Abstract
Shoot fly (Atherigona naqvii) is one of the major insects affecting spring maize in North India and can cause yield loss up to 60 per cent. The genetics of insect resistance is complex as influenced by genotypic background, insect population and climatic conditions. Therefore, quantitative trait loci (QTL) mapping is a highly effective approach for studying genetically complex forms of insect resistance. The objective of the present study was to dissect the genetic basis of resistance and identification of genomic regions associated with shoot fly resistance. A total of 107 F2 population derived from the cross CM143 (resistant) x CM144 (susceptible) was genotyped with 120 SSR markers. Phenotypic data were recorded on replicated F2:3 progenies for various component traits imparting resistance to shoot fly at different time intervals. Resistance to shoot fly was observed to be under polygenic control as evidenced by the identification of 19 putative QTLs governed by overdominance to partial dominance and additive gene actions. The major QTLs conditioning shoot fly resistance viz., qDH9.1 (deadheart) and qEC9.1 (oviposition) explaining 15.03 and 18.89 per cent phenotypic variance, respectively were colocalized on chromosome 9. These QTLs are syntenic to regions of chromosome 10 of sorghum which were also accounted for deadheart and oviposition suggesting that the same gene block may be responsible for shoot fly resistance. The candidate genes such as cysteine protease, subtilisin-chymotrypsin inhibitor, cytochrome P450 involved in synthesis of alleochemicals, receptor kinases, glossy15 and ubiquitin-proteasome degradation pathway were identified within the predicted QTL regions. This is the first reported mapping of QTLs conferring resistance to shoot fly in maize, and the markers identified here will be a valuable resource for developing elite maize cultivars with resistance to shoot fly.
1. Introduction
Globally maize is the third most important cereal crop after wheat and rice in terms of area and production having diversified uses as food, feed and a range of industrial products. In India, it was cultivated on an area of 9.63 million hectares with annual production of 25.90 million metric tonnes and average productivity of 2.69 metric tonnes per hectare during 2018 (www.indiastat.com). However, maize production is limited by insect pests [1] at different crop growth stages, thus hampering with the realization of yield potential. The continuous planting of maize throughout the year has led to increased incidence of shoot fly (Atherigona species) at seedling stage [2]. Sixteen shoot fly species have been reported on maize in Africa and Asia [3], of which A. naqvii Steyskal (Muscidae: Diptera) is most prevalent in North India [4] and reported to cause a loss of about 28–45 per cent in grain yield during spring season in the Indian Punjab [2].
The A. naqvii adult female lays eggs singly or in small groups on the stem above the ground or on/in cracks and crevices around the plants in the soil and on the under surface of the cotyledonary or first leaf of young seedlings. The maggots of shoot fly attack the whorl leaves of emerging seedlings causing deadheart while curled and distorted leaves are formed in bigger plants. The soil application of carbofuran 3 G @ 12.5 kg and phorate 10 G @ 10.0 kg per hectare at sowing time [5] or seed treatment with imidacloprid 600 FS @ 6 ml per kg seed one day before sowing has been found effective and recommended for management of shoot fly [6]. However, the intensive usage of insecticides leads to environmental pollution, kills natural enemies of the target pest, may also result in development of insecticide resistance in shoot fly populations. Besides, shoot fly is not easily exposed to insecticides in maize as the larvae feed inside the leaf whorls.
Genetic resistance is the most viable and sustainable strategy for shoot fly management. Low to moderate levels of resistance have been identified against shoot fly in the maize germplasm [7]. However, the genetics of shoot fly resistance in maize has not been investigated in details and no known source of cultivated maize accession is reported to confer absolute resistance to shoot fly. Plant resistance to Atherigona spp. is a complex trait and it depends on the interplay of several component characters [2]. Studies conducted in sorghum revealed that resistance to shoot fly was quantitative in nature [8, 9], with predominantly additive gene effects [10]. A series of past studies indicated that the resistance to different insect pests in maize is polygenic in nature. Various types of gene actions such as additive, dominant, and non-additive along with significant genotype by environment interactions have been reported in maize for resistance to storage pests [11] and stem borer [12, 13].
Substantial yield reduction caused by shoot fly incidence in maize has prompted maize workers to accelerate breeding research on shoot fly resistance in India. Conventional breeding for resistance to insect pest is a challenge due to complex inheritance [14]. The availability of molecular markers and development of linkage maps have led to the identification of genomic regions harbouring quantitative trait loci (QTL) for the traits of interest. Once marker-trait association is established, marker-assisted selection (MAS) could be followed to transfer resistance gene/QTLs in elite susceptible backgrounds. To date, several QTL mapping studies are available for storage pest species such as maize weevil [11, 15] and stem borer species such as European corn borer [16, 17], sugarcane borer [18], Southwestern corn borer [19, 20], Mediterranean corn borer [21] in maize. To the best of our knowledge, no report on the molecular mapping of QTL conferring shoot fly resistance in maize is available. Therefore, a detailed study of the underlying genetic basis of resistance to shoot fly in maize is central for designing more effective breeding strategies. Thus, the present study aims (i) to study the genetics and mapping of resistance to shoot fly in maize (ii) to compare the previously published genomic regions involved in different maize insect-pest resistance (iii) to identify syntenic regions associated with resistance to shoot fly in maize and sorghum and (iv) to understand the mechanism of shoot fly resistance.
We present here the first report on mapping QTLs conferring resistance to shoot fly in maize. A number of potential candidate genes were identified spanning within QTL regions which were involved directly or indirectly in shoot fly resistance. Syntenic regions for shoot fly resistance between maize and sorghum were found. Also, it was observed that some of the detected genomic regions were common for various insect-pests resistance in maize.
2. Materials and methods
2.1 Plant material
The experimental plant material consisted of two parental inbred lines viz. CM143 and CM144 and 107 F2 individuals & their F2:3 families. CM143 moderately resistant to shoot fly, is a well-adapted inbred line, whereas CM144 is susceptible to shoot fly. CM143 and CM144 are the parental lines of a popular hybrid, JH3459 recommended for cultivation in North-Western plains of India. The 107 F2:3 families along with parents (CM143 and CM144) were raised in randomized complete block design in two replications at Punjab Agricultural University, Ludhiana (S1 Fig). Each F2:3 family was planted in one row of three-meter length accommodating 16 plants with a plant to plant and row to row distance of 20 cm and 60 cm, respectively. Standard agronomical practices were followed for raising the crop. The F2:3 families were self-fertilized to generate F3:4 families.
2.2 Phenotypic evaluation
The fish-meal technique was used for increasing the shoot fly pressure under field conditions [7]. The moistened fish meal was applied @50 g/m2 one day after the seedling emergence by broadcasting to screen F2:3 families against shoot fly. The data on shoot fly infestation were recorded from ten plants of each F2:3 family at different time intervals. Phenotypic data on various traits such as egg count (oviposition, EC), leaf injury (LI), deadheart (DH), leaf glossiness (LG), seedling vigor (SV), leaf sheath pigmentation (LSP), leaf surface wetness (LSW), leaf length (LL), leaf width (LW), leaf area (LA), and stem girth (SG) were recorded. Ovipositional count as total number of eggs on ten random plants of each F2:3 family was recorded (S2 Fig). The mean number of eggs per family was calculated on 5, 10, and 15 days after emergence (DAE). The mean values of LI and DH per cent were calculated as number of plants with leaf injury or dead hearts / total number of plants × 100 at 7, 14, and 21 DAE. Data on LI and DH were recorded at different time intervals as these are expressed at early seedling stage to V5-V6 stage (21 days of germination). The percentages of the LI and DH were converted into different scales of 1–5 and 1–7, respectively as given in Table 1. The LI is the initial symptom and DH are formed later. However, the extent of DH formation depends on the host genotype irrespective of initial LI. Hence, different scales were used for these traits. The scale used was modified from Sharma et al [22]. The mean value of EC at 15 DAE was pursued for QTL analysis. The average values of LI and DH per cent recorded on 21 DAE were used for QTL identification. Leaf glossiness was visually scored on a scale of 1–5 at 5-leaf stage when there was maximum reflection of light from the leaf surfaces in the early morning hours [23]. The ranking of SV, LSP, and LSW was rated at 5-leaf stage on a 1–5 scale (S2 Fig) and was used to classify the families into different categories (Table 1). The LL (cm), LW (cm), LA (cm2), and SG (cm) was also recorded from seedlings at 5-leaf stage. Leaf width was recorded from upper, middle and lower portion of each leaf with measuring scale and its mean was calculated. The observations for LSP, SV, LG, LSW and leaf dimensions were recorded only once at the appropriate stage for influencing shoot fly oviposition and damage behaviour.
