Abstract
Background
Pediatric astrocytoma constitutes a majority of malignant pediatric brain tumors. Previous studies that investigated pediatric cancer predisposition have primarily been conducted in tertiary referral centers and focused on cancer predisposition genes. In this study, we investigated the contribution of rare germline variants to risk of malignant pediatric astrocytoma on a population level.
Methods
DNA samples were extracted from neonatal dried bloodspots from 280 pediatric astrocytoma patients (predominantly high grade) born and diagnosed in California and were subjected to whole-exome sequencing. Sequencing data were analyzed using agnostic exome-wide gene-burden testing and variant identification for putatively pathogenic variants in 175 a priori candidate cancer-predisposition genes.
Results
We identified 33 putatively pathogenic germline variants among 31 patients (11.1%) which were located in 24 genes largely involved in DNA repair and cell cycle control. Patients with pediatric glioblastoma were most likely to harbor putatively pathogenic germline variants (14.3%, N = 9/63). Five variants were located in tumor protein 53 (TP53), of which 4 were identified among patients with glioblastoma (6.3%, N = 4/63). The next most frequently mutated gene was neurofibromatosis 1 (NF1), in which putatively pathogenic variants were identified in 4 patients with astrocytoma not otherwise specified. Gene-burden testing also revealed that putatively pathogenic variants in TP53 were significantly associated with pediatric glioblastoma on an exome-wide level (odds ratio, 32.8, P = 8.04 × 10−7).
Conclusion
A considerable fraction of pediatric glioma patients, especially those of higher grade, harbor a putatively pathogenic variant in a cancer predisposition gene. Some of these variants may be clinically actionable or may warrant genetic counseling.
Keywords: pediatric glioma, Li–Fraumeni syndrome, glioblastoma, germline variant, exome sequencing
Key Points.
Putative pathogenic TP53 variants are associated with pediatric glioblastoma on a population level.
A considerable fraction of pediatric glioma patients harbors a putatively pathogenic variant and these patients may potentially be candidates for genetic counseling.
Importance of the Study.
Pediatric astrocytomas constitute a majority of malignant pediatric brain tumors.
Previous studies investigating pediatric cancer predisposition have primarily been conducted in tertiary referral centers and focused on cancer predisposition genes. In this ethnically diverse population study, 33 putatively pathogenic germline variants in cancer predisposition genes were identified among 31 out of 280 astrocytoma patients (11.1%). Five variants were located in TP53, of which 4 were identified among patients with glioblastoma (6.3%, N = 4/63). The next most frequently mutated gene was NF1 (N = 4). Patients with glioblastoma were most likely to harbor putatively pathogenic germline variants (14.3%, N = 9/63). Gene-burden testing revealed that putatively pathogenic variants in TP53 were significantly associated with glioblastoma on an exome-wide level (odds ratio, 32.8, P = 8.04 × 10−7). Therefore, a considerable fraction of pediatric glioma patients, especially those of higher grade, harbor a putatively pathogenic variant in a cancer predisposition gene. Some of these variants may be clinically actionable and or may warrant genetic counseling.
Pediatric brain tumors are the second most common cancer and the leading cause of cancer-related mortality in children.1,2 Pediatric astrocytomas are a heterogeneous group of tumors that comprise approximately 38% of all pediatric brain tumors.3 Mortality and morbidity vary greatly within histopathologic and molecular subtypes.4 For instance, 10-year survival can be as low as 13% for pediatric patients with glioblastoma.3 Pediatric astrocytoma is biologically distinct from adult astrocytoma in terms of both intracranial site and somatic driver genes, as well as clinical behavior.5,6 Although considerable progress has been made in our understanding of the somatic landscape of pediatric astrocytoma, much remains unknown regarding the etiology of these tumors.
Pediatric astrocytomas occur at increased rates in children with one of several cancer predisposition syndromes.7,8 For instance, constitutional mismatch repair deficiency (cMMRD) and Li–Fraumeni syndrome (LFS) have been linked to pediatric glioblastoma incidence.9–11 Similarly, neurofibromatosis type 1 (NF1) can present with optic pathway gliomas and pilocytic astrocytomas.12–16 Previous pan-cancer studies of rare variants have suggested that up to 10% of pediatric patients with glioma may harbor a pathogenic germline predisposition variant, with patients with high-grade glioma most likely to have an underlying genetic predisposition.7,8 These studies, however, are limited by the number of patients with pediatric glioma included, evaluation of only known cancer predisposition genes, lack of comparison with controls, and ascertainment biases when conducted in the setting of family-based pedigree analysis. Furthermore, clinic-based rather than population-based patient recruitment may lead to variant estimates that are not representative of the broader childhood glioma patient population.
To address these issues and explore genomic predisposition to pediatric astrocytoma, we applied an agnostic exome-wide approach to a population-based sample of 280 astrocytoma patients and publicly available controls, in addition to a focused assessment of putatively pathogenic variants in a set of candidate cancer predisposition genes. We also applied the focused analysis of putatively pathogenic variants to a series of 39 patients with exceedingly rare glioma histological pathologies.
