312. Dr. Glide-Hurst reports research agreements with Modus Medical, ViewRay Inc., and Philips Healthcare unrelated to the current work.
Contributor Information
Eric D. Morris, Department of Radiation Oncology, Henry Ford Cancer Institute, Detroit, MI, USA Department of Radiation Oncology, Wayne State University School of Medicine, Detroit, MI, USA.
Ahmed I. Ghanem, Department of Radiation Oncology, Henry Ford Cancer Institute, Detroit, MI, USA Department of Clinical Oncology, Alexandria University, Alexandria, Egypt.
Ming Dong, Department of Computer Science, Wayne State University, Detroit, MI, USA.
Milan V. Pantelic, Department of Radiology, Henry Ford Cancer Institute, Detroit, MI, USA
Eleanor M. Walker, Department of Radiation Oncology, Henry Ford Cancer Institute, Detroit, MI, USA
Carri K. Glide-Hurst, Department of Radiation Oncology, Henry Ford Cancer Institute, Detroit, MI, USA; Department of Radiation Oncology, Wayne State University School of Medicine, Detroit, MI, USA.
REFERENCES
- 1.Ng AK. Review of the cardiac long-term effects of therapy for Hodgkin lymphoma. Br J Haematol. 2011;154:23–31. [DOI] [PubMed] [Google Scholar]
- 2.Beukema JC, van Luijk P, Widder J, et al. Is cardiac toxicity a relevant issue in the radiation treatment of esophageal cancer? Radiother Oncol. 2015;114:85–90. [DOI] [PubMed] [Google Scholar]
- 3.Hardy D, Liu C-C, Cormier J, et al. Cardiac toxicity in association with chemotherapy and radiation therapy in a large cohort of older patients with non-small-cell lung cancer. Ann Oncol. 2010;21:1825–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darby SC, Ewertz M, McGale P, et al. Risk of ischemic heart disease in women after radiotherapy for breast cancer. N Engl J Med. 2013;368:987–998. [DOI] [PubMed] [Google Scholar]
- 5.Bradley JD, Paulus R, Komaki R, et al. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage iiia or iiib non-small-cell lung cancer (rtog 0617): a randomised, two-by-two factorial phase 3 study. Lancet Oncol. 2015;16:187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Movsas B, Hu C, Sloan J, et al. Quality of life analysis of a radiation dose-escalation study of patients with non-small-cell lung cancer: a secondary analysis of the radiation therapy oncology group 0617 randomized clinical trial. JAMA Oncol. 2016;2:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gagliardi G, Constine LS, Moiseenko V, et al. Radiation dose-volume effects in the heart. Int J Radiat Oncol Biol Phys. 2010;76:S77–S85. [DOI] [PubMed] [Google Scholar]
- 8.van den Bogaard V, Ta B, van der Schaaf A, et al. Validation and modification of a prediction model for acute cardiac events in patients with breast cancer treated with radiotherapy based on three-dimensional dose distributions to cardiac substructures. J Clin Oncol. 2017;35:1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vivekanandan S, Landau D, Counsell N, et al. The impact of cardiac radiation dosimetry on survival after radiation therapy for non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2017;99:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yusuf SW, Sami S, Daher IN. Radiation-induced heart disease: a clinical update. Cardiol Res Pract. 2011;2011:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieder C, Schill S, Kneschaurek P, et al. Influence of different treatment techniques on radiation dose to the lad coronary artery. Radiat Oncol. 2007;2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel S, Mahmood S, Nguyen T, et al. Comparing whole heart versus coronary artery dosimetry in predicting the risk of cardiac toxicity following breast radiation therapy. Int J Radiat Oncol Biol Phys. 2018;102:S46. [Google Scholar]
- 13.Morris ED, Ghanem AI, Pantelic MV, et al. Cardiac substructure segmentation and dosimetry using a novel hybrid magnetic resonance and computed tomography cardiac atlas. Int J Radiat Oncol Biol Phys. 2019;103:985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou R, Liao Z, Pan T, et al. Cardiac atlas development and validation for automatic segmentation of cardiac substructures. Radiother Oncol. 2017;122:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaderka R, Gillespie EF, Mundt RC, et al. Geometric and dosimetric evaluation of atlas based auto-segmentation of cardiac structures in breast cancer patients. Radiother Oncol. 2018;131:215–220. [DOI] [PubMed] [Google Scholar]
- 16.Zhuang X, Shen J. Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Med Image Anal. 2016;31:77–87. [DOI] [PubMed] [Google Scholar]
- 17.van den Bogaard VA, van Dijk LV, Vliegenthart R, et al. Development and evaluation of an auto-segmentation tool for the left anterior descending coronary artery of breast cancer patients based on anatomical landmarks. Radiother Oncol. 2019;136:15–20. [DOI] [PubMed] [Google Scholar]
