Abstract
Glaucoma, a multifactorial neurodegenerative disease characterized by progressive loss of retinal ganglion cells and their axons in the optic nerve, is a leading cause of irreversible vision loss. Intraocular pressure (IOP) is a risk factor for axonal damage, which initially occurs at the optic nerve head (ONH). Complex cellular and molecular mechanisms involved in the pathogenesis of glaucomatous optic neuropathy remain unclear. Here we define early molecular events in the ONH in an inherited large animal glaucoma model in which ONH structure resembles that of humans. Gene expression profiling of ONH tissues from rigorously phenotyped feline subjects with early-stage glaucoma and precisely age-matched controls was performed by RNA-sequencing (RNA-seq) analysis and complementary bioinformatic approaches applied to identify molecular processes and pathways of interest. Immunolabeling supported RNA-seq findings while providing cell-, region-, and disease stage–specific context in the ONH in situ. Transcriptomic evidence for cell proliferation and immune/inflammatory responses is identifiable in early glaucoma, soon after IOP elevation and prior to morphologically detectable axon loss, in this large animal model. In particular, proliferation of microglia and oligodendrocyte precursor cells is a prominent feature of early-stage, but not chronic, glaucoma. ONH microgliosis is a consistent hallmark in both early and chronic stages of glaucoma. Molecular pathways and cell type–specific responses strongly implicate toll-like receptor and NF-κB signaling in early glaucoma pathophysiology. The current study provides critical insights into molecular pathways, highly dependent on cell type and sub-region in the ONH even prior to irreversible axon degeneration in glaucoma.
Keywords: Glaucoma, Optic nerve head, Neuroinflammation, NF-κB, Toll-like receptor, Microglia, Oligodendrocyte, RNA-seq
Introduction
Glaucoma represents a spectrum of complex and multifactorial ocular disorders that are unified by an optic neuropathy with characteristic loss of retinal ganglion cells (RGCs) and their axons. Glaucoma remains a leading cause of irreversible blindness and visual impairment worldwide [1]. Intraocular pressure (IOP) is the most consistent and currently the only modifiable risk factor for glaucoma progression [2, 3], although individual susceptibility to IOP varies considerably [4, 5]. Current treatment strategies targeting the reduction of IOP often fail to prevent disease progression [6–8]. There is a critical need for deeper understanding of the cellular and molecular pathology of glaucoma, in order to develop more effective therapeutic strategies for patients with this common neurodegenerative disease.
In humans, RGC axons pass through “pores” between robust, multilayered, collagenous plates that form the lamina cribrosa (LC) of the optic nerve head (ONH). Substantial evidence from human patients, and from animal models including non-human primates, cats, and rodents, indicate that the LC region is an important initial site for optic nerve damage in glaucoma [9, 10]. A number of distinct, but potentially interrelated, pathobiological mechanisms have been implicated in glaucoma, including oxidative stress, biomechanical, vascular, neuroinflammatory, cellular, extracellular matrix-associated, and excitotoxic processes. These may all contribute to death of RGC axons and somas and ultimately result in loss of integrity of visual pathways [11–14]. However, molecular events that initiate, or are protective against, damage to RGC axons at the ONH in glaucoma have not been fully elucidated, largely due to limitations of existing experimental models and inability to reliably identify and study early-stage disease effectively in human patients.
Gene expression profiling provides comprehensive insight into molecular changes associated with disease. Advances in RNA-sequencing (RNA-seq) technology allow us to investigate transcriptome-wide gene expression without a priori assumptions. The greater dynamic range of RNA-seq, relative to hybridization-based microarrays, enhances its power to identify small, but potentially biologically important, quantitative changes. This enhanced sensitivity is valuable when studying an early phase of a complex disease process such as glaucomatous optic neuropathy, since changes in expression of individual genes may berelativelymodest. There havebeen very few reported RNA-seq studies conducted to examine ONH gene expression in spontaneous glaucoma models [15]. Importantly, these studies have been confined to rodents, in which the ONH lacks the collagenous plates of the human LC. To our knowledge, there have been no published studies which comprehensively interrogate early ONH gene expression changes in a spontaneous glaucoma model in which the ONH and LC closely resembles that of humans.
We previously reported a spontaneously occurring, recessively inherited form of feline congenital glaucoma (FCG), which represents an ortholog of human primary congenital glaucoma at the GLC3D locus (OMIM: 613086) due to a mutation in LTBP2 [16, 17]. In this feline model, onset of significant IOP elevation is recognized at 10 weeks of age, but glaucomatous optic neuropathy is chronic and gradually progressive over several years, as seen in many forms of human glaucoma [16, 18].
Here, we illuminate pro-inflammatory pathways and innate immune responses as early molecular events in the ONH at an early stage of glaucoma, while identifying cell proliferation and downregulation of neuronal and oligodendrocyte genes, prior to morphologically detectable RGC axon loss, in a cohort of meticulously phenotyped young cats with genetic glaucoma. Analyses of cell-specific gene enrichment provided evidence of significant enrichment of genesets that have been associated with pro-inflammatory such as LPS-activated microglia. In subsequent immunolabeling experiments, we confirmed microglial activation in the ONH in situ in this young cohort with early-stage disease. Furthermore, we identified proliferating cells of both microglia/macrophage and oligodendrocyte-lineage in early glaucomatous ONHs, but not in ONHs of adult subjects with established glaucoma. Together, these findings highlight molecular pathways associated with altered gene expression early in the course of glaucoma and provide strong evidence supporting roles for microglial activation and ONH cell proliferation in the early pathobiology of glaucoma.
Methods and Materials
Animals
Animals were maintained under a consistent 12 h light-dark cycle and all tissues collected between 8 A.M. and 11 A.M. In total, 33 domestic cats (Felis catus) were studied. Twelve, 10–12-week-old, homozygous LTBP2 mutant cats with fully penetrant, recessively inherited FCG [16, 19], and 7 age-matched wild-type (WT) cats of both sexes (FCG 6 male and 6 female, WT 1 male and 6 female) provided optic nerve head tissues for gene expression profiling. Subsequently, archived tissues from 7 adult homozygous LTBP2 mutant cats, with chronic glaucoma and 7 age-matched WTcats, were used for between-age/between-stage comparisons.
Phenotyping and Tonometry
Clinical ophthalmic phenotype was confirmed for all cats by ophthalmic examination by a board-certified veterinary ophthalmologist (GJM). Ophthalmic examination included rebound tonometry and slit-lamp biomicroscopy and indirect ophthalmoscopy following mydriasis. IOP was measured three times per week from 8 weeks of age by TonoVet® rebound tonometry (Icare Oy, Finland), as previously validated, in awake, gently restrained cats [18]. To minimize circadian variation in IOP, all values were obtained between 8 A. M and 10 A. M [20, 21].
Electrophysiological Testing
Full-field electroretinography (ERG) and visual evoked potential (VEP) were performed following standard procedures in the lab. Peak amplitudes and implicit times of a- and b-waves in ERG were scored, and intensity response function (Naka-Rushton) was plotted for b-wave amplitudes. Cortical VEP responses were recorded and root mean square of VEP amplitudes of the early wavelets (RMS), and peak amplitudes and latencies of the late positive component (designated P2) of VEPs were calculated. See Supplemental materials and methods for detailed methods.
OCT
Spectral domain optical coherence tomography (SD-OCT) was performed following electrophysiological tests, ensuring IOPs had been measured at ≤ 30 mmHg for ≥ 90 min to limit confounding effects of tissue compliance on parameters measured. Three to five ONH cube scans and HD horizontal raster scans centered on the ONH were obtained using Cirrus SD-OCT (Carl Zeiss Meditec Inc., Dublin, CA). To evaluate in vivo morphological ONH alterations, we manually quantified the following ONH parameters (Supplemental Fig. S1) using ImageJ (NIH, version 3.1): cup depth (CD), pre-laminar tissue thickness (PLT), neural canal opening (NCO), and posterior displacement of the lamina (PLD). Values for each parameter calculated from three high-quality images per subject (signal strength > 8/10) were averaged for analyses.
ONH Tissue Dissection
Eyes were enucleated immediately following euthanasia at the conclusion of clinical testing. Optic nerves were dissected 2 mm posterior to the globe for microscopy as outlined below and in Supplemental data (see Quantification of Optic Nerve Axons). The ONH was trephined by 4 mm biopsy punch, and adjacent retina, meninges, and connective tissue were carefully removed under magnification with a stereomicroscope under nuclease free conditions. The dissected ONHs were immersed in RNA stabilization solution (RNAlater®, Invitrogen, Carlsbad, CA) within 10 min of enucleation to prevent RNA degradation, incubated at 4 °C overnight and then stored at − 80 °C until RNA extraction.
Quantification of Optic Nerve Axons
A semi-automated targeted sampling method was used to quantify axons as validated and published for feline normal and glaucomatous optic nerves [22]. See Supplemental materials and methods for detailed methods.
RNA Sequencing, Read Alignment, Expression Estimation, and Differential Expression Analysis
Each RNA library was generated from total RNA isolated from ONH tissues. Libraries were sequenced by the Illumina® HiSeq2000 (Illumina, Inc., San Diego, CA) to obtain 100 bp strand-specific paired-end reads. Generated raw reads were quality-controlled, aligned, and mapped to the Felis catus reference genome (ICGSC Felis_catus_8.0/felCat8) using STAR [23] followed by estimation of transcript abundances using RSEM [24]. Differential expression (DE) analyses between normal and glaucomatous biological conditions were performed using DEseq2 [25]. Two other DE analysis tools were used to confirm the results of DEseq2. Differentially expressed genes (DEGs) between groups were considered significant with FDR < 0.05. See Supplemental materials and methods for detailed methods.
GSEA
Gene Set Enrichment Analysis (GSEA) was performed by pre-ranked enrichment analysis with GSEA (ver 2.2.0), analysis parameters: 1000 gene permutations [26]. The ONH RNA-seq dataset was processed following recommendations of the GSEA tutorial for RNA-seq data (http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html). To rank the genes, Log2 fold change (FC) values provided by DESeq2 were used. Normalized enrichment scores (NES) were used as a measure of magnitude of enrichment. Publicly available datasets for cell specific analysis as well as microglia molecular signature were downloaded and implemented in the GSEA analyses (Supplemental Table S1).
WGCNA
Weighted Gene Co-expression Network Analysis (WGCNA) is a co-expression network analysis that has been widely used in large transcriptome datasets [27]. A signed co-expression network was constructed using R package WGCNA (ver. 1.47) with normalized gene expression values filtered and log-transformed. Gene modules were formed by unsupervised clustering of genes by hierarchical clustering. The hub genes in upper quartile in the modules were visualized using Cytoscape (ver 3.3) [28] and Enrichment map Cytoscape plug-in [29]. See Supplemental materials and methods for detailed methods.
IF Labeling
Contralateral globes to those used for RNA-seq were fixed in 4% paraformaldehyde overnight at 4 °C. ONHs were then trephined by 4 mm biopsy punch and dissected as described above. The dissected ONH tissues were washed in 0.1 M PBS and immersed in a graded series of sucrose up to 30%, prior to embedding in Tissue-Tek® O.C.T. Compound (Sakura Finetek, Torrance, CA), and cryo-sectioning at 10 μm. Sections on slides were stored at − 80 °C until use. For immunofluorescence (IF), sections were dried for 1 h at room temperature, washed in 0.01 M PBS, and incubated for 45 min at room temperature in blocking solution (0.01 M PBS with 4% normal donkey serum, 1% BSA, and 0.1% Triton X-100). The slides were incubated overnight with appropriate dilutions of primary antibodies (Supplemental Table S2) in 0.01 M PBS with 2% normal donkey serum and 0.05% Tween 20, after washing three times with 0.01 M PBS. The sections were washed 3 times in 0.01 M PBS and then incubated with appropriate Alexa fluor® 488, 568, and 647 conjugated secondary antibody for 1 h at room temperature; nuclei were counterstained with 1:1000 DAPI (Life technologies) for 3 min then sections mounted using ProLong Gold Antifade Mountant (Life technologies). For each run, appropriate feline tissues that express the target protein as positive controls, and negative controls (omitting primary antibody) were included. All IF experiments were repeated at least three times for each condition to confirm reproducibility of the results. Images were captured using an SP8 confocal microscope (Leica, Buffalo Grove, IL). Relative intensities of IF labeling of GFAP, normalized to negative controls, were compared in sub-regions of the ONH, designated pre-laminar (PL), LC, and retro-laminar (RL).