Methods
Study Subjects
Patients with pediatric glioma were identified using previously described methods.17 In brief, the California Biobank for neonatal dried bloodspots (DBS) was linked to the California Cancer Registry via the Vital Statistics Registry, which allowed for identification and analysis of DBS of patients with pediatric glioma up to 19 years of age born between 1988 and 2009. The study protocol was approved by the institutional review boards at the California Health and Human Services Agency, University of California (San Francisco and Berkeley), and the University of Southern California. The California Health and Human Services Agency has waived the requirement for informed consent for use of DBS for research.
The third edition of the International Classification of Diseases for Oncology (ICD-O-3 version 0) was used for identification of patients. A total of 3260 DBS from patients with pediatric glioma were identified for potential analysis, of which 280 were selected specifically for the main analyses in this study with the following subtypes: astrocytoma not otherwise specified (NOS) (ICD-O-3: 9400/3, N = 106), anaplastic astrocytoma (ICD-0–3 code: 9400/3, N = 111), and glioblastoma (ICD-0–3 codes: 9440/3 and 9441/3, N = 63). Self-reported race/ethnicity was obtained from the California Cancer Registry. We restricted to patients of self-reported Latino or non-Latino white race/ethnicity to facilitate comparisons with public whole-exome sequencing control datasets. For the purposes of this study, high-grade gliomas were oversampled from those available (but randomly with regard to age, sex, and birth year; Supplementary Table 1). Pilocytic astrocytoma (World Health Organization [WHO] grade I) patients were not included in these analyses.
Additionally, 6 rare subtypes of glioma were selected from the same 3260 DBS to be analyzed separately with the aim to discover rare variants unique to these subtypes: astroblastoma (ICD-O-3: 9430/3, N = 5), desmoplastic infantile astrocytoma (ICD-O-3: 9412/3, N = 6), fibrillary astrocytoma (ICD-O-3: 9420/3, N = 14), gemistocytic astrocytoma (ICD-O-3: 9411/3, N = 5), gliomatosis cerebri (ICD-O-3: 9381/3, N = 4), and protoplasmic astrocytoma (ICD-O-3: 9410/3, N = 5).
Whole-Exome Sequencing and Variant Calling
DNA was extracted from one-third sections of 1.3 cm2 DBS using Qiagen QIAamp DNA Investigator kits and DNA quality and quantity assessed using Nanodrop and Picogreen assays, respectively. Whole-exome sequencing was performed using the Personalis ACE Exome 5GB capture kit. Samples were checked for correct sex, and one sample was removed as the registry-reported sex did not match the genetically determined sex.
Analyses were conducted based on the Genome Analysis Tool Kit best practices guidelines for genetic data preprocessing and germline variant calling.18,19 In summary, the Burrows-Wheeler Aligner 0.7 was used to align the FASTQ files to human reference genome 38 (hg38).20 The Genome Analysis Tool Kit 4.0 was then used to mark duplicates and base quality score recalibration. Variant calling was performed using the HaplotypeCaller command in Genomic Variant Call Format (GVCF) mode and joint variant calling across the entire cohort was subsequently performed using the GenotypeGVCFs command. Variants with a total read depth <8 and genotype quality (GQ) <20, average GQ <35, or missingness >10% were removed.21 Variant Quality Score Recalibration (VQSR) was then applied with a truth sensitivity level of 99.5% for single nucleotide variations and 99.0% for indels, and variants that did not pass VQSR were removed. For variant identification, additional quality metrics including QD (variant call confidence normalized by allele depth) >2, alternative allele read depth >5, and alternative allele fraction >0.2 were subsequently applied for further filtering of spurious variant calls.
Ancestry Ascertainment
A principal component analysis was conducted using fasT and Robust Ancestry Coordinate Estimation (TRACE) 1.03 with the 1000 Genome (1KG) dataset as reference.22,23 The first 4 principal components from the 1KG dataset samples were used to construct a K-nearest neighbor model for prediction of admixed American (Latino), European, African, East Asian, and South Asian ancestry in R 3.6.0 using the caret package.23,24 Eighty percent of the 1KG dataset was used as training data and 20% as test data, which resulted in an accuracy of the model of 99.5%. This model was then applied to the patients with pediatric glioma to ascertain ancestry groups (henceforth referred to as Latino, European ancestry, African American, East Asian, and South Asian; Supplementary Figure 1). The ascertained ancestries were used for gene-burden testing by ethnicity (described below).
