Abstract
The chemokine CXCL14 is a highly conserved, homeostatic chemokine which is constitutively expressed in skin epithelia. Responsible for immune cell recruitment and maturation, as well as impacting epithelial cell motility, CXCL14 contributes to the establishment of immune surveillance within normal epithelial layers. Furthermore, CXCL14 is critical to upregulating major histocompatibility complex class I (MHC-I) expression on tumor cells. Given these important roles, CXCL14 is often dysregulated in several types of carcinomas including cervical, colorectal, endometrial, and head and neck cancers. Its disruption has been shown to limit critical antitumor immune regulation and is correlated to poor patient prognosis. However, other studies have found that in certain cancers, namely pancreatic and some breast cancers, overexpression of stromal CXCL14 correlates with poor patient survival due to increased invasiveness. Contributing to the ambiguity CXCL14 plays in cancer is that the native CXCL14 receptor remains uncharacterized, although several candidate receptors have been proposed. Despite the complexity of CXCL14 functions, it remains clear that this chemokine is a key regulatory factor in cancer and represents a potential target for future cancer immunotherapies.
Keywords: CXCL14, chemokine, antitumor immunity, HPV, immunotherapy
1. INTRODUCTION
Epithelial cells make up the highly abundant portion of human tissues. Carcinomas, which comprise the majority of human malignancies, are cancers that arise from these epithelial cells. However, various other cell types surrounding epithelial tumors, such as fibroblasts and immune cells, are necessary to form the tumor microenvironment (TME). The TME helps determine specific niches for cancer cell survival, proliferation, invasion, and migration. Small secreted extracellular molecules such as chemokines and bioactive lipids are essential for the coordination of cells necessary to establish and sustain the TME1,2.
Chemokines (chemotactic cytokines) are small signaling proteins (8–10 kDa) produced by all cell types in the body3. Chemokines function by inducing directed chemotaxis of nearby responsive cells and regulating homeostasis and inflammation in the local environment. CXCL14 is a relatively novel chemokine with characteristics distinct from other chemokines. CXCL14 is constitutively expressed by normal skin keratinocytes, recruits various immune cells to the local environment, and functions as an antimicrobial and antitumoral factor4,5. Thus, it has been suggested that CXCL14 plays an important role in homeostasis and routine immune surveillance of the skin and that its dysregulation might greatly influence the TME (summarized in Fig. 1).
Figure 1.
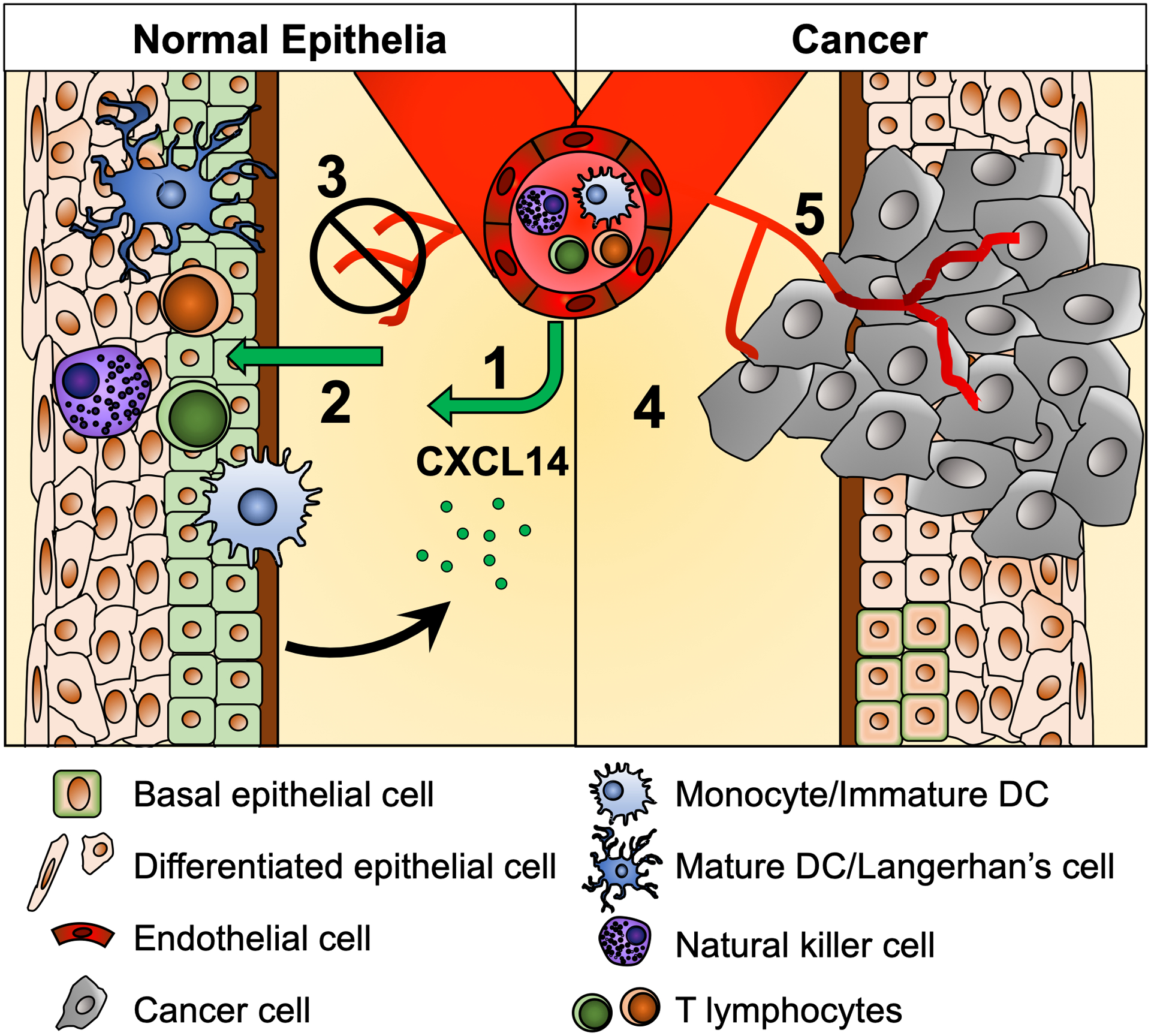
An overview of the CXCL14 functions in normal squamous epithelia (left) and the tumor microenvironment of head and neck cancer (right). In normal tissue, CXCL14 expression (green cell membrane) and secretion (green circle) (1) promotes immune cell recruitment to skin epithelia, (2) induces immune cell maturation and activation in skin layers, and (3) inhibits vascularization and epithelial cell motility, resulting in effective immune surveillance. In cancer tissue, the loss of CXCL14 expression (4) prevents immune cell recruitment and (5) enhances cancer cell invasion and angiogenesis, resulting in the immunosuppressive tumor microenvironment.
2. GENE AND PROTEIN STRUCTURE OF CXCL14
CXCL14 was first identified and cloned by Hromas et al. in 1999, showing 30% identity and 55% similarity with CXCL1, CXCL2, and CXCL8 which bind to the common chemokine receptor CXCR26 (Fig. 2A and 2B). Like other CXC chemokines, CXCL14 has four conserved cysteine residues that form disulfide bonds7. Also, in common, the first 22 amino acids of the N-terminus are strongly hydrophobic and act as a signal peptide, which is cleaved prior to secretion. Interestingly, despite the high degree of similarity to CXCL1, CXCL2, and CXCL8, CXCL14 is relatively unique among CXC chemokines and does not interact with their common receptor, CXCR2. Indeed, CXCL14 remains an orphan chemokine with its native receptor not yet fully characterized (Fig. 2B).
Figure 2.
