Abstract
Knowledge of structural details is very much essential from the drug-design perspective. In the systematic review, we systematically reviewed the structural basis of different target proteins of SARS-corona virus (CoV2) from a viral life cycle and from drug design perspective. We searched four literature (PubMed, EMBASE, NATURE, and Willey online library) databases and one structural database (RCSB.org) with appropriate keywords till April 18, and finally, 26 articles were included in the systematic review. The published literature mainly centered upon the structural details of “spike protein,” “main protease/M Pro/3CL pro,” “RNA-dependent RNA polymerase,” and “nonstructural protein 15 Endoribonuclease” of SARS-CoV-2. However, inhibitor bound structures were very less. We need better structures elucidating the interactions between different targets and their inhibitors which will help us in understanding the atomic level importance of different amino acid residues in the functionality of the target structures. To summarize, we need structures with fine resolution, co-crystallized structures with biologically validated inhibitors, and functional characterization of different target proteins. Some other routes of entry of SARS-CoV-2 are also mentioned (e.g., CD147); however, these findings are not structurally validated. This review may pave way for better understanding of SARS-CoV-2 life cycle from structural biology perspective.
Keywords: COVID-19, drug target, life cycle, SARS-corona virus-2, target structures
Introduction
SARS-corona virus (CoV-2) is the newest member of a large group of viruses coming under the order Nidovirales and family coronaviridae and genera beta coronavirus (group 2B).[1] SARS-CoV-2 consists of four basic structural proteins, which are club shape trimeric “spike protein (S)”, “membrane (M) protein”, “envelop (E) protein,” and helically symmetrical “nucleocapsid protein (N).”[2]
The molecular basis of transmission of coronavirus (CoV) is already explained in our previous systematic review.[3] The infection process starts with the binding of the spike protein S1-domain to the human host cell receptor angiotensin-converting enzyme 2 (ACEs), which leads to conformational change in the S1 and S2 domain of spike protein. These changes expose the “fusion peptide” of S2-domain, which mediates the fusion of the viral and host cell membranes. The RNA genome of the virus is then subsequently released into the host cell. The virus uses host-cell machinery to start the translation process to synthesize necessary polyproteins such as a pp1a, pp1ab which are further processed by proteases to release the nonstructural viral proteins (NSPs). The structural proteins (spike, E, N, and M protein) are translated from their respective location in the viral genome. The synthesized structural proteins, nonstructural proteins, and RNA genome assembles, which is then transported outside the cell by exocytosis.[4]
Coming to the presently available pharmacotherapeutic options for the treatment of COVID-19, chloroquine, hydroxychloroquine,[5] interferon-α, ribavirin, corticosteroids,[3] plasma therapy,[6] intravenous immune-globulins,[7] lopinavir/ritonavir, etc., are the mainstream treatment options; however, most of the agents are being used without major clinical evidence of efficacy and safety. Although many therapeutic options are under evaluation in different settings, for example, in silico, in vitro, preclinical, and clinical studies, however, definite evidence of efficacy of any of the agents in clinical settings is still not clear.[2,3,5,8]
SARS-CoV-2 is 70%–80% similar to SARS-CoV at the genomic level.[9] Recent studies highlighted many important genomic variations in SARS-CoV-2, for example, the absence of 8a protein, changes in number of amino acid in 3c and 8b protein.[4] The spike protein is also modified by the homologous recombination process.[10] As compared to SARS-CoV, a mutation (N501T) in the spike protein of SARS-CoV-2 confers higher-binding affinity of SARS-CoV2 to the ACE2 receptor.[11]
Better understanding the viral life cycle in terms of structural biology perspectives can help us in identifying druggable targets, which can be approached from drug design perspective. Here, we systematically reviewed the available target protein structures of SARS-CoV2 in relation to the viral life cycle and drug-design perspective.
Materials and Methods
We screened five literature database (PubMed, Embase, Nature, Willey online library, and RCSB database), using the keywords “crystal structure,” “NMR structure,” “X-Ray structure,” “COVID-19,” 2019-nCoV, SARS-CoV-2, “2019 novel corona virus” till April 18. At first titles and abstracts were screened, which was followed by full-text screening of the relevant articles as per the predefined inclusion/exclusion criteria for possible inclusion.
Inclusion-exclusion criteria
We included peer-reviewed published studies evaluating target protein three-dimensional (3D) structure (nuclear magnetic resonance, X-ray crystallography and cryo-electron microscopy structure) with or without bound ligand, with structures deposited and published in RCSB.org. Among unpublished studies with structure published in RCSB.org, we included only inhibitor bound structures.
Results and Discussion
A total of 206 nonduplicate studies were found after the preliminary screening of the databases. After title and abstract screening, a total of 55 studies were selected for further full-text screening. Among these 55 articles, 29 articles were excluded (review articles = 6, articles addressing SARS-CoV = 7, articles addressing MERS-CoV = 11, guideline = 1, in silico drug designing studies without intricate details of SARS-CoV-2 = 4). A total of 26 studies fulfilling the inclusion/exclusion criteria were included in the final review. The PRISMA flowchart of the study is shown in Figure 1. Details of published studies with important structural and functional target proteins are summarized in Table 1. Details of the important inhibitor-bound target structures without a peer-reviewed publication of the respective study are shown in Table 2.
