Abstract
Major recent advances in genetics and genomics present unique opportunities for enhancing our understanding of human physiology and disease predisposition. Here I demonstrate how analysis of genomic information can provide new insights into endocrine systems, using the human growth hormone (GH) signaling pathway as an illustrative example. GH is essential for normal postnatal growth in children, and plays important roles in other biological processes throughout life. GH actions are mediated by the GH receptor, primarily via the JAK2 protein tyrosine kinase and the STAT5B transcription factor, and inactivating mutations in this pathway all lead to impaired somatic growth. Variation in GH signaling genes has been evaluated using DNA sequence data from the Exome Aggregation Consortium, a compendium of information from >60,000 individuals. Results reveal many potential missense and other alterations in the coding regions ofGH1,GHR,JAK2, andSTAT5B, with most changes being uncommon. The total number of different alleles per gene varied by ~threefold, from 101 forGH1 to 338 forJAK2. Several known disease-linked mutations inGH1,GHR, andJAK2 were present but infrequent in the population; however, three amino acid changes inGHR were sufficiently prevalent (~4% to 44% of chromosomes) to suggest that they are not disease causing. Collectively, these data provide new opportunities to understand how genetically driven variability in GH signaling and action may modify human physiology and disease.
Extensive population variation in GH signaling proteins predicted in a human genome database provides new opportunities to understand the range of normal GH physiology and pathophysiology.
Major advances in molecular biology and in genetics and genomics during the last several years now present unique opportunities for enhancing our understanding of human physiology and disease predisposition. From the endocrinologist’s perspective, there is the potential not only for new hormones to be discovered and characterized, but also the unprecedented possibility of gaining novel insights into endocrine pathophysiology and disease mechanisms. A first step in taking advantage of these opportunities is to access publicly available databases. A wealth of information is available in these repositories; however, the volume of data is potentially overwhelming, and the ability to extract, catalog, evaluate the quality, and assess and interpret the information can be challenging. Despite these difficulties, the value of the data is potentially high because new research questions can be shaped and defined, leading to insights that may be actionable in terms of addressing important biomedical problems. For example, genome sequencing on a large scale has the potential to provide novel ideas about human disease susceptibility, and to shed light on human evolution and population variation (1–4).
In this mini-review, I demonstrate the power of close examination and analysis of genomic information in providing new understanding of endocrine systems. My illustrative example is the human growth hormone (GH) signaling pathway for the reasons subsequently outlined.
GH Signaling and Actions
GH plays a pivotal role in multiple physiologic processes in humans and other mammals. It is not only essential for somatic growth during childhood through its induction of insulinlike growth factor-1 (IGF1), but additionally it is an important regulator of intermediary metabolism and tissue repair throughout life (5,6). GH actions also have a dark side because they have been implicated in the development of certain cancers and in the pathophysiology associated with aging (7–9). Like related hormones and cytokines, GH acts after binding to a transmembrane receptor (10). Ligand binding to the growth hormone receptor (GHR) activates several signaling pathways (5,10,11). Among the most important is the just another kinase (JAK)–signal transducer and activator of transcription (STAT) cascade, in which induction of tyrosine kinase activity of JAK2 (5,10,11) causes phosphorylation of tyrosine residues on the intracellular part of the GHR (5,11), leading to recruitment of several signaling molecules, including STATs. The seven members of the STAT family are inducible transcription factors that function as effectors of activated cytokine and growth factor receptors (12,13). STAT5B in particular is a critical component of GH actions, being responsible for GH-stimulated production of IGF1 by promoting its gene transcription (14–16).
Decreased activity through the GH-IGF1 axis causes growth defects in humans that result in short stature (17–19). A variety of human genetic abnormalities have been shown to impair GH messenger RNA expression or protein biosynthesis [specifically theGH1 gene (20,21)], and mutations in theGHR have been described that reduce its synthesis or biological actions (22). A few individuals also have been reported with growth deficiency and inactivating mutations in theIGF1 orIGF1 receptor genes (23,24), or inSTAT5B (25).
Database Mining and Analysis
DNA sequence information from the Exome Aggregation Consortium (ExAC) (26–30) comprises the primary data source for this mini-review (http://exac.broadinstitute.org/), and consists of results of exome sequencing from 60,706 different individuals. Other information was obtained from the following resources: human messenger RNAs and genes from the Ensemble Genome Browser, genome assembly GRCh38 (www.ensemble.org) (31); and human protein sequences from the Uniprot browser (http://www.uniprot.org/), and from the National Center for Biotechnology Information Consensus CDS Protein Set (https://www.ncbi.nlm.nih.gov/CCDS/). The mutations in humanGH,GHR,JAK2, andSTAT5B reported inTables 2 and3 are from Online Mendelian Inheritance in Man (https://www.omim.org/), which is a compendium of human genes and genetic conditions (and has an excellent online tutorial for those who wish to learn how to use it), and the Growth Genetics Consortium (http://www.growthgenetics.com/). Among other useful resources for studying genetic connections with endocrine and other diseases are the following: the GWAS catalog (https://www.ebi.ac.uk/gwas/) contains a manually curated collection of published genome-wide association studies, and is searchable by disease, gene name, author, single nucleotide polymorphism, or human chromosomal location. COSMIC (http://cancer.sanger.ac.uk/cosmic) consists of a compendium of cancer mutations, and can be searched by gene name, cancer type, cancer site, molecule, author, single nucleotide polymorphism, and other topics.
