Abstract
Electronic health record phenotyping is the use of raw electronic health record data to assert characterizations about patients. Researchers have been doing it since the beginning of biomedical informatics, under different names. Phenotyping will benefit from an increasing focus on fidelity, both in the sense of increasing richness, such as measured levels, degree or severity, timing, probability, or conceptual relationships, and in the sense of reducing bias. Research agendas should shift from merely improving binary assignment to studying and improving richer representations. The field is actively researching new temporal directions and abstract representations, including deep learning. The field would benefit from research in nonlinear dynamics, in combining mechanistic models with empirical data, including data assimilation, and in topology. The health care process produces substantial bias, and studying that bias explicitly rather than treating it as merely another source of noise would facilitate addressing it.
INTRODUCTION
A phenotype is a specification of an observable, potentially changing state of an organism, as distinguished from the genotype, which is derived from an organism’s genetic makeup. The term phenotype can be applied to patient characteristics inferred from electronic health record (EHR) data.1,2 Researchers have been carrying out EHR phenotyping since the beginning of informatics, from both structured data3 and narrative data.4 The goal is to draw conclusions about a target concept based on raw EHR data, claims data, or other clinically relevant data. Phenotype algorithms – ie, algorithms that identify or characterize phenotypes – may be generated by domain exerts and knowledge engineers, including recent research in knowledge engineering,5–11 or through diverse forms of machine learning, with recent research in active learning,12 use of surrogate training sets,13–16 and deep learning,17 to generate novel representations of data.
In this paper, we discuss how to improve the fidelity of phenotype algorithms by addressing their richness and freedom from bias. Richness moves beyond the binary presence or absence of a condition to timing, degree, severity, cause, and relationship to factors like behavior, etc. We discuss temporal processing specifically, and we describe new ways to infer phenotypes that are not directly observable (eg, physiologic rate constants). We discuss phenotype bias inherent in the health care process and how to deconvolve the truth from these effects.
RICHNESS
Much of the work in phenotyping today answers the binary question of whether a condition is present or absent. For example, 2 large-scale phenotyping efforts, the Electronic Medical Records and Genomics (eMERGE) network18 and the Observational Health Data Sciences and Informatics (OHDSI) network,19 place a strong emphasis on identifying cohorts of patients to carry out association studies. Investigators may also seek to ascertain the probability, degree, severity, or level of a condition or parameter. Taking type 2 diabetes as an example (here and in further sections), one could pursue the probability of having the disease, the severity of the disease in terms of required interventions or sequelae, or the estimation of related parameters like glucose and hemoglobin A1c (HA1c). These, in turn, could feed more sophisticated analyses than are possible with just binary assignment, because they convey more information. For example, one could better assess treatment effects and disease progression and do a finer adjustment for diabetes comorbidity in other studies.
Manual queries usually produce binary answers or stated degrees or levels. Machine learning approaches often produce probabilities or scores (which can be converted to binary results with a threshold), although recent use of mechanistic models and data assimilation has produced continuous estimates of target concepts like HA1c.20 Even binary data points (eg, diagnosis codes) can be converted to continuous or ordinal values based on aggregating over many observations of the same patient.
Richer input streams will feed richer phenotypes. There are many data types beyond EHR data that will be useful for phenotyping.21 Mobile computing and self-monitoring produce streams of data that provide increased resolution. For example, depression symptoms have been inferred from mobile phone usage,22 and mobile applications are being used to capture behavioral information to characterize treatment response in type 2 diabetes.23 Linking disparate sources, such as EHR data with registries and national statistics, can lead to improved phenotyping.24 Phenotype algorithms can draw from complex concepts, such as those in narrative data, and produce similarly complex concepts. Narrative data are clearly becoming the basis of much phenotyping,25 as further evidenced by eMERGE.26,27 We refer readers to the many excellent reviews on narrative data.28–31
We recommend that phenotyping research move beyond the emphasis on binary phenotypes and address richer phenotypes. While much advanced phenotyping research is occurring, such as research using deep learning, these advanced methods are often then used to produce traditional binary classifications.32 More basic research on nonbinary targets is needed. For example, one area relates to the distribution of target concepts. The target concept may be not a single value, but a distribution. While distributions are often summarized with a mean and variance, recent work has shown that distributions with more than 2 parameters, such as the extreme value distribution for glucose measurement,33 may better reflect the complexity of human physiology.
