Abstract
Purpose of Review:
An increasing body of evidence suggests that the gut microbiome influences the pathogenesis of insulin resistance and type 2 diabetes (T2D). In this review, we will discuss the latest findings regarding the mechanisms linking the gut microbiome and microbial metabolites with T2D and therapeutic approaches based on the gut microbiota for the prevention and treatment of T2D.
Recent Findings:
Alterations in the gut microbial composition are associated with the risk of T2D. The gut microbiota can metabolize dietary- and host-derived factors to produce numerous microbial metabolites, which are involved in metabolic processes modulating nutrition and energy harvest, gut barrier function, systemic inflammation, and glucose metabolism.
Summary:
Microbial metabolites are important mediators of microbial-host crosstalk impacting host glucose metabolism. Furthermore, microbiome-based interventions may have beneficial effects on glycemic control. Future research is required to develop personalized T2D therapy based on microbial composition and/or metabolites.
Keywords: Gut microbiome, Microbial metabolites, T2D, Insulin resistance, Short-chain fatty acids, Branched-chain amino acids
Introduction
T2D is a highly prevalent metabolic disorder characterized by insulin resistance and insufficient compensatory circulating insulin. In the US, 13% of adults aged 18 or older have diabetes, of which 90–95% is T2D [1]. Though the prevalence of T2D in children and adolescents is substantially lower (up to 0.03%) [1], T2D is an increasing concern in this age group. The incidence of T2D in individuals aged 10 to 19 increased from 9.0 per 100,000 in 2002–2003 to 13.8 per 100,000 in 2014–2015 [2]. Although genetic factors play a critical role in the etiology of T2D, the remarkable increase in T2D prevalence in recent years has been driven by dramatic changes in non-genetic factors, including diet and physical activity. Recently, mounting evidence supports a critical role of the gut microbiota as a factor (beneficial or harmful) in the development of T2D. The gut microbiota exerts a great variety of functions impacting human physiology, including modulation of host nutrition and energy harvest, vitamin synthesis, fermentation of indigestible carbohydrates, regulation of the intestinal mucosal barrier, development of the host immune system, and protection against pathogens [3].
The human gastrointestinal (GI) tract contains a dynamic community of >1014 microbial cells, which in total is comprised of ~ 1,000 species including bacteria, archaea, viruses, and eukaryotes [4]. The collective genomes of the gut microbiota (metagenome) represent >100-fold more genes than encoded in the human genome, indicating the vast metabolic features provided to the host [5]. The human gut microbiota is composed of 6 main phyla: Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Verrucomicrobia, and Proteobacteria. Over 90% of the bacterial species present in the gut of adults belong to Firmicutes (mainly gram-positive) and Bacteroidetes (gram-negative) [6]. Initially influenced by mode of birth, the gut microbiome is established by age 2–3, resembling an adult microbiome, and remains relatively stable during adulthood [7]; yet it is responsive to dietary and medication alterations [8, 9].
The recent advances in high-throughput metagenomic sequencing (MGS) technologies have increased our knowledge of the symbiotic relationship between the gut microbiome and its host [4]. MGS enables researchers to identify pathogenic mechanisms of the microbiome related to host disease and provides insights into modulation of the gut microbial community for preventative and therapeutic purposes. In this review, we discuss the role of the gut microbiome on the development of T2D through microbial metabolites including short-chain fatty acids (SCFAs), branched-chain amino acids (BCAAs), bile acids, trimethylamine-N-oxide (TMAO), indole derivatives and imidazole propionate (ImP). Therapeutic approaches of gut microbiota manipulation to prevent and treat T2D are also discussed.
Gut microbiota and type 2 diabetes
Disruptions in the balance of gut microbial populations are linked with risk of obesity and T2D [10]. Reduced intestinal bacterial diversity (bacterial species number or richness) is associated with increased insulin resistance, adiposity, lipid levels, and inflammation [11]. The movement of gram-negative bacteria (e.g., Proteobacteria) out of the intestinal lumen and into the tissues of the host precedes the onset of T2D and abdominal adiposity [12]. Gut microbial metabolites can also slow intestinal transit, which may contribute to obesity risk by increasing energy absorption [10].
Several studies (Table 1) have performed large-scale microbiome profiling in T2D cases and controls. In 2010, Larsen et al. conducted one of the first studies in humans comparing the gut microbiota between individuals with T2D and healthy controls; T2D was associated with compositional changes in the gut microbiota at the phylum level using principal component analysis [13]. They found that phylum Firmicutes and class Clostridia were less abundant in the diabetic group compared to the control group, whereas class Betaproteobacteria was more enriched in diabetic persons and positively correlated with plasma glucose level. Many butyrate-producing bacteria in the human colon belong to the Firmicutes phylum and in particular class Clostridia [14, 15]. Thereafter, in two large MGS studies conducted in Chinese and European cohorts respectively, a common observation was that butyrate-producing bacteria (e.g., Roseburia intestinalis and Faecalibacterium prausnitzii), known for their anti-inflammatory properties [16], were less abundant in subjects with T2D [17, 18]. In a combined analysis of these two datasets, depletion of butyrate-producing taxa in T2D remained even after controlling for the microbiome-altering effects of metformin [19]. In another study, an increased level of F. prausnitzii was found to be associated with an anti-diabetic effect observed immediately after gastric bypass surgery, decoupled from significant weight loss [20].
Table 1.