Table 2.
Disease-Associated Mutations inGH1 andGHR
| Mutation | Population Variant | ExAC Prevalence |
|---|---|---|
| GH1 | ||
| W20stop | None | — |
| E58K | None | — |
| C79S | C79fs | 1 Allele |
| R103C | R103H | 1 Allele |
| D138G | None | — |
| G157fs | G157D | 13 Alleles |
| I205M | I205M | 56 Alleles |
| R209H | None | — |
| GHR | ||
| W4stop | None | — |
| W34stop | None | — |
| C56stop | C56stop | 1 Allele |
| R61stop | R61stop, R61G, R61L, R61Q | 2, 1, 1, 10 Alleles |
| Q83stop | Q83L | 1 Allele |
| W98stop | None | — |
| C101stop | None | — |
| Y113stop | None | — |
| V143Lfs | V143I | 2 Alleles |
| L159stop | None | — |
| W175stop | None | — |
| E198splice | None | — |
| V199-M206del | V199A, V199I | 6, 1 Alleles |
| E201stop | E201G | 2 Alleles |
| M206-M207ins | None | — |
| M207Ifs | M207V | 2 Alleles |
| R235stop | R235stop | 1 Allele |
| E242stop | E242D, E242K | 3, 7 Alleles |
| I293Kfs | I293Kfs | 1 Allele |
| I297Kfs | None | — |
| V301Sfs | V301I | 1 Allele |
| I328Pfs | I328T | 2 Alleles |
| A442Sfs | None | — |
| C56S | None | — |
| S58L | None | — |
| E60K | E60K | 1 Allele |
| E62K | E62K | 3 Alleles |
| W68R | None | — |
| R89K | None | — |
| Y104D | None | — |
| C112S | None | — |
| F118S | None | — |
| Q148P | None | — |
| V162I | V162I, V162F | 166, 17 Alleles |
| H168Q | H168Q, H168P | 1, 2 Alleles |
| D170H | None | — |
| I171T | None | — |
| Q172P | None | — |
| V173G | None | — |
| R179C | R179C, R179H | 496, 36 Alleles |
| Y226C | Y226F | 1 Allele |
| R229H, G | R229H, R229C, R229L | 159, 3, 1 Alleles |
| S244I | None | — |
| D262N | None | — |
| L544I | L544I | 67,843 Alleles |
Table 3.
Disease-Associated Mutations inJAK2 andSTAT5B
| Mutation | Population Variant | ExAC Prevalence |
|---|---|---|
| JAK2 | ||
| K539L | None | — |
| K607N | None | — |
| V617F, I | V617F | 82 Alleles |
| R683G, K, S | None | — |
| STAT5B | ||
| L142Rfs | None | — |
| R152stop | R152P, R152Q | 1, 6 Alleles |
| Q368fs | Q368K, Q368Pfs, Q368Rfs | 2, 91, 41 Alleles |
| N398fs | None | — |
| E561Rfs | None | — |
| A630P | None | — |
| F646S | None | — |
ExAC contains DNA sequencing results from the exons of genes of 60,706 people, and has been available publicly for less than a year (26). These individuals live in various parts of the world, and have different ethnic backgrounds. Their overall health status is unknown (26). Among the general conclusions from the initial published analysis of these 121,412 alleles is that variation within the coding regions of human genes is extensive (26). It appears however that most of the detected modifications are uncommon, with over half being seen in a single allele, and >99% being found in <1% of the ExAC population (26). Also, most the observed differences among people studied appear to consist of synonymous nucleotide changes or amino acid substitutions (26), with only the latter altering protein sequence. However, it seems likely that the extent of variation may differ significantly for individual human genes, and more importantly, that there may be substantial variability among components of pathways in which certain of the encoded proteins act. Hence, the analysis of the human GH signaling cascade presented here, which serves as an illustrative example of how such publicly available databases can be assessed to gain new insights into human endocrine physiology.