TIME
Time is important, because health is not static. Fundamentally, exploiting time involves capturing the progression of data over time to develop a more accurate estimate of whether a condition was present (ie, using time as an input) and reporting back when it was (ie, using time as an output). Time is essential, for example, in OHDSI, because phenotype definition elements are specifically related to the time of an index event.34 In type 2 diabetes, time is essential as an input to diagnose the disease, as an output at short time scales to aid in management such as nutrition, and as an output at long time scales to assess disease progression. The latter could be represented as a numeric severity scale that is dependent upon metrics like HA1c, required treatment, and complications, or it could be represented as an ordinal level of severity. Patients could then be graded in terms of stability: how quickly their diabetes is changing over time and what might be causing it.
Temporal concepts from related fields like physics are beginning to see use in biomedical informatics. For example, the extent to which patients persist in a particular physiologic state is called stationarity. Many algorithms require stationarity to work properly, but patients are often not stationary: they become ill and, hopefully, become healthy again. Albers et al. aggregated short sequences of data across patients and assessed their heterogeneity and bias,35,36 and applied this and related approaches to creatinine data,37 glucose data,38,39 and seizures in the neurological intensive care unit.40 Jung and Shah41 assessed the consequences of stationarity in predicting wound healing, showing that the relative performance of algorithms can be miscalculated and that simpler algorithms may be better in the setting of nonstationarity. Although it has rarely been employed in a health care setting, nonstationary can be addressed by applying a moving window to analyses, assuming there are enough data to resolve the window sizes, or by using advanced techniques like temporally adaptable Markov chain Monte Carlo models42 that estimate changing parameters over time. Sometimes nonstationarity is due to the health care process rather than physiology, and that is covered below under “Health Care Process Bias.”
Our field has certainly embraced time – for example, time has been used to generate phenotype algorithms and related correlations43–48 and to detect phenotypic patterns49–51 and temporal abstractions,52–57 with related research dating back to the 1980s. We recommend that this emphasis continue and that temporal methods from other fields like nonlinear dynamics be pulled in.
ABSTRACT REPRESENTATIONS AND INFERRED VARIABLES
Some phenotypes are directly reported in the EHR, although generally with low fidelity and mediocre, highly variable accuracy; diagnoses recorded as billing codes serve as one example of suboptimal performance.58 Therefore, for many phenotypes, the goal is to identify a state that is seemingly apparent (the diagnosis) but actually known only indirectly. By triangulating to the state, such as combining billing, laboratory, medication, and narrative data, one can arrive at a more reliable identification. This is the basis for most eMERGE phenotype algorithms.27
Other times, the target concept is latent, such that it is in no way directly measured. We include 3 examples in this section. Some learning techniques generate abstract representations that may serve as useful phenotypes themselves in the sense that they can be the input to subsequent learning or prediction algorithms. Other techniques, such as data assimilation and topology, support more interpretable inferred variables.
Abstract representations, including deep learning
The EHR is a high-dimensional space, and there are a number of techniques to reduce the dimensions to produce a more useful and sometimes more understandable representation. Traditional phenotyping employs feature selection based on clinical knowledge, but this is difficult to scale to many phenotypes, and it is difficult to know if the solution is in any way optimal. Bengio and colleagues59 provide an excellent review of learning representations, including principle component analysis, autoencoders, and manifolds. Similarly, topic modeling, such as through latent Dirichlet allocation,60,61 produces latent explanatory variables that are combinations of measured variables. Tensor factorization was used to generate latent groups of high-order features.62,63 If the goal of phenotyping is to provide input for a subsequent automated process, then these abstract representations may provide superior input. For example, Miotto et al.32 used deep learning layers to generate abstract representations of EHRs and found that predictions based on them were superior to other representation techniques. Our field is embracing technologies like deep learning, with examples like Deep Patient,32 and we recommend that this continue.