Figure 1.
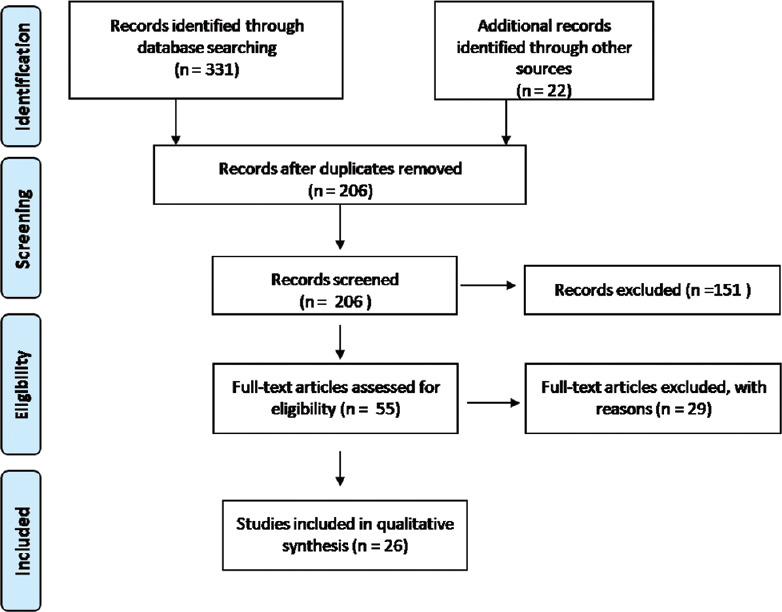
PRISMA flow chart of the study
Table 1.
Important PDB structures with published study details in peer-reviewed journal
| Target | PDB ID | Resolution | Method | Sequence length | Co-crystallized ligand structure name | Details of interaction between inhibitor and target | Important details |
|---|---|---|---|---|---|---|---|
| Spike protein S1 | 6VSB | 3.46Å | EM | 1288 | - | Nil | S trimer in perfusion conformation |
| 6M0J | 2.45 Å | XRD | 229 | - | Nil | Structure of spike protein RBD bind with ACE2 | |
| 6M17 | 2.9 Å | EM | 654 | Nil | The 2019-nCoV RBD/ACE2-B0AT1 complex | ||
| 6W41 | 3.08 Å | XRD | 231 | Human Ab C3022 | Available | Structure of RBD SARS coronavirus 2 | |
| 6VW1 | 2.68Å | XRD | 597 | Nil | Chimeric RBD bind with ACE2 structure in SARS CoV-2 | ||
| 6VXX | 2.8Å | EM | 1281 | Nil | Spike glycoprotein in close state structure of SARS CoV-2 | ||
| 6VYB | 3.2 Å | EM | 1281 | Nil | Spike ectodomain in open state structure of SARS-CoV-2 | ||
| Spike protein S2 | 6LXT | 2.9Å | XRD | 138 | Nil | Nil | Postfusion core of 2019-nCoV S2 subunit |
| Main protease/M Pro/3CL | 6Y2E | 1.75Ao | XRD | Nil | Nil | - | |
| 6Y2G | 2.2Å | XRD | 306 | α-ketoamide inhibitor | Available | Crystal structure (orthorhombic form) | |
| 6Y2F | 1.95Å | XRD | 306 | α-ketoamide inhibitor | Available | Crystal structure (monoclinic form) | |
| 6LU7 | 2.16 Å | XRD | 306 | Inhibitor N3 | Available | SARS-CoV-2 Main protease complexes with inhibitor | |
| RNA dependent RNA polymerase |
6M71 | 2.90 Å | EM | NSP12-942 NSP7-83 NSP8-198 |
Nil | Structure of RdRp with cofector | |
| NSP15 endoribonuclease | 6VWW | 2.20 Å | XRD | 371 | Nil | Nil | Structure of NSP15 from SARS CoV 2 |
RBD=Receptor-binding domain, ACE2=Angiotensin-converting enzyme 2, NSP=Nonstructural protein, RdRP=RNA-Dependent RNA-Polymerase, SARS CoV-2=Severe Acute Respiratory Syndrom Corona virus-2, PDB=Protein Data Bank, XRD=X-ray diffraction, EM=Electron Microscopy
Table 2.