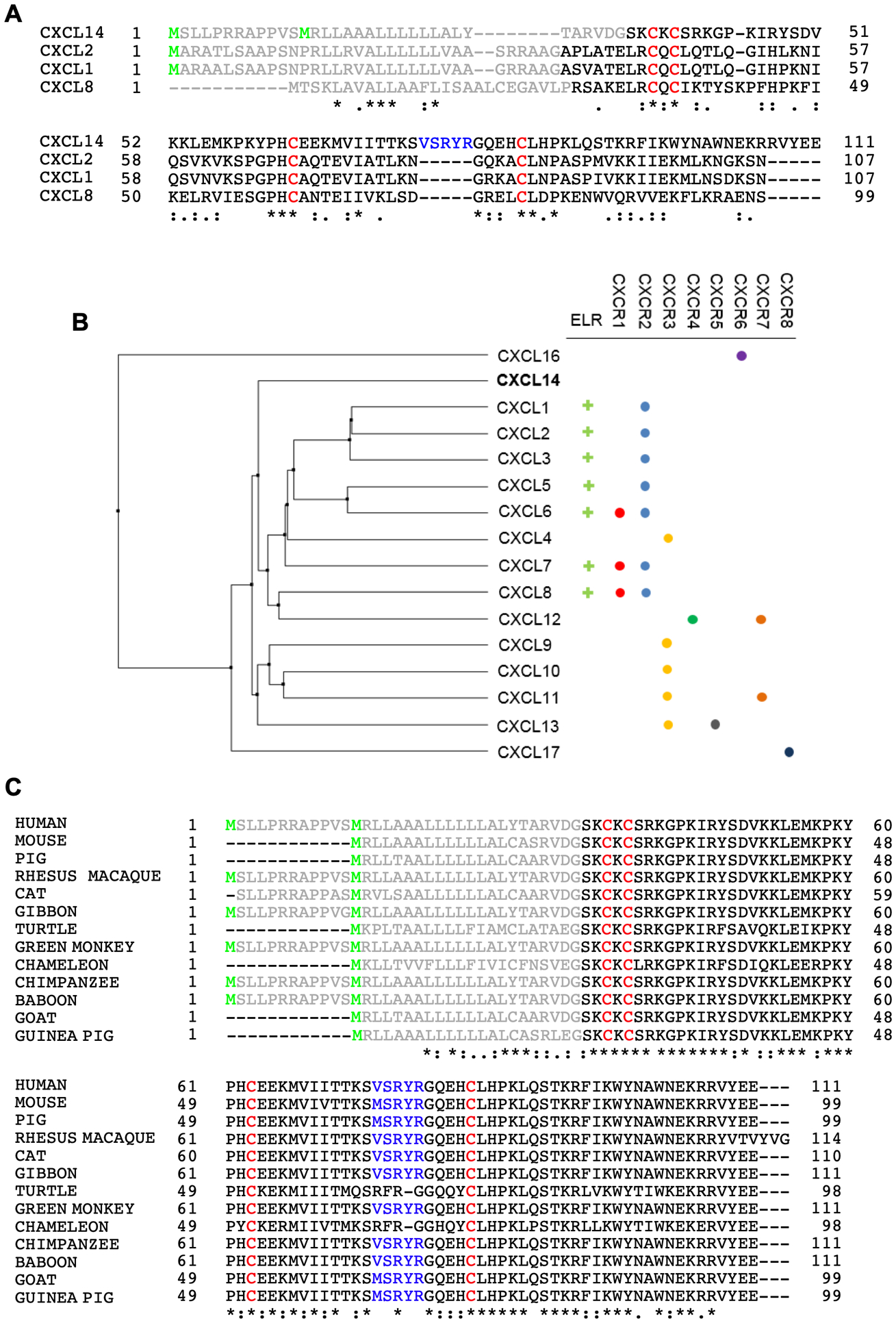
Multiple sequence alignment of human CXCL14 protein with CXCL1, CXCL2, and CXCL8 proteins (A), phylogram of human CXC ligands along with their receptors (B), and CXCL14 proteins of other animal species (C). In panels (A) and (C), start methionines (green), the signal peptides (grey), four conserved cysteine residues (red), and the unique five amino acid insertion (V/M-S-R-Y-R) (blue) are indicated. CXC ligand alignments (B) were performed using Clustal W2140 and the phylogram was generated in Jalview 2 using the BLOSUM62 matrix and UPGMA algorithm141.
CXCL14 is one of the most evolutionarily conserved chemokines, exhibiting remarkable similarity across a wide range of taxa including mammals and reptiles (Fig. 2C). In fact, mature human CXCL14 differs from murine CXCL14 at only two amino acids6. Among mammals, CXCL14 has a unique five amino acid insertion (V/M-S-R-Y-R) which is not present in in reptilian CXCL14 nor any other CXC chemokines (Fig. 2B)6,8. This VSRYR motif is recognized as a destruction box that is necessary and sufficient for ubiquitination and proteasomal degradation of CXCL148. Interestingly, CXCL14 is efficiently degraded by ubiquitin-mediated proteolysis in cancer and immortalized cells, but not in normal epithelial cells8. However, it is unknown if the deletion of the destruction box affects biological activity of CXCL14.
Unlike other chemokines, CXCL14 encodes a very short amino terminus (Ser-Lys) prior to the invariant CXC motif6. Another unique facet is that primate CXCL14 has two methionine start codons with the longer isoform encoding 12 additional amino acids prior to the 22 amino acid signal sequence (Fig. 2C). However, as of now, the molecular characteristics and functions of these two CXCL14 variants remain completely unknown.
3. REGULATION OF CXCL14 EXPRESSION
Unlike many other chemokines, CXCL14 is constitutively expressed in several normal tissues including adipose, brain, breast, cervix, lung, kidney, and skin (Fig. 3A). Squamous epithelia in particular expresses high levels of CXCL144 (Fig. 3B), supporting the idea that CXCL14 plays a homeostatic role in the skin layer. By analyzing human patient tissue samples and cell culture models, we have previously shown that CXCL14 expression is dramatically decreased during human papillomavirus (HPV)-driven progression of cervical (CxCa) and head and neck (HNC) cancers9,10 (Fig. 3B). Indeed, downregulation of CXCL14 expression is frequently observed in many cancers including cervical, prostate, lung, pancreatic, gastric, and oral cancers (Table 1)4,6,11–15. In contrast, several studies have shown that CXCL14 expression is upregulated in other cancers. While some of the studies found increased CXCL14 expression in epithelial cells, others identified cell types such as tumor-associated fibroblasts and infiltrating lymphocytes as common sources of CXCL14 overexpression4,16–20.
Figure 3.
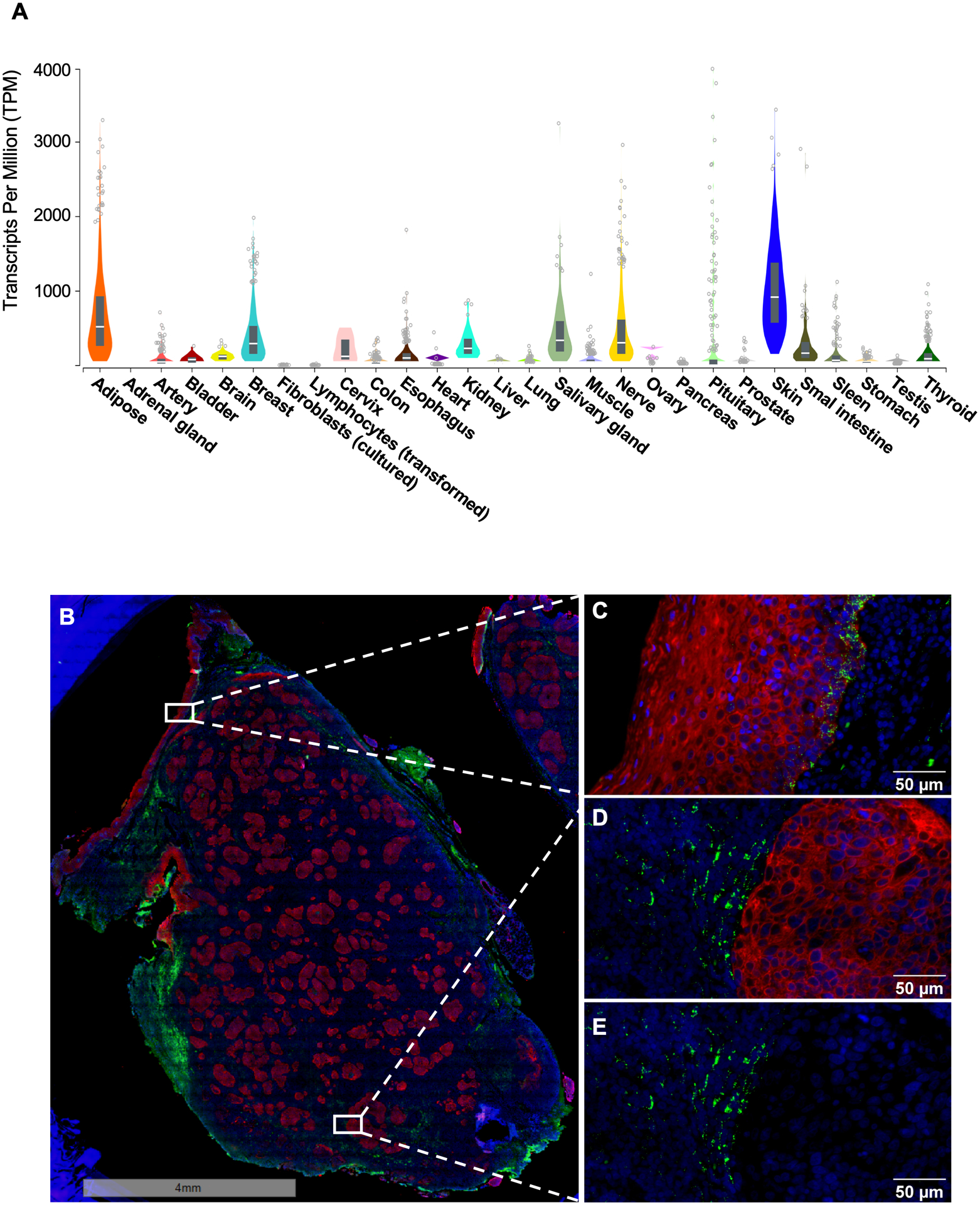
CXCL14 expression in normal human tissues in different organs (A) and RNA ISH of an HPV-positive tonsil tumor. The graph in (A) was generated in the Genotype-Tissue Expression (GTEx) Portal (gtexportal.org). The data used for the analyses were obtained from the GTEx Portal on 01/22/2020. The images of whole HNC tissue (B) and the magnified areas (C-E) show epithelial cells by cytokeratin immunostaining (red), CXCL14 RNA transcripts by in-situ hybridization (green), and nuclei by DAPI staining (blue). The basal layer of adjacent normal epithelium (C) and the tumor stroma (D and E) exhibit robust CXCL14 expression, while the tumor tissue (D and E) show a lack of CXCL14 expression.
Table 1.