Combining mechanistic knowledge and empirical data through data assimilation
Data assimilation64 is a technique that combines mechanical knowledge encoded in a mathematical model, eg, physiology, with empirical data to overcome the limitations of each. It has been used to predict weather, to fly jets, to run the power grid, and to create artificial pancreases for type 1 diabetes patients.65 Not many biomedical problems have the volume and accuracy of data required for accurate prediction from purely empirical models; instead, clinical data are often sparse, irregularly spaced, and inaccurate. Likewise, most biomedical mechanistic models are limited by our imperfect understanding and, without data and error analysis machinery, are unlikely to be quantitatively relatable to human data. Data assimilation is the machinery for combining mechanistic models with data and managing their errors and uncertainty. Data assimilation typically updates a model iteratively by making a forecast, comparing the forecast to observations, and updating the model parameters to improve future forecasts. The mechanistic nature of the model exploits knowledge to effectively constrain the search space so that far fewer data elements are needed to train the model than would otherwise be required. And the data optimize the parameters of the model and keep it anchored to reality.
The technique can be used to predict, smooth, and control, but most important for increasing phenotyping fidelity is its ability to estimate unmeasurable variables, represented as model parameters. Consider the setting of glucose metabolism. A mechanistic model could be used to generate estimates of interstitial space, insulin secretion rates, hepatic insulin extraction, glomerular filtration rate, and other things that could, in theory, be of interest but would not be measurable without advanced laboratory equipment. For example,Figure 1 shows the optimization of glucose-related parameters based on sparse, irregular finger-stick glucose measurements and meal estimates in normal and type 2 diabetes subjects.20 We recommend that phenotyping researchers consider ways of adding mechanistic knowledge to their empirical methods, including the use of data assimilation.
Figure 1.
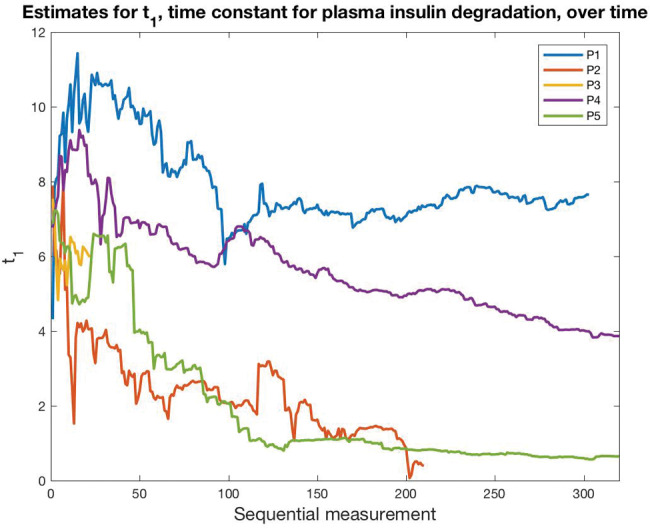
Data assimilation to find latent phenotypes. Data assimilation on a mechanistic glucose model produces estimates for a set of physiologic parameters, including plasma insulin degradation, shown here. Starting with the same initial value but based on 5 different patients’ data, data assimilation evolves the parameter to a different value for each patient (P1–P5). This represents a latent phenotype.
Topology
Topology66 is the study of properties that are preserved under the continuous transformation of an object – such as stretching but not tearing – in some real or conceptual space. It is therefore robust to normalizations, measurement distortions, and changes in units. Topology provides the machinery needed to draw statistical inferences about the shape of the clusters. Features of interest must be mapped to some space, such as illustrated inFigure 2. Topology was used to carry out unsupervised clustering of breast cancer patients based on genomic analysis.67 A new cancer subtype with 100% survival that was hidden from classical clustering techniques was found. In that study, the genomic data were mapped to a space that signified normal tissue and deviation from normal tissue. Similarly, Li et al.68 identified 3 type 2 diabetes subgroups through topological analysis. In another example, topological properties had a direct biological interpretation: shape was used to determine the number and rate of recombination events in viruses, as distinct from classic evolution through mutation.69 Topology should be applicable to clinical informatics, especially in these cases:
Where the space represents the state of a system, and that system has a continuously varying state rather than a set of distinct states that jump from one to another.