Microbiome-wide studies in T2D and prediabetes
| Ref | Sample size | Method | Selected key findings |
|---|---|---|---|
| Larsen et al. 2010 [13] | 36 men (18 T2D, 18 controls) | qPCR & 16S rRNA seq | Reduced Firmicutes and Clostridia in T2D. Betaproteobacteria enriched in T2D. Bacteroidetes/Firmicutes ratio (and other ratios) correlated positively with glucose levels. Relative enrichment for gram-negative bacteria in T2D. |
| Qin et al. 2012 [17] | 145 and 200 Chinese (two stage) | Shotgun sequencing ~2.6 Gb depth | Moderate dysbiosis in T2D. T2D enriched for opportunistic pathogens. Controls enriched for butyrate-producing bacteria (Clostridiales sp. SS3/4, Roseburia sp, F prausnitzii). Microbiota profile could classify T2D. |
| Karlsson et al. 2013 [18] | 145 European women, age 70 | Shotgun sequencing ~3.1 Gb depth | T2D enriched for four Lactobacillus sp. and depleted for five Clostridium sp. which correlated with glucose and insulin levels. Patterns of taxa distinguished T2D from controls better than BMI or existing diabetes risk scores. |
| Zhang et al. 2013 [22] | 121 Chinese (44 normal, 64 PD, 13 T2D) | 16S rDNA seq | Butyrate producing bacteria (e.g., A. muciniphilia, F. prausnitzii) more abundant in controls. Reduced abundance of Bacteroides and Verrucomicrobiae in prediabetes and T2D. |
| Sato et al. 2014 [93] | 50 Japanese T2D, 50 controls | RT-qPCR | In T2D, Clostridium coccoides group, Atopoium cluster, Prevotella decreased; Lactobacillus (L. reuteri, L. plantarum) increased. Gut bacteria detected in blood more often in T2D than in controls (28% vs 4%). |
| Forslund et al. 2015 [19] | 784 (includes some subjects from prior studies[17, 18] | Shotgun sequencing ~0.7 Gb depth | Controlling for metformin treatment, found a unified signature of depletion of butyrate-producing taxa (e.g., Roseburia sp) in T2D and microbial mediation of therapeutic effects of metformin via SCFA production. Accuracy of gut microbial signature to predict T2D [17, 18] was confounded by metformin use. |
| Egshatyan et al. 2016 [94] | 24 PD, 20 T2D, 48 controls | 16S rRNA seq | Blautia and Serratia genus enriched in T2D. |
| Candela et al. 2016 [95] | 40 T2D, 13 controls | 16S rRNA seq | In T2D, Enterobacteriaceae, Collinsella, Streptococcus, Lactobacillus increased; depletion of SCFA producers (e.g., Bacteroides, Prevotella). |
| Pedersen et al. 2016 [21] | 75 Danish T2D, 277 controls | qPCR & 16s rDNA seq | Insulin-resistant individuals had higher levels of serum BCAAs, associated with enriched BCAA-producing bacteria (Prevotella copri and Bacteroides vulgatus) and reduced potential for BCAA transport into bacterial cells. Butyrate-producing bacteria including A. muciniphila, F. prausnitzii, Firmicutes sp. and Clostridia sp. were negatively correlated with insulin resistance. |
| de la Cuesta-Zuluaga et al. 2017 [96] | 28 with diabetes, 84 controls | 16S rRNA seq | Diabetes without metformin (n=14) had higher Clostridiaceae 02d06 and a distinct OTU of Prevotella and lower Enterococcus casseliflavus than controls. Those on metformin had a different profile. |
| Wang et al. 2017 [97] | 40 Chinese minority ethnic groups (20 Uygurs and 20 Kazaks) | 16S rRNA seq | In Kazaks, Veillonellaceae was highly enriched in T2D, while families Planococcaceae and Coriobacteriaceae were more enriched in normal subjects. In Uygurs, T2D subjects had decreased levels of Erysipelotrichaceae. |
| Org et al. 2017 [98] | 352 PD, 164 controls | 16S rRNA seq | Pre-diabetic subjects had higher abundances of Anaerostipes and lower abundances of OTU from families Ruminococcaceae and Christencenellacea and genus Methanobrevibacter. |
| Sedighi et al. 2017 [99] | 18 T2D, 18 controls | RT-qPCR | Lactobacillus was significantly more enriched in T2D; Bifidobacterium was more frequent in the healthy subjects. |
| Allin et al. 2018 [100] | 134 PD, 134 controls | 16S rRNA seq | In prediabetes, Clostridium genus and A. muciniphilia were decreased and genera Dorea, Sutterella, and Streptococcus were increased. Several butyrate-producing bacteria decreased in prediabetes. |
| Salamon et al. 2018 [101] | 23 T2D, 23 controls | 16S rRNA seq | The ratio of Firmicutes/Bacteroidetes and phylum Verrucomicrobia increased in T2D. Lower relative percentages of SCFA-producing bacteria from genus Roseburia and genus Faecalibacterium were found in T2D. |
| Zhao et al. 2019 [24] | 65 T2D, 35 controls | 16S rRNA seq | In T2D, the abundance of Proteobacteria and the ratio of Firmicutes/Bacteroidetes were higher; the SCFAs, bile acids and lipids were disordered; the abundance of SCFA-producing bacteria (Lachnospiraceae and Ruminococcaceae etc.) increased, while the fecal SCFAs concentrations were decreased. |
PD, prediabetes; OTU, operational taxonomic unit; seq, sequencing; T2D, type 2 diabetes
A study including 277 non-diabetic Danish individuals showed that people with insulin resistance had increased levels of serum BCAAs, resulting from increased production of BCAA and reduced transport into bacterial cells [21]. Prevotella copri and Bacteroides vulgatus were identified as the main species driving the biosynthesis of BCAAs. They also demonstrated that P. copri could induce insulin resistance and elevate circulating levels of BCAAs in mice, suggesting that intestinal microbiota could be an important source of increased levels of BCAAs and play a key role in insulin resistance.
Using 16S ribosomal DNA (rDNA)-based sequencing, Zhang et al. found a lower abundance of Akkermansia muciniphila in individuals with prediabetes and newly diagnosed T2D, indicating that low abundance of this bacteria could be considered as a biomarker of glucose intolerance [22]. A. muciniphila is a human intestinal mucin-degrading bacterium that represents 3–5% of the human gut microbial community. Dao et al. showed an association between high abundance of A. muciniphila and healthier metabolic status in overweight/obese adults [23]. This study also demonstrated that higher abundance of A. muciniphila at baseline was associated with better clinical outcomes including glucose homoeostasis, blood lipids and body composition after calorie restriction. Metformin treatment has also been observed to increase levels of A. muciniphilia, which might mediate some of its metabolic benefits [9].
Recent studies discovered that the Firmicutes/Bacteroidetes ratio was significantly higher in patients with T2D than in healthy controls [24]. In contrast, other studies in Chinese, European and Danish cohorts found a lower Firmicutes/Bacteroidetes ratio in individuals with T2D [17, 18, 21]. The inconsistency between studies is probably due to confounding factors such as different sequencing techniques, study populations, diet and medication use. More controlled studies considering these confounding variables are needed. Variables not related to the disease of interest can considerably affect microbiome results [25].
In summary, the results to date have identified an association between gut microbiota and T2D; some rodent stool transplant studies even suggest causality of the gut microbiome on metabolic traits. Given variation between studies in humans, a common T2D microbiome signature has not emerged; however, several common themes have been observed regarding key microbial-derived metabolites, described below.
Microbial metabolites and type 2 diabetes
Several microbial metabolites are involved in the regulation of host metabolism and gut integrity, making them important links between the gut microbiome and the development of insulin resistance and T2D. Candidates for metabolically beneficial metabolites include SCFA, bile acids, sulfur-containing amino acids, indole derivatives, and vitamins (e.g., folate) while potentially harmful metabolites include BCAA, lipopolysaccharide (LPS), phenol, p-cresol, ammonia, amines, and methane [26]. Below, we highlight several metabolites of interest (Figure 1). Of note, while the balance of literature may suggest a particular metabolite is beneficial or harmful, it is often possible to find papers that come to opposite conclusions, depending on the experimental system used.