Statistics
For quantitative values, data from one eye per cat were compared between groups by two-tailed unpaired t test or ANOVA with Tukey’s multiple comparison test, after confirming normal distribution of data by Kolmogorov-Smirnov test. Where appropriate, to account for multiple hypothesis testing, Benjamini-Hochberg false discovery rate (FDR) procedures were applied. Unless noted otherwise in the text, descriptive statistics are presented as mean and standard error of the mean (SEM), and values of P < 0.05 considered significant. GraphPad InStat software (ver 3.06) and GraphPad Prism (ver 8.1.2) were used to conduct the statistical analyses. For RNA-seq DE analyses and functional enrichment analyses, an FDR < 0.05 was used, as implemented in DESeq2 software.
See Supplemental materials and methods for detailed methods as well as information on functional enrichmentanalysis and RT-qPCR.
Results
Functional Deficits and ONH Remodeling Precede Histopathologically Detectable RGC Axon Damage
Modest but statistically significant IOP elevation, first identified inFCG at 10 weeks of age relative to age-matched normal cats (Fig. 1a), was associated with subtle functional deficits characterized by reduced amplitude of VEP (Fig. 1b, c). Although VEP can reflect functional abnormalities anywhere in the visual pathway between distal retina and visual cortex, concurrent full-field flash ERG identified no significant differences in retinal responses between groups (Supplemental Fig. S2). Analysis of OCT-derived ONH structural parameters in vivo identified evidence of early ONH remodeling in FCG at 10–12 weeks of age (Fig. 1d, e). However, axon quantification and light microscopic evaluation of optic nerve morphology, which are established indicators of glaucomatous damage and stage of disease, revealed no quantitative or qualitative evidence of axonal damage (Fig. 1f–l). Collectively, in vivo and histologic findings represented an early stage of glaucoma in this cohort of young FCG cats, prior to significant irreversible axon loss.
Fig. 1.
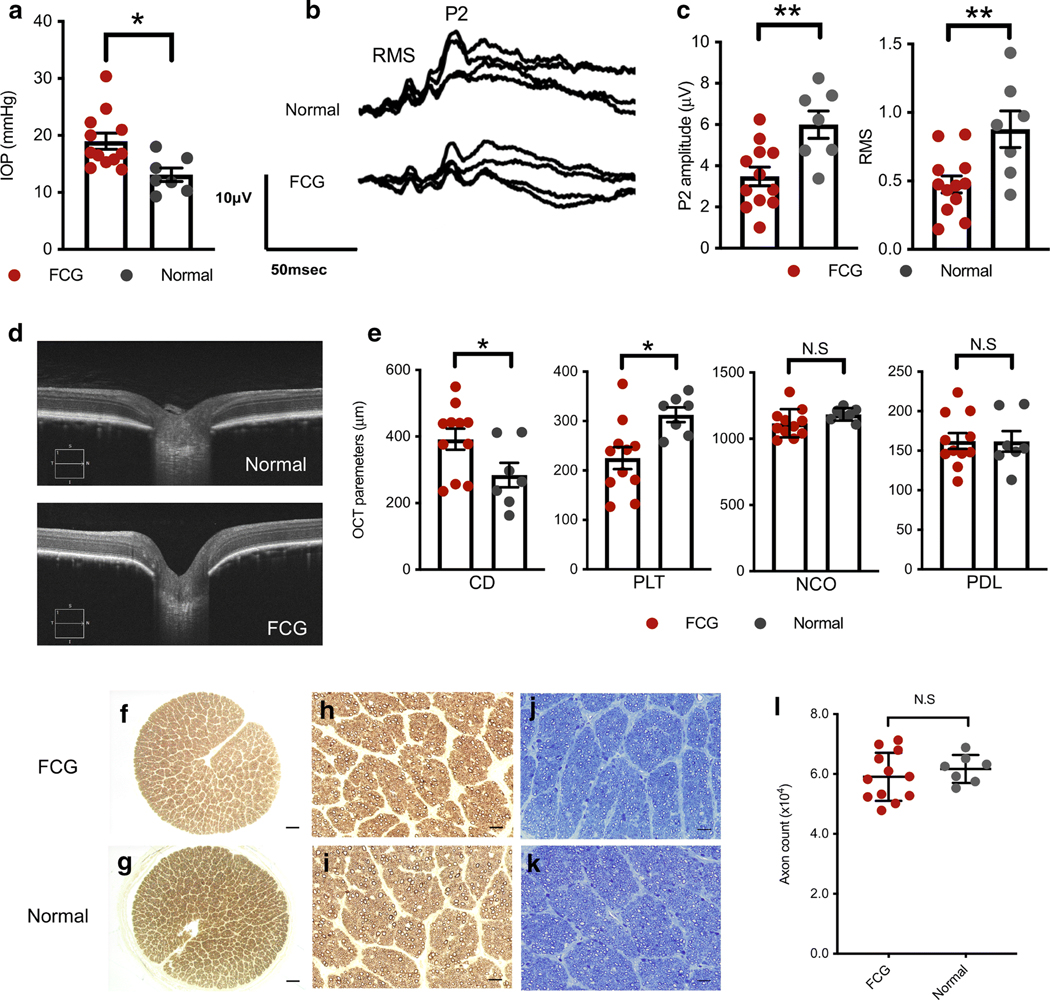
Optic nerve head (ONH) remodeling and functional deficits are evident soon after onset of IOP elevation, prior to axon loss in feline congenital glaucoma (FCG). Mean and SEM are presented throughout; all comparisons between normal (n = 7) and FCG (n = 12) were by unpaired 2-tailed t test. * P < 0.05; ** P < 0.01. a Intraocular pressure (IOP)was significantly higher in FCG than in normal subjects at 10 weeks of age (rebound tonometry; P = 0.01). b, c Peak amplitude of the late positive component (P2) and root mean square (RMS) of the early wavelets of the visual evoked potential (VEP) were significantly lower (P = 0.0058 and 0.0064, respectively) in FCG than controls at 10–12 weeks of age. d Representative ONH spectral domain-optical coherence tomography (SD-OCT) in vivo images of 12-week-old FCG (IOP 15 mmHg, scan quality 10/10) and normal cat (IOP 16 mmHg, scan quality 9/10). e Comparison of SD-OCT-derived quantitative ONH parameters between FCG and normal subjects revealed significantly increased optic cup depth (CD) and reduced pre-laminar tissue thickness (PLT) in FCG (P = 0.044 and 0.011, respectively), whereas width of the neural canal opening (NCO) and posterior displacement of the lamina (PDL) were not significantly different between groups. f–k Axon loss is not an early pathological feature in FCG. Representative optic nerve cross sections from 10- to 12-week-old FCG (f) and normal cats (g). No morphological evidence of axonal damage was observed in FCG (f, h, j) compared to normal control nerves (g, i, k) in either p-phenylenediamine (which stains myelin sheaths) (h, i) or Richardson’s stained sections (j, k). Scale bars = 200 μm (f, g); 20 μm (h–k). l Mean optic nerve axon count in FCG (n = 12) was not significantly different from normal (n = 7; P = 0.447, unpaired 2-tailed t test)
Cell Proliferation and Inflammatory Related Genes Are Upregulated in the ONH in Early Glaucoma
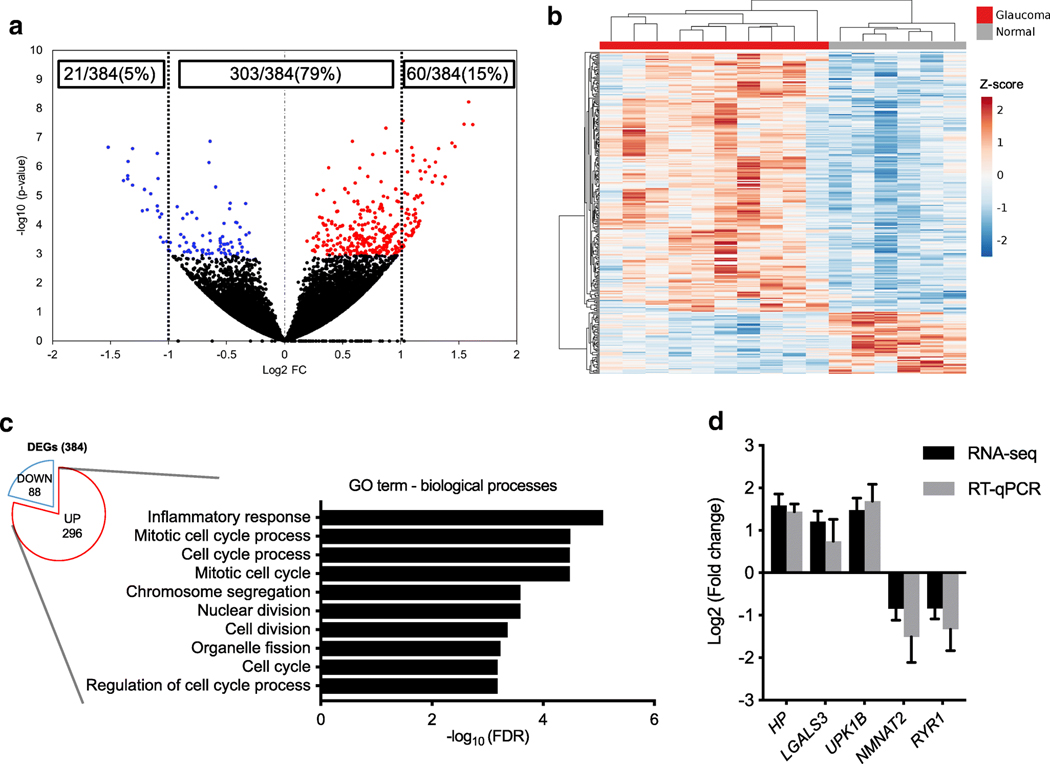
To interrogate early glaucomatous ONH transcriptomic changes in an unbiased comprehensive manner, we carried out RNA-seq of 10 ONHs from 10 to 12-week-old FCG cats and 6 age-matched controls. We identified 384 genes that were differentially expressed in the ONH of FCG relative to normal cats using DEseq2. Of these DEGs, 77% (296/384) were upregulated in FCG, whereas 23% (88/384) were downregulated, and 79% (303/384) expression changes were twofold or less (Fig. 2a). DEG expression profiles were remarkably consistent between subjects within either disease or control groups (Fig. 2b). The most highly up- and downregulated DEGs are presented in Supplemental Tables S3 and Supplemental Table S4, respectively (a list of all DEGs identified is also provided in Supplemental data 1). To provide greater confidence in the DEGs identified, we used two other differential expression analysis tools: edgeR and EBSeq. Over 92% (140/151) and 82% (153/185) of the DEGs identified by edgeR and EBSeq, respectively, overlapped with those identified by DEseq2. DEG expression profiles were remarkably consistent between subjects within groups, clustered by disease status (Fig. 2b). To provide a functional context for the DEGs identified, we utilized functional enrichment analyses by gene ontology (GO) terms and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Inflammatory response and mitotic cell process were highly over-represented GO terms identified as significantly enriched within the upregulated DEGs in FCG by homology-based GO term assignment and enrichment analysis (Fig. 2c). No significantly enriched GO term was identified in the downregulated DEGs. Cell cycle and infectious diseases, such as tuberculosis and leishmaniasis that activate immune/inflammatory responses, were highly enriched pathways identified on KEGG pathway analysis (FDR < 0.05). RT-qPCR for selected, representative DEGs (HP, LGALS3, UPK1B, NMNAT2, and RYR1) yielded directions and magnitude of gene expression differences between FCG and normal control ONH tissues consistent with and thereby validating RNA-seq results (Fig. 2d).