Where a space can be engineered so that critical (eg, invariant) features map to topological structures and nonessential features map to deformable parts that are ignored in the topological analyses.
Where adjacent interactions are more important than differences in objects on opposite ends of the problem space.
Where a problem can be mapped to a space, but the dimensions of that space cannot easily be normalized to produce a well-defined distance metric.
Figure 2.
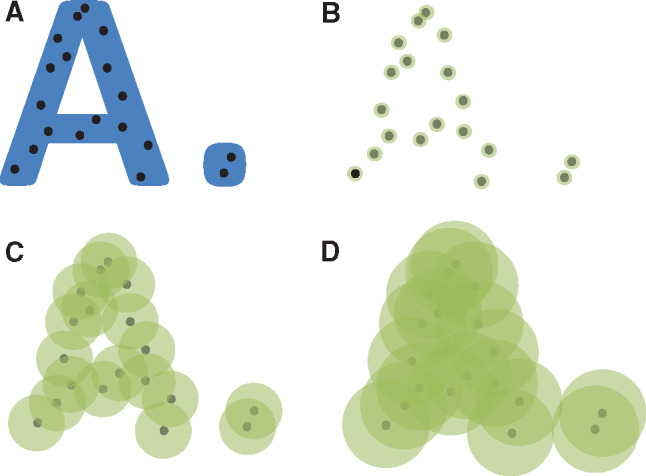
Topology. (A) Based on an underlying, unknown space that is shown in blue, a sample of black points are drawn. (B) We attempt to recreate the underlying space by creating green neighborhoods of radius epsilon around each point and joining touching neighborhoods. (C) As epsilon grows, we see features of the underlying space recreated, such as 2 distinct groups where one of them is a ring. (D) As epsilon grows further, all the points become joined. Arithmetic topology supplies the tools needed to infer properties of the underlying space based on properties of the neighborhoods as epsilon varies over its range.
We recommend that phenotyping researchers consider topological methods for their phenotyping research.
HEALTH CARE PROCESS BIAS
The health care process places its mark on phenotyping data.1,70 Measurements are made when a patient is ill, measurements are modified according to external goals, and a feed-forward loop is formed in which measurement affects state and state affects measurement. For example, laboratory tests show strong diurnal variation37,71 and other bias72; while this may reflect physiologic variation, we posit that overnight measurements, which are generally of sicker patients, might also play a role. Lasko et al.45 used time series analysis methods with unsupervised learning to address noisy, sparse, irregular health record data, applying it to uric acid laboratory data to distinguish clinical context. Weber and Kohane73 looked specifically at the time between laboratory tests to automatically determine normal ranges in a manner that exploits more medical intelligence than merely tallying standard deviations; it also identifies abnormal subpopulations and test overordering. In related work, physicians were found to oversample to correct for increases in a patient’s variance,74 but perhaps not enough.75
Many other effects arise; for example, ICD9-CM and ICD10-CM codes, which are used for billing and reflect financial incentives, may not reflect clinical state well.58 Such codes vary in granularity, with some representing heterogeneous collections of diseases as “not elsewhere classified,” and those definitions change over time as diseases are added elsewhere in the hierarchy. The clinical context of data collection can directly bias study results.76 One approach is to measure the effect of the health care process on clinical variables and group them by the effect,70 with the hope that the total phenotyping definition workload can be reduced by treating similarly affected variables with similar heuristics. For example, for chronic but not acute conditions, one can look for a longitudinal pattern of billing codes. Temporal information can help to decipher physiology from health care process effects, such as what is shown in a study of lagged linear correlation of EHR variables.44 More sophisticated lagged regression models, such as vector autoregression, can be used to correct for health care covariates.77
In our type 2 diabetes example, we must account for biased sampling of glucose78 – such as during an emergency department visit for an infection – when we attempt to assess a person’s chronic diabetes state, and we must realize that type 1 and 2 diabetes diagnosis codes are often interchanged. We recognize that phenotyping researchers are well aware of EHR bias, but we recommend that health care process bias be addressed as an area of research in itself and that its findings and techniques be shared, rather than merely treating the bias as one more detail to handle in the larger phenotyping process. Put another way, the EHR is actually a measure of the health care process, and that process must be understood well to infer details about the underlying patient; treating the EHR as a direct measure of the patient with noise misses the opportunity to correct the bias instead of just quantifying it.