All available XRD structure with Co-crystallized ligand which are not published till date
| Target protein name | XRD ID | Resolution | Method | Amino acid | Co-crystallized ligand structure name | Target protein name | XRD ID | Resolution | Method | Amino acid | Co-crystallized ligand structure name |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NSP15 | 6w01 | 1.90Å | XRD | 371 | Citrate | Main protease | 5RE7 | 1.79 Å | XRD | 306 | Z30932204 |
| NSP3 | 6W02 | 1.50 Å | XRD | 173 | ADP ribose | 5RE6 | 1.87 Å | Z54571979 | |||
| 6WCF | 1.06 Å | XRD | 170 | 2-(N-morpholino)- ethanesulfonic acid | 5RFB | 1.48 Å | Z1271660837 | ||||
| 5RFA | 1.52 Å | Z2643472210 | |||||||||
| 6W6Y | 1.45 Å | XRD | 170 | AMP | 5RFD | 1.41 Å | Z126932614 | ||||
| Spike protein | 6YLA | 2.42 Å | XRD | 213 | CR3022 Fab | 5RFC | 1.4 Å | Z979145504 | |||
| Main protease | 5R84 | 1.83 Å | XRD | 306 | Z31792168 | 5RFF | 1.78 Å | PCM-0102704 | |||
| 5R83 | 1.58 Å | Z44592329 | 5RFE | 1.46Å | Z509756472 | ||||||
| 5R7Y | 1.65 Å | Z45617795 | 5RFH | 1.58Å | PCM-0102277 | ||||||
| 5R80 | 1.93 Å | Z18197050 | 5RFG | 2.32 Å | PCM-0102372 | ||||||
| 5R82 | 1.31 Å | Z219104216 | 5REY | 1.96 Å | PCM-0102911 | ||||||
| 5r81 | 1.95 Å | Z1367324110 | 5REX | 2.07 Å | PCM-0102287 | ||||||
| 5REA | 1.63 Å | Z31432226 | 5RF9 | 1.43Å | Z217038356 | ||||||
| 5R7Z | 1.59 Å | Z1220452176 | 5REZ | 1.79Å | POB0129 | ||||||
| 5RGI | 1.57 Å | Z369936976 | 5RF2 | 1.53 Å | Z1741969146 | ||||||
| 5REC | 1.73 Å | Z1587220559 | 5REP | 1.81 Å | PCM-0102201 | ||||||
| 5REB | 1.68 Å | Z2856434899 | 5RFZ | 1.68 Å | PCM-0102274 | ||||||
| 5REE | 1.77 Å | Z2217052426 | 5RGF | 1.7 Å | Z1619978933 | ||||||
| FRFS | 1.7 Å | PCM-0102739 | 5REF | 1.61 Å | Z24758179 | ||||||
| 5red | 1.47 Å | Z2856434865 | 5Re9 | 1.72 Å | Z2856434836 | ||||||
| 5REG | 1.67 Å | Z1545313172 | 5RG0 | 1.72 Å | PCM-0102535 | ||||||
| 5REN | 2.15Å | PCM-0102425 | 5RE8 | 1.81 Å | Z2737076969 | ||||||
| 5RGG | 2.26 Å | Z2856434890 | 5REF | 2.07 Å | Z33545544 | ||||||
| FRF0 | 1.65Å | POB0073 | 5RE4 | 1.88 Å | Z1129283193 | ||||||
| 5REO | 1.88 Å | PCM-0102578 | 5REM | 1.96 Å | PCM-0103016 | ||||||
| 5RFL | 1.64 Å | PCM-0102389 | 5RFR | 1.71 Å | PCM-0102169 | ||||||
| 5RFK | 1.75 Å | PCM-0102575 | 5RG3 | 1.58 Å | NCL-00025412 | ||||||
| 5RFP | 2.03 Å | XRD | 306 | PCM-0102190 | 5RFQ | 1.76 Å | XRD | 306 | PCM-0102179 | ||
| 5REL | 1.62Å | PCM-0102340 | 5RG2 | 1.63 Å | NCL-00025058 | ||||||
| 5REK | 1.74 Å | PCM-0102327 | 5RFT | 1.58 Å | PCM-0102432 | ||||||
| 5REH | 1.8 Å | Z111507846 | FRFS | 1.7 Å | PCM-0102739 | ||||||
| 5REJ | 1.72 Å | PCM-0102241 | 5RFV | 1.48 Å | PCM-0102306 | ||||||
| 5RG1 | 1.65 Å | NCL-00024905 | 5RFU | 1.53 Å | PCM-0102121 | ||||||
| 5RFO | 1.83 Å | PCM-0102972 | 5RFX | 1.55 Å | PCM-0102254 | ||||||
| 5RF7 | 1.54 Å | Z316425948 | 5RFW | 1.43 Å | PCM-0102243 | ||||||
| 5REV | 1.6 Å | PCM-0103072 | 5RFJ | 1.8 Å | PCM-0103067 | ||||||
| 5REW | 1.55 Å | PCM-0102275 | 5RFI | 1.69 Å | PCM-0102353 | ||||||
| 5RF5 | 1.74 Å | Z3241250482 | 5RFL | 1.64 Å | PCM-0102389 | ||||||
| 5RET | 1.68 Å | PCM-0102269 | 5REI | 1.82 Å | Z2856434856 | ||||||
| 5RF6 | 1.45 Å | Z1348371854 | 5RGS | 1.72 Å | Z1259086950 | ||||||
| 5REU | 1.69 Å | PCM-0102395 | 5RGR | 1.41 Å | Z328695024 | ||||||
| 5RF3 | 1.5 Å | Z1741970824 | 5RGK | 1.43 Å | Z1310876699 | ||||||
| 5RER | 1.88 Å | PCM-0102615 | 5RGJ | 1.34 Å | Z1401276297 | ||||||
| 5RF4 | 1.61 Å | Z1741982125 | 5RGM | 2.04 Å | PCM-0102142 | ||||||
| 5RES | 1.65 Å | PCM-0102281 | 5RGL | 1.76 Å | PCM-0102962 | ||||||
| 5RF1 | 1.73 Å | NCL-00023830 | 5RGO | 1.74 Å | PCM-0102248 | ||||||
| 5RFY | 1.9 Å | PCM-0102974 | 5RGN | 1.86 Å | PCM-0102759 | ||||||
| 5RF8 | 1.44 Å | Z271004858 | 5RGQ | 2.15 Å | Z1849009686 | ||||||
| 5RFN | 1.80Å | PCM-0102868 | 6W63 | 2.10 Å | Noncovalent inhibitor X77 | ||||||
| 5RFM | 2.06 Å | PCM-0102539 | 6M2N | 2.20 Å | XRD | 306 | Novel inhibitor | ||||
| 5RGP | 2.07 Å | PCM-0102628 |
NSP=Nonstructural protein, XRD=X-ray diffraction
Target structures of COVID-19
