Abstract
The efficacy of probiotics as alternatives to antibiotics has been defined as one of the potential strategies to prevent Salmonella spp. infection in poultry. The purpose of this study was to isolate probiotic native Lactic acid bacteria (LAB) with high compatibility to intestinal tract and prevention of Salmonella typhimurium from broiler chicken feces. Thirty-seven samples of chicken feces were collected from seven broiler chicken farms in Northern Iran. The isolates identification was carried out with morphological and biochemical tests. Agar diffusion methods were used to evaluate the antimicrobial activities against Escherichia coli and S. typhimurium. The primary probiotic characteristics such as resistance to acid and bile and adhesion to Caco-2 cells were studied. Indeed, the ability of LAB isolates to inhibit adhesion of S. typhimurium to Caco-2 cells was evaluated by exclusion, competition and displacement assays. Among 42 isolates, S08, S01 and S06 isolates which showed appropriate probiotics characteristics were selected. Isolates S08 and S01 showed to be able to adhere strongly and also S06 was adhered moderately. In the exclusion assay, the isolates S08, S01 and S06 significantly hampered adhesion of S. typhimurium cell, in the competition assay, the isolates S08, S01 showed significant level of competition activity against S. typhimurium adherence to Caco-2 cells and isolate S08 showed the greatest displacement activity. The 16S rDNA sequence revealed that S08, S01, and S06 isolates were 99.00% similar to Lactobacillus salivarius, Lactobacillus johnsonii, and Pediococcus acidilactici, respectively. The result of this study suggested that LAB isolated from broiler chicken feces could be a remarkable reservoir for identification of probiotic to inhibit the pathogenic bacteria growth.
Key Words: Antimicrobial activity, Broiler chicken, Probiotic
Introduction
In intensive poultry production, there is no opportunity for newly hatched chickens to keep in touch with their mothers, which would lead to slow colonizing of microbial flora in their intestine tract. As a result, this condition makes them susceptible to infections of pathogenic microorganisms such as S. typhimurium, E. coli and Clostridium perfringens.1
Salmonellosis is one of the most widespread zoonosis all over the world and in charge of outbreaks of foodborne disease in humans through contamination and consumption of undercooked meats.2 It has been estimated that approximately 29.00% of chicken meats are contaminated with Salmonella spp. annually in Iran.3
Today the widespread of resistance to antibiotics among bacteria occurs due to increased consumption of antibiotics. Also, accumulation of antibiotic residues in animal carcasses and its transmission to consumers are major concerns influencing human health.4
Control of these foodborne enteric pathogens is a real challenge for the food industry and public health agency. Moreover, it is very difficult to protect the safety of food chains due to the resurgence of multidrug-resistant strains of foodborne pathogens. Therefore, the application of lactic acid bacteria as decontamination of foodborne pathogens might demonstrate antagonism towards pathogenic bacteria in meat preservation.5 The bio- compound from LAB could synergistically enhance the antimicrobial activity of eukaryotic peptides against Gram-negative bacteria and this would be another useful mechanism of action for LAB antimicrobial substances in the gastrointestinal tract.6
Indigenous LAB isolates were screened from broiler chicken feces grown in Golestan farms with a high potential poultry production in north of Iran. This is because of the appropriate climate, unique geophysical zones, biodiversity at microbial levels and pristine ecosystems that would be considered as a great area for isolating more varieties of indigenous bacteria with significant probiotic properties characteristic. These isolates might have more biocompatibility with a physicochemical chicken body and tolerate better acidic and bile salt conditions because of being originated from microflora of the gastrointestinal tract of Golestan chicken. Hence, indigenous LAB probiotic with the prior presence towards other organisms and increasing host resistance to Salmonella spp. infection by high competitive exclusion could be an effective intervention strategy for prevention of salmonellosis.7
It should be taken into consideration that there is no similar study in this area. Therefore, it is worthwhile to investigate more on native isolates of this region that would improve broiler chicken health by protecting them from intestinal pathogens and maintain the natural balance of intestinal microflora during antibiotic treatments.8,9
The present study was carried out to isolate, characterize and select the most suitable LAB isolated from poultry feces with inhibitory effects on the growth of E. coli and S. typhimurium for in vivo experiments as chicken probiotic adjuncts.
Materials and Methods
Isolation of LAB. To isolate LAB, Thirty-seven samples of Ross 308 broiler chicken feces (each sample was 1 g) were collected randomly in peptone water from seven farms with a capacity of rearing 20,000 birds fed with antibiotic-free diet from Golestan province (north of Iran) and then were transferred to the laboratory. To reduce the sampling error, the floor of each farm was divided into 20 areas and one sample was collected from each area. All samples from a farm were mixed and finally, five or six samples were collected from different parts of the mixed samples. The samples were homogenized and transferred to de Man, Rogosa and Sharpe broth (MRS; Merck, Darmstadt, Germany) and then incubated in anaerobic jars (Sharif Azmayeshgahi Co., Tehran, Iran) for 72 hr at 37.00 ˚C. After incubation, the isolates were spread on MRS agar plates. The plates were incubated at 37.00 ˚C for 48 hr in aerobic conditions. The isolates were characterized as LAB based on the Gram staining, catalase, oxidase, and hemo-lysis tests on blood agar. Only Gram- positive, catalase, oxidase, and hemolysis negative LAB was selected.10
Acid tolerance. The acid tolerance assay was tested according to Ehrmann et al. with modifications.11 The LAB isolates were grown overnight in MRS broth at 37.00 ˚C, adjusted to a final concentration of 7.00 to 8.00 log CFU mL-1, inoculated (1.00%, v/v) into sterile phosphate-buffered saline (PBS) (Sigma-Aldrich, USA) adjusted to pH 4.00 and 2.50 with 1.00 N HCl (Merck) (acidic condition) and PBS with normal pH 7.00 (control), and incubated anaerobically for 3 hr at 37.00 ˚C. After incubation, tenfold serial dilutions (up to 10−7) of each bacterial strain were prepared using PBS. Then 100 μL of 10−4 to 10−7 dilutions from each sample was spread plated on MRS agar and incubated anaerobically at 37.00 ˚C for 48 hr. After incubation, colonies on the plates were counted and enumerated as CFU mL-1. Tolerance to acidic condition was estimated by comparing viable cell counts after exposure to acidic and normal (control) conditions. The experiments were carried out in triplicate for each strain. The number of counts should be at least 106 CFU mL-1.
