Abstract
Background:
Microglia are implicated in the neuropathogenesis of HIV. Tetraspanin 2 (Tspan2) is closely related to CD9 and CD81 proteins, and are expressed on microglia cells. They have been implicated in cell fusion and adhesion and in the immune response, and neuroinflammation. Developing therapeutics that target microglia remains a challenge as these therapeutics must cross the Blood-Brain Barrier (BBB). Our goal was to use microglia derived exosomes as a vehicle to deliver siRNA across the BBB to target human telomerase reverse transcriptase immortalized human microglial cells (HTHU) latently infected by HIV (HTHU-HIV) and to evaluate if the knockdown of Tspan2 gene expression in changes the activation state of microglia cells, thereby modulating the neuroinflammatory response.
Methods:
A blood brain barrier (BBB) model that closely mimics and accurately reflects the characteristics and functional properties of the in vivo BBB was used to examine HTHU microglia exosome effects on BBB permeability, and their ability to migrate across the and delivery small interfering RNA (siRNA) to cells on the CNS side of the BBB model. Exosomes were loaded with Texas-Red control siRNA (20 pmol) or Cy5-Tspan2 siRNA and then placed in the apical side of the BBB model, 24 hr after incubation, HTHU-HIV cells microglial cells on the lower chamber were either imaged for siRNA uptake or analyzed for gene expression induced modifications.
Results:
HTHU exosomes transmigrate from the apical side of the BBB to deliver Texas-Red control siRNA or Cy5-Tspan2 siRNA to HTHU-HIV microglia cells on the CNS side of the BBB model. A dose dependent (5-40 pmol) increase in Cy5-Tspan2 uptake with a corresponding decrease in gene expression for Tspan2 occurred in HTHU-HIV microglia. A decrease in Tspan2 gene expression as a consequence of knockdown with Tspan2 siRNA at both 20 and 40 pmol concentrations resulted in a significant decrease in C-X-C motif chemokine 12 (CXCU12) and C-X-C chemokine receptor type 4 (CXCR4) gene expression in HTHU-HIV microglia. Furthermore, a decrease in the gene expression levels of the Interleukins, IL-13 and IL-10 and an increase in the gene expression levels for the Fc gamma receptor 2A(FCGR2A) and TNF-α occurred in HTHU-HIV microglial cells
Conclusions:
These data demonstrate that HTHU exosomes cross the BBB and are efficient delivery vehicles to the CNS. Moreover, modifying the expression levels of Tspan2, has downstream consequences that includes alterations in cytokines and microglia biomarkers.
Keywords: exosomes, microglia, HIV, siRNA
Graphical Abstract
Microglia-derived exosomes loaded with Tspan2 siRNA transmigrate across the BBB and knockdown Tspan2 gene expression in human microglial cells latently infected by HIV. This knockdown increases CXCL12, CXCR4, FCGR2A and TNF-α while decreasing IL-13 and IL-10 gene expression in HTHU-HIV microglial cells. Modulating Tspan2 modulates microglia cytokines and phenotype biomarkers.
1.0. Introduction
Neurocognitive disorders are among the most common and clinically important complications of human immunodeficiency virus (HIV-1) infection (1, 2). HIV-1 associated neurocognitive disorders (HAND) remain frequent and occur in nearly 60% of HIV-1 patients. These disorders, which include but are not limited to confusion, behavioral changes, headaches, cognitive motor impairment and anxiety disorders, have become a burden on the health care system (1, 2). Microglia are implicated in the neuropathogenesis of HIV and several other neurological disorders. Microglial cells are infected early in the course of HIV disease and can be a source of continuous infection throughout HIV/AIDS progression. Microglia act as viral reservoirs of active and latent HIV infections, thus posing a challenge to antiretroviral therapy and allow for HIV persistence in the brain (3–5)(6, 7). Targeting the neuro-inflammatory process altering microglial activation levels may be an effective strategy to decrease neurodegeneration in patients with HIV.
Tetraspanins are a 33-member family of 4-span transmembrane proteins that regulate cell motility, signaling and protein trafficking (8). Tetraspanin 2 (Tspan2) is one of the less well-characterized members of the tetraspanin superfamily. Tspan2 is closely related to CD9 and CD81 proteins, which have been implicated in biological functions related to cell fusion and adhesion and in processes such as the immune response, and neuroinflammation (9–11). Evidence suggests that the primary function of Tetraspanins such as Tspan2 is to control the trafficking of their partner proteins such as C-X-C motif chemokine 12 (CXCL12) and HIV receptor C-X-C chemokine receptor type 4 (CXCR4) whose altered function can affect neuronal repair and exacerbate HIV neuropathogenesis (12). We speculate that Tspan2 may be involved in the neuroinflammatory process by playing a role in microglial activation, thereby contributing to HIV neuropathogenesis. Thus, strategies to block or decrease Tspan2 activity in microglia cells may prevent or reduce microglia activation. Developing therapeutics that target microglia remains a challenge as these therapeutics must cross the Blood-Brain Barrier (BBB), a highly selective permeable barrier. This complex physiological checkpoint separates the circulating blood from the brain extracellular fluid and inhibits the free diffusion of circulating molecules from the blood into the brain (13). Only a limited number of drugs, i.e. small, lipophilic compounds (< 400–500 Da) can cross the BBB making delivery of agents across the BBB very challenging and warrants further research (14, 15). Hence, much effort has been channelled into improving drug transport across the BBB and into the central nervous system (CNS). Exosomes, membrane-contained vesicles that originate from the endocytic pathway or from plasma membrane, have all the desired attributes of effective drug delivery vehicles (16–19). They have been shown to diffuse across the plasma membranes of host cells including the BBB (20, 21). They are well tolerated, and can be loaded with various therapeutic agents such as small interfering RNA (siRNA) (22–25). Our goal was to use microglia derived exosomes to delivery siRNA across the BBB to target HTHU-HIV Microglia cells to evaluate if: 1) exosomes containing 2 siRNA were able to cross the BBB, 2) Tspan2 siRNA exosomes were incorporated by the microglia in sufficient quantity to enable a significant change in Tspan2 expression and 3) if knockdown of Tspan2 expression in the microglia results in a change in the microglia cell expression of activation markers, thereby modulating the neuroinflammatory response.