Allelic Variation in GH Signaling Molecules in Humans
Examination of the genes for GH signaling molecules in ExAC reveals a wide range of coding variation, with most changes inGH1,GHR,JAK2, andSTAT5B being missense mutations (94% to 97%) (Table 1). Second most common were alterations in the protein reading frame, including addition of truncating stop codons (~2% to 4%) (Table 1). The total number of different alleles per gene varied by more than threefold, from 101 forGH1 to 338 forJAK2. When corrected for protein length, protein modifications also varied by a factor of 3, from 0.14 nonsynonymous changes/codon forSTAT5B, to 0.47 forGH1 (Table 1). In terms of prevalence in the study population, 98% of missense alleles were found in <0.1% of individuals, and 99.6% were detected in <1.0%. These results show that overall variation in GH signaling proteins is low in this study group, and is consistent with general conclusions from the initial analysis of ExAC data (26).
Table 1.
Human Population Variation inGH1,GHR,JAK2, andSTAT5B
| Protein | No. of Codons a | Missense and In-Frame Insertions-Deletions | Frame Shifts, Stop Codons | Splicing Site Changes | Loss of Start Codon | Loss of Stop Codon | Total No. of Different Changes | Variants per Codon | Total Variant Alleles in Population (%) |
|---|---|---|---|---|---|---|---|---|---|
| GH | 217 | 96 | 4 | 1 | 0 | 0 | 101 | 0.47 | 1.4 |
| GHR | 638 | 229 | 8 | 6 | 1 | 0 | 244 | 0.38 | 53.9 |
| JAK2 | 1132 | 327 | 6 | 5 | 0 | 0 | 338 | 0.29 | 2.6 |
| STAT5B | 787 | 104 | 4 | 0 | 0 | 0 | 108 | 0.14 | 0.4 |
Based on transcripts used in ExAC database. One predicted variant does not correspond with the amino acid sequence of GHR and is not included in the compiled data.
Splicing changes at exon-intron and intron-exon junctions, reading frame alterations caused by insertions or deletions of DNA, and nucleotide changes that lead to the addition of stop codons each can contribute to loss of protein expression. The number of alleles showing these changes was very low among GH signaling genes, comprising a handful of different instances that were rare in the study population (0.007% to 0.11% allelic frequency). Copy number variation, in which a gene is absent or is present in more than one copy per chromosome, was similarly infrequent, and ranged from no instances forGH1 andGHR, to two forSTAT5B and 42 forJAK2.
Population Variation in GH
GH1 resides on chromosome 17q23.3 in a cluster of five related genes that include two genes coding for placental lactogen (chorionic somatomammotropin), one encoding a placental lactogen analog, and the other encoding the GH gene variant,GH2 (32), which unlikeGH1, is minimally expressed in the pituitary gland (32). The first characterized mutations inGH1 were gene deletions in individuals with familial GH deficiency (33), and multiple instances of gene and partial locus deletions have been described subsequently (34,35). Later studies identified individuals with short stature, who had presumptive inactivating amino acid changes inGH1 exons, or who had frame shift mutations or stop codons that prevented full-length GH from being synthesized in pituitary somatotrophs (34,35). Listed inTable 2 are eight such previously characterized mutations, and their prevalence in the ExAC population. Alterations at four of these sites are present in ExAC, but are rare, because only one substitution, I205>M, is detected as frequently as ~0.04% of the population (56 alleles), and the other three are found in <1 in 10,000 chromosomes (Table 2).
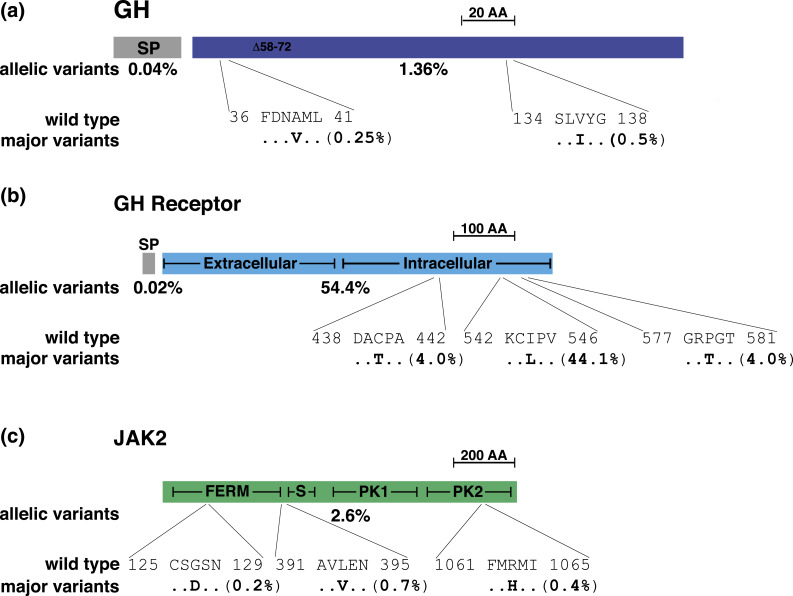
Two predicted alterations in mature GH, A39>V and V136>I, account for over half of all population changes among ExAC alleles [Fig. 1(a);Table 1]. Remarkably A39>V maps to site 1, one of two three-dimensional recognition domains that mediate binding of one molecule of GH to two molecules of the GHR (36) [site 1 comprises amino acids F36 to M40, F80 to Q94, and D195 to V206, and site 2 comprises amino acids F27 to R34 and D142 to E145 (36,37)]. Experiments now may be performed to address whether the valine side chain at position 39 could alter hormone-receptor recognition, and thus influence GH-mediated receptor activation. There are other predicted amino acid substitutions in the GH molecule, such as those associated with growth defects as previously noted (Table 2), but these occur with allelic frequencies that seem too low (from 1 in 2500 to 1 in 120,000) to have an appreciable population impact on human physiology.