Bile tolerance. The bile tolerance assay was tested according to Jacobsen et al. with modifications.12 The LAB isolates were grown overnight in MRS broth, adjusted to a final concentration of 7.00 to 8.00 log CFU mL-1, then inoculated (1.00%, v/v) into 10 mL of fresh MRS broth with or without (control) 0.30% (w/v) oxgall (Fluka Biochemika, Buchs, Switzerland) and incubated anaerobically at 37.00 ˚C for 3 hr. Bacterial optical density (OD) was measured at 650 nm using a Multimodal micro-plate reader (BioTek Instruments, Winooski, Vermont, USA). The experiments were carried out in triplicate for each strain. Bile tolerance was estimated, by calculating the coefficient of inhibition (Cinh), according to the formula described by Gopal et al.: 13
where, ∆T8 – T0 represents the difference in absorbance at time zero (T0) and after 8 hr (T8). The Cinh of less than 0.40 was considered significant, for the isolates to be considered as a suitable probiotic candidate.
In Vitro assay trypsin and pepsin tolerances. These assays evaluated the tolerance of isolate to the simulated gastric and intestinal juices. The bacterial suspensions were prepared with sterile PBS containing 2.00 g L-1 of trypsin at pH 7.00. Pepsin tolerance was determined using bacterial suspension in sterile PBS containing 2.00 g L-1 of pepsin at pH 3.00. After 3 hr incubation at 30.00 ˚C, 0.10 mL of each bacterial suspension was spread on TSA for the viable count.14 The assays were carried out in triplicate. The number of counts should be at least 106 CFU mL-1.
Antibiotic susceptibility minimal inhibitory concentrations test (MIC). The sensitivity of the bacteria to antibiotics was determined using the disc diffusion method according to the recommendations of the National Committee for Clinical Laboratory Standards (NCCLS) and European Food Safety Authority (EFSA).15 The Lacto-bacillus bacterial suspension (1.50 × 108 CFU mL-1; McFarland standard 0.50), was cultured overnight diluted 1:100, then 200 µL was seeded on Mueller-Hinton (MH) agar plates (Merck). Antibiotic-impregnated discs (Tadbir Fan Azma Co., Tehran, Iran) were placed on seeded plates and the zone of growth inhibition was measured after 24 hr of incubation at 37.00 ˚C. Inhibition zone diameters were measured in mm. MIC values were determined using serial antibiotic dilution procedure in the MH broth.16
Antagonistic activity against pathogens. The antagonistic activities of the LAB isolates against E. coli and S. typhimurium were evaluated using the agar spot test described by Touré et al. with modifications.17 Anti-bacterial activity was examined by spotting 10.00 µL of an overnight culture of each LAB strain (final concentration of 7.00 log CFU mL-1) on an overlaid agar plate inoculated with the indicator bacterium. Overlaid agar plates were prepared by pouring a soft agar layer (0.70% m/v) inoculated with a liquid culture (2.90% v/v) of an exponentially growing indicator bacterium (final concentration of 7.00 log CFU mL-1) on top of the agar medium. Plates were investigated for inhibition zones after incubation at 37.00 ˚C overnight. Inhibition zones more than 20.00 mm, 10.00-20.00 mm and less than 10.00 mm were determined as strong, intermediate and low inhibition, respectively. The experiments were carried out in triplicate for each strain.
Characterization of antimicrobial substances. The selected probiotics were assayed for the production of antimicrobial substances such as bacteriocins, hydrogen peroxide and organic acids using the agar well diffusion technique described by Touré et al. with modifications.17 For bacteriocins assay, the supernatant (5.00 mL) was treated with 1.00 mg mL-1 pronase. For organic acids assay, the supernatant was adjusted to pH 7.00 using 1.00 N NaOH and for hydrogen peroxide assay, the supernatant (5.00 mL) was treated with 0.50 mg mL-1 catalase. Treated supernatant was placed into 7.00 mm diameter wells, the plates were seeded with 1.00% (v/v) overnight culture of each indicator strain (test pathogen) and incubated for 24 hr at 37.00 ˚C. The diameter of the inhibition zones was measured.