Table 1. Scale used to record the data on different parameters from CM143, CM144 and 107 F2:3 families derived from the cross of CM143 × CM144.
| Scalea | % Leaf Injury (LI) | % Deadheart (DH) | Leaf surface wetnessb (LSW) | Seedling vigour (SV) | Leaf glossinessc (LG) | Leaf sheath pigmentation (LSP) |
|---|---|---|---|---|---|---|
| 1 (R)d | 0–15 | 0–5 | Entire leaf blade densely covered with water droplets | Highly vigorous (plants showing maximum height, more number of fully expanded leaves, good adaptation and robust seedlings) | Highly glossy (light green, shining, narrow and erect leaves) | Leaf sheath with dark pink pigment |
| 2 (MR) | 15.1–25 | 5.1–10 | Water droplets spread all over the leaf blade | Vigorous (good plant height, good number of fully expanded leaves, good adaptation and seedling growth) | Glossy (light green, less shining, narrow and erect leaves) | Leaf sheath with fair pink pigment |
| 3 (MR) | 25.1–35 | 10.1–15 | Leaf blade near mid rib covered with water droplets | Moderately vigorous (moderate plant height with moderate number of fully expanded leaves and fairly good seedling growth) | Moderate glossy (fair green, light shining, medium leaf width and less drooping leaves) | Leaf sheath with light pink pigment |
| 4 (MS) | 35.1–45 | 15.1–20 | Leaf blade with sparsely placed few water droplets | Less vigorous (less plant height with poor leaf expansion and poor adaptation) | Moderate nonglossy (green, pseudo-shine, broad and drooping leaves) | Leaf sheath with very light pink pigment |
| 5 (S) | 45.1–55 | 20.1–25 | Leaf blade without water droplets | Poor seedling vigour (plants showing poor growth and weak seedlings) | Nonglossy (dark green, dull, broad and drooping leaves) | Leaf sheath with green colour |
| 6 (S) | - | 25.1–30 | - | - | - | - |
| 7 (S) | - | 30.1–35 | - | - | - | - |
aThe 1–5 scale for scoring shoot fly damage in maize was calculated by converting percentages of the leaf injury into the rating score whereas the percentage of the dead hearts was classified into scale of 1–7. Leaf injury and deadheart counts were recorded thrice at an interval of 7 days i.e. at 7 days after emergence (DAE), 14 DAE and 21 DAE and were expressed in terms of percentage. The data on leaf glossiness, leaf sheath pigmentation, seedling vigor, and leaf surface wetness were recorded at 5 leaf stage of seedlings on the scale of 1–5
bThe observations on leaf surface wetness were recorded between 7.00 to 7:30 A.M.
cLeaf glossiness was evaluated in the early morning hours when there was maximum reflection of light from the leaf surfaces
dThe scale of 1–5 was categorized as: R-resistant, MR-moderately resistant, MS-moderately susceptible, S-susceptible
2.3 Molecular marker analysis
Genomic DNA was isolated from young seedlings of 107 F2 individuals and parents (CM143 and CM144) using the standard CTAB procedure [24]. In vitro amplification using polymerase chain reaction (PCR) was performed in a 96-well microplate in an EppendorfTM Master Cycler in 10 μl reaction volume as described by Kaur et al [25]. The PCR products were resolved in 3% 0.5X TBE agarose gel and the bands were visualized under the UVP gel documentation system. A total of 701 simple sequence repeats (SSR) markers curated from maize database (http://www.maizegdb.org), covering all regions of 10 linkage groups spanning all bins were selected for documentation of polymorphism between the parental lines. The F2 population was genotyped employing 199 polymorphic SSR markers and genetic map was constructed using 120 SSR markers.
2.4 Statistical analysis
Field data were subjected to analysis of variance as per standard procedure of randomized complete block design. Pearson correlation coefficients were calculated to establish association between different component traits. Linkage map was constructed with a threshold value of LOD score of 3.0 and recombination fraction of 0.3 using Mapdisto version 1.7.7 [26]. The QTLs were identified by composite interval mapping (CIM) using both forward and backward regression method with Windows QTL cartographer version 2.5 [27]. The threshold LOD was calculated using 1,000 permutations in each case with 5 per cent level of significance. The proportion of observed phenotypic variance explained by a QTL was estimated using coefficient of determination (R2) using maximum likelihood for CIM. Gene action for each QTL was calculated as per Stuber et al [28]. Briefly, values of 0 to 0.20 were taken for additive gene action, 0.21 to 0.80 as partial dominance, 0.81 to 1.20 as dominance and >1.20 as over dominance. The source of resistance allele was designated as per Jampatong et al [29]. Positive and negative additive values depicted that alleles were contributed by resistant parent CM143 and susceptible parent CM144, respectively.
2.5 In silico analysis
The potential candidate genes within the detected QTLs were fetched out from genome sequence of Zea mays (https://www.maizegdb.org/). Functional annotation of the downloaded genes was retrieved from maizegdb and the published literature. Blast2Go software was used for gene ontology analysis. The QTL-bearing sequences associated with shoot fly resistance revealed in the present study were compared to sorghum genome using NCBI blast to identify the syntenic regions between two genomes. Syntenic regions were plotted using CIRCOS program [30].
3. Results and discussion
3.1 Genetic basis of shoot fly resistance
3.1.1 Inheritance of shoot fly resistance
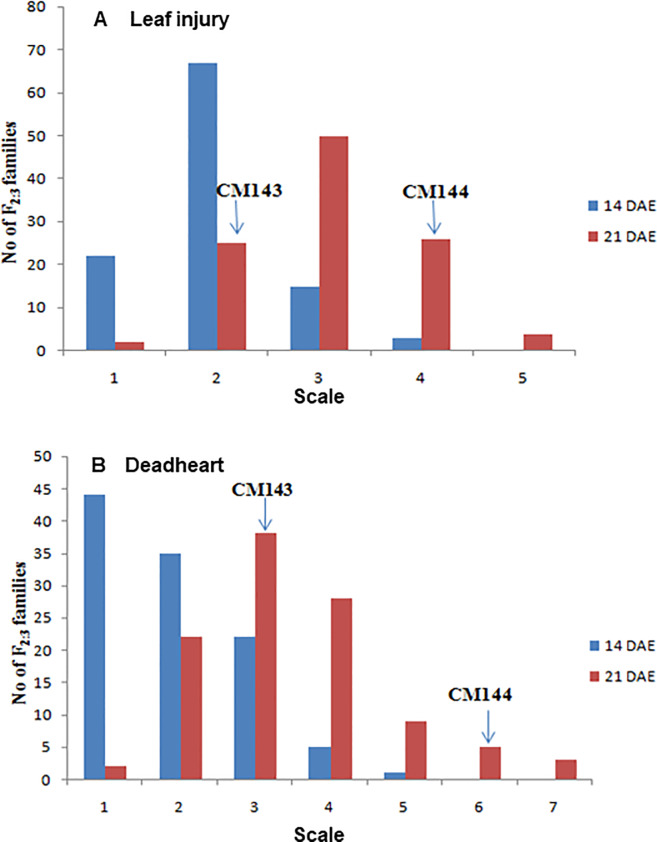
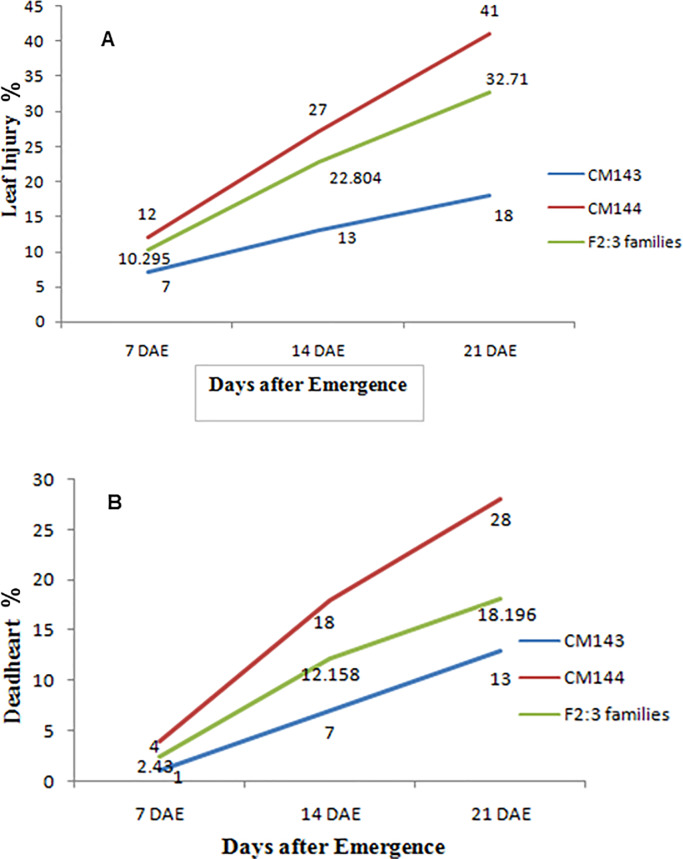
Plant morphology has a strong impact on shoot fly damage, especially seedling characteristics that physically reduce feeding, oviposition, and shelter. The phenotypic trait means of the parental lines and their F2:3 families for various component traits are presented in Tables 2 and 3. All the component traits showed significant differences between the parental lines and among the F2:3 families. Resistant parent CM143 recorded mean EC of 6.0, 13.0 and 20.0, whereas susceptible parent CM144 registered an average EC of 9.5, 21.0 and 38.5 at 5, 10 and 15 DAE, respectively indicating that a greater number of eggs was laid on susceptible parent CM144. The distribution of LI and DH data at 21 DAE revealed that the response of F2:3 families followed a normal curve with transgressive segregation in both directions (Fig 1). Moreover, different genotypes may vary in expression of injury depending upon their inherit ability to tolerate the damage. Similar observations were made for other traits. This indicated that variables for shoot fly resistance exhibited polygenic inheritance. The shoot fly severity progressed with time as indicated from LI and DH data (Fig 2). It could be inferred that the insect attack severity increases at 21 DAE and continues till the favourable conditions persists.