2.0. Methods
2.1. Cell culture
A). Primary cells:
Human telomerase reverse transcriptase immortalized human microglial cells (HTHU) and HTHU-HIV Microglia cells (HTHU-HIV, latently infected by HIV) (26) were used (generously provided by Dr. Jonathan Karn, CWRU, Cleveland, OH). The cell lines were maintained in Dulbecco’s Minimal Essential Medium (DMEM)/F12 (50:50) supplemented with 5% fetal bovine serum (FBS, exosome depleted), 5% Fetal Plex, 1X Non-Essential Amino Acid (NEAA) Cell Culture Supplement, 1X anti mycotic, and 1X penicillin/streptomycin. Additional cell lines used were primary cultures of human brain microvascular endothelial cells (BMVEC) (Cat# ACBRI-376) and Normal Human astrocytes (NHA) (Cat# ACBRI-371). BMVEC were cultured in Mesenchymal Stem Cell Chondrogenic Differentiation Medium media containing 50 mg/10 ml endothelial mitogen, heparin (10 mg/ml), 2.5% FBS (exosome depleted), 2.5% Fetal Plex, 1X NEAA, 1X anti mycotic, 1X glutamate, and 1X penicillin/streptomycin. NHA were cultured were cultured in DMEM containing 1X B-27, 2.5% FBS (exosome depleted), 2.5% Fetal Plex, 1X NEAA, 1X anti mycotic, 1X glutamate, and 1X penicillin/streptomycin. All cells were incubated in an atmosphere of 5% CO2 at 37° C.
B). Blood Brain Barrier Model:
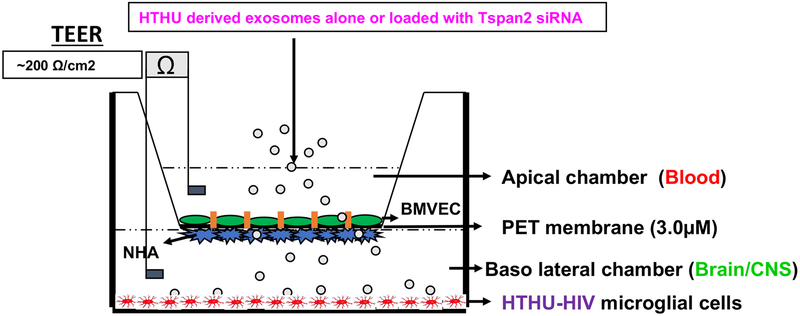
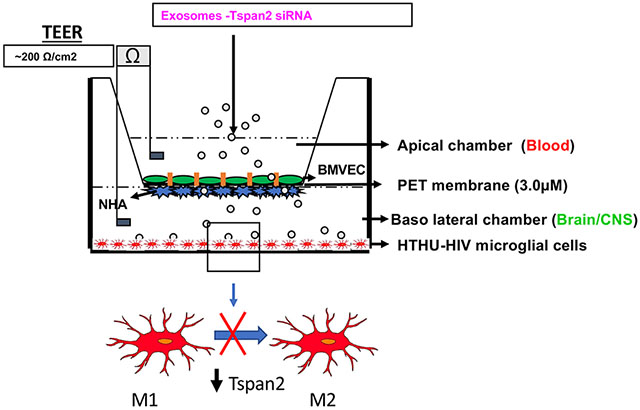
The BBB model that closely mimics and accurately reflects the characteristics and functional properties of the in vivo BBB was used. It is a well validated two-dimensional BBB in vitro co-culture system transwell model (27). The two cell types that are known to constitute the human BBB, HBMVECs and NHAs were used in this system. The NHAs were cultured on the underside of a polyethylene terephthalate (PET) transwell insert (basal end represents Brain/CNS side) and BMVECs were cultured on the inside of the PET insert (apical end represents blood side) and allowed to form monolayers and differentiate where the astrocytic feet extend to the BMVEC monolayer and together they form a tight barrier that effectively mimics the in vivo BBB (Figure 1). HTHU-HIV microglia were cultured as a monolayer on the CNS side of the cell culture 2D BBB cell culture system (Figure 1) to examine the uptake of exosomes and phenotypic changes in these microglia. Trans-endothelial electrical resistance (TEER) across the in vitro BBB was measured using an ohm meter Millicell ERS system (Millipore, Bedford, MA Cat # MERS 000 01). Electrodes were sterilized using 95% alcohol and rinsed in distilled water prior to measurement. A constant distance of 0.6 cm was maintained between the electrodes at all times during TEER measurement.
Figure 1. In vitro Blood Brain Barrier Model.
The BBB consists of BMVEC grown on the apical side of the membrane with NHA on the basolateral side of the membrane. HTHU-HIV microglial cells are grown in the lower chamber.
2.2. Exosomes
A). Isolation:
HTHU microglia cells were cultured containing 10% exosome-depleted FBS at 37°C for 2 days. Supernatant from HTHU microglia cells was collected and exosomes were isolated by differential centrifugation (2000 x g for 10 min, 10, 000 x g for 30 min, 100, 000 x g for 60 min, and 100, 000 x g for 60 min). Exosome pellets were resuspended in phosphate buffered saline (PBS).
B). Characterization:
Exosomes isolated from HTHU microglia cells were analyzed for their diameter and size using a Horiba nanopartica SZ-100 particle size analyzer (n=3). To determine morphology, exosome suspensions were then absorbed onto a 400-mesh carbon-coated copper grids, rinsed in filtered PBS, then stained with 2% phosphotungstic acid in aqueous suspension. Exosomes were visualized using transmission electron microscopy (TEM).
C). Labeling of siRNA:
Tspan2 siRNA (SMARTpool: siGENOME TSPAN2 siRNA- D017266-01-0020 Dharmacon) was labeled with Cy5 using the Label IT® siRNA Tracker Cy5™ Kit (Mirus Bio LLC, Madison, WI, Cat # 7213, red color, excitation of 649 emission of 670) according to manufactures instructions.
