Abstract
The mating-type switching endonuclease HO plays a central role in the natural life cycle of Saccharomyces cerevisiae, but its evolutionary origin is unknown. HO is a recent addition to yeast genomes, present in only a few genera close to Saccharomyces. Here we show that HO is structurally and phylogenetically related to a family of unorthodox homing genetic elements found in Torulaspora and Lachancea yeasts. These WHO elements home into the aldolase gene FBA1, replacing its 3' end each time they integrate. They resemble inteins but they operate by a different mechanism that does not require protein splicing. We show that a WHO protein cleaves Torulaspora delbrueckii FBA1 efficiently and in an allele-specific manner, leading to DNA repair by gene conversion or NHEJ. The DNA rearrangement steps during WHO element homing are very similar to those during mating-type switching, and indicate that HO is a domesticated WHO-like element.
Research organism: S. cerevisiae, Other
eLife digest
In the same way as a sperm from a male and an egg from a female join together to form an embryo in most animals, yeast cells have two sexes that coordinate how they reproduce. These are called “mating types” and, rather than male or female, an individual yeast cell can either be mating type “a” or “alpha”. Every yeast cell contains the genes for both mating types, and each cell’s mating type is determined by which of those genes it has active.
Only one mating type gene can be ‘on’ at a time, but some yeast species can swap mating type on demand by switching the corresponding genes ‘on’ or ‘off’. This switch is unusual. Rather than simply activate one of the genes it already has, the yeast cell keeps an inactive version of each mating type gene tucked away, makes a copy of the gene it wants to be active and pastes that copy into a different location in its genome. To do all of this yeast need another gene called HO. This gene codes for an enzyme that cuts the DNA at the location of the active mating type gene. This makes an opening that allows the cell to replace the ‘a’ gene with the ‘alpha’ gene, or vice versa. This system allows yeast cells to continue mating even if all the cells in a colony start off as the same mating type. But, cutting into the DNA is risky, and can damage the health of the cell. So, why did yeast cells evolve a system that could cause them harm?
To find out where the HO gene came from, Coughlan et al. searched through all the available genomes from yeast species for other genes with similar sequences and identified a cluster which they nicknamed “weird HO” genes, or WHO genes for short. Testing these genes revealed that they also code for enzymes that make cuts in the yeast genome, but the way the cell repairs the cuts is different. The WHO genes are jumping genes. When the enzyme encoded by a WHO gene makes a cut in the genome, the yeast cell copies the gene into the gap, allowing the gene to ‘jump’ from one part of the genome to another. It is possible that this was the starting point for the evolution of the HO gene. Changes to a WHO gene could have allowed it to cut into the mating type region of the yeast genome, giving the yeast an opportunity to ‘domesticate’ it. Over time, the yeast cell stopped the WHO gene from jumping into the gap and started using the cut to change its mating type.
Understanding how cells adapt genes for different purposes is a key question in evolutionary biology. There are many other examples of domesticated jumping genes in other organisms, including in the human immune system. Understanding the evolution of HO not only sheds light on how yeast mating type switching evolved, but on how other species might harness and adapt their genes.
Introduction
Mating-type switching is an important process in the natural life cycle of many budding yeast species. If an uninhabitable environment improves and becomes habitable, any yeast spores that germinate earlier than others will have a competitive advantage, provided that they have a backup mechanism to prevent death if they germinate too early. Mating-type switching provides this backup (Hanson and Wolfe, 2017). Yeast spores are survival capsules, and mating-type switching enables the microcolony formed from a newly-germinated spore to sporulate again after just a few cell divisions if necessary, preventing its extinction (Herskowitz, 1988). Across the phylogenetic tree of budding yeasts, mating-type switching has arisen independently at least 11 times, indicating strong natural selection in favor of switching (Krassowski et al., 2019).
In Saccharomyces cerevisiae, HO is the central gene in the mating-type switching process. It was one of the first yeast genes ever discovered, because haploid strains with a functional HO gene can switch their mating type and hence auto-diploidize and form visible spores, whereas ho mutants cannot (Winge and Roberts, 1949; Oshima, 1993). HO codes for an endonuclease that makes a double-strand DNA break at the mating type (MAT) locus, and is essential for efficient switching (Kostriken et al., 1983; Russell et al., 1986). Much of our knowledge about how eukaryotes repair double-strand DNA breaks in their chromosomes comes from studies that used HO as a model system (Haber, 2016). But despite the comprehensive functional and genetic characterization of HO, its evolutionary origin remains mysterious (Keeling and Roger, 1995; Haber and Wolfe, 2005; Koufopanou and Burt, 2005; Muller et al., 2007). The HO gene is a relatively recent evolutionary addition into the yeast genome, because it is found only in a few genera closely related to Saccharomyces (Butler et al., 2004; Hanson and Wolfe, 2017). The ‘three-locus’ system for mating-type switching, involving an active MAT locus and silent HML and HMR loci, pre-dates the origin of HO. An outgroup species, Kluyveromyces lactis, also has a three-locus system but has no HO gene, and employs alternative mechanisms to create a double-strand break at the MAT locus to initiate switching (Barsoum et al., 2010; Rajaei et al., 2014). Some more distantly related budding yeasts switch mating types using ‘two-locus’ flip/flop inversion systems, and again do not have an HO gene (Hanson and Wolfe, 2017; Krassowski et al., 2019).
As well as being a recent evolutionary innovation, HO endonuclease also has an unusual protein domain structure that begs the question of where it came from. It resembles inteins, but is not an intein itself. Inteins are mobile genetic elements that are completely protein-coding and occur as in-frame fusions within a host gene (Novikova et al., 2014). After the host gene is transcribed and translated, the intein is excised post-translationally and the host protein is assembled by protein splicing, making a peptide bond between its N- and C-terminal parts (exteins). HO has highest sequence similarity to the VDE intein of budding yeasts, which is the only intein in S. cerevisiae (Koufopanou and Burt, 2005; Green et al., 2018). The host gene for VDE is VMA1, which codes for a subunit of vacuolar H+-ATPase (Gimble and Thorner, 1992; Anraku et al., 2005). The excised VDE intein has endonuclease activity and can cleave empty (inteinless) alleles of VMA1, enabling the intein to spread through the population by homing – a selfish, super-Mendelian mode of inheritance (Burt and Koufopanou, 2004; Burt and Trivers, 2006). The VMA1 genes of several species in the budding yeast family Saccharomycetaceae are polymorphic for the presence/absence of VDE, due to active homing and interspecies spread of the intein (Koufopanou et al., 2002; Okuda et al., 2003). Homing of VDE into empty alleles of VMA1 occurs during meiosis in diploids that are heterozygotes for intein-containing and empty alleles of VMA1 (Gimble and Thorner, 1992). The VDE protein has two domains (Moure et al., 2002): a protein splicing domain that enables the host protein Vma1 to be made, and a homing endonuclease domain that enables the VDE DNA sequence to home into empty alleles of VMA1.
Most inteins are found in bacteria and archaea, not yeasts (Poulter et al., 2007; Novikova et al., 2014; Green et al., 2018). In phylogenetic and other sequence similarity analyses, the two yeast proteins HO and VDE were found to be each other’s closest relatives (Dalgaard et al., 1997; Koufopanou and Burt, 2005; Green et al., 2018). Although HO is related to inteins, and more distantly related to other homing endonucleases in the LAGLIDADG superfamily (Chevalier and Stoddard, 2001), it is an independently expressed standalone gene, whereas inteins and other homing endonucleases are self-splicing entities embedded within their host genes (Belfort et al., 2005; Belfort, 2017). HO does not undergo protein splicing and has no exteins. HO also has a unique zinc finger domain at its C-terminus that is not present in other intein-like proteins or homing endonucleases. In S. cerevisiae HO, amino acid residues essential for cleavage of the MAT locus are located both in the zinc finger and in the endonuclease domain of the intein-like region (Meiron et al., 1995; Bakhrat et al., 2004; Bakhrat et al., 2006), and the endonuclease has a stringent requirement for zinc ions (Jin et al., 1997).
A key feature differentiating HO from true homing endonucleases is that it does not propagate its own DNA sequence – in other words, it does not home. HO has become a normal cellular gene, and is an example of a domesticated mobile genetic element (Volff, 2006; Rusche and Rine, 2010). However, until now it has been unclear what type of mobile element HO originated from. Here, we show that HO is related to a large and diverse family of intein-zinc finger fusion proteins (WHO proteins) that occur mostly in the yeast genus Torulaspora. WHO proteins are encoded by a newly discovered homing genetic element, whose genomic organization is different from all other known homing elements, and whose host is the aldolase gene FBA1. The similarities between WHO and HO proteins, and between the DNA rearrangement steps that occur during WHO element homing and mating-type switching, show how the HO-catalyzed system of mating-type switching originated.
Results
WHO genes code for a family of intein-zinc finger fusion proteins similar to HO
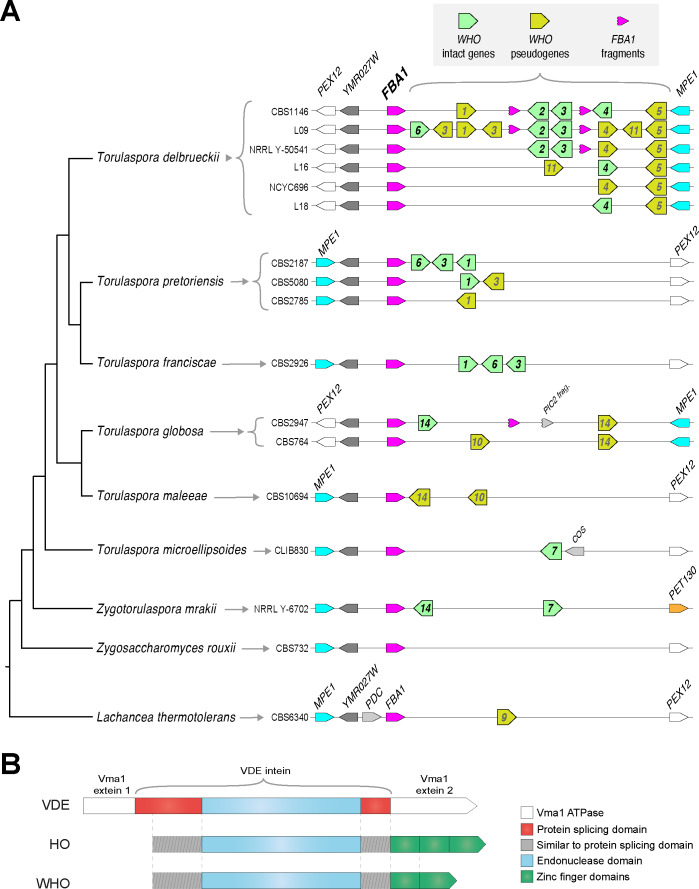
In the genome sequence of the type strain of Torulaspora delbrueckii (CBS1146; Gordon et al., 2011), we identified a cluster of five genes (TDEL0B06670 to TDEL0B06710) spanning 14 kb that have sequence similarity to HO. We renamed these genes WHO1 to WHO5, for ‘weird HO’ (Figure 1A). Two of them are pseudogenes, with a single frameshift in WHO1 and more extensive damage in WHO5. The WHO gene cluster is located downstream of the FBA1 gene encoding fructose-1,6-bisphosphate aldolase, an enzyme that functions bidirectionally in glycolysis and gluconeogenesis (Schwelberger et al., 1989). Amino acid sequence identity among the inferred WHO proteins is unusually low for a tandem gene cluster, ranging from 55% (Who2 vs. Who4) down to 24% (Who3 vs. Who4). T. delbrueckii also has an HO gene (TDEL0A00850) elsewhere in its genome, orthologous and syntenic with the HO gene of S. cerevisiae. The five inferred WHO proteins have only 22–25% identity to T. delbrueckii HO (BLASTP E-values in the range 1e-6 to 3e-30).