Figure 1.
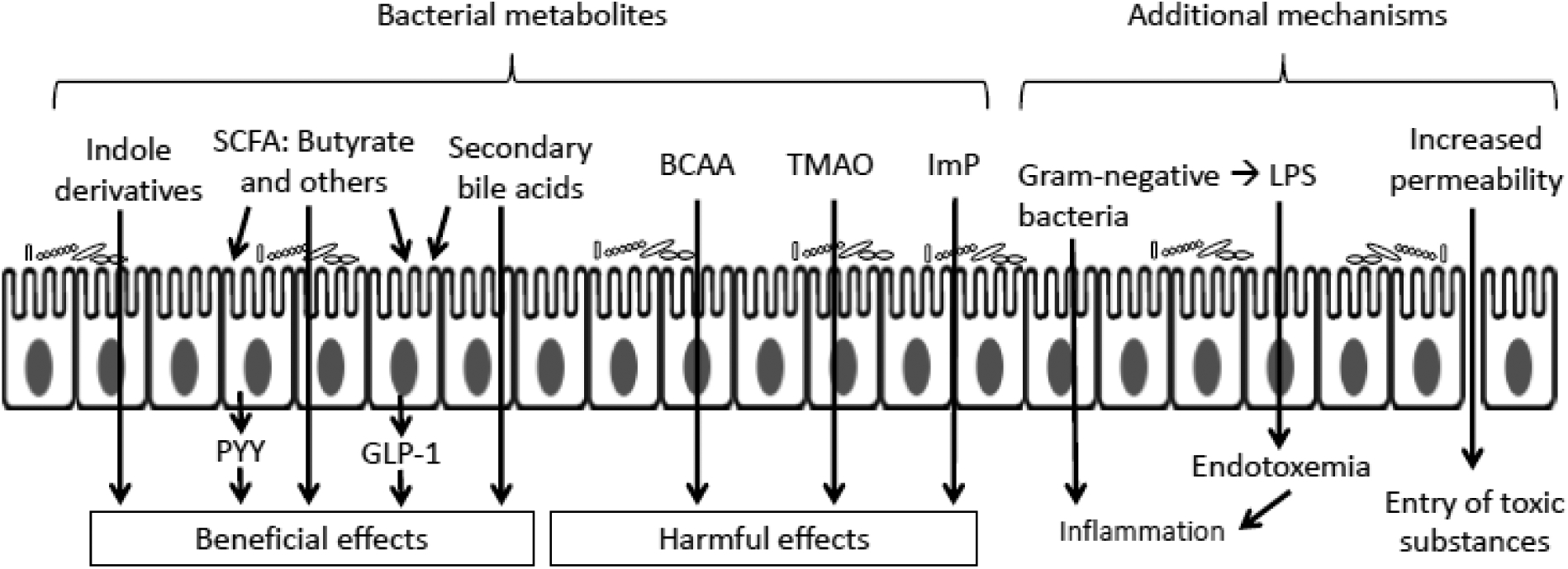
Metabolites linking the gut microbiota and T2D. SCFAs regulate host glucose homeostasis in part by stimulating the secretion of PYY and GLP-1 through binding to the receptors on intestinal epithelial cells. Indole derivatives have beneficial effects on insulin sensitivity. Bile acids may promote GLP-1 secretion and improve insulin sensitivity. BCAA, TMAO and ImP impair host glucose metabolism. In addition, increased gut permeability facilitates endotoxemia, whereby LPS released by the death of gram-negative bacteria crosses the epithelial barrier and enters the circulation, inducing inflammation, which impairs insulin sensitivity. SCFA, short-chain fatty acid; PYY, peptide YY; GLP-1, glucagon-like peptide-1; BCAA, branched-chain amino acid; TMAO, trimethylamine-N-oxide; ImP, imidazole propionate; LPS, lipopolysaccharide.
Short-chain fatty acids
The fermentation of non-digestible carbohydrates in the colon by the microbial community yields SCFAs. Acetate, propionate and butyrate are the most abundant SCFAs produced by this process. Butyrate is locally consumed in the gut as the primary energy source for colonocytes; propionate is utilized in the liver for gluconeogenesis while substantial amounts of acetate are utilized in the peripheral tissues [27]. SCFAs have been extensively reported to improve glucose homeostasis and metabolism in adipose, muscle, and liver [28]. SCFAs bind to G protein-coupled receptors GPR41 (Free Fatty Acid Receptor 3 or FFAR3) and GPR43 (Free Fatty Acid Receptor 2 or FFAR2) [29], which are expressed in a variety of cells including enteroendocrine cells, intestinal epithelial cells and the pancreatic islets [30]. GPR41 activation stimulates the secretion of peptide YY (PYY), which increases satiety [31]. Activation of GPR43 promotes the release of glucagon-like peptide-1 (GLP-1), which improves insulin secretion and inhibits glucagon secretion and also promotes satiety [32]. SCFAs, via GPR41, can also stimulate the production by adipose tissue of leptin, a hormone that regulates long-term food intake and energy expenditure [33]. Kimura et al. found that mice lacking GPR43 exhibited obesity on a normal diet, whereas transgenic mice with adipose-specific overexpression of GPR43 remained lean even when consuming a high-fat diet [34]. They further showed that SCFA-mediated activation of GPR43 suppressed adipose tissue-specific insulin signaling, which inhibited fat accumulation in white adipose tissue and enhanced energy utilization in other tissues, thereby maintaining metabolic homeostasis. The study found that although GPR43 suppressed insulin signaling in adipose tissue, systemic insulin sensitivity was enhanced in mice overexpressing GPR43.
SCFA have insulin sensitizing effects [35], and can improve metabolism by activating intestinal gluconeogenesis [36]. Butyrate inhibits histone deacetylase (HDAC); HDAC inhibition has been shown to promote β-cell development, proliferation, differentiation and function and inhibit apoptosis [37]. SCFAs have been shown to reduce mucosal and chronic systemic inflammation, probably due to suppression of proinflammatory cytokines such as tumor necrosis factor-alpha (TNF-α) and interleukin-6 (IL-6) [38], induction of anti-inflammatory cytokines [39], and reduction of immune cell infiltration into adipose tissue [40]. Butyrate is linked to enhanced gut integrity and intestinal barrier function through upregulation of tight junction protein claudin-1 expression and through redistribution of zonula occludens-1 (ZO-1) and occludin in cellular membranes [41].
