Abstract
Background
Inflammation is an important driver of malignant glioma disease. Inflammatory mediators are not only produced by immune cells in the tumor microenvironment, but also by glioblastoma (GBM) cells themselves creating a mutually reinforcing loop. We here aimed at identifying an “anti-inflammatory switch” that allows to dampen inflammation in GBM.
Methods
We used human GBM specimens, primary cultures, and cell lines. The response of GBM cells toward inflammatory stimuli was tested by incubation with supernatant of stimulated human immune cells. Expression levels were measured by whole transcriptome microarrays and qRT-PCR, and protein was quantified by LUMINEX and SDS-PAGE. MicroRNA binding to 3′UTRs was analyzed by luciferase assays. Proliferation rates were determined by flow cytometry, and invasion and angiogenesis were studied using migration and endothelial tube formation assays.
Results
We demonstrated GBM cells to secrete high amounts of proinflammatory mediators in an inflammatory microenvironment. We found miR-93 as a potential “anti-inflammatory tumor suppressor” dramatically downregulated in GBM. Concordantly, cytokine secretion dropped after miR-93 re-expression. Transfection of miR-93 in GBM cells led to down-regulation of hubs of the inflammatory networks, namely, HIF-1α and MAP3K2 as well as IL-6, G-CSF, IL-8, LIF, IL-1β, COX2, and CXCL5. We showed only COX2 and CXCL5 to be indirectly regulated by miR-93 while all other genes are true targets. Phenotypically, re-expression of miR-93 in GBM cells substantially suppressed proliferation, migration, and angiogenesis.
Conclusions
Alleviating GBM-derived inflammation by re-expression of miR-93 may be a powerful tool to mitigate these tumors’ aggressiveness and holds promise for new clinical approaches.
Key Points.
Inflammation is an important driver of malignant glioma disease and inflammatory mediators are also produced by glioblastoma (GBM) cells themselves.
MiR-93 strongly dampens the production of inflammatory mediators by GBM impeding the shaping of a proinflammatory microenvironment.
Importance of the Study.
Inflammation is an important driver of malignant glioma disease. Inflammatory mediators are not only produced by immune cells in the tumor microenvironment, but also by glioblastoma (GBM) cells themselves creating a mutually reinforcing loop. In this study, we uncover a new regulator of GBM-derived inflammation: MiR-93 is capable to strongly dampen the production of inflammatory mediators by GBM cells, thus impeding the shaping of a proinflammatory microenvironment by the tumor itself. We show that miR-93 exerts its effects by simultaneous targeting of inflammatory mediators and of central regulatory hubs within the networks of inflammation. In human GBM, miR-93 expression is strongly repressed. Its re-expression inhibited GBM cell proliferation, migration, and angiogenesis and induces a more benign phenotype. Dampening of GBM-derived inflammation by re-expression of miR-93 may mitigate tumors’ aggressiveness and holds promise for new clinical approaches.
Glioblastomas (GBMs) are devastating tumors with poor prognosis. Current multimodal treatment concepts mainly aim at reducing or destroying the tumor cells by application of surgical and radiation therapies, chemotherapy, and targeted agents such as epidermal growth factor receptor1 or cytokine inhibitors,2–4 however, with limited success.5 Thus, new approaches to improve this dissatisfying status quo are urgently needed.
Currently, inflammation—as a central driver of cancer6—has increasingly gained attention. GBM cells are surrounded by an inflammatory microenvironment mostly driven by the innate immunity, consisting of stimulated immune cells and inflammatory mediators, eg, cytokines, chemokines, and growth factors.4 The resulting immune “cocktail” activates signaling pathways enhancing angiogenesis, migration, and invasion of GBM cells thus consolidating their highly aggressive phenotype.7 Simultaneously, signs of paralysis of the adaptive immunity occur, such as enhanced recruitment of Tregs and attenuation of CD8 cytotoxicity.8–10 Due to this multilayered nature of the tumor microenvironment, therapeutic access based on immuno-modulation is challenging.
It is important to note, however, that the complex network of inflammation is not sufficiently covered by this model, as mainly the “external” side of the coin, ie, the tumor microenvironment, has been addressed. In fact, the tumor compartment itself mounts strong inflammatory activity and has to be regarded as an important constituent of the inflammatory process: Activated by the highly stimulating microenvironment, GBM cells produce considerable amounts of immuno-modulating mediators thereby strongly amplifying the inflammatory cascade.11,12 Thus, by interrupting this feed-forward mechanism, identification of “anti-inflammatory tumor suppressors” that regulate inflammatory signaling hubs within tumor cells could open up new therapeutic perspectives.
In this scenario, miRNAs (miRs) might gain attention. In the human immune system, potent miRs have been discovered that play pivotal roles in the regulation of inflammatory gene expression.13 These “immuno-miRs” usually regulate whole signaling networks with only one miR being able to target numerous genes located within functionally related cellular pathways, thereby amplifying their regulatory capacity. The role of these immune-miRs as regulators of inflammatory targets in tumor cells, however, remains grossly unclear so far.
In this study, we provide evidence of immuno-miR-93 acting as a tumor suppressor that is able to put a brake on tumor-derived inflammation. We report sharp downregulation of miR-93 in GBM tissue and in primary GBM cell lines. Re-expression of miR-93 strongly ameliorated GBM cell proliferation, migration, and angiogenesis and induced a more benign phenotype by direct targeting of central regulatory hubs within the networks of inflammation.
Our study helps to better understand the tumor-immunity axis in GBM and might provide new impulses for future therapeutic approaches.
Materials and Methods
Human Tissue Samples
Human GBM samples (n = 51) were obtained and processed as described previously.14 All GBM tumor specimens were obtained by stereotactic biopsy technique only from vital tumor areas. For quality check, samples of the direct vicinity (1 mm steps) were analyzed within the operating room by an experienced neuropathologist. Thereafter, a more detailed histopathology was performed using (1) light microscopy, (2) immunohistochemical staining for GFAP, MAP, and Ki-67, and (3) analysis of Isocitrate Dehydrogenas and O-methylguanine-DNA methyltransferase (MGMT) status. Please see Supplementary Table S2 for clinical and histological data. Normal brain (n = 9) was obtained from epilepsy surgery. One additional normal brain sample mRNA was purchased from Ambion. The study protocol was reviewed and approved by the institutional review board of the Ludwig Maximilians University, Munich, Germany (approval number: 216/14). Written informed consent was collected from all patients.
Purification of Peripheral Blood Mononuclear Cells
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood of 10 healthy volunteers by density centrifugation (Ficoll-Histopaque 1077; Sigma-Aldrich). Cell number and viability were assessed on a Vi-Cell (Beckman-Coulter). PBMCs were stimulated with Dynabeads Human T-Activator CD3/CD28 (Life Technologies).