D). Loading of exosomes with siRNA:
Exosomes were loaded using Exo-Fect Exosome Transfection Kit (System Biosciences SBI). Briefly, 50 μl of exosomes/PBS suspension (50 μg) were mixed with 10 μl of Exo-Fect solution and 20 μl of Cy5-labeled Tspan2 siRNA (SMARTpool: siGENOME TSPAN2 siRNA-D017266-01-0020 Dharmacon) concentration ranging from 10-40 pmol or mixed with 20 pmol Texas-Red control siRNA (red color, excitation of 585/29 emission of 628/32, System Biosciences).
2.3. BBB studies
A). BBB Integrity/permeability:
TEER across the BBB was measured before and after (48 hr) addition of exosomes alone or exosomes were loaded with Tspan2 siRNA (n=3). All TEER measurements were done in triplicate and the baseline TEER value of the PET insert alone without the cells was subtracted from the TEER values obtained when NHA and BMVEC cells were co-cultured.
B). BBB transmigration of loaded exosomes:
Exosomes were loaded with Texas-Red control siRNA (20 pmol) and then placed in the apical side of the BBB model, 24 hr after incubation, HTHU-HIV cells microglial cells on the lower chamber were fixed Texas-Red control siRNA (n=3). To determine that exosomes loaded with Texas-Red control siRNA transmigrated across the BBB to HTHU-HIV cells, cells were imaged for the presence of siRNA (red color, excitation of 585/29 emission of 628/32) using the EVOS® FL Cell Imaging System, Invitrogen. Cells were countered stained with 4’,6-diamidino-2-phenylindole (DAPI) (blue color, excitation of 357/44 nm, emission of 447/60 nm) to determine nuclear staining. Fluorescent intensity was measured using Image J (NIH) to quantify Texas-Red control siRNA and confirm transmigration.
C). Efficacy through of loaded exosomes:
Exosomes were loaded with scrambled siRNA or Cy5 labeled Tspan2 siRNA (10 to 40 pmol) and then placed in the apical side of the BBB model, 24 hr after incubation, HTHU-HIV cells microglial cells on the lower chamber (basolateral) were either fixed Texas-Red control siRNA (n=3) or harvested for RNA (n=3). Fixed HTHU-HIV cells were counter stained with DAPI and imaged using the EVOS® FL Cell Imaging System. Fluorescent intensity (red color) was measured using Image J software (NIH). RNA was extracted from harvested cells by an acid guanidinium-thiocyanate-phenol-chloroform method using Trizol reagent (Invitrogen - Life Technologies, Carlsbad, CA), and the amount of RNA was quantitated using a Nano-Drop ND-1000 spectrophotometer (Nano-Drop, Wilmington, DE). The extracted RNA was reverse transcribed to cDNA using a reverse transcriptase kit (Promega Inc, Madison, WI) by following the manufacturer’s instruction. The relative abundance of each mRNA species was quantitated by normalizing the expression levels of each mRNA to the endogenous β-actin levels. Q-PCR was done using specific primers to Tspan2, CXCL12, CXCR4, Interleukin-13 (IL-13), Interleukin-10 (IL-10), The Fc gamma receptor 2A (FCGR2A), TNF-α, and specific microglial activation/resting phenotypic markers, HLA-DQB1, HLA-DRA and HLA-DRB (GeneQuery™ Human Microglial Polarization Markers qPCR Array Kit (GQH-MPM) from ScienCell) (Table 1). All primers were validated for efficiency and specificity by qPCR with melt curve analysis, and gel electrophoresis. Brilliant SYBR green Q-PCR master mix (Stratagene Inc., La Jolla, CA) was used. Q-PCR was performed in a reaction volume of 25 μL. Briefly, 12.5 μL of master mix, 2.5 μL of assay primers (10×), and 10 μL of template cDNA (100 ng) were added to each well. After a short centrifugation, the PCR plate was subjected to 35 cycles under the following conditions: (i) PCR activation at 95°C for 5 min, (ii) denaturation at 95°C for 5s, and (iii) annealing/extension at 60°C for 10s. All samples and controls were run in triplicate on a Stratagene MX3000P real-time PCR system. To provide a precise quantification of the initial target in each PCR reaction, the amplification plot was examined and the data were calculated as described. Relative expression of mRNA species was calculated using the comparative threshold cycle number (CT) method (28). Briefly, for each sample, a difference in CT values (ΔCT) was calculated for each mRNA by taking the mean CT of duplicate tubes and subtracting the mean CT of the duplicate tubes for the reference RNA (β-actin) measured on an aliquot from the same RT reaction. The ΔCT for the treated sample was then subtracted from the ΔCT for the untreated control sample to generate a ΔΔCT. The mean of these ΔΔCT measurements was then used to calculate the levels in the targeted cytoplasmic RNA relative to the reference gene and normalized to the control as follows: relative levels or transcript accumulation index (TAI) = 2 − ΔΔCT. This calculation assumes that all PCR reactions are working with 100% efficiency. All PCR efficiencies were found to be >95%; therefore, this assumption introduced minimal error into the calculations.
TABLE 1:
QPCR Primer Sequence
| Gene | Forward Primer | Reverse Primer | Amplicon size |
|---|---|---|---|
|
Tspan2 Sequence ID: AF054839.1 |
GCT CAT TGG AAT TGT CGG TAT TG | TCA CAT CTC GTG AGT TTC GTA TC | 101 |
|
IL-10 Sequence ID: AF418271.1 |
GCA GAC TAC TCT TAC CCA CTT C | TGT GTT CCA GGC TCC TTT AC | 115 |
|
IL-13 Sequence ID: U10307.1 |
GTT GCA CAG ACC AAG GTA GT | CTA CGG GCC TGT TTC CTA ATC | 101 |
|
TNF-α Sequence ID: HQ201306.2 |
CCA GGG ACC TCT CTC TAA TCA | TCA GCT TGA GGG TTT GCT AC | 106 |
|
FCGR2A Sequence ID: AH002832.2 |
GGC ACC TAC TGA CGA TGA TAA A | CAA GCT GAG AGT ATG ACC ACA T | 111 |
|
HLADBQ1 Sequence ID: AY656681.1 |
TGG CCT TAT CAT CCG TCA AAG | AAG CAG GCA TCA CAG AAG AG | 100 |
|
HLADRA Sequence ID: AH001506.2 |
GCT CCT TTA CAG CCC ACT ATT | TCT GAG GTT CAA CAC TCT CAA C | 97 |
|
HLADRB1 Sequence ID: MK032407.1 |
CTC TAC GTC TGA GTG TCA TTT CTT | GCA CGT ACT CCT CTT GGT TAT AG | 86 |
|
CXCL12 (SDF-1) Sequence ID: AY644456.1 |
AAC TGT GCC CTT CAG ATT GT | CAG GTA CTC CTG AAT CCA CTT TAG | 90 |
|
CXCR4 Sequence ID: AY728138.1 |
CAT CCT CAT CCT GGC TTT CTT | CAC ACC CTT GCT TGA TGA TTT C | 98 |
2.4. Statistical analysis
Data was analyzed by the ANOVA followed by post-hoc analysis using Bonferroni corrections (Graph Pad Prism 7).