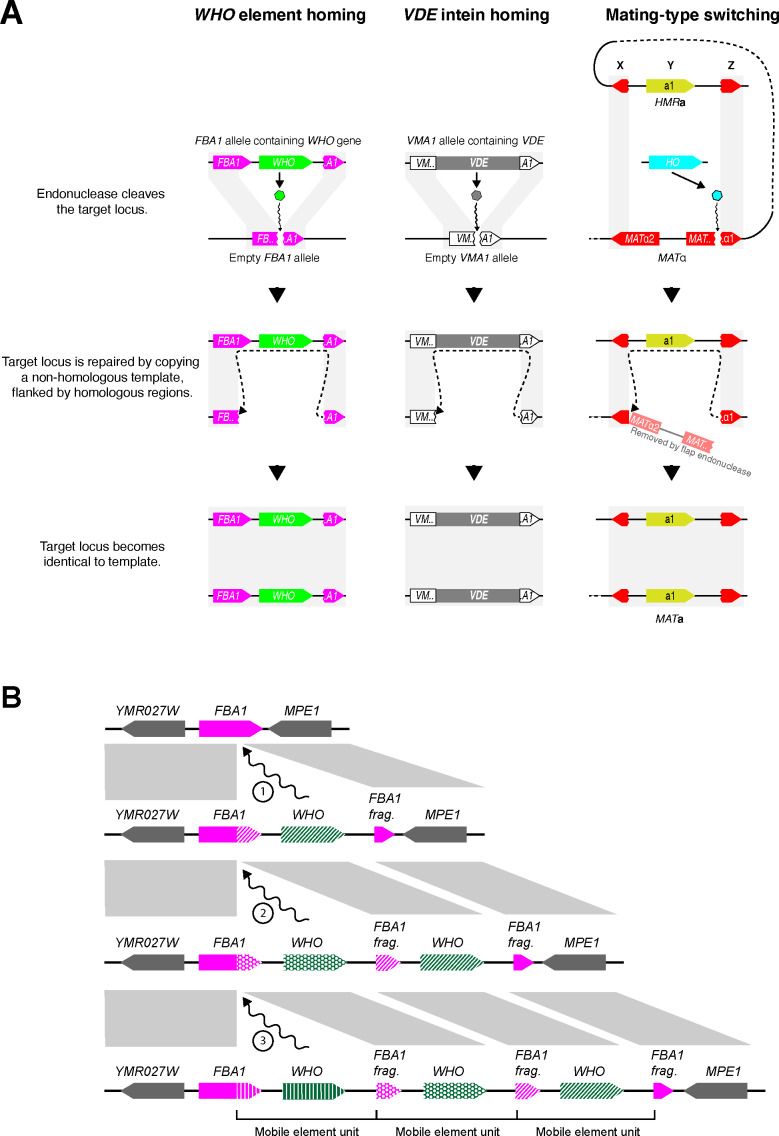
Figure 1. Genomic organization and domain structure of WHO genes.
(A) Polymorphic clusters of WHO genes and pseudogenes downstream of FBA1 in Torulaspora species. Multiple alleles are shown for T. delbrueckii, T. pretoriensis, and T. globosa. WHO genes are indicated by their family number. Fragments of the 3’ end of the FBA1 gene are marked. Genomic views are schematic and not drawn to scale. The phylogenetic tree is based on Shen et al. (2018). (B) Domain structure of HO, VDE and WHO proteins. The protein splicing domain is formed from two regions of the protein that flank the endonuclease domain (Moure et al., 2002).
Figure 1—figure supplement 1. Genomic organization and domain structure of WHO genes in Lachancea species.
The length and content of the WHO gene cluster is polymorphic among natural isolates of T. delbrueckii. This species has a primarily haploid (haplontic) life cycle (Kurtzman, 2011), so there is only one allele per strain. We found six different allelic WHO cluster arrangements among 15 strains we examined, with content ranging from 2 to 9 WHO genes and pseudogenes (Figure 1A; Supplementary file 1). The largest cluster (18 kb) is in strain L09, with three intact genes (WHO6, WHO2, WHO3) and six WHO pseudogenes.
As well as WHO genes and pseudogenes, some of the T. delbrueckii clusters contain one or two duplicated fragments of the 3’ end of FBA1, interspersed with WHO genes (Figure 1A). The FBA1 fragments are about 400 bp long (full-length FBA1 is 1080 bp). Some FBA1 fragments contain frameshifts or internal stop codons, and others are intact.
The WHO proteins consist of an intein-like region followed by a zinc finger domain (Figure 1B). This structure is similar to HO, but distinct from VDE which has no zinc finger. WHO and HO are the only intein-zinc finger fusion proteins known in any organism. WHO and HO both have no exteins, and their intein domains both lack an amino acid motif that is normally found at the C-terminal end (motif G; Pietrokovski, 1994). Similar to HO, the zinc finger domains of the WHO proteins contain variable numbers (3-5) of a Cys-X-X-Cys motif. By BLAST searches, we found that the most similar zinc finger domains in other yeast proteins occur in orthologs of S. cerevisiae Ash1 (a regulator of HO transcription), which is an atypical type of GATA zinc finger domain (Scazzocchio, 2000; Münchow et al., 2002), though the level of sequence identity is low (maximally 38% over 76 residues).
WHO gene/pseudogene clusters downstream of FBA1 are common in the genera Torulaspora and Lachancea
WHO gene/pseudogene clusters are present downstream of FBA1 in all five other species of the genus Torulaspora that we examined, and they again show within-species polymorphism in their gene content. We found three allelic WHO cluster arrangements among 9 T. pretoriensis strains, and two allelic arrangements in 2 T. globosa strains (Figure 1A). A pair of WHO genes is also present downstream of FBA1 in Zygotorulaspora mrakii, which is closely related to Torulaspora. There are no WHO genes in the next most closely related genus, Zygosaccharomyces.
By database searches, we found that WHO genes also occur in the genus Lachancea, but they are absent from almost every other sequenced budding yeast genome. Eleven of the 12 sequenced Lachancea species have clusters of WHO genes or pseudogenes downstream of FBA1, and five of these species have intact WHO genes (Figure 1—figure supplement 1). The WHO clusters in four Lachancea species also contain duplicated fragments of the 3’ end of FBA1. These structures in Lachancea are remarkably similar to the ones seen in Torulaspora, especially considering that these two genera are not closely related to each other, and there are no WHO genes in most other yeast genera. In addition, a few Lachancea genomes contain other WHO genes or pseudogenes at loci separate from FBA1 (Figure 1—figure supplement 1).
In summary, WHO genes resemble homing endonuclease genes (Gimble, 2000). Like HO, but unlike any other homing endonucleases, the endonuclease domain is fused to a zinc finger domain. The structure of WHO genes, and their occurrence in clusters that are mixtures of intact genes and pseudogenes, suggests that they are part of a mobile genetic element.
A WHO protein cleaves the T. delbrueckii FBA1 gene in an allele-specific manner
We hypothesized that WHO proteins are homing endonucleases whose target is the FBA1 gene. This hypothesis was motivated by two observations. First, WHO genes with very diverse sequences occur in tandem clusters downstream of FBA1, in both Torulaspora and Lachancea, suggestive of repeated integrations of different members of a mobile element family into the same target locus. Second, fragments of FBA1 are present within the WHO gene clusters in both Torulaspora and Lachancea (Figure 1 and Figure 1—figure supplement 1). All the FBA1 fragments consist of only the 3’ end of the gene, and many of them begin at approximately the same position (base 670–680 in the gene sequence), which we hypothesized could indicate a possible endonuclease cleavage site in FBA1.
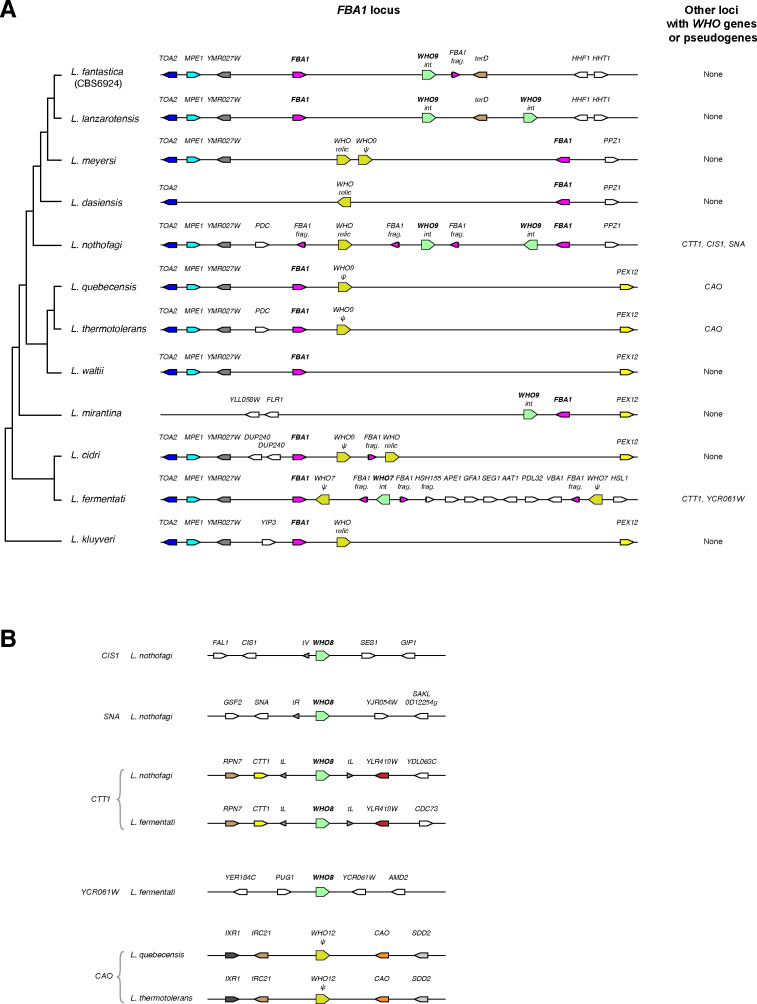
To test the hypothesis that WHO proteins target the FBA1 gene, we carried out experiments in S. cerevisiae because few tools exist for genetic manipulation of Torulaspora or Lachancea. We chose T. delbrueckii WHO6 for these experiments because it is intact, present in only a minority of the T. delbrueckii isolates we examined (3 of 15), and it is located at the end of the cluster closest to FBA1 (Figure 1A). Together, these features suggested that WHO6 could be the most recently-inserted WHO gene in the cluster in the strains that contain it, and therefore that WHO6 is a good candidate for a homing element that is currently active and spreading through the T. delbrueckii population.