Figure 1.
Population variation in human GH signaling molecules. (a) The human GH precursor consists of a 26-amino acid (AA) signal peptide (SP) and the 191-residue mature protein. The 20-kDa variant GH found in the blood lacks AAs 58 to 72 (Δ58-72 in the diagram). The overall population prevalence of variant alleles for each segment of pre-GH is listed below the map, and the location and prevalence of the two most common variants are depicted in single letter AA code. The scale bar represents 20 AAs. (b) The human GHR precursor consists of an 18-residue SP and the 620-AA mature protein. Major parts of the mature receptor are indicated. The overall population prevalence of variant alleles for the SP and for mature GHR is listed below the map, and the location and prevalence of the three most common variants is depicted in single letter AA code. The scale bar represents 100 AAs. (c) Diagram of 1132-AA human JAK2, with different domains indicated. The overall population prevalence of variant alleles is listed below the map, and the location of the three most common variants is depicted in single letter AA code. The scale bar represents 200 AAs. FERM, residues 37 to 380; PK1, pseudoprotein kinase domain, residues 545 to 809; PK2, protein kinase domain, AAs 849 to 1124; S, SH2 domain, AAs 401 to 482.
Variability in the GHR
The GHR was the first member of the cytokine receptor family to be characterized, and was among the first receptors to be shown to engage the JAK-STAT signaling cascade (5,10,11,38). Many different mutations in theGHR gene have been associated with human growth deficiency syndromes (17,22,39). Even though the first subject to be characterized molecularly had a complex rearrangement within theGHR locus on chromosome 5p13-p12 (40), most mutations identified to date are splicing alterations, protein truncating stop codons, or amino acid substitutions (17,22,39) (Table 2). Of the 23 different disease-associated frame shifts, stop codons, and splicing mutations listed inTable 2, only four are found in the ExAC database, with all being present at very low frequencies (C56stop, one allele; R61stop, two alleles; R235stop, one allele; I293>K-frame shift, one allele). In addition, amino acid substitutions at the same locations as the frame shifts and stop codons in theGHR gene also are rare, being detected at allelic frequencies of <0.01% in the population (Table 2).
Of 22 different disease-associated amino acid substitution locations in the GHR, only eight are present in the ExAC database (Table 2). Most changes that are connected to GHR dysfunction and growth defects map to the extracellular domain of the receptor (17,22,39), and five of these disease-linked alterations are very uncommon in the general population, being found in only one or three alleles of >121,000 surveyed (Table 2). However, two changes are present in ~0.1% of chromosomes (V162>I and R229>H, 166 and 149 alleles, respectively) and one in ~0.4% (R179>C, 496 alleles). The high prevalence of these three substitutions suggests either that the modifications may not be disease causing or that potentially subtle differences in the ability of the GHR to bind GH may be polymorphic in the human population. Further, one presumptive amino acid change in the intracellular region of the GHR that has been associated with growth impairment, L544>I, is actually the most common allele in ExAC, being found in 56% of all chromosomes sequenced (67,843 alleles) (Table 2). The other polymorphic allele accounts for the remaining 44% of chromosomes in the population [Fig. 1(b)]. Similarly, two other amino acid substitutions in the intracellular part of the GHR that were each first identified in an individual with a Laron-type syndrome of short stature and lack of response to GH treatment, C440>T and P579>T (41), are found in 4% of the population [Fig. 1(b)]. Thus, it seems likely that these latter modifications are also polymorphic variants. Of note, none appears to map to segments of the GHR that have been found previously to be critical for signal transduction (42–44). Experiments now may be designed to assess their effects on receptor function.