Fig. 2.
Cell proliferation and inflammatory-related genes are upregulated in the ONH in early glaucoma. a Volcano plot illustrating ONH gene expression differences between FCG (n = 10) and age-matched normal cats (n = 6). Each dot represents an individual gene. Red dots represent significantly upregulated, and blue dots represent significantly downregulated differentially expressed genes (DEGs) with thresholds at FDR < 0.05. Of the DEGs, 79% (303/384) showed less than two-fold changes in expression. b Hierarchical clustered heatmap depicts individual sample and gene expression differences of 384 DEGs between the ONHs from FCG on left (n = 10) and normal cats on right (n = 6). Scale: z –value. c In the upregulated DEGs, GO:0006954 inflammatory response (FDR = 8.3×10−6 ) and GO:1903047 mitotic cell cycle process (FDR = 3.22 × 10−5) were highly over-represented. d RT-qPCR performed for selected DEGs identified differences in gene expression between groups (n = 5 biological replicates per group) with comparable magnitude and direction of gene regulation to RNA-seq. Error bars in RNA-seq and RT-qPCR data represent standard error
Microglia and Oligodendrocyte Precursor Cells Proliferate in Early Spontaneous Glaucoma but Proliferating Cell Populations Vary by ONH Sub-region
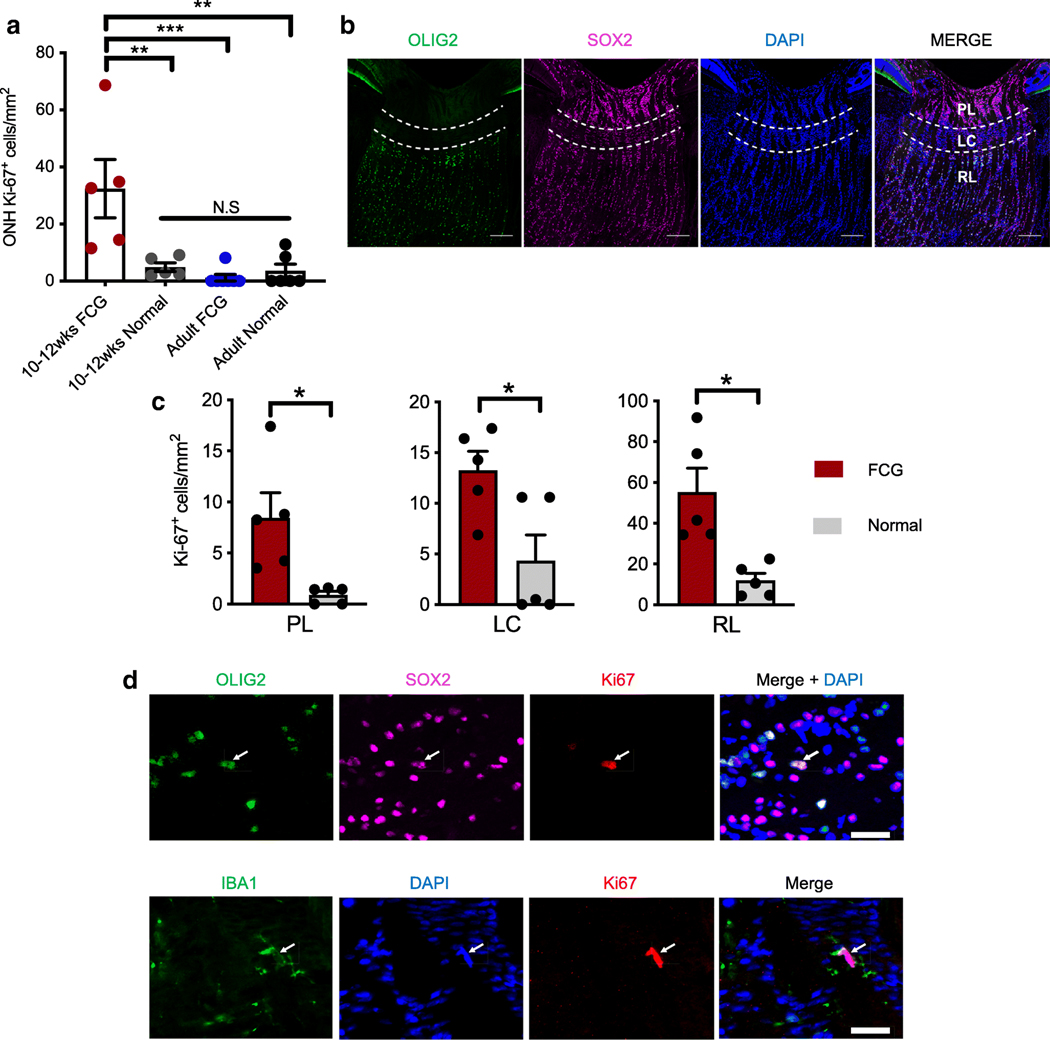
As we identified significant upregulation of genes associated with the cell cycle in the ONH of early FCG relative to agematched controls, we conducted a series of experiments to establish the degree and nature of cell proliferation in the ONH of cats with early FCG. Density of cells expressing Ki67, a mitotic cell marker specific to proliferating cells, was significantly increased in the ONH of 10–12 week-old cats with early FCG (Fig. 3a). The vast majority of cells in the ONH are glia, including astrocytes, microglia, and oligodendrocytes, in addition to axons of RGCs [30–33]. However, distribution of cell types, tissue structure, and biomechanical and physiological stresses are not uniform throughout the ONH [34]. We examined cell proliferation within three distinct sub-regions of the ONH: pre-laminar (PL), lamina cribrosa (LC), and retro-laminar (RL) regions (Fig. 3b, c) [30, 33, 35, 36]. Densities of proliferating cells were significantly greater in each of the ONH sub-regions in FCG compared to age-matched, normal ONHs (Fig. 3c), though cell proliferation was most pronounced in the RL region of the glaucomatous ONH. Next, we sought to determine the identity of these proliferating cells using markers for ONH astrocytes (SOX2 and GFAP), microglia/macrophages (IBA1), and cells of oligodendrocyte lineage (OLIG2) (Fig. 3d). In PL and LC regions of the early FCG ONH, almost all Ki67-labeling colocalized with IBA1, indicating that most proliferating cells in these regions are of microglia/macrophage lineage. In contrast, in the RL region, OLIG2+/SOX2+-immunopositive cells predominated and are presumed to be proliferating oligodendrocyte precursor cells (OPCs) (Fig. 3d) [37–39]. We subsequently addressed the question of whether glial cell proliferation remains a consistent hallmark of more chronic stages of glaucoma in FCG by immunolabeling ONHs from adult, 1–2-year-old, FCG and normal cats as described for younger animals. The young adult animal cohort was selected in an unbiased manner based solely on age and thus presented various degrees of axonal damage, although significant, chronic IOP elevation was consistently identified in all of these adult FCG cats (Supplemental Fig. S3). In contrast to younger kittens, no significant cell proliferation was identified in the adult ONH in FCG (Fig. 3a). In the much smaller population of proliferating cells in the adult ONH, the limited immunolabeling for Ki67 was largely restricted to microglia/macrophages, both in FCG and normal cats. This finding suggests that significant ONH glial cell proliferation is a feature that may be restricted to early-stage glaucoma.
Fig. 3.
Glial cell proliferation in the ONH is a feature of early disease in FCG. a Early glaucomatous ONHs had significantly increased density of proliferating (Ki67-immunopositive) cells compared to ONHs from agematched normal, adult FCG, and adult normal cats (**P < 0.01; ***P < 0.001; ANOVA followed by Tukey’s post-test for multiple comparisons; mean and SEM presented). In contrast, density of proliferating cells in the ONH was not significantly different between groups of adult cats (P = 0.78, n = 6 per group). b Photomicrographs of normal 10–12-week-old feline ONH showing three sub-regions (prelaminar [PL]; lamina cribrosa [LC], and retro-laminar [RL; extending 200 μm posterior to LC]) that are distinguishable based on their morphology and cell populations. OLIG2 oligodendrocyte-lineage marker (green); SOX2 astrocyte and glial progenitor marker (magenta); DAPI nuclear counter staining (blue). Scale bar = 200 μm. c In 10–12week-old cats with FCG, increased density of proliferating (Ki67-positive) cells was observed in each of the three ONH sub-regions, compared to age-matched normal controls (n = 5 per group; *P < 0.05, unpaired t test with Welch’s correction). d In the retrolaminar region of the ONH, Ki67 expression colocalized with SOX2 (magenta; top row) and OLIG2(green;toprow) (top;arrow). Dispersed throughout the ONH, other Ki67-positive cells expressed the microglia/macrophage marker IBA1 (bottom; arrow). Scale bar = 20 μm
As proliferation of various ONH cell types in early-stage FCG suggested cell type–specific responses to IOP elevation and initial glaucomatous damage, we next asked whether the ONH transcriptome in early FCG is enriched with cell typespecific genes, by conducting GSEA on the ONH transcriptome with cell type-specific genesets. Normalized enrichment scores (NES), a standardized metric that accounts for both differences in geneset size, and correlations between genesets and the expression dataset were used to compare and prioritize analysis results across gene sets [26]. Consistent with our immunolabeling results, we identified pronounced upregulation of microglia- and OPC-specific genes in the ONH in early glaucoma. Conversely, neuron- and myelinated oligodendrocyte–specific genes were significantly downregulated (Fig. 4a, b). Notably, the upregulation of astrocyte-specific genes was relatively modest compared to the other cell types in the ONH, suggesting more subtle molecular changes in astrocytes at this early stage of disease. Consistent with this finding, there was nosignificantchangein glial fibrillary acidic protein (GFAP) immunolabeling intensity in early FCG (Fig. 4c, d). Furthermore, no differences in normalized gene expression levels of astrocyte markers (GFAP, VIM, and AQP4) were observed between FCG and controls in this young cohort with early-stage disease (Fig. 4e).
Fig. 4.
