Artificial intelligence (AI) and machine learning (ML) methods have begun to reveal that complex imaging patterns can provide individualized biomarkers for diagnosis and prognosis. However, AI methods have been challenged by insufficient training, heterogeneity of imaging protocols across hospitals, and lack of generalization to new patient data. These challenges prompted the development of the ReSPOND (Radiomics Signatures for PrecisiON Diagnostics) consortium on glioblastoma (GBM). This collaboration of over 10 institutions, across 3 continents, is positioned to pool, harmonize, and analyze brain MRIs from more than 3300 de novo GBM patients who underwent the Stupp protocol, in addition to datasets from The Cancer Imaging Archive (TCIA).1 ReSPOND aims to further develop and test AI-based biomarkers for individualized prediction and prognostication, by moving from single-institution studies to generalized, well-validated predictive biomarkers in the following 4 areas:
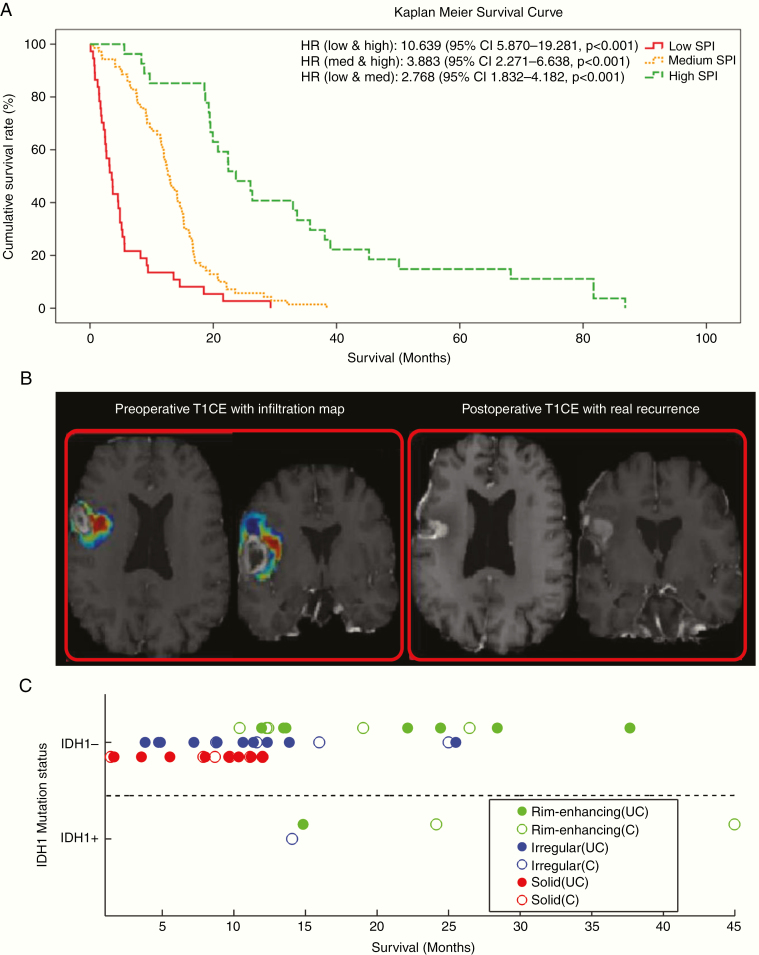
1. Prior work has shown that informative preoperative predictors of patient survival can be constructed using imaging-based ML methods2 (Fig. 1A). These individualized prognostic biomarkers may assist in targeted enrollment and enrichment of clinical trials. They also have the potential to support patient management by identifying poor-prognosis patients who may benefit from early initiation of alternative (to standard) or additional treatments.
Fig. 1.
Examples of ML-derived imaging signatures for diagnosis and prognosis for GBM. (A) Preoperative survival prediction index (SPI): survival curves of 3 subgroups formed according to preoperative prognostic SPI index in de novo GBM patients2 (reprinted with permission from: “Imaging Patterns Predict Patient Survival and Molecular Subtype in Glioblastoma via Machine Learning Techniques,” Neuro-Oncology, 18 (3), 2015, pp. 417–425, https://doi.org/10.1093/neuonc/nov127). (B) ML-based imaging signature of peritumoral infiltration predicts future recurrence; regions with higher probability of recurrence (red), as calculated from preoperative MRIs of de novo GBM patients (left), were more than 10 times more likely to present recurrence at follow-up (right)4 (reprinted with permission from: “Radiomic Signature of Infiltration in Peritumoral Edema Predicts Subsequent Recurrence in Glioblastoma: Implications for Personalized Radiotherapy Planning,” J Medical Imaging, 5(2), 021219 (2018), https://doi.org/10.1117/1.JMI.5.2.02121). (C) Imaging subtype offers predictive value beyond IDH1 mutation status. Three imaging subtypes were identified in preoperative MRIs of de novo GBM patients (marked by green, red, and blue). Among IDH1 non-mutant tumors, one of the GBM subtypes (green) has significantly better prognosis, which was comparable to IDH1-mutant tumors7 (reprinted from: “Radiomic MRI Signature Reveals Three Distinct Subtypes of Glioblastoma with Different Clinical and Molecular Characteristics, Offering Prognostic Value Beyond IDH1,” Sci Rep 8, 5087 (2018) doi:10.1038/s41598-018-22739-2; reuse permitted under Creative Commons license).