FBA1 is an essential gene in most growth conditions (Lobo, 1984; Schwelberger et al., 1989; Boles and Zimmermann, 1993), so we reasoned that if it is the natural target of WHO endonuclease cleavage, then strains of T. delbrueckii that contain a WHO6 gene should contain alleles of FBA1 that are resistant to cleavage by Who6 endonuclease, whereas other T. delbrueckii strains might contain alleles that are sensitive to Who6. We constructed haploid strains of S. cerevisiae that contain the open reading frame (ORF) of T. delbrueckii FBA1 (TdFBA1) integrated into the ADE2 gene on chromosome XV. These strains also have the native S. cerevisiae FBA1 gene on chromosome XI. We used two different alleles of TdFBA1: one from a T. delbrueckii isolate (strain L09) that has a WHO6 gene downstream of it, and one from an isolate (CBS1146) that has no WHO6 gene (Figure 1A). We then introduced a high copy-number panARS plasmid (pWHO6-HA) on which WHO6 was expressed from the constitutive T. delbrueckii TDH3 promoter (Figure 2A), with a 3xHA epitope tag at its 3’ end. As described below, we found that this plasmid induces cleavage of the allele from strain CBS1146, but not of the allele from strain L09. Hence we designated the CBS1146 allele TdFBA1-S (for sensitivity to cleavage by Who6), and the L09 allele TdFBA1-R (for resistance). We also found that the plasmid does not induce cleavage of S. cerevisiae FBA1.
Figure 2. WHO6 induces allele-specific DNA cleavage of the T. delbrueckii FBA1 gene, with DNA repair by gene conversion or NHEJ.
(A) Summary of the experiment. Haploid S. cerevisiae strains, containing a non-expressed T. delbrueckii FBA1 ORF (TdFBA1-S or TdFBA1-R alleles) integrated at ADE2, were transformed multiple independent times with plasmids expressing WHO6 or WHO6-HA. In all transformations of strains containing the TdFBA1-S allele, the only colonies that survived expression of WHO6 were ones in which TdFBA1-S underwent DNA cleavage and repair by gene conversion or imprecise NHEJ, changing its sequence and making it resistant to further cleavage. (B) Gene conversion and imprecise NHEJ events in TdFBA1-S. The reference DNA sequence (uppercase) shows the 3’ end of the TdFBA1-S allele from T. delbrueckii strain CBS1146. Survivors 1–5 are transformants in which TdFBA1-S was cleaved by Who6 and repaired by imprecise NHEJ near position 668 (green box and triangle; survivors 1 and 2 have a 1 bp insertion, and survivors 3–5 have a 1 bp deletion, relative to the sequence TTT in the reference). Survivors 6–12 are transformants in which TdFBA1-S was cleaved by Who6 and partially overwritten by gene conversion with the endogenous S. cerevisiae FBA1 gene. Gene conversion regions are highlighted with pink backgrounds. The TdFBA1-R allele from T. delbrueckii strain L09, which is the natural host of WHO6, is also shown; this allele acquired no sequence changes among 10 independent pWHO6 transformants examined. A putative Who6 recognition site (yellow) and cleavage site with 4 bp 3’ overhang (underlined) are marked. Survivors 10 and 11 are from transformations with pWHO6-HA; all other survivors are from transformations with pWHO6. The complete TdFBA1 gene was sequenced from all transformants but only positions 616 to 975 are shown; there were no changes outside this region.
Our experiment is similar to one carried out by Moore and Haber (1996) who overexpressed HO so that it continually cleaved the MAT locus in haploid S. cerevisiae cells that had no HML/HMR loci. They found that the only cells that survived HO overexpression were ones in which inaccurate DNA repair ligated the chromosome back together but modified the target site sequence in such a way that HO could no longer cleave it, because chromosomes with accurate repairs were re-cleaved by HO. Similarly, in our experiment, the only haploid S. cerevisiae cells containing TdFBA1-S that survived overexpression of Who6 were ones in which the TdFBA1-S sequence became modified, either by gene conversion or by imprecise non-homologous end joining (NHEJ) (Figure 2A).
By genome sequencing, we found that in two independent experiments where pWHO6-HA was introduced into S. cerevisiae strains containing TdFBA1-S, this T. delbrueckii FBA1 allele was modified by gene conversion with the native S. cerevisiae FBA1 gene. In contrast, no gene conversion was seen when pWHO6-HA was introduced into S. cerevisiae strains containing the TdFBA1-R allele (two independent transformants), nor in control transformations in which a similar plasmid expressing 3xHA-tagged Green Fluorescent Protein (pGFP-HA) was transformed into S. cerevisiae strains containing TdFBA1-S or TdFBA1-R. Apart from the gene conversions at the TdFBA1-S transgene, no other nucleotide changes were detected in the genomes of the strains transformed with pWHO6-HA, and the native S. cerevisiae FBA1 gene remained unchanged in this experiment.
To verify that the gene conversions were not caused by the 3xHA tag, we carried out additional experiments in which S. cerevisiae strains containing either TdFBA1-S or TdFBA1-R were each independently transformed 10 times with either pWHO6 (a plasmid expressing Who6 with no 3xHA tag) or pGFP-HA as a control. A single colony was chosen from each transformation, and its ade2::TdFBA1 locus was amplified by PCR and sequenced. Approximately 100-fold fewer colonies were obtained from the transformations of pWHO6 into the TdFBA1-S strain than in any of the other three combinations of plasmid (pWHO6 or pGFP-HA) and allele (TdFBA1-S or TdFBA1-R). Evidence of cleavage of the T. delbrueckii FBA1 ORF was observed in all 10 independent transformants of pWHO6 into the TdFBA1-S strain: five transformants showed gene conversion between TdFBA1-S and the native S. cerevisiae FBA1 gene, and the other five had single-nucleotide insertions or deletions in TdFBA1-S consistent with cleavage and repair by imprecise NHEJ (Figure 2B). In contrast, no sequence changes at ade2::TdFBA1 were detected in the 10 independent transformants of pWHO6 into the TdFBA1-R strain, nor (as expected) in any of the 20 transformants with pGFP-HA.
From these experiments we conclude that the T. delbrueckii isolate (L09) that contains the WHO6 gene also contains, 588 bp upstream, an allele of FBA1 (TdFBA1-R) that is resistant to cleavage by the Who6 endonuclease. It can therefore stably maintain this endonuclease gene in its genome. In contrast, an isolate (CBS1146) that has no WHO6 gene contains an FBA1 allele (TdFBA1-S) that is sensitive to cleavage by Who6.
The WHO endonuclease cleavage site in FBA1
Similar to the HO overexpression survivors in Moore and Haber (1996), our transformants that contained the sensitive TdFBA1-S allele of T. delbrueckii FBA1 and survived overexpression of Who6 have acquired mutations that can be inferred to have damaged the Who6 recognition or cleavage sites, making the cells resistant to Who6. These mutations enable us to identify the approximate location of the Who6 recognition and cleavage sites in T. delbrueckii FBA1.
The five transformants showing evidence of imprecise NHEJ (survivors 1–5 in Figure 2B) each sustained a single 1 bp insertion or deletion in the TdFBA1-S sequence, at the same site in the gene (positions 667–669: TTT→TTTT, or TTT→TT). Like other LAGLIDADG endonucleases, HO and VDE both make a staggered double-strand break with 4 bp 3’ overhangs when they cleave DNA, and they have large (~24 bp) degenerate recognition sequences that span the cleavage site (Nickoloff et al., 1990; Gimble and Thorner, 1993; Taylor et al., 2012). We therefore infer that the overhang made by Who6 must include some or all of the TTT sequence centered on position 668 (Figure 2B). The HO recognition site at the MAT locus is moderately conserved between S. cerevisiae and other budding yeasts, and has at its core the sequence CGCAACA, where the 4 bp overhang is underlined. By analogy, we suggest that the core of the Who6 recognition site in the sensitive TdFBA1-S allele of T. delbrueckii FBA1 is CGCATTT (positions 663–669). In the resistant TdFBA1-R allele, 3 of these seven bases are different (CagcTTT). In S. cerevisiae FBA1, which is also resistant to cleavage by Who6, 3 of 7 bases are different (tGCtTTc) (Figure 2B).
The seven transformants showing evidence of gene conversion (survivors 6–12, including two from pWHO6-HA transformations and five from pWHO6 transformations) each replaced a section of the T. delbrueckii FBA1-S sequence with the corresponding section of the S. cerevisiae FBA1 gene from chromosome XI (Figure 2B). The T. delbrueckii and S. cerevisiae genes have 84% nucleotide sequence identity overall. The gene conversion tracts are asymmetrical: they extend rightwards (towards the stop codon of FBA1) from the cleavage site for 106–306 bp, whereas they extend leftwards for only 5–14 bp. This asymmetry suggests that one side of the WHO-induced double-strand break is more active in recombination than the other. Similarly, during mating-type switching in S. cerevisiae, the DNA on one side of the HO-induced break (the Z-side) participates in exchange with HML/HMR, whereas DNA on the Y-side remains inert until it is eventually clipped off (Lee and Haber, 2015).
In summary, the pattern of NHEJ events and gene conversions in S. cerevisiae cells that contain TdFBA1-S and survive continuous expression of a WHO protein is very similar to the pattern in cells that contain MAT and survive continuous expression of HO (Moore and Haber, 1996). We infer that WHO endonucleases cleave FBA1 genes at approximately base 668, which is slightly upstream of the 5’ ends of many of the FBA1 fragments seen in Torulaspora and Lachancea species. The core of the putative recognition site of a WHO endonuclease is also similar to that of HO, with the sequence 5’-CGC-3’ adjacent to the overhang.
Repeated homing continually replaces the 3’ end of FBA1 and builds WHO clusters
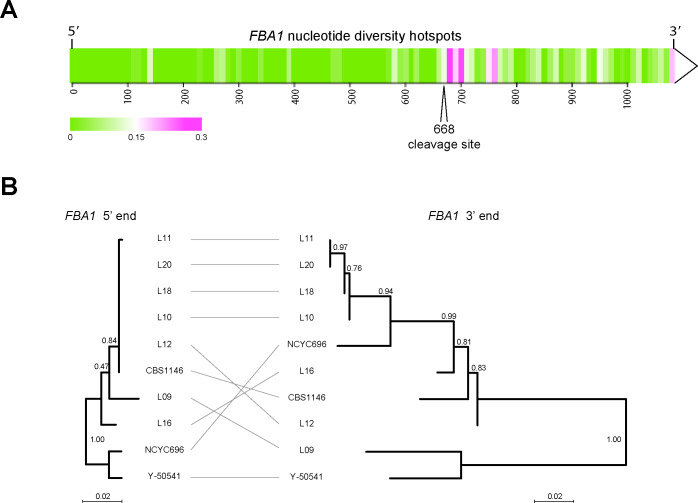
The TdFBA1-S and TdFBA1-R alleles have only 85% nucleotide sequence identity downstream of base 668, which is remarkably low for two alleles from the same species. In contrast, they have 99% identity upstream of this position. More generally, among the full-length FBA1 genes of the 15 T. delbrueckii isolates we analyzed, nucleotide sequence diversity is much higher in the 3’ part of the gene (Figure 3A). Moreover, phylogenetic trees constructed from the 5’ and 3’ parts of TdFBA1 (upstream and downstream of position 668) have contradictory topologies (Figure 3B). The heterogeneous evolution of the two ends of FBA1 in T. delbrueckii, and the presence of 3’ FBA1 fragments in its genome, suggest a mechanism for how the homing genetic element containing WHO genes operates.
Figure 3. Different evolutionary dynamics of the 5’ and 3’ parts of T. delbrueckii FBA1.