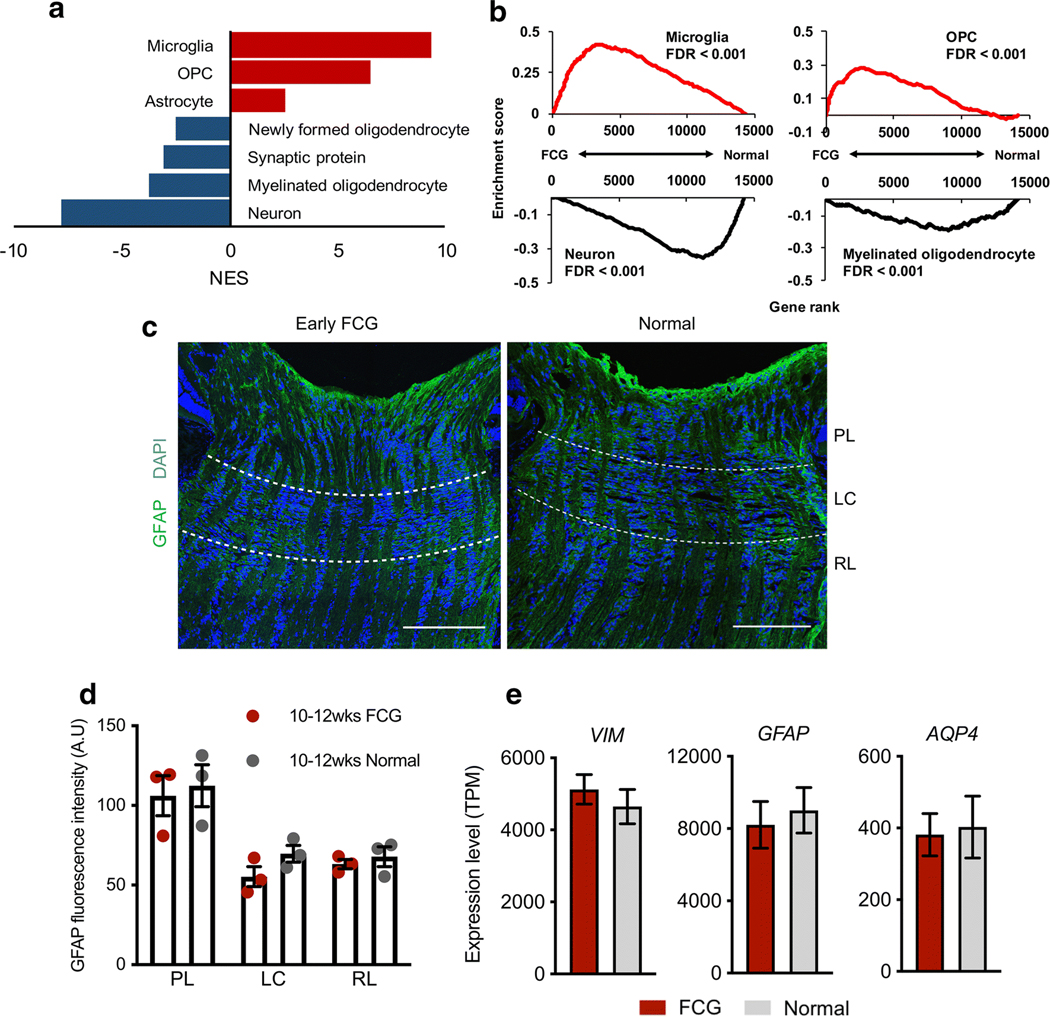
Highly cell type–specific enrichments were identified in the ONH transcriptome in FCG. a Normalized enrichment scores (NES) prioritize enrichment of different cell-specific genesets in the early FCG transcriptome. Microglia and oligodendrocyte precursor cell (OPC) and, to a lesser extent, astrocyte genesets showed upregulation in FCG, while downregulation of neuron, synaptic protein, and oligodendrocyte genesets was demonstrated. b GSEA-identified microglia- and OPC-specific gene sets exhibited statistically significant, coordinated gene upregulation in FCG (NES = 9.37 and 6.48, respectively; FDR < 0.001), whereas the neuron and myelinated oligodendrocyte gene sets were significantly downregulated (NES = − 7.80 and − 3.77, respectively; FDR < 0.001). Note that upregulation of astrocyte-specific genes is modest compared to the other ONH cell types (NES = 2.58; FDR < 0.001). Enrichment plots demonstrate cell type–specific geneset expression across the whole transcriptome. Pattern (c) and intensity (d) of immunofluorescent-labeling for the astrocyte marker GFAP in the optic nerve head were broadly similar between 10- and 12-week-old FCG and age-matched normal feline subjects (FCG: n = 3, normal: n = 3). e Normalized mRNA expression (transcript per million: TPM) of general astrocyte markers (GFAP, vimentin, aquaporin-4) in FCG from our RNA-seq dataset was not significantly increased relative to normal subjects (FDR = 0.98, 0.45, and 0.97, respectively). FCG: n = 10, normal: n = 6. Data presented as mean and SEM
Microglial Activation Is a Prominent Feature of Optic Neuropathy in Both Early-Stage and Chronic FCG
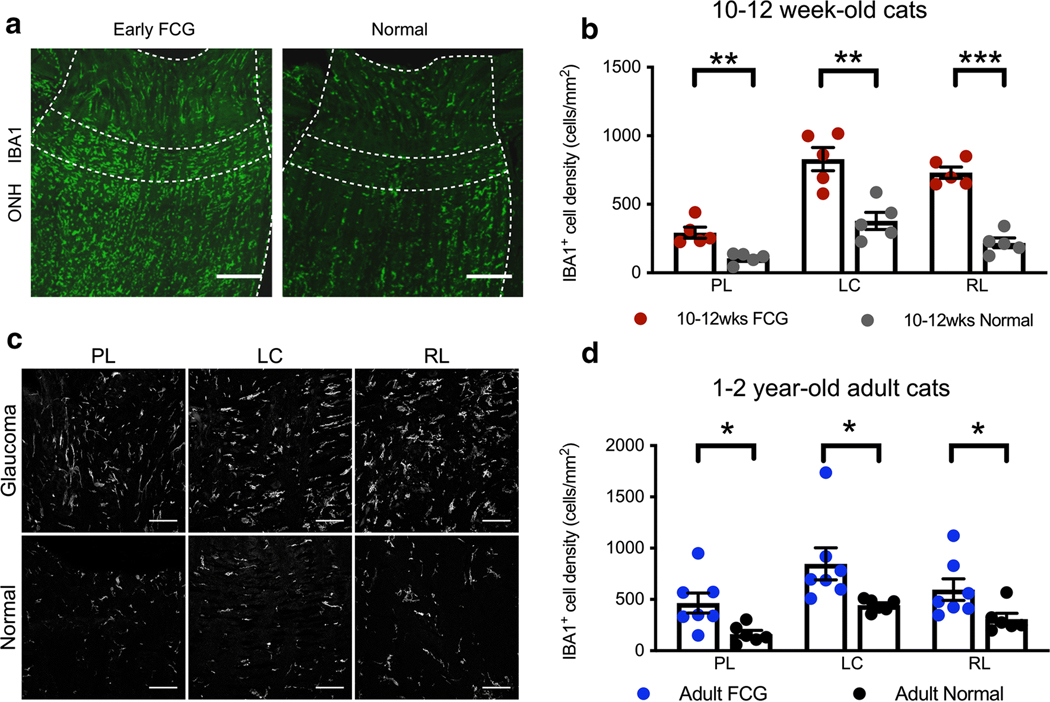
Guided by the pronounced enrichment of microglia specific genes and significant upregulation of immune/inflammation-related genes we observed in the ONH in early FCG, we next focused our attention on determining microglial cellular and molecular states. In all ONH sub-regions in early glaucomatous cats, there was a significant increase in the density of IBA1+ cells, compared to controls (Fig. 5a, b), providing further evidence for microglial/macrophage activation throughout the ONH early in glaucoma, prior to axon degeneration. We next sought to determine whether microglia in the ONH become activated early in the course of FCG, soon after onset of IOP elevation. To directly assess microglial activation state in the ONH, we examined cell morphology in addition to the number of microglia, in each of the three ONH subregions in situ (Fig. 5c). Although IBA1+ cells show diverse morphology in each of these regions, and there are clearly limitations imposed by the assessment of three-dimensional structure in two-dimensional tissue sections, IBA1+ cells appear to have more activated shapes (rod-like, bushy, or ameboid) in glaucomatous ONH, compared to age-matched normal controls. To assess molecular microglial activation, we first examined whether the ONH transcriptome in early FCG demonstrates enrichment of microglia genesets representative of various microglial cell states. GSEA identified significant enrichment of LPS-activated microglia genesets, which was a predominant molecular signature over other enriched microglial genesets in the ONH in early glaucoma (Supplemental Fig. S4). IBA1 expression colocalized with P2RY12 immunoreactivity (a microglia specific marker) in the majority of the IBA1+cell population in the PL and RL of normal ONH, but not in the LC region of young FCG and age-matched normal subjects (Supplemental Fig. S5). Thus, with the possible exception of the LC region, ONH resident microglial activation contributes to microgliosis in early stage of glaucoma. Furthermore, IBA1+ cell types and/or their phenotype may vary between sub-regions. However, as in early FCG, density of IBA1+ cells was increased in all three regions of the adult glaucomatous ONHs, relative to age-matched controls, and microglial and potentially macrophage activation appears to be a consistent feature across stages of disease in this glaucoma model (Fig. 5d).
Fig. 5.
Glaucomatous ONH pathobiology is characterized by microglial/ macrophage activation in both early and chronic FCG. a Photomicrographs illustrate the distribution of IBA1+ microglia in immunolabeled longitudinal sections of early FCG and age-matched control ONHs. Microglia are distributed throughout the ONH in both groups (scale bar = 100 μm). b The density of IBA1+ microglia was significantly increased in all 3 regions of the ONH in early FCG, compared to control subjects (n = 5 per group) (unpaired t test: PL, **P < 0.01; LC, **P < 0.01; RL, ***P < 0.001, respectively). FDR < 0.01 in all three tests. Data presented as mean and SEM. c Representative photomicrographs illustrating IBA1+ cells in prelaminar (PL), lamina cribrosa (LC), and retro-laminar (RL) regions in early glaucomatous and normal age-matched ONH sections. Microgliosis was evident in all sub-regions of the ONH in early FCG (scale bar = 25 μm). d The density of IBA1+ microglia was also increased in all 3 regions of in the ONH in 1–2-year-old adult cats with chronic FCG, relative to sections from age-matched control subjects (FCG; n = 7 and normal; n = 6) (*P < 0.05, unpaired t test. FDR < 0.05 in all three tests. Data presented as mean and SEM)
Early OPC Proliferation and Late Oligodendrocyte Loss Are Features of Spontaneous Glaucoma
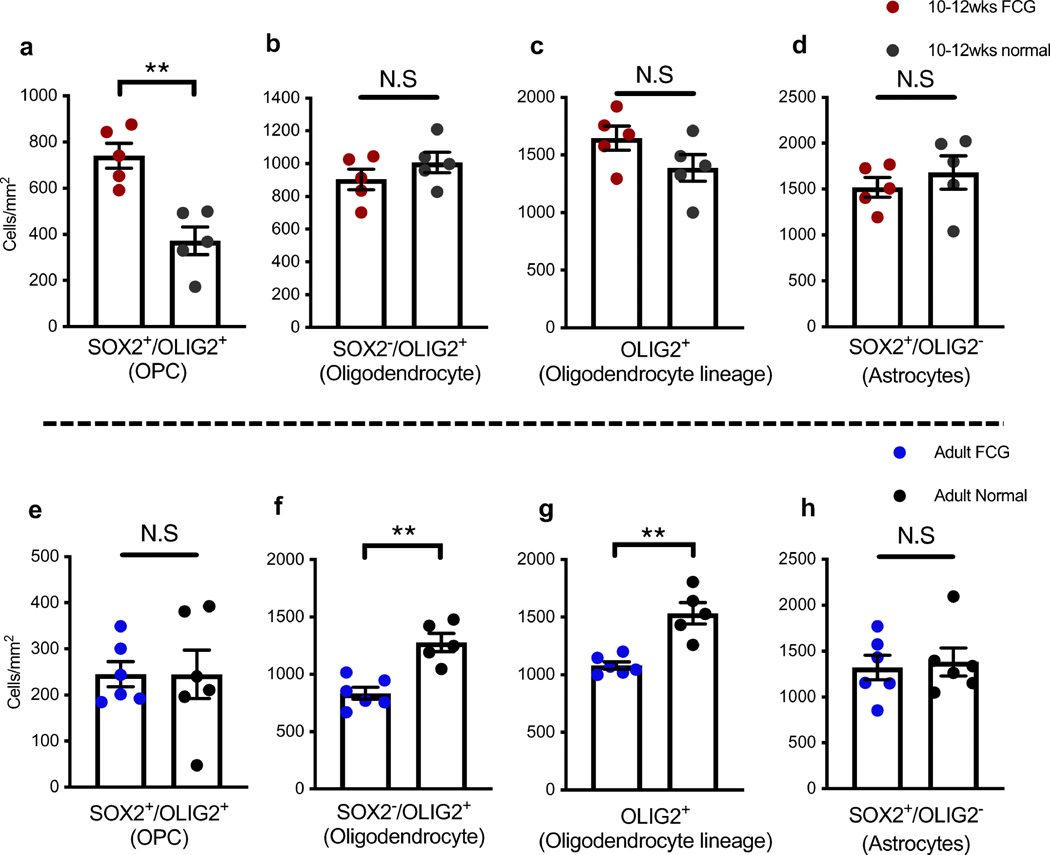
Increased density of OPCs (OLIG2+/SOX2+) was identified in the RL region of the ONH in early FCG in comparison with age-matched controls (Fig. 6a). In contrast, densities of differentiated oligodendrocytes (OLIG2+/SOX2−), oligodendrocyte-lineage cells (OLIG2+), or astrocytes (OLIG2−/SOX2+) were not significantly different at this early disease stage relative to age-matched controls(Fig.6b–d). Next, we investigated whether this increase in density of OPCs persisted in chronic disease by examining archived ONH tissues from adult cats with chronic FCG together with additional, appropriate age-matched normal cats. No significant difference in density of OPCs or astrocytes (OLIG2−/SOX2+) was observed between adult FCG cats and normal controls (Fig. 6e, h), while the density of OLIG2+/SOX2− differentiated oligodendrocytes was significantly lower in adult FCG ONHs (Fig. 6f). Together, this resulted in an overall decrease in the number of OLIG2+ oligodendrocyte-lineage cells in chronic glaucoma (Fig. 6g). Collectively, our experiments indicated that increase in density of OPCs with cell proliferation is a feature of the early glaucomatous ONH, whereas loss of mature oligodendrocytes occurs later, in chronic disease.