Population Aspects of JAK2 and STAT5B
JAK2 is one of four members of a nonreceptor tyrosine kinase family that also includes JAK1, JAK3, and Tyk2 (45). All these molecules function as obligate protein kinases for multiple cytokine receptors (46,47). Seven different amino acid substitution mutations in JAK2 have been described in individuals with blood dyscrasias, including polycythemia vera, thrombocythemia, and myelofibrosis, and several classes of leukemia (48–52), but no abnormalities in JAK2 have been associated with growth disorders in humans. Of these disease-linked alterations, only V617>F is detected in ExAC, and surprisingly is present in 0.07% of chromosomes (82 alleles) (Table 3), perhaps indicating that a predisposition to one of these precancerous diseases is prevalent among different human populations.
Overall, variant alleles are found inJAK2 in 2.6% of the population (Table 1), with three substitutions, G127>D, L393>A, and R1063>H, accounting for half of the changes [Fig. 1(c)]. The biological significance of these three modifications is unknown. Although they are found in different domains of the protein [Fig. 1(c)], none map to segments of the molecule involved in signaling with either the GHR or other cytokine receptors (11,53).
STAT5B, a 787-amino acid protein (54), is a critical transcription factor for GH-activatedIGF1 gene expression (14–16), and mutations in human STAT5B phenocopy GH and GHR deficiencies regarding growth deficits; however, most of the handful of individuals found to have inactivatingSTAT5B gene alterations also presented to medical attention with evidence of immune system defects (25). Compared with the other genes studied here,STAT5B is an outlier because its extent of variation in the population (0.4%, 0.14 changes per codon) was much lower than was detected forGH1,GHR, orJAK2 (Table 1). Additionally, no amino acid substitution or frame shift mutation was present in >0.08% of chromosomes (Table 3). Although speculative, perhaps the minimal level of variability in theSTAT5B gene reflects its multifactorial role as a signal transducer for GH, prolactin, erythropoietin, and multiple other cytokines (13,46,54).
As noted, a small number of humans have been identified withSTAT5B gene mutations (19,25). In all cases, these individuals have been ascertained because of severe growth failure associated with evidence of immune deficiency (19,25), and of these, two are single amino acid changes and five are frame shift mutations or stop codons (Table 3). Alterations at two of these sites are present in the ExAC database, with predicted amino acid substitutions at the Q368 frame shift location being present in ~0.1% of chromosomes in the population (132 alleles) (Table 3). The other mutations are either very rare or absent in ExAC (Table 3).
Limitations and Strengths of Population-Based Genome Sequence Data in Understanding GH Actions in Humans
As potentially expected with any large-scale DNA sequencing project, the ExAC database contains extensive material for novel biological insights, and both ambiguities and errors. From the perspective of GH signaling and actions, potential problems include the presence of at least one presumptive polymorphic coding variant that cannot be mapped to theGHR gene or protein. Other limitations of the data include the potential skewing of the study population. Although several different groups are represented, >60% of samples derive from Europeans, ~20% are East or South Asian, and only ~8% are either of Hispanic or African origin (26). Thus, the true rate and extent of variation among proteins in humans may not be established yet. In addition, there is a likely error rate associated with the many nucleotide changes that appear only once in 121,412 chromosomes evaluated.
Despite these limitations, these data provide a multitude of new opportunities to understand and reconsider what is normal GH physiology and to redefine the extent of its pathophysiology in humans. GH actions are critical for normal somatic growth in children, with growth rates functioning as a possible readout for dynamic interactions between genetic and environmental factors (55). These interactions, and the range of outcomes labeled normal, now may be reassessed in the context of many versions of GH and the GHR in the population. GH actions also have been postulated to be involved in aging (9) and in cancer pathogenesis (7,8), and the hypothesis now may be considered that some variants of either GH or the GHR, or specific combinations with different types of JAK2 or STAT5B, may enhance disease susceptibility, whereas others may be protective. A similar perspective on variation in insulinlike growth factor signaling and action, and its roles in human physiology and disease, has recently been published (56).
The extensive variability captured by ExAC should be traceable to our ancestors, including extinct populations (57,58). Modern humans contain marks in their DNA of past contacts with these populations, including both Denisovans and Neanderthals, and the legacy of these interactions still influences some traits and probably some disease susceptibilities (58). New hypotheses inspired by the data from ExAC and other genome sequencing projects could lead to novel insights about how the complex biology of GH actions has been shaped over millennia. Similar opportunities to develop new research questions and to define new physiologic paradigms exist for other areas of endocrinology, and should incentivize investigators to extract, critically evaluate, and interpret these data.
Acknowledgments
I thank my colleagues for their comments on the manuscript.
Disclosure Summary: The author has nothing to disclose.
Abbreviations:
- ExAC
Exome Aggregation Consortium
- GH
growth hormone
- GHR
growth hormone receptor
- IGF1
insulinlike growth factor-1
- JAK
just another kinase
- STAT
signal transducer and activator of transcription.