CONCLUSIONS
Phenotyping is already expanding in scope in biomedical informatics, supporting richer phenotypes and addressing bias. Biomedical informatics can draw from other fields, such as deep learning, data assimilation, and topology, to further expand its treatment. Health care process bias should be addressed explicitly. Combinations of methods, such as applying temporal deep learning to address health care process bias, may bear fruit.
CONTRIBUTORS
Both authors made substantial contributions to the conception and design of the work, drafted the work or revised it critically for important intellectual content, had final approval of the version to be published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
FUNDING
This work was funded by grants from the National Institutes of Health: R01 LM006910 “Discovering and applying knowledge in clinical databases” and U01 HG008680 “Columbia GENIE (GENomic Integration with EHR).”
COMPETING INTERESTS
None.
REFERENCES
- 1. Hripcsak G,Albers DJ. Next-generation phenotyping of electronic health records.J Am Med Inform. Assoc. 2013;20:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pathak J,Kho AN,Denny JC. Electronic health records–driven phenotyping: challenges, recent advances, and perspectives.J Am Med Inform Assoc. 2013;20(e2):e206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warner HR. Knowledge sectors for logical processing of patient data in the HELP system.Proc Annu Symp Comput Appl Med Care. 1978:401–04. [Google Scholar]
- 4. Hripcsak G,Friedman C,Alderson PO,DuMouchel W,Johnson SB,Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing.Ann Intern Med. 1995;122:681–88. [DOI] [PubMed] [Google Scholar]
- 5. Newton KM,Peissig PL,Kho AN,et al. Validation of electronic medical record–based phenotyping algorithms: results and lessons learned from the eMERGE network.J Am Med Inform Assoc. 2013;20(e1):e147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rasmussen LV,Thompson WK,Pacheco JA,et al. Design patterns for the development of electronic health record–driven phenotype extraction algorithms.J Biomed Inform. 2014;51:280–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rasmussen LV,Kiefer RC,Mo H,et al. A modular architecture for electronic health record–driven phenotyping.AMIA Jt Summits Transl Sci Proc. 2015;147–51. [PMC free article] [PubMed] [Google Scholar]
- 8. Rea S,Pathak J,Savova G,et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project.J Biomed Inform. 2012;45:763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conway M,Berg RL,Carrell D,et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms.AMIA Annu Symp Proc. 2011;274–83. [PMC free article] [PubMed] [Google Scholar]
- 10. Overby CL,Pathak J,Gottesman O,et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury.J Am Med Inform Assoc. 2013;20:e243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morley KI,Wallace J,Denaxas SC,et al. Defining disease phenotypes using national linked electronic health records: a case study of atrial fibrillation.PLoS One. 2014;9:e110900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y,Carroll RJ,McPeek Hinz ER,Shah A,Eyler AE,Denny JC,Xu H. Applying active learning to high-throughput phenotyping algorithms for electronic health records data.J Am Med Inform Assoc. 2013;20:e253–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Halpern Y,Choi Y,Horng S,Sontag D. Using anchors to estimate clinical state without labeled data.AMIA Annu Symp Proc. 2014;2014:606–15. [PMC free article] [PubMed] [Google Scholar]
- 14. Agarwal V,Lependu P,Podchiyska T,Barber R,Boland M,Hripcsak G,Shah N. Using narratives as a source to automatically learn phenotype models.1st Workshop on Data Mining for Medical Informatics: Electronic Phenotyping, Washington, DC;2014. [Google Scholar]