Annotation
Annotation was performed using ANNOVAR,25 which incorporated information for Gencode v26,26 Genome Aggregation Database (gnomAD) 2..2 exome allelic frequencies,27 and ClinVar (07–01–2018).28 BCFtools 1.929 was used to annotate for Trans-Omics for Precision Medicine (TOPMed) program Freeze 530 allelic frequencies, CADD scores (version 1.4),31 and mean gnomAD exonic coverage.27 Identified splice site variants were evaluated using Human Splicing Finder 3.1 for their effect on pre-mRNA splicing.32
Gene-Burden Test
We performed exome-wide gene-based burden testing to identify genes mutated at a significantly higher frequency in patients with pediatric glioma compared with publicly available controls (from gnomAD, N = 123 126) using the Test Rare vAriants with Public Data (TRAPD) method.33 The gnomAD database is a large collection of whole-exome and genome sequencing data obtained from African American, Latino, Ashkenazi Jewish, East Asian, Finnish, Non-Finnish European, South Asian, and other ethnicities for which allelic frequencies are available (N = 123 126 for version 2.2).34 Only the 280 samples from patients with astrocytoma NOS, anaplastic astrocytoma, and glioblastomas were included in the gene-burden testing. TRAPD utilizes variant-level summary statistics from publicly available control data for case-control rare variant gene-burden testing, and allows for application of both dominant and recessive models.33 For the gene-burden test, the gnomAD variant call format (VCF) file was converted to hg38 using liftOver and annotated in the same fashion as described above. Only variants with PASS in the Filter field in the gnomAD VCF were used. To ensure a non-inflated gene-burden test, variants were only included if coverage was >10 in at least 90% of samples and QD >5 in both our cohort and gnomAD. Genomic inflation was evaluated by λ Δ95, as previously described.33 Variants were included in the gene-burden test if the gnomAD population maximum allele frequency was ≤0.001 and had a CADD score >20 and/or were annotated as loss-of-function. Genes on sex chromosomes were not evaluated. We also performed separate gene-burden tests stratified by brain tumor subgroup (glioblastoma [N = 63], high-grade gliomas (WHO grades III and IV [N = 187]), low-grade glioma (WHO grade II [N = 93]), and all non-glioblastoma glioma [N = 217]), limiting to loss-of-function variants only. Additional gene-burden tests were stratified by genetically ascertained ancestry (limiting to the largest groups: European ancestry [N = 155] and Latino [N = 122]), which were compared with the Non-Finnish European (N = 55 860) and Latino (N = 16 791) populations, respectively, within gnomAD. A P-value <2.5 × 10−6 was considered exome-wide significant (Bonferroni correction based on approximately 20 000 genes).33 Variants in genes that reached exome-wide significance were visually inspected using Integrative Genomics Viewer version 2.4 and excluded if deemed an artifact.35 Quantile-quantile (q-q) plots were constructed in R (v3.6.0).
Identification of Putatively Pathogenic Variants in Cancer-Predisposition Genes
To identify putatively pathogenic variants, we removed variants with gnomAD exonic or TOPMed allelic frequency >0.0001.36 Furthermore, only exonic variants were included that were annotated as loss-of-function (stop gain, stop loss, or frameshift insertion/deletion) or as “Pathogenic” or “Likely pathogenic” in ClinVar, or deemed to result in alternative splicing using the Human Splicing Finder. In an analysis with less strict filtering criteria, variants were also included if they had a CADD score >20, regardless of functional annotation or ClinVar annotation. Variants that were annotated as “benign” or “likely benign” based on ClinVar annotation were removed from both analyses. We limited variants to those in 162 cancer genes associated with dominant or recessive pediatric cancer predisposition, based on a list used by Gröbner et al,7 as well as 13 additional genes of interest in glioma risk, including: shelterin complex genes previously implicated in familial glioma (ACD, POT1, TERF1, TERF2, TERF2IP, TINF237), genes potentially related to pediatric gliomagenesis (ATRX, DAXX, H3F3A,38IDH139, NOTCH2, and NOTCH2NL40), and a gene identified in family segregating multiple glioma cases (CASP941; Supplementary Table 2). Variants in tumor protein 53 (TP53) were also evaluated for their predicted function in the International Agency for Research on Cancer (IARC) TP53 germline database.42 Compound heterozygosity was evaluated for mismatch repair genes MSH2, MSH6, PMS2, and MLH1 to evaluate presence of cMMRD.43 All putatively pathogenic variants were visually inspected using the Integrative Genomics Viewer.35 Samples with rare glioma subtypes described above were analyzed in the same fashion for discovery of putatively pathogenic variants unique to these subtypes.
Data Availability
This study used biospecimens from the California Biobank Program. Any uploading of genomic data and/or sharing of these biospecimens or individual data derived from these biospecimens has been determined to violate the statutory scheme of the California Health and Safety Code Sections 124980(j), 124991(b), (g), (h), and 103850 (a) and (d), which protect the confidential nature of biospecimens and individual data derived from biospecimens. Certain aggregate results may be available from the authors by request.