References
- 1. Acuna-Hidalgo R,Veltman JA,Hoischen A. New insights into the generation and role of de novo mutations in health and disease.Genome Biol.2016;17(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katsanis N.The continuum of causality in human genetic disorders.Genome Biol.2016;17(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Quintana-Murci L.Understanding rare and common diseases in the context of human evolution.Genome Biol.2016;17(1):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amr SS,Al Turki SH,Lebo M,Sarmady M,Rehm HL,Abou Tayoun AN. Using large sequencing data sets to refine intragenic disease regions and prioritize clinical variant interpretation.Genet Med.2017;19(5):496–504. [DOI] [PubMed] [Google Scholar]
- 5. Lanning NJ,Carter-Su C. Recent advances in growth hormone signaling.Rev Endocr Metab Disord.2006;7(4):225–235. [DOI] [PubMed] [Google Scholar]
- 6. Rosenfeld RG,Hwa V. The growth hormone cascade and its role in mammalian growth.Horm Res.2009;71(Suppl 2):36–40. [DOI] [PubMed] [Google Scholar]
- 7. Perry JK,Liu DX,Wu ZS,Zhu T,Lobie PE. Growth hormone and cancer: an update on progress.Curr Opin Endocrinol Diabetes Obes.2013;20(4):307–313. [DOI] [PubMed] [Google Scholar]
- 8. Beckwith H,Yee D. Insulin-like growth factors, insulin, and growth hormone signaling in breast cancer: implications for targeted therapy.Endocr Pract.2014;20(11):1214–1221. [DOI] [PubMed] [Google Scholar]
- 9. Berryman DE,Christiansen JS,Johannsson G,Thorner MO,Kopchick JJ. Role of the GH/IGF-1 axis in lifespan and healthspan: lessons from animal models.Growth Horm IGF Res.2008;18(6):455–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waters MJ.The growth hormone receptor.Growth Horm IGF Res.2016;28:6–10. [DOI] [PubMed] [Google Scholar]
- 11. Waters MJ,Brooks AJ. JAK2 activation by growth hormone and other cytokines.Biochem J.2015;466(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levy DE,Darnell JEJ Jr. Stats: transcriptional control and biological impact.Nat Rev Mol Cell Biol.2002;3(9):651–662. [DOI] [PubMed] [Google Scholar]
- 13. Schindler C,Plumlee C. Inteferons pen the JAK-STAT pathway.Semin Cell Dev Biol.2008;19(4):311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rotwein P.Mapping the growth hormone--Stat5b--IGF-I transcriptional circuit.Trends Endocrinol Metab.2012;23(4):186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alzhanov D,Mukherjee A,Rotwein P. Identifying growth hormone-regulated enhancers in the Igf1 locus.Physiol Genomics.2015;47(11):559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukherjee A,Alzhanov D,Rotwein P. Defining human insulin-like growth factor I gene regulation.Am J Physiol Endocrinol Metab.2016;311(2):E519–E529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosenfeld RG,Belgorosky A,Camacho-Hubner C,Savage MO,Wit JM,Hwa V. Defects in growth hormone receptor signaling.Trends Endocrinol Metab.2007;18(4):134–141. [DOI] [PubMed] [Google Scholar]
- 18. Feigerlova E,Hwa V,Derr MA,Rosenfeld RG. Current issues on molecular diagnosis of GH signaling defects.Endocr Dev.2013;24:118–127. [DOI] [PubMed] [Google Scholar]
- 19. Wit JM,Oostdijk W,Losekoot M,van Duyvenvoorde HA,Ruivenkamp CA,Kant SG. Mechanisms in endocrinology: novel genetic causes of short stature.Eur J Endocrinol.2016;174(4):R145–R173. [DOI] [PubMed] [Google Scholar]
- 20. Alatzoglou KS,Webb EA,Le Tissier P,Dattani MT. Isolated growth hormone deficiency (GHD) in childhood and adolescence: recent advances.Endocr Rev.2014;35(3):376–432. [DOI] [PubMed] [Google Scholar]
- 21. Giordano M.Genetic causes of isolated and combined pituitary hormone deficiency.Best Pract Res Clin Endocrinol Metab.2016;30(6):679–691. [DOI] [PubMed] [Google Scholar]
- 22. Janecka A,Kołodziej-Rzepa M,Biesaga B. Clinical and molecular features of Laron syndrome, a genetic disorder protecting from cancer.In Vivo.2016;30(4):375–381. [PubMed] [Google Scholar]
- 23. Netchine I,Azzi S,Le Bouc Y,Savage MO. IGF1 molecular anomalies demonstrate its critical role in fetal, postnatal growth and brain development.Best Pract Res Clin Endocrinol Metab.2011;25(1):181–190. [DOI] [PubMed] [Google Scholar]