Dysregulation of CXCL14 expression and its correlations to patient survival.
| Cancer | CXCL14 Gene Expression | Prognostic Correlation | Re-expression Effect in vivo | References |
|---|---|---|---|---|
| Breast | Up/Downregulated* | Positive** | Tumor Suppression** | 4,6,16,131,132, Fig.4 |
| Cervical | Downregulated | Positive | NA | 4,9, Fig.4 |
| Colorectal | Up/Downregulated | Positive | NA | 4,25,133, Fig.4 |
| Endometrial | Downregulated | Positive | NA | 134, Fig.4 |
| Gastric | Downregulated | No Correlation | NA | 14,22,27 |
| Glioblastoma | Upregulated | Negative | NA | 135 |
| Head & Neck | Downregulated | Positive | Tumor Suppression | 4,10,12,14,28, Fig.4 |
| Kidney | Downregulated | No correlation | NA | 4 |
| Liver | Downregulated | No correlation | Tumor Suppression | 15,26 |
| Lung | Downregulated | No correlation | Tumor Suppression | 24,52 |
| Melanoma | NA | Negative | Tumor Suppression | 52,136, Fig. 4 |
| Osteosarcoma | Upregulated | Negative | NA | 137 |
| Ovarian | Not changed | No correlation | NA | 4 |
| Pancreatic | Upregulated | No correlation | NA | 13 |
| Prostate | Up/Downregulated | Positive | NA | 21,49,138 |
| Testis | NA | No correlation | NA | Data not shown |
| Thyroid | Upregulated | No correlation | NA | 139 |
| Urothelial | NA | No correlation | NA | Data not shown |
NA: no published data available
Upregulated in stromal cells and downregulated in epithelial cells
Not in agreement
Mechanistically, promoter hypermethylation has been identified as a common mechanism for downregulating CXCL14 expression in a variety of cancers10,21,22. For example, in a mouse model of acute myeloid leukemia, CXCL14 promoter hypermethylation occurs in the early stages of cancer progression, appearing in the preleukemic or early leukemic stage23. Hypermethylation of CXCL14 is also common in lung adenocarcinoma where decitabine treatment, a DNA methyltransferase inhibitor, restores CXCL14 expression24. In agreement to these findings, our study shows that CXCL14 promoter methylation is induced by the HPV oncoprotein E7 in virus-infected normal keratinocytes. This virus-mediated DNA hypermethylation is accumulated through low- and high-grade cervical lesions to invasive cancer10. In addition, CXCL14 expression and methylation status are highly correlated in HNC patients, while CXCL14 levels are restored in cervical cancer cells treated with the demethylating agent decitabine10. These findings suggest that CXCL14 promoter hypermethylation is the key mechanism in downregulation of CXCL14 expression in CxCa and HNC. Finally, promoter hypermethylation has been implicated in the downregulation of CXCL14 in multiple other cancers, including prostate, colorectal, gastrointestinal, liver, and oral cancers21,22,25–28.
An additional mechanism of CXCL14 loss was demonstrated via an atypical Rho family small GTPase, RhoBTB2. RhoBTB2 is frequently mutated, deleted or silenced in breast and lung cancers and leads to the downregulation of CXCL14 expression in primary epithelial cells29. Nevertheless, RhoBTB2 expression can also be downregulated in breast, liver, and bladder cancers by promoter hypermethylation30–33, thus DNA hypermethylation may be involved in the downregulation of CXCL14 by reducing RhoBTB2 expression.
CXCL14 expression can also be regulated by a microRNA in cancer-associated fibroblasts (CAFs). Downregulation of miR-29b, an important tumor suppressor34, in CAFs activates p38-STAT1 signaling and upregulates CXCL14 expression in the stroma of breast cancer35. CXCL14 expression in interneurons also fully depends on the zinc finger transcription factor Sp8 showing that Sp8-deficient mice completely abrogate CXCL14 expression in caudal ganglionic eminence-derived cells36.
Activation of epidermal growth factor receptor (EGFR) signaling also downregulates CXCL14 expression. Treatment with the EGFR inhibitors, cetuximab and gefitinib, increases CXCL14 expression up to 50-fold in oral squamous carcinoma cells37. Inhibition of the extracellular signal-regulated kinase (ERK) signaling by EGFR activation increased CXCL14 expression in human tongue squamous carcinoma cells, but not in cervical adenocarcinoma cells37. Consistently, an earlier study showed that reactive oxygen species (ROS) reduce the expression of CXCL14 by activating EGFR-ERK signaling38.
Beyond cancer, regulation of CXCL14 expression by promoter methylation is also associated with immune regulation in viral and bacterial infections39,40, autoimmune diseases such as inflammatory bowel disease41, and even fetal development42,43. Although these studies, are outside the scope of this review, it exemplifies the importance of CXCL14 expression in both normal and diseased tissues.
4. CXCL14 EXPRESSION AND CANCER PATIENT SURVIVAL
To assess the relationship between CXCL14 expression and patient survival, we analyzed TCGA data obtained through cBioPortal and the Human Protein Atlas for a variety of cancers. Our TCGA analysis showed that the overall survival rates of CxCa and HNC patients are significantly higher with high CXCL14 expression levels (Fig. 4). Further, the positive correlations between CXCL14 expression levels and overall patient survival are also found in colorectal, breast, and endometrial cancers (Fig. 4). In contrast, melanoma is only cancer showing that high CXCL14 expression level correlates to poor patient survival (Fig. 4), while there are no significant associations in glioma, thyroid, lung, gastric, liver, pancreatic, renal, urothelial, testis, and ovarian cancers (data not shown). Previous studies have shown similar results (Table 1). Additionally, CXCL14 along with other chemokines (CCL13 and CCL27) is highly upregulated in the tissue samples of patients without tumor recurrence/perineural invasion compared to those with tumor recurrence44. It has also been reported that high CXCL14 expression levels are associated with the overall survival of breast cancer patients and progression-free survival of ovarian cancer patients45,46. Furthermore, a negative correlation exists between CXCL14 expression and lymph node metastasis. In contrast, other studies have shown that high stromal, but not epithelial, CXCL14 expression is significantly linked to poor survival in breast cancer and non-small-cell lung carcinoma47,48. In addition, CXCL14 expression levels are positively correlated with aggressive prostate cancer49. These findings suggest that the role of CXCL14 is dependent on the context and interconnection with other factors in the various TME.
Figure 4.
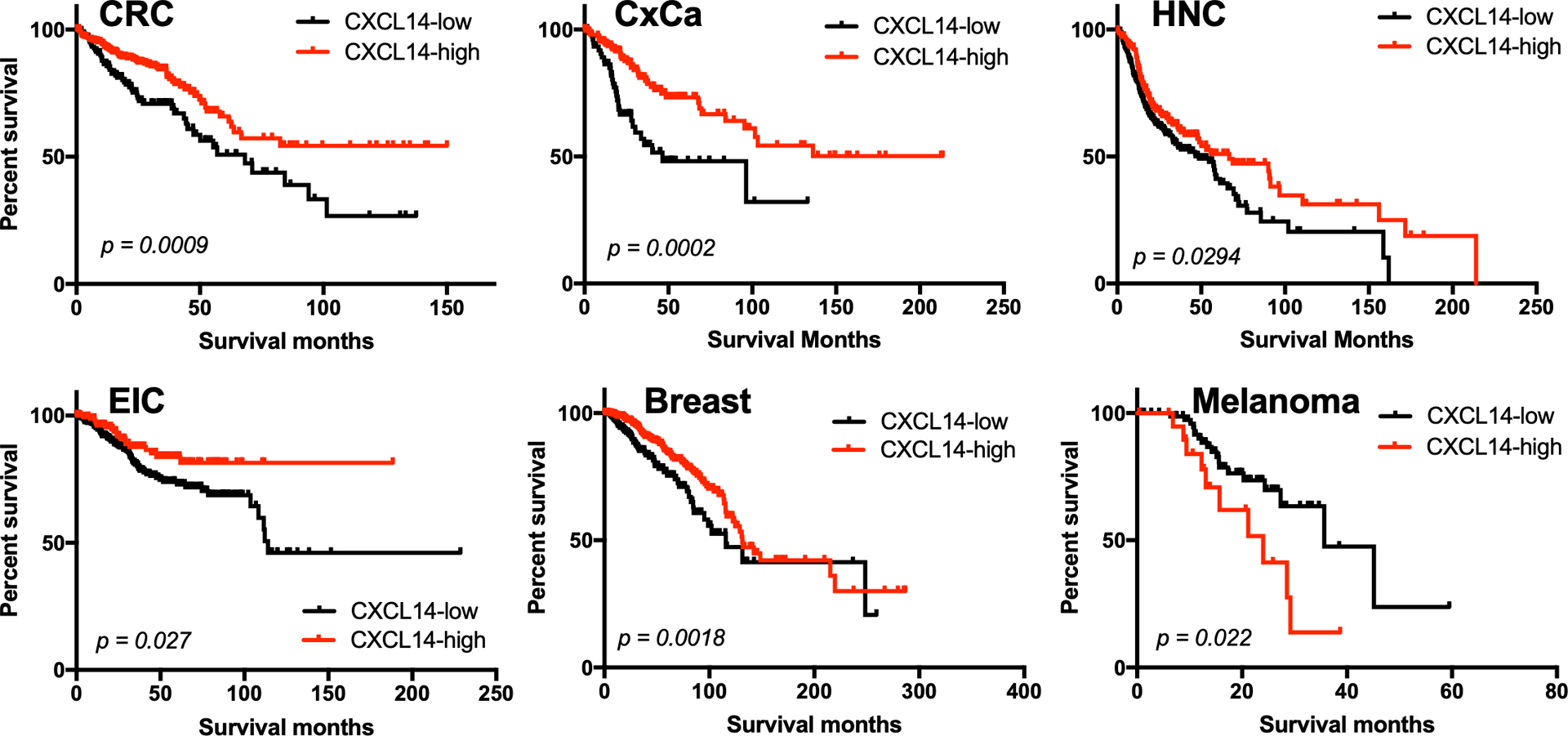
Correlations of CXCL14 expression with overall survival in patients with colorectal cancer (CRC), cervical cancer (CxCa), head and neck cancer (HNC), endometrial intraepithelial carcinoma (EIC), breast, and melanoma. The Cancer Genome Atlas (TCGA) RNA sequencing and patient survival data were obtained through the Human Protein Atlas (proteinatlas.org). Patient survival rates were analyzed using a Kaplan–Meier estimator and log-rank p-values were calculated as we previously described74. Patients of each cancer type were classified into high- and low-expression groups based on the best expression cut-off, which refers to the FPKM value that yields the lowest log-rank p-value.