Increased acetate production due to an altered gut microbiota in rodents leads to activation of the parasympathetic nervous system, resulting in increased glucose-stimulated insulin secretion and increased ghrelin secretion. This caused a positive feedback loop leading to hyperphagia, increased fat storage, and the associated sequelae of obesity [42]. In mice dietary supplementation with butyrate and propionate can protect against high-fat diet-induced obesity and insulin resistance [43]. Long-term colonic propionate delivery significantly reduced body weight gain and intra-abdominal fat accumulation and prevented deterioration in insulin sensitivity in overweight adults [44]. In a bidirectional Mendelian randomization (MR) study including 952 normoglycemic individuals, Sanna et al. discovered that host genetic variation resulting in increased production of butyrate was associated with improved insulin response (P = 9.8 × 10−5), whereas increased propionate levels were causally related to an increased risk of T2D (P = 0.004) [45]. Recently, Tirosh et al. showed that propionate increased glucagon and the adipokine fatty acid-binding protein 4 (FABP4) production, which stimulated glycogenolysis and hyperglycemia, leading to insulin resistance in mice and humans [46]. In summary, while the balance of literature suggests butyrate has beneficial effects, data are more mixed for propionate and acetate.
Branched-chain amino acids
BCAAs (leucine, isoleucine, and valine), characterized by non-linear aliphatic side-chains, are among the essential amino acids synthesized by gut microbiota. BCAAs have emerged as biomarkers of insulin resistance and predictors of T2D and cardiovascular diseases [47, 48]. Human studies have demonstrated that elevated levels of plasma BCAAs are associated with insulin resistance and an increased risk of T2D [21, 49]. Reduced dietary intake of BCAAs restores metabolic health with improved glucose tolerance and insulin sensitivity in obese mice, even as they continue to be fed a high-fat, high-sugar diet [50]. One mechanism underlying altered BCAA levels is that insulin resistance may cause reduced suppression of proteolysis [51]. Another mechanism may be that impaired adiponectin signaling contributes to reductions in BCAA catabolism in peripheral tissues in T2D [52].
Given the positive correlation between fasting plasma BCAA levels and homeostasis model assessment of insulin resistance (HOMA-IR), investigators conducted a bidirectional MR study to interrogate causality [53]. While the genetic risk score (GRS) for circulating BCAA levels was not associated with HOMA-IR or fasting insulin levels, the GRS for insulin resistance traits was significantly associated with increased plasma BCAA levels. These results suggest that higher BCAAs do not have a causal role on insulin resistance while insulin resistance has a causal effect on higher circulating BCAA levels. While more data are needed to conclusively establish the cause and effect relationships between BCAAs and insulin resistance, the current evidence suggests BCAA as a promising target for risk stratification and possible treatment of obesity and insulin resistance.
Bile acids
Bile acids (cholic acid and chenodeoxycholic acid) are steroid molecules produced in hepatocytes from cholesterol and then further processed into secondary bile acids by the intestinal microbiota, facilitating absorption and transport of dietary lipids. Taurocholic acid, glycocholic acid, taurochenodeoxycholic acid and glycochenodeoxycholic acid are the major secondary bile acids in humans [54]. Microbiota can shape the bile acid pool; bile acids, in turn, can also regulate the gut microbiota composition due to their antimicrobial activity [55]. By binding to the nuclear farnesoid X receptor (FXR) and G protein-coupled bile acid receptor-1 (Takeda G protein-coupled receptor-5 or TGR5), bile acids (e.g., lithocholic acid) can stimulate secretion of the incretin hormone GLP-1 by intestinal L cells, thus regulating glucose metabolism and improving insulin sensitivity [56, 57]. Bile acids also decrease hepatic gluconeogenesis, promote glycogen synthesis, increase energy expenditure, stimulate insulin secretion, and attenuate inflammation [57]. Administration of oral vancomycin significantly reduced gut microbiota diversity in male subjects with metabolic syndrome, with a decrease in gram-positive bacteria from the Firmicutes phylum and an increase in gram-negative bacteria from the Proteobacteria phylum [8]. Vancomycin decreased plasma secondary bile acids deoxycholic acid, lithocholic acid and iso-lithocholic acid but increased the primary bile acids cholic acid and chenodeoxycholic acid. Moreover, administration of vancomycin decreased peripheral insulin sensitivity, with reduction in fecal secondary bile acids correlating with reduction in insulin sensitivity. Given the generally beneficial effects of bile acids described above, it seems paradoxical that treatment of diabetic patients with the bile acid sequestrant colesevelam leads to improved glycemic control [58]. By interrupting enterohepatic recirculation and depleting the bile acid pool, colesevelam results in upregulation of cholesterol 7-α-hydroxylase, increasing the conversion of cholesterol to bile acids, which leads to enhanced expression and activity of HMG-CoA reductase and increased expression of LDL-cholesterol receptors on hepatocytes [59]. Given that reduced HMG-CoA reductase activity has been causally linked to dysglycemia [60], we propose that the effect of colesevelam to increase HMG-CoA reductase activity is responsible for its glucose lowering effect (rather than a mechanism involving bile acids).
Trimethylamine N-oxide
Trimethylamine (TMA) is an amine synthesized exclusively by the gut microbiota from dietary nutrients including choline, betaine, lecithin, and carnitine, which are enriched in red meat and liver and other animal products. TMA is subsequently oxidized in the liver by flavin-containing monooxygenase 3 (FMO3) into TMAO [61]. A recent dietary intervention study found that chronic red meat intake significantly increased systemic TMAO levels through enhanced production and reduced elimination of TMAO, compared to isocaloric white meat and non-meat diets [62]. Higher plasma TMAO was associated with an increased risk of newly diagnosed T2D in a case-control study of 2,694 participants [63]. In a recent MR study with 149,821 subjects by Jia et al., TMAO did not have a causal effect on T2D, whereas T2D was causally associated with increased TMAO levels [64]. Dietary TMAO increased fasting insulin levels and HOMA-IR and exacerbated impaired glucose tolerance in high fat diet-fed mice by interfering with the hepatic insulin signaling pathway and inducing inflammation in adipose tissue [65]. Silencing FMO3 in mice decreased circulating TMAO levels as well as glucose and insulin, whereas FMO3 overexpression in a human hepatoma cell line resulted in increased glucose secretion and insulin resistance [66]. In the POUNDS Lost trial, reduction in TMAO, choline and L-carnitine due to dietary changes were associated with improved insulin sensitivity [67].