Results
Baseline Characteristics
A total of 280 patients with pediatric gliomas were identified, sequenced, and included in the analysis. The mean age of diagnosis was 5.8 years (SD: 4.1) and 134 samples were from females (47.9%). Most self-identified as non-Latino white (N = 175 (62.5%), followed by Latino (N = 105, 37.5%, Table 1). Anaplastic astrocytoma was the most common tumor in our cohort (N = 111, 9.6%), followed by astrocytoma NOS (N = 106, 37.9%). There was a relatively large number of glioblastomas as a result of oversampling (N = 63, 22.5%). The majority of patients had high-grade gliomas, defined as WHO grade III or IV (N = 190, 67.9%). The number of grade IV patients was greater than the number of glioblastomas (N = 64) likely due to diffuse intrinsic pontine gliomas and similar tumors being coded as grade IV prior to 2016. The ascertained ancestry groups based on principal component analysis were discordant with self-reported race/ethnicity in 8.6% of patients (N = 24; Supplementary Table 3, Supplementary Figure 1).
Table 1.
Baseline characteristics of 280 pediatric glioma patients
| Characteristic | Overall, N (%) | |
|---|---|---|
| Total | 280 | |
| Age, y, mean (SD) | 5.83 (4.12) | |
| Sex | Female | 134 (47.9) |
| Male | 146 (52.1) | |
| Ethnicity (self-identified) | Latino | 105 (37.5) |
| Non-Latino white | 175 (62.5) | |
| Pathology (ICD-O-3.0) | Astrocytoma NOS (9400/3) | 106 (37.9) |
| Astrocytoma, anaplastic (9401/3) | 111 (39.6) | |
| Glioblastoma (9440/3, 9441/3) | 63 (22.5) | |
| Location (ICD-O-3.0 topographical location) | Cerebrum (C710-C714) | 127 (45.4) |
| Cerebellum (C716) | 25 (8.9) | |
| Ventricle (C715) | 14 (5.0) | |
| Brainstem (C717) | 61 (21.8) | |
| Overlapping/ unspecified (C718-C719) | 53 (18.9) | |
| Grade (WHO) | I | 0 (.0) |
| II | 90 (32.1) | |
| III | 115 (41.1) | |
| IV | 75 (26.8) |
Agnostic Exome-Wide Gene-Burden Test
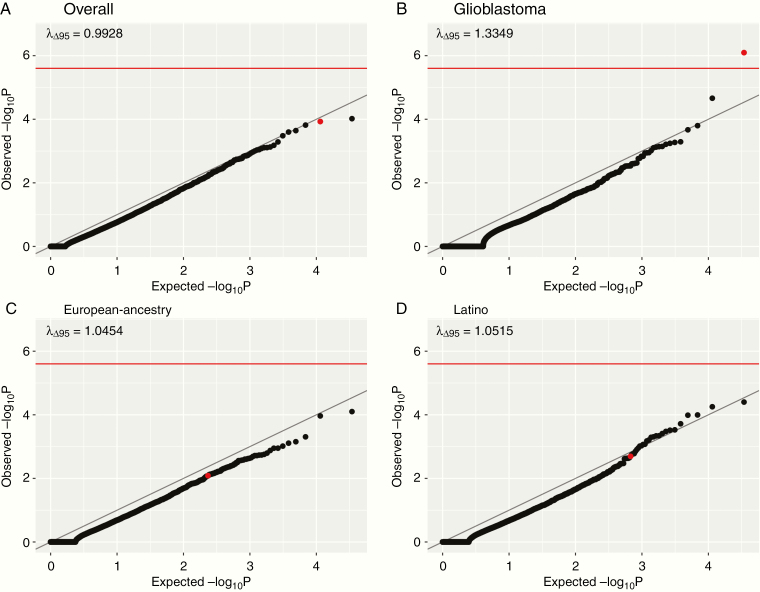
Exome-wide gene-burden tests were carried out to compare the frequency of putatively pathogenic germline variants per gene between the patients with pediatric astrocytomas and gnomAD controls. The analysis across all subjects was not inflated (λ Δ95 = 0.99) but identified no genes associated with glioma on an exome-wide significance level (Figure 1A). When limited to glioblastomas, TP53 was significantly associated at an exome-wide significance level (odds ratio [OR], 32.8, 95% CI: 10.2–81.7, P-value: 8.04 × 10−7; Figure 1B). We did not find any significantly overrepresented genes in subgroup analyses stratified by race/ethnicity (Figure 1C and D), limited to high-grade gliomas (WHO grades III and IV; Supplementary Figure 2A), limited to low-grade gliomas (WHO grade II; Supplementary Figure 2B), with glioblastomas excluded (Supplementary Figure 2C), or with only loss-of-function variants (Supplementary Figure 2D; all P > 2.5 × 10−6). Furthermore, no significant associations were identified using recessive models (all P > 2.5 × 10−6).
Fig. 1.
Q-Q plots for gene-burden test using a dominant model for overall (A), glioblastomas only (B), ascertained European ancestry (C), and ascertained Latinos (D). The red line on the y-axis indicates exome-wide significance at P = 2.5 × 10−6. The λ Δ95 indicates the amount of genomic inflation. TP53 is indicated by red dots.