After reviewing the full text of the articles, we could find published papers with 3D structures of only four targets, which are spike protein, main protease (also known as main protease [Mpro] and 3CLpro), RNA-Dependent RNA-Polymerase (RdRP), and NSP15 of SARS CoV-2.
Spike protein
The spike proteins are trimeric fusion proteins, with two main domains, the S1 and the S2 domain. The structure the spike protein of SARS-CoV2 is around 77% conserved when compared to SARS-CoV.[12] Root mean square deviation (RMSD) between SARS-CoV receptor-binding domain (RBD) and SARS-CoV-2 RBD is around 1.2 angstrom and RMSD between receptor-binding motif (RBM) of SARS-CoV and RBM of SARS-CoV-2 is around 1.3 angstrom, implicating high similarity between these two structures.[13] The affinity of interaction between ectodomain of SARS-CoV-2 and ACE-2 is very high (10–20 fold) as compared to SARS-CoV, and thus, it contributes to the high infectivity SARS-CoV-2.[14]
The spike proteins are considered as a class-1 viral fusion-protein, and it requires two-step protease cleavages for activation. The site between S1 and S2 is the location of priming cleavage; however, the activation cleavage occurs solely on S2. Different protease, for example, trans-membrane protease serine protease-2 (TMPRSS-2), TMPRSS-4, human airway trypsin (HAT) like protease, cathepsins, etc., are involved in these proteolytic activation process. In cell-based experiments (293/hACE2 cells), the expression of different TMPRSS iso-forms enhanced SARS-CoV-2 spike protein-mediated cell-cell fusion.[15]
Importance of S1 domain
The S1-RBD-mediated recognition and attachment to the host receptor (ACE2) is a prime-initiating event, which leads to viral and host-cell fusion and other subsequent events. This highlights the importance of detailed understanding of the interactions between the S1 domain of spike protein and ACE2.
Details of S1 domain and its interaction with angiotensin-converting enzyme 2
The RBD of the S1-spike protein of SARS-CoV-2 consists of a core region and the receptor-binding motif (RBM).[16] The Core region consists of 5 beta-pleated sheets (1, 2 3, 4, and 7), which are organized in the antiparallel mode. The RBM consists of two short beta-pleated sheets (β5 and β6), loops and alpha helices (alpha 4 and 5). A total of three cysteine residue pairs provide stability to the core and another cysteine residue pair helps in connecting the distal end of RBM.[13] However, the interacting surface between the S1 RBD and ACE2 is quite huge (total buried surface area of 1687 angstraon[2] with around 800 Å each on the S1 RBD side and ACE2 side).[13] Compared to SARS-CoV, many variations were seen in the SARS-CoV RBD region, for example, in the N terminal, the variations ARG426 to ASN 439, TYR484 to GLN498, and THR487 to ASN50 were observed between SARS-CoV and novel SARS-CoV2.[17]
S1 domain of spike protein and its interaction with peptidase domain of ACE2
ACE2 also serves as a chaperone to the amino acid transporter BoAt1 (SLC6A19). A full length structure of ACE2-BoAt1 complex is already reported. The complex represent a homodimer (2× [ACE2-BoAt1]) of two heterodimers (ACE2-BoAt1), exhibit closed or open conformation due to shift of the peptidase domain (PD) of ACE2. However, the homo-dimerization is mediated by the collectrin like domain (CLD).[18] Interestingly, the open close conformation of the complex is governed by the state of the peptidase domain of the ACE2. The peptidase domain also binds to the S1 domain of spike protein. A dimeric ACE2 complex can accommodate two S proteins together. These interactions may play an important role in membrane invagination during endocytosis. However, ACE2 may stay as homodimer even in the absence of Bo AT1.[18] PDB I. D. 6M17 represents a cryo-EM Structure ACE-2 (amino acids 814) in the presence of the transporter B0AT1 with and without RBD of “SARS-COV-2-spike protein” (resolution of 2.9 angstroms).[18] The structure of “spike-protein RBD” in complex with “ACE2-BoAT complex” is shown in Figure 2.