5. CXCL14 FUNCTIONS IN CANCER PROGRESSION
5.1. CXCL14 as a Tumor Suppressor
We and other groups have revealed tumor suppressor functions of CXCL14 involved in immune responses, cell motility, and angiogenesis. Several studies have consistently shown that restoration of CXCL14 expression in cancer cells suppresses tumor growth in mouse models of HNC, melanoma, colon, liver, and lung cancers10,26,50–53. CXCL14 is expressed at a high level in normal oral epithelial cells, but frequently absent in oral carcinoma cells8,54. However, human oral squamous carcinoma cells, in which CXCL14 expression is restored by transfection of a CXCL14 expression vector, showed significantly slower tumor growth in athymic nude or severe combined immunodeficiency (SCID) mice compared to vector-transfected cells50,51. Similarly, our previous study showed that HPV-positive mouse HNC cells engineered to express physiological levels of CXCL14 considerably suppressed tumor growth in immunocompetent syngeneic mice10. Restoration of CXCL14 expression by an EGFR inhibitor also demonstrates the antitumor effect on HNC53.
In addition to HNC cancers, the development of melanoma and chronic colitis-associated colon cancer is suppressed in CXCL14 transgenic mice through NK cell activity52. The metastasis of melanoma cells is synergistically inhibited in CXCL14 transgenic mice treated with α-galactosyl ceramide, a strong immunostimulatory agent, showing a significant increase in mouse survival rate after injection of melanoma cells52. However, this is contradictory to the poor patient survival correlated to CXCL14 expression levels (Fig. 4). CXCL14 overexpression in colorectal carcinoma cells inhibits cell proliferation, while low CXCL14 expression correlates to increased lymph node metastasis and poor patient survival55.
CXCL14 has also been shown to play a crucial role as a tumor suppressor in hepatocellular cell carcinoma (HCC). CXCL14 expression is significantly downregulated in hepatitis B virus (HBV)-positive HCC patient tissues and CXCL14 polymorphisms are associated with disease progression of HBV infection56. Thus, it has been suggested to use re-expression or upregulation of CXCL14 as a novel strategy to treat HCC patients26. Stable expression of CXCL14 in lung adenocarcinoma cells induces tumor cell necrosis in vitro and dramatically suppresses tumor growth in athymic nude mice in vivo24. In addition, apoptosis of clear cell renal cell carcinoma cells is decreased by knockdown of CXCL14, subsequently enhancing tumorigenesis57. These findings suggest that CXCL14 is a potent tumor suppressing chemokine constitutively expressed in epithelial cells.
5.2. CXCL14 as a Tumor Promoter
Although our analysis shows better overall survival of breast cancer patients with high CXCL14 expression (Fig. 4), a separate study found a contrasting result that poor recurrence-free survival rates are correlated with high CXCL14 expression47. CXCL14 increases cell proliferation, migration, and resistance to paclitaxel in in vitro assays using breast cancer cells35. Interestingly, CXCL14 expression is highly upregulated in breast cancer containing a deletion of the gene hypermethylated in cancer 1 (HIC1), which is a tumor suppressor frequently hypermethylated in human tumors58–60. HIC1 downregulation activates CXCL14 promoter activity and increases CXCL14 expression. Increased CXCL14 expression in the absence of HIC1 induces hyperplasia of the mammary gland and breast cancer cell migration through the activation of stromal fibroblasts in vivo. Since all invasive ductal carcinomas and breast cancer cells expressing CXCL14 were obtained from young, premenopausal patients, Allinen et al. suggests a potential connection of CXCL14 expression with female hormone effects61. Interestingly, the negative impact CXCL14 exhibits differs in different cancers. As is observed in breast cancer, CXCL14 augments pancreatic cancer cell invasion but dissimilarly does not have an impact on cell viability and drug resistance13. Furthermore, CXCL14 secreted by surrounding fibroblasts increases the expression of mesenchymal markers and induces epithelial-mesenchymal transition (EMT) and lung metastasis16. These results suggest that CXCL14 functions as a tumor promoter in certain contexts.
5.3. Discrepancy of CXCL14 functions in tumor growth
To a large extent the contexts and factors that determine if CXCL14 acts as a tumor suppressor or promoter remain unknown. In addition to the technical limitations and variations (further discussed in the Conclusion section), the source of CXCL14 may be important. In breast cancer, CXCL14 expression in stromal fibroblasts is a poor prognostic marker47. Likewise, while it is broadly understood that estrogen receptor (ER) signaling is required for cervical cancer progression and maintenance62–64, our study has shown that expression of estrogen receptor α (ERα), an essential component of ER, is frequently absent in cancer epithelial tissues of cervical cancer patients9. Instead, ERα expression in stromal fibroblasts of cancer tissues is highly increased and essential for chemokine dysregulation and reprogramming the TME during cervical cancer progression9,65. Furthermore, in breast cancer epithelial CXCL14 expression was significantly associated with ERα-positive tumors and lower proliferation status47. These findings suggest that communications between cancer cells and CAFs may be important for the roles of CXCL14 in tumor suppression and promotion.
6. MOLECULAR FUNCTIONS OF CXCL14
6.1. Angiogenesis
CXCL14 was discovered as a potent inhibitor of angiogenesis, which is stimulated by CXCL8, fibroblast growth factor 2 (FGF2), and vascular endothelial growth factor (VEGF)54. Mechanistically, CXCL14 inhibits chemotaxis of endothelial cells by directly binding to IL-8 and FGF2, thus hindering their interaction with high-affinity receptors on human vascular endothelial cells54. Rivera et al. also showed that treatment of the antiangiogenic agent, sorafenib, induces a high level of CXCL14 expression in Gr1+ myeloid cells in a pancreatic neuroendocrine tumor, and neutralization of CXCL14 by antibody abrogates angiogenesis inhibition by sorafenib66, indicating a dominant angiostatic effect of CXCL14.
CXC chemokines containing an ELR (glutamic acid-leucine-arginine) motif at the amino-terminus such as CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, and CXCL8 show a potent angiogenic activity that induces endothelial chemotaxis and neovascularization67,68. In contrast, CXCL14 does not contain the ELR motif6 (Fig. 2B). However, a previous study has shown that CXCL14 can enhance angiogenesis. Fibroblasts overexpressing CXCL14 increase tumor angiogenesis and macrophage infiltration in prostate cancer xenografts17. This result suggests that CXCL14 secreted from fibroblasts may function in a completely different manner. Thus, depending on its source cell type, epithelial cells or fibroblasts, CXCL14 may exhibit opposite effects resulting in either tumor suppression or promotion, respectively.
6.2. Immune responses
Like other chemokines, CXCL14 broadly modulates chemotaxis, differentiation, and activation of various types of immune cells. Re-expression of CXCL14 in HNC cells induces chemotaxis of dendritic cells (DCs) and increases DC infiltration into the TME. Further enhancing the efficacy of DCs, treatment with recombinant human CXCL14 facilitates DC maturation through NF-κB signaling activation12. Circulating CD14+ DC precursors and monocytes are recruited by CXCL14 secreted from the epidermis and differentiated to tissue-resident macrophages and Langerhans cells for antigen presentation69,70. The number of Langerhans cells and their activation is impaired in HPV16 E7 expressing epidermis71. This may be explained by our previous finding of low CXCL14 expression in HPV-positive cancer cells caused by HPV16 E7-mediated hypermethylation of the CXCL14 promoter10. As tissue-resident macrophages and Langerhans cells are critical for immune defense in the skin layer, these findings support the homeostatic functions of CXCL14 in immune surveillance of skin epithelia.
CXCL14 was previously found to provoke the migration of activated human natural killer (NK) cells72 and CXCL14-deficient mice show significantly lower NK cell numbers in the uterus of pregnant mice73. Suppression of tumor growth and metastasis by CXCL14 is abolished when NK cells are depleted in mice with melanoma52, suggesting that the NK cell activity enhanced by CXCL14 is key for tumor suppression in melanoma. Our studies have also shown that NK cells are recruited by re-expression of CXCL14 in HPV-positive HNC cells, and NK cell depletion reverted CXCL14-mediated tumor suppression10,74. In addition, our immunocompetent syngeneic HNC model has revealed that CXCL14 re-expression also induces chemotaxis of CD4+ and CD8+ T cells. While depletion of CD4+ T cells shows minimal effects, all mice with a compromised CD8+ T cell repertoire grow tumors, indicating that CXCL14-mediated tumor suppression is reliant on CD8+ T cells74. Further contributing to the tumor suppression, we have discovered that CXCL14 increases MHC-I expression on the tumor cell surface and activates antigen-specific CD8+ T cells to kill HPV-positive HNC cells74. These results suggest a potent tumor suppressor function of CXCL14, which could be used as an immunotherapeutic agent to treat cancers.
6.3. Epithelial cell proliferation and migration
In addition to immune cells, CXCL14 also modulates epithelial cell function and mobility. Our study has shown that restoration of CXCL14 expression in human CxCa cells and mouse HNC cells consistently inhibits cell migration and wound healing in vitro. Inhibition of the CXCL12-CXCR4 signaling axis by CXCL14 has been suggested as one of the mechanisms by which CXCL14 inhibits cell migration75. This inhibitory effect of CXCL14 on cell migration may explain the correlation between CXCL14 expression and decreased metastasis in melanoma, colorectal cancer (CRC), and breast cancers45,52. In contrast, other studies have reported that CXCL14 enhances cancer cell migration and invasion. Treatment with recombinant human CXCL14 accelerates cell migration and invasion of CRC and breast cancer cells61,76.