Putatively Pathogenic Variants in Cancer Predisposition Genes
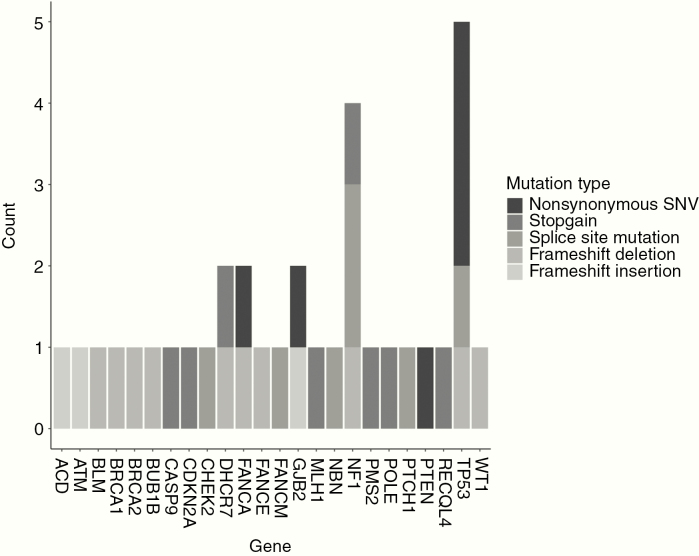
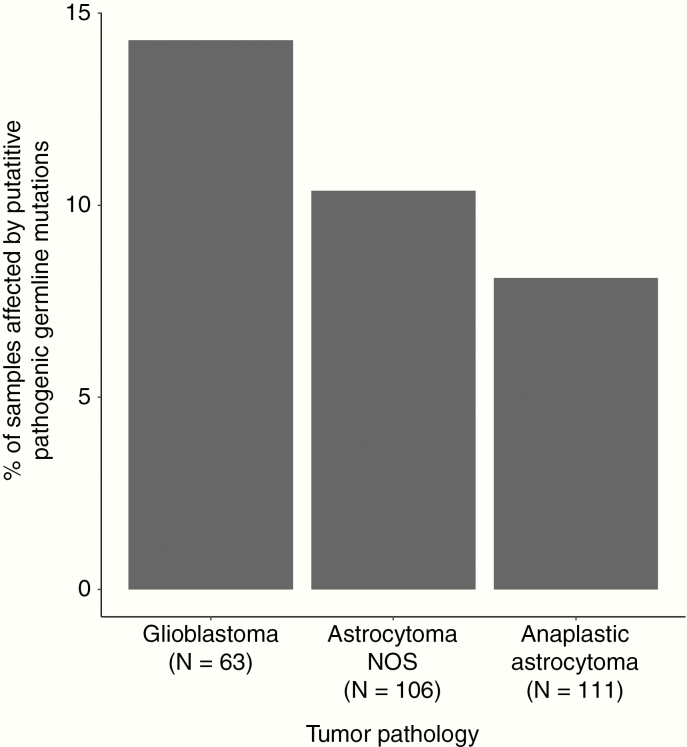
Thirty-three variants in a priori cancer predisposition genes were identified that were either loss-of-function variants or predicted functional splice site variants (Supplementary Table 4) or had a pathogenic/likely pathogenic ClinVar annotation (Figure 2, Table 2). In total, 31 patients (11.1%) harbored one or more likely causal germline variants in a cancer predisposition gene. A higher proportion of patients with glioblastoma (total glioblastoma: 9/63, 14.3%) carried a likely causal germline variant compared with other subtypes (22/217, 10.1%; Figure 3).
Fig. 2.
Prevalence of putatively pathogenic germline variants in cancer predisposition genes by variant type based on strict filtering criteria. Bars are shaded by variant type. Variants were deemed putatively pathogenic if deemed nonsense, frameshift, altering splice site, or annotated as “Pathogenic” or “Likely pathogenic” based on ClinVar and had an allele frequency ≤0.0001 in both TOPMed and gnomAD.
Table 2.