Primary GBM Cell Isolation
Tissue samples were collected from open GBM resections and processed with the Brain Tumor Dissociation Kit, P (Miltenyi) according to the manufacturer’s instructions. Sampling was approved by the institutional review board of the Ludwig Maximilians University, Munich, Germany (approval number: 216/14). In order to determine the neural origin, immunohistochemical staining for GFAP and MAP was carried out. Cells were used up to a cell culture passage number of 10.
Human Umbilical Vein Endothelial Cells Isolation
Primary human umbilical vein endothelial cells (HUVECs) were isolated from umbilical cords of healthy neonates directly after cesarean delivery at the Department of Gynecology and Obstetrics, University Hospital, LMU Munich, Germany. Written informed consent from the mother was obtained before donating umbilical cords in accordance with the Declaration of Helsinki. Cells of one single cord at passage 2–4 were used for each of the independent experiments.
Cell Culture
The human GBM cell lines U87 and T98G,15 as well as HEK293 cells (all from ATCC), were maintained according to the vendor’s specifications. Primary GBM cells were cultured in MACS Neuro Medium (Miltenyi), supplied with 2% MACS Neuro Brew-21 w/o Vit. A (Miltenyi), 10% FCS, 2% Penicillin-Streptomycin, and 2% l-Glutamine. HUVECs were cultured in 200PRF (Thermo Fisher) with 2% LSGS (Thermo Fisher). All cell lines were cultured at 37°C, 5% CO2 in a tissue culture incubator (Heraeus). For experiments under hypoxic conditions, cells were incubated in 5% O₂ and 40 mmHg CO₂ in an air-tight hypoxia chamber (Billups-Rothenberg) at 37°C. For stimulation experiments, cells were incubated with supernatant of stimulated PBMCs or IL-1β (10 ng/mL, Miltenyi) for 24 h. Unstimulated controls were present in all stimulation experiments (Supplementary Figure S2).
Vector Construction
3′ UTRs were PCR amplified using genomic DNA and specific primers (Supplementary Table S1, Metabion). PCR products were ligated into the pSCB-amp/kan vector (Stratagene) according to the manufacturer’s protocol and subcloned into the psiCHECK-2 plasmid using restriction endonucleases NotI, XhoI, and PmeI. All constructs were sequence verified by MWG Biotech (Ebersberg, Germany).
Transfections
Transfections were performed by electroporation (Neon Transfection System, Life Technologies) using 50 nM synthetic hsa-miR-93 miRNA Precursor (premiR-93) or 50 nM of premiR Precursor Negative Control #1 (both from Life Technologies). For RNA interference experiments, 100 nM of SMARTpool siRNA (Dharmacon) or non-targeting siRNA control was transfected. Subsequently, cells were incubated for 48 h before being harvested. Co-transfection of luciferase reporter constructs and pre-miRs was performed using 100 000 HEK-293 or U87 cells, 1 µg of PsiCHECK-2 plasmid and 50 nM pre-miRs. All transfection experiments were performed in triplicates.
Reporter Gene Assays
Co-transfected cells were harvested and luminescence was measured using the FilterMaxF3 (Molecular Devices) and the Dual-Glo-Luciferase Assay system (Promega) according to the manufacturer’s protocol. Renilla luciferase activity was normalized to firefly luciferase activity. All experiments were performed in triplicates.
Migration Assay
About 70 000 U87 cells per well were seeded in 2-well culture inserts (Ibidi) and cultured at 37°C, 5% CO2 in a cell culture incubator (Heraeus). After incubation overnight, inserts were removed, and cells were washed twice. Cells were then incubated with supernatant of transfected U87 cells stimulated with IL-1β and incubated in 5% O₂ overnight. Cells were photographed using an inverted microscope (Carl Zeiss).
Tube Formation Assay
About 10 µL of Geltrex Basement Membrane Matrix (Thermo Fisher) was transferred to a µ-slide (Ibidi) and incubated for 45 min at 37°C, 5% CO2. HUVECs were harvested and 50 µL of a cell suspension (1 × 106 cells/mL) was applied to each well. After 2 h, cell culture medium was discarded and supernatant of pre-miR-transfected U87 cells, stimulated with IL-1β and incubated in 5% O₂, was applied to HUVECs. Phase contrast microscopy was performed after 4 h of incubation. The resulting pictures of 7 independent experiments were analyzed using the 2 publicly available programs ImageJ (version 1.51k) Angiogenesis Analyzer16 and Angiotool version 0.6a (National Cancer Institute).17
RNA Isolation and cDNA Synthesis
RNA was extracted using the RNAqueous Total RNA-Isolation Kit or, for quantification of miR expression, the mirVana™ miRNA Isolation Kit (Ambion) according to the manufacturer’s protocols. After treatment with Turbo DNAse (Ambion), RNA purity was assessed on a NanoDrop2000 spectrometer (Thermo Scientific). cDNA was generated by reverse transcription of equal amounts of RNA using the SuperScript III Reverse Transcriptase Kit, RNase OUT (Invitrogen), Oligo-dT Primers, and Random Hexamers (Qiagen).
Quantitative Real-Time PCR
Relative quantification of mRNA levels was performed using a LightCycler 480 (Roche Diagnostics) as previously described.18 Primers (Metabion) and UPL Probes (Roche Diagnostics) are provided in Supplementary Table S1. All assays were designed intron spanning. Tata-box-binding protein (TBP) and succinate dehydrogenase complex, subunit A (SDHA) served as reference genes. Quantification cycle (Cq) values were calculated employing the “second derivative maximum” method computed by the LightCycler software.
Quantification of MiR Expression
MiR-93 levels were measured using TaqMan MicroRNA Assays (Applied Biosystems) according to the manufacturer’s protocol. U47 served as an endogenous control. MiR-93 and reference gene expression were measured in duplicate. All samples were calibrated using a sample of normal brain tissue. The following cycling conditions were applied: 95°C, 10 min; 50 cycles: 95°C, 15 s, 60°C, 60 s; 40°C, 30 s.
Preparation of Protein Extracts and SDS-PAGE
Cells were lysed in cell lysis buffer containing protease and phosphatase inhibitors (Cell Signaling Technologies). Protein concentrations were assessed by BCA assays (Thermo Fisher Scientific) according to the manufacturer’s instructions. About 40 µg of protein was separated on 10% SDS-PAGE gels and electroblotted on PVDF membranes. Nonspecific binding was blocked using 5% bovine serum albumin (BSA) (Sigma) in TBS-Tween-20 (TBST) (Sigma). MAP3K2 (MEKK2), HIF-1α, and COX2 protein were detected using MEKK2 (#19607, Cell Signaling Technology), Purified Mouse Anti-Human HIF-1α Clone 54/HIF-1α (RUO) (BD Biosciences), and Cox-2 (clone H-3, #sc-376861, Santa Cruz Biotech) antibodies. β-Actin (8H10D10) Mouse mAb (#3700, Cell Signaling Technology) served as loading control. Antibodies were diluted in TBST with 1% BSA. Immunoreactivity was assessed using horseradish peroxidase-labeled antibodies (Cell Signaling Technology).