- 18.Ronneberger O, Fischer P, Brox T.U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention Springer. 2015. pp. 234–241. [Google Scholar]
- 19.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 20.Payer C, Štern D, Bischof H, et al. Multi-label whole heart segmentation using CNNs and anatomical label configurations In: Pop M, Sermesant M, Jodoin P-M, Lalande A, Zhuang X, Yang G, Young A, Bernard O, eds. Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges. Cham. Springer International Publishing; 2018:190–198. [Google Scholar]
- 21.Mortazi A, Burt J, Bagci U. Multi-planar deep segmentation networks for cardiac substructures from MRI and CT In: Pop M, Sermesant M, Jodoin P-M, Lalande A, Zhuang X, Yang G, Young A, Bernard O, eds. Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges Cham: Springer International Publishing; 2018:199–206. [Google Scholar]
- 22.Zhang Z, Yang L, Zheng Y. Translating and segmenting multimodal medical volumes with cycle- and shape-consistency generative adversarial network. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2018. pp. 9242–9251 [Google Scholar]
- 23.Haq R, Hotca A, Apte A, et al. Cardio-pulmonary substructure segmentation of ct images using convolutional neural networks In: Nguyen D, Xing L, Jiang S, eds. Artificial intelligence in radiation therapy. Cham. Springer International Publishing; 2019:162–169. [Google Scholar]
- 24.Kamnitsas K, Ledig C, Newcombe VF, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- 25.Pluim JP, Maintz JA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging. 2003;22:986–1004. [DOI] [PubMed] [Google Scholar]
- 26.Feng M, Moran JM, Koelling T, et al. Development and validation of a heart atlas to study cardiac exposure to radiation following treatment for breast cancer. Int J Radiat Oncol Biol Phys. 2011;79:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baughman DR, Liu YA. Neural networks in bioprocessing and chemical engineering. San Diego, CA: Academic press; 2014. [Google Scholar]
- 28.Isensee F, Kickingereder P, Wick W, et al. Brain tumor segmentation and radiomics survival prediction: contribution to the brats 2017 challenge In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, eds. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham: Springer International Publishing; 2018:287–297. [Google Scholar]
- 29.Lee C-Y, Xie S, Gallagher P, et al. Deeply-supervised nets. Artificial intelligence and statistics. 2015;562–570. [Google Scholar]
- 30.Zeng G, Yang X, Li J, et al. 3D u-net with multi-level deep supervision: fully automatic segmentation of proximal femur in 3D MR images In: Wang Q, Shi Y, Suk H-I, Suzuki K, eds. Machine learning in medical imaging. Cham: Springer International Publishing; 2017:274–282. [Google Scholar]
- 31.Mao X, Shen C, Yang Y-B. Image restoration using very deep convolutional encoder-decoder networks with symmetric skip connections. Adv Neural Inf Process Syst. 2016;29:2802–2810. [Google Scholar]
- 32.Men K, Chen X, Zhang Y, et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol. 2017;7:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milletari F, Navab N, Ahmadi S-A.V-net: Fully convolutional neural networks for volumetric medical image segmentation. 3D Vision (3DV), 2016 Fourth International Conference on IEEE. 2016. pp. 565–571. [Google Scholar]
- 34.Nair AA, Tran TD, Reiter A, et al. A deep learning based alternative to beamforming ultrasound images. 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE. 2018. pp. 3359–3363. [Google Scholar]
- 35.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv. 2014. [Google Scholar]
- 36.Emami H, Dong M, Nejad-Davarani SP, et al. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med Phys. 2018;45:3627–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition 2015. pp. 3431–3440 [DOI] [PubMed] [Google Scholar]
- 38.Larsson M, Alvén J, Kahl F. Max-margin learning of deep structured models for semantic segmentation In: Sharma P, Bianchi FM, eds. Image Analysis. Cham. Springer International Publishing; 2017:28–40. [Google Scholar]
- 39.Krähenbühl P, Koltun V. Efficient inference in fully connected CRFs with gaussian edge potentials. Adv Neural Inf Process Syst. 2011;24:109–117. [Google Scholar]
- 40.Paris S, Durand F. A fast approximation of the bilateral filter using a signal processing approach In: Leonardis A, Bischof H, Pinz A, editors. Computer Vision – ECCV 2006. Berlin, Heidelberg: Springer, Berlin Heidelberg; 2006:568–580. [Google Scholar]
- 41.Shotton J, Winn J, Rother C, et al. Textonboost for image understanding: multi-class object recognition and segmentation by jointly modeling texture, layout, and context. Int J Comput Vision. 2009;81:2–23. [Google Scholar]