Identified putatively pathogenic germline variants in cancer-predisposition genes in 280 pediatric glioma patients
| Chromosome | Position (hg38) | Ref. | Alt. | Gene | Exonic Predicted Function | ClinVar | Pathology | Ancestry (ascertained) |
|---|---|---|---|---|---|---|---|---|
| 1 | 15518335 | G | A | CASP9 | Stop gain | NA | Astrocytoma NOS | Latino |
| 3 | 37047578 | G | A | MLH1 | Stop gain | Pathogenic | Glioblastoma | Latino |
| 6 | 35459689 | TCAAA | T | FANCE | Frameshift deletion | NA | Anaplastic astrocytoma | European ancestry |
| 7 | 5987468 | T | A | PMS2 | Stop gain | Pathogenic | Glioblastoma | European ancestry |
| 8 | 89971291 | C | G | NBN | Splicing | NA | Glioblastoma | Latino |
| 8 | 144512940 | G | A | RECQL4 | Stop gain | NA | Anaplastic astrocytoma | Latino |
| 9 | 21971149 | G | A | CDKN2A | Stop gain | Uncertain significance | Astrocytoma NOS | European ancestry |
| 9 | 95446392 | C | T | PTCH1 | Splicing | NA | Astrocytoma NOS | European ancestry |
| 10 | 87933130 | G | A | PTEN | Nonsynonymous SNV | Likely pathogenic | Anaplastic astrocytoma | European ancestry |
| 11 | 32435209 | GC | G | WT1 | Frameshift deletion | Likely pathogenic | Anaplastic astrocytoma | European ancestry |
| 11 | 71437905 | C | T | DHCR7 | Stop gain | NA | Anaplastic astrocytoma | European ancestry |
| 11 | 71442257 | GGCTACCTGCAGGAGT CACGGCCCCCTCCTGGAT | G | DHCR7 | Frameshift deletion | Likely pathogenic | Glioblastoma | Latino |
| 11 | 108272812 | C | CATCA | ATM | Frameshift insertion | NA | Astrocytoma NOS | European ancestry |
| 12 | 132635978 | G | A | POLE | Stop gain | Uncertain significance | Astrocytoma NOS | European ancestry |
| 13 | 20189312 | T | TA | GJB2 | Frameshift insertion | Pathogenic/Likely pathogenic | Anaplastic astrocytoma | European ancestry |
| 13 | 20189538 | T | G | GJB2 | Nonsynonymous SNV | Pathogenic/Likely pathogenic | Astrocytoma NOS | European ancestry |
| 13 | 32337521 | CAAAAG | C | BRCA2 | Frameshift deletion | Pathogenic | Astrocytoma NOS | European ancestry |
| 14 | 45189363 | G | T | FANCM | Splicing | NA | Astrocytoma NOS | European ancestry |
| 15 | 40213359 | AC | A | BUB1B | Frameshift deletion | NA | Anaplastic astrocytoma | Latino |
| 15 | 90754838 | CAGAAA | C | BLM | Frameshift deletion | Likely pathogenic | Glioblastoma | European ancestry |
| 16 | 67660397 | T | TCCCGCT GGTCCAC | ACD | Frameshift insertion | NA | Anaplastic astrocytoma | Latino |
| 16 | 89746890 | T | C | FANCA | Nonsynonymous SNV | Pathogenic | Astrocytoma NOS | European ancestry |
| 16 | 89792033 | GCCAA | G | FANCA | Frameshift deletion | Pathogenic | Glioblastoma | Latino |
| 17 | 7670700 | G | A | TP53 | Nonsynonymous SNV | Pathogenic | Glioblastoma | European ancestry |
| 17 | 7673533 | AC | A | TP53 | Splicing | Pathogenic | Glioblastoma | European ancestry |
| 17 | 7673776 | G | A | TP53 | Nonsynonymous SNV | Pathogenic | Anaplastic astrocytoma | Latino |
| 17 | 7676082 | GA | G | TP53 | Frameshift deletion | NA | Glioblastoma | Latino |
| 17 | 7674250* | C | T | TP53 | Nonsynonymous SNV | Conflicting reports in Clinvar | Glioblastoma | Latino |
| 17 | 31159008 | A | G | NF1 | Splicing | NA | Astrocytoma NOS | Latino |
| 17 | 31225251 | G | T | NF1 | Splicing | NA | Astrocytoma NOS | Latino |
| 17 | 31327535 | C | T | NF1 | Stop gain | Pathogenic | Astrocytoma NOS | European ancestry |
| 17 | 31327696 | GT | G | NF1 | Frameshift deletion | NA | Astrocytoma NOS | European ancestry |
| 17 | 43092848 | GTT | G | BRCA1 | Frameshift deletion | Pathogenic | Anaplastic astrocytoma | European ancestry |
| 22 | 28710060 | C | T | CHEK2 | Splicing | Likely pathogenic | Glioblastoma | Latino |
Abbreviations: Alt, alternative allele; NA: not available; NOS: not otherwise specified; Ref: reference allele: *identified through the IARC TP53 Database (http://p53.iarc.fr); SNV, single nucleotide variation.
Fig. 3.
Percentage of pediatric astrocytoma patients affected by putatively pathogenic germline variants by tumor type.
Five pathogenic TP53 variants were identified, of which 4 occurred in patients with glioblastoma (Table 2). Four NF1 variants were identified. Three occurred in patients with a WHO grade II tumor and none were optic pathway gliomas. DHCR7, GJB2, and FANCA variants were each identified in 2 patients. One variant each was discovered in ATM, ACD, BLM, BRCA1, BRCA2, BUB1B, CASP9, CDKN2A, CHEK2, FANCE, FANCM, MLH1, NBN, PMS2, POLE, PTCH1, PTEN, RECQL4, and WT1. One patient with a glioblastoma harbored both a CHECK2 and a DHCR7 variant and 1 patient with an astrocytoma NOS harbored both a GJB2 and an NF1 variant. No patients with cMMRD were identified in analysis of compound heterozygosity.