Indole derivatives
Indole is a signaling molecule produced from the essential amino acid tryptophan by the bacterial enzyme tryptophanase [68]. Indole influenced GLP-1 secretion by intestinal L-cells in vitro, with effects differing based on duration of exposure [69]. Indolepropionic acid, a microbial metabolite from tryptophan, was associated with lower risk of developing T2D and better insulin sensitivity and negatively correlated with low-grade inflammation [70]. The beneficial effects of indolepropionic acid might relate to the interplay between dietary fiber intake and inflammation or by a direct effect on β-cell function [71]. Indole-3-acetic acid (IAA) was shown to ameliorate insulin resistance, lipid dysmetabolism, oxidative stress and inflammation, protecting against liver injury in mice fed with high-fat diet [72].
Imidazole propionate
ImP is a metabolite produced from histidine by the gut microbiota. Koh et al. recently demonstrated that the concentrations of ImP were higher in subjects with versus without T2D [73]. They showed that the fecal microbiota from patients with T2D had increased capacity of producing ImP in an in vitro gut simulator (a stabilized fermenter with microbial community in an anaerobic and reducing environment mimicking that of the human gut). Administration of ImP impaired glucose tolerance in mice. They further showed that ImP inhibited insulin signaling at the level of insulin receptor substrates (IRS, both IRS1 and IRS2) through activation of a signaling pathway involving mechanistic target of rapamycin complex 1 (mTORC1). Their findings indicated that the microbial metabolite ImP may play a role in the pathogenesis of T2D.
Microbiome-based therapeutic applications in T2D
Fecal microbiota transplant
Fecal microbiota transplant (FMT) is the process of transferring the fecal microbes of a healthy donor into a recipient. FMT has been effective in the treatment of recurring Clostridium difficile infection. Recently, its potential in treating other diseases including metabolic disorders has been explored. Vrieze et al. demonstrated that FMT from lean donors to adult males with metabolic syndrome resulted in improvements in peripheral insulin sensitivity and an increase in butyrate-producing bacteria in the recipient’s fecal microbiota [16]. The beneficial effects of lean donor FMT may be influenced by the baseline fecal microbiota composition (diversity) of the recipients [74]. Current evidence for FMT as a therapeutic tool to improve insulin sensitivity is limited by small sample sizes and absence of data on glycemic control; hence, more studies will be required to explore the effects and potential risks of such treatment and to eliminate the threat of transplantation of pathogenic bacteria.
Dietary interventions
The richness and composition of the gut microbial community can be altered by diet. An animal-based diet was shown to decrease the abundance of Firmicutes that metabolize plant polysaccharides, leading to reduced production of beneficial SCFAs [75]. A recent meta-analysis reported that dietary interventions modulated the gut microbiota and improved glucose control, as represented by HbA1c, while no improvement in fasting blood glucose, fasting insulin, or HOMA-IR was found [76]. In another clinical study, high dietary fiber intake selected promoted a group of SCFA-producing bacteria and improved HbA1c levels, partly via increased GLP-1 production. Promotion of these SCFA producers also decreased producers of detrimental metabolites such as indole and hydrogen sulfide [77].
Inulin (naturally occurring soluble dietary fiber) treatment was found to reduce fasting blood glucose and decrease insulin resistance associated with increased serum GLP-1 level in diabetic rats [78]. In these rats, treatment elevated levels of Lactobacillus and SCFA-producing bacteria Lachnospiraceae, Phascolarctobacterium, and Bacteroides, whereas the abundance of Desulfovibrio, which produces LPS, was decreased. A human study showed that while treatment with only inulin did not reduce fasting blood glucose, administration of inulin with sodium butyrate significantly lowered fasting blood glucose [79]. A recent meta-analysis suggested that inulin-type carbohydrate intervention might reduce fasting plasma glucose, fasting insulin, HbA1c and HOMA-IR, with little effect on BMI [80].
The response to dietary intervention can vary substantially among individuals due to interindividual differences in gut microbiota composition [81]. Analysis of the gut microbial composition can be used to identify individuals who would benefit from dietary interventions and to customize personalized dietary interventions according to bacterial composition. Synergistic therapies involving both bacteria and prebiotics may also be a promising future approach for the prevention and treatment of T2D.
Probiotics
Probiotics are live microorganisms that confer health benefits on the host when administered in proper amounts. Lactobacillus paracasei, L. rhamnosus and Bifidobacterium animalis attenuated weight gain and significantly improved glucose-insulin homeostasis and hepatic steatosis when individually administered to high fat diet-fed mice [82]. Furthermore, statistical analysis of fecal bacterial 16S rRNA genes showed that the supplemented strains shifted the overall microbiota structure of the high fat diet-fed mice toward that of lean mice fed a normal diet. Oral administration of Lactobacillus casei Shirota suppressed plasma levels of lipopolysaccharide-binding protein (LBP, a marker of endotoxemia) and improved insulin resistance in diet-induced obese mice [83]. L. casei Zhang administration improved glucose tolerance in high fructose-induced hyperinsulinemic rats [84]. Dietary supplementation with Bifidobacterium pseudocatenulatum CECT 7765 was found to reduce serum triglyceride, hepatic fat, cholesterol and glucose levels and improve insulin sensitivity in obese mice [85]. Administration of A. muciniphila reversed high fat diet-induced metabolic disorders, restored gut barrier function and reduced inflammation in mice [86]. Another study showed that administration of a purified membrane protein from A. muciniphila or the pasteurized bacteria reduced fat mass and improved insulin resistance and dyslipidemia in obese and diabetic mice [87].
In contrast, results from human studies are less clear. A meta-analysis has shown that probiotics reduce glucose, HbA1c, insulin and HOMA-IR in participants with diabetes, but not in participants with other associated risk factors [88]. In a randomized controlled trial (RCT), supplementation with L. acidophilus La5 and B. animalis subsp lactis Bb12 for 6 weeks did not affect glycemic control in overweight subjects [89]. In another RCT, supplementation with L. reuteri DSM 17938 for 12 weeks did not affect HbA1c, liver steatosis, or adiposity in patients with T2D. However, it did improve insulin sensitivity in a subset of participants likely due to high diversity of their gut microbiota at baseline [90].
Apart from naturally occurring bacteria, genetically modified strains have also been developed to promote glycemic control. Oral administration of a recombinant Lactococcus lactis strain expressing GLP-1 resulted in increased insulin secretion and improved glucose tolerance in rats with T2D [91]. Engineering bacterial vectors with customized gene products may be a useful approach in future T2D therapy.
