Abstract
Fusobacterium nucleatum (F. nucleatum), which has been associated with colorectal carcinogenesis, can impair anti-tumour immunity and actively invade colon epithelial cells. Considering the critical role of autophagy in host defence against microorganisms, we hypothesised that autophagic activity of tumour cells might influence the amount of F. nucleatum in colorectal cancer tissue. Using 724 rectal and colon cancer cases within the Nurses’ Health Study and the Health Professionals Follow-up Study, we evaluated autophagic activity of tumour cells by immunohistochemical analyses of BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression. We measured the amount of F. nucleatum DNA in tumour tissue by quantitative PCR. We conducted multivariable ordinal logistic regression analyses to examine the association of tumour BECN1, MAP1LC3, and SQSTM1 expression with the amount of F. nucleatum, adjusting for potential confounders, including microsatellite instability status, CpG island methylator phenotype, long-interspersed nucleotide element-1 methylation, and KRAS, BRAF and PIK3CA mutations. Compared with BECN1-low cases, BECN1-intermediate and BECN1-high cases were associated with lower amounts of F. nucleatum with odds ratios (for a unit increase in three ordinal categories of the amount of F. nucleatum) of 0.54 (95% confidence interval, 0.29–0.99) and 0.31 (95% confidence interval, 0.16–0.60), respectively (Ptrend < 0.001 across ordinal BECN1 categories). Tumour MAP1LC3 and SQSTM1 levels were not significantly associated with the amount of F. nucleatum (Ptrend > 0.06). Tumour BECN1, MAP1LC3, and SQSTM1 levels were not significantly associated with patient survival (Ptrend > 0.10). In conclusion, tumour BECN1 expression is inversely associated with the amount of F. nucleatum in colorectal cancer tissue, suggesting a possible role of autophagy in the elimination of invasive microorganisms.
Keywords: colorectal neoplasms, immunology, microbiology, microbiome, molecular pathological epidemiology, tumour microenvironment
Introduction
Accumulating evidence supports the importance of the gut microbiota in intestinal carcinogenesis through modulation of tumour immune microenvironment [1–5]. Fusobacterium nucleatum (F. nucleatum) has emerged as a potentially influential driver of colorectal carcinogenesis due to its contribution to the formation of proinflammatory condition [6–8]. Studies have shown an enrichment of F. nucleatum in colorectal cancer tissue compared with the adjacent normal colon [7–9], and revealed an association of high F. nucleatum levels with distinct clinical and tumour molecular features including proximal tumour location, high-level microsatellite instability (MSI), lower levels of tumour-infiltrating CD3+ cells, and poor patient prognosis [9–12]. Given incomplete knowledge of the interaction between microbes and the host immune system, a better understanding of mechanism through which specific microbes can localise in colorectal cancer tissue would inform efforts to develop cancer prevention and treatment strategies [13–15].
Autophagy involves the lysosomal degradation and recycling of intracellular components. The BECN1 (beclin 1) and MAP1LC3 (LC3) proteins are key positive regulators of autophagy that initiate autophagosome formation, while the SQSTM1 (p62) protein plays a crucial role in lysosomal degradation of ubiquitinated substrate [16–19]. This homeostatic process adjusts cellular biomass and function in response to diverse stimuli, including metabolic stress, inflammation, and pathogen infection. Autophagic activity of tumour cells may enhance anti-tumour immune response by presenting tumour antigens [16–18,20,21]. Autophagy also plays a key role in determining microbial composition in the microenvironment. Further evidence suggests that activated autophagic activity may eliminate intracellular microorganisms [22–28]. Therefore, we hypothesised that autophagic activity of tumour cells might influence tumour bacterial load including levels of F. nucleatum in colorectal cancer tissue.
Even the best experimental model could not recapitulate the complexity of human tumour microenvironment, which is influenced by genetic and epigenetic alterations, environmental exposures, gut microbiota, and host factors [29,30]. Thus, the importance of large-scale human studies on tumour molecular characteristics and microbiota cannot be overemphasised. To test our hypothesis, we utilised a molecular pathological epidemiology database of colorectal cancer cases within two large U.S. prospective cohort studies, with data on demographic, clinical, pathological, and tumour molecular characteristics. This comprehensive dataset enabled us to examine the association of autophagic activity of tumour cells with tumoural bacterial load including the amount of F. nucleatum as well as Bifidobacterium genus in the tumour tissue, taking into account the distinct features of microbial species.
Materials and methods
Study population
We collected data from two prospective cohort studies in the U.S., the Nurses’ Health Study (NHS, 121,701 women aged 30–55 years followed since 1976) and the Health Professionals Follow-up Study (HPFS, 51,529 men aged 40–75 years followed since 1986) [31]. Study participants have been followed with biennial questionnaires on lifestyle factors and newly-diagnosed diseases including colorectal cancer. The response rate has been more than 90% for each follow-up questionnaire cycle in both cohort studies. The National Death Index was used to ascertain deaths of study participants and identify unreported lethal colorectal cancer cases. Participating physicians, who were blind to exposure data, reviewed medical records of identified colorectal cancer cases to confirm the disease diagnosis and to collect data on clinical characteristics including tumour size, tumour anatomical location, and disease stage based on the American Joint Committee on Cancer TNM classification.
Among 724 patients with available data on the amount of F. nucleatum, we analysed 628 patients with available data on tumour BECN1 expression, 689 patients with available data on tumour MAP1LC3 expression, and 674 patients with available data on tumour SQSTM1 expression in colorectal cancer tissues diagnosed up to 2008. We included both colon and rectal carcinomas based on the colorectal continuum model [12,32]. Patients were followed until death or the end of follow-up (June 30, 2014 for the NHS; and January 1, 2014 for the HPFS), whichever came first. We collected formalin-fixed paraffin-embedded (FFPE) tumour tissue blocks from hospitals throughout the U.S. where colorectal cancer patients had undergone surgical resection. A single pathologist (S.O.), blinded to other data, reviewed haematoxylin and eosin-stained tissue sections from all collected blocks and recorded pathological features. Tumour differentiation was categorised into well/moderate vs. poor (> 50% vs. ≤ 50% gland formation, respectively).
Informed consent was obtained from all study participants at enrollment. This study was approved by the institutional review boards of Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health (Boston, MA, USA), and those of participating registries as required.
Immunohistochemical evaluation of BECN1, MAP1LC3, SQSTM1, and CTSB
We constructed tissue microarrays of colorectal cancer cases with sufficient tissue materials, including up to four tumour cores from each case in one tissue microarray block [33]. Immunohistochemical studies of BECN1, MAP1LC3, and SQSTM1 were performed to evaluate autophagic activity of tumour cells. The BECN1 and MAP1LC3 expression have been positively associated with autophagic activity, while SQSTM1, degraded by autophagy, has been inversely associated with autophagic activity [16–18]. As secondary analysis, we evaluated tumour cell expression of CTSB (cathepsin B), which is one of the lysosomal enzymes involved in autophagic degradation and various other intracellular proteolytic processes [34].