- 15. Halpern Y,Horng S,Choi Y,Sontag D. Electronic medical record phenotyping using the anchor and learn framework.J Am Med Inform Assoc. 2016;23:731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agarwal V,Podchiyska T,Banda JM,et al. Learning statistical models of phenotypes using noisy labeled training data.J Am Med Inform Assoc. 2016;23:1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LeCun Y,Bengio Y,Hinton G. Deep learning.Nature. 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- 18. Gottesman O,Kuivaniemi H,Tromp G,et al. ;eMERGE Network. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future.Genet Med. 2013;1510:761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hripcsak G,Duke JD,Shah NH,et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers.MEDINFO’15; August 19–23, São Paulo, Brazil;2015. [PMC free article] [PubMed] [Google Scholar]
- 20. Albers DJ,Levine ME,Gluckman BJ,Ginsberg H,Hripcsak G,Mamykina L. Personalized glucose forecasting for type 2 diabetics using data assimilation.PLOS Comput Biol. 2017;134:e1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weber GM,Mandl KD,Kohane IS. Finding the missing link for big biomedical data.JAMA. 2014;311:2479–80. [DOI] [PubMed] [Google Scholar]
- 22. Hung GC,Yang PC,Chang CC,Chiang JH,Chen YY. Predicting negative emotions based on mobile phone usage patterns: an exploratory study.JMIR Res Protoc. 2016;53:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mamykina L,Levine ME,Davidson PG,Smaldone AM,Elhadad N,Albers DJ. Data-driven health management: reasoning about personally generated data in diabetes with information technologies.J Am Med Inform Assoc. 2016;23:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denaxas SC,George J,Herrett E,et al. Data resource profile: cardiovascular disease research using linked bespoke studies and electronic health records (CALIBER).Int J Epidemiol. 2012;41:1625–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pivovarov R,Perotte AJ,Grave E,Angiolillo J,Wiggins CH,Elhadad N. Learning probabilistic phenotypes from heterogeneous EHR data.J Biomed Inform. 2015;58:156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. PheKB: a Knowledgebase for Discovering Phenotypes from Electronic Health Records.https://phekb.org. Accessed June 4, 2017. [Google Scholar]
- 27. Kirby JC,Speltz P,Rasmussen LV,et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability.J Am Med Inform Assoc. 2016;23:1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shivade C,Raghavan P,Fosler-Lussier E,Embi PJ,Elhadad N,Johnson SB,Lai AM. A review of approaches to identifying patient phenotype cohorts using electronic health records.J Am Med Inform Assoc. 2014;21:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pathak J,Bailey KR,Beebe CE,et al. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium.J Am Med Inform Assoc. 2013;20(e2):e341–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao KP,Cai T,Savova GK,et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing.BMJ. 2015;350:h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu S,Liao KP,Shaw SY,et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources.J Am Med Inform Assoc. 2015;22:993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miotto R,Li L,Kidd BA,Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records.Sci Rep. 2016;6:26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Albers D,Pivovarov R,Schmidt JM,Elhadad N,Hripcsak G. Model selection for EHR laboratory tests preserving healthcare context and underlying physiology (abstract).Proc AMIA Symp.2015;1–2. [Google Scholar]
- 34. Observational Health Data Sciences and Informatics (OHDSI).www.ohdsi.org. Accessed June 4, 2017. [Google Scholar]