Caco-2 cell cultures and adherence assay. The Caco-2 cells were obtained from the Cellular Bank of the Pasteur Institute of Iran and Roswell Park Memorial Institute (RPMI) medium -1640 medium (Merck) supplemented with 10.00% (v/v) inactivated (56.00 ˚C for 36 min) fetal bovine serum (FBS; Merck), 2.00% of 200 mM (v/v) L-glutamine and 1.00% unnecessary amine acids (v/v). The 1.00% Strep (10.00 mg mL-1) antibiotic was added to prevent contamination and incubated with 5.00% CO2 at 37.00 ˚C. The culture was changed daily until Caco-2 monolayers were at 80.00-90.00% confluence. For adherence of LAB isolates to Caco-2, 1.00 mL isolates (108 CFU mL-1) and 1.00 mL of cell line culture medium were added to each well of the culture plate and incubated at 37.00 ˚C in 5.00% CO2 After 2 hr of incubation, monolayers were washed four times, fixed, Gram-stained and examined microscopically. An adherence index was determined from 20 random microscope fields of adhering LAB per 100 Caco-2 cells.18
Test of inhibition of S. typhimurium adhesion to Caco-2. The ability of lactobacilli to inhibit S. typhimurium adhesion to Caco-2 cells was assayed following methods reported by Candela et al. with some modifications.19 In brief, approximately 3.00 × 105 Caco-2 cells per well were seeded in a 12-well plate (Corning Inc., Corning, USA), the culture medium was changed daily and Caco-2 monolayers at 80.00-90.00% confluence were washed twice with PBS (pH 7.40) before experiments. In the exclusion assay, Caco-2 monolayer were inoculated with 300 µL of LAB isolates suspension (107 CFU per well) in RPMI-1640 medium and incubated for 1 hr at 37.00 ˚C in 5.00% CO2. The Caco-2 monolayers were then inoculated with 100 µL of S. typhimurium suspension (107 CFU per well) in RPMI-1640 medium, incubated for 1 hr at 37.00 ˚C in 5.00% CO2. In a competition assay, Caco-2 monolayer were inoculated with 400 µL of LAB isolates suspensions (107 CFU per well) and S. typhimurium suspension (107 CFU per well) in RPMI-1640 medium and incubated for 1 hr at 37.00 ˚C in 5.00% CO2. In the displacement assay, Caco-2 monolayer were inoculated with 300 µL of S. typhimurium in RPMI-1640 medium suspension (107 CFU per well) and incubated for 1 hr at 37.00 ˚C in 5.00% CO2. The Caco-2 monolayer were then inoculated with 100 µL of LAB isolates suspension (107 CFU per well) in RPMI-1640 medium and incubated for 1 hr at 37.00 ˚C in 5.00% CO2. Wells containing S. typhimurium alone served as control. In all experiments, non-adhering cells were removed by washing four times with sterile PBS and then treated with 0.50 mL of 0.50% (v/v) Triton X-100 (Amresco, Solon, USA) for 5 min in an ice-water bath. The cells were then serially diluted (101 - 105) and a 5-fold dilution plated onto TSA plates for counting S. typhimurium. The ability of lactobacilli to exclude, compete and displace S. typhimurium was determined by comparing adhesion of S. typhimurium in the presence of lactobacilli to that of S. typhimurium alone. The experiments were carried out in triplicate for each strain.
Assay for the activity of digestive enzymes. All the seven isolates selected as a result of the primary screening tests were checked for presence of enzyme (i.e., amylase, phosphatase, and protease) activities according to Kim et al. with minor modifications.20 For amylase activity, all of seven isolates were subcultured on modified MRS (including 0.25% starch) broth and spot-inoculated on to relevant medium that consisted of meat peptone (0.50%), yeast extract (0.70%), NaCl (0.20%), starch (2.00%), and agar (1.50%) and then incubated anaerobically for 48 hr at 37.00 ˚C. After incubation, the halo zone surrounding each colony was measured with a caliper. For detecting the clear zones of amylase activity, Lugolʼs solution was poured over the plates.
Phosphatase activity was measured using calcium phytate. The selected LAB were subcultured in the MRS broth that contained 0.25% calcium phytate (Merck) and then spot-inoculated onto relevant medium that consisted of glucose (1.50%), calcium phytate (0.50%), NH4NO3 (0.50%), KCl (0.05%), MgSO4・7H2O (0.05%), MnSO4・7H2O (0.02%), FeSO4・7H2O (0.001%), and agar (1.50; adjusted to pH 7.00) and then incubated anaerobically for 48 hr at 37.00 ˚C. After incubation, the halo zone surrounding each colony was measured with a caliper. For detection of protease activity, the isolates were cultured on MRS broth and after anaerobic incubation for 24 hr at 37.00 ˚C, 30.00 μL of culture supernatant was transferred onto a disc placed over a medium consisting of skim milk (1.00%) and agar (1.50%). Incubated anaerobically for 48 hr at 37.00 ˚C. After incubation, the halo zone surrounding each colony was measured with the caliper.