While we found no change in cell proliferation rates by CXCL14 expression in human CxCa and mouse HNC cells, a study has shown that CXCL14 expression inhibits the proliferation of oral cancer cells and CRC cells by arresting the cell cycle in the G1 phase55,77. CXCL14 also functions as a negative regulator of skeletal muscle regeneration through cell cycle arrest and differentiation of myoblasts78. These findings suggest a potential role of CXCL14 in cell cycle arrest of cancer epithelial cells.
6.4. Antimicrobial activity
Like several chemokines expressed by mucosal skin such as CXCL9, CXCL10, CXCL17, CCL20, CCL25, and CCL2879–84, CXCL14 also shows antimicrobial activity7,85. CXCL14 effectively clears infection of Streptococcus pneumoniae, but not Pseudomonas aeruginosa, in respiratory tracts7. Interestingly, the N-terminal 13 amino acids of CXCL14 (SKCKCSRKGPKIR) are sufficient to enact the full antimicrobial activity, comparable to the activity of full-length CXCL147. These results demonstrate that CXCL14 plays an important multifunctional role in immune surveillance in normal epithelial tissues86.
7. CXCL14 CANDIDATE RECEPTORS
7.1. CXCR4
CXCR4, the receptor of CXCL12, was the first receptor identified having direct interaction with CXCL1475,87. Previous studies have shown that CXCL14 may function as a decoy ligand of CXCR4 and inhibit activation of CXCR4 signaling by CXCL12. Mechanistically, CXCL14 binds to CXCR4 competitively with CXCL12 and internalizes CXCR4, interfering with receptor signaling. Further, a dimerized CXCL14 peptide (amino acids 51–77) is sufficient to bind to CXCR4 and inhibit CXCL12-CXCR4 signaling activation by triggering CXCR4 internalization88. The CXCL12-CXCR4 signaling regulates organogenesis, hematopoiesis, and angiogenesis during gestational development and lymphocyte development89–92 as well as the typical chemokine functions for the recruitment of immune cells such as dendritic cells and lymphocytes93,94. The CXCL12-CXCR4 axis is well known to promote cancer development and metastasis by enhancing angiogenesis, cell migration, and epithelial-mesenchymal transition95–97. Thus, CXCL14 inhibition of CXCL12 binding to CXCR4 may be a pertinent mechanism of CXCL14-mediated tumor suppression.
Interestingly, however, another group has shown a contrary result that CXCL14 synergistically activates CXCR4 in the presence of CXCL12. While CXCL14 alone does not activate CXCR4 signaling despite the high-affinity interaction, CXCL14 synergizes the sub-optimal concentration of CXCL12 to activate CXCR4 signal transduction98. CXCL14 binding to CXCR4 changes the receptor conformation in the cell surface membrane. Another study has shown that CXCL14 is highly expressed and colocalized with CXCR4 in idiopathic pulmonary fibrosis99, and supports the role of CXCL14 in CXCR4 signaling activation.
Finally, another group refutes any associations between CXCL14 and CXCR4, showing that CXCL14 does not affect CXCL12-induced CXCR4 phosphorylation, G protein coupling, internalization, or downstream mitogen-activated protein kinase (MAPK) signaling activation100. The conflicting results of all three possibilities (inhibition, activation, and no effect on CXCR4) suggest that the mechanisms by which CXCL14 functions are complex and may depend on effector vs. target cell types, the context of the tissue microenvironment, and combinations of other cytokines5. These discrepant functions of CXCL14 even on the identical receptor may well represent both tumor suppressor and promoter activities in different cancers described above.
7.2. GPR85
CXCL14 secreted by HIC1-deleted breast cancer cells binds to a member of the G protein-coupled receptor (GPCR) family, GPR85 (also known as Super Conserved Receptor Expressed In Brain 2, or SREB2), expressed on fibroblasts58. The CXCL14 and GPR85 interaction activates ERK1/2, AKT, neddylation signaling and induces CCL17 production. CCL17 secreted by the activated fibroblasts returns and binds to CCR4 on breast cancer cells and induces breast cancer cell migration58. GPR85 is also known to be expressed on macrophages101,102 and can be targeted by miR-29, which also targets CXCL14 mRNA102. However, GPR85 expression has not been previously observed on fibroblasts or epithelial cells nor shown to be associated with any cancer. Thus, it would be interesting to investigate the regulations of chemokine expression in the TME by GPR85 signaling.
7.3. ACKR2
A recent study has shown that CXCL14 also binds another GPCR named Atypical Chemokine Receptor 2 (ACKR2). ACKR2 is known as a chemokine decoy receptor that interacts with the majority of inflammatory CC chemokines, degrades them, and thus inhibits inflammation103–105 ACKR2 is expressed in lymphatic tissues at high levels, particularly on lymphatic endothelial cells within non-inflamed tissues106,107. ACKR2 expression is induced by VEGF-D, transforming growth factor-β (TGFβ), IL-6, interferon (IFN)-α/β, and IFN-γ108. It has been well known that ACKR2 plays a central role in the regulation of chemokine levels near afferent lymphatic vessels and the elimination of proinflammatory chemokines from inflamed tissues109,110.
Interestingly, ACKR2 expression in tumor cells is protective against cancer progression by inhibiting proinflammatory chemokines and infiltration of immune cells such as M2 macrophages111–114. High ACKR2 expression in human breast cancer is associated with low levels of axillary lymph node metastasis, and a single nucleotide polymorphism of ACKR2 is associated with lymph node metastasis and decreased patient recurrence-free survival115–117. These results suggest that increased ACKR2 expression hinders cancer progression by blocking inflammatory chemokines, inhibiting angiogenesis, and limiting the infiltration of immunosuppressive immune cells. Surprisingly, however, expression of CXCL14 and ACKR2 in fibroblasts induces EMT, tumor cell invasion, and metastasis in breast cancer16. These results imply that CXCL14 and ACKR2 function completely different ways depending on their expression in different cell types.
8. CONCLUSIONS
As described, previous studies on CXCL14 have shown contradictory functions of CXCL14: tumor suppression vs. promotion, increased vs. decreased cell migration, correlations with better vs. worse patient survival rates, angiogenesis vs. angiostasis, and CXCR4 inhibition vs. activation vs. no effect. The mechanisms that regulate these multi-faced functions of CXCL14 are mysterious and potentially defined by the cell and tissue types that produce and respond to CXCL14 as well as by other chemokines and cytokines that interacts with CXCL145. Generally, while CXCL14 secreted by epithelial cells has largely been shown to suppress tumor growth, CXCL14 produced from CAFs promotes tumor growth and metastasis. While CXCL14-mediated tumor suppression and expression correlates with better patient survival in HNC, CRC, and HCC, CXCL14-mediated tumor promotion frequently occurs in breast and pancreatic cancers. CXCL14 induces chemotaxis of DCs, NK cells, and T cells, but inhibits epithelial and endothelial cell migration.
In addition to the complexity and variations in biological mechanisms, experimental conditions may also contribute to these contradictory results. CXCL14 proteins used in the previous experiments were mostly synthesized or purified from bacterial expression. Our unpublished data show that glycosylation of CXCL14 protein, which is lacked from chemical synthesis and bacterial expression, may be important for its function in the inhibition of cell migration. In addition, human CXCL14, but not mouse CXCL14, has two potential start codons, but their effects and regulation have not been considered or determined in any studies to date. A further confounding factor is that the CXCL14 protein is extremely unstable, resulting in difficult purification and consistent treatments during ex vivo experiments. This instability is principally structurally driven, as CXCL14 contains a C-terminal degron (two glutamates) which mediate efficient protein interaction with E3 ubiquitin ligases to facilitate rapid protein degradation118. Another caveat is that a vast majority of the gene expression data from patient samples (e.g., TCGA) analyzed whole tumor tissues including both epithelial and stromal components. Our previous study has shown that the proportions of cancer epithelial cells in patient tissue samples are extremely variable, mixed with normal stromal cells119. As the source of the CXCL14 is highly associated with its impact on tumor growth, the variability of these samples may result in unclear findings. Thus, precise microdissection should be required for accurate gene expression analysis using tumor tissue samples.
Despite the complex functions of CXCL14 in cancer progression, there is a consensus on its tumor suppressor functions in HNC, particularly with HPV infection. Current immunotherapies using immune checkpoint inhibitors (ICI) have been effective in some cases, but over 70% of treated patients fail to respond120–122. These ICI-refractory cancers have markedly reduced expression of MHC-I antigen presentation, with little T cell infiltration123–126. Low MHC-I expression significantly correlates with poor survival rates in HNC patients127 and upregulation of MHC-I antigen presentation stimulates antitumor immunity responding to ICI in ICI-refractory cancer128. It has been suggested that modulating chemokines to recruit T cells may overcome this hurdle129,130. We have previously shown that restoring CXCL14 expression in HPV-positive HNC cells suffices to recruit NK and T cells in vitro and in vivo, and importantly that it also upregulates MHC-I in epithelial tumor cells, resulting in suppressed tumor growth10,74. Thus, further studies on CXCL14 may lead to novel immunotherapeutic strategies to treat HNC and other cancer patients, particularly non-responders to current immunotherapies that do not reverse the lack of MHC-I expression and T cell infiltration in the TME.
ACKNOWLEDGMENT
This work was supported in part by NIH R01 DE026125 (D. Pyeon and W.C. Spanos) and NIH T32 AI052066 (J.A. Westrich). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Funding information: National Institutes of Health, Grant/Award Number: R01 DE026125, T32 AI052066, and P20 GM103548
Footnotes
Conflicts of interest: The authors declare that there are is conflict of interests.