Figure 2.
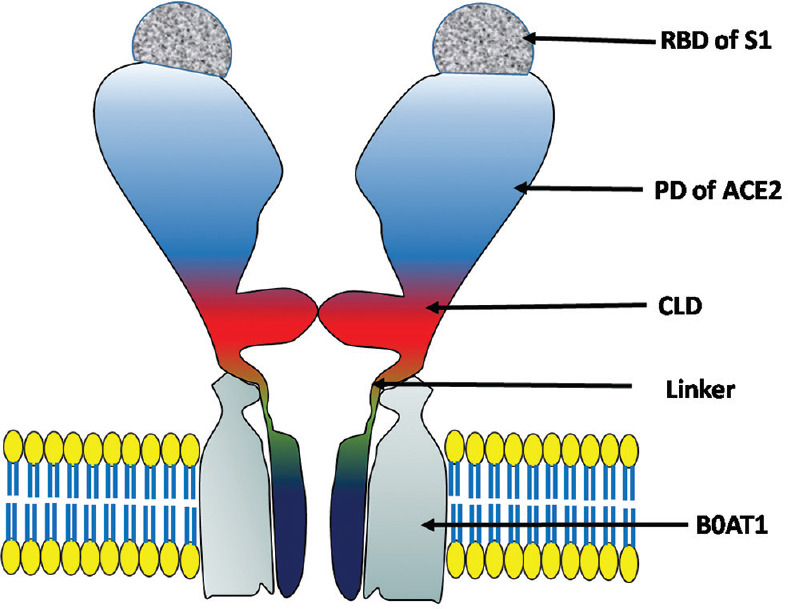
Structure of ACE-2 receptor with RBD region of S protein. Abbreviations: RBD = Receptor-binding domain of SARS-corona virus-2, PD = Peptidase domain of ACE2, CLT = Collectrin-like domain, BOAT1 = Sodium-dependent neutral amino acid transported B(o)AT1
The PD of ACE2 is the main region responsible for its interaction with the spike protein (dissociation constant [Kd] of ~15 nM).[17] The major interactions being H bond between RBD (Gln498, Thr500, and Asn501) with N terminal of ACE-2 (Tyr41, Gln42, Lys353, and Arg357), TYR453 and LYS417 of RBD region bind with middle of the bridge region of ACE2 (Asp30 and His34), hydrogen bond forms in between Gln474 residues of RBD and Gln24 residues of C terminal of ACE2. Phe486 of the RBD shows van-der Waal force interaction with Met 82 of ACE-2.[17]
S2 domain
The S2 subunit has a key role in the membrane fusion.[19] The homology between S2 segments of SARS-CoV and SARS-CoV-2 is around 89%.[20] The fusion process is initiated by the initial interactions between the RBD of spike S1 domain of SARS-CoV-2 and the PD of ACE2.[20] This process is followed by the interaction between heptad repeat1 (HR1) and heptad repeat2 (HR2) domain of S2 to form a six helix bundle fusion core, which fuses with the host cell membrane.[21] Compared to SARS-CoV-2, an eight residue sequence in the fusion region of S2 of SARS-CoV-2 imparts more stability to the six helix bundle core.[22] An already established pan CoV inhibitor EK1 was also found to be useful against SARS-CoV-2. By conjugating cholesterol with the EK1, a new molecule was designed (EK1C4), which showed more potent inhibitory action against SARS-CoV-2.[23]
Compared to SARS-CoV, the host membrane fusion ability is higher in the case of SARS-CoV-2. Few mutated amino acids in the HR1 results in enhanced interaction with the HR2 domain.[22] Details of interaction between HR1 and HR2 of S2 can be visualized in the PDB: 6 LXT.[22]
Role of fusion peptide of S2
The S2 domain contains two heptad repeats (HR1 and HR2) and one fusion peptide. In case of SARS-CoV, critical cleavage at R797 may expose the viral fusion peptide.[24] This fusion peptide-associated fusion platform (bipartite) alters the membrane integrity in the presence of calcium.[25] A proteolytic cleavage in the S2 exposes the fusion peptide. In virology, fusion peptides are used as the mediators for the fusion process as they help lipid membranes to fuse and permeabilize. In case of SARS-CoV-2, two fusion peptides are recognized that is SARSww-I and SARSww-II at the N terminal region of spike protein of nCoV. However, among these, only SARSww-I peptide robustly divided into the membranes of large unilamellar vesicles and induced the fusion of lipid vesicles causing vesicular aggregation and induced the leakage of lipid vesicles (an indicator of the propensity to perturb membrane integrity).[26]
In another experiment, treatment with bafilomycin or ammonium chloride decreased the SARS-CoV-entry to 293/hACE2 cells indicating endocytosis as the possible route of establishment of cellular infection. Some phosphoinositides may play an important role in this endocytosis process, for example, phosphatydleinositol 3-phosphate 5 kinase (PIPKIN) which plays a major role in the synthesis of phosphotydilinositol 3,5 bis phosphate in early endosome. Inhibiting PIPKIN with apilimod resulted in the decreased entry of SARS-CoV-2 spike pseudo-virion into 293/hACE2 cells. However, in the downstream, TCP2 was found to be another major determinant in the SARS-CoV-2 endocytosis process.[15]
Details of published target structures of S1 and S2