3.0. Results
3.1. Characterization of exosomes derived from HTHU-microglia:
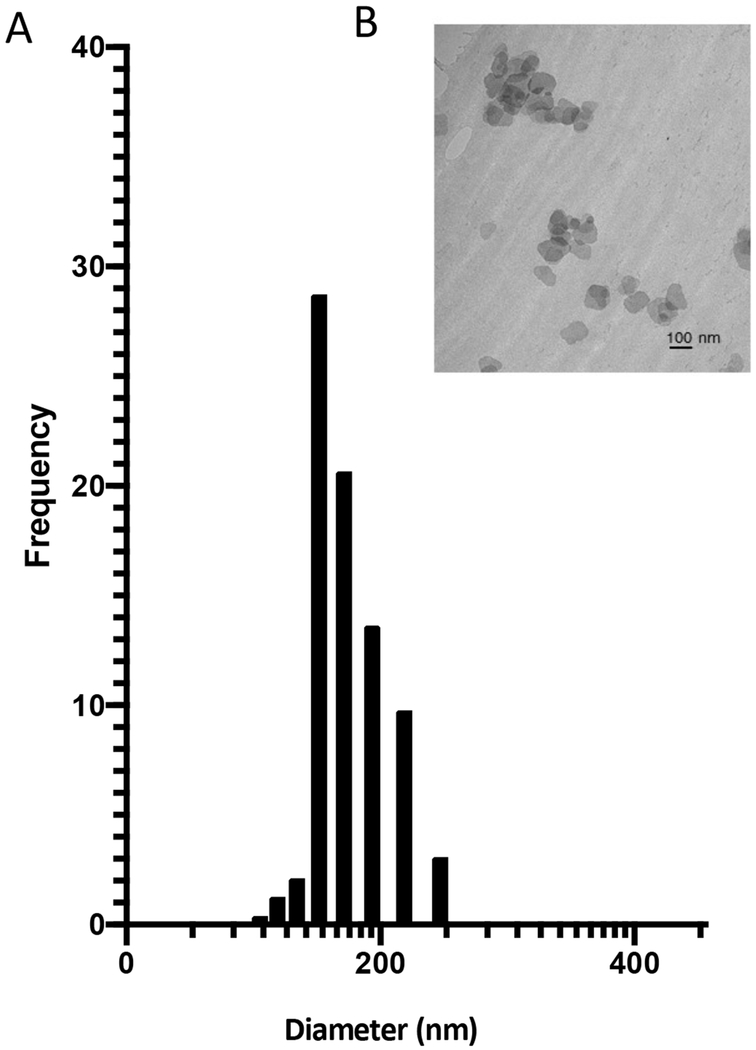
DLS analysis demonstrates that isolated exosomes range in diameter/size from 93 to 218 nm with a predominant peak at 142 nm (Figure 2A). A representative TEM image (Figure 2B) demonstrates that exosomes have a vesicular morphology. After successful isolation of exosomes, determining if exosomes derived from HTHU-microglia could be used as therapeutic delivery vehicles to the CNS was next examined.
Figure 2. Characterization of exosomes.
A) Analysis of exosome diameter/size using dynamic light scattering (DLS). B) Representative TEM image of exosomes demonstrating morphology (n=3).
3.2. Exosome effects on BBB integrity and ability to transmigrate BBB:
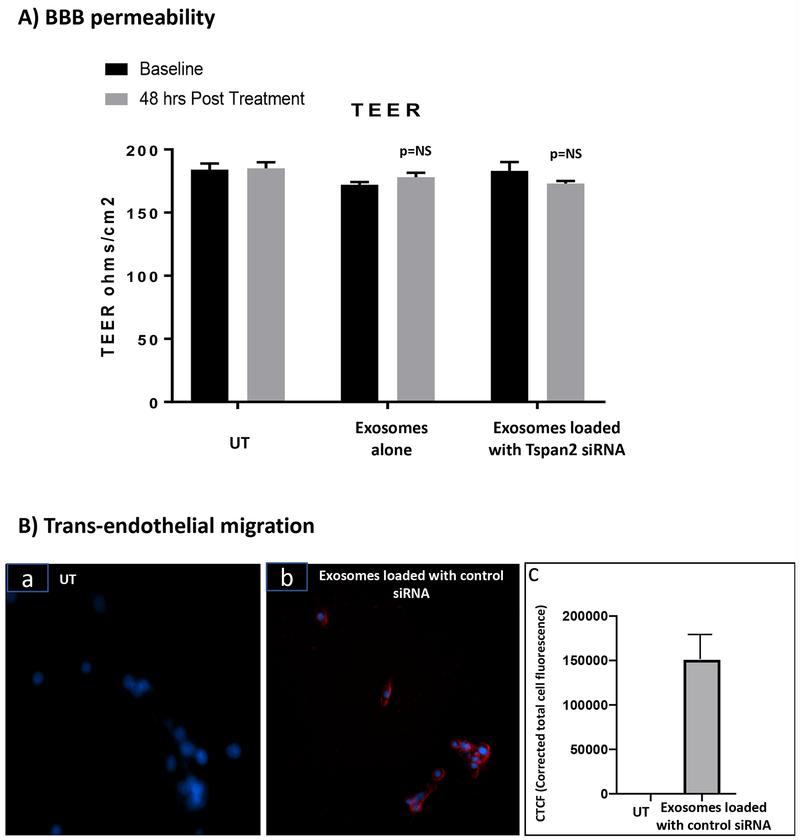
To determine the effect of HTHU exosomes on BBB integrity, BBB TEER both before and 48 hr post treatment was evaluated. There was no statistically significant difference (Figure 3A) in TEER before and after treatment with HTHU exosomes alone or with HTHU exosomes loaded with Tspan2 siRNA. The average TEER value at baseline prior to addition of exosomes was 184.0 ± 4.9, 188.0 ± 5.5 and 175.0 ± 2.1 for the untreated control, HTHU exosomes alone and HTHU exosomes loaded with Tspan2 siRNA respectively, while the corresponding TEER values 48 hr post treatment were 183.0 ± 4.5, 184.0 ± 8.8 and 173.0 ± 2.2 respectively.