REFERENCES
- 1.Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller MC, Mayo KH. Chemokines from a Structural Perspective. Int J Mol Sci. 2017;18(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frederick MJ, Henderson Y, Xu X, et al. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am J Pathol. 2000;156(6):1937–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang XY, Ozawa S, Kato Y, et al. C-X-C Motif Chemokine Ligand 14 is a Unique Multifunctional Regulator of Tumor Progression. Int J Mol Sci. 2019;20(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hromas R, Broxmeyer HE, Kim C, et al. Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem Biophys Res Commun. 1999;255(3):703–706. [DOI] [PubMed] [Google Scholar]
- 7.Dai C, Basilico P, Cremona TP, et al. CXCL14 displays antimicrobial activity against respiratory tract bacteria and contributes to clearance of Streptococcus pneumoniae pulmonary infection. J Immunol. 2015;194(12):5980–5989. [DOI] [PubMed] [Google Scholar]
- 8.Peterson FC, Thorpe JA, Harder AG, Volkman BF, Schwarze SR. Structural determinants involved in the regulation of CXCL14/BRAK expression by the 26 S proteasome. J Mol Biol. 2006;363(4):813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Boon JA, Pyeon D, Wang SS, et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A. 2015;112(25):E3255–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cicchini L, Westrich JA, Xu T, et al. Suppression of Antitumor Immune Responses by Human Papillomavirus through Epigenetic Downregulation of CXCL14. MBio. 2016;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito S, Ozawa S, Ikoma T, Yajima N, Kiyono T, Hata R. Expression of a chemokine BRAK/CXCL14 in oral floor carcinoma cells reduces the settlement rate of the cells and suppresses their proliferation in vivo. Biomed Res. 2010;31(3):199–206. [DOI] [PubMed] [Google Scholar]
- 12.Shurin GV, Ferris RL, Tourkova IL, et al. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. Journal of immunology (Baltimore, Md : 1950). 2005;174:5490–5498. [DOI] [PubMed] [Google Scholar]
- 13.Wente MN, Mayer C, Gaida MM, et al. CXCL14 expression and potential function in pancreatic cancer. Cancer Lett. 2008;259(2):209–217. [DOI] [PubMed] [Google Scholar]
- 14.Kou Y, Zhao Y, Bao C, Wang Q. Comparison of Gene Expression Profile Between Tumor Tissue and Adjacent Non-tumor Tissue in Patients with Gastric Gastrointestinal Stromal Tumor (GIST). Cell Biochem Biophys. 2015;72(2):571–578. [DOI] [PubMed] [Google Scholar]
- 15.Niu J, Lin Y, Liu P, Yu Y, Su C, Wang X. Microarray analysis on the lncRNA expression profile in male hepatocelluar carcinoma patients with chronic hepatitis B virus infection. Oncotarget. 2016;7(46):76169–76180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sjoberg E, Meyrath M, Milde L, et al. A Novel ACKR2-Dependent Role of Fibroblast-Derived CXCL14 in Epithelial-to-Mesenchymal Transition and Metastasis of Breast Cancer. Clin Cancer Res. 2019. [DOI] [PubMed] [Google Scholar]
- 17.Augsten M, Hagglof C, Olsson E, et al. CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth. Proc Natl Acad Sci U S A. 2009;106(9):3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Augsten M, Sjoberg E, Frings O, et al. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer Res. 2014;74(11):2999–3010. [DOI] [PubMed] [Google Scholar]
- 19.Jia G, Chandriani S, Abbas AR, et al. CXCL14 is a candidate biomarker for Hedgehog signalling in idiopathic pulmonary fibrosis. Thorax. 2017;72(9):780–787. [DOI] [PubMed] [Google Scholar]
- 20.Eiro N, Fernandez-Gomez J, Sacristan R, et al. Stromal factors involved in human prostate cancer development, progression and castration resistance. J Cancer Res Clin Oncol. 2017;143(2):351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song EY, Shurin MR, Tourkova IL, Gutkin DW, Shurin GV. Epigenetic mechanisms of promigratory chemokine CXCL14 regulation in human prostate cancer cells. Cancer Research. 2010;70:4394–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fotouhi O, Adel Fahmideh M, Kjellman M, et al. Global hypomethylation and promoter methylation in small intestinal neuroendocrine tumors: an in vivo and in vitro study. Epigenetics. 2014;9(7):987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnet M, Claus R, Becker N, et al. Early aberrant DNA methylation events in a mouse model of acute myeloid leukemia. Genome Med. 2014;6(4):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tessema M, Klinge DM, Yingling CM, Do K, Van Neste L, Belinsky Sa. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene. 2010;29:5159–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao B, Yang Y, Pan Y, et al. Epigenetic silencing of CXCL14 induced colorectal cancer migration and invasion. Discov Med. 2013;16(88):137–147. [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Huang P, Zhang L, et al. Antitumor efficacy of C-X-C motif chemokine ligand 14 in hepatocellular carcinoma in vitro and in vivo. Cancer Sci. 2013;104(11):1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu C, Lin F, Zhu G, et al. Abnormal hypermethylation of promoter region downregulates chemokine CXC ligand 14 expression in gastric cancer. Int J Oncol. 2013;43(5):1487–1494. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama R, Arikawa K, Bhawal UK. The epigenetic regulation of CXCL14 plays a role in the pathobiology of oral cancers. J Cancer. 2017;8(15):3014–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinnon CM, Lygoe KA, Skelton L, Mitter R, Mellor H. The atypical Rho GTPase RhoBTB2 is required for expression of the chemokine CXCL14 in normal and cancerous epithelial cells. Oncogene. 2008;27(54):6856–6865. [DOI] [PubMed] [Google Scholar]
- 30.Tang W, Wang C, Fu F, Chen Q. RhoBTB2 gene in breast cancer is silenced by promoter methylation. Int J Mol Med. 2014;33(3):722–728. [DOI] [PubMed] [Google Scholar]
- 31.Han L, Hou L, Song J, et al. Decreased expression of the DBC2 gene and its clinicopathological significance in breast cancer: correlation with aberrant DNA methylation. Biotechnol Lett. 2013;35(8):1175–1181. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y, Chen JY, Yang J, Li B, Chen ZH, Xiao CG. DBC2 gene is silenced by promoter methylation in bladder cancer. Urol Oncol. 2008;26(5):465–469. [DOI] [PubMed] [Google Scholar]
- 33.Ye C, Tao R, Cao Q, et al. Whole-genome DNA methylation and hydroxymethylation profiling for HBV-related hepatocellular carcinoma. Int J Oncol. 2016;49(2):589–602. [DOI] [PubMed] [Google Scholar]
- 34.Yan B, Guo Q, Fu FJ, et al. The role of miR-29b in cancer: regulation, function, and signaling. Onco Targets Ther. 2015;8:539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Zhang J, Sun X, Su Q, You C. Down-regulation of miR-29b in carcinoma associated fibroblasts promotes cell growth and metastasis of breast cancer. Oncotarget. 2017;8(24):39559–39570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei S, Du H, Li Z, et al. Transcription factors Sp8 and Sp9 regulate the development of caudal ganglionic eminence-derived cortical interneurons. J Comp Neurol. 2019;527(17):2860–2874. [DOI] [PubMed] [Google Scholar]
- 37.Kondo T, Ozawa S, Ikoma T, et al. Expression of the chemokine CXCL14 and cetuximab-dependent tumour suppression in head and neck squamous cell carcinoma. Oncogenesis. 2016;5(7):e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maehata Y, Ozawa S, Kobayashi K, et al. Reactive oxygen species (ROS) reduce the expression of BRAK/CXCL14 in human head and neck squamous cell carcinoma cells. Free Radic Res. 2010;44(8):913–924. [DOI] [PubMed] [Google Scholar]
- 39.Mukherjee S, Vipat VC, Chakrabarti AK. Infection with influenza A viruses causes changes in promoter DNA methylation of inflammatory genes. Influenza Other Respir Viruses. 2013;7(6):979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sajjanar B, Trakooljul N, Wimmers K, Ponsuksili S. DNA methylation analysis of porcine mammary epithelial cells reveals differentially methylated loci associated with immune response against Escherichia coli challenge. BMC Genomics. 2019;20(1):623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karatzas PS, Mantzaris GJ, Safioleas M, Gazouli M. DNA methylation profile of genes involved in inflammation and autoimmunity in inflammatory bowel disease. Medicine (Baltimore). 2014;93(28):e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheong CY, Chng K, Lim MK, et al. Alterations to DNA methylation and expression of CXCL14 are associated with suboptimal birth outcomes. J Hum Genet. 2014;59(9):504–511. [DOI] [PubMed] [Google Scholar]
- 43.Xu N, Barlow GM, Cui J, et al. Comparison of Genome-Wide and Gene-Specific DNA Methylation Profiling in First-Trimester Chorionic Villi From Pregnancies Conceived With Infertility Treatments. Reprod Sci. 2017;24(7):996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mays AC, Feng X, Browne JD, Sullivan CA. Chemokine and Chemokine Receptor Profiles in Metastatic Salivary Adenoid Cystic Carcinoma. Anticancer Res. 2016;36(8):4013–4018. [PubMed] [Google Scholar]
- 45.Gu XL, Ou ZL, Lin FJ, et al. Expression of CXCL14 and its anticancer role in breast cancer. Breast Cancer Res Treat. 2012;135(3):725–735. [DOI] [PubMed] [Google Scholar]