- 35. Albers DJ,Hripcsak G. Using time-delayed mutual information to discover and interpret temporal correlation structure in complex populations.Chaos. 2012;22:013111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Albers DJ,Hripcsak G. Estimation of time-delayed mutual information and bias for irregularly and sparsely sampled time-series.Chaos, Solutions, Fract. 2012;45;853–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Albers DJ,Hripcsak G. A statistical dynamics approach to the study of human health data: resolving population scale diurnal variation in laboratory data.Phys Lett A. 2010;374:1159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Albers DJ,Hripcsak G,Schmidt M. Population physiology: leveraging electronic health record data to understand human endocrine dynamics.PLoS One. 2012;712:e48058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Albers DJ,Elhadad N,Tabak E,Perotte A,Hripcsak G. Dynamical phenotyping: using temporal analysis of clinically collected physiologic data to stratify populations.PLoS One. 2014;96:e96443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Claassen J,Albers D,Schmidt JM,et al. Nonconvulsive seizures in subarachnoid hemorrhage link inflammation and outcome.Ann Neurol. 2014;75:771–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jung K,Shah NH. Implications of non-stationarity on predictive modeling using EHRs.J Biomed Inform. 2015;58:168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hagar Y,Albers D,Pivovarov R,Chase H,Dukic V,Elhadad N. Survival analysis with electronic health record data: experiments with chronic kidney disease.Statistical Analy Data Mining. 2014;7:385–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Warner JL,Zollanvari A,Ding Q,Zhang P,Snyder GM,Alterovitz G. Temporal phenome analysis of a large electronic health record cohort enables identification of hospital-acquired complications.J Am Med Inform Assoc. 2013;20:e281–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hripcsak G,Albers DJ,Perotte A. Exploiting time in electronic health record correlations.J Am Med Inform Assoc. 2011;18 (Suppl 1): i109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lasko TA,Denny JC,Levy MA. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data.PLoS One. 2013;8:e66341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hauskrecht M,Visweswaran S,Cooper G,Clermont G. Data-driven identification of unusual clinical actions in the ICU.Annual American Medical Informatics Association Symposium, Washington, DC;2013;580. [Google Scholar]
- 47. Liu Z,Hauskrecht M. Sparse linear dynamical system with its application in multivariate clinical time series.NIPS 2013 Workshop on Machine Learning for Clinical Data Analysis and Healthcare,December 2013; preprint available online athttps://arxiv.org/abs/1311.7071. Accessed October 4, 2017. [Google Scholar]
- 48. Liu Z,Hauskrecht M. Clinical time series prediction with a hierarchical dynamical system.The 14th Conference on Artificial Intelligence in Medicine, Murcia, Spain;2013;227–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang F,Lee N,Hu J,Sun J,Ebadollahi S. Towards heterogeneous temporal clinical event pattern discovery: a convolutional approach. In:KDD’12,August 12–16, 2012, Beijing, China;2012:453–61. [Google Scholar]
- 50. Batal I,Valizadegan H,Cooper GF,Hauskrecht M. A pattern mining approach for classifying multivariate temporal data. In:Proc IEEE Int Conf Bioinformatics Biomed. 2011;358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Noren GN,Hopstadius J,Bate A,Star K,Edwards IR. Temporal pattern discovery in longitudinal electronic patient records.Data Min Knowl Discov. 2010;20:361–87. [Google Scholar]
- 52. Shahar Y. A framework for knowledge-based temporal abstraction.Artif Intell. 1997;90(1–2):79–133. [Google Scholar]
- 53. Stacey M,McGregor C. Temporal abstraction in intelligent clinical data analysis: a survey.Artif Intell Med. 2007;39:1–24. [DOI] [PubMed] [Google Scholar]
- 54. Moskovitch R,Peek N,Shahar Y. Classification of ICU patients via temporal abstraction and temporal patterns mining.Notes of the Intelligent Data Analysis in Medicine and Pharmacology (IDAMAP 2009) Workshop. Verona, Italy;2009;35–40. [Google Scholar]