Table 2. Means and range of parents and F2:3 families derived from cross of CM143 × CM144 at different days after emergence (DAE) for egg count, leaf injury (%) and deadheart (%) after shoot fly infestation.
| Parents/ Population | Oviposition (Egg count, EC)a | |||||
|---|---|---|---|---|---|---|
| 5 DAEa | 10 DAE | 15 DAE | ||||
| Range | Mean | Range | Mean | Range | Mean | |
| CM143 | 0–8 | 6.0±0.12 | 0–15 | 13.0±0.23 | 10–25 | 20.0±0.21 |
| CM144 | 0–13 | 9.5±0.22 | 5–25 | 21.0±0.11 | 20–50 | 38.50±0.19 |
| F2:3 families | 0–11 | 7.64±0.13 | 10–25 | 17.59±0.08 | 17–43 | 28.36±0.16 |
| CDd | 5.186 | CVe | 9.3 | |||
| Leaf injury% (LI)b | ||||||
| 7 DAE | 14 DAE | 21 DAE | ||||
| CM143 | 0–10 | 7.0±0.114 | 0–15 | 13.0±0.167 | 15.0–25.0 | 18.0±0.104 |
| CM144 | 0–15.0 | 12.0±0.126 | 25.0–35.0 | 27.0±0.118 | 35.0–45.0 | 41.0±0.234 |
| F2:3 families | 0–20.0 | 10.295±0.132 | 10–45.0 | 22.804±0.23 | 15.0–55.0 | 32.71±0.194 |
| CD | 12.492 | CV | 19.24 | |||
| Deadheart% (DH)c | ||||||
| 7 DAE | 14 DAE | 21 DAE | ||||
| CM143 | 0–5.0 | 1.0±0.216 | 5.0–10 | 7.0±0.109 | 10.0–15.0 | 13.0±0.134 |
| CM144 | 0–10.0 | 4.0±0.109 | 15.0–20.0 | 18.0±0.094 | 25.0–30.0 | 28.0±0.122 |
| F2:3 families | 0–10.0 | 2.425±0.301 | 5.0–25.0 | 12.158±0.134 | 10.0–35.0 | 18.92±0.137 |
| CD | 10.734 | CV | 31.97 | |||
aEgg count was recorded at an interval of 5 days i.e. at 5 days after emergence (DAE), 10 DAE and 15 DAE
b&cLeaf injury and deadheart were recorded at an interval of 7 days i.e. at 7 DAE, 14 DAE and 21 DAE and were expressed in terms of percentage
dCD: Critical difference
eCV: Coefficient of variation
Table 3. Mean values of parents and F2:3 families derived from the cross of CM143 × CM144 for various component traits after shoot fly infestation.
| Traitsa/ Lines | Seedling vigor (SV) | Leaf glossiness (LG) | Leaf sheath pigmentation (LSP) | Leaf surface wetness (LSW) | Leaf length (LL; cm) | Leaf width (LW; cm) | Leaf area (LA; cm2) | Stem girth (SG; cm) |
|---|---|---|---|---|---|---|---|---|
| CM143 | 1.5±0.137d | 2.0±0.216 | 1.0±0.122 | 2.0±0.164 | 11.56±0.112 | 1.69±0.198 | 19.53±0.13 | 2.07±0.205 |
| CM144 | 4.25±0.836 | 4.5±0.228 | 4.0±0.202 | 4.5±0.181 | 13.04±0.166 | 1.87±0.12 | 24.38±0.20 | 1.76±0.08 |
| F2:3 families | 2.18±0.195 | 2.71±0.058 | 1.66±0.311 | 3.28±0.115 | 12.001±0.175 | 1.711±0.123 | 20.53±0.124 | 1.98±0.177 |
| CDb | 1.333 | 1.028 | 1.001 | 1.094 | 2.098 | 0.379 | 8.157 | 4.76 |
| CVc | 30.78 | 19.15 | 30.22 | 16.82 | 7.54 | 10.56 | 15.64 | 11.54 |
aThe data on SV, LG, LSP, LSW, LL, LW, LA and SG were recorded at 5th leaf stage of seedlings. The data on SV, LG, LSP and LSW were recorded on the scale of 1–5. LG was evaluated in the early morning hours when there was maximum reflection of light from the leaf surfaces whereas the observations on LSW were recorded between 7.00 to 7:30 A.M
bCD: Critical difference
cCV: Coefficient of variation
d± Value is standard error of difference
Fig 1. Distribution of leaf injury and deadheart at 14 and 21 days after emergence (DAE) for shoot fly resistance among F2:3 families derived from cross of CM143 × CM144.
The 1–5 scale for shoot fly damage in F2:3 families was calculated by converting percentages of the leaf injury into the rating score (A), whereas the percentage of the dead hearts was classified into scale of 1–7 (B). Leaf injury scale- 1: 0–15%, 2: 15.1–25%, 3: 25.1–35%, 4: 35.1–45%, 5: 45.1–55%. Deadheart scale- 1: 0–5%, 2: 5.1–10%, 3: 10.1–15%, 4: 15.1–20%, 5: 20.1–25%, 6: 25.1–30%, 7: 30.1–35%. The position of the average scores of the parental types, CM143 and CM144, are indicated (Refer Table 2).
Fig 2.
Leaf injury % (A) and deadheart % (B) progress curve of CM143, CM144 and F2:3 families derived from cross of CM143 × CM144 at 7, 14 and 21 days after emergence (DAE) after shoot fly infestation. The values represent the mean value of the parents and F2:3 families (Refer Table 2).
High degree of positive association between EC and DH (r = 0.936); LI and EC (r = 0.891); LI and DH (r = 0.824); and between LG and LSW (r = 0.83) were observed (Table 4). The present results suggested that LI and DH are the major contributors to shoot fly damage because the eggs are laid by shoot fly on the leaves of emerging maize seedlings and subsequently growing tip is damaged leading to DH. The ability to recover from leaf injury is key for expression of resistance in a genotype. Both these traits are directly associated with the EC of shoot fly which is related or dependent on LG, and SV. It has been reported earlier that higher values of LL, LW, LA, and SG were associated with susceptibility to shoot fly in maize [2]. Similar results have been observed in the present study that the traits like LL, LW, LA, and SG were negatively associated with resistance (Table 4). Also, LL, LW, LA, and SG were non-significantly related with other component traits for shoot fly resistance. The leaf glossiness (light green and shiny leaves), leaf surface wetness (reduces the movement of freshly hatched larvae) and leaf sheath pigmentation (dark pink) at seedling stage has a strong influence on the oviposition of shoot fly and these traits were positively associated with oviposition and deadheart in the present work which are in agreement with Dhillon et al [22] but differ with the results of Satish et al [9] in sorghum. Also, association of seedling vigor with oviposition and deadheart was positive in present study but negatively correlated in sorghum as reported by Satish et al [9]. This suggests that the genetic relationship between these traits depends on the germplasm being evaluated and the differences may exist at genus level. Also, the inherent mechanisms of tolerance of plants drive the reaction either towards resistance or susceptibility.
Table 4. Correlation coefficients among various component traits for shoot fly resistance among F2:3 families derived from the cross of CM143 × CM144.