For immunohistochemistry, deparaffinised tissue sections were heated in a microwave using a pressure cooker for 17 minutes in Antigen Retrieval Citra Solution, pH 6 (BioGenex Laboratories, San Ramon, CA, USA). Tissue sections were incubated with a dual endogenous enzyme block (Dako, Glostrup, Denmark) for 30 minutes and then serum-free protein block (Dako) for 10 minutes. Slides were incubated for 16 hours at 4 °C with a primary antibody against BECN1 [rabbit polyclonal antibody (ab55878), Abcam, Cambridge, MA, USA; dilution, 1:600], MAP1LC3A and MAP1LC3B [rabbit polyclonal antibody (ab58610), Abcam; dilution, 1:100], and SQSTM1 [mouse monoclonal antibody, clone 2C11, Abnova, Taipei, Taiwan; dilution, 1:1500]. The EnVision HRP-labeled polymer (Dako) was then applied to the sections for 30 minutes, followed by visualization with 3,3-diaminobenzidine and counterstaining with haematoxylin. Sections processed with the replacement of the primary antibody with Tris-buffered saline were used as negative controls.
The cytoplasmic expression levels (intensity) of BECN1, MAP1LC3, and SQSTM1 were recorded as low, intermediate, or high (Figure 1, Figure 2, and Figure 3). The immunostained slides were interpreted by the blinded study pathologists (S.A.K. for BECN1, Z.R.Q. for MAP1LC3, and Y.M. for SQSTM1). A subset of cases (N = 118 for BECN1, N = 111 for MAP1LC3, and N = 143 for SQSTM1) was scored independently by the second blinded pathologists (Y.M. for BECN1 and MAP1LC3, and A.d.S. for SQSTM1). The weighted κ values for the ratings of the two observers were 0.76 for BECN1, 0.69 for MAP1LC3, and 0.63 for SQSTM1 [20], indicating reasonably good interobserver agreement (all P < 0.001).
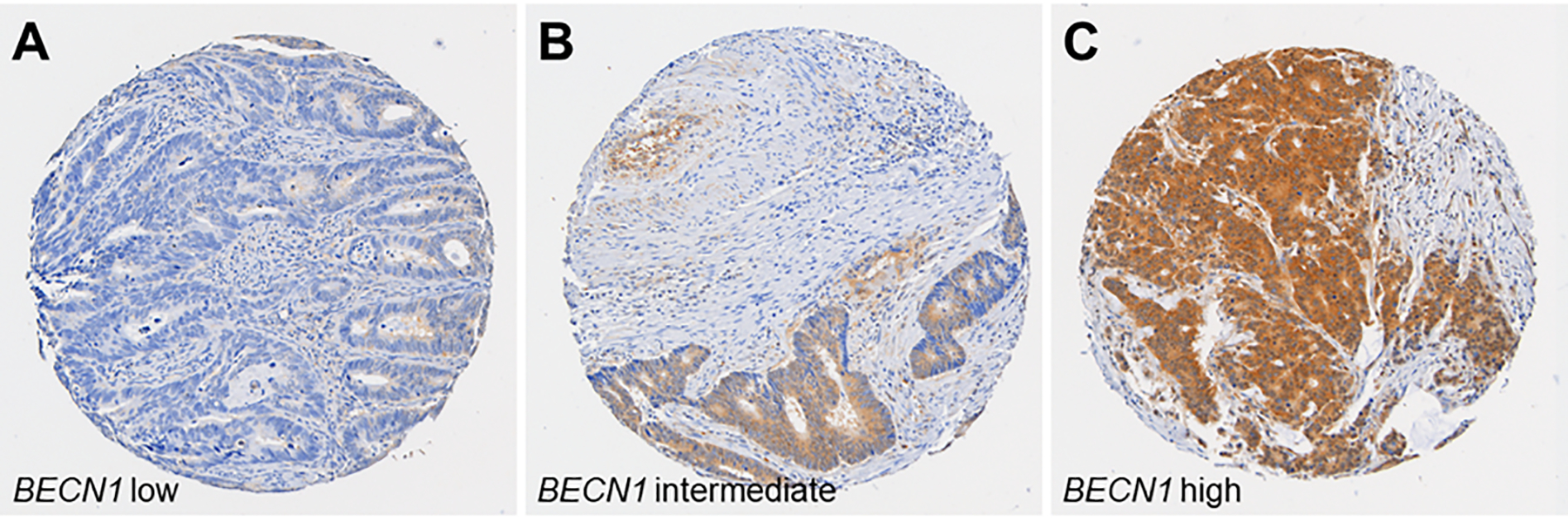
Figure 1.

Tumour BECN1 (beclin 1) expression in colorectal cancer. Tumour BECN1 expression was scored as low (A), intermediate (B), or high (C), according to cytoplasmic expression level of BECN1.
Figure 2.

Tumour MAP1LC3 (LC3) expression in colorectal cancer. Tumour MAP1LC3 expression was scored as low (A), intermediate (B), or high (C), according to cytoplasmic expression level of MAP1LC3.
Figure 3.
Tumour SQSTM1 (p62) expression in colorectal cancer. Tumour SQSTM1 expression was scored as low (A), intermediate (B), or high (C), according to cytoplasmic expression level of SQSTM1.
As previously described [35], we conducted immunohistochemical study of CTSB using an antibody against CTSB [goat polyclonal antibody (S-12), Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA; 1:1250 dilution]. The cytoplasmic expression of CTSB was recorded as absent, weak, moderate, or strong, as previously described [35].
Quantitative polymerase chain reaction (PCR) for Fusobacterium nucleatum and Bifidobacterium genus
Genomic DNA was extracted from archival FFPE tissue sections of colorectal cancer using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). As previously described [9,36], we performed a quantitative polymerase chain reaction (PCR) assay to measure the amount of F. nucleatum and Bifidobacterium genus DNA in the tumour tissue. Cases with any detectable F. nucleatum and Bifidobacterium genus DNA were categorised as low vs. high based on the median cut point amount of F. nucleatum or Bifidobacterium genus, while cases without detectable F. nucleatum and Bifidobacterium genus were categorised as negative.
Evaluation of tumour molecular characteristics
Tumour MSI status was analysed using PCR of 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487), and MSI-high was defined as presence of instability in ≥ 30% of the markers, as previously described [32]. Using bisulfite-treated DNA, methylation statuses of eight CpG island methylator phenotype (CIMP)-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) and long-interspersed nucleotide element-1 (LINE-1) was determined as previously described [32]. CIMP-high was defined as ≥ 6 methylated promoters of eight promoters, and CIMP-low/negative as 0–5 methylated promoters as previously described [32]. PCR and pyrosequencing were performed for KRAS (codons 12, 13, 61, and 146), BRAF (codon 600), and PIK3CA (exons 9 and 20), as previously described [32].