2. Prior work using ML-based imaging signatures has shown promise for identifying tumor infiltration beyond the visible margins and into peritumoral edematous (“bright-FLAIR”) tissue3,4 (Fig. 1B). These imaging signatures have been found to identify tissue that is 10 times more likely to present early recurrence, and hence could help establish aggressive, yet targeted, treatments of GBM via peritumoral dose escalation and extensive resection.
3. ML-based imaging signatures have been found to differentiate between treatment-related pseudoprogression and progressive disease.5,6
4. Unsupervised clustering methods applied to rich imaging features have previously identified 3 imaging subtypes of GBM with divergent prognosis and molecular compositions. These imaging subtypes could help refine World Health Organization classifications, as they appear to offer prognostic information complementary to isocitrate dehydrogenase 1 (IDH1) mutation status7 (Fig. 1C)
To support further development, generalization, and clinical translation in these areas, the ReSPOND consortium will use MRIs to conduct rigorously designed biomarker development and validation studies. The following institutions and organizations constitute the founding members of ReSPOND and have agreed to contribute MRI datasets (sample size in parentheses), along with demographic and basic clinical information: University of Pennsylvania (826), University of Pittsburgh (450), Tata Memorial (396), Catalan Institute of Oncology (301), Thomas Jefferson University (300), University Hospitals of Cleveland/Case Western Reserve University (250), Washington University (250), Yonsei University (211), TCIA (135), Henry Ford Hospital (100), Kings College London (100). Of these datasets, approximately 43% will have only conventional MRI (T1, T1-gadolinium, T2, fluid attenuated inversion recovery [FLAIR]), and 57% will have advanced MRI with at least diffusion-weighted imaging or diffusion tensor imaging or dynamic susceptibility contrast‒MRI. To support diagnostic and prognostic marker development, clinical data will also be collected and harmonized. It is expected that approximately 60% of the cases will have O6-methylguanine-DNA methyltransferase methylation status known and 58% will have IDH1 status available. Over 80% of the cases have recurrence information and follow-up scans from the time of first recurrence will be additionally collected. The matched image sets support development of pseudoprogression imaging signatures and refinement of preoperative estimates of patient survival, based on the time and imaging characteristics of first recurrence.
In summary, ReSPOND will develop a large database of harmonized brain MRIs, along with up-to-date clinical annotations, from a diverse pool of GBM patients across multiple institutions. In addition, while we have described the current composition of the consortium, ReSPOND aims to engage the broader community of researchers investigating AI-based tools for GBM, and hence welcomes additional contributors.
Funding
This work was supported in part by NIH R01NS042645, U24CA189523–01A, Penn Center for Precision Medicine (PCPM), Marato Tv3 Project 665/C/2013, ISCIII (Project PI18/01062), Peter D.Cristal Chair, the Kimble Family Fund, the Ferry Family Fund, the Center of Excellence for Translational Neuro-Oncology at UH, the Seidman Cancer Center, NCI/ITCR U01 - U01 CA242871 01, Sidney Kimmel Cancer Center at Jefferson Health, NCI/NIH Award Number P30CA056036, a Challenge Award from the Prostate Cancer Foundation, the Wellcome/Engineering and Physical Sciences Research Council Center for Medical Engineering (WT 203148/Z/16/Z), and Funding for Precision Radiation Therapy through the Abramson Cancer Center.
Conflict of interest statement
No conflicts declared.
References
- 1. Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macyszyn L, Akbari H, Pisapia JM, et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol. 2016;18(3):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akbari H, Macyszyn L, Da X, et al. Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery. 2016;78(4):572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rathore S, Akbari H, Doshi J, et al. Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning. J Med Imaging (Bellingham). 2018;5(2):021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elshafeey N, Kotrotsou A, Giniebra Camejo D, et al. Multicenter Study to Demonstrate Radiomic Texture Features Derived from MR Perfusion Images of Pseudoprogression Compared to True Progression in Glioblastoma Patients. American Society of Clinical Oncology; 2017;35(Supp15):2016. [Google Scholar]
- 6. Prasanna P, Nayate A, Gupta A, et al. Distinguishing radiation necrosis from brain tumor recurrence on routine MRI: a preliminary human-machine reader comparison study. Paper presented at: Neuro-Oncology. 2016;18(Supp 6):vi139–vi140. [Google Scholar]
- 7. Rathore S, Akbari H, Rozycki M, et al. Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1. Sci Rep. 2018;8(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]