After searching the RCSB database, we could find eight structures of spike protein S1 and S2 domain, of which respective study was also published in a peer reviewed journal. However, we could find few structures, of which details were not available in the peer reviewed domain. However, among these structures, we could find one structure (PDB I. D. 6W41) in which RBD of S1 of SARS-CoV-2 was co-crystallized with a neutralizing antibody (isolated from convalescent SARS-CoV patients).[12] The PDB I. D. 6VW1 represents SARS-CoV-2 chimeric RBD crystallization structure complex.[16] The structures 6VXX and 6VYB represent the SARS-CoV-2 S ectodomain trimer both in both closed and open states. The PDB I. D. 6VSB represents a high-resolution structure (cryo EM) of the spike protein (3.46 Å) with sequence length of 1288 amino acids.[14] The PDB I. D. 6M0J represents a crystal structure of RBD region of spike protein (S) bound to the ACE2 receptor at the resolution of 2.45 Å.[13] Again, some of the polyclonal antibodies against spike protein of SARS-CoV also neutralized SATRS-CoV-2, indicating that the conserved regions of the S epitopes can be targeted for developing a vaccine.[27] However, three SARS-CoV specific antibodies failed to bind against the spike protein of SARS-COV-2.[14]
Protease
The replicase gene of SARS-CoV-2 encodes two polyproteins (pp1a and pp1ab), which are necessary for replication/transcription process of the virus.[20,28,29] The Main protease and the papain-like protease are the two major proteases, which processes these two polyproteins to release 16 nonstructural proteins (NSPs) of polyproteins which results from translation of viral RNA.[28]
Main protease/MPro/3CL pro
The structure typically contains two protomers (protomer I and II), each of which consisting of 3 domains (domain I, II and III), with around 100 amino acids in each domain. The substrate-binding site residues in the gap between I and II domain. One interesting feature is that the substrate binding site is highly conserved among all SARS-CoV Main protease and the most variable domain being helical domain III.[30] In domain III, five helices cluster is located which implicated in dimerization of Mpro through a salt bridge interaction (Arg-4 of one protomer and Glu-290 of the other protomer). For the catalytic activity, the dimerization is necessary because the N-finger of each protomer interacts with the other protomer through its interaction with residues Glu166 and thus helps to form the shape of the substrate binding site.[28] The structure of the main protease is represented in Figure 3.
Figure 3.
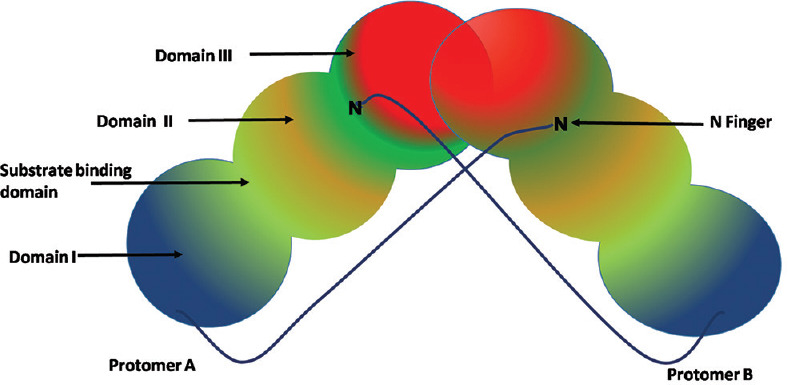
Structure of main protease. Each protomer consists of three domains containing approximately 100 amino acid residues in each. The substrate binding site resides in the cleft between domain I and II. Domain II and III is linked by a long loop
M pro has ability to cleave the larger pp1ab at particular 11 sites; The recognition site being leu-gln*-*ser-ala-gly in most of the cases.[28]
The Mpro crystal structure with an inhibitor (N3) is already reported (PDB I. D. 6 LU7).[30] Structure of SARS-CoV-2 Mpro is reported with α-ketoamide inhibitor (PDB: 6Y2E,6Y2G,6Y2F).[28]
Nonstructural proteins
The genomic organization of COVID-19 contains 5' UTR, orf-1ab, S, E, M, N gene and UTR 3'. Among them, ORF1ab cover the largest part of viral genome and is responsible for coding of 16 NSP nonstructural-proteins (1-16).[31] NSP3 and NSP 5 encodes the “papain like protease (PL pro)” and “chymotrypsin-like protease (CL Pro or M Pro)” respectively, while NSP15 shows endo-ribonulease activity.[32]
ORF1b encoded NSPs (12–16) are involved in RNA replication/transcription process. These NSPs e.g., RdRp and helicase complex comprises the replication transcription complex (RTC).[32,33] These RTC are typically anchored in endoplasmic reticulum (ER) originated double membrane containing vesicles which are further integrated into convoluted membranes (network of modified ER membrane).[33]
Nonstructural protein 15
Among the all SARS-CoV-2 NSP, the only available crystal-structure of is of NSP15 (PDB I. D. 6VWW, 371 amino acid, 2.20 Å).[34] However, none of the structures of NSP 15 were inhibitor bound. NSP15 belongs to the endoribonuclease (EndoU) family,[35] which play role in RNA processing. NSP15 is an essential element in the coronavirus life cycle.[36] Nsp15 is an endoribonuclease nidoviral RNA uridylate-specific (NendoU), belonging to the EndoU family. The C-terminal-NendoU domain is responsible for the catalytic-function of Nsp15. The active site present in between 2 B sheet, which comprises of six conserved residues HIS235, LY290, HIS250, THR341, SER294, TYR343.[36] NendoU activity of the Nsp15 can induce protein-interference in the innate-immune response, however, this effect may be independent of the endonuclease activity also. NSP-15 is a necessary element in the life cycle of for coronavirus.[36]