| Traits | Leaf glossiness | Leaf surface wetness (LSW) | Leaf sheath Pigmentation | Leaf length (LL) | Leaf width (LW) | Leaf area (LA) | Stem girth (SG) | Leaf injury (LIa) | Deadheart (DHb) | Oviposition (EC15c) |
|---|---|---|---|---|---|---|---|---|---|---|
| (LG) | ||||||||||
| (LSP) | ||||||||||
| Seedling vigor (SV) | 0.752 | 0.739 | 0.709 | -0.103 | -0.087 | 0.008 | -0.008 | 0.669 | 0.669 | 0.668 |
| (< 0.0001)d | (< 0.0001) | (< 0.0001) | (0.289) | (0.373) | (0.932) | (0.935) | (< 0.0001) | (< 0.0001) | (< 0.0001) | |
| Leaf glossiness (LG) | 0.831 | 0.725 | -0.068 | -0.079 | 0.042 | 0.088 | 0.737 | 0.692 | 0.738 | |
| (< 0.0001) | (< 0.0001) | (0.489) | (0.417) | (0.664) | (0.368) | (< 0.0001) | (< 0.0001) | (< 0.0001) | ||
| Leaf surface wetness (LSW) | 0.694 | -0.121 | -0.166 | -0.023 | 0.079 | 0.769 | 0.701 | 0.752 | ||
| (< 0.0001) | (0.215) | (0.088) | (0.810) | (0.419) | (< 0.0001) | (< 0.0001) | (< 0.0001) | |||
| Leaf sheath pigmentation (LSP) | -0.165 | -0.183 | -0.099 | 0.040 | 0.702 | 0.658 | 0.702 | |||
| (0.090) | (0.060) | (0.309) | (0.684) | (< 0.0001) | (< 0.0001) | (< 0.0001) | ||||
| Leaf length (LL) | 0.736 | 0.854 | 0.342 | -0.027 | 0.022 | -0.019 | ||||
| (< 0.0001) | (< 0.0001) | (0.001) | (0.783) | (0.819) | (0.849) | |||||
| Leaf width (LW) | 0.855 | 0.318 | -0.035 | -0.010 | -0.036 | |||||
| (< 0.0001) | (0.001) | (0.720) | (0.918) | (0.712) | ||||||
| Leaf area (LA) | 0.371 | 0.113 | 0.131 | 0.100 | ||||||
| (< 0.0001) | (0.248) | (0.180) | (0.307) | |||||||
| Stem girth (SG) | 0.217 | 0.148 | 0.204 | |||||||
| (0.024) | (0.129) | (0.035) | ||||||||
| Leaf injury (LI) | 0.824 | 0.891 | ||||||||
| (< 0.0001) | (< 0.0001) | |||||||||
| Deadheart (DH) | 0.936 | |||||||||
| (< 0.0001) |
aEC: Egg count data at 15 days after emergence (DAE) was used for analysis
bLI: Leaf injury data at 21 DAE was permutated for analysis
cDH: Deadheart data at 21 DAE was analyzed
dThe value in parenthesis indicates the significance probability associated with the statistic
3.1.2 QTL mapping
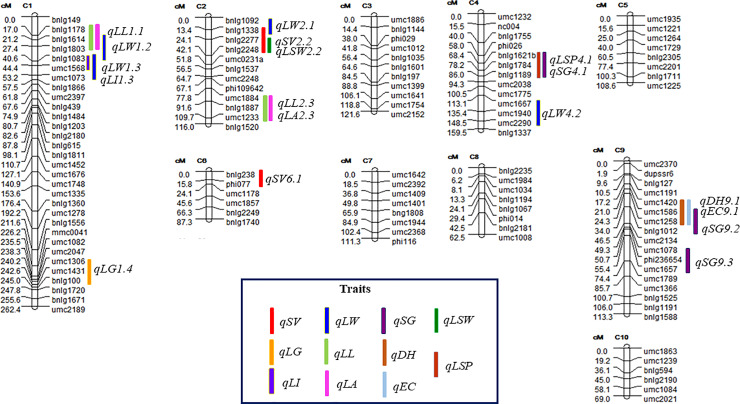
A total of 701 maize SSR markers were surveyed for parental polymorphism and 228 (32.52%) markers were found to be polymorphic. A set of 199 polymorphic SSR markers were genotyped on F2 population. Significant deviation from expected Mendelian segregation was observed for 74 SSR markers (37.18%); 21 and 18 SSR markers were skewed towards CM143 and CM 144, respectively. Whereas, 30 markers had banding pattern more towards heterozygosity. Segregation distortion might be due to gamete abortion or the selective fertilization of particular gametic genotypes, presence of segregation-distortion loci (SDL) in the vicinity of markers, epistasis, enhanced recombination [31]. In the present investigation five of the SSR markers showed banding pattern resembling either of the parents in the whole population. This might be due to the presence of ‘cold spot regions’ showing little or no recombination [32] or due to a gamete elimination system [33]. All these markers were excluded from data analysis. Five of the SSR markers did not show linkage with their respective linkage group during the development of genetic linkage map. The relative position and genetic distances of 120 SSR markers along the respective chromosome are presented in Fig 3. The genetic map had a total length of 1211.5 cM with an average distance of 10.1 cM between markers. The order of the markers on the linkage map agreed with the corresponding positions on the IBM 2008 map (https://www.maizegdb.org/) except four markers (umc1452, bnlg1866, bnlg615, and umc2248) whose chromosomal location agreed but not specified according to the bin position.
Fig 3. Genetic linkage map of maize representing 19 quantitative trait loci (QTL) for shoot fly resistance identified in F2:3 families derived from cross of CM143 × CM144.
Putative QTL are designated by the corresponding chromosome in which they are found. The map distance is given on the left in centimorgans (cM) from the top of each chromosome and marker name is represented on the right side of the chromosome. Different color bars are used to indicate QTL for each trait. LL: Leaf length, LW: Leaf width, LA: Leaf area, LI: Leaf injury, LSW: Leaf surface wetness, LSP: Leaf sheath pigmentation, LG: Leaf glossiness, SV: Seedling vigor, SG: Stem girth, EC: Oviposition, DH: Deadheart.
Numerous studies have been conducted to map QTLs for resistance to European corn borer, Mediterranean corn borer, Asian corn borer, sugarcane borer, maize weevil, Southwestern corn borer, and fall army worm [34]. However, no information is available about the number, mode of action, and location of QTLs governing shoot fly resistance in maize. A total of 19 putative QTLs associated with various component traits governing shoot fly resistance were detected on chromosomes 1, 2, 4, 6, and 9 (Table 5). It has been observed that resistant as well as susceptible parent contributed alleles for shoot fly resistance as none of the parents was completely resistant or susceptible to shoot fly. Nine (qLW4.2, qLL2.3, qLA2.3, qLSW2.2, qSV2.2, qEC9.1, qDH9.1, qSG9.2, and qSG9.3) of the 19 QTLs alleles conferring resistance to the shoot fly in the present investigation were contributed by the susceptible parent, CM144, whereas positive alleles of other 10 QTLs were contributed by the resistant parent, CM143. Other authors have also reported QTL alleles for insect resistance that came from a highly susceptible parent in maize [11, 16], rice [35] and cotton [36]. It indicates that susceptible parent also harbor some favorable alleles, which can be employed in gene pyramiding.
Table 5. Marker intervals showing putative QTL for shoot fly resistance component traits in F2 population from cross of CM143 × CM144.
| Trait | QTLa | Marker interval | Binb | LOD scorec | Phenotypic varianced (%) | Additive effecte | Dominance effectf | Gene Actiong |
|---|---|---|---|---|---|---|---|---|
| Leaf width | qLW1.2 | bnlg1614-bnlg1083 | 1.02 | 3.68 | 9.32 | 0.0526 | 0.0314 | PD |
| qLW1.3 | bnlg1083-umc1073 | 1.02–1.03 | 4.27 | 9.94 | 0.0595 | 0.0194 | PD | |
| qLW2.1 | bnlg1092-bnlg1338 | 2.01 | 2.65 | 8.16 | 0.05 | 0.0031 | A | |
| qLW4.2 | umc1667-umc2290 | 4.08 | 3.18 | 11.26 | -0.0457 | 0.0312 | PD | |
| Leaf length | qLL1.1 | bnlg1178-bnlg1803 | 1.02 | 4.67 | 4.25 | 0.1162 | 0.2853 | OD |
| qLL2.3 | bnlg1884-umc1233 | 2.05 | 2.94 | 10.85 | -0.1465 | 0.0236 | A | |
| Leaf area | qLA1.1 | bnlg1178-bnlg1803 | 1.02 | 6.28 | 8.48 | 0.3731 | 0.6358 | OD |
| qLA2.3 | umc1884-umc1233 | 2.05 | 2.55 | 9.82 | -0.3392 | 0.1158 | PD | |
| Leaf injury | qLI1.3 | bnlg1083-umc1568 | 1.02 | 3.14 | 11.96 | 0.1143 | - | A |
| Leaf surface wetness | qLSW2.2 | bnlg2277-bnlg2248 | 2.02–2.03 | 3.22 | 7.30 | -0.018 | - | A |
| Leaf glossiness | qLG1.4 | umc1306-bnlg100 | 1.09 | 2.62 | 12.98 | 0.0501 | -0.0614 | D |
| Leaf sheath pigmentation | qLSP4.1 | bnlg1621b-bnlg1189 | 4.07 | 3.08 | 7.58 | 0.1207 | 0.0439 | OD |
| Seedling vigour | qSV2.2 | bnlg1338-bnlg2248 | 2.01–2.03 | 2.73 | 9.8 | -0.0432 | 0.1402 | OD |
| qSV6.1 | bnlg238-phi077 | 6.00–6.01 | 2.80 | 9.7 | 0.0340 | -0.1229 | OD | |
| Oviposition | qEC9.1 | umc1420-umc1258 | 9.03 | 4.09 | 18.49 | -2.881 | 0.8197 | OD |
| Dead heart | qDH9.1 | umc1420-umc1258 | 9.03 | 3.49 | 15.03 | -0.0393 | 0.0040 | A |
| Stem girth | qSG9.2 | umc1586-bnlg1012 | 9.05 | 2.65 | 7.26 | -0.1722 | -0.2282 | OD |
| qSG9.3 | umc1078-umc1657 | 9.06 | 4.27 | 10.5 | -0.1118 | -0.5880 | OD | |
| qSG4.1 | bnlg1621b-bnlg1189 | 4.07 | 3.47 | 9.8 | 0.0670 | -0.3669 | OD |
aPutative QTL are designated by the corresponding chromosome in which they are found
LL: Leaf length, LW: Leaf width, LA: Leaf area, LI: Leaf injury, LSW: Leaf surface wetness, LSP: Leaf sheath pigmentation, LG: Leaf glossiness, SV: Seedling vigor, SG: Stem girth, EC: Oviposition, DH: Deadheart
bChromosome bin location of QTL peak of the maize genome. Bins divide the genetic map into 100 approximately equal segments of approximately 20 centiMorgans between two fixed Core Marker. The segments are designated with the chromosome number followed by a two-digit decimal (e.g., 1.00, 1.01, 1.02, etc)
cThe maximum LOD score associated with each QTL
dR2 estimates the proportion of phenotypic variance (%) explained by individual QTL
eThe additive genetic effect of the putative QTL. A positive number indicates that the alleles for resistance are derived from CM143 and a negative number means that the alleles for resistance are derived from CM144
fThe dominant genetic effect of the putative QTL
gGene action displayed by a QTL: A (additive) = > 0.20, PD (partial dominance) = 0.21 to 0.80, D (dominance) = 0.81 to 1.20, OD (over dominance) = > 1.20
Four QTLs for leaf width (qLW1.2, qLW1.3, qLW2.1 and qLW4.2) depicting partial-dominance and additive gene action were identified on short arm of chromosome 1 (bins 1.02–1.03), chromosome 2 (bin 2.01) and long arm of chromosome 4 (bin 4.08) explaining 38.68% of total phenotypic variance among F2:3 families. Most of the alleles associated with leaf width were contributed by the resistant parent, CM143. The QTLs for leaf length and leaf area present on short arm of chromosome 1 (qLL1.1 & qLA1.1) revealed over dominance gene action whereas, the qLL2.3 and qLA2.3 present on long arm of chromosome 2 displayed partial dominance and additive gene action, respectively.