ELISA and Multiplex Analyses
Cell culture supernatant was immediately stored at −80°C. Cytokine levels were assessed using LEGEND MAX Human IL-6, CXCL5, IL-8 ELISA Kits (BioLegend) and a custom-made LUMINEX Multiplex Assay (ProcartaPlex Mix&Match Human Kit, Thermo Fisher) according to the manufacturer’s protocol. Cytokine concentrations were determined using the FilterMax F3 MultiMode Microplate Reader (ELISA) and Luminex 200 and xPONENT quantification software (LUMINEX assays).
Microarray
An Affymetrix GeneChip PrimeView Human Gene Expression Microarray was performed by IMGM Laboratories (Martinsried). Bioinformatics was performed by the Anesthesia & Critical Care Informatics & Data Analysis Group (Department of Anesthesiology). Microarray Data is accessible at ArrayExpress (https://www.ebi.ac.uk/arrayexpress/, accession number E-MTAB-8395).
Immunofluorescence
Pre-miR or siRNA-transfected U87 cells (1 × 106 cells/mL) were seeded in µ-Slides (Ibidi), incubated for 24 h at 37°C and 5% CO2 in antibiotics-free medium, and stimulated with IL-1β (10 ng/mL) in 5% O2 for 24 h. Cells were fixed with chilled Methanol (−20°C) for 10 min, washed with PBS, and blocked with 5% BSA for 1 h. Primary antibody incubation (Mouse anti-Human Cox-2, clone H-3, #sc-376861, Santa Cruz Biotech, final dilution: 1:200) was performed overnight at 4°C. Slides were washed and stained with secondary Goat anti-Mouse IgG-Alexa Fluor® 488 (final dilution: 1:400, Invitrogen). Pictures of 10 randomly chosen microscopic fields at ×200 magnification were obtained (microscope: IX51 [Olympus], camera: Diagnostic Instruments Model #3.2.0 color ccd).
Flow Cytometry and Intracellular Staining
Cellular proliferation was assessed by intracellular staining of Ki67 protein. U87 and T98G cells (0.5 × 106) were transfected as previously described and stimulated with IL-1β (10 ng/mL). After 48 h, cells were permeabilized, fixed, and stained (PE anti-human Ki-67 Antibody, BioLegend, Cat. #350504) according to the manufacturer’s instructions. Analysis was performed on a FACSCanto II flow cytometer applying single-cell gating. Unstained samples and PE Mouse IgG1κ isotypes were used to determine autofluorescence and nonspecific binding.
Statistics and Bioinformatics
Statistical significance was defined as P ≤ .05 (*P ≤ .05; **P ≤ .01). Data were tested for normal distribution using the Shapiro–Wilk test. Differences between groups were analyzed using paired or unpaired Student’s t-test (two-tailed) unless stated otherwise. All data are presented as mean ± SEM, unless stated otherwise. GraphPad Prism 6 software (GraphPad Software, Inc.) was used for all statistical analyses.
Potential MiR-93 target genes were identified using the prediction programs TargetScanHuman 6.2 (http://www.targetscan.org/) and PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html). Involvement of target genes in tumor-associated pathways was evaluated using the KEGG database (www.genome.jp/kegg/pathway.html).
GBM datasets from The Cancer Genome Atlas (TCGA) with determined MGMT promoter methylation status and miR-93 expression were downloaded19 and assessed for use as a validation dataset.
Results
A Proinflammatory Microenvironment Induces an Inflammatory Gene Expression Profile in GBM Cell Lines
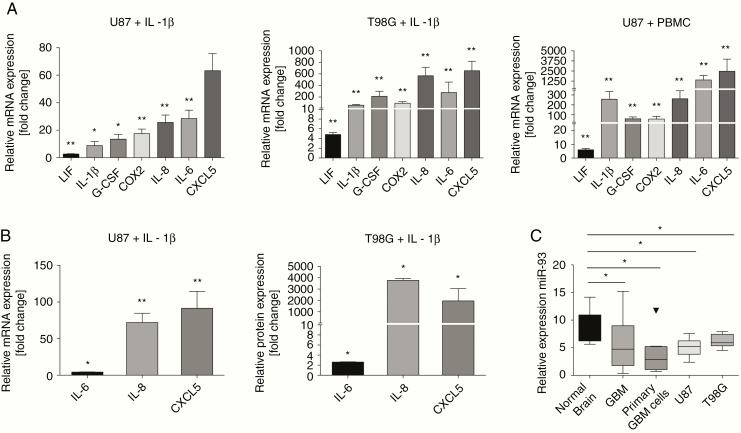
We first aimed at characterizing the response of GBM cells toward inflammatory stimuli. To this end, we incubated U87 or T98G cells with the proinflammatory “master” cytokine IL-1β 20,21 or with cell culture supernatant of stimulated PBMCs and assessed the expression levels of inflammatory mediators known to promote GBM tumor progression.22–31 Both approaches resulted in a marked induction of leukemia inhibitory factor (LIF), IL-1β, COX2, IL-8, IL-6, CXCL5, and G-CSF mRNA levels (Figure 1A). Accordingly, IL-6, IL-8, and CXCL5 protein levels were significantly increased in GBM cell culture supernatants after stimulation with IL-1β (Figure 1B). These results provide evidence that GBM cells are highly susceptible to inflammatory stimulation and respond by secreting a considerable amount of immunomodulatory mediators.
Figure 1.
An inflammatory microenvironment induces proinflammatory gene expression in GBM cells. Immuno-miR-93 is repressed in GBM. (A) mRNA expression of inflammatory genes in U87 (left panel, n = 3) and T98G (middle panel, n = 5) cells after stimulation with either IL-1β or supernatant of stimulated PBMCs (right panel, n = 3). (B) Protein expression of inflammatory genes after stimulation with IL-1β in U87 (left panel, n = 3) and T98G (right panel, n = 3) cells. All experiments were performed in triplicates; *P < .05; **P < .01. (C) Normalized expression levels of miR-93 in GBM biopsies (n = 51), primary GBM cell lines (n = 8), U87, T98G cells (n = 6), and normal brain tissue (n = 9). Endogenous reference: U47; *P < .05.
MiR-93 Is Repressed in Human Gliomas and its Re-expression Impairs GBM-Derived Cytokine Secretion
To identify an immuno-miR that may ameliorate GBM-derived inflammation, we used bioinformatics analyses and literature research and selected miR-93 out of the group of immuno-miRs as its expression is strongly linked to an anti-inflammatory phenotype in a variety of tissue types other than immune cells, eg, cardiomyocytes, hepatocytes, and neural cells.32–34 To obtain a first clue of whether miR-93 may also play a role in GBM, we analyzed tumor tissues (n = 42), primary GBM cell lines (n = 8), U87 and T98G cell lines (n = 6 each) for miR-93 levels as compared to normal brain tissue (n = 9) and found a sharp downregulation (Figure 1C). This led us to hypothesize that miR-93 indeed may be an “anti-inflammatory tumor suppressor” and thus is repressed in GBM tissue.