Fig. 6.
Early oligodendrocyte precursor cell (OPC) activation and late oligodendrocyte loss in FCG. a The density of OPCs (SOX2 and OLIG2 co-labeled cells) in the retrolaminar ONH of 10–12 week-old cats with early-stage glaucoma was significantly greater than in agematched control ONHs (n = 5 per group). b–d Based on patterns of SOX2 and OLIG2 immunolabeling, density of oligodendrocytes (b), cells of oligodendrocyte-lineage (c), and astrocytes (d) did not significantly differ between early FCG and age-matched normal ONHs. e–h Densities of OPCs (e) and astrocytes (h) were not significantly different in adult cats with chronic FCG compared to age-matched normal cats. In contrast, in the ONH of adult FCG cats, densities of both oligodendrocytes (f) and cells of oligodendrocyte lineage(g) weresignificantly decreased in comparison to normal adult cats (a–d n = 5 for each group, e–h n = 6 for FCG and n = 5 for normal; **P < 0.01; unpaired t test)
Consistent, Coordinated Gene Expression Changes Are Identifiable in Early Glaucoma by Complementary Computational Approaches
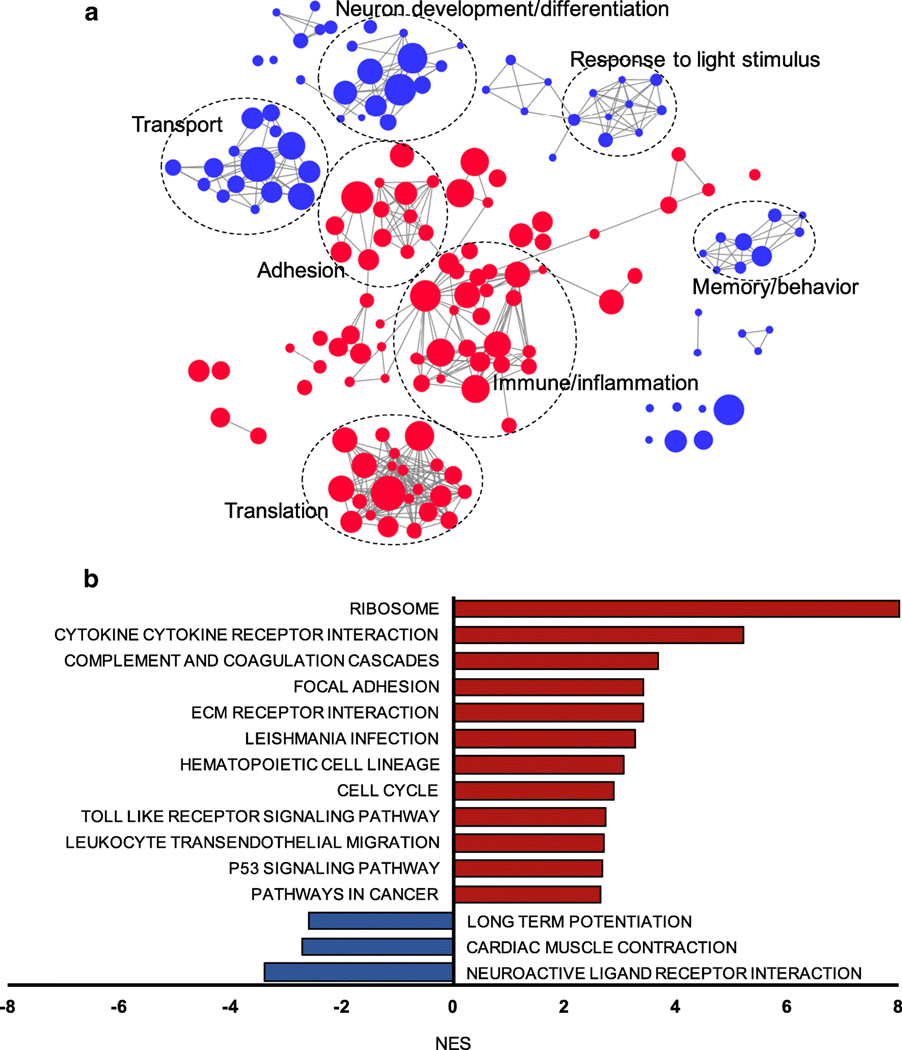
Expression levels of single genes may not change dramatically, early in the course of a slowly progressive neurodegenerative disease such as glaucoma. Our data showed that, relative to age-matched controls, changes in ONH gene expression in early FCG were less than twofold for nearly 80% of the DEGs identified. To account for limitations this imposes on the detection of differentially expressed genes and pathways by conventional pairwise comparison and pre-determined cut-off values, we utilized GSEA as a more powerful bioinformatics tool to identify disease-relevant molecular pathways and functions that may play a role in earlystage glaucoma [26]. Our results provided a number of biologically meaningful insights into the pathogenesis of glaucoma. Importantly, the molecular pathways/functions we identified were not detectable in conventional pairwise gene to gene DE analyses. For GO functional analysis, we employed enrichment network analysis to highlight the most enriched molecular functions and reduce redundancy of enriched GO gene sets. This provided a summary of up- and downregulated GO biological processes in early glaucomatous ONH (Fig. 7a). In the upregulated GO biological processes in early FCG, immune/inflammatory responses, adhesion, and translation functions were significantly enriched, whereas downregulated GO terms were enriched in processes associated with neuron development/differentiation, transport, responses tolight stimulus, and memory/behavior. Utilizing KEGG pathway curated genesets, GSEA identified 38 significantly enriched upregulated pathways and 3 downregulated molecular pathways in early glaucoma, including significantly upregulated ribosome, cytokine-cytokine receptor interaction, complementcoagulation cascades, and extracellular matrix (ECM)-receptor interaction pathways. Incontrast, neuroactive ligand receptor interaction, cardiac muscle contraction, and long potentiation pathways were downregulated in early FCG (Fig. 7b).
Fig. 7.
Coordinated changes in gene expression associated with functions/pathways are identifiable in early-stage FCG. a Enrichment map based on the GSEA results of GO biological processes was generated using Enrichmentmap plug-in for Cytoscape. Red nodes represent genesets that are positively enriched and over-represented in the ONH in early FCG, while blue nodes represent genesets that are negatively enriched and under-represented. Node size is proportional to the size of the functional GO term gene sets. Only gene sets that were enriched with FDR < 0.001 and the top 100 enriched GO biological processes (both upregulated and downregulated) in FCG are depicted. Genesets with common biological function are grouped by cluster and are labeled with representative functions. b Significantly up- (FDR < 0.001) and downregulated KEGG pathways in FCG (FDR < 0.05) support many cellular and molecular mechanisms previously implicated in glaucoma pathogenesis. Red bars represent significantly upregulated KEGG pathways, whereas blue bars represent downregulated KEGG pathways
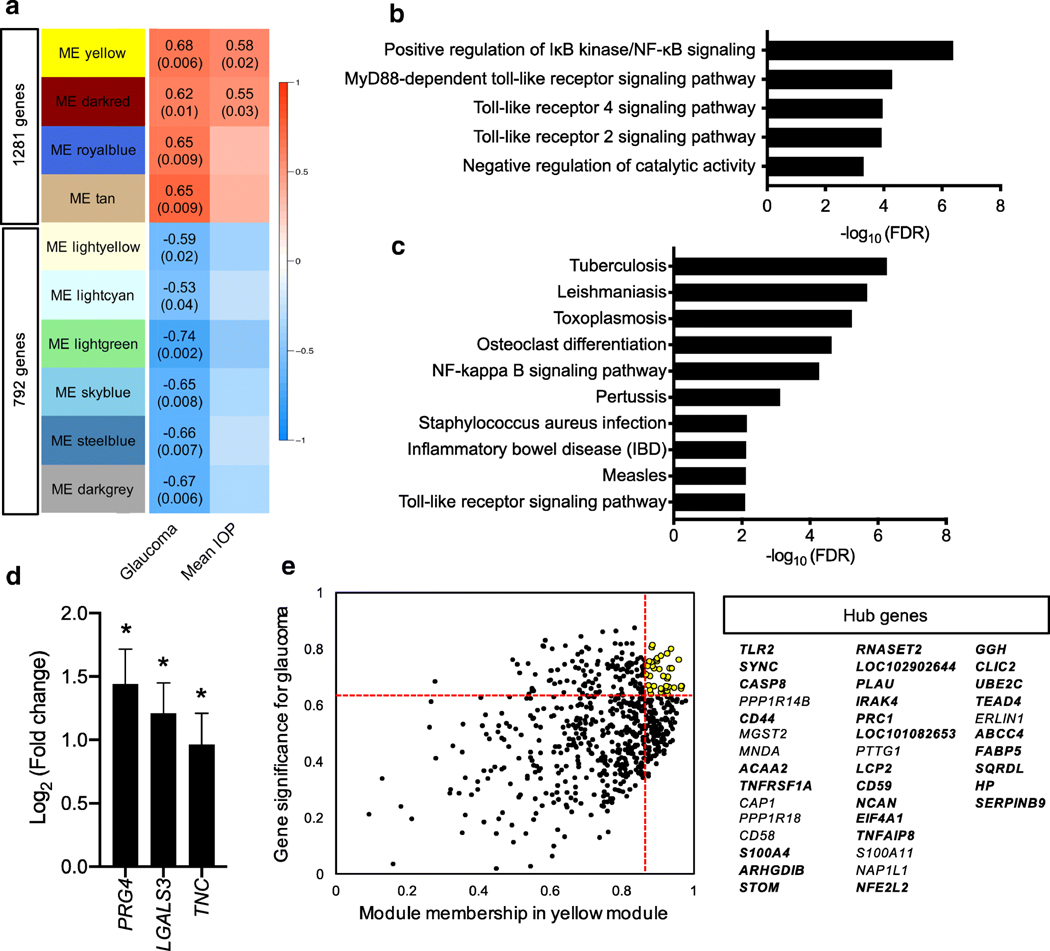
Expanding our characterization of the ONH transcriptome in early FCG, we applied weighted gene co-expression network analysis (WGCNA) to explore gene modules (gene clusters) that are associatedwith early FCG. The fourmodules that positively correlated with glaucoma genotype contained a total of 1281 genes, whereas the six modules that were negatively correlated with glaucoma genotype comprised 792 genes (Fig. 8a; and list of genes belonging to the modules provided in Supplemental data 2). Importantly, irrespective of LTBP2 genetic background, mean IOP was significantly positively correlated with two modules (P < 0.05), supporting an association between magnitude of IOP and ONH gene expression. Functional GO enrichment analysis of the modules identified distinctive biological processes associated with early FCG. One of the identified modules was significantly enriched for positive regulation of NF-κB signaling and MyD88-dependent toll-like receptor (TLR) signaling pathway (Fig. 8b). With the KEGG genesets, this demonstrated significantly over-represented pathways, again featuring TLR receptor signaling and NF-κB signaling pathways (Fig. 8c). Of note, genes encoding damage-associated molecular patterns (DAMPs) such as LGALS3, PRG4, and TNC [40], which are endogenous ligands for the TLR receptor signaling pathway, were included in the module, and these genes were also significantly upregulated in early FCG by conventional DE analysis (Fig. 8d). These results further support TLR receptor signaling and NF-κB signaling as molecular features of the early pathobiology ofglaucoma. Theother module wasenrichedfor proteasomal ubiquitin-independent protein catabolic process, and KEGG pathways showed an enrichment in the proteasome (Supplemental Fig. S6A). Setting thresholds with upper quintile for membership and gene significance in glaucoma, we identified 40 hub genes in the modules, likely to play central roles in the network [27]. In one module that was significantly and positively correlated with IOP, we further filtered the genes within the upper-quintile for gene significance in IOP and identified 30 genes as hub genes in one of the implicated gene modules (Fig. 8e). These included genes involved in toll-like receptor signaling (TLR2 and IRAK4), tumor necrosis factor receptor (TNFRSF1A), and apoptosis and NF-κB induction (CASP8). Filtering of the darkred module revealed 14 genes as hub genes, 5 of which were also hub genes in IOP (Supplemental Fig. S6B). Overall, our analyses confirmed orchestrated gene expression changes, at the level of molecular pathways in early-stage glaucoma and, importantly, underscored the relationship between magnitude of IOP and initial molecular responses in this common neurodegenerative disease.