Molecular identification of LAB isolates. The DNA (deoxyribonucleic acid) of selected LAB was extracted using the lysozyme digestion according to Araújo et al. 21 The specific universal primer pairs of 27F (5'_AGAGTTTG ATCMTGGCTCAG_3') and 1492R (5'_TACGGYTACCTTGTTA CGACTT_3') were used for amplification of a 1500 bp fragment from 16S rDNA region (Metabion, Martinsried, Germany). The 25.00 μL PCR reaction mixture consisted of 150 ng template DNA, 2.50 μL of PCR 10x Buffer, 200 μM dNTP, 1.50 mM MgCl2, 10.00 pM of each primer, 1.00 U of Taq DNA Polymerase enzyme and distilled water. All PCR chemicals were obtained from Cinnagen (Tehran, Iran). The thermal cycling (FFG-05TUD; Techne Flexigene, Minneapolis, USA) included primary denaturation at 94.00 ˚C for 8 min, followed by 30 cycles of denaturation at 94.00 ˚C for 30 sec, annealing of primer at 58.00 ˚C for 30 min, extension at 72.00 ˚C for 30 sec, and final extension at 94.00 ˚C for 5 min. Then, the PCR products were electro-phoresed on 1.50% agarose gel and stained by Rima Sight DNA stain and were detected with UV light. The molecular weight marker (SinaClon, Tehran, Iran) was used as a molecular marker. After purification of the PCR product from the agarose gel, the samples were sent for sequencing. (Bioneer Inc., Daejeon, South Korea).
Statistical and bioinformatics analysis. The results were analyzed by GraphPad Prism (version 7.0; GraphPad Software Inc., San Diego, USA) for selected LAB. The statistical analyses were done at a 95.00% confidence level (p < 0.05), and results were reported as mean ± SD. The alignment of 16S rDNA gene sequences of LAB isolates was done by the BLASTn bioinformatics tool of the EZ TAXON database and a phylogenetic tree was drawn using MEGA software (version 4.0; Biodesign Institute, Tempe, USA).22
Results
Bacterial isolates. From the screening of 67 isolates from chicken feces samples, only 42 isolates were identified as Gram-positive, catalase, oxidase, and hemolysis negative.
Test for resistance to bile salts and acid. The results of the present study showed that seven selected isolates could tolerate bile salt with a concentration of 0.30% and acid conditions (pH 2.00, 4.00). However, the degrees of tolerance varied among the seven isolates, three isolates S08, S01 and S06 showed the highest growth and compatibility with acid with loss in cell viability of only 0.27 to 0.62 and bile salt conditions.
Test for trypsin and pepsin tolerance. All studied LAB isolates could survive after 3 hr of exposure to pepsin (3.00 g L-1) at pH 2.00 and trypsin (3.00 g L-1) at pH 7.00 that showing a model simulating of the composition of gastric juices. The viable LAB cell numbers were decreased by approximately 1.00 log CUF mL-1 for most isolates. S05, S06, S08 showed the lowest sensitivity to gastric juices.
Antibiotic susceptibility test. According to EFSA, the results of MIC values for antibiotic susceptibility of LAB isolates against seven tested antibiotics showed that all the seven LAB isolates exhibited MIC values the same or lower than the MIC breakpoint values recommended for LAB. In addition, since none of the strains were resistant to the tested antibiotics, no further studies on their antibiotic resistance were needed.
Antagonistic effects. The results of antagonistic effects of LAB bacteria isolates were subjected against the indicator microorganisms E. coli, S. typhimurium (Table 1). All seven isolates showed antagonistic effects against all indicator microorganisms tested, however, degrees of antagonism varied among them. The results revealed that all the isolated Lactobacillus strains, exhibited the average inhibition zones of more than 10.00 mm on the growth of test pathogens, however, the isolates S8, S1, and S06 were the most effective noticeable isolates in inhibiting the growth of the test pathogens (16.00-27.00 mm).
Table 1.
Antagonistic activity of LAB isolates against test pathogens (diameter of observed holes in mm). Values are expressed as of the independent experiments, each in triplicate
| Bacteria | S. typhimurium | E. coli |
| S01* | 26.71 ± 0.15 | 24.53 ± 0.85 |
| S04 | 18.05 ± 0.23 | 16.22 ± 0.29 |
| S05 | 8.40 ± 0.41 | 10.60 ± 0.60 |
| S06* | 20.41 ± 0.16 | 16.04 ± 0.55 |
| S07 | 10.74 ± 0.07 | 4.82 ± 0.60 |
| S08* | 44.22 ± 0.38 | 40.21 ± 0.81 |
| S12 | 12.20 ± 0.61 | 14.25 ± 1.34 |
* Strains that showed higher antagonistic activities than other strains against most of the tested pathogenic strains.
Characterization of inhibitory substance. The antimicrobial substances provided by the isolates were characterized by agar well diffusion assay against the test pathogens. The results showed that culture supernatants of all seven isolated treated with pronase (1.00 mg mL-1) did not have any inhibitory activities against the indicator strains. This indicated that the inhibitory effect of LAB isolates were not due to bacteriocins production. Culture supernatants treated with catalase also did not affect the inhibitory activities of the LAB isolates against the test pathogens. This showed that inhibition by the LAB isolates were not due to hydrogen peroxide production. Neutralized supernatant (pH 6.50) of all seven LAB isolates did not have any inhibitory activity effects. Therefore, it could be concluded that the inhibitory effects of the LAB isolates were due to their organic acid productions.