- 46.Ito S, Takahara Y, Hyodo T, et al. The roles of two distinct regions of PINCH-1 in the regulation of cell attachment and spreading. Mol Biol Cell. 2010;21(23):4120–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sjoberg E, Augsten M, Bergh J, Jirstrom K, Ostman A. Expression of the chemokine CXCL14 in the tumour stroma is an independent marker of survival in breast cancer. Br J Cancer. 2016;114(10):1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji X, Shen Z, Zhao B, Yuan X, Zhu X. CXCL14 and NOS1 expression in specimens from patients with stage I-IIIA nonsmall cell lung cancer after curative resection. Medicine (Baltimore). 2018;97(10):e0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams KA, Lee M, Hu Y, et al. A systems genetics approach identifies CXCL14, ITGAX, and LPCAT2 as novel aggressive prostate cancer susceptibility genes. PLoS Genet. 2014;10(11):e1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozawa S, Kato Y, Komori R, Maehata Y, Kubota E, Hata R. BRAK/CXCL14 expression suppresses tumor growth in vivo in human oral carcinoma cells. Biochem Biophys Res Commun. 2006;348(2):406–412. [DOI] [PubMed] [Google Scholar]
- 51.Ozawa S, Kato Y, Kubota E, Hata R. BRAK/CXCL14 expression in oral carcinoma cells completely suppresses tumor cell xenografts in SCID mouse. Biomed Res. 2009;30(5):315–318. [DOI] [PubMed] [Google Scholar]
- 52.Hata R, Izukuri K, Kato Y, et al. Suppressed rate of carcinogenesis and decreases in tumour volume and lung metastasis in CXCL14/BRAK transgenic mice. Sci Rep. 2015;5:9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozawa S, Kato Y, Ito S, et al. Restoration of BRAK / CXCL14 gene expression by gefitinib is associated with antitumor efficacy of the drug in head and neck squamous cell carcinoma. Cancer Sci. 2009;100(11):2202–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shellenberger TD, Wang M, Gujrati M, et al. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Research. 2004;64:8262–8270. [DOI] [PubMed] [Google Scholar]
- 55.Lin K, Zou R, Lin F, Zheng S, Shen X, Xue X. Expression and effect of CXCL14 in colorectal carcinoma. Mol Med Rep. 2014;10(3):1561–1568. [DOI] [PubMed] [Google Scholar]
- 56.Lin Y, Chen BM, Yu XL, Yi HC, Niu JJ, Li SL. Suppressed Expression of CXCL14 in Hepatocellular Carcinoma Tissues and Its Reduction in the Advanced Stage of Chronic HBV Infection. Cancer Manag Res. 2019;11:10435–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyu XJ, Li HZ, Ma X, et al. Elevated S100A6 (Calcyclin) enhances tumorigenesis and suppresses CXCL14-induced apoptosis in clear cell renal cell carcinoma. Oncotarget. 2015;6(9):6656–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Weng X, Wang L, et al. HIC1 deletion promotes breast cancer progression by activating tumor cell/fibroblast crosstalk. J Clin Invest. 2018;128(12):5235–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujii H, Biel MA, Zhou W, Weitzman SA, Baylin SB, Gabrielson E. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 1998;16(16):2159–2164. [DOI] [PubMed] [Google Scholar]
- 60.Zheng J, Wang J, Sun X, et al. HIC1 modulates prostate cancer progression by epigenetic modification. Clin Cancer Res. 2013;19(6):1400–1410. [DOI] [PubMed] [Google Scholar]
- 61.Allinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6(1):17–32. [DOI] [PubMed] [Google Scholar]
- 62.Brake T, Lambert PF. Estrogen contributes to the onset, persistence, and malignant progression of cervical cancer in a human papillomavirus-transgenic mouse model. Proc Natl Acad Sci U S A. 2005;102(7):2490–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chung SH, Franceschi S, Lambert PF. Estrogen and ERalpha: culprits in cervical cancer? Trends Endocrinol Metab. 2010;21(8):504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Son J, Park JW, Lambert PF, Chung SH. Requirement of estrogen receptor alpha DNA-binding domain for HPV oncogene-induced cervical carcinogenesis in mice. Carcinogenesis. 2014;35(2):489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spurgeon ME, den Boon JA, Horswill M, et al. Human papillomavirus oncogenes reprogram the cervical cancer microenvironment independently of and synergistically with estrogen. Proc Natl Acad Sci U S A. 2017;114(43):E9076–E9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rivera LB, Meyronet D, Hervieu V, Frederick MJ, Bergsland E, Bergers G. Intratumoral myeloid cells regulate responsiveness and resistance to antiangiogenic therapy. Cell Rep. 2015;11(4):577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strieter RM, Polverini PJ, Kunkel SL, et al. The functional role of the ELR motif in CXC chemokine-mediated angiogenesis. J Biol Chem. 1995;270(45):27348–27357. [DOI] [PubMed] [Google Scholar]
- 68.Strieter RM, Belperio JA, Phillips RJ, Keane MP. CXC chemokines in angiogenesis of cancer. Semin Cancer Biol. 2004;14(3):195–200. [DOI] [PubMed] [Google Scholar]
- 69.Schaerli P, Willimann K, Ebert LM, Walz A, Moser B. Cutaneous CXCL14 targets blood precursors to epidermal niches for Langerhans cell differentiation. Immunity. 2005;23(3):331–342. [DOI] [PubMed] [Google Scholar]
- 70.Kurth I, Willimann K, Schaerli P, Hunziker T, Clark-Lewis I, Moser B. Monocyte selectivity and tissue localization suggests a role for breast and kidney-expressed chemokine (BRAK) in macrophage development. J Exp Med. 2001;194(6):855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abd Warif NM, Stoitzner P, Leggatt GR, Mattarollo SR, Frazer IH, Hibma MH. Langerhans cell homeostasis and activation is altered in hyperplastic human papillomavirus type 16 E7 expressing epidermis. PLoS One. 2015;10(5):e0127155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Starnes T, Rasila KK, Robertson MJ, et al. The chemokine CXCL14 (BRAK) stimulates activated NK cell migration: implications for the downregulation of CXCL14 in malignancy. Exp Hematol. 2006;34(8):1101–1105. [DOI] [PubMed] [Google Scholar]
- 73.Cao Q, Chen H, Deng Z, et al. Genetic deletion of Cxcl14 in mice alters uterine NK cells. Biochem Biophys Res Commun. 2013;435(4):664–670. [DOI] [PubMed] [Google Scholar]
- 74.Westrich JA, Vermeer DW, Silva A, et al. CXCL14 suppresses human papillomavirus-associated head and neck cancer through antigen-specific CD8(+) T-cell responses by upregulating MHC-I expression. Oncogene. 2019;38(46):7166–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanegashima K, Suzuki K, Nakayama Y, et al. CXCL14 is a natural inhibitor of the CXCL12-CXCR4 signaling axis. FEBS Lett. 2013;587(12):1731–1735. [DOI] [PubMed] [Google Scholar]
- 76.Zeng J, Yang X, Cheng L, et al. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J Transl Med. 2013;11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sato K, Ozawa S, Izukuri K, Kato Y, Hata R. Expression of tumour-suppressing chemokine BRAK/CXCL14 reduces cell migration rate of HSC-3 tongue carcinoma cells and stimulates attachment to collagen and formation of elongated focal adhesions in vitro. Cell Biol Int. 2010;34(5):513–522. [DOI] [PubMed] [Google Scholar]
- 78.Waldemer-Streyer RJ, Reyes-Ordonez A, Kim D, Zhang R, Singh N, Chen J. Cxcl14 depletion accelerates skeletal myogenesis by promoting cell cycle withdrawal. NPJ Regen Med. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burkhardt AM, Tai KP, Flores-Guiterrez JP, et al. CXCL17 is a mucosal chemokine elevated in idiopathic pulmonary fibrosis that exhibits broad antimicrobial activity. J Immunol. 2012;188(12):6399–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu B, Wilson E. The antimicrobial activity of CCL28 is dependent on C-terminal positively-charged amino acids. Eur J Immunol. 2010;40(1):186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hieshima K, Ohtani H, Shibano M, et al. CCL28 has dual roles in mucosal immunity as a chemokine with broad-spectrum antimicrobial activity. J Immunol. 2003;170(3):1452–1461. [DOI] [PubMed] [Google Scholar]
- 82.Yang D, Chen Q, Hoover DM, et al. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J Leukoc Biol. 2003;74(3):448–455. [DOI] [PubMed] [Google Scholar]
- 83.Marshall A, Celentano A, Cirillo N, Mignogna MD, McCullough M, Porter S. Antimicrobial activity and regulation of CXCL9 and CXCL10 in oral keratinocytes. Eur J Oral Sci. 2016;124(5):433–439. [DOI] [PubMed] [Google Scholar]
- 84.Crawford MA, Fisher DJ, Leung LM, et al. CXC Chemokines Exhibit Bactericidal Activity against Multidrug-Resistant Gram-Negative Pathogens. mBio. 2017;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maerki C, Meuter S, Liebi M, et al. Potent and broad-spectrum antimicrobial activity of CXCL14 suggests an immediate role in skin infections. J Immunol. 2009;182(1):507–514. [DOI] [PubMed] [Google Scholar]
- 86.Wolf M, Moser B. Antimicrobial activities of chemokines: not just a side-effect? Front Immunol. 2012;3:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hara T, Tanegashima K. CXCL14 antagonizes the CXCL12-CXCR4 signaling axis. Biomol Concepts. 2014;5(2):167–173. [DOI] [PubMed] [Google Scholar]
- 88.Tanegashima K, Tsuji K, Suzuki K, Shigenaga A, Otaka A, Hara T. Dimeric peptides of the C-terminal region of CXCL14 function as CXCL12 inhibitors. FEBS Lett. 2013;587(23):3770–3775. [DOI] [PubMed] [Google Scholar]
- 89.Nagasawa T, Hirota S, Tachibana K, et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382(6592):635–638. [DOI] [PubMed] [Google Scholar]
- 90.Tachibana K, Hirota S, Iizasa H, et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393(6685):591–594. [DOI] [PubMed] [Google Scholar]
- 91.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393(6685):595–599. [DOI] [PubMed] [Google Scholar]