Figure3. Exosome effects on BBB permeability and ability for trans-endothelial migration.
A) TEER measurements of the BBB pre and post incubation (48 hr) with exosomes isolated from HTHU cells alone or with exosomes isolated from HTHU cells loaded with Tspan2 siRNA in the apical side of BBB model (n=3). B) Trans-endothelial migration (24 hr) of HTHU exosomes loaded with Texas-red control siRNA to HTHU-HIV microglial cells, a) untransfected (UT) HTHU-HIV cells, b) HTHU exosomes loaded with Texas-Red control siRNA. The blue fluorescence arises from DAPI staining of nuclei, and the red fluorescence arises from Texas-Red control siRNA. Representative 20X magnification image. C) Quantification of fluorescence intensity Texas-Red control siRNA using Image J software (total of n=3 experiments).
To determine if HTHU exosomes transmigrate across the BBB, HTHU exosomes loaded Texas-Red control siRNA were applied to the apical side of the BBB, 24-hr later, HTHU-HIV microglial cells that were cultured at the bottom of the 2D BBB cell culture system were imaged. Fluorescent control siRNA was observed in HTHU-HIV microglial cells (Figure 3B–a&b, a: transfected control; b: exosomes loaded with Texas-Red control siRNA (red coloring)) and quantitated using Image J software (Figure 3B–c). These data demonstrate that exosomes transmigrated from the apical side of BBB to deliver Texas-Red control siRNA to HTHU-HIV microglia cells on the CNS side of the BBB model demonstrating that HTHU exosomes cross the BBB and are efficient delivery vehicles to the CNS.
3.3. Efficacy of knock-down of Tspan2 gene expression.
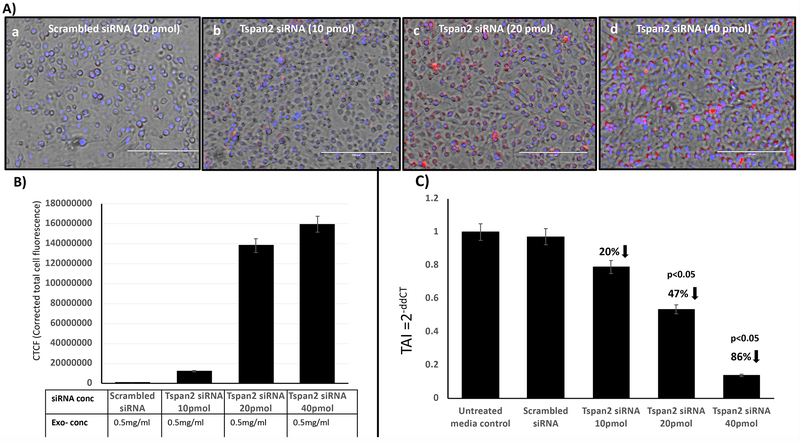
Based on data from above that exosomes isolated from HTHU-HIV microglia cells efficiently transmigrate across the BBB and deliver therapeutics, examining if HTHU exosomes loaded with Cy5-labeled Tspan2 siRNA transmigrate across the BBB and inhibit gene expression for Tspan2 in HTHU-HIV microglial cells at the CNS side of the BBB model was done. Data demonstrating transmigration and delivery are shown in Figure 4. Of the, exosomes that transmigrated across the BBB, a range of concentrations of Tspan2 siRNA (10, 20 & 40 pmol, Figure 4A–b through d) loaded exosomes showed a significant dose response increase in Tspan2 siRNA uptake as compared to the untreated control (Figure 4A–a). Quantification of cell fluorescence is shown in Figure 4B, demonstrating a dose response in uptake. A corresponding significant decrease in Tspan2 gene expression with 20 (47% decrease; p<0.05) and 40 pmol (86% decrease; p<0.05) of Tspan2 siRNA-exosomes is shown in Figure 4C.
Figure 4. Therapeutic effect of exosomes on Tspan2 gene expression.
HTHU exosomes loaded with Cy5 labeled Tspan2 siRNA were applied to the apical side of the BBB, 24 hr later, HTHU-HIV microglial cells on the basolateral side were imaged or analyzed for Tspan2 gene expression. A) Representative 20X images of HTHU-HIV cells following transmigration of exosomes loaded with increasing concentrations of Cy5-Tspan2 siRNA (red color). B) Corresponding quantification of cell fluorescence. C) Corresponding gene expression levels for Tspan2 quantified by q-PCR (total of n=3 experiments with 2 replicates). TAI= transcript accumulation index.