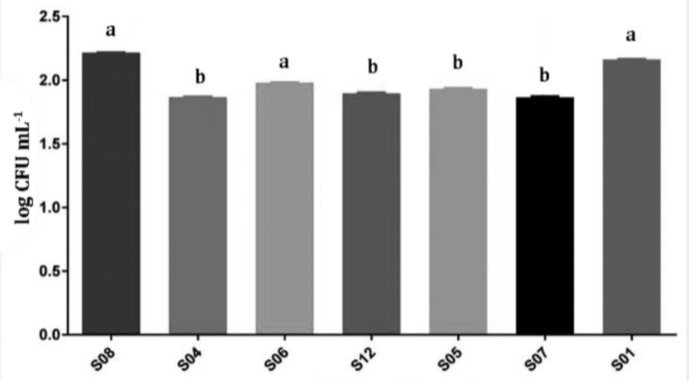
Attachment test. The ability of LAB attachment to Caco-2 cells was significantly different among strains which are shown in Figure 1. A moderate attachment to Caco-2 differentiated cells were observed in S04, S12, S05, S06, S05, S07 strains, among which the S06 strains showed higher attachment ability. Also, a high level of attachment ability was observed in strains S08 and S01, in which the S08 strain showed the highest attachment rate among all seven isolates (Fig. 2).
Fig. 1.
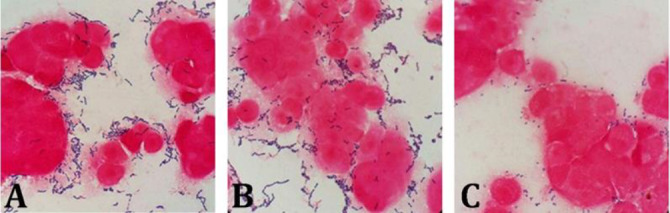
Attachment ability of lactic acid bacteria isolates to intestinal epithelial cells A: S01/ B: S08/ C: S06)
Fig. 2.
The adherence activity of lactic acid bacteria to intestinal epithelial cells
ab Different letters indicate significant differences (p < 0.05).
Test of competitive elimination of Salmonella attachment to intestinal epithelial cells. In the exclusion experiment, the seven selected LAB isolates showed different effects on S. typhimurium adhesion to Caco-2 cells. The isolates S08, S01, and S06 significantly hampered adhesion of S. typhimurium cell (p ≤ 0.05), (Table 2), as summarized in Table 2 and the other four isolates failed to display any significant differences in their capacity to exclude S. typhimurium from Caco-2 cells (p ˃ 0.05). In the competition experiment, LAB isolates and S. typhimurium were provided with an equal opportunity for adherence to Caco-2 cells. The isolates S08, S01 showed significant level of competition activity (p ≤ 0.05). The other isolates produced a non-significant level of competitive activity. With regard to the displacement of previously adhering S. typhimurium, the isolates S08 significantly displaced previously adhered S. typhimurium (p ≤ 0.05). The isolate S08 showed the greatest displacement activity. No significant difference was found for the other isolates.
Table 2.
Exclusion, Competition and displacement of adhering S. typhimurium from Caco-2 cells by the lactic acid bacteria isolates. Data are expressed as mean (×105 CFU per well) ± standard deviation of three independent experiments
|
Exclusion experiment
|
Competition experiment
|
Displacement experiment
|
||||
|---|---|---|---|---|---|---|
| Strains | S . typhimurium* |
S
.
typhimurium
adherence inhibited |
S . typhimurium* | S . typhimurium adherence inhibited | S . typhimurium* | S . typhimurium adherence inhibited |
| Control | - | 2.46 ± 0.18a | - | 2.73 ± 0.37a | - | 2.59 ± 0.42a |
| S01 | 1.38 ± 0.15b | 1.08 ± 0.12b | 2.07 ± 0.32b | 0.66 ± 0.06b | 2.12 ± 0.33a | 0.47 ± 0.07a |
| S04 | 2.19 ± 0.38a | 0.27 ± 0.07a | 2.59 ± 0.36a | 0.14 ± 0.02a | 2.23 ± 0.11a | 0.36 ± 0.01a |
| S05 | 1.98 ± 0.33a | 0.48 ± 0.10a | 2.53 ± 0.34a | 0.20 ± 0.02a | 2.46 ± 0.21a | 0.13 ± 0.01a |
| S06 | 1.73 ± 0.17b | 0.73 ± 0.12b | 2.38 ± 0.27a | 0.35 ± 0.03a | 2.38 ± 0.33a | 0.21 ± 0.02a |
| S07 | 2.07 ± 0.38a | 0.39 ± 0.09a | 2.49 ± 0.28a | 0.24 ± 0.04a | 2.32 ± 0.16a | 0.27 ± 0.01a |
| S08 | 1.02 ± 0.14b | 1.44 ± 0.16b | 1.76 ± 0.16b | 0.97 ± 0.08b | 2.01 ± 0.22b | 0.58 ± 0.06 b |
| S12 | 1.81 ± 0.24a | 0.65 ± 0.06a | 2.46 ± 0.39a | 0.27 ± 0.05a | 2.27 ± 0.19a | 0.32 ± 0.02a |
ab indicate significant difference compared to control at p < 0.05.
Evaluation of the ability to produce enzymes by LAB isolates. All the isolates were investigated for enzymatic activity. The size of the halo zones surrounding the colonies gave an approximate indication of extracellular enzyme activity. None of seven bacterial isolates produced α-amylase and protease enzymes, however, all of them showed phosphatase activity.