- 92.McGrath KE, Koniski AD, Maltby KM, McGann JK, Palis J. Embryonic expression and function of the chemokine SDF-1 and its receptor, CXCR4. Dev Biol. 1999;213(2):442–456. [DOI] [PubMed] [Google Scholar]
- 93.Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. [DOI] [PubMed] [Google Scholar]
- 94.Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2(2):123–128. [DOI] [PubMed] [Google Scholar]
- 95.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107(5):1761–1767. [DOI] [PubMed] [Google Scholar]
- 96.Sun Y, Mao X, Fan C, et al. CXCL12-CXCR4 axis promotes the natural selection of breast cancer cell metastasis. Tumour Biol. 2014;35(8):7765–7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang P, Hu Y, Zhou Q. The CXCL12-CXCR4 signaling axis plays a key role in cancer metastasis and is a potential target for developing novel therapeutics against metastatic cancer. Curr Med Chem. 2019. [DOI] [PubMed] [Google Scholar]
- 98.Collins PJ, McCully ML, Martinez-Munoz L, et al. Epithelial chemokine CXCL14 synergizes with CXCL12 via allosteric modulation of CXCR4. FASEB J. 2017;31(7):3084–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rodriguez LR, Emblom-Callahan M, Chhina M, et al. Global Gene Expression Analysis in an in vitro Fibroblast Model of Idiopathic Pulmonary Fibrosis Reveals Potential Role for CXCL14/CXCR4. Sci Rep. 2018;8(1):3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Otte M, Kliewer A, Schutz D, Reimann C, Schulz S, Stumm R. CXCL14 is no direct modulator of CXCR4. FEBS Lett. 2014;588(24):4769–4775. [DOI] [PubMed] [Google Scholar]
- 101.Lattin JE, Schroder K, Su AI, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Franceschetti T, Kessler CB, Lee SK, Delany AM. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J Biol Chem. 2013;288(46):33347–33360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bonecchi R, Locati M, Galliera E, et al. Differential recognition and scavenging of native and truncated macrophage-derived chemokine (macrophage-derived chemokine/CC chemokine ligand 22) by the D6 decoy receptor. J Immunol. 2004;172(8):4972–4976. [DOI] [PubMed] [Google Scholar]
- 104.Graham GJ. D6/Ackr2. Front Immunol. 2015;6:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mabuchi T, Hwang ST. ACKR2: Nature’s Decoy Receptor Lures Unsuspecting Chemokines in Psoriasis. J Invest Dermatol. 2017;137(1):7–11. [DOI] [PubMed] [Google Scholar]
- 106.Nibbs RJ, Kriehuber E, Ponath PD, et al. The beta-chemokine receptor D6 is expressed by lymphatic endothelium and a subset of vascular tumors. Am J Pathol. 2001;158(3):867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bonavita O, Mollica Poeta V, Setten E, Massara M, Bonecchi R. ACKR2: An Atypical Chemokine Receptor Regulating Lymphatic Biology. Front Immunol. 2016;7:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McKimmie CS, Singh MD, Hewit K, et al. An analysis of the function and expression of D6 on lymphatic endothelial cells. Blood. 2013;121(18):3768–3777. [DOI] [PubMed] [Google Scholar]
- 109.Nibbs R, Graham G, Rot A. Chemokines on the move: control by the chemokine “interceptors” Duffy blood group antigen and D6. Semin Immunol. 2003;15(5):287–294. [DOI] [PubMed] [Google Scholar]
- 110.Locati M, Torre YM, Galliera E, et al. Silent chemoattractant receptors: D6 as a decoy and scavenger receptor for inflammatory CC chemokines. Cytokine Growth Factor Rev. 2005;16(6):679–686. [DOI] [PubMed] [Google Scholar]
- 111.Nibbs RJ, Gilchrist DS, King V, et al. The atypical chemokine receptor D6 suppresses the development of chemically induced skin tumors. J Clin Invest. 2007;117(7):1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vetrano S, Borroni EM, Sarukhan A, et al. The lymphatic system controls intestinal inflammation and inflammation-associated Colon Cancer through the chemokine decoy receptor D6. Gut. 2010;59(2):197–206. [DOI] [PubMed] [Google Scholar]
- 113.Savino B, Caronni N, Anselmo A, et al. ERK-dependent downregulation of the atypical chemokine receptor D6 drives tumor aggressiveness in Kaposi sarcoma. Cancer Immunol Res. 2014;2(7):679–689. [DOI] [PubMed] [Google Scholar]
- 114.Bonecchi R, Savino B, Caronni N, Celesti G, Mantovani A, Locati M. Atypical chemokine receptor 2: a brake against Kaposi’s sarcoma aggressiveness. Oncoimmunology. 2014;3(12):e955337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu KD, Wang X, Yang C, Zeng XH, Shao ZM. Host genotype and tumor phenotype of chemokine decoy receptors integrally affect breast cancer relapse. Oncotarget. 2015;6(28):26519–26527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zeng XH, Ou ZL, Yu KD, et al. Absence of multiple atypical chemokine binders (ACBs) and the presence of VEGF and MMP-9 predict axillary lymph node metastasis in early breast carcinomas. Med Oncol. 2014;31(9):145. [DOI] [PubMed] [Google Scholar]
- 117.Yang C, Yu KD, Xu WH, et al. Effect of genetic variants in two chemokine decoy receptor genes, DARC and CCBP2, on metastatic potential of breast cancer. PLoS One. 2013;8(11):e78901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Koren I, Timms RT, Kula T, Xu Q, Li MZ, Elledge SJ. The Eukaryotic Proteome Is Shaped by E3 Ubiquitin Ligases Targeting C-Terminal Degrons. Cell. 2018;173(7):1622–1635 e1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pyeon D, Newton Ma, Lambert PF, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Research. 2007;67:4605–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gildener-Leapman N, Ferris RL, Bauman JE. Promising systemic immunotherapies in head and neck squamous cell carcinoma. Oral Oncol. 2013;49:1089–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. The New England journal of medicine. 2013;369:134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wolchok JD, Kluger H, Callahan MK, et al. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hamid O, Schmidt H, Nissan A, et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med. 2011;9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schoppy DW, Sunwoo JB. Immunotherapy for Head and Neck Squamous Cell Carcinoma. Hematol Oncol Clin North Am. 2015;29(6):1033–1043. [DOI] [PubMed] [Google Scholar]
- 126.Ugurel S, Spassova I, Wohlfarth J, et al. MHC class-I downregulation in PD-1/PD-L1 inhibitor refractory Merkel cell carcinoma and its potential reversal by histone deacetylase inhibition: a case series. Cancer Immunol Immunother. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yoo SH, Keam B, Ock CY, et al. Prognostic value of the association between MHC class I downregulation and PD-L1 upregulation in head and neck squamous cell carcinoma patients. Sci Rep. 2019;9(1):7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Luo N, Nixon MJ, Gonzalez-Ericsson PI, et al. DNA methyltransferase inhibition upregulates MHC-I to potentiate cytotoxic T lymphocyte responses in breast cancer. Nat Commun. 2018;9(1):248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gajewski TF. The Next Hurdle in Cancer Immunotherapy: Overcoming the Non-T-Cell-Inflamed Tumor Microenvironment. Semin Oncol. 2015;42(4):663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Spranger S, Spaapen RM, Zha Y, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5(200):200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Doebar SC, Sieuwerts AM, de Weerd V, Stoop H, Martens JWM, van Deurzen CHM. Gene Expression Differences between Ductal Carcinoma in Situ with and without Progression to Invasive Breast Cancer. Am J Pathol. 2017;187(7):1648–1655. [DOI] [PubMed] [Google Scholar]
- 132.Li J, Liu C, Chen Y, et al. Tumor Characterization in Breast Cancer Identifies Immune-Relevant Gene Signatures Associated With Prognosis. Front Genet. 2019;10:1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li X, Zhong Q, Luo D, Du Q, Liu W. The prognostic value of CXC subfamily ligands in stage I-III patients with colorectal cancer. PLoS One. 2019;14(4):e0214611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van der Horst PH, Wang Y, Vandenput I, et al. Progesterone inhibits epithelial-to-mesenchymal transition in endometrial cancer. PLoS One. 2012;7(1):e30840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fazi B, Proserpio C, Galardi S, et al. The Expression of the Chemokine CXCL14 Correlates with Several Aggressive Aspects of Glioblastoma and Promotes Key Properties of Glioblastoma Cells. Int J Mol Sci. 2019;20(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Izukuri K, Suzuki K, Yajima N, et al. Chemokine CXCL14/BRAK transgenic mice suppress growth of carcinoma cell transplants. [corrected]. Transgenic Res. 2010;19(6):1109–1117. [DOI] [PubMed] [Google Scholar]
- 137.Liang W, Yang C, Peng J, Qian Y, Wang Z. The Expression of HSPD1, SCUBE3, CXCL14 and Its Relations with the Prognosis in Osteosarcoma. Cell Biochem Biophys. 2015;73(3):763–768. [DOI] [PubMed] [Google Scholar]
- 138.Schwarze SR, Luo J, Isaacs WB, Jarrard DF. Modulation of CXCL14 (BRAK) expression in prostate cancer. Prostate. 2005;64(1):67–74. [DOI] [PubMed] [Google Scholar]
- 139.Oler G, Camacho CP, Hojaij FC, Michaluart P, Jr., Riggins GJ, Cerutti JM. Gene expression profiling of papillary thyroid carcinoma identifies transcripts correlated with BRAF mutational status and lymph node metastasis. Clin Cancer Res. 2008;14(15):4735–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- 141.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]