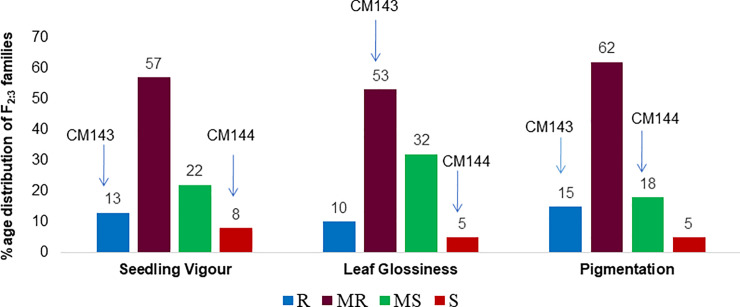
The QTL for LI and LG were detected in different genomic regions of chromosome 1 (short arm, bin 1.02 and long arm, bin 1.09) accounting for 11.96 and 12.98% phenotypic variation, respectively (Fig 3 and Table 5). Gene action effects were additive and dominant for LI and LG, respectively. Two QTL, qSV2.2 and qSV6.1, for seedling vigor exhibiting overdominance were detected on short arm of both chromosomes 2 and 6 with favourable alleles contributed by CM144 and CM143, respectively. Overdominance gene action was exhibited by all the three QTL for SG present on long arm of chromosome 4 (qSG4.1) and 9 (qSG9.2, qSG9.3) accounting for 27.56% phenotypic variance together (Table 5). Alleles associated with reduced SG were contributed by susceptible parent, CM144. Most of the QTLs for insect resistance in maize showed additive gene action [16, 21]. However, present study depicted that maize shoot fly resistance was governed by overdominance (nine QTLs), dominance (one QTL), partial dominance (four QTLs) and additive (five QTLs) type of gene action. It indicates that genetics of shoot fly resistance is complex and involves different types of gene action for different component traits. Similar results were reported by Garcia-Lara et al [11] that genetic effects were mainly dominant and additive for maize weevil resistance. Our results also support that leaf glossiness was positively correlated with the level of resistance as 53 per cent of F2:3 families were moderately resistant and the allelic contribution for leaf glossiness was inherited from the female parent (CM143) in dominant manner. Similarly, leaf sheath pigmentation and seedling vigor traits showed overdominance expression as specified from maximum number of F2:3 families were resistant and moderately resistant (Fig 4).
Fig 4. Per cent distribution of F2:3 families derived from cross of CM143 × CM144 into different categories with respect to seedling vigor, leaf glossiness and leaf sheath pigmentation.
The data on these traits were recorded at 5 leaf stage of seedlings on the scale of 1–5. The scale of 1–5 was categorized as: R-resistant = 1, MR-moderately resistant = 2, 3, MS-moderately susceptible = 4, S-susceptible = 5. The number on the bar indicates the percentage of F2:3 families into that category. The position of the average scores of the parental types, CM143 and CM144, are indicated.
The detected QTLs for shoot fly component traits co-localized in seven genomic regions located on chromosomes 1, 2, 4 and 9 (Fig 3). Two QTLs (present on chromosome bins 1.02 and 2.05) each for leaf length (qLL1.1, qLL2.3) and leaf area (qLA1.1, qLA2.3) were co-localized (Fig 3) as both traits are positively correlated. The QTL qLW1.3 present in the genomic region bnlg1083-umc1073 overlapped with qLI1.3. Likewise, leaf sheath pigmentation QTL, qLSP4.1, was co-localized to stem girth QTL, qSG4.1, on chromosome 4 whereas seedling vigor QTL, qSV2.2 (R2 = 9.8%) was overlapped with leaf sheath pigmentation QTL, qLSW2.2 (R2 = 7.3%).
The QTL for deadheart (qDH9.1) spanning the marker interval of umc1420 and umc1258 (9.03) explained 15.03% of phenotypic variance. This putative QTL was co-localized with oviposition QTL (qEC9.1) with 18.89% of phenotypic variance (S3 Fig). This QTL was residing near the centromeric region of chromosome 9. Our results are in concordance to Satish et al [9] and Apotikar et al [37] regarding co-localization of EC and DH QTLs in sorghum chromosome 10. So, this region could be further dissected to identify the candidate genes for shoot fly resistance in maize. Co-localization of QTLs for different traits might be result of tight linkage of several genes (cluster of genes are present in the form of multigene family controlling different traits) or pleiotropic effect of a locus [9].
The previous studies concluded that disease and insect resistance genes in maize appear in “clusters” [38]. The detected bins in the experiment (1.02–03, 1.09, 2.05, 6.00–6.01, 9.03 and 9.05–06) have been reported in different studies for insect resistance especially to European corn borer, fall army worm and maize weevil as presented in S1 Table. Based on meta QTL analysis, Badji et al [34] highlighted the presence of combined insect resistance genomic regions in maize and thus, could form the basis for multiple pest’s resistance breeding. This clearly supports the hypothesis that there are certainly some common genomic regions that confer resistance against insect pests along with specified genomic regions unique to specificity for each of the pests. Hence, these common genomic regions are key for further studies to dissect the resistance genes and might enable for deciphering the common metabolic pathway conferring insect pest resistance.
3.2 Resistance mechanism
Plant resistance to insect herbivory can be classified as antixenosis (non-preference of host plants), antibiosis, and tolerance [1]. Non-preference by insects is often projected as a property of the plant to render it unattractive for oviposition, feeding, and shelter. The antixenosis for oviposition by shoot fly was not observed in maize genotypes and the tolerance was mainly dependent on the ability of plant to recover from injury [2, 7]. Similar observations were recorded in the present investigation. However, the differences in oviposition exists that may be due to aggregation distribution of eggs at high density or may be due to antixenosis. Scanty information is available on the mechanism of antibiosis for resistance to shoot fly.
The QTLs for leaf dimensions and leaf injury were detected in bin 1.02 in the present study. In an earlier investigation by Barriere et al [39], cell wall-bound phenolic compounds (p-coumaric acid, esterified ferulic acid etc.) were found to be associated in bin 1.01/1.02. Similarly, the identified QTL in bin 1.09, 2.01, 2.05, 4.08, 6.01, 9.03, 9.05–9.06 also included QTL for different metabolites (maysin, chlorogenic acid, Feuric acid, p-coumaric acid, DIMBOA and diferuloyl putrescine) as detected in other experiments [40–42]. It is well known that the cell wall constituents act as a barrier to feeding insects and its proximal co-localization with shoot fly component trait QTLs validates the importance of cell wall structure and composition in resistance. It has been reported earlier that QTLs for leaf feeding & stem feeding co-localized with cell wall constituents in 29 maize bins [1]. It could be concluded that antibiosis increases the mortality and hampers the growth and feeding of larvae on the host plant. As there is close liaison among QTLs linked with shoot fly resistance and cell wall components, it will be a rewarding exercise to map QTLs for these metabolites using the same mapping population.
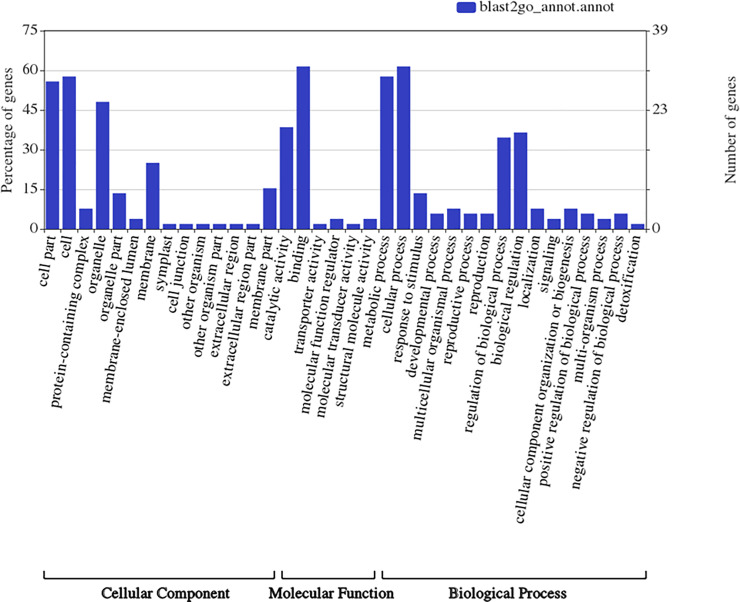
In the present study, the number of genes detected in the QTL regions varied from 91 to 295. The putative 58 candidate genes were short-listed from a previous study by Satish et al [9] and on the basis of their role in plant stress response. These genes were categorized into receptor kinases (leucine-rich repeat receptor kinase, serine/threonine protein kinases and sucrose non-fermenting related protein kinases 1), signalling molecules or secondary messengers, metabolic enzymes, transcription factors and some proteins acting as chaperons (S2 Table). Most of the candidate genes are involved in jasmonic signalling pathway, lignin biosynthesis, meristem growth, and biotic and abiotic stress tolerance. These genes were classified into three categories namely, cellular function, molecular function and biological process according to gene ontology (GO) using Blast2Go (Fig 5). GO analysis of cellular components showed that genes were associated with all parts of the cell. In the molecular function category, catalytic activity and various binding activities genes were enriched. In biological process, most of the genes were associated with cellular process, metabolic process, biological regulation and regulation of biological process. It can be foreseen that crosstalk might be existing among different pathways involved in shoot fly resistance.