- 42.Zheng S, Jayasumana S, Romera-Paredes B, et al. Conditional random fields as recurrent neural networks. Proceedings of the IEEE international conference on computer vision 2015. pp. 1529–1537 [Google Scholar]
- 43.Zhao X, Wu Y, Song G, et al. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal. 2018;43:98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Nie H. A post-processing method based on fully connected CRFs for chronic wound images segmentation and identification. NUDT.2018. [Google Scholar]
- 45.Lustberg T, van Soest J, Gooding M, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126(2):312–317. [DOI] [PubMed] [Google Scholar]
- 46.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 47.Thomson D, Boylan C, Liptrot T, et al. Evaluation of an automatic segmentation algorithm for definition of head and neck organs at risk. Radiat Oncol. 2014;9:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumarasiri A, Siddiqui F, Liu C, et al. Deformable image registration based automatic ct-to-ct contour propagation for head and neck adaptive radiotherapy in the routine clinical setting. Med Phys. 2014;41:121712. [DOI] [PubMed] [Google Scholar]
- 49.Aljabar P, Gooding M. The cutting edge: delineating contours with deep learning. Mach Learn. 2001;2005:2013. [Google Scholar]
- 50.Ben-Cohen A, Klang E, Amitai MM, et al. Anatomical data augmentation for cnn based pixel-wise classification. Biomedical Imaging (ISBI 2018), 2018 IEEE 15th International Symposium on IEEE. 2018. pp. 1096–1099. [Google Scholar]
- 51.Frid-Adar M, Diamant I, Klang E, et al. Gan-based synthetic medical image augmentation for increased cnn performance in liver lesion classification. Neurocomputing. 2018;321:321–331. [Google Scholar]
- 52.Kjerland ∅. Segmentation of coronary arteries from ct-scans of the heart using deep learning: MS Thesis. NTNU. 2017. [Google Scholar]
- 53.Crum WR, Camara O, Hill DL. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans Med Imaging. 2006;25:1451–1461. [DOI] [PubMed] [Google Scholar]
- 54.Sudre CH, Li W, Vercauteren T, et al. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations Deep learning in medical image analysis and multimodal learning for clinical decision support In Proceedings of the MICCAI workshop on Deep Learning in Medical Image Analysis (DLMIA) Vol. 10553. Cham: Springer; 2017:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu Y, Mazur TR, Wu X, et al. A novel MRI segmentation method using CNN-based correction network for MRI-guided adaptive radiotherapy. Med Phys. 2018;45:5129–5137. [DOI] [PubMed] [Google Scholar]
- 56.Rajchl M, Lee MCH, Oktay O, et al. Deepcut: Object segmentation from bounding box annotations using convolutional neural networks. IEEE Trans Med Imaging. 2017;36:674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamnitsas K, Bai W, Ferrante E, et al. Ensembles of multiple models and architectures for robust brain tumour segmentation In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, eds. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham. Springer International Publishing; 2018:450–462. [Google Scholar]
- 58.Jiang H, Guo Y. Multi-class multimodal semantic segmentation with an improved 3D fully convolutional networks. Neurocomputing. 2019. 10.1016/j.neucom.2018.11.103 [DOI] [Google Scholar]
- 59.Liu Z, Li X, Luo P, et al. Semantic image segmentation via deep parsing network. Proceedings of the IEEE international conference on computer vision 2015. pp. 1377–1385 [Google Scholar]
- 60.Trullo R, Petitjean C, Ruan S, et al. Segmentation of organs at risk in thoracic ct images using a sharpmask architecture and conditional random fields. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) IEEE. 2017. pp. 1003–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu J-Y, Park T, Isola P, et al. Unpaired image-to-image translation using cycle-consistent adversarial networks. Proceedings of the IEEE international conference on computer vision 2017. pp. 2223–2232 [Google Scholar]
- 62.Huang X, Li Y, Poursaeed O, et al. Stacked generative adversarial networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017. pp. 5077–5086 [Google Scholar]
- 63.Tan W, Xu L, Wang X, et al. Estimation of the displacement of cardiac substructures and the motion of the coronary arteries using electrocardiographic gating. Onco Targets Ther. 2013;6:1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kataria T, Bisht SS, Gupta D, et al. Quantification of coronary artery motion and internal risk volume from ECG gated radiotherapy planning scans. Radiother Oncol. 2016;121:59–63. [DOI] [PubMed] [Google Scholar]
- 65.Li Q, Tong Y, Yin Y, et al. Definition of the margin of major coronary artery bifurcations during radiotherapy with electrocardiograph-gated 4d-ct. Physica Med. 2018;49:90–94. [DOI] [PubMed] [Google Scholar]