- 55. Sohn S,Savova GK. Mayo Clinic smoking status classification system: extensions and improvements.AMIA Annu Symp Proc. 2009;2009:619–23. [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou L,Hripcsak G. Temporal reasoning with medical data – a review with emphasis on medical natural language processing.J Biomed Inform. 2007;40:183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hripcsak G,Elhadad N,Chen C,Zhou L,Morrison FP. Using empirical semantic correlation to interpret temporal assertions in clinical texts.J Am Med Inform Assoc. 2009;16:220–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hogan WR,Wagner MM. Accuracy of data in computer-based patient records.J Am Med Inform Assoc. 1997;4:342–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bengio Y,Courville A,Vincent P. Representation learning: a review and new perspectives.IEEE T Pattern Anal Mach Intell. 2013;35:1798–828. [DOI] [PubMed] [Google Scholar]
- 60. Blei DM,Ng AY,Jordan MI. Latent Dirichlet allocation.J Machine Learning Res. 2003;3:993–1022. [Google Scholar]
- 61. Pivovarov R,Perotte AJ,Grave E,Angiolillo J,Wiggins CH,Elhadad N. Learning probabilistic phenotypes from heterogeneous EHR data.J Biomed Inform. 2015;58:156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Luo Y,Wang F,Szolovits P. Tensor factorization toward precision medicine.Brief Bioinform. 2017;18:511–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Luo Y,Xin Y,Hochberg E,Joshi R,Uzuner O,Szolovits P. Subgraph augmented non-negative tensor factorization (SANTF) for modeling clinical narrative text.J Am Med Inform Assoc. 2015;22:1009–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Law K,Stuart A,Zygalakis K. Data Assimilation. Cham, Switzerland: Springer;2015. [Google Scholar]
- 65. Kovatchev B,Breton M,Man C,Cobelli C. In silico preclinical trials: a proof of concept in closed-loop control of type 1 diabetes.J Diabetes Sci Technol. 2009;3:44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Carlsson G. Topology and data.Bull Am Mathematical Soc. 2009;462:255–308. [Google Scholar]
- 67. Nicolau M,Levine AJ,Carlsson G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival.Proc Natl Acad Sci USA. 2011;10817:7265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li L,Cheng WY,Glicksberg BS,et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity.Sci Transl Med. 2015;7:311ra174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cámara PG,Levine AJ,Rabadán R. Inference of ancestral recombination graphs through topological data analysis.PLoS Comput Biol. 2016;128:e1005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hripcsak G,Albers DJ. Correlating electronic health record concepts with healthcare process events.J Am Med Inform Assoc. 2013;20:e311–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Albers DJ,Hripcsak G. An information-theoretic approach to the phenome (abstract).AMIA Summit on Translational Bioinformatics,March 15–17; San Francisco;2009. [Google Scholar]
- 72. Pivovarov R,Albers DJ,Sepulveda JL,Elhadad N. Identifying and mitigating biases in EHR laboratory tests.J Biomed Inform. 2014;51:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weber GM,Kohane IS. Extracting physician group intelligence from electronic health records to support evidence based medicine.PLoS One. 2013;8:e64933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hripcsak G,Albers DJ,Perotte A. Parameterizing time in electronic health record studies.J Am Med Inform Assoc. 2015;224:794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lasko TA. Nonstationary Gaussian process regression for evaluating clinical laboratory test sampling strategies.Proc 29th AAAI Conference on Artificial Intelligence 2015;1777–83. [PMC free article] [PubMed] [Google Scholar]
- 76. Hripcsak G,Knirsch C,Zhou L,Wilcox A,Melton GB. Bias associated with mining electronic health records.J Biomed Discov Collab. 2011;6:48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Levine ME,Albers DJ,Hripcsak G. Comparing lagged linear correlation, lagged regression, Granger causality, and vector autoregression for uncovering associations in EHR data.Proc AMIA Symp. 2017;2016:779–88. [PMC free article] [PubMed] [Google Scholar]
- 78. Albers DJ,Elhadad N,Tabak E,Perotte A,Hripcsak G. Dynamical phenotyping: using temporal analysis of clinically collected physiologic data to stratify populations.PLoS One. 2014;9:e96443. [DOI] [PMC free article] [PubMed] [Google Scholar]