Fig 5. Gene ontology classification of the genes identified within the detected QTL regions for shoot fly resistance.
The insect oral components act as elicitors recognized by the plant to regulate the wounding response [43]. The endogenous resistance mechanism to shoot fly wounding is induced by the synthesis of proteinase inhibitors as evident from the QTLs spanning LG and SV traits harbour candidate genes for cysteine protease and subtilisin-chymotrypsin inhibitor on chromosome 2 and 6, respectively (S2 Table). The accumulation of the 33-kDa cysteine protease in the maize midwhorl was associated with a significant reduction in larval growth due to diminished nutrient utilization [44]. Similarly, subtilisin/chymotrypsin inhibitor gene (Zm00001d035683) detected on chromosome 6 also plays a role of natural defense against attacks of pests and pathogens [45]. This might be responsible for high SV associated with shoot fly resistance. The induced direct defenses also involve the production of toxic or repellent secondary metabolites and volatiles that attract natural enemies of the herbivore to the plant as indirect defense. The presence of putative genes for cytochrome P450s superfamily in the present study indicates that maize plants are involved in the production of allelochemicals, as P450s mediate the synthesis of allelochemicals [46]. The nature and type of allelochemical for the shoot fly resistance is yet to be elucidated.
The identified genes for GH10 family of glycoside hydrolases and extracellular sulfatases in the QTL interval of DH are present in extracellular space and have been implicated in the defense response by mobilization of energy reserves through degradation of starch and proteoglycan metabolism, respectively. These enzymes regulate signalling pathways by regulating the binding of signal ligands and receptor kinase [47, 48]. The genes for membrane bound receptor protein kinase have been identified within the QTL region of DH and EC, SG, and SV. All these have been implicated in initiating various signalling pathways, including meristem function, brassinosteroid perception, overall plant morphology, and plant defense [49]. These are further under regulation of phosphatases (PP2Cs) and ubiquitin/proteasome system. The genes for protein deubiquitination, ubiquitylation, protein-protein interaction, and proteasomal degradation (PP2C, Ubx2, ubiquitin C-terminal hydrolase, Speckle-Type Poz Protein (SPOP), and Ring finger zinc protein) have been localized within the QTL region of LG, SV, SG, and DH (S2 Table).
It has been reported that there is co-activation of numerous intracellular signalling cascades (Table 5). Jasmonic acid (JA) levels increase after wounding or attack by herbivores [50]. Fatty acid desaturase has been shown to trigger the JA defense signalling pathway which in turn regulates the induction of cysteine protease [51]. The gene for fatty acid desaturase identified in the present study is likely to be involved in imparting tolerance to shoot fly as gene for cysteine protease was also detected. Several studies have also shown that Ca2+ is an important messenger in many biotic and abiotic signals. Most of the Ca2+ sensors are proteins with atleast one highly conserved Ca2+ binding helix-turn-helix structures (EF-hands) and members of the MYB class of TFs [52]. The candidate genes for EF-hand MYB related transcription factors identified in the QTL interval of SV and LG traits provide a clue that Ca2+ signaling may play a role in shoot fly tolerance. Also, the gene for plasma membrane localized cyclic nucleotide gated channels (CNGCs) on chromosome 6 (qSV6.1; bnlg238-phi077) identified in the present work, has been reported to be involved in uptake of Ca2+ and other cations [53]. Likewise, a gene for fatty acid binding proteins present in QTL interval of qSV6.1 have been identified in response to wound, it could be inferred that oral secretions of shoot fly might contain fatty acid conjugates which is sensed by the plants. It indicates that fatty acid binding proteins may be involved not only in intracellular signalling but also play a role in long-distance lipid signalling and may function as ‘chaperones’ which move lipids into and/or throughout the phloem [54].
In the present investigation, the genes identified for proline oxidase, nudix hydrolase, peroxidise, γ-glutamylcysteine, glutathione S-transferases and TETRATRICOPEPTIDE THIOREDOXIN-LIKE (TTL) have been reported to be involved in response to oxidative stress, redox balance regulation, and to modulate the levels of their substrates to maintain physiological homeostasis [55–58]. The identified genes for some of the transcription factors, including members of the C2H2 zinc finger protein (ZFP), HDZIP, NAC, MYB, Knotted1-like and bHLH families participate in plant stress response [59]. Most of the members of the Knox (Knotted1-like) class are associated with the maintenance and growth of the shoot meristems [60]. Both HD-ZIP class I (HAT7) and HD-ZIP class II (ATHB4, HAT14 & HAT22) respond to illumination sensing and phytochome-mediated shoot morphogenesis, respectively. Some of the HDZip I and II genes are involved in mediating the effects of external conditions to regulate plant growth and development [61].
Glossy phenotypes have less deposition of epicuticular wax and usually impart tolerance to insect damage. The QTL spanning LG in chromosome bin 1.09 contain some putative genes belonging to MYB transcription factors. Some MYB family transcription factors regulate the genes involved in wax biosynthesis and transport of cuticular components [62]. The candidate gene for MYB transcription factor identified for LG in the present study might be acting as negative regulator for wax biosynthesis. The previous studies demonstrated that wax biosynthesis genes were down regulated in the glossy mutant of Brassica [63] and by AP2/ERF type transcription factor in Arabidopsis [64]. The gl15 (glossy 15) gene present in bin 9.03 was also reported as a candidate gene for fall army worm, south western corn borer and European corn borer resistance [19]. It alters the standard shift from vegetative to adult plant growth stage. This could be also one of the probable candidate genes associated with maize shoot fly resistance. From the present investigation, it could be inferred that tolerance in conjunction with antibiosis mechanism exists for shoot fly resistance in maize. Further studies are required to explore the metabolic signalling pathways for deciphering shoot fly resistance mechanism.
3.3 Syntenic relationships
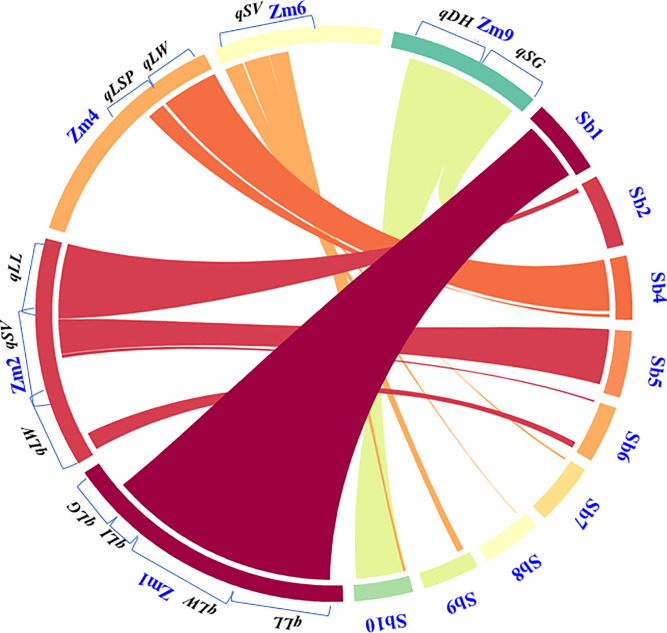
We investigated the syntenic relationships of the QTLs identified in the present study with those of reported in sorghum associated with shoot fly resistance [9]. Two major QTLs associated with DH between markers Xnhsbm1044-Xnhsbm1013 and Xnhsbm1033-Xcup16 on chromosome 10 explaining 15.0 and 11.4 per cent phenotypic variance, respectively in sorghum were found to be syntenic to bins 9.02–9.03 of maize chromosome 9 on which QTL for DH and EC were localized in the present study (Fig 6). The SV QTL located on maize chromosome 2 was found to be syntenic to SV QTL on chromosome 6 in sorghum. Similarly, SV QTL present on chromosome 6 of maize was coinciding to small genomic regions of sorghum chromosome 7, 8, 9 and 10. The QTLs (qLL1.1, qLW1.2, qLI1.3) identified on maize chromosome 1 and QTLs (qSG9.2, qSG9.3) located on chromosome 9 were syntenic to sorghum chromosome 1 where QTL for trichome density on lower leaf surface was detected by Satish et al [9]. Similarly, chromosome 4 (4.07–4.09) of maize harbouring QTLs for LW, SG, and LSP corresponded to sorghum chromosome 4 accounting for trichome density on lower leaf surface QTL. The foregoing results suggests that same gene blocks may be responsible for shoot fly resistance in both the cereal crops, sorghum and maize.