Fig. 8.
Weighted gene co-expression network analysis (WGCNA) identified disease-relevant gene clusters. a WGCNA identified 34 nonoverlapping gene clusters (modules) that are significantly correlated with glaucoma. Heat map (correlation matrix) exhibits the correlation of gene co-expression modules. Ten modules significantly correlated with glaucoma genotype are shown. Numbers on the heat map in each module denote Pearson correlation coefficients (top) and P values (below, in parentheses). Two modules (yellow and dark red) are positively correlated with both glaucoma and mean IOP. The yellow module consisted of 665 genes including 41% (122/294) of the upregulated DEGs identified (r = 0.68, P = 0.006), and the darkred module comprised 151 genes (r = 0.62, P = 0.01). b The top five overrepresented GO terms in the “Yellow module” indicated that positive regulation of IκB kinase/NF-κB signaling (GO:0043123, FDR < 4.19×10−7 ) and MyD88-dependent toll-like receptor pathway related genes (GO:0002775, FDR < 0.05) are over-expressed in early glaucoma in the FCG model. c The top 10 over-represented KEGG pathways in the “Yellow module” includes NF-κB signaling and toll-like receptor signaling as well as inflammatory diseases associated with innate immune responses (FDR < 0.05). d Bar graph illustrates three differentially expressed damage associated pattern recognition (DAMP) genes (TNC, LGALS3, PRG4) that were significantly upregulated in early FCG relative to age-matched normal controls, based on RNA-seq data. Error bars represent estimated standard errors for the estimated coefficients of log2 (fold change) values. *FDR < 0.05. e Scatter plot demonstrates module membership and gene significance in the yellow module for FCG. Each dot represents a single gene in the modules. Genes that have high module membership (vertical red line; upper quartile) and high gene significance for glaucoma (horizontal red line; upper quartile) are considered hub genes (yellow dots). The hub genes in both glaucoma genotype and IOP are highlighted in bold print
Discussion
As for many other neurodegenerative diseases, the pathogenesis of glaucoma is complex, and changes in single molecules and molecular pathways alone do not fully explain disease pathogenesis. Moreover, glaucoma is typically a slowly progressive disease, and molecular changes associated with disease pathology may not be dramatic, especially early in the disease process. However, to develop effective therapeutic strategies that target reversible processes in glaucoma, an understanding of pathobiology of early glaucoma will be critical. In this study, we performed transcriptomic profiling to gain comprehensive and non-biased molecular insights into early pathobiology of glaucomatous optic neuropathy at a stage of the disease when phenotypic heterogeneity and complexity are limited in our model. RNA-seq affords a large dynamic range, which enabled us to identify modest but important gene expression changes in contrast to previous gene expression studies that investigated early changes using microarrays [41–43]. While inherited rodent glaucoma models, which include the DBA/2J mouse that shares similarities to human pigmentary glaucoma [44], are established and widely used, structural differences between mice and humans, including the murine lack of a collagenous LC and small eye size, may limit the translational value of this model. Given that the feline ONH more closely resembles that of humans than the ONH of rodents, our identification of changes in the ONH transcriptome in our glaucoma model prior to histological evidence of irreversible axon degeneration provides valuable insight into the early molecular events likely involved in the pathogenesis of glaucoma in human patients.
We identified significant upregulation of genes involved in cell proliferation and immune/inflammation responses inearly spontaneous glaucoma, relative to precisely age-matched control subjects. In this respect, our findings are consistent with ONH gene expression changes reported in adult DBA/2J mice [41] and rats with experimental ocular hypertension [42] that had no, or minimal, evidence of axonal damage. Thus, our results are supported by a body of evidence that points to cell proliferation and immune responses/inflammation as common molecular features of early glaucoma.
An upregulation of cell cycle–associated genes has been reported previously in rodent glaucoma models, both in ONHs from DBA/2J mice and from rats with experimentally induced IOP elevation [32, 33, 36]. However, the identities and the in situ localization of the majority of these proliferating cells within the ONH were not definitively established in most of these studies and have not been reported in spontaneous glaucoma models that have ONH structure and cellular organization comparable to humans, although a recent study has shown that astrocyte proliferation predominates in the ONH of experimental rat models of glaucoma [45]. Here, we demonstrated that proliferating cells are present throughout the ONH soon after onset of IOP elevation in cats with FCG. Importantly, the proliferating cell types varied by ONH sub-region. Microglia/macrophages dominate this response in the PL and LC sub-regions of the ONH, whereas cells of oligodendrocyte lineage, in particular OPCs, are the predominant glial cell type proliferating in the RL region. In contrast, ONHs from adult FCG subjects with more chronic disease did not show evidence of cell proliferation to the extent seen in early disease, suggesting that this molecular profile may be restricted to early-stage glaucoma. Although proliferation of microglia and OPCs has been implicated in a wide variety of neurodegenerative diseases with axon degeneration[46, 47],it remains unclear whether the glial cell proliferation we observed in the glaucomatous ONH in early disease is beneficial or detrimental. It is likely that the role played by glial cell proliferation differs with stage of disease.
Collectively, our data provide unique insight into coordinated gene expression changes in glial and neural cell-specific genes in response to initial glaucomatous damage. In early FCG with no histologically detectable damage, we identified cell type–specific upregulation of microglia and OPC genes and, to a lesser extent, upregulation of astrocyte genes, whereas neuronal and mature oligodendrocyte specific genes were downregulated. These changes in tissue expression of cell-specific genes could indicate an absolute alteration in cell number, cell composition, and/or cell functional state, which could all contribute to the observed changes in gene expression [48, 49]. Through subsequent immunolabeling and microscopy, we confirmed that microglia and OPCs in the ONH both proliferate and become activated, with increase in number of these cells observed in early glaucoma, prior to histological evidence of axon damage and loss. Although astrocytes are one of the major cell types in the ONH, our experiments showed that enrichment and activation of ONH astrocytes were less prominent features of early disease. This may reflect reversible rapid and dynamic reactivity in astrocytes, which could occur without concomitant changes in gene expression after a short-term elevation of IOP, as previously reported in experimental rodent models of acute ocular hypertension [50, 51]. Our findings in early FCG are in sharp contrast to pronounced gliosis dominated by astrocytes, which we have observed much later in the course of disease in the ONH of adult cats with chronic, established glaucoma, and that others have reported in the ONH in rodent experimental glaucoma with advanced ONH injury [45] and of human donors with chronic stages of glaucoma [11, 52, 53].
Although we identified downregulation of mature oligodendrocyte and neuronal genes, no significant change in the number of mature oligodendrocytes or of RGC axons was observed at this early stage of glaucoma. We therefore propose that downregulation of these genes reflects changes in cell states rather than loss of cells. As there are no neuronal somas in the ONH, the downregulation of neuronal specific genes may indicate pathologic RGC axonal mRNA alterations. For instance, expression of NMNAT2 was significantly reduced in early FCG ONHs. This gene, which encodes nicotinamide/nicotinic acid mononucleotide adenylyltransferase 2, has been postulated to play a significant role in axonal health and degeneration processes [54], is significantly more abundant in adult mouse RGC axons than in RGC somas [55], and is also downregulated in RGCs of DBA/2Jmicewith minimal axonal damage [56]. Similarly, RGC axonal mRNAs and oligodendrocyte genes have been shown to be downregulated in a rodent optic nerve crush model [57], as well as in other neurodegenerative diseases, including multiple sclerosis [58].
Profound microglial/macrophage activation is a consistent hallmark of glaucomatous optic neuropathy, identified not just during the early stages of disease in our model, prior to axon loss, but also shown to persist in more advanced, chronic stages of disease. Microglial/macrophage activation has been implicated in a variety of neurodegenerative diseases, including glaucoma as identified in donor ONH tissue from human glaucoma patients and in retinal, ONH, and optic nerve tissues in rodent models of glaucoma [31, 59–62]. As in our cohort of young cats with FCG, in DBA/2J mice, ONH and retinal microglial activation precedes detectable axon loss [59, 63]. It is noteworthy that transcriptome data in early FCG provided evidence for robust enrichment in pro-inflammatory phenotypes, such as LPS activated microglia, in the ONH over other microglial molecular phenotypes. Emerging experimental evidence suggests LPS-activated microglia direct astrocytes to a neurotoxic state by production of pro-inflammatory cytokines [64]. Therefore, the microglial activation that we observed in early-stage glaucoma could initiate signaling detrimental to other, intimately associated, adjacent cells in the ONH including astrocytes, oligodendrocytes, and RGC axons. Microglia-specific markers such as P2RY12 have been used to distinguish microglia from macrophages [65]. Unfortunately, these markers are generally expressed by microglia in homeostatic states and their expression is downregulated upon activation in disease states [66]. Thus, in the absence of robust, consistently expressed microglia-specific markers for immunolabeling, and a practical means to specifically label microglia or macrophages in our model in vivo, it was not possible within the scope of the present studies for us to establish whether activated IBA1+ cells were resident ONH microglia or macrophages or whether they represented infiltrating monocytes that have been proposed to migrate into the glaucomatous mouse ONH [67, 68]. Regardless of their origins however, we propose that these activated immune cells are likely to contribute to prodegenerative molecular pathways in glaucoma that in turn promote disease progression and axon loss. Emerging evidence from experimental rat models showed positive correlation between IBA1+ cell density and optic nerve injury grade, further supporting critical roles played by myeloid cells in glaucoma progression [45].
OPCs represented a large subset of the proliferating cells in the RL region of the ONH, contributing to a significant increase in the number of ONH OPCs in early glaucoma, prior to axon degeneration and/or significant loss of oligodendrocytes. We cannot fully exclude the possibility that OPC proliferation was related to a specific stage of optic nerve development, particularly given that myelination of the retrolaminar optic nerve is a post-natal process in cats. However, proliferating OPCs were identified in significantly greater numbers in the ONH of FCG subjects than in the normal, precisely age-matched cats. Additionally, proliferation of OPCs and chronic loss of mature oligodendrocytes have been reported in the myelinated orbital ON of aged, glaucomatous DBA/2J mice and of rats with experimentally induced ocular hypertension [69, 70]. As OPCs can be differentiated to new oligodendrocytes, their activation in early glaucoma may be protective [71] and may help maintain integrity of the myelin sheath of the optic nerve. In contrast to early-stage FCG, OPC activation and proliferation were not features of later, chronic stages of FCG, in which overall oligodendrocyte loss was evident, consistent with findings in established experimental glaucoma in rats. Together, our findings support a role for OPCs in glaucoma pathophysiology. Further studies are needed to determine whether other features of OPC biology, besides their capability of generating new oligodendrocytes, may also contribute to disease progression or neuroprotection.