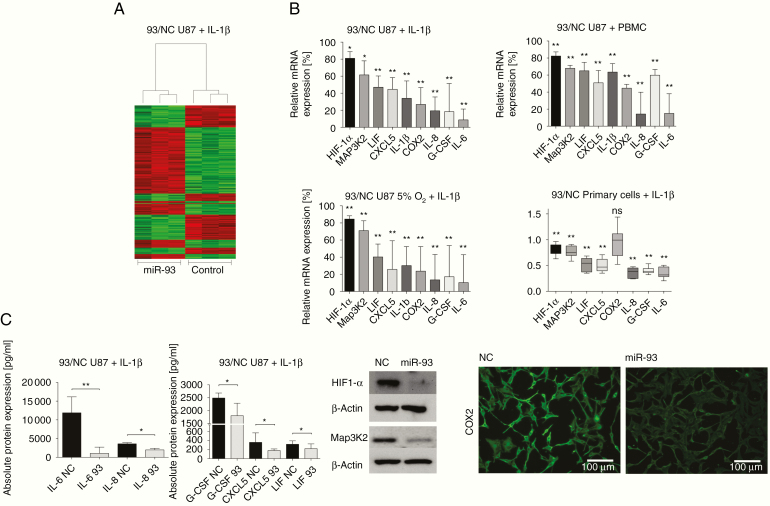
We next transiently re-expressed miR-93 in U87 GBM cells, incubated with IL-1β, and conducted microarray analyses to identify genes regulated by miR-93. This approach discovered a number of regulated genes with a clear emphasis on those involved in the control of inflammatory or/and hypoxic pathways (Figure 2A). Particularly the signaling hubs HIF-1α and Map3K2 were significantly downregulated, which was subsequently validated by qRT-PCR (Figure 2B). Notably, mRNA levels of IL-6, G-CSF, IL-8, COX2, IL-1β, CXCL5, and LIF, the same genes induced by inflammation, were found to be significantly repressed after re-expression of miR-93 in both IL-1β-stimulated (Figure 2B) and unstimulated (Supplementary Figure S2) U87 GBM cells. Above all, transfection of primary GBM cells with miR-93 resulted in significantly reduced mRNA levels for all mediators except COX2, stressing the substantial impact of miR-93 on the expression of inflammatory genes even more (Figure 2B, lower right panel). Impaired cytokine secretion upon miR-93 transfection, as measured by Multiplex protein assays (Figure 2C, upper left panel), further emphasized these results.
Figure 2.
MiR-93 re-expression produces an anti-inflammatory cell phenotype and impairs GBM-derived cytokine secretion. (A) Microarray analysis showing differentially expressed genes in U87 cells transfected with either miR-93 or scrambled control (NC) (n = 3). Green: induction, red: repression of mRNA levels. (B) Cytokine mRNA levels in U87 cells transfected with either miR-93 or scrambled control (NC) and stimulated with IL-1β (upper left panel, n = 5), IL-1β, and 5% O2 (lower left panel, n = 4) or supernatant of stimulated PBMCs (upper right panel, n = 3). Lower right panel: mRNA expression of cytokines in primary GBM cell lines (n = 6) transfected with either miR-93 or scrambled control (NC), stimulated with IL-1β. All values represent expression relative to scrambled control. Experiments were performed in triplicates; *P < .05; **P < .01. (C) Upper left panel: Protein expression of cytokines in U87 cell culture supernatant after transfection with either miR-93 or scrambled control (NC) and stimulation with IL-1β. Data were quantified using multiplex protein analyses. Values represent expression relative to scrambled control (n = 5); **P < .01. Upper right panel: Immunodetection of HIF-1α and Map3K2 in U87 cell extracts after transfection of miR-93 or scrambled control (NC), stimulation with IL-1β, and incubation in 5% O2. A representative example of 3 independent experiments is shown. Lower panel: Immunofluorescence of COX2 in U87 cells transfected with miR-93 or scrambled control (NC), stimulated with IL-1β, and incubation in 5% O2. A representative example of 3 independent experiments is shown.
Recent publications have discussed both oxygen deficiency35,36 and inflammation as central enhancers of GBM progression and extensively studied their close interrelation.37–40 To mimic the situation within the vital tumor microenvironment where the majority of viable immune cells is located, we assessed the impact of miR-93 during presence of both moderate hypoxia and inflammation. In line with our previous results, and even after being prone to a high level of inflammatory stress, transfection of miR-93 once again resulted in a significant decrease of IL-6, IL-8, IL-1β, LIF, G-CSF, COX2, and CXCL5 (Figure 2B, lower left panel). Similarly, Map3K2 and HIF-1α mRNA and protein levels were significantly reduced (Figure 2B, lower left panel; Figure 2C, right panel). COX2 protein expression was downregulated as shown by immunofluorescence microscopy (Figure 2C, right panel). Taken together, these data suggest that miR-93 attenuates the secretion of GBM-derived tumorigenic cytokines in an inflammatory and hypoxic microenvironment. Moreover, we were able to show that key components of inflammatory and hypoxic signaling, Map3K2 and HIF-1α, respectively, are repressed by miR-93.
Target Validation Identifies LIF, IL-6, G-CSF, Map3K2, and HIF-1α as Novel miR-93 Targets
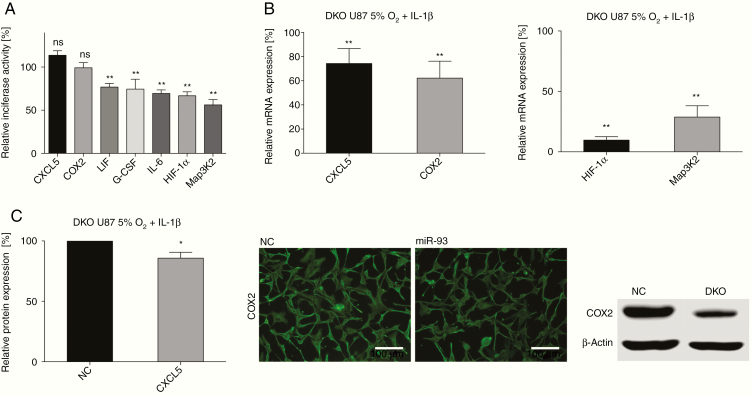
To identify direct interactions between miR-93 and the 3′ UTRs of the differentially regulated genes, we created vector constructs containing the 3′ UTRs of either LIF, HIF-1α, IL-6, G-CSF, Map3K2, COX2, or CXCL5 and performed luciferase reporter gene assays. Direct targeting of IL-8 by miR-93 has previously been found in GBM and leiomyomas.41,42 Co-transfection of miR-93 and reporter vectors significantly quenched luciferase activity of IL-6, LIF, G-CSF, HIF-1α, and Map3K2 reporter constructs (Figure 3A). However, reporter gene activities of COX2 and CXCL5 were unaltered by miR-93 transfection. We thus assumed that the observed downregulation of COX2 and CXCL5 may be conducted indirectly, presumably through miR-93-mediated, direct inhibition of genes that induce COX2 and CXCL5 gene expression.