Statistical analysis
All statistical analyses were conducted using SAS software (version 9.4, SAS Institute, Cary, NC, USA), and all P values were two-sided. We used the two-sided α level of 0.005 [37]. Our primary hypothesis testing was an assessment of the association of tumour BECN1, MAP1LC3, and SQSTM1 expression levels (low, intermediate, and high; as an ordinal predictor variable) with the amount of F. nucleatum DNA (negative, low, and high; as an ordinal outcome variable). All other tests, including analyses of Bifidobacterium genus, assessment of stratum-specific risk estimates, and survival analyses, represented secondary analyses.
We performed multivariable ordinal logistic regression analyses to control for potential confounders. The multivariable ordinal logistic model initially included sex (female vs. male), age at diagnosis (continuous), year of diagnosis (continuous), family history of colorectal cancer in any first-degree relative (present vs. absent), tumour location (proximal colon vs. distal colon vs. rectum), MSI status (MSI-high vs. non-MSI-high), CIMP status (high vs. low/negative), LINE-1 methylation level (continuous), KRAS mutation (mutant vs. wild-type), BRAF mutation (mutant vs. wild-type), and PIK3CA mutation (mutant vs. wild-type). A backward elimination was conducted with a threshold P of 0.05 to select variables for the final models. Cases with missing data [family history of colorectal cancer in a first-degree relative (1.1%), tumour location (0.3%), MSI (2.8%), CIMP (8.3%), KRAS (2.9%), BRAF (2.5%), and PIK3CA mutation (7.9%)] were included in the majority category of a given categorical covariate to limit the degrees of freedom of the models. For the cases with missing data on LINE-1 methylation (2.5%), we assigned a separate indicator variable. We confirmed that excluding the cases with missing information in any of the covariates did not substantially alter results (data not shown). We assessed the proportional odds assumption in an ordinal logistic regression model, which was generally satisfied (P > 0.1).
To assess the association between ordinal categories of the amount of F. nucleatum DNA and other categorical variables (except for tumour differentiation and pT stage, for which Fisher’s exact test was performed), the chi-square test was performed. To compare continuous variables, an analysis of variance assuming equal variances was performed.
We estimated cumulative survival probabilities using the Kaplan-Meier method, and compared the differences between categories using the log-rank test. For analyses of colorectal cancer-specific mortality, deaths as a result from other causes were censored. To control for potential confounders, Cox proportional hazards regression analysis was performed and hazard ratio was calculated for mortality. The multivariable Cox proportional hazards regression models initially included tumour differentiation (well to moderate vs. poor), disease stage (I/II vs. III/IV) in addition to the same set of variables as in multivariable ordinal logistic regression analysis. A backward elimination was conducted with a threshold P of 0.05 to select variables for the final models. In secondary analyses, we assessed the statistical interaction between BECN1. MAP1LC3, and SQSTM1 expression (high vs. low/intermediate) and F. nucleatum status in colorectal cancer tissue (positive vs. negative) in relation to colorectal cancer-specific or overall mortality. We used the Wald test for the cross-product in multivariable-adjusted Cox proportional hazards regression models. We estimated hazard ratios for colorectal cancer and overall mortality comparing cases with high expression of BECN1, MAP1LC3, and SQSTM1 to cases with low/intermediate expression in strata of F. nucleatum status using re-parameterization of the interaction term in a single regression model [38]. Cases with missing data on tumour differentiation (0.1%) and disease stage (7.7%) were included in the majority category, and the other covariates were dealt as in the multivariable ordinal logistic regression models. The assumption of proportional hazards was generally satisfied using the assessment of a time-varying covariate, which was the cross-product of log-transformed survival time and the level of BECN1, MAP1LC3, and SQSTM1 expression in the whole population or in strata of F. nucleatum status (P > 0.08).
In ordinal logistic and Cox regression analyses, the inverse probability weighting (IPW) method was applied to reduce the potential bias due to the availability of tumour tissue [39–41]. Using the multivariable logistic regression model for the entire dataset of colorectal cancer cases (regardless of available tissue), we estimated the probability of the availability of tumour tissue, as previously described [36]. Each patient with complete data was weighted by the inverse probability. Weights greater than the 95th percentile were truncated and set to the value of the 95th percentile to reduce outlier effects [41]. We confirmed that results without weight truncation did not change substantially (data not shown). The logistic and Cox regression analyses without IPW yielded similar results to the IPW-adjusted model (Supplementary Table S2 and S8).
Results
Among the 724 colorectal cancer cases from the two prospective cohort studies, F. nucleatum DNA was detected using a quantitative PCR assay in 99 (14%) cases. Tumour BECN1 (beclin 1) expression levels were low, intermediate, and high in 102 (16%), 289 (46%), and 237 (38%) cases, respectively. Tumour MAP1LC3 expression levels were low, intermediate, and high in 201 (29%), 221 (32%), and 267 (39%) cases, respectively. Tumour SQSTM1 (p62) expression levels were low, intermediate, and high in 158 (23%), 309 (46%), and 207 (31%) cases, respectively. Table 1 and Supplementary Table S1 show clinical, pathological, and molecular features of colorectal cancer cases according to the amount of F. nucleatum DNA and Bifidobacterium genus DNA, respectively. As previously reported [9,11], a greater amount of F. nucleatum DNA was associated with poor tumour differentiation, higher pT stage, MSI-high status, and CIMP-high status (P < 0.005; with the α level of 0.005). Tumour BECN1 expression inversely correlated with the amount of F. nucleatum DNA (P < 0.001, by Spearman’s correlation test with the α level of 0.005), while tumour MAP1LC3 or SQSTM1 expression was not significantly correlated with the amount of F. nucleatum DNA (P > 0.061, by Spearman’s correlation test). There was no significant correlation between any of tumour BECN1, MAP1LC3, and SQSTM1 expression and the amount of Bifidobacterium genus DNA (P > 0.13).
Table 1.