Previously published gene expression studies in early stages of rodent glaucoma models have mainly focused on pairwise comparison of expression of single genes between groups, with or without hierarchical clustering [15, 41, 42, 67, 72, 73]. However, in very early pathology of slowly progressive neurodegenerative disease such as glaucoma, expression levels of single genes may not change dramatically. In our study, changes in gene expression were less than twofold for nearly 80% of the DEGs identified in the ONH in early FCG. Similarly, in DBA/2J mice with no histologically detectable evidence of glaucoma, over 94% of DEG expression changes were less than twofold [41]. By applying a series of complementary bioinformatics approaches, we were able to identify a number of molecular pathways which may play a role in glaucoma progression but were not detectable by conventional pairwise single gene comparison, due to small magnitude of individual gene expression changes. Coordinated gene upregulation in molecular pathways that include cytokine-cytokine interactions, complement pathways, TLR signaling, extracellular matrix (ECM) receptor pathways, focal adhesion, and cell cycle was identified by GSEA in our FCG model at an early stage in the disease process. These pathways have been implicated previously in glaucoma pathobiology [11, 15, 41, 45, 68]. In contrast, molecular pathways such as neuroactive ligand receptor interactions, cardiac muscle contraction, and long-term potentiation were significantly downregulated. These downregulated pathways have not previously been specifically reported in the glaucomatous ONH. Combined with our finding that neuron-specific gene expression was downregulated, it is likely that these particular function/pathwaywide alterations reflect molecular changes in RGCs, in particular within their axons.
Our results highlighted molecular hallmarks of the innate immune response particularly associated with the TLR and NF-κB signaling pathway. Glial cells and neurons in the mammalian CNS are known to express TLRs and TLR-signaling molecules that modulate innate immunity, through dynastic activation of the NF-kB pathway, and resulting production of cytokines, including proinflammatory cytokines such as TNF-α, IL-1β, and IL-6 [74, 75]. Consistent with our findings, upregulation of TLR signaling genes in the ONH and retina has been reported previously in DBA/2J mice [41], and proteomic and immunohistochemical analyses of human donor retinae have shown increased expression of TLR proteins in astrocytes and microglia in glaucomatous tissue [76]. Furthermore, an association between TLR polymorphisms and normal tension glaucoma has been identified in human patients [77]. Together, these data strongly suggest an important role for TLR in the complex pathophysiology of glaucomatous optic neuropathy. Our dataset also indicated significant upregulation of DAMP genes in early FCG, further implicating TLR signaling as an early and consistent feature of glaucoma pathophysiology. Upregulation of the tenascin-C gene (TNC), for example,could contribute to TLR activation in the ONH in our subjects with early FCG, as highlighted by prior studies involving ONH tissues from human glaucoma patients, DBA/2J mice, and rats with experimental ocular hypertension [41, 67, 72, 78], as well as many other neurodegenerative diseases including Huntington disease, Alzheimer’s disease, and multiple sclerosis [79]. Importantly, in our studies, magnitude of gene expression in these molecular pathways correlated not only with FCG genotype but also with IOP phenotype. Our results therefore provide the first direct evidence of upregulation of immune/inflammation genes in the ONH as an early cellular and molecular response to elevation in IOP in spontaneous glaucoma, reinforcing the long-recognized role of IOP in susceptibility to glaucomatous damage of RGCs and their axons.
Conclusions
The current study provides powerful evidence that a number of molecular pathways are regulated in the ONH prior to axon degeneration in glaucoma and these pathways are highly dependent on cell type and sub-region within the ONH. Results of pathway analyses support microglial proliferation and activation to a pro-inflammatory phenotype as key features of early disease. As neuroinflammation and innate immune responses, modulated by TLR and NF-κB pathways, have emerged as consistent features of glaucomatous optic neuropathy across multiple studies, ongoing studies in our lab seek to leverage strengths of the FCG model to interrogate changes in the transcriptome of distinct populations of cells in different ONH sub-regions across different stages of disease. Pathways identified correlate with IOP exposure and enhance our understanding of early cellular and molecular pathology of glaucoma. These pathways and processes may offer promising therapeutic targets to limit subsequent neurodegeneration in glaucoma.
Supplementary Material
Acknowledgments
The authors would like to thank Akihiro Ikeda, Robert Nickells, Dale Bjorling, and Colin Dewey for their advice on the conduct of these studies; Satoshi Kinoshita for preparation of cryosections; Ben August for preparation of semi-thin optic nerve sections; Carol A. Rasmussen and Elizabeth A. Hennes-Beean for technical support and acquisition of OCT images and electrophysiology data, respectively; Youngwoo Park and Jaesang Ahn for assistance with tissue dissection, OCTanalysis, and axon counting; UW-Madison’s Biotechnology Center for library preparation, sequencing, and the support of bioinformatics analyses; and the student trainees and laboratory assistants in the McLellan lab who assisted with animal procedures and collation of data.
Funding Information The studies were supported in part by NIH Grants K08 EY018609 and R01 EY027396 (GJM); S10 OD018221, P30 EY0016665, and a CTSA award from UW-Madison’s Institute for Clinical and Translational Research through NCATS grant UL1TR000427 (GJM); a National Glaucoma Research Grant from the BrightFocus Foundation (GJM); a Grant-in-Aid Award from Fight For Sight (GJM); the University of Wisconsin–Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation (GJM); and an unrestricted grant to the University of Wisconsin-Madison Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness. Additional support was provided by the Center for Integrated Animal Genomics, Iowa State University (NME and GJM); a Battelle General Platform and Infrastructure Award (NME); and a JASSO scholarship (awarded to KO). Tissue sectioning was performed by the University of Wisconsin Translational Research Initiatives in Pathology laboratory (TRIP), supported by the UW Department of Pathology and Laboratory Medicine, UWCCC (P30 CA014520) and the Office of The Director- NIH (S10OD023526).
Footnotes
Data Availability The datasets generated and/or analyzed during the current study are available the NCBI’s Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo; accession number: GSE110019).
Compliance with Ethical Standards
All animal procedures were conducted in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research, the NIH Guide for the Care and Use of Laboratory Animals, and in compliance with protocols approved by the University of Wisconsin-Madison’s IACUC.
Competing Interests The authors declare that they have no competing interests.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s12035–020-01910–9) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tham Y-C, Li X, Wong TY, Quigley HA, Aung T, Cheng C- Y (2014) Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 121(11):2081–2090 [DOI] [PubMed] [Google Scholar]
- 2.Kass MA, Heuer DK, Higginbotham EJ, Johnson CA, Keltner JL, Miller JP et al. (2002) The Ocular Hypertension Treatment Study: a randomized trial determines that topical ocular hypotensive medication delays or prevents the onset of primary open-angle glaucoma. Arch Ophthalmol 120(6):701–713 discussion 829–30 [DOI] [PubMed] [Google Scholar]
- 3.Anderson DR, Drance SM, Schulzer M (1998) The effectiveness of intraocular pressure reduction in the treatment of normal-tension glaucoma. Collaborative Normal-Tension Glaucoma Study Group. Am J Ophthalmol 126(4):498–505 [DOI] [PubMed] [Google Scholar]
- 4.Drance S, Anderson DR, Schulzer M, Collaborative Normal-Tension Glaucoma Study Group. Risk factors for progression of visual field abnormalities in normal-tension glaucoma. Am J Ophthalmol 2001;131(6):699–708. [DOI] [PubMed] [Google Scholar]
- 5.Anderson DR, Drance SM, Schulzer M, Collaborative NormalTension Glaucoma Study Group (2001) Natural history of normal-tension glaucoma. Ophthalmology 108(2):247–253 [DOI] [PubMed] [Google Scholar]
- 6.Heijl A, Leske MC, Bengtsson B, Hyman L, Bengtsson B, Hussein M et al. (2002) Reduction of intraocular pressure and glaucoma progression: results from the Early Manifest Glaucoma Trial. Arch Ophthalmol 120(10):1268–1279 [DOI] [PubMed] [Google Scholar]
- 7.Susanna R Jr, De Moraes CG, Cioffi GA, Ritch R (2015) Why do people (still) go blind from Glaucoma? Transl Vis Sci Technol 4(2): 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland MV, Ervin A-M, Friedman DS, Jampel HD, Hawkins BS, Vollenweider D, Chelladurai Y, Ward D et al. (2013) Comparative effectiveness of treatments for open-angle glaucoma: a systematic review for the U.S. Preventive Services Task Force. Ann Intern Med 158(4):271–279 [DOI] [PubMed] [Google Scholar]
- 9.Quigley HA, Anderson DR (1977) Distribution of axonal transport blockadeby acute intraocular pressure elevation in theprimate optic nerve head. Invest Ophthalmol Vis Sci 16(7):640–644 [PubMed] [Google Scholar]
- 10.Howell GR, Libby RT, Jakobs TC, Smith RS, Phalan FC, Barter JW, Barbay JM, Marchant JK et al. (2007) Axons of retinal ganglion cells are insulted in the optic nerve early in DBA/2J glaucoma. J Cell Biol 179(7):1523–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez MR, Miao H, Lukas T (2008) Astrocytes in glaucomatous optic neuropathy. Prog Brain Res 173:353–373 [DOI] [PubMed] [Google Scholar]
- 12.Nickells RW, Howell GR, Soto I, John SWM (2012) Under pressure: cellular and molecular responses during glaucoma, a common neurodegeneration with axonopathy. Annu Rev Neurosci 35(1): 153–179 [DOI] [PubMed] [Google Scholar]
- 13.Burgoyne CF, Downs JC, Bellezza AJ, Suh J-KF, Hart RT (2005) The optic nerve head as a biomechanical structure: a new paradigm for understanding the role of IOP-related stress and strain in the pathophysiology of glaucomatous optic nerve head damage. Prog Retin Eye Res 24(1):39–73 [DOI] [PubMed] [Google Scholar]
- 14.Williams PA, Marsh-Armstrong N, Howell GR, Bosco A, Danias J, Simon J et al. (2017) Neuroinflammation in glaucoma: a new opportunity. Exp Eye Res 157:20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harder JM, Braine CE, Williams PA, Zhu X, MacNicoll KH, Sousa GL et al. (2017) Early immune responses are independent of RGC dysfunction in glaucoma with complement component C3 being protective. Proc Natl Acad Sci U S A 114(19):E3839–E3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuehn MH, Lipsett KA, Menotti-Raymond M, Whitmore SS, Scheetz TE, David VA, et al. (2016) A mutation in LTBP2 causes congenital glaucoma in domestic cats (Felis catus). Chidlow G, editor. PLoS ONE.11(5):e0154412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali M, McKibbin M, Booth A, Parry DA, Jain P, Riazuddin SA, Hejtmancik JF, Khan SN et al. (2009) Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet 84(5):664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adelman S, Shinsako D, Kiland JA, Yaccarino V, Ellinwood NM, Ben-Shlomo G et al. (2018) The post-natal development of intraocular pressure in normal domestic cats (Felis catus) and in feline congenital glaucoma. Exp Eye Res 166:70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutz-Mendicino MM, Snella EM, Jens JK, Gandolfi B, Carlson SA, Kuehn MH, McLellan G, Ellinwood NM (2011) Removal of potentially confounding phenotypes from a Siamese-derived feline glaucoma breeding colony. Comparative Medicine 61(3):251–257 [PMC free article] [PubMed] [Google Scholar]