RNA-dependent RNA-polymerase
NSP12 also known as a RdRp is a key component in viral life cycle, which plays an essential role in synthesis of viral RNA and transcription/replication. NSP7 and NSP8 act are required cofactors for this machinery.[37]
The NSP12 structures comprises of the NiRAN domain towards the N terminal end and the RdRp domain toward the C terminal end.[38] NiRAN domain and polymerase domain are Linked by an interface domain.[39] The complex structure of RdRP is depicted in Figure 4.
Figure 4.
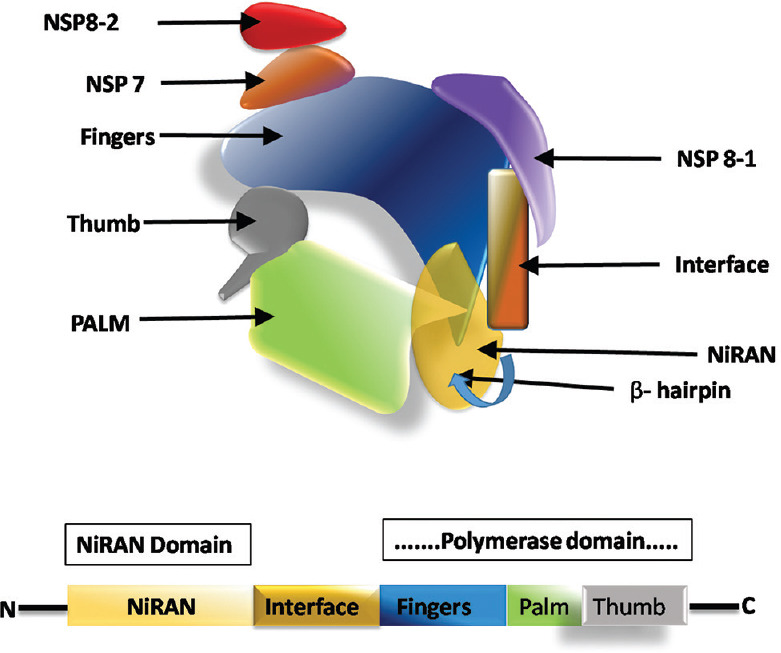
Structure of RNA-dependent RNA-polymerase in complex with nonstructural protein -7 and 8. Nonstructural protein 8 and nonstructural protein 7 are the necessary co-factors of nonstructural protein -12. The NiRAN domain and the polymerase domain of the nonstructural protein 12 complex are connected by an interface. A beta-hairpin structure resides in the N terminal domain
Nsp12 is thought to be a primary target of nucleotide analogues such as remdesvir (antiviral inhibitor), which has the ability to treat SARS-CoV-2 viral infection.[40] A cryo EM structure of NSP12 with remdesvir is reported in complex with triphosphate form of remdesvir, NSP-7 and NSP-8 (PDB I. D. 7BV2, 2.9-Å resolution),[41] in presence and absence of DTT.[39]
Conclusion
Detailed knowledge biological interactions are of utmost importance while designing new drugs or in case of virtual screening of ligand databases against particular target. We have structures of most of the SARS-CoV-2 targets; however, we don't have crystallized structure of few NSPs and few structural proteins yet. We need more structural details to understand the atomic level importance of different targets proteins and their interactions with inhibitors, which may pave way for the development of safe and effective anti-COVID-19 therapy.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Fehr AR, Perlman S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prajapat M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, et al. Drug targets for corona virus: A systematic review. Indian J Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarma P, Prajapat M, Avti P, Kaur H, Kumar S, Medhi B. Therapeutic options for the treatment of 2019-novel coronavirus: An evidence-based approach. Indian J Pharmacol. 2020;52:1–5. doi: 10.4103/ijp.IJP_119_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–8. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarma P, Kaur H, Kumar H, Mahendru D, Avti P, Bhattacharyya A, et al. Virological and Clinical Cure in Covid-19 Patients Treated with Hydroxychloroquine: A Systematic Review and Meta-Analysis. J Med Virol. 2020 doi: 10.1002/jmv.25898. doi:101002/jmv25898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Díez J-M, Romero C, Gajardo R. Currently available intravenous immunoglobulin contains antibodies reacting against severe acute respiratory syndrome coronavirus 2 antigens. Immunotherapy. 2020 doi: 10.2217/imt-2020-0095. DOI: 102217/imt-2020-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarma P, Shekhar N, Prajapat M, Avti P, Kaur H, Kumar S, et al. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain) J Biomol Struct Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1753580. doi:101080/0739110220201753580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceccarelli M, Berretta M, Rullo EV, Nunnari G, Cacopardo B. Editorial – Differences and Similarities between Severe Acute Respiratory Syndrome (SARS)-CoronaVirus (CoV) and SARS-CoV-2? Would a Rose by Another name Smell as Sweet Eur Rev Med Pharmacol Sci. 2020;24(5):2781–3. doi: 10.26355/eurrev_202003_20551. doi:10.26355/eurrev_202003_20551. [DOI] [PubMed] [Google Scholar]
- 10.Discovery of Bat Coronaviruses through Surveillance and Probe Capture-Based Next-Generation Sequencing mSphere. [Last accessed on 2020 Apr 26]. Available from: https://msphereasmorg/content/5/1/e00807-19abstract . [DOI] [PMC free article] [PubMed]