(A) Heatmap showing regions of nucleotide sequence diversity (π) among 15 sequenced FBA1 alleles from T. delbrueckii isolates, plotted in 10 bp windows. (B) Inconsistency of phylogenetic trees obtained from the 5’ and 3’ ends of FBA1 alleles from different T. delbrueckii strains (5’ end: bases 1–667; 3’ end: bases 669–1083). The alleles from strains CBS1146 and L09 are also called TdFBA1-S and TdFBA1-R, respectively. The trees are drawn to the same scale and were generated from nucleotide sequences using PhyML as implemented in Seaview v4.5.0 (Gouy et al., 2010) with default parameters. Bootstrap support from 1000 replicates is shown.
We propose that WHO elements home into the FBA1 locus by using a mechanism that involves replacing the 3’ end of FBA1, thereby converting a sensitive FBA1 allele into one that is resistant to the particular WHO protein encoded by the element (Figure 4A, left column). During meiosis in a heterozygous diploid cell, the WHO protein encoded by the donor allele cleaves the sensitive full-length FBA1 gene of the recipient allele at position 668. The double-strand break is then repaired by using the WHO-containing chromosome as a template. The 3’ region of the cleaved full-length FBA1 gene interacts with the FBA1 fragment in the template, resulting in incorporation of the WHO gene into the previously empty allele. After homing, the recipient chromosome contains a WHO gene located between a resistant full-length FBA1 gene (a chimera of the recipient’s previous 5’ end and a copy of the donor’s 3’ end) and a new FBA1 fragment formed from the recipient’s previous 3’ end. In this model, the FBA1 fragment downstream of the donor’s WHO gene is an essential part of the WHO element because it provides a region of homology that acts as a recombination site (Figure 4A). The fragment is not part of the expressed FBA1 gene so it does not need to maintain an open reading frame.
Figure 4. Proposed mechanism of WHO element homing.
(A) Similarity of mechanisms of action of WHO, VDE and HO. The mechanism we propose for WHO elements integrating at FBA1 is compared to the known mechanisms for VDE integrating into VMA1 and for HO-mediated switching of the MAT locus (Gimble and Thorner, 1992; Lee and Haber, 2015). WHO and VDE homing occur between allelic chromosomes in a diploid cell, whereas mating-type switching occurs between MAT and HML/HMR loci in a haploid cell. Gray rectangles indicate regions of sequence identity. The column on the right shows mating-type switching from MATα to MATa in S. cerevisiae. Switching from MATa to MATα occurs by an identical mechanism; the core of the HO recognition site (CGCAACA) is the first 7 nucleotides of the Z region, which is present in both of the MAT alleles even though it is part of the MATα1 gene sequence. The HO gene is on a different chromosome than MAT-HML-HMR. (B) Model for WHO cluster formation by successive integration of WHO elements. Every time a WHO element integrates into the locus, the 3’ end of the full-length FBA1 gene is replaced. The previous 3’ end is pushed rightwards, together with any older WHO genes, after which they can decay into pseudogenes. The complete WHO mobile element unit consists of a WHO gene and the upstream 3’ end of FBA1, which confers resistance to it.
While the modified FBA1 is now resistant to the newly acquired WHO gene, it may still be sensitive to other WHO genes. Repeated homing of multiple different WHO elements into the same chromosome will build tandem clusters of WHO genes and FBA1 fragments, with the most recent elements being located closest to the full-length FBA1 gene (Figure 4B). Each homing event replaces the 3’ end of the full-length FBA1 gene with the sequence from a different allele, causing its rapid evolution and discordant phylogenies. Over time, the WHO genes and FBA1 fragments can decay into pseudogenes or become deleted, because they are not required for the aldolase function of FBA1. The functional unit of a WHO element can be defined as a WHO gene and the resistance-conferring FBA1 3’ region upstream of it (Figure 4B).
In summary, T. delbrueckii WHO genes are part of a homing genetic element that targets FBA1. Our model for its mechanism of action explains how WHO clusters and FBA1 fragments are formed, and the unusual chimeric mode of evolution of FBA1.
Phylogeny of WHO genes shows an FBA1-associated backbone and multiple transpositions to other genomic loci
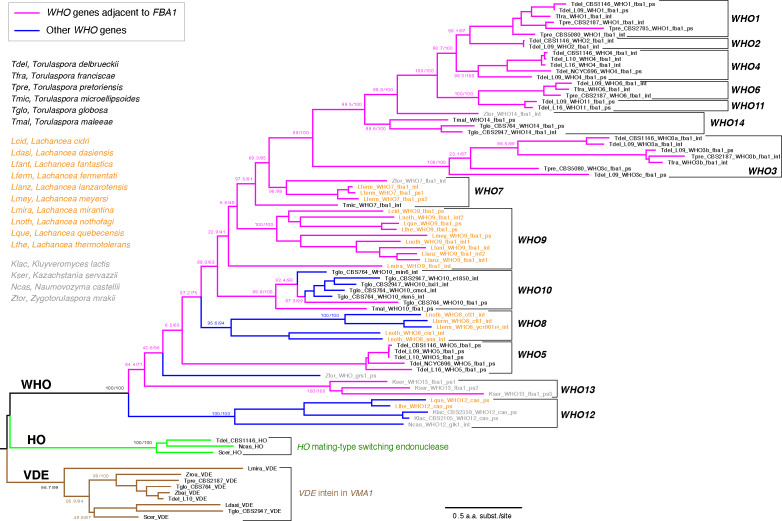
We investigated the phylogenetic relationship among WHO, HO, and VDE genes, using amino acid sequences inferred from intact genes and from some of the less-damaged WHO pseudogenes. In view of the high divergence among the sequences, the tree topology may not be fully accurate, but it permits identification of approximately 14 families of WHO genes (Figure 5). The WHO families form a monophyletic group, separate from HO and VDE. Most of the families are either Torulaspora-specific or Lachancea-specific, indicative of recent gene duplications within each genus. The WHO2, WHO4, WHO5 and WHO11 families are specific to T. delbrueckii so they must be young. Overall, the tree indicates a dynamic history of extensive WHO gene duplication and frequent formation of pseudogenes, consistent with the ‘cycle of degeneration’ expected for a homing genetic element (Burt and Koufopanou, 2004).
Figure 5. Families of WHO genes and their phylogenetic relationship to HO and VDE.
Magenta branches indicate WHO genes that are located at the FBA1 locus, and blue branches indicate WHO genes that are not beside FBA1. 14 WHO families are marked by brackets. Individual WHO gene names are colored by their source genus (black, Torulaspora; orange, Lachancea; gray, other genera). WHO gene names indicate the source species and strain number (if multiple strains were analyzed), WHO family (in uppercase), the name of a neighboring gene in the genome (in lowercase), and the suffix ‘int’ for intact WHO genes or ‘ps’ for WHO pseudogenes. Protein sequences were aligned using MUSCLE and filtered with Gblocks as implemented in Seaview v4.5.0 (Gouy et al., 2010). Badly degraded pseudogenes (relics) were not included. The tree was constructed by maximum likelihood using IQ-TREE v1.6.12 (Trifinopoulos et al., 2016), utilizing the built-in model finder option. Numbers on branches show support values from SH-aLRT and 1000 ultrafast bootstraps, separated by a slash (Trifinopoulos et al., 2016). The tree was rooted using VDE because WHO and HO share a zinc finger domain.
Figure 5—figure supplement 1. Some Kazachstania species have an intron in FBA1 at a location corresponding to the WHO cleavage site in Torulaspora.
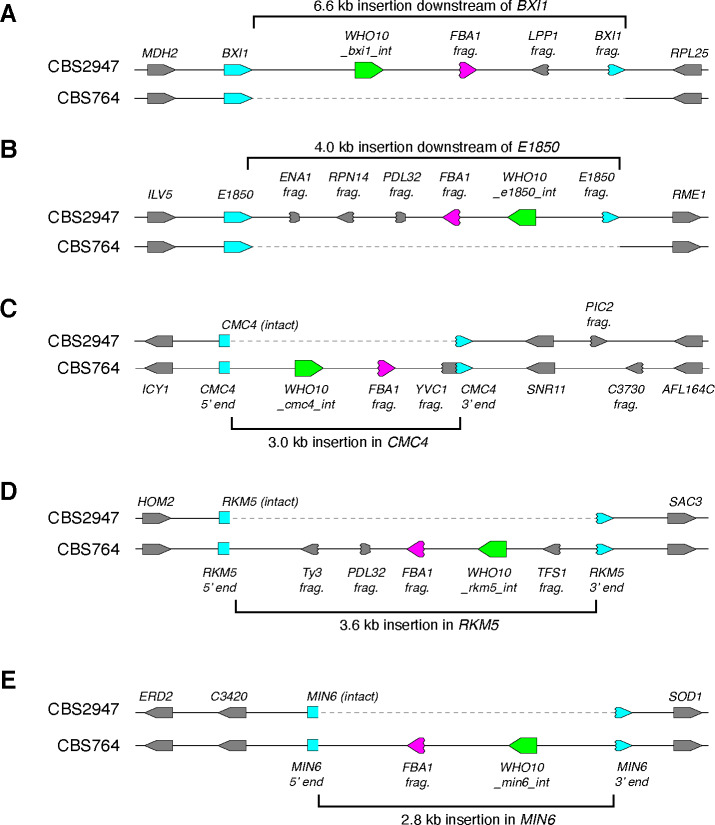
Although most WHO genes and pseudogenes are located downstream of FBA1 genes (magenta branches in Figure 5), a few of them are not. It is striking that these non-FBA1-associated genes fall into a small number of clades (blue branches in Figure 5). The WHO genes in these clades seem to have lost their target specificity for FBA1 and transposed to other places in the genome, and several of these WHO genes are intact. Most notably, the WHO10 family includes five intact genes from T. globosa that are located at five different places in the genomes of the two isolates we sequenced (Figure 6). There is a WHO10 pseudogene beside FBA1 in one T. globosa isolate and the sister species T. maleeae (Figure 1A), indicating that WHO10 was originally associated with FBA1. Another WHO clade unlinked to FBA1 is WHO8, which is present only in two species of Lachancea (Figure 5).
Figure 6. WHO10 is an active mobile genetic element in T. globosa and integrates into loci other than FBA1.
Each panel shows a pair of allelic regions from T. globosa strains CBS2947 and CBS764T. Intact WHO10 genes are shown in green, FBA1 fragments in magenta, and host genes in blue. (A-E) Five strain-specific insertions of WHO10 elements into different host loci. At the BXI1 and E1850 loci of CBS2947 (A,B), the 3’ end of the host gene became duplicated, whereas at the CMC4, RKM5 and MIN6 loci of CBS764 (C-E) the host gene was disrupted by the insertion and no part of it became duplicated. All the integrations contain a fragment of FBA1 immediately downstream of WHO10, and most also contain fragments of other genes. Genes are named after their S. cerevisiae orthologs where possible. E1850 is the T. globosa ortholog of T. delbrueckii TDEL0E01850, a gene with no homolog in baker’s yeast.