Conclusion
Available evidence suggests that the gut microbiota plays an important role in the development of insulin resistance and T2D. Gut microbes affect host glucose metabolism through microbial metabolites, which are involved in diverse metabolic pathways. The gut microbial composition is significantly different between patients with T2D and healthy subjects. In many studies, the gut microbiota in subjects with T2D is less abundant in SCFA butyrate-producing bacteria. This may reflect dietary habits given that individuals with T2D tend to consume a high-fat, high-glycemic index, low-fiber dietary pattern compared to healthy individuals [92]. We have highlighted several metabolites of interest. Future efforts are needed to integrate knowledge of these metabolites with the MGS studies, looking for effects on production and clearance of these metabolites. Prospective studies are needed to identify pathogenic metabolites, given that MR studies have suggested that some metabolite alternations are consequences of, rather than causes of, insulin resistance and T2D [64, 53].
Microbiota manipulation including FMT, dietary interventions and probiotic administration in experimental and clinical studies has led to beneficial effects on glucose homeostasis. However, there are still several issues to consider before we can apply these approaches to clinical practice in T2D. First, most of the positive metabolic effects were transient. Repeated treatments would be needed to maintain the effect, and the long-term efficacy of these treatments still needs to be assessed. Second, there are potential risks of introducing pathogenic bacteria when using the FMT approach. Further research needs to be conducted to eliminate this risk. Third, it has been shown that responses to interventions are different among individuals due to different host microbial composition. Therefore, customized therapy based on personal microbiota profile may be an effective treatment approach for T2D in the future.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Human and animal rights: All reported studies/experiments with human or animal subjects performed by the authors have been previously published and complied with all applicable ethical standards (including the Helsinki declaration and its amendments, institutional/national research committee standards, and international/national/institutional guidelines).
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Centers for Disease Control and Prevention. National Diabetes Statistics Report, 2020. Atlanta, GA: Centers for Disease Control and Prevention, U.S. Dept of Health and Human Services; 2020. [Google Scholar]
- 2.Divers J, Mayer-Davis EJ, Lawrence JM, Isom S, Dabelea D, Dolan L, et al. Trends in Incidence of type 1 and type 2 diabetes among youths - selected counties and Indian reservations, United States, 2002–2015. MMWR Morb Mortal Wkly Rep. 2020;69(6):161–5. doi: 10.15585/mmwr.mm6906a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions Science. 2012;336(6086):1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 4.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome Nature. 2012;486(7402):207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60(4):824–31. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 9.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. Population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63(5):727–35. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 10.Moreno-Indias I, Cardona F, Tinahones FJ, Queipo-Ortuno MI. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–6. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 12.Amar J, Serino M, Lange C, Chabo C, Iacovoni J, Mondot S, et al. Involvement of tissue bacteria in the onset of diabetes in humans: evidence for a concept. Diabetologia. 2011;54(12):3055–61. doi: 10.1007/s00125-011-2329-8. [DOI] [PubMed] [Google Scholar]
- 13.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5(2):e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 15.Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7(5):949–61. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–6 e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 17.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 19.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–6. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59(12):3049–57. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.••.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–81. doi: 10.1038/nature18646. This study found that human insulin resistance was associated with increased serum BCAA levels, with the association mainly driven by Prevotella copri and Bacteroides vulgatus. Their experiment in mice suggested that microbial interventions may have the potential to improve insulin resistance and thus reduce the risk of T2D and cardiovascular disease.
- 22.Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8(8):e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65(3):426–36. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 24.Zhao L, Lou H, Peng Y, Chen S, Zhang Y, Li X. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine. 2019. doi: 10.1007/s12020-019-02103-8. [DOI] [PubMed] [Google Scholar]
- 25.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–4. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 26.Khan MT, Nieuwdorp M, Backhed F. Microbial modulation of insulin sensitivity. Cell Metab. 2014;20(5):753–60. doi: 10.1016/j.cmet.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–45. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 28.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11(10):577–91. doi: 10.1038/nrendo.2015.128. [DOI] [PubMed] [Google Scholar]
- 29.Stoddart LA, Smith NJ, Milligan G. International Union of Pharmacology. LXXI. free fatty acid receptors FFA1,−2, and−3: pharmacology and pathophysiological functions. Pharmacological Reviews. 2008;60(4):405–17. doi: 10.1124/pr.108.00802. [DOI] [PubMed] [Google Scholar]
- 30.Priyadarshini M, Navarro G, Layden BT. Gut microbiota: FFAR reaching effects on islets. Endocrinology. 2018;159(6):2495–505. doi: 10.1210/en.2018-00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larraufie P, Martin-Gallausiaux C, Lapaque N, Dore J, Gribble FM, Reimann F, et al. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci Rep. 2018;8(1):74. doi: 10.1038/s41598-017-18259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61(2):364–71. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong Y, Miyamoto N, Shibata K, Valasek MA, Motoike T, Kedzierski RM, et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci U S A. 2004;101(4):1045–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puddu A, Sanguineti R, Montecucco F, Viviani GL. Evidence for the gut microbiota short-chain fatty acids as key pathophysiological molecules improving diabetes. Mediators Inflamm. 2014;2014:162021. doi: 10.1155/2014/162021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1–2):84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 37.Khan S, Jena G. The role of butyrate, a histone deacetylase inhibitor in diabetes mellitus: experimental evidence for therapeutic intervention. Epigenomics. 2015;7(4):669–80. doi: 10.2217/epi.15.20. [DOI] [PubMed] [Google Scholar]
- 38.Roelofsen H, Priebe MG, Vonk RJ. The interaction of short-chain fatty acids with adipose tissue: relevance for prevention of type 2 diabetes. Benef Microbes. 2010;1(4):433–7. doi: 10.3920/BM2010.0028. [DOI] [PubMed] [Google Scholar]
- 39.Saemann MD, Bohmig GA, Osterreicher CH, Burtscher H, Parolini O, Diakos C, et al. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000;14(15):2380–2. doi: 10.1096/fj.00-0359fje. [DOI] [PubMed] [Google Scholar]
- 40.Meijer K, de Vos P, Priebe MG. Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Current Opinion in Clinical Nutrition and Metabolic Care. 2010;13(6):715–21. doi: 10.1097/MCO.0b013e32833eebe5. [DOI] [PubMed] [Google Scholar]
- 41.Wang HB, Wang PY, Wang X, Wan YL, Liu YC. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig Dis Sci. 2012;57(12):3126–35. doi: 10.1007/s10620-012-2259-4. [DOI] [PubMed] [Google Scholar]
- 42.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534(7606):213–7. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin HV, Frassetto A, Kowalik EJ Jr., Nawrocki AR, Lu MM, Kosinski JR, et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;7(4):e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SE, et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64(11):1744–54. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.••.Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nature Genetics. 2019;51(4):600–5. doi: 10.1038/s41588-019-0350-x. This bidirectional MR study supports a causal effect of the gut microbiome on metabolic traits. They found that host-genetics predicted increase in gut production of butyrate was associated with improved insulin sensitivity, whereas increased propionate levels were causally related to an increased risk of T2D.