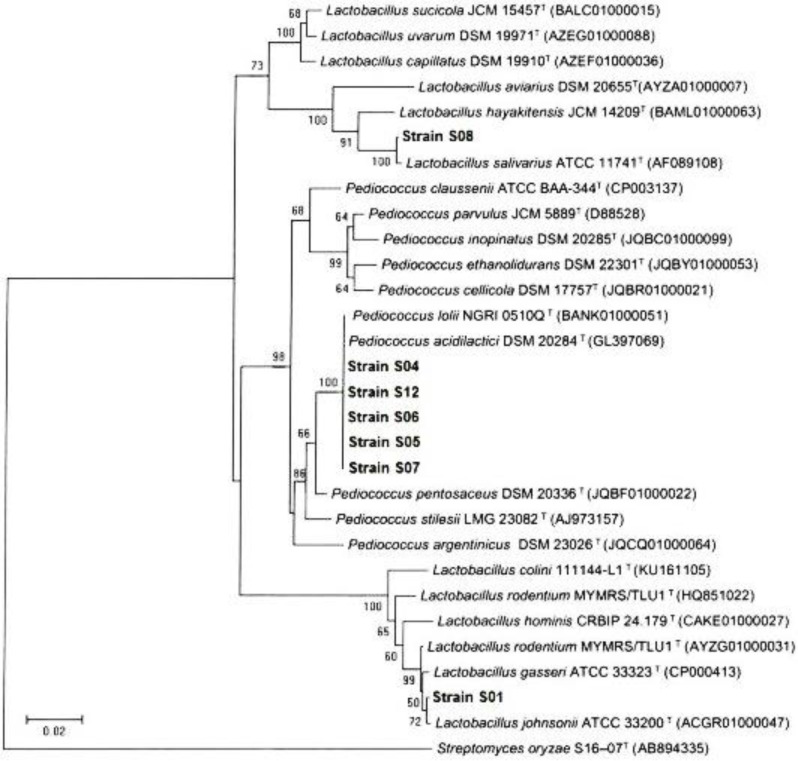
Identification of LAB. Genotypically, the selected strains of LAB with probiotic features were identified based on sequence analysis of 16S rDNA gene and showed that DNA fragments of amplification products were about 1500 bp. The analysis of 16S rDNA gene of isolates has been successfully sequenced, aligned and compared. The Streptomyces oryzae was used as an out-group. the results showed that the isolates S04, S12, S06, S05, and S07 were 99.00% similar to Pediococcus acidilactici strain DSM 20284, the S01 isolate was 99.00% similar to L. johnsonii strain ATCC33200 and the S08 isolate was 99.00% similar to L. salivarius strain ATCC11741 (Fig. 3).
Fig. 3.
The Phylogenetic tree of LAB bacteria based on the neighbor-joining method of 16S rRNA gene sequences
Discussion
Selecting the most beneficial organism in vitro as a cheap and fast assay is more efficient than in vivo. Although all the in situ conditions of the intestinal ecosystem cannot be provided in in vitro condition, it is still a powerful method for the rapid screening of high potential strains, allowing extensive study of a large number of isolates in order to discover the specific characteristics of probiotic strains.11,23
The potential probiotic LAB strains are expected to tolerate the stress conditions of digestive tract in order to improve the health status of hosts. The ability to tolerate acid, bile salts, pancreatic fluid and production of antimicrobial agents are considered as good indicators for the survival of strains in the gastrointestinal tract.
Shokryazdan et al. have suggested that culture medium with pH 3.00 and a level of 0.10-0.30% bile salt was considered as a standard medium for the evaluation of acid and bile tolerance in probiotic bacteria.24
Ehrmann et al. have reported that strains of L. reuteri, L. salivarius, and L. animalis were able to tolerate pH 3.00 for 4 hr, however, the degrees of tolerance varied among the strains.11 Earlier, Charteris et al. have pointed out in their review that most Lactobacillus spp. were able to tolerate pH 4.00 for 1 hr, however, the percentage of cell viability varied considerably among different strains.25 It is also apparent from the results of the current study that the highest viability of the three isolates S08; S01and S06 with the acidic and bile salt conditions might be as a result of strain-specific and natural habitat in proventriculus.
Antimicrobial activity against pathogens is another important potential feature considered in the selection of potential probiotic strains for maintaining a healthy microbial balance in the gastrointestinal tract. The antagonistic activity has mostly been attributed to the production of antimicrobial substances or metabolites such as organic acids, bacteriocins, bacteriocin-like components and hydrogen peroxide by the probiotic.5 In this study, although inhibition zones varied between seven isolates, however, all of them showed inhibitory activity against Escherichia coli and Salmonella spp. The most inhibitory activity was for S08, S01 and S06 isolates with an inhibition zone 16.00-27.00 mm which were found to be due to their organic acid production, not hydrogen peroxide or bacteriocins. Species- and strain-specific antimicrobial activities of lactobacilli have also been reported by Corsetti et al. which were consistent with the results of the present Study.26 This activity, together with the mechanism of competitive exclusion, in which pro-biotic strains competed with pathogens for nutrients and attachment sites, prevented the pathogens colonization in the intestine. It was indicated that not only antimicrobial metabolites were produced by LAB, but the presence of specific adhesion molecules and receptors could have significant factors in preventing the pathogen adhesion. Therefore, the effect of the in vitro inhibitory activity of probiotic on pathogen adhesion has to be confirmed in vivo.27 It was suggested that lactobacilli isolated from chicken feces has a strain-dependent ability to attach to the intestinal epithelial cells.28
Studies have shown that the stronger attachment of bacteria to the intestinal epithelial cells might have a higher ability to inhibit intestinal pathogenic bacteria such as S. typhimurium and E. coli, therefore, this had a protective role in improving the chicken gastro-intestinal tract health. In this study, LAB isolates had attachment ability to Caco-2 cells from 1.80 to 2.20 log CFU per well. It has also been determined that the highest attachment was for to S08 and S01 isolates with the high ability in the competitive elimination of Salmonella spp. attachment to intestinal cells. Lim et al. have shown that in vitro condition, attachment to Caco-2 cell is significantly dependent on the strain (strain-specific).29
This investigation exhibited a significant difference between two methods of exclusion and competition in preventing Salmonella spp. attachment to Caco-2 cells in the presence of S01 and S08 strains.