- 11.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS Coronavirus? J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. DOI: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan M, Wu NC, Zhu X, Lee CD, So RTY, Lv H, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–3. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–20. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 14.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–3. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–4. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–8. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan R, Zhang Y, Li Y, Xia L, Zhou Q. Structure of dimeric full-length human ACE2 in complex with B0AT1. bioRxiv. 2020 Preprint. [Google Scholar]
- 19.Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–21. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) Infection Inhibition Using Spike Protein Heptad Repeat-Derived peptides PNAS. [Last accessed on 2020 Apr 26]. Available from: https://wwwpnasorg/content/101/22/8455 . [DOI] [PMC free article] [PubMed]
- 22.Inhibition of SARS-CoV-2 (previously 2019-nCoV) Infection by a Highly Potent Pan-Coronavirus Fusion Inhibitor Targeting its Spike Protein that Harbors a high Capacity to Mediate Membrane Fusion Cell Research. [Last accessed on 2020 Apr 20]. Available from: https://wwwnaturecom/articles/s41422-020-0305-x . [DOI] [PMC free article] [PubMed]
- 23.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–55. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madu IG, Belouzard S, Whittaker GR. SARS-coronavirus spike S2 domain flanked by cysteine residues C822 and C833 is important for activation of membrane fusion. Virology. 2009;393:265–71. doi: 10.1016/j.virol.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai AL, Millet JK, Daniel S, Freed JH, Whittaker GR. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J Mol Biol. 2017;429:3875–92. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sainz B, Jr, Rausch JM, Gallaher WR, Garry RF, Wimley WC. Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J Virol. 2005;79:7195–206. doi: 10.1128/JVI.79.11.7195-7206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–920. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–12. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A New Coronavirus Associated with Human Respiratory Disease in China Nature. [Last accessed on 2020 Apr 26]. Available from: https://wwwnaturecom/articles/s41586-020-2008-3 .
- 30.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. doi: 101038/s41586-020-2223-y [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 31.da Silva SJ, da Silva CT, Mendes RP, Pena L. Role of nonstructural proteins in the pathogenesis of SARS-CoV-2 –. Silva J Med Virol. 2020 doi: 10.1002/jmv.25858. doi: 101002/jmv25858 [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Astuti I, Ysrafil Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response? Diabetes Metab Syndr. 2020;14:407–12. doi: 10.1016/j.dsx.2020.04.020. doi:10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagemeijer MC, Verheije MH, Ulasli M, Shaltiël IA, de Vries LA, Reggiori F, et al. Dynamics of coronavirus replication-transcription complexes. J Virol. 2010;84:2134–49. doi: 10.1128/JVI.01716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.RCSB PDB-6VWW: Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. [Last accessed on 2020 Apr 17]. Available from: https://wwwrcsborg/structure/6VWW .
- 35.Deng X, Baker SC. An “Old” protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018;517:157–63. doi: 10.1016/j.virol.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim Y, Jedrzejczak R, Maltseva NI, Wilamowski M, Endres M, Godzik A, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020 doi: 10.1002/pro.3873. 101002/pro3873 doi:101002/pro3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, Decroly E, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A. 2014;111:E3900–9. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehmann KC, Gulyaeva A, Zevenhoven-Dobbe JC, Janssen GM, Ruben M, Overkleeft HS, et al. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–34. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–82. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–71. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin W, Mao C, Luan X, Shen D-D, Shen Q, Su H, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. eabc1560. doi: 101126/scienceabc1560 [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]