Fig 6. Syntenic relationship of maize genomic regions associated for shoot fly resistance component traits with sorghum genome.
Zm1refers to Zea mays chromosome 1 and so on. Sb1 refers to Sorgum bicolor chromosome 1 and so on. LL: Leaf length, LW: Leaf width, LI: Leaf injury, LSP: Leaf sheath pigmentation, LG: Leaf glossiness, SV: Seedling vigor, SG: Stem girth, DH: Deadheart.
Conclusion
Till date, all published QTL mapping studies on maize insect resistance has involved stem borers and storage pests. To the best of our knowledge, there is no report on the identification of QTLs for shoot fly tolerance in maize. We identified 19 QTLs on chromosomes 1, 2, 4, 6, and 9 in bins 1.02–1.03, 1.09, 2.01, 2.05, 4.07, 4.08, 6.00–01, 9.03 and 9.05–9.06 conferring tolerance to shoot fly. Many of the QTLs for component traits were found to be either overlapped or co-localized suggesting tight linkage of several genes. Also, the identification of co-localized QTL could be utilized for improvement of more than one trait at a time using the same linked markers. Some of the identified maize QTLs were observed to be syntenic with those of sorghum, thus confirming their association with shoot fly resistance. The transgressive segregants from the present material have been advanced and are being used in maize breeding programs. The identified regions of the QTLs need to be saturated with an additional set of markers, which will facilitate fine mapping of the QTLs. The present study has provided an insight in understanding the genetic basis of shoot fly resistance in maize.
Supporting information
A: CM143 (moderately resistant) and B: CM144 (susceptible), infested with shoot fly after the seedling emergence using fish meal technique. C: Field view of F2:3 families derived from the cross of CM143 × CM144 (C).
(TIF)
The data was recorded at different time intervals for different traits. A: Egg count on seedlings, B: Symptoms of leaf injury, C1: Highly vigorous seeding, C5: Poor seedling vigor, D1: Highly glossy, D5: Non-glossy, E1: Leaf sheath with dark pink pigment, E5: Leaf sheath with green color, F: Deadheart formation (refer Table 1).
(TIF)
The vertical axes in graph indicate LOD scores, and the horizontal line indicate the empirically derived LOD threshold for calling a QTL position. Small triangles on the x-axes denote the position of mapped SSR markers in the population and number represent the genetic distance in cM. One triangle may represent one or more markers in the case of very closely linked markers.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
Funding provided by the Punjab Agricultural University is gratefully acknowledged. Mention of a specific product by Punjab Agricultural University does not constitute an endorsement and does not imply recommendation of products.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding provided by the Punjab Agricultural University is gratefully acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Meihls LN, Kaur H, Jander G. Natural variation in maize defense against insect herbivores. Symposia on quantitative biology. Cold Spring Harbor Laboratory Press. 2012; 77:269–283. 10.1101/sqb.2012.77.014662 [DOI] [PubMed] [Google Scholar]
- 2.Jindal J. Incidence of insect pests and management of shoot fly, Atherigona spp. in spring sown maize. Ph.D Thesis, Punjab Agricultural University, Ludhiana, India. 2013.
- 3.Panwar VPS, Sarup P. Distribution and host plant of shoot fly species attacking maize in different parts of the world. J Ent Res. 1985; 9:207–217. [Google Scholar]
- 4.Sandhu GS, Kaushal KK. Occurrence and biology of maize shoot fly in India. Entomol Res. 1976; 88:27. [Google Scholar]
- 5.Kanta U, Singh DP, Sekhon SS. Chemical control of shoot fly, Atherigona naqvii Steyskal on spring maize in Punjab. J Insect Sci. 2006; 19:105–106. [Google Scholar]
- 6.Jindal J, Hari NS. Efficacy of seed treatments against shoot fly, Atherigona naqvii Steyskal on spring maize in Punjab. J Insect Sci. 2011; 24:86–90. [Google Scholar]
- 7.Jindal J, Hari NS, Grewal MS, Chawla JS. Evaluation of different genotypes against shoot fly, Atherigona naqvii Steyskal in spring sown maize. Crop Improv. 2007; 34:160–162. [Google Scholar]
- 8.Hallali MS, Gowda BTS, Kulkarni KA, Gould JV. Inheritance of resistance to shoot fly (Atherigona soccata Rond.) in sorghum. SABRAO J. 1982; 14:165–170. [Google Scholar]
- 9.Satish K, Srinivas G, Madhusudhana R, Padmaja PG, Reddy RN, Mohan SM, et al. Identification of quantitative trait loci for resistance to shoot fly in sorghum [Sorghum bicolour (L.) Moench]. Theor App Genet. 2009; 119:1425–1439. [DOI] [PubMed] [Google Scholar]
- 10.Nimbalkar VS, Bapat DR. Inheritance of shoot fly resistance in sorghum. J Maharashtra Agric Univ. 1992; 17:93–96. [Google Scholar]
- 11.García-Lara S, Khairallah MM., Vargas M and Bergvinson DJ. Mapping of QTL associated with maize weevil resistance in tropical maize. Crop Sci. 2009; 49:139–149. [Google Scholar]
- 12.André AM, van Rensburg JBJ, Labuschagne MT. Inheritance of resistance in maize to the African stalk borer, Busseolafusca (Fuller) (Lepidoptera: Noctuidae). South Afr J Plant Soil. 2003; 20:64–71. 10.1080/02571862.2003.10634910 [DOI] [Google Scholar]
- 13.Barros J, Malvar RA, Butrón A, Santiago R. Combining abilities in maize for the length of the internode basal ring, the entry point of the Mediterranean corn borer larvae. Plant Breed 2011; 130:268–270. [Google Scholar]
- 14.Murenga M, Derera J, Mugo S, Tongoona P. A review of genetic analysis and response to selection for resistance to Busseola fusca and Chilo partellus, stem borers in tropical maize germplasm: a Kenyan perspective. Maydica. 2016; 61:1–11. [Google Scholar]
- 15.Castro-Álvarez FF, Manilal Á, Bergvinson DJ, García-Lara S. Genetic mapping of QTL for maize weevil resistance in a RIL population of tropical maize. Theor Appl Genet. 2015; 128: 411–419. 10.1007/s00122-014-2440-6 [DOI] [PubMed] [Google Scholar]
- 16.Bohn M, Schulz B, Kreps R, Klein D, Melchinger AE. QTL mapping for resistance against the European corn borer (Ostrinia nubilalis H.) in early maturing European dent germplasm. Theor Appl Genet. 2000; 101:907–917. [Google Scholar]
- 17.Cardinal AJ, Lee M, Guthrie WD, Bing J, Austin DF, Veldboom LR, et al. Mapping of factors for resistance to leaf-blade feeding by European corn borer (Ostrinia nubilalis) in maize. Maydica. 2006; 51:93–102. [Google Scholar]
- 18.Groh S, Gonzalez-de-Leon D, Khairallah MM, Jiang C, Bergvinson D, Bohn M, et al. QTL Mapping in Tropical Maize: III. genomic regions for resistance to Diatraea spp. and associated traits in two RIL populations. Crop Sci. 1998; 38:1062–1072. [Google Scholar]
- 19.Brooks TD, Bushman BS, Williams WP, McMullen MD, Buckley PM. Genetic basis of resistance to fall armyworm (Lepidoptera: Noctuidae) and southwestern corn borer (Lepidoptera: Crambidae) leaf-feeding damage in maize. J Econ Entomol. 2007; 100:1470–1475. 10.1603/0022-0493(2007)100[1470:gbortf]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- 20.Brooks TD, Willcox MC, Williams WP, Buckley PM. Quantitative trait loci conferring resistance to fall armyworm and southwestern corn borer leaf feeding damage. Crop Sci. 2005; 45:2430–2434. [Google Scholar]
- 21.Jiménez-Galindo JC, Ordás B, Butron A, Samayoa LF, Malvar RA. QTL Mapping for yield and resistance against Mediterranean corn borer in maize. Front Plant Sci. 2017; 8:698 10.3389/fpls.2017.00698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma HC, Taneja SL, Leuschner K, Nwanze KF. Techniques to screen sorghum for resistance to insect pests: Information Bull No. 32. 1992. pp. 48 International Crops Research Institute for the Semi-Arid Tropics, Patancheru, Andhra Pradesh, India. [Google Scholar]
- 23.Dhillon MK, Sharma HC, Singh R, Naresh JS. Mechanisms of resistance to shoot fly, Atherigona soccata in sorghum. Euphytica. 2005; 144:301–312. [Google Scholar]
- 24.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acid Res. 1980; 8:4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaur M, Vikal Y, Kaur H, Lalit, Kaur K, Chawla JS. Mapping quantitative trait loci associated with southern leaf blight resistance in maize (Zea mays L.). J Phytopathol. 2019; 167:591–600. 10.1111/jph.12849 [DOI] [Google Scholar]
- 26.Lorieux M (2012) MapDisto: Fast and efficient computation of genetic linkage maps. Mol Breed 30:1231–1235. 10.1007/s11032-012-9706-y [DOI] [Google Scholar]
- 27.Basten CJ, Weir BS, Zeng ZB. QTL Cartographer: A reference manual and tutorial for QTL mapping Centre for Quantitative Genetics, NCSU, USA: statgen.ncsu.edu/qtlcart. 2005. [Google Scholar]
- 28.Stuber CW, Edwards MD, Wendel JF. Molecular marker facilitated investigations of quantitative trait loci in Maize. II. Factors influencing yield and its component traits. Crop Sci. 1987; 27:639–648. [Google Scholar]