- 24. Wit JM,Oostdijk W,Losekoot M. Spectrum of insulin-like growth factor deficiency.Endocr Dev.2012;23:30–41. [DOI] [PubMed] [Google Scholar]
- 25. Hwa V.STAT5B deficiency: impacts on human growth and immunity.Growth Horm IGF Res.2016;28:16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lek M,Karczewski KJ,Minikel EV,Samocha KE,Banks E,Fennell T,O’Donnell-Luria AH,Ware JS,Hill AJ,Cummings BB,Tukiainen T,Birnbaum DP,Kosmicki JA,Duncan LE,Estrada K,Zhao F,Zou J,Pierce-Hoffman E,Berghout J,Cooper DN,Deflaux N,DePristo M,Do R,Flannick J,Fromer M,Gauthier L,Goldstein J,Gupta N,Howrigan D,Kiezun A,Kurki MI,Moonshine AL,Natarajan P,Orozco L,Peloso GM,Poplin R,Rivas MA,Ruano-Rubio V,Rose SA,Ruderfer DM,Shakir K,Stenson PD,Stevens C,Thomas BP,Tiao G,Tusie-Luna MT,Weisburd B,Won HH,Yu D,Altshuler DM,Ardissino D,Boehnke M,Danesh J,Donnelly S,Elosua R,Florez JC,Gabriel SB,Getz G,Glatt SJ,Hultman CM,Kathiresan S,Laakso M,McCarroll S,McCarthy MI,McGovern D,McPherson R,Neale BM,Palotie A,Purcell SM,Saleheen D,Scharf JM,Sklar P,Sullivan PF,Tuomilehto J,Tsuang MT,Watkins HC,Wilson JG,Daly MJ,MacArthur DG;Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans.Nature.2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruderfer DM,Hamamsy T,Lek M,Karczewski KJ,Kavanagh D,Samocha KE,Daly MJ,MacArthur DG,Fromer M,Purcell SM;Exome Aggregation Consortium . Patterns of genic intolerance of rare copy number variation in 59,898 human exomes.Nat Genet.2016;48(10):1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zou J,Valiant G,Valiant P,Karczewski K,Chan SO,Samocha K,Lek M,Sunyaev S,Daly M,MacArthur DG. Quantifying unobserved protein-coding variants in human populations provides a roadmap for large-scale sequencing projects.Nat Commun.2016;7:13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kruger RP.Knockout! Knockout! Who’s not there?.Cell.2016;167(2):289–291. [DOI] [PubMed] [Google Scholar]
- 30. Bahcall OG.Genetic variation: ExAC boosts clinical variant interpretation in rare diseases.Nat Rev Genet.2016;17(10):584. [DOI] [PubMed] [Google Scholar]
- 31.Ensembl 2016 Ensembl release 86. Available atwww.ensembl.org. Accessed 1 May 2017.
- 32. Chen EY,Liao YC,Smith DH,Barrera-Saldaña HA,Gelinas RE,Seeburg PH. The human growth hormone locus: nucleotide sequence, biology, and evolution.Genomics.1989;4(4):479–497. [DOI] [PubMed] [Google Scholar]
- 33. Procter AM,Phillips JA III,Cooper DN. The molecular genetics of growth hormone deficiency.Hum Genet.1998;103(3):255–272. [DOI] [PubMed] [Google Scholar]
- 34. Mullis PE.Genetics of isolated growth hormone deficiency.J Clin Res Pediatr Endocrinol.2010;2(2):52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miletta MC,Lochmatter D,Pektovic V,Mullis PE. Isolated growth hormone deficiency type 2: from gene to therapy.Endocr Dev.2012;23:109–120. [DOI] [PubMed] [Google Scholar]
- 36. Fuh G,Cunningham BC,Fukunaga R,Nagata S,Goeddel DV,Wells JA. Rational design of potent antagonists to the human growth hormone receptor.Science.1992;256(5064):1677–1680. [DOI] [PubMed] [Google Scholar]
- 37. Cunningham BC,Ultsch M,De Vos AM,Mulkerrin MG,Clauser KR,Wells JA. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule.Science.1991;254(5033):821–825. [DOI] [PubMed] [Google Scholar]
- 38. Waters MJ,Hoang HN,Fairlie DP,Pelekanos RA,Brown RJ. New insights into growth hormone action.J Mol Endocrinol.2006;36(1):1–7. [DOI] [PubMed] [Google Scholar]
- 39. Rosenfeld RG,Rosenbloom AL,Guevara-Aguirre J. Growth hormone (GH) insensitivity due to primary GH receptor deficiency.Endocr Rev.1994;15(3):369–390. [DOI] [PubMed] [Google Scholar]
- 40. Godowski PJ,Leung DW,Meacham LR,Galgani JP,Hellmiss R,Keret R,Rotwein PS,Parks JS,Laron Z,Wood WI. Characterization of the human growth hormone receptor gene and demonstration of a partial gene deletion in two patients with Laron-type dwarfism.Proc Natl Acad Sci USA.1989;86(20):8083–8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kou K,Lajara R,Rotwein P. Amino acid substitutions in the intracellular part of the growth hormone receptor in a patient with the Laron syndrome.J Clin Endocrinol Metab.1993;76(1):54–59. [DOI] [PubMed] [Google Scholar]