3.4. Downstream effects of changes in Tspan2 gene expression
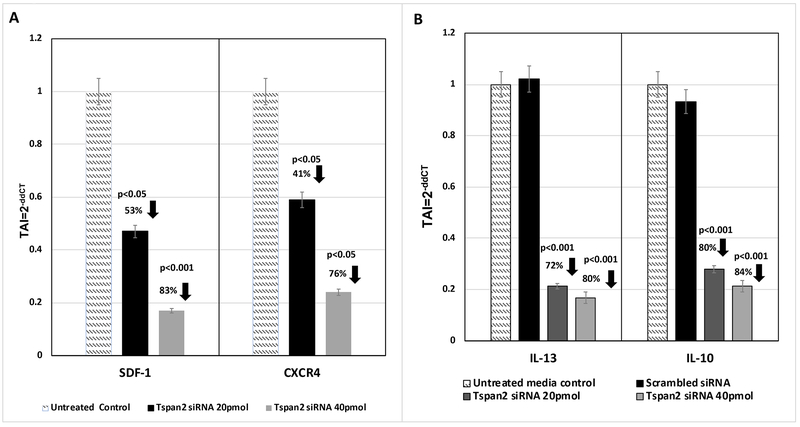
After determining that exosomes transmigrate across the BBB and are efficacious in knocking down gene expression in HTHU-HIV microglia cells, the downstream effects of modifications in Tspan2 gene expression were examined. A decrease in Tspan2 gene expression as a consequence of knockdown with Tspan2 siRNA at both 20 and 40 pmol concentrations resulted in a significant decrease in CXCL12 (TAI = 0.5; 53% decrease; p<0.05 and TAI = 0.2; 83% decrease; p<0.001 respectively) and CXCR4 gene expression (TAI = 0.6; 41% decrease; p<0.05 and TAI = 0.2; 76% decrease; p<0.05 respectively) as compared to the untreated control (TAI = 1.0) (Figure 5A). A decrease in the gene expression levels of M2 phenotypic markers IL-13 (TAI = 0.3; 72% decrease; p<0.001) and IL-10 (TAI = 0.2; 80% decrease; p<0.001) in HTHU-HIV microglial cells with uptake of 20 pmol Tspan2 siRNA exosomes as compared to the untreated control microglia (TAI = 1.0) was observed (Figure 5B). Increasing Tspan2 siRNA concentration to 40 pmol resulted in a comparable decrease in IL-10 (TAI = 0.2; 84% decrease; p<0.001) and IL-13 (TAI = 0.2; 80% decrease; p<0.001) gene expression when compared to the untreated control (TAI = 1.0) (Figure 5B).
Figure 5. Downstream effects of changes in Tspan2 gene expression.
HTHU exosomes loaded with Cy5 labeled Tspan2 siRNA were applied to the apical side of the BBB, 24 hr later, HTHU-HIV microglial cells on the basolateral side were imaged or analyzed for gene expression. A) CXCL12 and CXCR4 gene expression in the presence or absence of Tspan2 siRNA in HTHU-HIV (total of n=3 experiments with 2 replicates). B) I1-13 and I1-10 gene expression in the presence or absence of Tspan2 siRNA in HTHU-HIV (total of n=3 experiments with 2 replicates).
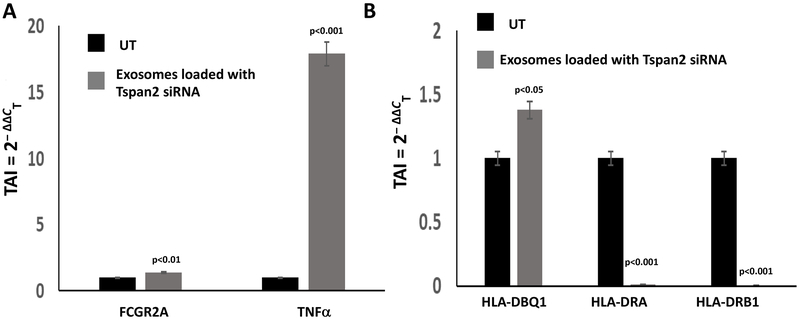
Gene expression levels of key microglial biomarkers in the HTHU-HIV microglial cells cultured at the bottom of the 2D BBB cell culture system were examined. FCGR2A, HLA-DQB1, HLA-DRA, HLA-DRB (Figure 6). An increase in the gene expression levels of FCGR2A (TAI = 1.4, 40.2% increase; p<0.0-1) and TNF-α (TAI = 17.9% (179% increase; p<0.001) occurred in microglial cells that had uptake of Tspan2 siRNA exosomes as compared to the untreated control microglia (TAI = 1.0) (Figure 6A). Additionally, HLA-DBQ1 (TAI = 1.4; 38% increase; p<0.05), HLADRA (TAI = 0.01; 99% decrease; p<0.001) and HLADRB1 (TAI = 0.002; 99% increase; p<0.001) (Figure 6B) were significantly different between the microglial cells treated with Tspan2 siRNA exosomes as compared to the untreated control microglia (TAI = 1.0). These data demonstrate that modifying the expression levels of Tspan2, has downstream consequences that includes alterations in cytokines and microglia biomarkers.
Figure 6. Downstream effects on microglia markers.
HTHU exosomes loaded with Cy5 labeled Tspan2 siRNA were applied to the apical side of the BBB, 24 hr later, HTHU-HIV microglial cells on the basolateral side were imaged or analyzed for gene expression. A) FCFR2A and TNFα gene expression in the presence or absence of Tspan2 siRNA in HTHU-HIV (total of n=3 experiments with 2 replicates). B) HLA-DBQ1, HLA-DRA and HLA-DRB1 gene expression in the presence or absence of Tspan2 siRNA in HTHU-HIV (total of n=3 experiments with 2 replicates).
4.0. Discussion:
Microglia, which are the innate immune cells of CNS, largely rely on mobile-vesicles to propagate cytokines-mediated inflammatory response across distant brain regions (29). Microglial activation in the CNS occurs in response to a pro-inflammatory stimulus and microglial phenotypic transformation can occur. Depending on the phenotypes activated, microglia can produce either cytotoxic or neuroprotective effects. Activated microglia exhibit migratory, proliferative, and phagocytic properties as well as the capacity to release chemokines, cytokines, neurotrophic factors and to present antigens (6, 30). Targeting the neuro-inflammatory process early by preventing microglial activation may thus be an effective strategy to stall the neurodegeneration in patients with HIV-associated dementia.