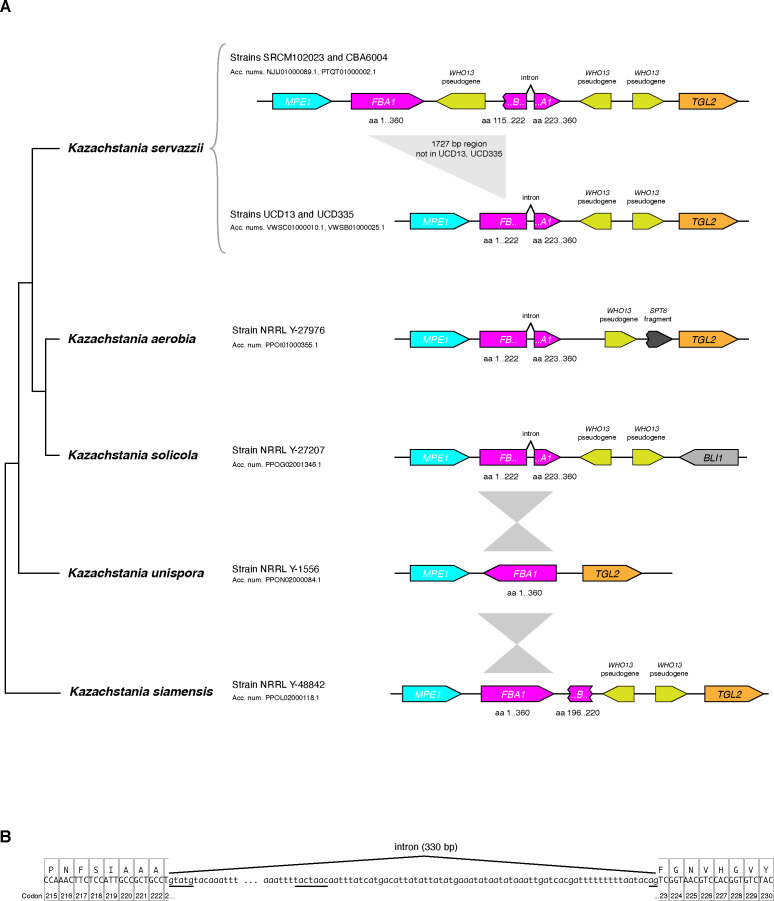
We detected only a few WHO sequences in species other than Torulaspora (or Zygotorulaspora) and Lachancea in BLAST searches against the NCBI database, which includes genome sequences from hundreds of yeast species including members of almost every genus in the family Saccharomycetaceae (Shen et al., 2018). Thus the WHO family has a very limited phylogenetic distribution, and occurs mostly in two genera that are not sisters of each other (Shen et al., 2018). The few WHO sequences outside Torulaspora and Lachancea all lie in the WHO13 and WHO12 families, which are outgroups to the other WHO families (Figure 5), and most of them are pseudogenes. WHO13 is FBA1-associated but WHO12 is not. The WHO13 sequences are all pseudogenes and were detected only in a small clade of Kazachstania species, downstream of FBA1. Interestingly, these species have an intron in FBA1 at precisely the inferred WHO endonuclease cleavage site (Figure 5—figure supplement 1), and this is the only intron in any budding yeast FBA1 gene. In other eukaryotes, evolutionarily novel introns are gained at sites of double-strand DNA breakage (Li et al., 2009).
The position of WHO12 as an outgroup to all the other WHO families (Figure 5) raises the possibility that the common ancestor of all the WHO families might have used a different gene as its original target, before changing target to FBA1 after the WHO12 family separated from the others. The only intact WHO12 gene occurs in Naumovozyma castellii, where it is located beside GLK1 (glucokinase). We sequenced the genomes of four strains of this species and found two with intact WHO12, and two with frameshifted WHO12 pseudogenes. There was no structural polymorphism or high sequence divergence at this locus in N. castellii, and no GLK1 gene fragments, so no evidence of active homing. The other WHO12 sequences are pseudogenes in K. lactis (Fabre et al., 2005) and two Lachancea species (Figure 1—figure supplement 1), beside a CAO copper amine oxidase gene in all three cases. The K. lactis WHO12 pseudogene lies between full-length CAO and a damaged fragment of CAO. It is therefore possible that WHO12 is an active element targeting CAO, and that CAO pre-dates FBA1 as the target of the whole WHO superfamily, but given the rarity of WHO12 it seems more likely that FBA1 is the ancestral target and WHO12 is a family that lost its specificity for FBA1 more recently, similar to WHO10.
In summary, the phylogenetic tree indicates that WHO genes have been located beside FBA1 throughout most of their diversification. The WHO genes that are not now located beside FBA1 are descended from FBA1-linked ancestors. The patchy taxonomic distribution of WHO genes suggests that they are native to the genus Torulaspora and/or Lachancea and have probably been transmitted between these genera by horizontal gene transfer. Horizontal transfer among budding yeast species has previously been inferred for the VDE intein (Koufopanou et al., 2002; Okuda et al., 2003). The split between the zinc finger-containing proteins HO and WHO pre-dates the diversification of the WHO families.
Recent transposition of WHO10 genes in Torulaspora globosa confirms the mobility of WHO elements
The WHO10 family has become amplified in T. globosa. In the two strains that we sequenced, strain-specific insertions of intact WHO10 genes are present at five loci not linked to FBA1 (Figure 6). The five Who10 proteins have only 72–80% amino acid sequence identity to one another. The T. globosa WHO10 genes are located within regions of inserted DNA 2.8–6.6 kb long. In two cases, the inserted DNA includes a duplicated fragment of the 3’ end of a host gene (BXI1 and E1850) at one end, so that the host gene was not disrupted (Figure 6A,B). These duplications resemble the FBA1 fragments seen downstream of FBA1 in many species (Figure 1A). In the other three cases, the WHO10-containing insertion interrupts a host gene without forming a duplication, probably inactivating it (Figure 6C,D,E).
All five T. globosa insertions include an FBA1 fragment immediately downstream of the WHO10 gene, even though the insertions are not near the FBA1 locus. Some insertions also contain fragments of various other genes, mostly from their 3’ ends (Figure 6). The structure and variable location of the DNA insertions indicate that WHO10 genes are part of a mobile genetic element that is active in T. globosa. The mobile element consists of WHO10 and the gene fragments, which may be molecular fossils of previous host sites into which the element inserted.
The genomic evidence indicates that most WHO genes function as part of a homing genetic element that targets the FBA1 locus (Figure 4). However, in T. globosa the WHO10 family has lost its specificity for FBA1 and become a more general mobile element rather than a homing element, resulting in the proliferation of intact WHO10 genes to multiple other sites in the genome. How the WHO10 genes become integrated into the non-FBA1 sites in T. globosa is not clear, because there are no homologous flanking sequences to guide integration of WHO10 into a double-strand break.
Discussion
We have shown that WHO elements are homing genetic elements in the budding yeast genera Torulaspora and Lachancea, that primarily target the aldolase gene FBA1. They have diversified into a large family with very divergent endonuclease genes. Our model proposes that WHO elements home into sensitive alleles of FBA1 by using a duplicated fragment of the 3’ end of FBA1 as a second region of homology downstream of the WHO gene (Figure 4A, left column). Homing replaces the 3’ end of FBA1, making it resistant to cleavage by the element’s WHO endonuclease. The DNA manipulation steps in WHO’s homing mechanism are identical to those that occur during VDE intein homing into VMA1, but the gene organization of WHO elements and their relationship to the host gene differ substantially from VDE (Figure 4A, first two columns). Resistance to endonuclease cleavage in FBA1 comes from allelic sequence differences, whereas resistance in VMA1 comes directly from interruption of the cleavage site by the VDE element (Gimble and Thorner, 1992).
FBA1 can be described as the host gene for the WHO element, even though the element lies downstream of FBA1 rather than interrupting it. This structural organization makes WHO elements different from the two currently recognized classes of homing genetic elements, which are inteins and intron-encoded homing endonucleases (Belfort et al., 2005; Belfort, 2017). In both of these other classes the homing element is a self-splicing entity, transcribed as an internal part of the host gene, that must be removed (by mRNA or protein splicing) in order to express the mature host protein. In contrast, WHO genes are transcribed independently of FBA1 (some of them are in the opposite orientation to FBA1; Figure 1A), and FBA1 will remain functional even if the WHO gene becomes a pseudogene. WHO elements therefore constitute a third structural class of homing element, and the only one with a propensity to form clusters. The mechanism of action of WHO elements has altered the evolutionary trajectory of their host gene FBA1, disrupting the normal vertical inheritance of this gene and leading to a chimeric mode of evolution in which the two ends of the gene have different histories (Figure 3).
The reason why WHO elements chose FBA1 as their host gene is probably that aldolase is absolutely required for spore formation, due to its role in gluconeogenesis (Dickinson and Williams, 1986). Meiosis and sporulation require cells to be grown on a non-fermentable carbon source such as acetate, and in these conditions gluconeogenesis is necessary to make the glucose monomers used for synthesis of the polysaccharide layers of the spore wall, a late stage in the meiosis-sporulation pathway (Neiman, 2005; Walther et al., 2014). S. cerevisiae fba1 mutants cannot make spores (Lobo, 1984; Dickinson and Williams, 1986) but they should not be blocked in meiosis, which is when WHO element homing is expected to occur. It is unlikely that the FBA1 genes in either the donor or the recipient chromosome can be transcribed at the same time as DNA cleavage and recombination is occurring during homing. By temporarily inactivating FBA1, the WHO element may be able to delay the cell from progressing from meiosis into sporulation until homing has finished. Homing is likely to be a slow process, because mating-type switching takes more than an hour (Lee and Haber, 2015).
During mating-type switching (Figure 4A, right column), HO initiates a series of DNA manipulation steps that closely resemble the steps that occur during homing of WHO elements and the VDE intein. Together with the sequence similarity among the three proteins, this similarity of the molecular mechanisms indicates a shared evolutionary origin of the three processes. While the mechanisms of WHO and VDE homing are essentially identical, the mechanism of HO action at the MAT locus has diverged from them in two critical ways. First, the HO gene is not part of the template used for DNA repair. Second, switching occurs in haploids, whereas homing occurs in diploids during meiosis. There is no homologous chromosome for the cleaved MAT locus to interact with, so instead it interacts with HML or HMR.
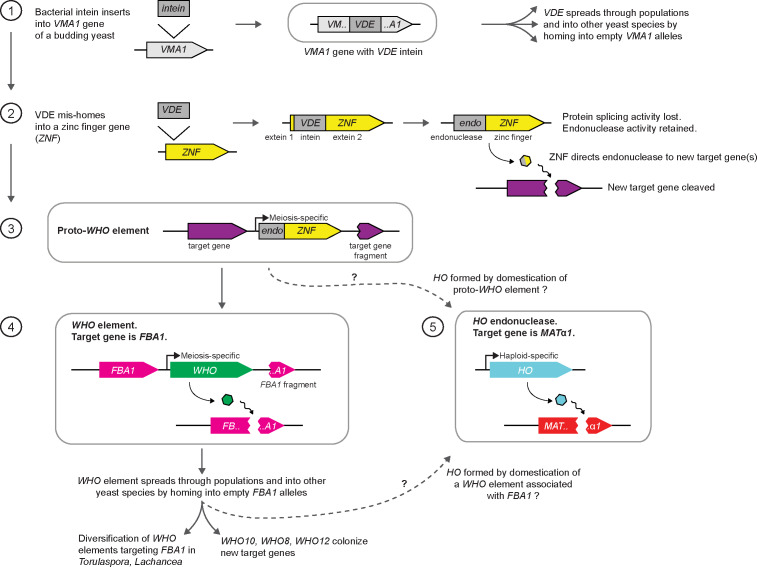
Our results finally illuminate the origin of HO endonuclease. Based on the fact that WHO and HO share features that are otherwise unique – the presence of a zinc finger domain and the absence of exteins – we propose the following evolutionary model (Figure 7). (1) An intein from a bacterial source invaded the VMA1 gene of an early budding yeast species to become VDE. (2) VDE subsequently duplicated and mis-homed into a zinc finger protein gene (possibly a paralog of ASH1), close to the 5’ end, to make a fusion gene that was the common ancestor of WHO and HO. The zinc finger directed the endonuclease to new target gene(s) in the genome. (3) The fusion gene became located between the target gene and a duplicated fragment of the target gene, forming a proto-WHO element. This step resembles some of the WHO10 insertion sites seen in T. globosa. The target gene may have been FBA1, or possibly a different, unknown, gene. To function as a homing element, the proto-WHO element must have had a meiosis-specific promoter. (4) The proto-WHO element diversified and spread through yeast populations and into additional species, with FBA1 as its main target. Occasional mis-homing events spread the element into new targets such as the WHO10 locations in T. globosa. (5) At an early stage of diversification, a WHO endonuclease developed an ability to cleave MATα1, in a species that already contained a three-locus MAT/HML/HMR mating-type switching system, and became domesticated as HO. During domestication, the transcriptional regulation of the gene must have changed from meiosis-specific expression to haploid-specific expression, as well as gaining cell lineage and cell cycle constraints (Stillman, 2013). The boundary between the Y and Z regions of the MAT locus, which was previously variable among species, became permanently fixed at the site where the endonuclease cleaved MATα1 (Figure 4A; Hanson and Wolfe, 2017).