Figure 3.
MiR-93-induced changes in GBM-derived cytokine secretion are elicited via direct targeting of LIF, IL-6, G-CSF, Map3K2, and HIF-1α. (A) Luciferase reporter activity of cells co-transfected with constructs containing the 3′ UTR of COX2 (n = 3), CXCL5 (n = 3), LIF (n = 4), G-CSF (n = 5), IL-6 (n = 5), HIF-1α (n = 9), or Map3K2 (n = 11) and either miR-93 or scrambled control. All Luciferase experiments were performed in triplicates; ns = not significant, **P < .01. (B) Analysis of GBM cells incubated in 5% O2 and stimulated with IL-1 after siRNA-mediated double knockdown of HIF-1α and Map3K2 (DKO). Left panel: qRT-PCR analysis of COX2 and CXCL5 (n = 8); **P < .01. Right panel: Knockdown efficiency of HIF-1α and Map3K2 siRNA transfection (qRT-PCR, n = 3, **P < .01). (C) Left panel: CXCL5 protein expression after double knockdown of HIF-1α and Map3K2 as measured by ELISA; n = 6, *P < .05. Middle and right panel: COX-2 protein expression after double knockdown of HIF-1α and Map3K2, measured by immunofluorescence and SDS-PAGE. A typical example of 3 experiments is shown.
These results provide evidence that IL-6, LIF, G-CSF, HIF-1α, and MAP3K2 genes are direct targets of miR-93, whereas COX2 and CXCL5 are regulated indirectly.
Repression of HIF-1α and MAP3K2 Reduces COX2 and CXCL5 Levels
Expression levels of COX2 and CXCL5 are efficiently induced by both inflammation and moderate hypoxia.43–45 Since Map3K2 and HIF-1α are hubs in both inflammatory and hypoxic signaling,43,46,47 we reasoned that miR-93-mediated regulation of Map3K2 and HIF-1α may indirectly affect COX2 and CXCL5 expression. To investigate this assumption, we conducted gene-specific knockdown of Map3K2 and HIF-1α in U87 cells stimulated with IL-1β and incubated in 5% O2. Single knockdown of HIF and MAP3K2 resulted in only a weak, not significant decrease of COX2 and CXCL5 expression (data not shown). As miR-93 affects both genes simultaneously, we performed double-knockdown experiments. Here, COX2 and CXCL5 mRNA levels were significantly decreased by −37.8% ± 14.0% and −25.7% ± 12.4% compared to controls (Figure 3B, left panel). These results were confirmed on protein level by western blot, immunofluorescence, or ELISA (Figure 3C). Taken together, our results show that knockdown of Map3K2 and HIF-1α is able to evoke changes in COX2 and CXCL5 levels similar to those after overexpression of miR-93. Thus, targeting of Map3K2 and HIF-1α by miR-93 accounts for the reduction of COX2 and CXCL5 levels in miR-93-transfected GBM cells.
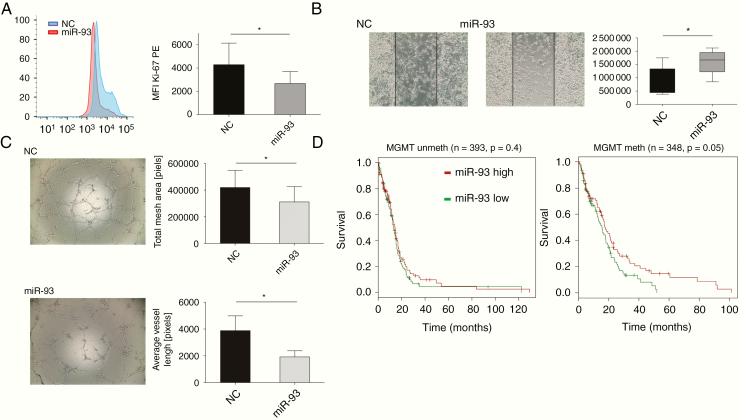
MiR-93 Inhibits U87 Cell Proliferation and Migration and Attenuates Angiogenesis
Having shown that overexpression of miR-93 strongly inhibits the expression and secretion of GBM-derived cytokines known to augment tumor progression, we next assessed the functional impact of these reduced cytokine levels on proliferation, migration, and angiogenesis, all hallmarks of GBM tumor progression. To this end, untreated GBM cells were incubated with cell culture supernatant from miR-93-transfected GBM cells incubated with 5% O2 and stimulated with IL-1β. As depicted in Figure 4 and Supplementary Figure S3, Ki-67 expression was significantly reduced indicating decreased cell proliferation (Figure 4A), and 2D migration assays showed a decrease in cellular spreading (Figure 4B; −47% ± 21.8%). In addition, angiogenic potential of HUVECs was markedly impaired upon incubation with supernatant of miR-93-transfected GBM cells, as measured by HUVEC tube formation assays (Figure 4C; average vessel length: −49.5% ± 11.8%, total mesh area: −25.7% ± 27.43%).
Figure 4.
Functional impact of miR-93 on migration, angiogenesis, and proliferation. (A) Intracellular flow-cytometric staining of Ki-67 protein in GBM cells transfected with miR-93 or scrambled control (NC). Left panel: Representative histogram of 3 independent experiments is shown. Right panel: Mean Fluorescence Intensity (MFI). (B and C) GBM cells were transfected with miR-93 or scrambled control (NC), incubated in 5% O2, and stimulated with IL-1. Resulting cell culture supernatant was applied to GBM cells or HUVECs (B). Left panel: 2D migration assay of U87 cells after incubation with U87 cell culture supernatant (n = 4, P < .05). Depicted are start state and after 24 h incubation. Lines mark the cell-free area. A typical example of 3 similar experiments is shown. Right panel: Quantitation of cell migration based on open-area analysis. (C) HUVEC tube formation assay after incubation with cell supernatants of miR-93-transfected GBM cells. Left panel: A typical example of 7 experiments is shown. Right panel: total mesh area (top, P = .021, t-test) and average vessel length (bottom, P = .023, ratio paired t-test). (D) TCGA data analysis of miR-93 expression and overall survival rate. Left panel: unmethylated MGMT promoter (n = 217; P = .4). Right panel: methylated MGMT promotor (n = 170, P = .05).
Taken together, these results provide evidence that the suppression of tumor-promoting cytokine secretion by miR-93 substantially suppresses proliferation, migration, and angiogenesis of GBM.
MiR-93 Expression Correlates With Survival in MGMT-Methylated GBM
In order to validate whether the effects of miR-93 observed in vitro also play a role in a clinical setting, we analyzed TCGA datasets. We found a negative correlation of all target genes with miR-93 expression, which was particularly pronounced for LIF (Supplementary Table S4; n = 566, r: −0.24, P = .004). Moreover, survival was lower for tumors expressing higher levels of miR-93. This difference was significant in case of a methylated MGMT promoter (P = .05, n = 170; Figure 4D and Supplementary Figure S4). These data suggest that high expression of miR-93 is associated with prolonged survival in the cohort of MGMT-methylated GBM patients.