Clinical, pathological, and molecular characteristics of colorectal cancer cases according to the amount of Fusobacterium nucleatum (F. nucleatum) DNA in tumour tissue
| Amount of F. nucleatum DNA in colorectal cancer tissue | |||||
|---|---|---|---|---|---|
| Characteristic* | All cases (N = 724) |
Negative (N = 625) |
Low (N = 50) |
High (N = 49) |
P value† |
| Sex | 0.14 | ||||
| Female (NHS) | 423 (58%) | 361 (58%) | 27 (54%) | 35 (71%) | |
| Male (HPFS) | 301 (42%) | 264 (42%) | 23 (46%) | 14 (29%) | |
| Mean age ± SD (years) | 69.3 ± 8.9 | 69.3 ± 8.9 | 70.5 ± 8.7 | 68.0 ± 8.5 | 0.38 |
| Year of diagnosis | 0.17 | ||||
| 1995 or before | 223 (31%) | 200 (32%) | 8 (16%) | 15 (31%) | |
| 1996–2000 | 222 (31%) | 191 (31%) | 16 (32%) | 15 (31%) | |
| 2001–2008 | 279 (39%) | 234 (37%) | 26 (52%) | 19 (39%) | |
| Family history of colorectal cancer in first-degree relative(s) | 0.37 | ||||
| Absent | 574 (80%) | 492 (79%) | 43 (88%) | 39 (81%) | |
| Present | 142 (20%) | 127 (21%) | 6 (12%) | 9 (19%) | |
| Tumour location | 0.020 | ||||
| Cecum | 129 (18%) | 102 (16%) | 11 (22%) | 16 (33%) | |
| Ascending to transverse colon | 244 (34%) | 209 (34%) | 18 (36%) | 17 (35%) | |
| Descending to sigmoid colon | 197 (27%) | 180 (29%) | 7 (14%) | 10 (20%) | |
| Rectum | 152 (21%) | 132 (21%) | 14 (28%) | 6 (12%) | |
| Tumour differentiation | <0.001 | ||||
| Well to moderate | 660 (91%) | 581 (93%) | 42 (86%) | 37 (76%) | |
| Poor | 63 (8.7%) | 44 (7.0%) | 7 (14%) | 12 (24%) | |
| pT stage | 0.003 | ||||
| pT1 | 55 (8.2%) | 52 (9.0%) | 1 (2.1%) | 2 (4.3%) | |
| pT2 | 135 (20%) | 124 (22%) | 9 (19%) | 2 (4.3%) | |
| pT3 | 443 (66%) | 373 (65%) | 33 (70%) | 37 (79%) | |
| pT4 | 37 (5.5%) | 27 (4.7%) | 4 (8.5%) | 6 (13%) | |
| pN stage | 0.29 | ||||
| pN0 | 396 (61%) | 349 (62%) | 23 (51%) | 24 (53%) | |
| pN1 | 158 (24%) | 132 (24%) | 15 (33%) | 11 (24%) | |
| pN2 | 95 (15%) | 78 (14%) | 7 (16%) | 10 (22%) | |
| M stage | 0.11 | ||||
| M0 | 572 (86%) | 493 (86%) | 43 (93%) | 36 (78%) | |
| M1 | 96 (14%) | 83 (14%) | 3 (6.5%) | 10 (22%) | |
| AJCC disease stage | 0.008 | ||||
| I | 150 (22%) | 141 (24%) | 6 (13%) | 3 (6.5%) | |
| II | 223 (33%) | 187 (32%) | 16 (35%) | 20 (43%) | |
| III | 199 (30%) | 165 (29%) | 21 (46%) | 13 (28%) | |
| IV | 96 (14%) | 83 (14%) | 3 (6.5%) | 10 (22%) | |
| MSI status | < 0.001 | ||||
| Non-MSI-high | 588 (84%) | 527 (87%) | 32 (67%) | 29 (59%) | |
| MSI-high | 116 (16%) | 80 (13%) | 16 (33%) | 20 (41%) | |
| CIMP status | < 0.001 | ||||
| Low/negative | 546 (82%) | 484 (84%) | 35 (78%) | 27 (59%) | |
| High | 118 (18%) | 89 (16%) | 10 (22%) | 19 (41%) | |
| Mean LINE-1 methylation level ± SD (%) | 62.4 ± 9.6 | 62.1 ± 9.6 | 62.7 ± 10.0 | 65.0 ± 9.2 | 0.12 |
| KRAS mutation | 0.16 | ||||
| Wild-type | 414 (59%) | 363 (60%) | 21 (46%) | 30 (62%) | |
| Mutant | 289 (41%) | 246 (40%) | 25 (54%) | 18 (38%) | |
| BRAF mutation | 0.009 | ||||
| Wild-type | 597 (85%) | 523 (86%) | 40 (83%) | 34 (69%) | |
| Mutant | 109 (15%) | 86 (14%) | 8 (17%) | 15 (31%) | |
| PIK3CA mutation | 0.66 | ||||
| Wild-type | 566 (85%) | 493 (85%) | 36 (84%) | 37 (80%) | |
| Mutant | 101 (15%) | 85 (15%) | 7 (16%) | 9 (20%) | |
| BECN1 (beclin 1) expression‡ | < 0.001 | ||||
| Low | 102 (16%) | 77 (14%) | 14 (31%) | 11 (28%) | |
| Intermediate | 289 (46%) | 251 (46%) | 20 (44%) | 18 (46%) | |
| High | 237 (38%) | 216 (40%) | 11 (24%) | 10 (26%) | |
| MAP1LC3 (LC3) expression‡ | 0.062 | ||||
| Low | 201 (29%) | 163 (27%) | 17 (36%) | 21 (46%) | |
| Intermediate | 221 (32%) | 199 (33%) | 12 (26%) | 10 (22%) | |
| High | 267 (39%) | 234 (39%) | 18 (38%) | 15 (33%) | |
| SQSTM1 (p62) expression‡ | 0.83 | ||||
| Low | 158 (23%) | 131 (22%) | 15 (33%) | 12 (28%) | |
| Intermediate | 309 (46%) | 278 (48%) | 18 (39%) | 13 (30%) | |
| High | 207 (31%) | 176 (30%) | 13 (28%) | 18 (42%) | |
Percentage indicates the proportion of patients with a specific clinical, pathologic, or molecular characteristic among all patients or in strata of the amount of F. nucleatum DNA.
To compare categorical data between subgroups classified by the amount of F. nucleatum DNA (except for tumour differentiation and pT stage, for which the Fisher’s exact test was performed), the chi-square test was performed, unless otherwise noted. To compare continuous variables, an analysis of variance was performed.
To assess associations between the amount of F. nucleatum DNA in colorectal cancer tissue (negative, low, and high) and tumour BECN1, MAP1LC3, and SQSTM1 expression level (low, intermediate, and high) and, the Spearman’s correlation test was performed.
Abbreviations: AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-up Study; LINE-1, long-interspersed nucleotide element-1; MSI, microsatellite instability; NHS, Nurses’ Health Study; SD, standard deviation.
In our primary hypothesis testing, we used an ordinal logistic regression analysis to assess the association of tumour BECN1, MAP1LC3, or SQSTM1 expression levels with the amount of F. nucleatum DNA (Table 2, and Supplementary Table S3, S4, and S5). In the multivariable analyses, compared with BECN1-low cases, multivariable odd ratios (ORs) for the amount of F. nucleatum DNA were 0.54 [95% confidence interval (CI), 0.29–0.99] for BECN1-intermediate cases and 0.31 (95% CI, 0.16–0.60) for BECN1-high cases (Ptrend < 0.001; with the α level of 0.005). We did not observe a statistically significant association of tumour MAP1LC3 or SQSTM1 expression levels with the amount of F. nucleatum DNA in colorectal cancer (Ptrend > 0.061; with the α level of 0.005).
Table 2.
Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome)
| Amount of F. nucleatum DNA in colorectal cancer tissue | ||
|---|---|---|
| Univariable OR (95% CI)* |
Multivariable OR (95% CI)*,† |
|
| Tumour BECN1 expression (N = 628) | ||
| Low | 1 (referent) | 1 (referent) |
| Intermediate | 0.48 (0.26–0.88) | 0.54 (0.29–0.99) |
| High | 0.27 (0.14–0.52) | 0.31 (0.16–0.60) |
| Ptrend‡ | < 0.001 | < 0.001 |
| Tumour MAP1LC3 expression (N = 689) | ||
| Low | 1 (referent) | 1 (referent) |
| Intermediate | 0.46 (0.25–0.83) | 0.46 (0.25–0.85) |
| High | 0.55 (0.32–0.95) | 0.57 (0.33–0.99) |
| Ptrend‡ | 0.043 | 0.061 |
| Tumour SQSTM1 expression (N = 674) | ||
| Low | 1 (referent) | 1 (referent) |
| Intermediate | 0.54 (0.30–0.97) | 0.55 (0.30–1.01) |
| High | 0.88 (0.48–1.60) | 0.89 (0.48–1.65) |
| Ptrend‡ | 0.75 | 0.79 |
IPW was applied to reduce a bias due to the availability of tumour tissue after cancer diagnosis (see “Statistical analysis” subsection for details).
The multivariable ordinal logistic regression model initially included age, sex, year of diagnosis, family history of colorectal cancer, tumour location, microsatellite instability, CpG island methylator phenotype, long-interspersed nucleotide element-1 methylation level, and KRAS, BRAF, and PIK3CA mutations. A backward elimination with a threshold P of 0.05 was used to select variables for the final model. The variables which remained in the final models are shown in Supplementary Table S3, S4, and S5.
Ptrend was calculated by the linear trend across the ordinal categories of tumour BECN1, MAP1LC3, and SQSTM1 expression level (low, intermediate, and high, as an ordinal predictor variable) in the IPW-adjusted ordinal logistic regression model for the amount of F. nucleatum DNA (negative, low, and high, as an ordinal outcome variable).
Abbreviations: CI, confidence interval; IPW, inverse probability weighting; OR, odds ratio.
In secondary analysis using a subset of cases with available CTSB data, we did not observe a statistically significant association of tumour CTSB expression with the amount of F. nucleatum DNA (Ptrend = 0.74; with the α level of 0.005) (Supplementary Table S6 and S7).
As secondary analyses to assess the prognostic association of BECN1, MAP1LC3, and SQSTM1 expression, we conducted Kaplan-Meier analyses and Cox regression analyses, and did not observe a significant association of BECN1, MAP1LC3, or SQSTM1 levels with colorectal cancer-specific or overall mortality (Ptrend > 0.10; with the α level of 0.005) (Table 3, Supplementary Table S8, and Supplementary Figure S1, S2, and S3). We further examined whether prognostic association of BECN1, MAP1LC3, and SQSTM1 expression in strata of F. nucleatum status and did not observe a significant interaction between BECN1, MAP1LC3, or SQSTM1 expression and of F. nucleatum status in relation to colorectal cancer-specific or overall mortality (Pinteraction > 0.25; with the α level of 0.005) (Supplementary Table S9).
Table 3.
Tumour BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression level and patient survival with inverse probability weighting (IPW)
| Colorectal cancer-specific survival | Overall survival | ||||||
|---|---|---|---|---|---|---|---|
| No. of cases | No. of events | Univariable HR (95% CI)* |
Multivariable HR (95% CI)*,† |
No. of events | Univariable HR (95% CI)* |
Multivariable HR (95% CI)*,† |
|
| Tumour BECN1 expression | |||||||
| Low | 98 | 29 | 1 (referent) | 1 (referent) | 60 | 1 (referent) | 1 (referent) |
| Intermediate | 283 | 79 | 0.96 (0.61–1.51) | 0.80 (0.50–1.27) | 163 | 0.94 (0.66–1.34) | 0.89 (0.62–1.26) |
| High | 233 | 66 | 0.99 (0.63–1.57) | 0.74 (0.45–1.20) | 143 | 0.95 (0.67–1.36) | 0.85 (0.59–1.22) |
| Ptrend‡ | 0.99 | 0.26 | 0.85 | 0.39 | |||
| Tumour MAP1LC3 expression | |||||||
| Low | 196 | 47 | 1 (referent) | 1 (referent) | 103 | 1 (referent) | 1 (referent) |
| Intermediate | 218 | 65 | 1.33 (0.89–1.99) | 1.49 (1.00–2.22) | 136 | 1.18 (0.88–1.57) | 1.19 (0.90–1.58) |
| High | 261 | 77 | 1.30 (0.89–1.91) | 1.07 (0.72–1.58) | 164 | 1.18 (0.91–1.53) | 1.11 (0.84–1.45) |
| Ptrend‡ | 0.20 | 0.94 | 0.25 | 0.63 | |||
| Tumour SQSTM1 expression | |||||||
| Low | 152 | 50 | 1 (referent) | 1 (referent) | 98 | 1 (referent) | 1 (referent) |
| Intermediate | 304 | 83 | 0.80 (0.55–1.16) | 0.78 (0.54–1.12) | 183 | 0.97 (0.73–1.29) | 0.94 (0.71–1.23) |
| High | 204 | 54 | 0.82 (0.55–1.24) | 0.71 (0.47–1.08) | 119 | 0.94 (0.70–1.26) | 0.86 (0.63–1.17) |
| Ptrend‡ | 0.38 | 0.10 | 0.66 | 0.32 | |||
IPW was applied to reduce a bias due to the availability of tumour tissue after cancer diagnosis (see “Statistical analysis” subsection for details).
The multivariable Cox regression model initially included sex, age, year of diagnosis, family history of colorectal cancer, tumour location, tumour differentiation, disease stage, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and long-interspersed nucleotide element-1 methylation level. A backward elimination with a threshold P of 0.05 was used to select variables for the final models.
Ptrend value was calculated across the ordinal categories of tumour BECN1, MAP1LC3, and SQSTM1 expression levels (low, intermediate, and high) in the IPW-adjusted Cox regression model.
Abbreviations: CI, confidence interval; HR, hazard ratio; IPW, inverse probability weighting.
Discussion
Using two large prospective cohort studies in the U.S., we tested the hypothesis that autophagic activity of tumour cells might influence the amount of F. nucleatum in colorectal cancer tissue. Notably, we found an inverse association of tumour BECN1 expression level with the amount of F. nucleatum, but not with the amount of Bifidobacterium genus in colorectal cancer tissue. Our findings suggest a possible role of tumour autophagic activity in the elimination of a specific microorganism within the colorectal cancer microenvironment.