Furthermore, the experiment showed that Lactobacillus with high antimicrobial secretion and potential attachment has a great ability to prevent the attachment of Salmonella spp. to Caco-2.20 It can be concluded that the ability of LAB in controlling pathogenic bacteria is highly specific and related to the type of probiotic strains and pathogenic bacteria, as well as the time of bacterial communication with intestinal epithelial cells. It is supposed that the presence of receptors is not only important factor in attachment of bacteria to the cells but also the secretion of antimicrobial produced by the strains can play an important role in this regard.30,31
All the isolates were investigated for enzymatic activity. None of seven bacterial isolates produced α-amylase and protease enzymes, however, all of them displayed phosphatase activity. These results were no in agreement with the findings of Kim et al. that found LAB isolated from the broiler digestive tract had amylase and phytase activities.20 Phosphatase enzyme provides an interesting alternative to reduce the anti-nutritional effects of phytic acid and could prove to be a safe route for improving the digestion of phosphate in the chicken diet.32
In the present study, three strains of native LAB (Lactobacillus salivarius, Lactobacillus johnsonii and Pediococcus acidilactici) with probiotic properties were isolated from chicken feces in Golestan province of Iran. These isolates due to bearing resistance to acid and bile salts and creating the high levels of organic acids were more compatible with the poultry gastrointestinal tract which leads to increase growth performance and also improving the immune system. Also, these isolates showed the high ability to attach to intestinal epithelial cells and the competitive elimination of Salmonella spp. from colonization on intestinal cells. Therefore, the isolated LAB from the present study could be used to produce native probiotic products that are suitable for usage in poultry to reduce the consumption of antibiotics and the incidence of various diseases.
Acknowledgments
The authors would like to express their sincere gratitude to Takgene Zist Co. (Tehran, Iran) for providing processing facilities.
Conflict of interests
The authors declare that they have no conflict of interests regarding the publication of this manuscript.
References
- 1.Williams BA, Verstegen MW, Tamminga S. Fermentation in the large intestine of single-stomached animals and its relationship to animal health. Nutr Res Rev. 2001;14(2):207–228. doi: 10.1079/NRR200127. [DOI] [PubMed] [Google Scholar]
- 2.Mishu B, Griffin PM, Tauxe RV, et al. Salmonellaenteritidis gastroenteritis transmitted by intact chicken eggs. Ann Intern Med. 1991;115(3):190–194. doi: 10.7326/0003-4819-115-3-190. [DOI] [PubMed] [Google Scholar]
- 3.Sodagari HR, Mashak Z, Ghadimianazar A. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from retail chicken meat and giblets in Iran. J Infect Dev Ctries. 2015;9(5):463–469. doi: 10.3855/jidc.5945. [DOI] [PubMed] [Google Scholar]
- 4.Edens FW. An alternative for antibiotic use in poultry: Probiotics. Braz J Poultry Sci. 2003;5(2):75–97. [Google Scholar]
- 5.Hassanzadazar H, Ehsani A, Mardani K. Antibacterial activity of Enterococcus faecium derived from Koopeh cheese against Listeria monocytogenes in probiotic ultra-filtrated cheese. Vet Res Forum. 2014;5(3):169–175. [PMC free article] [PubMed] [Google Scholar]
- 6.Taghi-Zadeh A, Nejati F. Screening of lactic acid bacteria isolated from Iranian sourdoughs for antifungal activity: Enterococcusfaecium showed the most potent antifungal activity in bread. Appl Food Biotechnol. 2017;4(4):219–227. [Google Scholar]
- 7.Revolledo L, Ferreira AJ, Mead GC. Prospects in Salmonella control: Competitive exclusion, probiotics, and enhancement of avian intestinal immunity. J Appl Poult Res. 2006;15(2):341–351. [Google Scholar]
- 8.Rehman HU, Vahjen W, Awad WA. Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch Anim Nutr. 2007;61(5):319–335. doi: 10.1080/17450390701556817. [DOI] [PubMed] [Google Scholar]
- 9.Shalaei M, Hosseini SM, Zergani E. Effect of different supplements on eggshell quality, some characteristics of gastrointestinal tract and performance of laying hens. Vet Res Forum. 2014;5(4):277–286. [PMC free article] [PubMed] [Google Scholar]