There was no significant difference in the proportions of ascertained Latino patients (12.3%) versus patients of European ancestry (10.7%) affected by putatively pathogenic germline variants (P = 0.82 respectively [chi-square test]). There was no significant difference in the proportions of male (12.3%) versus female patients affected by a putatively pathogenic germline variants (9.0%, P = 0.47 [chi-square test]). Brainstem (14.8%) and cerebellar (12.0%) locations were not associated with increased proportions of putatively pathogenic germline variants compared with cerebral location (9.45%, P = 0.32 and P = 0.71, respectively [Fisher’s exact test]). High-grade tumors (WHO grades III and IV) were not associated with an increased number of putatively pathogenic variants compared with low-grade tumors (P = 0.64 [chi-square test]). There was no significant difference in age at diagnosis between patients who harbored a putatively pathogenic germline variant (6.8 years) and those who did not (5.7 years, P = 0.13). No differences in age at diagnosis were identified among various pathologies when patients with putatively pathogenic variants were compared with patients with the same diagnosis without a putatively pathogenic germline variant (glioblastoma: P = 0.07; astrocytoma NOS: P = 0.17; anaplastic astrocytoma: P = 0.49, all chi-square test).
Three hundred and eighteen variants were identified in our list of candidate cancer predisposition genes when only CADD scores (CADD >20) were used to identify variants (Supplementary Table 5). The most commonly mutated genes included COL7A1 (N = 14) and POLE (N = 13).
Putatively Pathogenic Variants in Rare Glioma Subtypes
Among 39 patients with rare subtypes of glioma, 5 putatively pathogenic variants were identified (Supplementary Table 6). Three TP53 variants were identified among a patient with a gemistocytic astrocytoma, a patient with a fibrillary astrocytoma, and a patient with desmoplastic infantile astrocytoma. The patient with the fibrillary astrocytoma also harbored a TSC2 variant. One astroblastoma patient harbored a putatively pathogenic RECQL4 variant.
Discussion
In this study, we performed whole-exome sequencing of germline DNA in 280 pediatric patients with astrocytoma to assess the contribution of rare germline variants to disease predisposition. This is the largest investigation of the prevalence of germline variants in pediatric patients with brain tumors to date, and the first such study to use a population-based approach that avoids bias-related case ascertainment at tertiary referral centers. Overall, we found that at least 10% of patients with high-grade pediatric glioma harbored a putatively pathogenic germline variant in a known cancer predisposition gene, similar to previous reports for pediatric glioma in 2 recent pan-cancer sequencing studies of children recruited at diagnosis, by Zhang et al8 (12/137, 8.8%) and Gröbner et al7 (22/223, 9.9%). We did not find differences in frequency of putatively pathogenic variants between ascertained Latinos versus patients from ascertained European ancestry, suggesting that the increased incidence of high-grade pediatric gliomas in patients from European ancestry is likelier explained by common low-penetrance risk alleles, environmental exposures, or a combination of the two.
Our analyses revealed that 14.3% of pediatric glioblastoma diagnoses in this sample are Potentially attributable to high-penetrance germline predisposition. In this patient subgroup, TP53 appears to account for the greatest burden of glioblastoma predisposition, as 6.4% of patients harbored a TP53 variant. We were, however, unable to assess whether carriers of these putatively pathogenic TP53 variants also had a clinical diagnosis of LFS. For lower-grade tumors, NF1 accounted for the greatest burden of cancer predisposition. This study also identified various loss-of-function and splice site variants hitherto undescribed in the evaluated cancer-predisposition genes. In addition, we identified putatively pathogenic variants in various genes, such as GJB2 and FANCA (Figure 2), that have not previously been implicated in pediatric glioma etiology, along with DHCR7, which was recently reported as a possible glioma predisposition allele within Smith-Lemli-Opitz syndrome.44 No cMMRD patients were identified in our cohort, likely due to the low frequency of consanguinity in a large population-based sample of Californians.9,10
Capitalizing on large, publicly available sequencing data from unselected individuals, we performed the first rare variant gene-burden test in a childhood cancer. This revealed significant enrichment of germline TP53 variants in pediatric patients with glioblastoma, conferring ~30-fold risk of disease. Germline variants in TP53 have previously been identified among patients with pediatric glioma through family-based studies of LFS, a cancer predisposition syndrome caused by germline TP53 variants.11,45 Other studies that evaluated large cohorts of pediatric patients with brain tumors identified germline TP53 variants among pediatric patients with higher-grade glioma, similar to our findings.7,8
NF1 germline variants have previously been identified among optic pathway gliomas and, to a lesser extent, pediatric pilocytic astrocytoma, neither of which were included in our study.7,13–16 Likely pathogenic germline NF1 variants have been reported in both lower-grade and higher-grade glioma patients,7,8 although we detected NF1 variants predominantly among patients with low-grade glioma (N = 3/4) and no putatively pathogenic NF1 variants in glioblastoma patients. Similar to previous studies in patients recruited at diagnosis, we identified various putatively pathogenic germline variants in a range of other cancer-predisposition genes. However, it remains unclear how these variants may contribute to risk of pediatric glioma or whether they may cooperate with additional risk factors considering that many controls also harbored rare putatively pathogenic germline variants, including in TP53.46
Currently, a pediatric glioma diagnosis is by itself not a recommendation for genetic counseling, with the exception of optic pathway gliomas because of the strong association with NF1.47 Our study, however, shows that the percentage of pediatric patients with glioblastoma presenting with a germline variant in a cancer predisposition gene is considerable. Despite this, not all variants identified in the various cancer predisposition genes may be clinically actionable. However, 6.4% of pediatric patients with glioblastoma may carry a putatively pathogenic variant in TP53 which could establish an LFS diagnosis and may have consequences for screening and treatment.47,48 With regard to other tumors evaluated in this study, the percentage of actionable germline variants was relatively low and the diagnosis of any of these tumors may not, therefore, form an indication for genetic testing or counseling.