There is growing interest in the role of the gut microbiota in cancer biology, especially F. nucleatum in colorectal cancer. A translational study has shown that F. nucleatum is associated with colorectal cancer recurrence through the promotion of chemoresistance by engaging autophagic activity [42]. F. nucleatum binds host epithelial CDH1 (E-cadherin) via the fusobacterial adhesin FadA and invades epithelial cells, resulting in the promotion of inflammation and tumour cell growth in transformed cells [6,43,44]. Moreover, invasive F. nucleatum is persistently associated with distant metastases from primary colorectal cancers [45]. In contrast, the members of Bifidobacterium genus have been considered to inhibit colorectal carcinogenesis through the prevention of enteropathogenic infection and the inhibition of secondary bile acid production [46–48]. Our previous population-based study has shown that intratumoural Bifidobacterium genus may reflect loss of intestinal barrier function in poorly differentiated colorectal cancer [36]. These distinct characteristics of F. nucleatum and Bifidobacterium genus are supported by our results of differing associations with autophagic activity. Our finding suggests the autophagy may target only highly virulent bacteria which can cause stress to the tumour cells, while autophagy is not associated with the presence of Bifidobacteria genus. From an immunological point of view, F. nucleatum may suppress the adaptive immune responses and inhibit T-cell- and natural killer cell-mediated immune response against colorectal cancer through the immune cell receptor TIGIT [6,49]. In line with these studies, our previous population-based studies have shown an inverse association of F. nucleatum in colorectal cancer tissue with CD3+ pan-T cell density and adaptive anti-tumour immune response in MSI-high tumours [9,50]. Hence, the elimination of F. nucleatum may potentiate the effect of current front-line immunotherapeutic treatments.
Invasion of bacterial pathogens into epithelial cells triggers various innate immune responses, such as proinflammatory signaling and cell-autonomous restriction of bacterial growth. Among these host defence mechanisms, autophagy plays a critical role in the targeting and degradation of intracellular bacteria [24,28,51]. The autophagic machinery targets intracellular bacterium and promotes protective immune and stress responses, as the invasion of bacterium triggers the rapid induction of the intracellular starvation state [52]. There are three main autophagic mechanisms that provide a series of barriers against invading microorganisms. The first anti-microbial function is xenophagy, which is the uptake of intracellular microorganisms into double membrane autophagosomes regulated by BECN1 and MAP1LC3 [24,25]. The second is MAP1LC3-associated phagocytosis which promotes phagosome fusion with lysosomes [26]. Finally, a group of autophagic adaptors, known as SQSTM1-like receptors, are involved in the elimination of microorganisms from the cytoplasm [27]. In this study, tumour BECN1 expression level was inversely associated with the amount of F. nucleatum and tumour MAP1LC3 expression had the same trend as that of BECN1, however there was no correlation of tumour SQSTM1 expression with the amount of F. nucleatum. Potential reason for the difference might be caused by distinct features of autophagy markers. BECN1 and MAP1LC3 are involved in the same phase of autophagy (i.e. autophagosome formation), while SQSTM1 contributes to a different phase and also regulates transcriptional factors, suggesting that F. nucleatum might be eliminated through xenophagy and MAP1LC3-phagocytosis [17,18]. Autophagic machineries are not only required to eliminate microorganisms but also contribute to major histocompatibility complex class II presentation of their derived antigens that stimulate the CD4+ T-cell adaptive response [22,23]. Moreover, autophagy is likely to influence the efficacy of immune checkpoint blockade therapy as well as gut microbiota [4,5,16,53]. Collectively, a better understanding of autophagy in relation to gut microbiota would have considerable therapeutic implications by its connection to tumour cells, host cells, and microorganisms in the tumour microenvironment. We did not observe significant association of tumour BECN1, MAP1LC3, and SQSTM1 expression levels with colorectal cancer mortality. Prognostic roles of tumour BECN1, MAP1LC3, and SQSTM1 in colorectal cancer remain controversial, possibly due to differences in study populations, designs and methods as well as a multifaceted nature of autophagic process [54–61].
We acknowledge potential limitations of the current study. First, we designed a cross-sectional study, by which we cannot exclude the possibility of reverse causation. While there is a possibility that F. nucleatum might modify autophagic activity of tumour cells, our specific hypothesis was based on several lines of experimental evidence indicating that autophagic activity eliminates microorganisms [22–28]. Second, our study only concentrated on a limited number of microbial species and genus. Accumulating evidence suggests that various species of microbiota and dysregulated microbial communities are involved in carcinogenesis and anti-tumour immune response [1,3–5]. Although F. nucleatum has garnered considerable attention as a potential cancer promoting microbe, other bacteria, such as Bacteroides fragilis, colibactin-producing Escherichia coli, and Peptostreptococcus stomatis, have been reported to be enriched in colorectal cancer and to be associated with colorecral cancer carcinogenesis [62,63]. Therefore, more comprehensive analyses including 16S rRNA gene sequencing and whole metagenomic shotgun sequencing would provide further insight into the relationship between microbiota and autophagy. Third, we evaluated the autophagic activity using a limited number of autophagic markers. Autophagy is a complex multistep process where more than 30 autophagy-related genes have been identified to make up the core machinery [16–19]. Although BECN1, MAP1LC3, and SQSTM1 have been widely used as immunohistochemistry-based autophagy markers in human FFPE tissues [54–61], a simultaneous multimarker evaluation of other components of the autophagic pathway might further enlighten the interactions between autophagy and microbiota. Finally, we used the quantitative PCR assay for F. nucleatum and Bifidobacterium genus in FFPE tissue specimens; therefore, the detection rates might have been influenced by histopathology procedures and storage conditions. However, our previous validation study using the quantitative PCR assay showed a good concordance in detection of F. nucleatum in paired FFPE and frozen tissue specimens [9]. We also observed high linearity and reproducibility of F. nucleatum and Bifidobacterium genus measurements in FFPE tissue specimens [9,36].
The current study has notable strengths, including the utilisation of a molecular pathological epidemiology database [14,15,64] derived from two U.S.-based large prospective cohort studies. The integrated data on autophagic activity, tumour molecular characteristics, pathological findings, and microbial features enabled us to comprehensively examine possible roles of autophagy in relation to microorganisms in colorectal cancers. Importantly, our study population was derived from a large number of hospitals located throughout the U.S., which increases the generalisability of our findings. Nevertheless, our findings need to be validated in independent studies.
In conclusion, we have shown an inverse association of tumour BECN1 expression level with the amount of F. nucleatum in colorectal cancer tissue allowing us to derive a possible role of autophagy in the elimination of invasive microorganisms such as F. nucleatum from the tumour microenvironment. Our population-based data, if validated, may inform future translational research on anti-microorganism therapy including treatment strategies targeting autophagy in colorectal cancer.
Supplementary Material
Supplementary material online
Supplementary Table S1. Clinical, pathological, and molecular characteristics of colorectal cancer cases according to the amount of Bifidobacterium genus DNA in tumour tissue
Supplementary Table S2. Ordinal logistic regression analysis to assess the association of tumour BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome) without inverse probability weighting
Supplementary Table S3. Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour BECN1 (beclin 1) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome) (Final model)
Supplementary Table S4. Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour MAP1LC3 (LC3) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome) (Final model)
Supplementary Table S5. Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour SQSTM1 (p62) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome) (Final model)
Supplementary Table S6. Distribution of colorectal cancer cases according to tumour CTSB (cathepsin B) expression level, and the amount of Fusobacterium nucleatum (F. nucleatum) or Bifidobacterium genus DNA in tumour tissue
Supplementary Table S7. Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour CTSB (cathepsin B) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome)
Supplementary Table S8. Tumour BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression level and patient survival without inverse probability weighting
Supplementary Table S9. Tumour BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression level and patient survival in strata of Fusobacterium nucleatum (F. nucleatum) status with inverse probability weighting (IPW)
Supplementary Figure S1. Inverse probability weighting (IPW)-adjusted Kaplan-Meier survival curves of colorectal cancer patients according to the levels of tumour BECN1 (beclin 1) expression.