- 29.Jampatong CMD, Barry BD, Darrah LL, Byrne PF, Kross H. Quantitative trait loci for first- and second-generation European corn borer resistance derived from the maize inbred Mo47. Crop Sci. 2002; 42:584–593. [Google Scholar]
- 30.Krzywinski MI, Jacqueline ES, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alheit KV, Reif JC, Maurer HP, Hahn V, Weissmann EA, Miedaner T, et al. Detection of segregation distortion loci in triticale (x triticosecale Wittmack) based on a high-density DArT marker consensus genetic linkage map. BMC Genomics. 2011; 12:380 10.1186/1471-2164-12-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vikal Y, Chawla H, Sharma R, Lore JS, Singh K. Mapping of bacterial blight resistance gene xa8 in rice (Oryza sativa L.). Indian J Genet. 2014; 74:589–595. [Google Scholar]
- 33.Baumbach J, Rogers JP, Slattery RA, Narayanan NN, Xu M, Palmer, et al. Segregation distortion in a region containing a male-sterility, female-sterility locus in soybean. Plant Sci. 2012; 195:151–156. 10.1016/j.plantsci.2012.07.003 [DOI] [PubMed] [Google Scholar]
- 34.Badji A, Otim M, Machida L, Odong T, Kwemoi DB, Okii D, et al. Maize combined insect resistance genomic regions and their co-localization with cell wall constituents revealed by tissue-specific QTL meta-analyses. Front Plant Sci. 2018; 9:895 10.3389/fpls.2018.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao N, Lee C-R, Semagn K, Sow M, Nwilene F, Kolade O et al. QTL mapping in three rice populations uncovers major genomic regions associated with African rice gall midge resistance. PLoS ONE. 2016; 11(8):e0160749 10.1371/journal.pone.0160749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Yuan Y, Wei Z, Guo X, Guo Y, Zhang S et al. Molecular mapping and validation of a major QTL conferring resistance to a defoliating isolate of Verticillium Wilt in cotton (Gossypium hirsutum L.). PLoS ONE. 2014; 9(4):e96226 10.1371/journal.pone.0096226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Apotipkar DB, Venkateswarlu D, Ghorade RB, Wadaskar RM, Patil JV, Kulwal PL. Mapping of shoot fly tolerance in sorghum using SSR markers. J Genet. 2011; 90:59–66. 10.1007/s12041-011-0046-1 [DOI] [PubMed] [Google Scholar]
- 38.McMullen MD, Simcox KD. Genomic organization of disease and insect resistance genes in maize. Mol Plant Microbe Interact. 1995; 8:811–815. [Google Scholar]
- 39.Barriere Y, Thomas J, Denouse D. QTL mapping for lignin content, lignin monomeric composition, p-hydroxycinnamate content, and cell wall digestibility in the maize recombinant inbred line progeny F838 × F286. Plant Sci. 2008; 175:585–595. [Google Scholar]
- 40.Butro´n A, Chen YC, Rottinghaus GE, McMullen MD. Genetic variation at bx1 controls DIMBOA content in maize. Theor Appl Genet. 2010; 120:721–734. 10.1007/s00122-009-1192-1 [DOI] [PubMed] [Google Scholar]
- 41.García-lara S, Burt AJ, Arnason JT, Bergvinson DJ. QTL mapping of tropical maize grain components associated with maize weevil resistance. Crop Sci. 2010; 50:815–825. [Google Scholar]
- 42.Szalma SJ, Snook ME, Bushman BS, Houchins KE, McMullen MD. Duplicate loci as QTL: The role of chalcone synthase loci in flavone and phenylpropanoid biosynthesis in maize. Crop Sci. 2002; 42:1679–1687. [Google Scholar]
- 43.Ferry N, Edwards MG, Gatehouse JA, Gatehouse AM. Plant–insect interactions: molecular approaches to insect resistance. Curr Opin Biotechnol. 2004; 15:155–161. 10.1016/j.copbio.2004.01.008 [DOI] [PubMed] [Google Scholar]
- 44.Pechan T, Ye L, Chang YM, Mitra A, Lin L, Davis FM, et al. A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell. 2000; 12(7):1031–1040. 10.1105/tpc.12.7.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamayo MC, Rufat M, Bravo JM, Segundo SB. Accumulation of a maize proteinase inhibitor in response to wounding and insect feeding, and characterization of its activity toward digestive proteinases of Spodoptera littoralis larvae. Planta. 2000; 211(1):62–71. 10.1007/s004250000258 [DOI] [PubMed] [Google Scholar]
- 46.Schuler MA. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996; 112:1411–1419. 10.1104/pp.112.4.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ai X, Kusche-Gullberg M, Lindahl U, Emerson CP. Remodeling of heparan sulfate sulfation by extracellular endosulfatases In: Garg HG, Linhardt RJ, CA Hales, editors. Chemistry and Biology of Heparin and Heparan Sulfate. Elsevier, New York: 2005. pp. 245–258. [Google Scholar]
- 48.Henrissat B, Davies GJ. Glycoside Hydrolases and Glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 2000; 124:1515–1519. 10.1104/pp.124.4.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becraft PW. Receptor kinase signalling in plant development. Annu Rev Cell Dev Biol. 2002; 18(1):163–192. [DOI] [PubMed] [Google Scholar]
- 50.Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 2008; 177:301–318. 10.1111/j.1469-8137.2007.02292.x [DOI] [PubMed] [Google Scholar]
- 51.Ankala A, Luthe DS, Williams WP, Wilkinson JR. Integration of ethylene and jasmonic acid signaling pathways in the expression of maize defense protein Mir1-CP. Mol Plant-Microbe Interact. 2009; 22:1555–1564. 10.1094/MPMI-22-12-1555 [DOI] [PubMed] [Google Scholar]
- 52.Reddy ASN, Ali GS, Celesnik H, Day IS. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011; 23:2010–2032. 10.1105/tpc.111.084988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finka A, Quendet AFH, Maathuis FJ, Saidi Y, Goloubinoff P. Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. The Plant Cell. 2012; 24(8):3333–3348. 10.1105/tpc.112.095844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guelette BS, Benning UF, Hoffmann-Benning S. Identification of lipids and lipid binding proteins in phloem exudates of Arabidopsis thaliana. J Exp Bot. 2012; 63:3603–3616. 10.1093/jxb/ers028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ge X, Xia Y. The role of AtNUDT7, a Nudix hydrolase, in the plant defense response. Plant Signal Behav. 2008; 3(2):119–120. 10.4161/psb.3.2.5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gullner G, Komives T, Király L, Schröder P. Glutathione S-Transferase enzymes in plant-pathogen interactions. Front Plant Sci. 2018; 9:1836 10.3389/fpls.2018.01836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lakhssassi N, Doblas VG, Rosado A, Valle AE, Pose D, Jimenez AJ, et al. The Arabidopsis TETRATRICOPEPTIDE THIOREDOXIN-LIKE gene family is required for osmotic stress tolerance and male sporogenesis. Plant Physiol. 2012; 158:1252–1266. 10.1104/pp.111.188920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller G, Honig A, Stein H, Suzuki N, Mittler R, Zilberstein A. Unravelling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem. 2009; 284:26482–26492. 10.1074/jbc.M109.009340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golldack D, Lüking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011; 30:1383–1391. 10.1007/s00299-011-1068-0 [DOI] [PubMed] [Google Scholar]
- 60.Bowman JL, Eshed Y. Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 2000; 5:110–115. 10.1016/s1360-1385(00)01569-7 [DOI] [PubMed] [Google Scholar]
- 61.Sessa G, Carabelli M, Possenti M, Morelli G, Ruberti I. Multiple links between HD-Zip proteins and hormone networks. Int J Mol Sci. 2018; 19:4047 10.3390/ijms19124047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008; 20:752–767. 10.1105/tpc.107.054858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pu Y, Gao J, Guo Y, Liu T, Zhu L, Xu P, et al. A novel dominant glossy mutation causes suppression of wax biosynthesis pathway and deficiency of cuticular wax in Brassica napus. BMC Plant Biol. 2013; 13:215 10.1186/1471-2229-13-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Go YS, Kim H, Kim HJ, Suh MC. Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-Type transcription factor. Plant Cell. 2014; 26:1666–1680. 10.1105/tpc.114.123307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: CM143 (moderately resistant) and B: CM144 (susceptible), infested with shoot fly after the seedling emergence using fish meal technique. C: Field view of F2:3 families derived from the cross of CM143 × CM144 (C).
(TIF)
The data was recorded at different time intervals for different traits. A: Egg count on seedlings, B: Symptoms of leaf injury, C1: Highly vigorous seeding, C5: Poor seedling vigor, D1: Highly glossy, D5: Non-glossy, E1: Leaf sheath with dark pink pigment, E5: Leaf sheath with green color, F: Deadheart formation (refer Table 1).
(TIF)
The vertical axes in graph indicate LOD scores, and the horizontal line indicate the empirically derived LOD threshold for calling a QTL position. Small triangles on the x-axes denote the position of mapped SSR markers in the population and number represent the genetic distance in cM. One triangle may represent one or more markers in the case of very closely linked markers.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.