- 20.Sigle KJ, Camaño-Garcia G, Carriquiry AL, Betts DM, Kuehn MH, McLellan GJ (2011) The effect of dorzolamide 2% on circadian intraocular pressure in cats with primary congenital glaucoma. Vet Ophthalmol 14(Suppl. 1):48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Sole MJ, Sande PH, Bernades JM, Aba MA, Rosenstein RE (2007) Circadian rhythm of intraocular pressure in cats. Vet Ophthalmol 10(3):155–161 [DOI] [PubMed] [Google Scholar]
- 22.Teixeira LBC, Buhr KA, Bowie O, Duke FD, Nork TM, Dubielzig RR, McLellan G (2014) Quantifying optic nerve axons in a cat glaucoma model by a semi-automated targeted counting method. Mol Vis 20:376–385 [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M et al. (2012) STAR: ultrafast universal RNAseq aligner. Bioinformatics. 29(1):15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102(43):15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P, Horvath S (2008) WGCNA: anR package for weighted correlation network analysis. BMC Bioinformatics 9(1):559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merico D, Isserlin R, Stueker O, Emili A, Bader GD (2010;) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. Ravasi T, editor. PLoS One 5(11): e13984–e13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye H, Hernandez MR (1995) Heterogeneity of astrocytes in human optic nerve head. J Comp Neurol 362(4):441–452 [DOI] [PubMed] [Google Scholar]
- 31.Neufeld AH (1999) Microglia in the optic nerve head and theregion of parapapillary chorioretinal atrophy in glaucoma. Arch Ophthalmol 117(8):1050–1056 [DOI] [PubMed] [Google Scholar]
- 32.Jennings AR, Carroll WM (2014) Oligodendrocyte lineage cells in chronic demyelination of multiple sclerosis optic nerve. Brain Pathol 25(5):517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balaratnasingam C, Kang MH, Yu P, Chan G, Morgan WH Cringle SJ, Yu DY (2014) Comparative quantitative study of astrocytes and capillary distribution in optic nerve laminar regions. Exp Eye Res 121:11–22 [DOI] [PubMed] [Google Scholar]
- 34.Stowell C, Burgoyne CF, Tamm ER, Ethier CR, Lasker/IRRF Initiative on Astrocytes and Glaucomatous Neurodegeneration Participants (2017) Biomechanical aspects of axonal damage in glaucoma: a brief review. Exp Eye Res 157:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balaratnasingam C, Morgan WH, Johnstone V, Pandav SS, Cringle SJ, Yu D-Y (2009) Histomorphometric measurements in human and dog optic nerve and an estimation of optic nerve pressure gradients in human. Exp Eye Res 89(5):618–628 [DOI] [PubMed] [Google Scholar]
- 36.Kang MH, Law-Davis S, Balaratnasingam C, Yu D-Y (2014) Sectoral variations in the distribution of axonal cytoskeleton proteins in the human optic nerve head. Exp Eye Res 128:141–150 [DOI] [PubMed] [Google Scholar]
- 37.Ossola B, Zhao C, Compston A, Pluchino S, Franklin RJM, Spillantini MG (2016) Neuronal expression of pathological tau accelerates oligodendrocyte progenitor cell differentiation. Glia 64(3):457–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer AJ, Zelinka C, Scott MA (2010) Heterogeneity ofglia inthe retina and optic nerve of birds and mammals. Koch K-W, editor. PLoS ONE. 5(6):e10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiwari S, Dharmarajan S, Shivanna M, Otteson DC, BeleckyAdams TL (2014) Histone deacetylase expression patterns in developing murine optic nerve. BMC Dev Biol 14(1):30–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thundyil J, Lim K-L (2015) DAMPs and neurodegeneration. Ageing Res Rev 24(Part A):17–28 [DOI] [PubMed] [Google Scholar]
- 41.Howell GR, Macalinao DG, Sousa GL, Walden M, Soto I, Kneeland SC, Barbay JM, King BL et al. (2011) Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J Clin Invest 121(4):1429–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson EC, Doser TA, Cepurna WO, Dyck JA, Jia L, Guo Y, Lambert WS, Morrison JC (2011) Cell proliferation and interleukin-6-type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Invest Ophthalmol Vis Sci 52(1):504–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kompass KS, Agapova OA, Li W, Kaufman PL, Rasmussen CA, Hernandez MR (2008) Bioinformatic and statistical analysis of the optic nerve head in a primate model of ocular hypertension. BMC Neurosci 9(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.John SW, Smith RS, Savinova OV, Hawes NL, Chang B, Turnbull D, Davisson M, Roderick TH et al. (1998) Essential iris atrophy, pigment dispersion, and glaucoma in DBA/2J mice. Invest Ophthalmol Vis Sci 39(6):951–962 [PubMed] [Google Scholar]
- 45.Lozano DC, Choe TE, Cepurna WO, Morrison JC, Johnson EC (2019) Early optic nerve head glial proliferation and Jak-stat pathway activation in chronic experimental glaucoma. Invest Ophthalmol Vis Sci 60(4):921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tauheed AM, Ayo JO, Kawu MU (2016) Regulation of oligodendrocyte differentiation: insights and approaches for the management of neurodegenerative disease. Pathophysiology 23(3):203–210 [DOI] [PubMed] [Google Scholar]
- 47.Gomez-Nicola D, Fransen NL, Suzzi S, Perry VH (2013) Regulation of microglial proliferation during chronic neurodegeneration. J Neurosci 33(6):2481–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C (2015) Defining cell types and states with single-cell genomics. Genome Res 25(10):1491–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM et al. (2010) Cell type-specific gene expression differences in complex tissues. Nat Methods 7(4):287–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun D, Qu J, Jakobs TC (2013) Reversible reactivity by optic nerve astrocytes. Glia. 61(8):1218–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tehrani S, Davis L, Cepurna WO, Choe TE, Lozano DC, Monfared A et al. (2016) Astrocyte structural and molecular response to elevated intraocular pressure occurs rapidly and precedes axonal tubulin rearrangement within the optic nerve head ina rat model. Cho KS, editor. PLoS One 11(11):e0167364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varela HJ, Hernandez MR (1997) Astrocyte responses in human optic nerve head with primary open-angle glaucoma. J Glaucoma 6(5):303–313 [PubMed] [Google Scholar]
- 53.Hernandez MR, Igoe F, Neufeld AH (1986) Extracellular matrix of the human optic nerve head. Am J Ophthalmol 102(2):139–148 [DOI] [PubMed] [Google Scholar]
- 54.Conforti L, Gilley J, Coleman MP (2014) Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat Rev Neurosci 15(6):394–409 [DOI] [PubMed] [Google Scholar]
- 55.Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, Amieux PS, Holt CE (2016) Dynamic axonal translation in developing and mature visual circuits. Cell. 166(1):181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams PA, Harder JM, Foxworth NE, Cochran KE, Philip VM, Porciatti V, Smithies O, John SW (2017) Vitamin B3 modulates mitochondrial vulnerability and prevents glaucoma in aged mice. Science. 355(6326):756–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu J, Jakobs TC (2013) The time course of gene expression during reactive gliosis in the optic nerve. Di Giovanni S, editor. PLoS One 8(6):e67094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evangelidou M, Karamita M, Vamvakas SS, Szymkowski DE, Probert L (2014) Altered expression of oligodendrocyte and neuronal marker genes predicts the clinical onset of autoimmune encephalomyelitis and indicates the effectiveness of multiple sclerosisdirected therapeutics. J Immunol 192(9):4122–4133 [DOI] [PubMed] [Google Scholar]
- 59.Bosco A, Steele MR, Vetter ML (2011) Early microglia activation in a mouse model of chronic glaucoma. J Comp Neurol 519(4): 599–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan L, Neufeld AH (2001) Activated microglia in the human glaucomatous optic nerve head. J Neurosci Res 64(5):523–532 [DOI] [PubMed] [Google Scholar]
- 61.Ebneter A, Casson RJ, Wood JPM, Chidlow G (2010) Microglial activation in the visual pathway in experimental glaucoma: spatiotemporal characterization and correlation with axonal injury. Invest Ophthalmol Vis Sci 51(12):6448–6413 [DOI] [PubMed] [Google Scholar]
- 62.Block ML, Hong J-S (2005) Microglia and inflammation-mediated neurodegeneration: multiple triggers with a common mechanism. Prog Neurobiol 76(2):77–98 [DOI] [PubMed] [Google Scholar]
- 63.Bosco A, Breen KT, Anderson SR, Steele MR, Calkins DJ, Vetter ML (2016) Glial coverage in the optic nerve expands in proportion to optic axon loss in chronic mouse glaucoma. Exp Eye Res 150: 34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L et al. (2017) Neurotoxic reactive astrocytes are induced by activated microglia. Nature 18:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B et al. (2013) Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat Neurosci 17(1):131–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Butovsky O, Weiner HL (2018) Microglial signatures and their role in health and disease. Nat Rev Neurosci 19(10):622–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howell GR, Soto I, Zhu X, Ryan M, Macalinao DG, Sousa GL et al. (2012) Radiation treatment inhibits monocyte entry into the optic nerve head and prevents neuronal damage in a mouse model of glaucoma. J Clin Invest 122(4):1246–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams PA, Braine CE, Foxworth NE, Cochran KE, John SWM (2017) GlyCAM1 negatively regulates monocyte entry into the optic nerve head and contributes to radiation-based protection in glaucoma. J Neuroinflammation 14(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Son JL, Soto I, Oglesby E, Lopez-Roca T, Pease ME, Quigley HA, Marsh-Armstrong N (2010) Glaucomatous optic nerve injury involves early astrocyte reactivity and late oligodendrocyte loss. Glia. 58(7):780–789 [DOI] [PubMed] [Google Scholar]
- 70.Nakazawa T, Nakazawa C, Matsubara A, Noda K, Hisatomi T, She H et al. (2006) Tumor necrosis factor-alpha mediates oligodendrocyte death and delayed retinal ganglion cell loss in a mouse model of glaucoma. J Neurosci 26(49):12633–12641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hill RA, Patel KD, Goncalves CM, Grutzendler J, Nishiyama A (2014) Modulation of oligodendrocyte generation during a critical temporal window after NG2 cell division. Nat Neurosci 17(11): 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson EC, Jia L, Cepurna WO, Doser TA, Morrison JC (2007) Global changes in optic nerve head gene expression after exposure to elevated intraocular pressure in a rat glaucoma model. Invest Ophthalmol Vis Sci 48(7):3161–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang DY, Ray A, Rodgers K, Ergorul C, Hyman BT, Huang W, Grosskreutz CL (2010) Global gene expression changes in rat retinal ganglion cells in experimental glaucoma. Invest Ophthalmol Vis Sci 51(8):4084–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol 11(5):373–384 [DOI] [PubMed] [Google Scholar]
- 75.Rosenberger K, Derkow K, Dembny P, Krüger C, Schott E, Lehnardt S (2014) The impact of single and pairwise Toll-like receptor activation on neuroinflammation and neurodegeneration. J Neuroinflammation 11(1):373–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo C, Yang X, Kain AD, Powell DW, Kuehn MH, Tezel G (2010) Glaucomatous tissue stress and the regulation of immune response through glial Toll-like receptor signaling. Invest Ophthalmol Vis Sci 51(11):5697–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takano Y, Shi D, Shimizu A, Funayama T, Mashima Y, Yasuda N et al. (2012) Association of Toll-like receptor 4 genepolymorphisms in Japanese subjects with primary open-angle, normal-tension, and exfoliation glaucoma. Am J Ophthalmol 154(5):825–832.e1 [DOI] [PubMed] [Google Scholar]
- 78.Pena JDO, Varela HJ, Ricard CS, Hernandez MR (1999) Enhanced tenascin expression associated with reactive astrocytes in human optic nerve heads with primary open angle glaucoma. Exp Eye Res 68(1):29–40 [DOI] [PubMed] [Google Scholar]
- 79.Bonneh Barkay D, Wiley CA (2009) Brain extracellular matrix in neurodegeneration. Brain Pathol 19(4):573–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.