Supplementary Figure S2. Inverse probability weighting (IPW)-adjusted Kaplan-Meier survival curves of colorectal cancer patients according to the levels of tumour MAP1LC3 (LC3) expression.
Supplementary Figure S3. Inverse probability weighting (IPW)-adjusted Kaplan-Meier survival curves of colorectal cancer patients according to the levels of tumour SQSTM1 (p62) expression.
Acknowledgments
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Funding: This work was supported by U.S. National Institutes of Health (NIH) grants (P01 CA87969 to M.J. Stampfer; UM1 CA186107 to M.J. Stampfer; P01 CA55075 to W.C. Willett; UM1 CA167552 to W.C. Willett; U01 CA167552 to W.C. Willett and L.A. Mucci; P50 CA127003 to C.S.F.; R01 CA118553 to C.S.F.; R01 CA169141 to C.S.F.; R01 CA137178 to A.T.C.; K24 DK098311 to A.T.C.; R35 CA197735 to S.O.; R01 CA151993 to S.O.; K07 CA190673 to R.N.; and K07 CA188126 to X.Z.); by Cancer Research UK’s Grand Challenge Initiative (C10674/A27140 to W.S.G., M.G., C.H., and S.O.); by Nodal Award (2016-02) from the Dana-Farber Harvard Cancer Center (to S.O.); by the Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17 to C.S.F. and M.G.), administered by the American Association for Cancer Research, a scientific partner of SU2C; and by grants from the Project P Fund, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance and SU2C. K.H. was supported by fellowship grants from the Uehara Memorial Foundation and the Mitsukoshi Health and Welfare Foundation. K.K. was supported by grants from Overseas Research Fellowship (JP2017-0775) from Japan Society for the Promotion of Science and JSPS Fujita Memorial Fund for Medical Research. K.A. was supported by a grant from Overseas Research Fellowship (JP2018-60083) from Japan Society for the Promotion of Science. J.B. was supported by a grant from the Australia Awards-Endeavour Scholarships and Fellowships Program. K.F. was supported by a fellowship grant from the Uehara Memorial Foundation. M.G. is supported by an ASCO Conquer Cancer Foundation Career Development Award. A.T.C. is a Stuart and Suzanne Steele MGH Research Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- AJCC
American Joint Committee on Cancer
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- FFPE
formalin-fixed paraffin-embedded
- HPFS
Health Professionals Follow-up Study
- IPW
inverse probability weighting
- LINE-1
long-interspersed nucleotide element-1
- MSI
microsatellite instability
- NHS
Nurses’ Health Study
- OR
odds ratio
- PCR
polymerase chain reaction
- SD
standard deviation
Footnotes
Disclosure of potential conflicts of interest: A.T.C. previously served as a consultant for Bayer Healthcare and Pfizer Inc.. R.N. is currently employed by Pfizer Inc.; she contributed to this study before she became an employee of Pfizer Inc. M.G. was on an advisory board for AstraZeneca and receives research funding from Bristol-Myers Squibb. C.S.F. previously served as a consultant for Agios, Bain Capital, Bayer, Celgene, Dicerna, Five Prime Therapeutics, Gilead Sciences, Eli Lilly, Entrinsic Health, Genentech, KEW, Merck, Merrimack Pharmaceuticals, Pfizer Inc, Sanofi, Taiho, and Unum Therapeutics; C.S.F. also serves as a Director for CytomX Therapeutics and owns unexercised stock options for CytomX and Entrinsic Health. This study was not funded by any of these commercial entities. No other conflicts of interest exist. The other authors declare that they have no conflicts of interest.
Use of Standardised official symbols
We use HUGO (Human Genome Organisation)-approved official symbols (or root symbols) for genes and gene products, including BECN1, BRAF, CACNA1G, CD3, CD4, CDKN2A, CDH1, CRABP1, CTSB, IGF2, KRAS, MAP1LC3A, MAP1LC3B, MLH1, NEUROG1, PIK3CA, RUNX3, SOCS1, SQSTM1, and TIGIT; all of which are described at www.genenames.org. The official symbols are italicised to differentiate from non-italicised colloquial names that are used along with the official symbols. This format enables readers to familiarise themselves with the official symbols for genes and gene products together with common colloquial names.
References
- 1.Tilg H, Adolph TE, Gerner RR, et al. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018; 33: 954–964. [DOI] [PubMed] [Google Scholar]
- 2.Goodman B, Gardner H. The microbiome and cancer. J Pathol 2018; 244: 667–676. [DOI] [PubMed] [Google Scholar]
- 3.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016; 16: 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018; 359: 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018; 359: 91–97. [DOI] [PubMed] [Google Scholar]
- 6.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013; 14: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012; 22: 292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012; 22: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol 2015; 1: 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale VL, Jeraldo P, Chen J, et al. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome Med 2018; 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016; 65: 1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mima K, Cao Y, Chan AT, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol 2016; 7: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajpoot M, Sharma AK, Sharma A, et al. Understanding the microbiome: Emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin Cancer Biol 2018; 52: 1–8. [DOI] [PubMed] [Google Scholar]
- 14.Ogino S, Nowak JA, Hamada T, et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018; 67: 1168–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada T, Nowak JA, Milner DA Jr., et al. Integration of microbiology, molecular pathology, and epidemiology: a new paradigm to explore the pathogenesis of microbiome-driven neoplasms. J Pathol 2019; 247: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov 2019; 9: 1167–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X, Overholtzer M, Thompson CB. Autophagy in cellular metabolism and cancer. J Clin Invest 2015; 125: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B, Wen X, Cheng Y. Survival or death: disequilibrating the oncogenic and tumor suppressive autophagy in cancer. Cell Death Dis 2013; 4: e892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol 2019; 247: 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosumi K, Masugi Y, Yang J, et al. Tumor SQSTM1 (p62) expression and T cells in colorectal cancer. Oncoimmunology 2017; 6: e1284720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy J, Cacheux W, Bara MA, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol 2015; 17: 1062–1073. [DOI] [PubMed] [Google Scholar]
- 22.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell 2014; 54: 224–233. [DOI] [PubMed] [Google Scholar]
- 23.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013; 13: 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mimouna S, Bazin M, Mograbi B, et al. HIF1A regulates xenophagic degradation of adherent and invasive Escherichia coli (AIEC). Autophagy 2014; 10: 2333–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubber A, Kubori T, Coban C, et al. Bacterial secretion system skews the fate of Legionella-containing vacuoles towards LC3-associated phagocytosis. Sci Rep 2017; 7: 44795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heath RJ, Goel G, Baxt LA, et al. RNF166 Determines Recruitment of Adaptor Proteins during Antibacterial Autophagy. Cell Rep 2016; 17: 2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Hu W, Cho CH, et al. Reduced lysosomal clearance of autophagosomes promotes survival and colonization of Helicobacter pylori. J Pathol 2018; 244: 432–444. [DOI] [PubMed] [Google Scholar]