Discussion
Inflammation is a major driver of malignancy. It is increasingly recognized that cytokines and other inflammatory mediators are not only produced by immune cells, but also by cancer cells themselves, thereby creating a mutually reinforcing loop.7,48 This context is currently gaining attention also in malignant glioma disease.22 In this study, we uncover a new central regulator of GBM-derived inflammation: miR-93, previously recognized as a classical “immuno-miR” that regulates immune cell functions,49–52 is capable to dampen down the production of inflammatory mediators by GBM cells, thus impeding the shaping of a proinflammatory microenvironment by the tumor itself. We show that miR-93 acts as an “anti-inflammatory tumor suppressor” that is repressed in human GBM. Its re-expression inhibits GBM cell proliferation, migration, and angiogenesis and induces a more benign phenotype. This is accomplished by simultaneous direct targeting of inflammatory mediators and of central regulatory hubs within the networks of inflammation.
Massive infiltration and unspecific activation of immune cells within the microenvironment of GBM have been shown to prompt GBM cells to secrete cytokines themselves.53 Yet, this inflammatory interplay has not been analyzed in detail. We here treated GBM cells with supernatants of stimulated immune cells to mimic the in vivo tumor microenvironment and found a strong production of the inflammatory mediators IL-1β, IL-6, IL-8, CXCL5, COX2, G-CSF, and LIF. All of these mediators are known to promote proliferation, migration, and invasion of cancer cells.22–31 Hence, GBMs are capable to create a tumor-promoting milieu themselves thereby strongly amplifying the vicious influence of “external” inflammation.
We next aimed at identifying an “anti-inflammatory switch” that allows to control inflammation in GBM cells by interrupting the detrimental feed-forward loop. We focused on miRs with a previously established role in the regulation of inflammatory mediators in immune cells. These immuno-miRs usually simultaneously target several genes within functionally related signaling pathways, thereby amplifying their regulative impact.13 Based on bioinformatics analyses and literature research, we selected miR-93 out of the group of immuno-miRs. MiR-93 is located with the 13th intron of its host gene MCM7. Bioinformatical data suggest that miR-93 does not possess a promoter by itself and thus might be cotranscribed with the host gene,54 but regulation of its expression currently remains elusive. MiR-93 levels are strongly linked to an anti-inflammatory phenotype in a variety of cell types.32–34,55 We hypothesized that it may also play an anti-inflammatory role in GBM. Fitting our hypothesis, we found the expression of miR-93 to be strongly repressed in GBM cell lines, primary GBM cell lines, and human GBM specimens.
Re-expression of miR-93 by in vitro transfection led to a pronounced repression of inflammatory cytokines and of genes involved in inflammatory or hypoxic signaling, as determined by whole transcriptome analyses. In particular, the inflammation-induced mediators IL-1β, IL-6, IL-8, CXCL5, COX2, G-CSF, and LIF, as well as the signaling hubs HIF-1α and Map3K2, were found to be significantly downregulated. Furthermore, we showed that HIF-1α and Map3K2 also mediate the repressive effect of miR-93 on COX2 and CXCL5, as both genes were significantly downregulated after concurrent knockdown of the 2 key molecules Map3K2 and HIF1α. Notably, this strong anti-inflammatory effect of transfected miR-93 was even preserved in an inflammatory and hypoxic microenvironment.
For IL-8, direct targeting by miR-93 has already been reported.41,50 Using reporter experiments, we were able to validate 5 new bona fide targets: LIF, G-CSF, IL-6 and, of utmost importance, HIF-1α and Map3K2. HIF-1α is considered the key transcription factor during cellular responses to oxygen deprivation and a major link between hypoxic and inflammatory signaling.56,57 Its role in GBM as a major player supporting invasion and progression is already well defined.58,59 Map3K2 is a crucial activator of NFKB transcriptional activity46 thus strongly involved in the activation of inflammatory signaling.
It was then up to provide the proof that the control of inflammatory mediator secretion of GBM cells by re-expression of miR-93, indeed, mitigates their aggressiveness. Using functional assays, we showed that GBM cell proliferation and migration were significantly inhibited upon incubation with supernatants of miR-93-transfected GBM cells. As expected, neovascularization as a hallmark of GBM malignancy60,61 was also affected. Supernatants of miR-93-transfected GBM cells considerably decreased the angiogenic potential of HUVECs, highlighting the important role of miR-93 as a central “anti-inflammatory tumor suppressor” in the context of GBM. Recent studies provided heterogeneous results, so far as tumor-promoting effects of miR-93 have been reported in cervical and prostate cancer.62,63 However, in lymphoma of the central nervous system, decreased expression has been found and antitumor effects have been suggested.64
Our in vitro results were further supported by TGCA analyses, which indicate a negative correlation of inflammatory target genes with miR-93 expression. Moreover, survival was longer for patients with methylated MGMT promoter and high miR-93 expression.
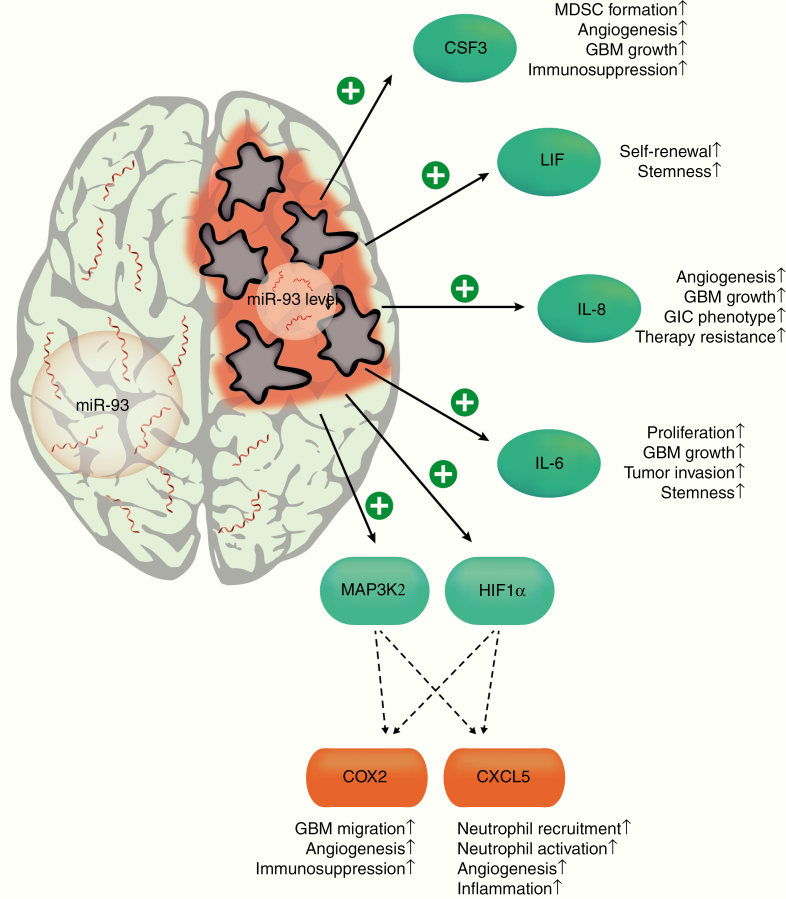
Taken together, we have identified and validated an inflammatory network in GBM cells consisting of the target genes IL-1β, IL-6, IL-8, CXCL5, COX-2, G-CSF, LIF, HIF-1α, and Map3K2, all regulated by miR-93 as a “central hub” (Figure 5). Dampening of GBM-derived inflammation by re-expression of miR-93 may be a powerful tool to alleviate these tumors’ aggressiveness and holds promise for new clinical approaches in the future.