Evidence suggests that the primary function of Tetraspanins such as Tspan2 is to control the trafficking of their partner proteins such CXCL12 and HIV receptor CXCR4 whose altered function can affect neuronal repair and exacerbate HIV neuropathogenesis. During neuroinflammation, elevated concentrations of CXCL12 due to its increased production by microglial and/or brain microvascular endothelial cells that constitute the BBB, could affect neuronal and neuroendocrine activities thereby brain pathology and/or neurotoxicity. Additionally, receptor-ligand interaction between CXCR4-CXCL12 and other neurotransmitter systems of the CNS, suggests that these may be key players of the neuroimmune interface that participates in shaping the brain function in response to changes in the CNS environment (12). Tspan2 can not only control the trafficking of CXCL12 but can also regulate the cell surface expression of CXCL12 and its binding efficacy to its ligand CXCR4 on targeted microglia and thereby modulate microglial activation and subsequent neuropathology. Evidence suggests that extracellular vesicles, containing siRNA or other payloads such pro-peptides, cytosolic proteins, microRNAs and mRNAs etc, represent an efficient way to transfer functional molecules from one cell to another. The exosome vesicular content can be delivered to targeted recipient cells (19, 29, 31, 32). Microglial cells provide constant neurotrophic support, microglia and neurons can communicate by releasing and receiving extracellular vesicles (33–36). We investigated this ability of the microglial derived exosomes to communicate across physiological barriers. However, the greatest challenge with using biologics to target the brain has been getting them across the BBB in sufficient amounts to provide effective treatment. In our study, we derived exosomes from microglial cells and demonstrate that exosomes can not only carry a siRNA payload across the BBB but can also be taken up efficiently by HTHU-HIV microglia and potentially modulate the transition of microglial phenotype. We observed a significant decrease in both the expression of CXCL12 and its receptor CXCR4 in HTHU-HIV microglia that were transfected with Tspan2 siRNA and had decreased gene expression for Tspan2. Furthermore, a decrease in Tspan2 gene resulted in a significant decrease the gene expression levels of IL-13 and IL-10 and an increase in the gene expression levels for FCGR2A and TNF-α. Blocking Tspan2 activity or knockdown Tspan2 gene expression in the brain may alter microglial activation and limit or stall the neuro-pathogenesis process. During chronic neuroinflammation, microglial cells are continually activated by proinflammatory stimuli and these chronically activated microglia will continue to produce inflammatory cytokines and reactive oxygen/nitrogen species, which leads to neuronal death. Pro- and anti-inflammatory cytokines can polarize microglia to distinct activation states, however the exact mechanism that underlies microglial polarization and the functional relevance of this process in both acute and chronic CNS disease remains to be investigated. In the context of HIV-1 infection, microglia exhibit various signs of activation including secretion of elevated levels of pro-inflammatory cytokines (7).
In the context of HIV-1 infection, microglia exhibit various signs of activation including secretion of elevated levels of pro-inflammatory cytokines (7). We observed significant changes in the expression levels of microglial activation markers and resting markers HLA-DBQ1, HLADRA and HLADRB1 between the microglial cells treated with Tspan2 siRNA exosomes as compared to the untreated control microglia. HLA-DRB markers belongs to the HLA class II beta chain paralogs which are heterodimers consisting of an alpha and a beta chain. These Class II molecules are expressed in antigen presenting cells. Analysis of MHC expression by the microglia is highly informative, as endothelial and perivascular cells constitutively express HLA class I molecules, and upregulated expression can be found in degenerative and/or inflammatory neurological disorders (37–39). Resting microglia may undergo rapid phenotypic changes with a marked increase in HLA class II expression that may be critical for antigen presentation (40, 41). Reactive microglia with increased class II expression have been reported in a number of neurological disorders with direct or indirect immune involvement (42).
In summary, microglia activation is complex and could result in various activation states such as the distinct M1 or M2 phenotypes or an intermediary phenotype that is as yet not completely distinct. During chronic neuroinflammation, microglial cells are continually activated by proinflammatory stimuli which leads to their producing inflammatory cytokines and reactive oxygen species, leading to neuronal death and neuropathology that underlies neuroHIV. The M1/M2 polarization of microglia contributes significantly to how neuroinflammation is modulated in the CNS (6, 7) and thus an exosome mediated therapeutic strategy that is explored in this study may be crucial step in limiting HIV associated neuroinflammation. Future investigations will include identifying the role of Tspan2 and the microglia-dependent NLR family pyrin domain containing 3 inflammasome and damage-associated molecular pattern molecules that play a significant role in the brain under neuroinflammatory conditions.
Acknowledgements
Research reported in this publication was supported by 1R01AI129649 (NIAID) (JR), 1R01DA047410 (NIDA) (SM) and the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR001412 (JR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Literature Cited
- 1.Hogan C, Wilkins E. Neurological complications in HIV. Clinical Medicine 2011; 11 (6):571–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valcour V, Sithinamsuwan P, Letendre S, Ances B. Pathogenesis of HIV in the central nervous system. Current HIV/AIDS reports. 2011;8(1):54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnell G, Joseph S, Spudich S, Swanstrom R. HIV-1 Replication in the Central Nervous System Occurs in Two Distinct Cell Types. PLoS Pathog 2001;7(10):e1002286 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sillman B, Woldstad C, Mcmillan J, Gendelman HE. Neuropathogenesis of human immunodeficiency virus infection. Handb Clin Neurol. 2018;152:21–40. [DOI] [PubMed] [Google Scholar]
- 5.Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R. HIV-1 target cells in the CNS. J Neurovirol 2015;21(3):276–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherry J, Olschowka J, O’Banion MK. Neuroinflammation and M2 microglia: the good, the bad, and the inflamed. J Neuroinflammation. 2014; 11:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y, Weidong L. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Molecular Neurobiology. 2016;53(2): 1181–94. [DOI] [PubMed] [Google Scholar]