- 46.Tirosh A, Calay ES, Tuncman G, Claiborn KC, Inouye KE, Eguchi K, et al. The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci Transl Med. 2019;11(489). doi: 10.1126/scitranslmed.aav0120. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Canela M, Guasch-Ferre M, Toledo E, Clish CB, Razquin C, Liang LM, et al. Plasma branched chain/aromatic amino acids, enriched Mediterranean diet and risk of type 2 diabetes: case-cohort study within the PREDIMED Trial. Diabetologia. 2018;61(7):1560–71. doi: 10.1007/s00125-018-4611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tobias DK, Lawler PR, Harada PH, Demler OV, Ridker PM, Manson JE, et al. Circulating branched-chain amino acids and incident cardiovascular disease in a prospective cohort of US women. Circ Genom Precis Med. 2018;11(4):e002157. doi: 10.1161/CIRCGEN.118.002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flores-Guerrero JL, Oste MCJ, Kieneker LM, Gruppen EG, Wolak-Dinsmore J, Otvos JD, et al. Plasma branched-chain amino acids and risk of incident type 2 diabetes: results from the PREVEND prospective cohort study. J Clin Med. 2018;7(12). doi: 10.3390/jcm7120513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cummings NE, Williams EM, Kasza I, Konon EN, Schaid MD, Schmidt BA, et al. Restoration of metabolic health by decreased consumption of branched-chain amino acids. J Physiol. 2018;596(4):623–45. doi: 10.1113/JP275075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giesbertz P, Daniel H. Branched-chain amino acids as biomarkers in diabetes. Curr Opin Clin Nutr Metab Care. 2016;19(1):48–54. [DOI] [PubMed] [Google Scholar]
- 52.Lian K, Du C, Liu Y, Zhu D, Yan W, Zhang H, et al. Impaired adiponectin signaling contributes to disturbed catabolism of branched-chain amino acids in diabetic mice. Diabetes. 2015;64(1):49–59. doi: 10.2337/db14-0312. [DOI] [PubMed] [Google Scholar]
- 53.Mahendran Y, Jonsson A, Have CT, Allin KH, Witte DR, Jorgensen ME, et al. Genetic evidence of a causal effect of insulin resistance on branched-chain amino acid levels. Diabetologia. 2017;60(5):873–8.doi: 10.1007/s00125-017-4222-6. [DOI] [PubMed] [Google Scholar]
- 54.Hofmann AF. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med. 1999;159(22):2647–58. doi: 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- 55.Fiorucci S, Distrutti E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med. 2015;21(11):702–14. doi: 10.1016/j.molmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Pathak P, Xie C, Nichols RG, Ferrell JM, Boehme S, Krausz KW, et al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology. 2018;68(4):1574–88. doi: 10.1002/hep.29857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shapiro H, Kolodziejczyk AA, Halstuch D, Elinav E. Bile acids in glucose metabolism in health and disease. J Exp Med. 2018;215(2):383–96. doi: 10.1084/jem.20171965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen M, Sonne DP, Mikkelsen KH, Gluud LL, Vilsboll T, Knop FK. Bile acid sequestrants for glycemic control in patients with type 2 diabetes: A systematic review with meta-analysis of randomized controlled trials. Journal of Diabetes and Its Complications. 2017;31(5):918–27. doi: 10.1016/j.jdiacomp.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Melian EB, Plosker GL. Colesevelam. Am J Cardiovasc Drugs. 2001;1(2):141–6; discussion 7–8. doi: 10.2165/00129784-200101020-00007. [DOI] [PubMed] [Google Scholar]
- 60.Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JE, Shah T, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015;385(9965):351–61. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17(1):49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Bergeron N, Levison BS, Li XS, Chiu S, Jia X, et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur Heart J. 2019;40(7):583–94. doi: 10.1093/eurheartj/ehy799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shan ZL, Sun TP, Huang H, Chen SJ, Chen LK, Luo C, et al. Association between microbiota-dependent metabolite trimethylamine-N-oxide and type 2 diabetes. American Journal of Clinical Nutrition. 2017;106(3):888–94. doi: 10.3945/ajcn.117.157107. [DOI] [PubMed] [Google Scholar]
- 64.Jia J, Dou P, Gao M, Kong X, Li C, Liu Z, et al. Assessment of causal direction between gut microbiota-dependent metabolites and cardiometabolic health: a bidirectional Mendelian randomization analysis. Diabetes. 2019;68(9):1747–55. doi: 10.2337/db19-0153. [DOI] [PubMed] [Google Scholar]
- 65.Gao X, Liu X, Xu J, Xue C, Xue Y, Wang Y. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J Biosci Bioeng. 2014;118(4):476–81. doi: 10.1016/j.jbiosc.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 66.Shih DM, Wang Z, Lee R, Meng Y, Che N, Charugundla S, et al. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J Lipid Res. 2015;56(1):22–37. doi: 10.1194/jlr.M051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heianza Y, Sun D, Li X, DiDonato JA, Bray GA, Sacks FM, et al. Gut microbiota metabolites, amino acid metabolites and improvements in insulin sensitivity and glucose metabolism: the POUNDS Lost trial. Gut. 2019;68(2):263–70. doi: 10.1136/gutjnl-2018-316155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li G, Young KD. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology-Sgm. 2013;159:402–10. doi: 10.1099/mic.0.064139-0. [DOI] [PubMed] [Google Scholar]
- 69.Chimerel C, Emery E, Summers DK, Keyser U, Gribble FM, Reimann F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 2014;9(4):1202–8. doi: 10.1016/j.celrep.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Mello VD, Paananen J, Lindstrom J, Lankinen MA, Shi L, Kuusisto J, et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci Rep. 2017;7:46337. doi: 10.1038/srep46337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tuomainen M, Lindstrom J, Lehtonen M, Auriola S, Pihlajamaki J, Peltonen M, et al. Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals. Nutr Diabetes. 2018;8(1):35. doi: 10.1038/s41387-018-0046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ji Y, Gao Y, Chen H, Yin Y, Zhang W. Indole-3-acetic acid alleviates nonalcoholic fatty liver disease in mice via attenuation of hepatic lipogenesis, and oxidative and inflammatory stress. Nutrients. 2019;11(9). doi: 10.3390/nu11092062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koh A, Molinaro A, Stahlman M, Khan MT, Schmidt C, Manneras-Holm L, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell. 2018;175(4):947–61 e17. doi: 10.1016/j.cell.2018.09.055. [DOI] [PubMed] [Google Scholar]
- 74.Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26(4):611–9. [DOI] [PubMed] [Google Scholar]
- 75.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Houghton D, Hardy T, Stewart C, Errington L, Day CP, Trenell MI, et al. Systematic review assessing the effectiveness of dietary intervention on gut microbiota in adults with type 2 diabetes. Diabetologia. 2018;61(8):1700–11. doi: 10.1007/s00125-018-4632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.••.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151–6. doi: 10.1126/science.aao5774. This study identified a group of acetate- and butyrate-producing bacteria selectively promoted by dietary fibers. Promotion of this group of SCFA producers not only had a beneficial effect on glucose homeostasis but also kept detrimental bacteria at bay. This study presents a potential novel approach for managing T2D by targeted restoration of specific SCFA producers with dietary fibers.