- 29.Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 2011; 60: 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol 2017; 14: 235–246. [DOI] [PubMed] [Google Scholar]
- 31.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 2013; 369: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012; 61: 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med 2007; 356: 2131–2142. [DOI] [PubMed] [Google Scholar]
- 34.Hu W, Chan H, Lu L, et al. Autophagy in intracellular bacterial infection. Semin Cell Dev Biol 2019. [DOI] [PubMed] [Google Scholar]
- 35.Chan AT, Baba Y, Shima K, et al. Cathepsin B expression and survival in colon cancer: implications for molecular detection of neoplasia. Cancer Epidemiol Biomarkers Prev 2010; 19: 2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosumi K, Hamada T, Koh H, et al. The Amount of Bifidobacterium Genus in Colorectal Carcinoma Tissue in Relation to Tumor Characteristics and Clinical Outcome. Am J Pathol 2018; 188: 2839–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamin DJ, Berger JO, Johannesson M, et al. Redefine statistical significance. Nat Hum Behav 2018; 2: 6–10. [DOI] [PubMed] [Google Scholar]
- 38.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One 2008; 3: e3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamada T, Cao Y, Qian ZR, et al. Aspirin Use and Colorectal Cancer Survival According to Tumor CD274 (Programmed Cell Death 1 Ligand 1) Expression Status. J Clin Oncol 2017; 35: 1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Nevo D, Nishihara R, et al. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol 2018; 33: 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seaman SR, White IR. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res 2013; 22: 278–295. [DOI] [PubMed] [Google Scholar]
- 42.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017; 170: 548–563 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abed J, Emgard JE, Zamir G, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016; 20: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 2013; 14: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017; 358: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011; 469: 543–547. [DOI] [PubMed] [Google Scholar]
- 47.Laforest-Lapointe I, Arrieta MC. Patterns of Early-Life Gut Microbial Colonization during Human Immune Development: An Ecological Perspective. Front Immunol 2017; 8: 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ubeda C, Djukovic A, Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunology 2017; 6: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015; 42: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamada T, Zhang X, Mima K, et al. Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol Res 2018; 6: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang J, Canadien V, Lam GY, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A 2009; 106: 6226–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tattoli I, Sorbara MT, Vuckovic D, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 2012; 11: 563–575. [DOI] [PubMed] [Google Scholar]
- 53.Noman MZ, Paggetti J, Moussay E, et al. Driving Natural Killer cells toward the melanoma tumor battlefield: Autophagy as a valuable therapeutic target. Oncoimmunology 2018; 7: e1452583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmitz KJ, Ademi C, Bertram S, et al. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, Beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J Surg Oncol 2016; 14: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang M, Zhao H, Guo L, et al. Autophagy-based survival prognosis in human colorectal carcinoma. Oncotarget 2015; 6: 7084–7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han Y, Xue XF, Shen HG, et al. Prognostic significance of Beclin-1 expression in colorectal cancer: a meta-analysis. Asian Pac J Cancer Prev 2014; 15: 4583–4587. [DOI] [PubMed] [Google Scholar]
- 57.Park JM, Huang S, Wu TT, et al. Prognostic impact of Beclin 1, p62/sequestosome 1 and LC3 protein expression in colon carcinomas from patients receiving 5-fluorouracil as adjuvant chemotherapy. Cancer Biol Ther 2013; 14: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo GF, Jiang WQ, Zhang B, et al. Autophagy-related proteins Beclin-1 and LC3 predict cetuximab efficacy in advanced colorectal cancer. World J Gastroenterol 2011; 17: 4779–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giatromanolaki A, Koukourakis MI, Harris AL, et al. Prognostic relevance of light chain 3 (LC3A) autophagy patterns in colorectal adenocarcinomas. J Clin Pathol 2010; 63: 867–872. [DOI] [PubMed] [Google Scholar]
- 60.Li BX, Li CY, Peng RQ, et al. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy 2009; 5: 303–306. [DOI] [PubMed] [Google Scholar]
- 61.Koukourakis MI, Giatromanolaki A, Sivridis E, et al. Beclin 1 over- and underexpression in colorectal cancer: distinct patterns relate to prognosis and tumour hypoxia. Br J Cancer 2010; 103: 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med 2019; 25: 968–976. [DOI] [PubMed] [Google Scholar]
- 63.Garrett WS. The gut microbiota and colon cancer. Science 2019; 364: 1133–1135. [DOI] [PubMed] [Google Scholar]
- 64.Ogino S, Nowak JA, Hamada T, et al. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol 2019; 14: 83–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material online
Supplementary Table S1. Clinical, pathological, and molecular characteristics of colorectal cancer cases according to the amount of Bifidobacterium genus DNA in tumour tissue
Supplementary Table S2. Ordinal logistic regression analysis to assess the association of tumour BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome) without inverse probability weighting
Supplementary Table S3. Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour BECN1 (beclin 1) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome) (Final model)
Supplementary Table S4. Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour MAP1LC3 (LC3) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome) (Final model)
Supplementary Table S5. Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour SQSTM1 (p62) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome) (Final model)
Supplementary Table S6. Distribution of colorectal cancer cases according to tumour CTSB (cathepsin B) expression level, and the amount of Fusobacterium nucleatum (F. nucleatum) or Bifidobacterium genus DNA in tumour tissue
Supplementary Table S7. Inverse probability weighting (IPW)-adjusted ordinal logistic regression analysis to assess the association of tumour CTSB (cathepsin B) expression level (Predictor) with the amount of Fusobacterium nucleatum (F. nucleatum) DNA (Outcome)
Supplementary Table S8. Tumour BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression level and patient survival without inverse probability weighting
Supplementary Table S9. Tumour BECN1 (beclin 1), MAP1LC3 (LC3), and SQSTM1 (p62) expression level and patient survival in strata of Fusobacterium nucleatum (F. nucleatum) status with inverse probability weighting (IPW)
Supplementary Figure S1. Inverse probability weighting (IPW)-adjusted Kaplan-Meier survival curves of colorectal cancer patients according to the levels of tumour BECN1 (beclin 1) expression.
Supplementary Figure S2. Inverse probability weighting (IPW)-adjusted Kaplan-Meier survival curves of colorectal cancer patients according to the levels of tumour MAP1LC3 (LC3) expression.
Supplementary Figure S3. Inverse probability weighting (IPW)-adjusted Kaplan-Meier survival curves of colorectal cancer patients according to the levels of tumour SQSTM1 (p62) expression.