Figure 5.
Role of miR-93 as an “anti-inflammatory tumor suppressor” in GBM. MiR-93 exerts its effects by simultaneous targeting of inflammatory mediators and of central regulatory hubs within the networks of inflammation. In human GBM, miR-93 expression is strongly repressed, which enables an unrestrained expression of its target genes, thereby promoting the aggressive phenotype of these tumors. CSF3, colony stimulating factor 3; LIF, leukemia inhibitory factor; IL-8, interleukin 8; IL-6, interleukin 6; HIF1α, hypoxia inhibitory factor 1α; MAP3K2, mitogen-activated protein kinase kinase kinase 2; COX2, cyclooxygenase 2; CXCL5, C-X-C motif chemokine 5.
Supplementary Material
Acknowledgments
We thank Niklas Thon for providing GBM patient samples. We thank Jessica Rink, Gabriele Gröger, and Gudrun Prangenberg for excellent technical assistance.
Funding
This study was funded by institutional grants of the LMU Munich.
Conflict of interest statement. The authors declare no conflict of interest.
Authorship Statement
M.H. designed, planned, and performed experiments; analyzed and interpreted data; and wrote the manuscript. N.M. planned and performed experiments; analyzed and interpreted data; and participated in writing the manuscript. D.E. performed experiments, analyzed data, and contributed artwork. C.L.H. analyzed data. T.W., M.B.M, and G.S. performed experiments. F.-W. K. contributed patient samples, interpreted data, and participated in writing the manuscript. S.K. designed experiments, interpreted data, wrote the manuscript, and supervised the study.
References
- 1. Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19(11):1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gilbert MR, Dignam JJ, Armstrong TS, et al. . A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chinot OL, Wick W, Mason W, et al. . Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):709–722. [DOI] [PubMed] [Google Scholar]
- 4. Yeung YT, McDonald KL, Grewal T, Munoz L. Interleukins in glioblastoma pathophysiology: implications for therapy. Br J Pharmacol. 2013;168(3):591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stupp R, Taillibert S, Kanner A, et al. . Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318(23):2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125(9): 3347–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, Breakefield XO. Multidimensional communication in the microenvirons of glioblastoma. Nat Rev Neurol. 2018;14(8):482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crane CA, Ahn BJ, Han SJ, Parsa AT. Soluble factors secreted by glioblastoma cell lines facilitate recruitment, survival, and expansion of regulatory T cells: implications for immunotherapy. Neuro Oncol. 2012;14(5):584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heimberger AB, Abou-Ghazal M, Reina-Ortiz C, et al. . Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin Cancer Res. 2008;14(16):5166–5172. [DOI] [PubMed] [Google Scholar]
- 10. Mathios D, Kim JE, Mangraviti A, et al. . Anti-PD-1 antitumor immunity is enhanced by local and abrogated by systemic chemotherapy in GBM. Sci Transl Med. 2016;8(370):370ra180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tarassishin L, Lim J, Weatherly DB, Angeletti RH, Lee SC. Interleukin-1-induced changes in the glioblastoma secretome suggest its role in tumor progression. J Proteomics. 2014;99:152–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tarassishin L, Casper D, Lee SC. Aberrant expression of interleukin-1β and inflammasome activation in human malignant gliomas. PLoS One. 2014;9(7):e103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirschberger S, Hinske LC, Kreth S. MiRNAs: dynamic regulators of immune cell functions in inflammation and cancer. Cancer Lett. 2018;431:11–21. [DOI] [PubMed] [Google Scholar]
- 14. Kreth S, Thon N, Eigenbrod S, et al. . O-methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One. 2011;6(2):e17156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patil V, Pal J, Somasundaram K. Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing. Oncotarget. 2015;6(41):43452–43471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carpentier G, Martinelli M, Courty J, Cascone I. Angiogenesis analyzer for ImageJ. 4th ImageJ User and Developer Conference Proceedings. 2012:198–201. [Google Scholar]
- 17. Zudaire E, Gambardella L, Kurcz C, Vermeren S. A computational tool for quantitative analysis of vascular networks. PLoS One. 2011;6(11):e27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hübner M, Hinske CL, Effinger D, et al. . Intronic miR-744 inhibits glioblastoma migration by functionally antagonizing its host gene MAP2K4. Cancers. 2018;10(11):400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11(8):633–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mills KH, Dunne A. Immune modulation: IL-1, master mediator or initiator of inflammation. Nat Med. 2009;15(12):1363–1364. [DOI] [PubMed] [Google Scholar]
- 22. Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fathima Hurmath K, Ramaswamy P, Nandakumar DN. IL-1β microenvironment promotes proliferation, migration, and invasion of human glioma cells. Cell Biol Int. 2014;38(12):1415–1422. [DOI] [PubMed] [Google Scholar]
- 24. West AJ, Tsui V, Stylli SS, et al. . The role of interleukin-6-STAT3 signalling in glioblastoma. Oncol Lett. 2018;16(4):4095–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCoy MG, Nyanyo D, Hung CK, et al. . Endothelial cells promote 3D invasion of GBM by IL-8-dependent induction of cancer stem cell properties. Sci Rep. 2019;9(1):9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hasan T, Caragher SP, Shireman JM, et al. . Interleukin-8/CXCR2 signaling regulates therapy-induced plasticity and enhances tumorigenicity in glioblastoma. Cell Death Dis. 2019;10(4):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai Z, Wu J, Chen F, et al. . CXCL5 promotes the proliferation and migration of glioma cells in autocrine- and paracrine-dependent manners. Oncol Rep. 2016;36(6):3303–3310. [DOI] [PubMed] [Google Scholar]
- 28. Feng X, Yu Y, He S, et al. . Dying glioma cells establish a proangiogenic microenvironment through a caspase 3 dependent mechanism. Cancer Lett. 2017;385:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quail DF, Bowman RL, Akkari L, et al. . The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352(6288):aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Butowski N, Colman H, De Groot JF, et al. . Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro Oncol. 2016;18(4):557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edwards LA, Li A, Berel D, et al. . ZEB1 regulates glioma stemness through LIF repression. Sci Rep. 2017;7(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J, Jiang M, Deng S, et al. . miR-93-5p-containing exosomes treatment attenuates acute myocardial infarction-induced myocardial damage. Mol Ther Nucleic Acids. 2018;11:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang B, Xuan L, Tang M, et al. . miR-93-3p alleviates lipopolysaccharide-induced inflammation and apoptosis in H9c2 cardiomyocytes by inhibiting toll-like receptor 4. Pathol Res Pract. 2018;214(10):1686–1693. [DOI] [PubMed] [Google Scholar]