Figure 7. Model for the origin of HO by domestication of a WHO element.
See Discussion for details. The dashed arrows indicate two possible routes to HO, from ancestral WHO elements that either were, or were not, specifically associated with FBA1.
Many examples are known of mobile genetic element genes that have been domesticated to take on a new role in the cell (Volff, 2006). In some of these examples, a domesticated endonuclease gene has retained its nucleolytic activity and functions in a programmed genome rearrangement process, such as the RAG1 gene in V(D)J recombination in the immune system of jawed vertebrates (Huang et al., 2016), and the PiggyMac gene in elimination of germline sequences during development of the macronucleus in Paramecium (Baudry et al., 2009). In other examples the ability to cleave DNA has been lost, such as in the bacterial DUF199/WhiA family which originated as a LAGLIDADG endonuclease but is now a regulator of transcription (Kaiser et al., 2009). HO has retained its endonuclease activity, and its origin from a homing element may help explain some unusual properties of this protein in vitro, such as its extreme catalytic inefficiency and its ability to attach to both ends of linear DNA molecules, forming loops visible by electron microscopy (Jin et al., 1997).
The domestication of a WHO element to become HO is similar to the domestication of the transposon-derived genes KAT1 and α3 to act as generators of double-strand breaks at the MAT locus during mating-type switching in K. lactis (Barsoum et al., 2010; Rusche and Rine, 2010; Rajaei et al., 2014). In all three cases, a mobile element gene was domesticated in a genome that already had a three-locus MAT/HML/HMR arrangement and probably switched mating types by a passive process based on homologous recombination without a specific mechanism for making a double-strand break at MAT. Why were mobile genetic elements repeatedly recruited into these switching systems? If we consider that, in any population of haploid cells, (1) mating-type switching can only increase a cell’s probability of mating, (2) mating leads to the formation of a diploid and inevitably to meiosis, even if many vegetative generations later, and (3) homing genetic elements can only home during meiosis, it logically follows that it is in a homing genetic element’s self-interest to increase the frequency of mating-type switching (Hanson and Wolfe, 2017). Thus, a WHO element could increase its rate of spread into empty FBA1 alleles in a population, if its WHO protein developed a secondary activity of cleaving the MAT locus as well as cleaving FBA1. Importantly, haplontic species such as Torulaspora and Lachancea require a nutritional signal to mate, so mating-type switching is unlikely to be followed by immediate mating of switched cells with their clonal relatives as occurs in S. cerevisiae. Instead, we expect that a switched cell in a haplontic species could go through many cycles of mitotic division before mating, which increases the opportunity for switched cell lineages to disperse and outbreed, and therefore increases the opportunity for homing elements to spread. Since frequent and accurate switching are probably favored by natural selection (Hanson et al., 2014), the subsequent steps that domesticated a WHO element to form the non-mobile and exquisitely regulated gene HO (Stillman, 2013) would also have been advantageous.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Torulaspora delbrueckii) | FBA1 | Gordon et al., 2011 | TDEL0B06660; NCBI: XM_003679958 | Strain CBS1146; TdFBA1-S allele |
| Strain, strain background (Saccharomyces cerevisiae) | IMX585 | Mans et al., 2015 | S. cerevisiae strain with integrated CAS9 gene | |
| Strain, strain background (Torulaspora delbrueckii) | L09; L10; L11; L12; L13; L15; L16; L18; L19; L20 | This paper | Strain collection of Lallemand, Inc | |
| Strain, strain background (Torulaspora delbrueckii) | NCYC696 | National Collection of Yeast Cultures (UK) | ||
| Strain, strain background (multiple Torulaspora species) | All CBS strains | Westerdijk Fungal Biodiversity Institute (Netherlands) | ||
| Strain, strain background (Torulaspora pretoriensis) | UWOPS 83–1046.2 | MA Lachance, University of Western Ontario (Canada) | ||
| Strain, strain background (Zygotorulaspora mrakii) | NRRL Y-6702 | CP Kurtzman, USDA Agricultural Research Service | ||
| Strain, strain background (Naumovozyma castellii) | Y056 (NRRL Y-12630T); Y174 (CBS4310); Y287 (CBS3006); Y668 (CBS1579) | Jure Piškur, Lund University (Sweden) | See Spírek et al., 2003 | |
| Recombinant DNA reagent | pMEL13 plasmid | Mans et al., 2015 | ||
| Sequence-based reagent | ADE2.Y sgRNA template | DiCarlo et al., 2013 | ||
| Recombinant DNA reagent | pIL75-KanMX plasmid | Liachko and Dunham, 2014 | ||
| Recombinant DNA reagent | pWHO6 | This paper | DNA sequence is in Supplementary file 3 | |
| Recombinant DNA reagent | pWHO6-HA | This paper | DNA sequence is in Supplementary file 3 | |
| Recombinant DNA reagent | pGFP-HA | This paper | DNA sequence is in Supplementary file 3 | |
| Sequence-based reagent | Sc ade2::TdFBA1-S repair template | This paper | DNA sequence is in Supplementary file 3 | |
| Sequence-based reagent | Sc ade2::TdFBA1-R repair template | This paper | DNA sequence is in Supplementary file 3 | |
| Sequence-based reagent | Primers for PCR of Sc ade2::TdFBA1 locus | This paper | 5’-TGACCACGTT AATGGCTCC-3’ and 5’-CACCAGCTCCA GCGATAATTG-3’ | |
| Software, algorithm | SPAdes assembler | Bankevich et al., 2012 | V3.11.1 | RRID:SCR_000131 |
| Software, algorithm | HGAP3 assembler | Chin et al., 2013 |
Yeast isolates and genome sequences
Genomes analyzed in this study are listed in Supplementary files 1 and 2. New genome sequences were obtained as follows. T. delbrueckii strains L09–L20 were from the strain collection of Lallemand Inc (Montréal, Canada), generously provided by Dr. Caroline Wilde. They were sequenced at the University of Missouri core facility (Illumina, SE 1 × 50 bp). T. delbrueckii strain NCYC696 data were downloaded from opendata.ifr.ac.uk/NCYC on 23-Feb-2017 as unassembled Illumina sequence reads (PE 2 × 100 bp). T. globosa strains CBS764T and CBS2947 were purchased from the Westerdijk Institute (Netherlands) and sequenced using both Illumina (PE 2 × 150 bp; BGI Tech Solutions, Hong Kong) and Pacific Biosciences Sequel technologies (1 SMRT cell; Earlham Institute, Norwich, UK). T. pretoriensis strain CBS2187T (Illumina, PE 2 × 100 bp + 6 kb library MP 2 × 100 bp) and T. franciscae strain CBS2926T (Illumina, PE 2 × 100 bp) were sequenced and assembled at INRAE Montpellier. Eight other T. pretoriensis strains (CBS2785, CBS5080, CBS9333, CBS11100, CBS11121, CBS11123, CBS11124 from the Westerdijk Institute, and UWOPS 83–1046.2 from M.A. Lachance, University of Western Ontario) were sequenced at the Earlham Institute using their proprietary LITE protocol for the Illumina platform. Zygotorulaspora mrakii strain NRRL Y-6702T was obtained from the USDA Agricultural Research Service (Peoria, IL, USA) and sequenced at the Earlham Institute using both Pacific Biosciences RSII (4 SMRT cells) and Illumina LITE methods. Naumovozyma castellii strains Y056, Y174, Y287 and Y668 were gifts from Prof. Jure Piškur (Lund University, Sweden) and were sequenced at the Earlham Institute using the Illumina LITE method.
Cultures were grown under standard rich-medium conditions. DNA for Illumina sequencing was harvested from stationary-phase cultures by homogenization with glass beads followed by phenol-chloroform extraction and ethanol precipitation. Purified DNA was concentrated with the Genomic DNA Clean and Concentrator-10 (Zymo Research, catalog D4010). DNA for PacBio sequencing was prepared as in Ortiz-Merino et al. (2017).
Illumina data were assembled using SPAdes version v3.11.1 (Bankevich et al., 2012). PacBio data were assembled using HGAP3 (Chin et al., 2013).
Other genome sequences used in this study were taken from the NCBI database. The previously published genome sequences for Torulaspora species are from Gordon et al. (2011), Gomez-Angulo et al. (2015), Tondini et al., 2018, Galeote et al. (2018) and Shen et al. (2018); Lachancea species are from Souciet et al. (2009), Sarilar et al. (2015), Vakirlis et al. (2016), Freel et al. (2016) and Kellis et al. (2004); and Kluyveromyces species are from Dujon et al. (2004) and Varela et al. (2019).
Construction of S. cerevisiae ade2::TdFBA1 strains
S. cerevisiae strains in which the coding region (ORF) of T. delbrueckii FBA1 was integrated into the S. cerevisiae ADE2 gene, in opposite orientation to ADE2 so that it is not functional, were constructed using CRISPR-Cas9 as follows. The ADE2-targeting sgRNA ADE2.Y from DiCarlo et al. (2013) was synthesized as a gene fragment by Integrated DNA Technologies, and inserted into the sgRNA plasmid pMEL13 from Mans et al. (2015) by restriction digestion and ligation. This plasmid was then transformed into S. cerevisiae strain IMX585 expressing Cas9 (Mans et al., 2015), together with a repair template containing the T. delbrueckii FBA1 ORF (TdFBA1-S or TdFBA1-R allele) flanked with homology to S. cerevisiae ADE2 in reverse orientation (bases 564456..564832 and 565952..566366 of S. cerevisiae chromosome XV). Sequences of the ade2::TdFBA1-S and ade2::TdFBA1-R constructs are given in Supplementary file 3. Transformants were selected on YPAD (YPD (Formedium) supplemented with 40 μg/ml adenine sulfate (Sigma)) containing 200 μg/ml G418. ADE2 knockouts were identified by formation of red colonies. Successful integrants were confirmed by PCR amplification of the ade2::TdFBA1 locus and Sanger sequencing (Eurofins). Two replicate ade2::TdFBA1-S strains were designated C1 and C4, and two replicate ade2::TdFBA1-R strains were designated L1 and L3.