- 10.Blajman J, Gaziano C, Zbrun MV, et al. In vitro and in vivo screening of native lactic acid bacteria toward their selection as a probiotic in broiler chickens. Res Vet Sci. 2015;101:50–56. doi: 10.1016/j.rvsc.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Ehrmann MA, Kurzak P, Bauer J, et al. Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J Appl Microbiol. 2002;92(5):966–975. doi: 10.1046/j.1365-2672.2002.01608.x. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen CN, Nielsen VR, Hayford AE, et al. Screening of probiotic activities of forty-seven strains of Lactobacillus spp by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65(11):4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopal PK, Prasad J, Smart J, et al. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their anta-gonistic activity against an enterotoxigenic Escherichia coli. Int J Food Microbial. 2001;67(3):207–216. doi: 10.1016/s0168-1605(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhang YC, Zhang LW, Tuo YF, et al. Inhibition of Shigella sonnei adherence to HT-29 cells by lactobacilli from Chinese fermented food and preliminary characterization of S-layer protein involvement. Res Microbial. 2010;161(8):667–672. doi: 10.1016/j.resmic.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Aquilina G, Bories G, Chesson A, et al. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012;10(6):2740. [Google Scholar]
- 16.Carasi P, Díaz M, Racedo SM, et al. Safety characterization and antimicrobial properties of kefir-isolated Lactobacillus kefiri. Biomed Res Int. 2014;2014:208974. doi: 10.1155/2014/208974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Touré R, Kheadr E, Lacroix C, et al. Production of antibacterial substances by bifidobacterial isolates from infant stool active against Listeria monocytogenes. J Appl Microbiol. 2003;95(5):1058–1069. doi: 10.1046/j.1365-2672.2003.02085.x. [DOI] [PubMed] [Google Scholar]
- 18.Chauviere G, Coconnier MH, Kerneis SO, et al. Adhesion of human Lactobacillusacidophilus strain LB to human enterocyte-like Caco-2 cells. Microbiology. 1992;138(8):1689–1696. doi: 10.1099/00221287-138-8-1689. [DOI] [PubMed] [Google Scholar]
- 19.Candela M, Perna F, Carnevali P, et al. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol. 2008;125(3):286–292. doi: 10.1016/j.ijfoodmicro.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Kim EY, Kim YH, Rhee MH, et al. Selection of Lactobacillus sp PSC101 that produces active dietary enzymes such as amylase, lipase, phytase and protease in pigs. J Gen Appl Microbial. 2007;53(2):111–117. doi: 10.2323/jgam.53.111. [DOI] [PubMed] [Google Scholar]
- 21.Araújo WL, Angellis DA, Azevedo JL. Direct RAPD evaluation of bacteria without conventional DNA extraction. Braz Arch Biol Technol. 2004;47(3):375–380. [Google Scholar]
- 22.Jannah SN, Dinoto A, Wiryawan KG, et al. Characteristics of lactic acid bacteria isolated from gastrointestinal tract of Cemani chicken and their potential use as probiotics. Media Peternakan. 2014;37(3):182–189. [Google Scholar]
- 23.Nemcova R. Criteria for selection of Lactobacillus for probiotic use. Vet Med (Praha) 1997;42(1):19–27. [PubMed] [Google Scholar]
- 24.Shokryazdan P, Sieo CC, Kalavathy R, et al. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed Res Int. 2014;2014:927268. doi: 10.1155/2014/927268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charteris WP, Kelly PM, Morelli L, et al. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J Appl Microbial. 1998;84(5):759–768. doi: 10.1046/j.1365-2672.1998.00407.x. [DOI] [PubMed] [Google Scholar]
- 26.Corsetti A, Gobbetti M, Smacchi E. Antibacterial activity of sourdough lactic acid bacteria: Isolation of a bacteriocin-like inhibitory substance from Lactobacillus sanfrancisco C57. Food Microbial. 1996;13(6):447–456. [Google Scholar]
- 27.Jankowska A, Laubitz D, Antushevich H, et al. Competition of Lactobacillus paracasei with Salmonella enterica for adhesion to Caco-2 cells. Biomed Res Int BioMed J Biomed Biotechnol. 2008:357964. doi: 10.1155/2008/357964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kizerwetter-Świda M, Binek M. Assessment of potentially probiotic properties of Lactobacillus strains isolated from chickens. Pol J Vet Sci. 2016;19(1):15–20. doi: 10.1515/pjvs-2016-0003. [DOI] [PubMed] [Google Scholar]
- 29.Lim SM, Ahn DH. Factors affecting adhesion of lactic acid bacteria to Caco-2 cells and inhibitory effect on infection of Salmonella typhimurium. J Microbiol Biotechnol. 2012;22(12):1731–1739. doi: 10.4014/jmb.1208.08049. [DOI] [PubMed] [Google Scholar]
- 30.Gueimonde M, Jalonen L, He F, et al. Adhesion and competitive inhibition and displacement of human enteropathogens by selected lactobacilli. Food Res Int. 2006;39(4):467–471. [Google Scholar]
- 31.Lee YK, Puong KY, Ouwehand AC, et al. Displacement of bacterial pathogens from mucus and Caco-2 cell surface by lactobacilli. J Med Microbial. 2003;52(10):925–930. doi: 10.1099/jmm.0.05009-0. [DOI] [PubMed] [Google Scholar]
- 32.Oh BC, Choi WC, Park S, et al. Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Appl Microbiol Biotechnol. 2004;63(4):362–372. doi: 10.1007/s00253-003-1345-0. [DOI] [PubMed] [Google Scholar]