- 8.Hantak M, Qing E, Earnest JT, Gallagher T. Tetraspanins: Architects of Viral Entry and Exit Platforms. J Virol 2019;93(9):01429–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krementsov DN, Weng J, Lambele M, Roy NH, Thali M. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology. 2009;6:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sims B, Farrow AL, Williams SD, Bansal A, Krendelchtchikov A, Matthews QL. Tetraspanin blockage reduces exosome-mediated HIV-1 entry. Arch Virol. 2018; 163(6): 1683–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaseen IH, Monk PN, Partridge LJ. Tspan2: a tetraspanin protein involved in oligodendrogenesis and cancer metastasis. Biochem Soc Trans. 2017;45(2):465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A G. CXCL12 chemokine and its receptors as major players in the interactions between immune and nervous systems. Front Cell Neurosci 2014;8(65):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serlin Y, Shelef I, Knyazer B, Friedman A. Anatomy and physiology of the blood-brain barrier. Semin Cell Dev Biol. 2015;38:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbott N Prediction of blood–brain barrier permeation in drug discovery from in vivo, in vitro and in silico models. Drug Discovery Today: Technologies. 2004;1(4):407–16. [DOI] [PubMed] [Google Scholar]
- 15.Abbott NJ. Dynamics of CNS Barriers: Evolution, Differentiation, and Modulation. Cellular and Molecular Neurobiology. 2005;25(1):5–23. [DOI] [PubMed] [Google Scholar]
- 16.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6(4):287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnology advances. 2013;31(5):543–51. [DOI] [PubMed] [Google Scholar]
- 18.van Dommelen SM, Vader P, Lakhal S, Kooijmans SA, van Solinge WW, Wood MJ, et al. Microvesicles and exosomes: opportunities for cell-derived membrane vesicles in drug delivery. J Control Release. 2012;161(2):635–44. [DOI] [PubMed] [Google Scholar]
- 19.Aryani A, Denecke B. Exosomes as a Nanodelivery System: a Key to the Future of Neuromedicine? Mol Neurobiol. 2016;53(2):818–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CC, Liu L, Ma F, Wong CW, Guo XE, Chacko JV, et al. Elucidation of Exosome Migration across the Blood-Brain Barrier Model In Vitro. Cell Mol Bioeng. 2016;9(4):509–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumoto J, Stewart T, Sheng L, Li N, Bullock K, Song N, et al. Transmission of alpha-synuclein-containing erythrocyte-derived extracellular vesicles across the blood-brain barrier via adsorptive mediated transcytosis: another mechanism for initiation and progression of Parkinson’s disease? Acta Neuropathol Commun. 2017;5(1):71-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood M. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature biotechnology. 2011. ;29(4):341–5. [DOI] [PubMed] [Google Scholar]
- 23.El Andaloussi S, Lakhal S, Mager I, Wood MJ. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65(3):391–7. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Chen L, Huang Y, Li K, Jinye A, Fan T, et al. Exosome-delivered TRPP2 siRNA inhibits the epithelial-mesenchymal transition of FaDu cells. Oncol Lett. 2019;17(2):1953–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Sui H, Zheng Y, Jiang Y, Shi Y, Liang J, et al. Curcumin-primed exosomes potently ameliorate cognitive function in AD mice by inhibiting hyperphosphorylation of the Tau protein through the AKT/GSK-3beta pathway. Nanoscale. 2019;11(15):7481–96. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Mesa Y, Jay TR, Checkley MA, Luttge B, Dobrowolski C, Valadkhan S, et al. Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. J Neurovirol. 2017;23(1):47–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan SD, Roy I, Xu G, Yong KT, Ding H, Aalinkeel R, et al. Enhancing the delivery of anti retroviral drug “Saquinavir” across the blood brain barrier using nanoparticles. Current HIV Research. 2010;8(5):396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmittgen TD, Livak K. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–8. [DOI] [PubMed] [Google Scholar]
- 29.Frühbeis C, Fröhlich D, Kuo WP, Krämer-Albers EM. Extracellular vesicles as mediators of neuron-glia communication. Front Cell Neurosci. 2013;7:182-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011;91:461–553. [DOI] [PubMed] [Google Scholar]
- 31.Andras I, Toborek M. Extracellular vesicles of the blood-brain barrier. Tissue Barriers. 2016;4(1):e1131804-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bang C, Thum T. Exosomes: new players in cell-cell communication. The international journal of biochemistry & cell biology. 2012;44(11):2060–4. [DOI] [PubMed] [Google Scholar]
- 33.Fitzner D, Schnaars M, Van Rossum D, Krishnamoorthy G, Dibaj P, Bakhit M, et al. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. Journal of Cell Science. 2011;124:447–58. [DOI] [PubMed] [Google Scholar]
- 34.Jonavice U, Tunaitis V, Kriauciunaite K, Jarmalaviciute A, Pivoriunas A. Extracellular vesicles can act as a potent immunomodulators of human microglial cells. J Tissue Eng Regen Med. 2019; 13(2):309–18. [DOI] [PubMed] [Google Scholar]
- 35.Paolicelli RC, Bergamini G, Rajendran L. Cell-to-cell Communication by Extracellular Vesicles: Focus on Microglia. Neuroscience. 2019;405:148–57. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Boza-Serrano A, Dunning CJR, Clausen BH, Lambertsen KL, Deierborg T. Inflammation leads to distinct populations of extracellular vesicles from microglia. J Neuroinflammation. 2018;15(1):168: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobel RA, Blanchette BW, Bhan AK, Colvin RB. The immunopathology of experimental allergic encephalomyelitis. II. Endothelial cell Ia increases prior to inflammatory cell infiltration. J Immunol. 1984;132:2402–7. [PubMed] [Google Scholar]
- 38.Lampson LA, Hickey WF. Monoclonal antibody analysis of MHC expression in human brain biopsies: tissue ranging from “histologically normal” to that showing different levels of glial tumor involvement. J Immunol. 1986;136:4054–62. [PubMed] [Google Scholar]
- 39.Sobel RA, Ames MB. Major histocompatibility complex molecule expression in the human central nervous system: immunohistochemical analysis of 40 patients. J Neuropath Exp Neurol. 1988;47:19–28. [DOI] [PubMed] [Google Scholar]
- 40.Hayes G, Woodroofe MN, Cuzner ML. Microglia are the major cell type expressing MHC class II in human white matter. J Neurol Sci 1987;80:25–37. [DOI] [PubMed] [Google Scholar]
- 41.Hickey W, Kimura H. Perivascular microglial cells of the CNS are bone marrow-derived and present antigen in vivo. Science. 1988;239:290–2. [DOI] [PubMed] [Google Scholar]
- 42.McGeer PL, Itagaki S, McGeer EG. Expression of the histocompatibility glycoprotein HLA-DR in neurological disease. Acta Neurpathol. 1988;76:550–55. [DOI] [PubMed] [Google Scholar]