- 78.Zhang Q, Yu H, Xiao X, Hu L, Xin F, Yu X. Inulin-type fructan improves diabetic phenotype and gut microbiota profiles in rats. PeerJ. 2018;6:e4446. doi: 10.7717/peerj.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roshanravan N, Mahdavi R, Alizadeh E, Jafarabadi MA, Hedayati M, Ghavami A, et al. Effect of butyrate and inulin supplementation on glycemic status, lipid profile and glucagon-like peptide 1 level in patients with type 2 diabetes: a randomized double-blind, placebo-controlled trial. Horm Metab Res. 2017;49(11):886–91. doi: 10.1055/s-0043-119089. [DOI] [PubMed] [Google Scholar]
- 80.Rao M, Gao C, Xu L, Jiang L, Zhu J, Chen G, et al. Effect of inulin-type carbohydrates on insulin resistance in patients with type 2 diabetes and obesity: a systematic review and meta-analysis. J Diabetes Res. 2019;2019:5101423. doi: 10.1155/2019/5101423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metabolism. 2015;22(6):971–82. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 82.Wang J, Tang H, Zhang C, Zhao Y, Derrien M, Rocher E, et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015;9(1):1–15. doi: 10.1038/ismej.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Naito E, Yoshida Y, Makino K, Kounoshi Y, Kunihiro S, Takahashi R, et al. Beneficial effect of oral administration of Lactobacillus casei strain Shirota on insulin resistance in diet-induced obesity mice. J Appl Microbiol. 2011;110(3):650–7. doi: 10.1111/j.1365-2672.2010.04922.x. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Wang L, Zhang J, Li Y, He Q, Li H, et al. Probiotic Lactobacillus casei Zhang ameliorates high-fructose-induced impaired glucose tolerance in hyperinsulinemia rats. Eur J Nutr. 2014;53(1):221–32. doi: 10.1007/s00394-013-0519-5. [DOI] [PubMed] [Google Scholar]
- 85.Cano PG, Santacruz A, Trejo FM, Sanz Y. Bifidobacterium CECT 7765 improves metabolic and immunological alterations associated with obesity in high-fat diet-fed mice. Obesity (Silver Spring). 2013;21(11):2310–21. doi: 10.1002/oby.20330. [DOI] [PubMed] [Google Scholar]
- 86.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(22):9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107–13. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 88.Sun J, Buys NJ. Glucose- and glycaemic factor-lowering effects of probiotics on diabetes: a meta-analysis of randomised placebo-controlled trials. Br J Nutr. 2016;115(7):1167–77. doi: 10.1017/S0007114516000076. [DOI] [PubMed] [Google Scholar]
- 89.Ivey KL, Hodgson JM, Kerr DA, Lewis JR, Thompson PL, Prince RL. The effects of probiotic bacteria on glycaemic control in overweight men and women: a randomised controlled trial. Eur J Clin Nutr. 2014;68(4):447–52. doi: 10.1038/ejcn.2013.294. [DOI] [PubMed] [Google Scholar]
- 90.Mobini R, Tremaroli V, Stahlman M, Karlsson F, Levin M, Ljungberg M, et al. Metabolic effects of Lactobacillus reuteri DSM 17938 in people with type 2 diabetes: A randomized controlled trial. Diabetes Obes Metab. 2017;19(4):579–89. doi: 10.1111/dom.12861. [DOI] [PubMed] [Google Scholar]
- 91.Agarwal P, Khatri P, Billack B, Low WK, Shao J. Oral delivery of glucagon like peptide-1 by a recombinant Lactococcus lactis. Pharm Res. 2014;31(12):3404–14. doi: 10.1007/s11095-014-1430-3. [DOI] [PubMed] [Google Scholar]
- 92.Pastorino S, Richards M, Pierce M, Ambrosini GL. A high-fat, high-glycaemic index, low-fibre dietary pattern is prospectively associated with type 2 diabetes in a British birth cohort. Br J Nutr. 2016;115(9):1632–42. doi: 10.1017/s0007114516000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sato J, Kanazawa A, Ikeda F, Yoshihara T, Goto H, Abe H, et al. Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care. 2014;37(8):2343–50. doi: 10.2337/dc13-2817. [DOI] [PubMed] [Google Scholar]
- 94.Egshatyan L, Kashtanova D, Popenko A, Tkacheva O, Tyakht A, Alexeev D, et al. Gut microbiota and diet in patients with different glucose tolerance. Endocr Connect. 2016;5(1):1–9. doi: 10.1530/EC-15-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Candela M, Biagi E, Soverini M, Consolandi C, Quercia S, Severgnini M, et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br J Nutr. 2016;116(1):80–93. doi: 10.1017/S0007114516001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40(1):54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 97.Wang Y, Luo X, Mao X, Tao Y, Ran X, Zhao H, et al. Gut microbiome analysis of type 2 diabetic patients from the Chinese minority ethnic groups the Uygurs and Kazaks. PLoS One. 2017;12(3):e0172774. doi: 10.1371/journal.pone.0172774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ, et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biology. 2017;18(1):70. doi: 10.1186/s13059-017-1194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sedighi M, Razavi S, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Mehrtash A, et al. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb Pathog. 2017;111:362–9. doi: 10.1016/j.micpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 100.•.Allin KH, Tremaroli V, Caesar R, Jensen BAH, Damgaard MTF, Bahl MI, et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia. 2018;61(4):810–20. doi: 10.1007/s00125-018-4550-1. This study found altered gut microbial composition in individuals with prediabetes with a decreased abundance of Clostridium and A. muciniphila. However, the prediabetic phenotype was not reproduced in mice that underwent human fecal microbiota transplantation.
- 101.Salamon D, Sroka-Oleksiak A, Kapusta P, Szopa M, Mrozinska S, Ludwig-Slomczynska AH, et al. Characteristics of gut microbiota in adult patients with type 1 and type 2 diabetes based on nextgeneration sequencing of the 16S rRNA gene fragment. Pol Arch Intern Med. 2018;128(6):336–43. doi: 10.20452/pamw.4246. [DOI] [PubMed] [Google Scholar]