WHO6 expression plasmid construction
Replicating plasmids constitutively expressing Who6, Who6-HA, or GFP-HA were constructed in the panARS replicating vector pIL75 (Liachko and Dunham, 2014) containing a KanMX marker. The nucleotide sequences of these plasmids (pWHO6, pWHO6-HA, pGFP-HA) are given in Supplementary file 3. In pWHO6, the WHO6 gene from T. delbrueckii strain L09 was placed under the control of the promoter and terminator of the T. delbrueckii glyceraldehyde-3-phosphate dehydrogenase gene TDH3 (TDEL0E04750). These regions were amplified by PCR from T. delbrueckii genomic DNA using high fidelity polymerase (New England Biolabs, M0492S) and inserted into pIL75 by restriction digestion and ligation. Plasmid pWHO6-HA is identical to pWHO6 except that its WHO6 gene is fused to a C-terminal 3xHA tag. A similar control plasmid (pGFP-HA) was made containing a GFP gene fused to 3xHA tag (GBlock made by Integrated DNA Technologies and inserted into pIL75 by restriction digestion and ligation), under the control of the T. delbrueckii TDH3 promoter and terminator.
Gene conversion assays
The S. cerevisiae ade2::TdFBA1 strains (C1, C4, L1, L3) as described above were transformed with the plasmids pWHO6, pWHO6-HA, or pGFP-HA. Multiple independent transformations were made to ensure that all gene conversion or NHEJ events recovered were independent. First, pWHO6-HA and pGFP-HA were each transformed into strains C1, C4, L1 and L3, and the whole genomes of these eight strains were sequenced, resulting in the unexpected discovery of gene conversion at the ade2::TdFBA1 locus when C1 and C4 were transformed with pWHO6-HA (called survivors 11 and 10, respectively, in Figure 2B). Genome sequencing was done by BGI Tech Solutions using a BGISEQ instrument with 50 bp single-end reads. Second, pWHO6 (no 3xHA tag) and pGFP-HA were each transformed 10 times into strains C4 and L1. The ade2::TdFBA1 locus from each transformant was amplified by PCR with a high-fidelity polymerase (New England Biolabs, M0492S) and Sanger sequenced. Primers for amplification were TGACCACGTTAATGGCTCC and CACCAGCTCCAGCGATAATTG.
For transformation, S. cerevisiae cells were grown overnight in liquid cultures of YPAD. Cultures were reinoculated in 50 ml and grown to mid-log phase. Cells were incubated with 1M LiAc, 50% PEG, salmon sperm DNA and plasmid DNA for 30 min, and then heat shocked at 42°C for 15 min. Transformants were selected on YPAD containing 200 μg/ml G418. Only one colony was picked from each transformation plate. The number of colonies obtained on the plates with the combination C4 + pWHO6 was dramatically lower (approximately 100-fold) than on the three other combinations. Individual colonies were grown in liquid YPAD overnight, before genomic DNA was harvested using a QIAamp DNA Mini kit (Qiagen).
Acknowledgements
We thank Caroline Wilde, Weilong Hao, Warren Albertin and Robert Mans for strains, Devin Scannell and Mike Eisen for preliminary data on T. globosa, and Raúl Ortiz-Merino and Amanda Lohan for assistance. We thank Sara Hanson, Geraldine Butler and Wolfe lab members for comments on the manuscript. We thank Jo Dicks for permission to use the NCYC696 sequence data, which were produced by the UK National Collection of Yeast Cultures, in partnership with The Earlham Institute, using funding awarded to the Institute of Food Research, Norwich, UK by the Biotechnology and Biological Sciences Research Council. This study was supported by Science Foundation Ireland (13/IA/1910) and the European Research Council (789341).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Kenneth H Wolfe, Email: kenneth.wolfe@ucd.ie.
Alan M Moses, University of Toronto, Canada.
Patricia J Wittkopp, University of Michigan, United States.
Funding Information
This paper was supported by the following grants:
Science Foundation Ireland 13/IA/1910 to Kenneth H Wolfe.
European Research Council 789341 to Kenneth H Wolfe.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Investigation, Writing - original draft, Writing - review and editing.
Investigation, Writing - review and editing.
Investigation.
Investigation.
Investigation.
Investigation.
Investigation.
Data curation, Investigation, Writing - review and editing.
Conceptualization, Formal analysis, Writing - original draft, Writing - review and editing.
Additional files
Data availability
Key nucleotide sequence data is provided in Supplementary File 3. New genome sequences have been deposited at NCBI. Their Bioproject numbers are in the dataset table and also in Supplementary files 1 and 2.
The following datasets were generated:
Coughlan AY, Lombardi L, Braun-Galleani S, Martos AAR, Galeote V, Bigey F, Dequin S, Byrne KP, Wolfe KH. 2020. Torulaspora franciscae genome sequencing. NCBI BioProject. PRJNA622240
Coughlan AY, Lombardi L, Braun-Galleani S, Martos AAR, Galeote V, Bigey F, Dequin S, Byrne KP, Wolfe KH. 2020. Torulaspora delbrueckii genome sequencing. NCBI BioProject. PRJNA623898
Coughlan AY, Lombardi L, Braun-Galleani S, Martos AAR, Galeote V, Bigey F, Dequin S, Byrne KP, Wolfe KH. 2020. Torulaspora delbrueckii strain:NCYC696 Genome sequencing. NCBI BioProject. PRJNA623891
Coughlan AY, Lombardi L, Braun-Galleani S, Martos AAR, Galeote V, Bigey F, Dequin S, Byrne KP, Wolfe KH. 2020. Torulaspora pretoriensis genome sequencing. NCBI BioProject. PRJNA623867
Coughlan AY, Lombardi L, Braun-Galleani S, Martos AAR, Galeote V, Bigey F, Dequin S, Byrne KP, Wolfe KH. 2020. Torulaspora globosa CBS764 genome sequencing and assembly. NCBI BioProject. PRJNA625704
Coughlan AY, Lombardi L, Braun-Galleani S, Martos AAR, Galeote V, Bigey F, Dequin S, Byrne KP, Wolfe KH. 2020. Torulaspora globosa CBS2947 genome sequencing and assembly. NCBI BioProject. PRJNA625705
Coughlan AY, Lombardi L, Braun-Galleani S, Martos AAR, Galeote V, Bigey F, Dequin S, Byrne KP, Wolfe KH. 2020. Zygotorulaspora mrakii strain:NRRL Y-6702 Genome sequencing and assembly. NCBI BioProject. PRJNA625702
Coughlan AY, Lombardi L, Braun-Galleani S, Martos AAR, Galeote V, Bigey F, Dequin S, Byrne KP, Wolfe KH. 2020. Naumovozyma castellii genome sequencing. NCBI BioProject. PRJNA623732
References
- Anraku Y, Mizutani R, Satow Y. Protein splicing: its discovery and structural insight into novel chemical mechanisms. IUBMB Life. 2005;57:563–574. doi: 10.1080/15216540500215499. [DOI] [PubMed] [Google Scholar]
- Bakhrat A, Jurica MS, Stoddard BL, Raveh D. Homology modeling and mutational analysis of ho endonuclease of yeast. Genetics. 2004;166:721–728. doi: 10.1534/genetics.166.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhrat A, Baranes K, Krichevsky O, Rom I, Schlenstedt G, Pietrokovski S, Raveh D. Nuclear import of ho endonuclease utilizes two nuclear localization signals and four importins of the ribosomal import system. Journal of Biological Chemistry. 2006;281:12218–12226. doi: 10.1074/jbc.M600238200. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum E, Martinez P, Aström SU. Alpha3, a transposable element that promotes host sexual reproduction. Genes & Development. 2010;24:33–44. doi: 10.1101/gad.557310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Bétermier M. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes & Development. 2009;23:2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Wood DW, Stoddard BL, Derbyshire V. Homing Endonucleases and Inteins. Springer-Verlag; 2005. [DOI] [Google Scholar]
- Belfort M. Mobile self-splicing introns and inteins as environmental sensors. Current Opinion in Microbiology. 2017;38:51–58. doi: 10.1016/j.mib.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E, Zimmermann FK. Saccharomyces cerevisiae phosphoglucose isomerase and fructose bisphosphate aldolase can be replaced functionally by the corresponding enzymes of Escherichia coli and Drosophila melanogaster. Current Genetics. 1993;23:187–191. doi: 10.1007/BF00351494. [DOI] [PubMed] [Google Scholar]
- Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Current Opinion in Genetics & Development. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Harvard University Press; 2006. [Google Scholar]
- Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe KH. Evolution of the MAT locus and its ho endonuclease in yeast species. PNAS. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Research. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ, Moser MJ, Holley WR, Chatterjee A, Mian IS. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Research. 1997;25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Research. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JR, Williams AS. A genetic and biochemical analysis of the role of gluconeogenesis in sporulation of Saccharomyces cerevisiae. Microbiology. 1986;132:2605–2610. doi: 10.1099/00221287-132-9-2605. [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wésolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Fabre E, Muller H, Therizols P, Lafontaine I, Dujon B, Fairhead C. Comparative genomics in Hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Molecular Biology and Evolution. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- Freel KC, Friedrich A, Sarilar V, Devillers H, Neuvéglise C, Schacherer J. Whole-Genome sequencing and intraspecific analysis of the yeast species Lachancea quebecensis. Genome Biology and Evolution. 2016;8:733–741. doi: 10.1093/gbe/evv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote V, Bigey F, Devillers H, Ortiz-Merino RA, Dequin S, Wolfe KH, Neuvéglise C. Genome sequence of Torulaspora microellipsoides CLIB 830T. Genome Announcements. 2018;6:e00615–e00618. doi: 10.1128/genomeA.00615-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble FS. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiology Letters. 2000;185:99–107. doi: 10.1111/j.1574-6968.2000.tb09046.x. [DOI] [PubMed] [Google Scholar]
- Gimble FS, Thorner J. Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature. 1992;357:301–306. doi: 10.1038/357301a0. [DOI] [PubMed] [Google Scholar]
- Gimble FS, Thorner J. Purification and characterization of VDE, a site-specific endonuclease from the yeast Saccharomyces cerevisiae. The Journal of Biological Chemistry. 1993;268:21844–21853. [PubMed] [Google Scholar]
- Gomez-Angulo J, Vega-Alvarado L, Escalante-García Z, Grande R, Gschaedler-Mathis A, Amaya-Delgado L, Arrizon J, Sanchez-Flores A. Genome sequence of Torulaspora delbrueckii NRRL Y-50541, Isolated from Mezcal Fermentation. Genome Announcements. 2015;3:e00438. doi: 10.1128/genomeA.00438-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Armisén D, Proux-Wéra E, ÓhÉigeartaigh SS, Byrne KP, Wolfe KH. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. PNAS. 2011;108:20024–20029. doi: 10.1073/pnas.1112808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Green CM, Novikova O, Belfort M. The dynamic intein landscape of eukaryotes. Mobile DNA. 2018;9:4. doi: 10.1186/s13100-018-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Wolfe K. Evolution and function of HO and VDE endonucleases in fungi. In: Belfort M, Wood D. W, Stoddard B. L, Derbyshire V, editors. Homing Endonucleases and Inteins. Berlin: Springer-Verlag; 2005. pp. 161–176. [DOI] [Google Scholar]
- Haber JE. A life investigating pathways that repair broken chromosomes. Annual Review of Genetics. 2016;50:1–28. doi: 10.1146/annurev-genet-120215-035043. [DOI] [PubMed] [Google Scholar]