- 34. Shang Y, Dai S, Chen X, Wen W, Liu X. MicroRNA-93 regulates the neurological function, cerebral edema and neuronal apoptosis of rats with intracerebral hemorrhage through TLR4/NF-κB signaling pathway. Cell Cycle. September 2019:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Eales KL, Hollinshead KE, Tennant DA. Hypoxia and metabolic adaptation of cancer cells. Oncogenesis. 2016;5:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khan N, Mupparaju S, Hou H, Williams BB, Swartz H. Repeated assessment of orthotopic glioma pO(2) by multi-site EPR oximetry: a technique with the potential to guide therapeutic optimization by repeated measurements of oxygen. J Neurosci Methods. 2012;204(1):111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma V, Dixit D, Koul N, Mehta VS, Sen E. Ras regulates interleukin-1β-induced HIF-1α transcriptional activity in glioblastoma. J Mol Med (Berl). 2011;89(2):123–136. [DOI] [PubMed] [Google Scholar]
- 38. Corcoran SE, O’Neill LA. HIF1α and metabolic reprogramming in inflammation. J Clin Invest. 2016;126(10):3699–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jensen RL. Brain tumor hypoxia: tumorigenesis, angiogenesis, imaging, pseudoprogression, and as a therapeutic target. J Neurooncol. 2009;92(3):317–335. [DOI] [PubMed] [Google Scholar]
- 40. Folco EJ, Sukhova GK, Quillard T, Libby P. Moderate hypoxia potentiates interleukin-1β production in activated human macrophages. Circ Res. 2014;115(10):875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chuang TD, Luo X, Panda H, Chegini N. miR-93/106b and their host gene, MCM7, are differentially expressed in leiomyomas and functionally target F3 and IL-8. Mol Endocrinol. 2012;26(6):1028–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fabbri E, Brognara E, Montagner G, et al. . Regulation of IL-8 gene expression in gliomas by microRNA miR-93. BMC Cancer. 2015;15:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Triner D, Shah YM. Hypoxia-inducible factors: a central link between inflammation and cancer. J Clin Invest. 2016;126(10):3689–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang LY, Tu YF, Lin YC, Huang CC. CXCL5 signaling is a shared pathway of neuroinflammation and blood-brain barrier injury contributing to white matter injury in the immature brain. J Neuroinflammation. 2016;13:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee JJ, Natsuizaka M, Ohashi S, et al. . Hypoxia activates the cyclooxygenase-2-prostaglandin E synthase axis. Carcinogenesis. 2010;31(3):427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmidt C, Peng B, Li Z, et al. . Mechanisms of proinflammatory cytokine-induced biphasic NF-kappaB activation. Mol Cell. 2003;12(5):1287–1300. [DOI] [PubMed] [Google Scholar]
- 47. Kesavan K, Lobel-Rice K, Sun W, et al. . MEKK2 regulates the coordinate activation of ERK5 and JNK in response to FGF-2 in fibroblasts. J Cell Physiol. 2004;199(1):140–148. [DOI] [PubMed] [Google Scholar]
- 48. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu Y, Jin H, Yang X, et al. . MicroRNA-93 inhibits inflammatory cytokine production in LPS-stimulated murine macrophages by targeting IRAK4. FEBS Lett. 2014;588(9):1692–1698. [DOI] [PubMed] [Google Scholar]
- 50. Fabbri E, Borgatti M, Montagner G, et al. . Expression of microRNA-93 and interleukin-8 during Pseudomonas aeruginosa-mediated induction of proinflammatory responses. Am J Respir Cell Mol Biol. 2014;50(6):1144–1155. [DOI] [PubMed] [Google Scholar]
- 51. Eissmann P, Evans JH, Mehrabi M, Rose EL, Nedvetzki S, Davis DM. Multiple mechanisms downstream of TLR-4 stimulation allow expression of NKG2D ligands to facilitate macrophage/NK cell crosstalk. J Immunol. 2010;184(12):6901–6909. [DOI] [PubMed] [Google Scholar]
- 52. Tian F, Yuan C, Hu L, Shan S. MicroRNA-93 inhibits inflammatory responses and cell apoptosis after cerebral ischemia reperfusion by targeting interleukin-1 receptor-associated kinase 4. Exp Ther Med. 2017;14(4):2903–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mantovani A, Barajon I, Garlanda C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol Rev. 2018;281(1):57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hinske LC, Dos Santos FRC, Ohara DT, Ohno-Machado L, Kreth S, Galante PAF. MiRIAD update: using alternative polyadenylation, protein interaction network analysis and additional species to enhance exploration of the role of intragenic miRNAs and their host genes. Database. 2017;2017. doi: 10.1093/database/bax053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiong L, Yu KH, Zhen SQ. MiR-93 blocks STAT3 to alleviate hepatic injury after ischemia-reperfusion. Eur Rev Med Pharmacol Sci. 2018;22(16):5295–5304. [DOI] [PubMed] [Google Scholar]
- 56. Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364(7):656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schito L, Semenza GL. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2(12):758–770. [DOI] [PubMed] [Google Scholar]
- 58. Tafani M, Di Vito M, Frati A, et al. . Pro-inflammatory gene expression in solid glioblastoma microenvironment and in hypoxic stem cells from human glioblastoma. J Neuroinflammation. 2011;8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Monteiro AR, Hill R, Pilkington GJ, Madureira PA. The role of hypoxia in glioblastoma invasion. Cells. 2017;6(4). doi: 10.3390/cells6040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 61. Colwell N, Larion M, Giles AJ, et al. . Hypoxia in the glioblastoma microenvironment: shaping the phenotype of cancer stem-like cells. Neuro Oncol. 2017;19(7):887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang X, Li F, Zhu L. Clinical significance and functions of microRNA-93/CDKN1A axis in human cervical cancer. Life Sci. 2018;209: 242–248. [DOI] [PubMed] [Google Scholar]
- 63. Choi N, Park J, Lee JS, et al. . miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget. 2015;6(27):23533–23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Takashima Y, Kawaguchi A, Iwadate Y, et al. . MicroRNA signature constituted of miR-30d, miR-93, and miR-181b is a promising prognostic marker in primary central nervous system lymphoma. PLoS One. 2019;14(1):e0210400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.