- 42. VanderKuur JA,Wang X,Zhang L,Campbell GS,Allevato G,Billestrup N,Norstedt G,Carter-Su C. Domains of the growth hormone receptor required for association and activation of JAK2 tyrosine kinase.J Biol Chem.1994;269(34):21709–21717. [PubMed] [Google Scholar]
- 43. Kelly PA,Edery M,Finidori J,Postel-Vinay MC,Gougon L,Ali S,Dinerstein H,Sotiropoulos A,Lochnan H,Ferrag F. Receptor domains involved in signal transduction of prolactin and growth hormone.Proc Soc Exp Biol Med.1994;206(3):280–283. [DOI] [PubMed] [Google Scholar]
- 44. Rowland JE,Lichanska AM,Kerr LM,White M,d’Aniello EM,Maher SL,Brown R,Teasdale RD,Noakes PG,Waters MJ. In vivo analysis of growth hormone receptor signaling domains and their associated transcripts.Mol Cell Biol.2005;25(1):66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ihle JN.The Janus kinase family and signaling through members of the cytokine receptor superfamily.Proc Soc Exp Biol Med.1994;206(3):268–272. [DOI] [PubMed] [Google Scholar]
- 46. Imada K,Leonard WJ. The Jak-STAT pathway.Mol Immunol.2000;37(1-2):1–11. [DOI] [PubMed] [Google Scholar]
- 47. Babon JJ,Lucet IS,Murphy JM,Nicola NA,Varghese LN. The molecular regulation of Janus kinase (JAK) activation.Biochem J.2014;462(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Percy MJ,McMullin MF. The V617F JAK2 mutation and the myeloproliferative disorders.Hematol Oncol.2005;23(3-4):91–93. [DOI] [PubMed] [Google Scholar]
- 49. Mullighan CG.The molecular genetic makeup of acute lymphoblastic leukemia.Hematology (Am Soc Hematol Educ Program).2012;2012:389–396. [DOI] [PubMed] [Google Scholar]
- 50. Stein BL,Oh ST,Berenzon D,Hobbs GS,Kremyanskaya M,Rampal RK,Abboud CN,Adler K,Heaney ML,Jabbour EJ,Komrokji RS,Moliterno AR,Ritchie EK,Rice L,Mascarenhas J,Hoffman R. Polycythemia vera: an appraisal of the biology and management 10 years after the discovery of JAK2 V617F.J Clin Oncol.2015;33(33):3953–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grinfeld J,Godfrey AL. After 10 years of JAK2V617F: disease biology and current management strategies in polycythaemia vera.Blood Rev.2016;31(3):101–118. [DOI] [PubMed] [Google Scholar]
- 52. Bose P,Verstovsek S. Prognosis of primary myelofibrosis in the genomic era.Clin Lymphoma Myeloma Leuk.2016;16(Suppl):S105–S113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brooks AJ,Dai W,O’Mara ML,Abankwa D,Chhabra Y,Pelekanos RA,Gardon O,Tunny KA,Blucher KM,Morton CJ,Parker MW,Sierecki E,Gambin Y,Gomez GA,Alexandrov K,Wilson IA,Doxastakis M,Mark AE,Waters MJ. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor.Science.2014;344(6185):1249783. [DOI] [PubMed] [Google Scholar]
- 54. Hennighausen L,Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B.Genes Dev.2008;22(6):711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Baron J,Sävendahl L,De Luca F,Dauber A,Phillip M,Wit JM,Nilsson O. Short and tall stature: a new paradigm emerges.Nat Rev Endocrinol.2015;11(12):735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rotwein P.Large scale analysis of variation in the insulin-like growth factor family in humans reveals rare disease links and common polymorphisms [published online ahead of print April 7, 2017].J Biol Chem. doi:jbc.M117.783639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jones ER,Gonzalez-Fortes G,Connell S,Siska V,Eriksson A,Martiniano R,McLaughlin RL,Gallego Llorente M,Cassidy LM,Gamba C,Meshveliani T,Bar-Yosef O,Müller W,Belfer-Cohen A,Matskevich Z,Jakeli N,Higham TF,Currat M,Lordkipanidze D,Hofreiter M,Manica A,Pinhasi R,Bradley DG. Upper palaeolithic genomes reveal deep roots of modern Eurasians.Nat Commun.2015;6:8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vattathil S,Akey JM. Small amounts of archaic admixture provide big insights into human history.Cell.2015;163(2):281–284. [DOI] [PubMed] [Google Scholar]