- Hanson SJ, Byrne KP, Wolfe KH. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. PNAS. 2014;111:E4851–E4858. doi: 10.1073/pnas.1416014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SJ, Wolfe KH. An evolutionary perspective on yeast Mating-Type switching. Genetics. 2017;206:9–32. doi: 10.1534/genetics.117.202036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiological Reviews. 1988;52:536–553. doi: 10.1128/MMBR.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, Le Petillon Y, Liu X, Chen S, Schatz DG, Xu A. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Binkowski G, Simon LD, Norris D. Ho Endonuclease Cleaves MAT DNA in vitro by an inefficient stoichiometric reaction mechanism. The Journal of Biological Chemistry. 1997;272:7352–7359. doi: 10.1074/jbc.272.11.7352. [DOI] [PubMed] [Google Scholar]
- Kaiser BK, Clifton MC, Shen BW, Stoddard BL. The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease. Structure. 2009;17:1368–1376. doi: 10.1016/j.str.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Roger AJ. The selfish pursuit of sex. Nature. 1995;375:283. doi: 10.1038/375283a0. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kostriken R, Strathern JN, Klar AJ, Hicks JB, Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983;35:167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Koufopanou V, Goddard MR, Burt A. Adaptation for horizontal transfer in a homing endonuclease. Molecular Biology and Evolution. 2002;19:239–246. doi: 10.1093/oxfordjournals.molbev.a004077. [DOI] [PubMed] [Google Scholar]
- Koufopanou V, Burt A. Degeneration and domestication of a selfish gene in yeast: molecular evolution versus site-directed mutagenesis. Molecular Biology and Evolution. 2005;22:1535–1538. doi: 10.1093/molbev/msi149. [DOI] [PubMed] [Google Scholar]
- Krassowski T, Kominek J, Shen XX, Opulente DA, Zhou X, Rokas A, Hittinger CT, Wolfe KH. Multiple reinventions of Mating-type switching during budding yeast evolution. Current Biology. 2019;29:2555–2562. doi: 10.1016/j.cub.2019.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. The Yeasts, a Taxonomic Study. Amsterdam: Elsevier; 2011. Torulaspora Lindner (1904) pp. 867–874. [Google Scholar]
- Lee C-S, Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Microbiology Spectrum. 2015;3:0013–2014. doi: 10.1128/microbiolspec.MDNA3-0013-2014. [DOI] [PubMed] [Google Scholar]
- Li W, Tucker AE, Sung W, Thomas WK, Lynch M. Extensive, recent intron gains in Daphnia populations. Science. 2009;326:1260–1262. doi: 10.1126/science.1179302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Dunham MJ. An autonomously replicating sequence for use in a wide range of budding yeasts. FEMS Yeast Research. 2014;14:364–367. doi: 10.1111/1567-1364.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo Z. Saccharomyces cerevisiae aldolase mutants. Journal of Bacteriology. 1984;160:222–226. doi: 10.1128/JB.160.1.222-226.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NG, van den Broek M, Daran-Lapujade P, Pronk JT, van Maris AJ, Daran JM. CRISPR/Cas9: a molecular swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Research. 2015;15:fov004. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiron H, Nahon E, Raveh D. Identification of the heterothallic mutation in HO-endonuclease of S. cerevisiae using HO/ho chimeric genes. Current Genetics. 1995;28:367–373. doi: 10.1007/BF00326435. [DOI] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1996;16:2164–2173. doi: 10.1128/MCB.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moure CM, Gimble FS, Quiocho FA. Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence. Nature Structural Biology. 2002;9:764–770. doi: 10.1038/nsb840. [DOI] [PubMed] [Google Scholar]
- Muller H, Hennequin C, Dujon B, Fairhead C. Ascomycetes: the Candida MAT locus: Comparing MAT in the genomes of hemiascomycetous yeasts. In: Heitman J, Kronstad J. W, Taylor J. W, Casselton L. A, editors. Sex in Fungi: Molecular Determination and Evolutionary Implications. Washington: ASM Press; 2007. pp. 247–263. [DOI] [Google Scholar]
- Münchow S, Ferring D, Kahlina K, Jansen RP. Characterization of Candida Albicans ASH1 in Saccharomyces cerevisiae. Current Genetics. 2002;41:73–81. doi: 10.1007/s00294-002-0286-y. [DOI] [PubMed] [Google Scholar]
- Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff JA, Singer JD, Heffron F. In vivo analysis of the Saccharomyces cerevisiae HO nuclease recognition site by site-directed mutagenesis. Molecular and Cellular Biology. 1990;10:1174–1179. doi: 10.1128/MCB.10.3.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova O, Topilina N, Belfort M. Enigmatic distribution, evolution, and function of inteins. Journal of Biological Chemistry. 2014;289:14490–14497. doi: 10.1074/jbc.R114.548255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda Y, Sasaki D, Nogami S, Kaneko Y, Ohya Y, Anraku Y. Occurrence, horizontal transfer and degeneration of VDE intein family in Saccharomycete yeasts. Yeast. 2003;20:563–573. doi: 10.1002/yea.984. [DOI] [PubMed] [Google Scholar]
- Ortiz-Merino RA, Kuanyshev N, Braun-Galleani S, Byrne KP, Porro D, Branduardi P, Wolfe KH. Evolutionary restoration of fertility in an interspecies hybrid yeast, by whole-genome duplication after a failed mating-type switch. PLOS Biology. 2017;15:e2002128. doi: 10.1371/journal.pbio.2002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y. The Early Days of Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 1993. Homothallism, mating-type switching, and the controlling element model in Saccharomyces cerevisiae; pp. 291–304. [Google Scholar]
- Pietrokovski S. Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Science. 1994;3:2340–2350. doi: 10.1002/pro.5560031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter RT, Goodwin TJ, Butler MI. The nuclear-encoded inteins of fungi. Fungal Genetics and Biology. 2007;44:153–179. doi: 10.1016/j.fgb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Rajaei N, Chiruvella KK, Lin F, Aström SU. Domesticated transposase Kat1 and its fossil imprints induce sexual differentiation in yeast. PNAS. 2014;111:15491–15496. doi: 10.1073/pnas.1406027111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Rine J. Switching the mechanism of mating type switching: a domesticated transposase supplants a domesticated homing endonuclease. Genes & Development. 2010;24:10–14. doi: 10.1101/gad.1886310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Jensen R, Zoller MJ, Burke J, Errede B, Smith M, Herskowitz I. Structure of the Saccharomyces cerevisiae HO gene and analysis of its upstream regulatory region. Molecular and Cellular Biology. 1986;6:4281–4294. doi: 10.1128/MCB.6.12.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarilar V, Devillers H, Freel KC, Schacherer J, Neuvéglise C. Draft genome sequence of Lachancea lanzarotensis CBS 12615T, an ascomycetous yeast isolated from grapes. Genome Announcements. 2015;3:e00292-15. doi: 10.1128/genomeA.00292-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C. The fungal GATA factors. Current Opinion in Microbiology. 2000;3:126–131. doi: 10.1016/S1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- Schwelberger HG, Kohlwein SD, Paltauf F. Molecular cloning, primary structure and disruption of the structural gene of aldolase from Saccharomyces cerevisiae. European Journal of Biochemistry. 1989;180:301–308. doi: 10.1111/j.1432-1033.1989.tb14648.x. [DOI] [PubMed] [Google Scholar]
- Shen XX, Opulente DA, Kominek J, Zhou X, Steenwyk JL, Buh KV, Haase MAB, Wisecaver JH, Wang M, Doering DT, Boudouris JT, Schneider RM, Langdon QK, Ohkuma M, Endoh R, Takashima M, Manabe RI, Čadež N, Libkind D, Rosa CA, DeVirgilio J, Hulfachor AB, Groenewald M, Kurtzman CP, Hittinger CT, Rokas A. Tempo and mode of genome evolution in the budding yeast subphylum. Cell. 2018;175:1533–1545. doi: 10.1016/j.cell.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souciet JL, Dujon B, Gaillardin C, Johnston M, Baret PV, Cliften P, Sherman DJ, Weissenbach J, Westhof E, Wincker P, Jubin C, Poulain J, Barbe V, Ségurens B, Artiguenave F, Anthouard V, Vacherie B, Val ME, Fulton RS, Minx P, Wilson R, Durrens P, Jean G, Marck C, Martin T, Nikolski M, Rolland T, Seret ML, Casarégola S, Despons L, Fairhead C, Fischer G, Lafontaine I, Leh V, Lemaire M, de Montigny J, Neuvéglise C, Thierry A, Blanc-Lenfle I, Bleykasten C, Diffels J, Fritsch E, Frangeul L, Goëffon A, Jauniaux N, Kachouri-Lafond R, Payen C, Potier S, Pribylova L, Ozanne C, Richard GF, Sacerdot C, Straub ML, Talla E, Génolevures Consortium Comparative genomics of protoploid Saccharomycetaceae. Genome Research. 2009;19:1696–1709. doi: 10.1101/gr.091546.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spírek M, Yang J, Groth C, Petersen RF, Langkjaer RB, Naumova ES, Sulo P, Naumov GI, Piskur J. High-rate evolution of Saccharomyces sensu lato chromosomes. FEMS Yeast Research. 2003;3:363–373. doi: 10.1016/S1567-1356(02)00204-0. [DOI] [PubMed] [Google Scholar]
- Stillman DJ. Dancing the cell cycle two-step: regulation of yeast G1-cell-cycle genes by chromatin structure. Trends in Biochemical Sciences. 2013;38:467–475. doi: 10.1016/j.tibs.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GK, Petrucci LH, Lambert AR, Baxter SK, Jarjour J, Stoddard BL. LAHEDES: the LAGLIDADG homing endonuclease database and engineering server. Nucleic Acids Research. 2012;40:W110–W116. doi: 10.1093/nar/gks365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondini F, Jiranek V, Grbin PR, Onetto CA. Genome Sequence of Australian Indigenous Wine Yeast Torulaspora delbrueckii COFT1 Using Nanopore Sequencing. Genome Announcements. 2018;6:00321-18. doi: 10.1128/genomeA.00321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakirlis N, Sarilar V, Drillon G, Fleiss A, Agier N, Meyniel JP, Blanpain L, Carbone A, Devillers H, Dubois K, Gillet-Markowska A, Graziani S, Huu-Vang N, Poirel M, Reisser C, Schott J, Schacherer J, Lafontaine I, Llorente B, Neuvéglise C, Fischer G. Reconstruction of ancestral chromosome architecture and gene repertoire reveals principles of genome evolution in a model yeast genus. Genome Research. 2016;26:918–932. doi: 10.1101/gr.204420.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela JA, Puricelli M, Ortiz-Merino RA, Giacomobono R, Braun-Galleani S, Wolfe KH, Morrissey JP. Origin of lactose fermentation in Kluyveromyces lactis by Interspecies Transfer of a Neo-functionalized Gene Cluster during Domestication. Current Biology. 2019;29:4284–4290. doi: 10.1016/j.cub.2019.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan-Martini A, Lachance MA, Kurtzman CP. The Yeasts, a Taxonomic Study. Amsterdam: Elsevier; 2011. Kazachstania Zubkova (1971) pp. 439–470. [Google Scholar]
- Volff J-N. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Walther T, Létisse F, Peyriga L, Alkim C, Liu Y, Lardenois A, Martin-Yken H, Portais JC, Primig M, François J. Developmental stage dependent metabolic regulation during meiotic differentiation in budding yeast. BMC Biology. 2014;12:60. doi: 10.1186/s12915-014-0060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge Ö, Roberts C. A gene for diploidization in yeasts. Comptes Rendus Des Travaux Du Laboratoire Carlsberg. Serie Physiologique. 1949;24:341–346. [Google Scholar]