This study has two major strengths: first, the use of population-based patients identified from the California Cancer Registry, which removes potential ascertainment bias; and second, the number of pediatric patients with glioma analyzed is, to our knowledge, the largest reported to date. There are, however, some caveats to this study. For example, we focused analyses on self-identified non-Latino white and Latino children only, who together are over 90% of births in California, and oversampled children with higher-grade tumors. More recent versions of ICD-O-3 coding have incorporated more specific codes for pediatric glioma pathologies, which were not available to us due to the inclusion period of this study. Further, tumor material was not available for investigation of “second hits,” as the DNA samples were derived from the DBS repository and are not linked to specific hospital records or pathology archives. Although we did apply a novel agnostic approach to identify genes associated with genetic predisposition to pediatric glioma, our gene-burden testing has various limitations. It did not result in identification of novel genes, even when ethnicities were evaluated separately, perhaps due to lack of statistical power. Comparison with a public dataset, rather than population-matched controls, may be problematic due to differences in population structure, sequencing methods, and especially differences in depth of sequencing.49 We did, however, endeavor to limit these potential issues by filtering for variants with adequate depth in both gnomAD and our cohort,33 using similar data analysis pipelines, and by evaluating genomic inflation. Moreover, our most significant finding was with TP53 in pediatric glioblastoma, which is unlikely to be a spurious finding and supports the validity of our methodology.
As rare germline variants appear unable to explain the majority of pediatric astrocytomas, future studies should explore other potential risk factors, including non-exonic variation, common variants, and environmental exposures. Future studies can also include more patients and additional ethnicities and study the effect of specific variants with respect to phenotype, function, and associated somatic alterations. Whole-genome sequencing may reveal additional non-coding predisposing germline variants—for example, in enhancer loci, as well as germline structural variants that may underlie pediatric glioma risk.50
In conclusion, this study provides further evidence that putatively pathogenic germline variants in cancer-predisposing genes contribute to the etiology of approximately 10% of pediatric astrocytomas. These variants were found predominantly among patients with high-grade glioma, with the highest burden of germline variants—almost half of which were located in TP53—observed in pediatric glioblastoma patients. Pediatric patients with glioblastoma and their families may be candidates for genetic testing and genetic counseling in certain circumstances such as a positive family history.
Supplementary Material
Acknowledgments
The biospecimens and/or data used in this study were obtained from the California Biobank Program at the California Department of Public Health, SIS request number 311, in accordance with Section 6555(b), 17 CCR. The authors acknowledge Robin Cooley and Steve Graham of the California Department of Public Health for their assistance providing banked specimens and record linkage services for this portion of the study. This study used birth data obtained from the Center for Health Statistics and Informatics, California Department of Public Health. The California Department of Public Health is not responsible for the analyses, results, interpretations, or conclusions drawn by the authors regarding the birth data used in this publication. The collection of cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; Centers for Disease Control and Prevention’s National Program of Cancer Registries, under cooperative agreement 5NU58DP003862–04/DP003862; the National Cancer Institute’s Surveillance, Epidemiology, and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the author(s) and do not reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their contractors and subcontractors. Computation for the work described in this paper was supported by the University of Southern California’s Center for High-Performance Computing (hpcc.usc.edu). Parts of the work described in this manuscript were presented at SNO Pediatric Neuro-Oncology Basic and Translational Research Conference in San Francisco (May 2019).
Funding
This study was supported by R01CA194189 from the NIH.
Conflict of interest statement
The authors report no conflicts of interest.
Authorship statement
Concept of study: ISM, AJD, KMW, CM, XM, JLW. Acquisition of data: HMH, LM, XM, KMW, XM, JLW. Analysis: ISM, CZ, AJD. Draft of manuscript: ISM, AJD, KMW JLW. Careful review of manuscript: AJD, KMW, CM, XM, JLW.
References
- 1. Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992–2004). Cancer. 2008;112(2):416–432. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 3. Ostrom QT, Gittleman H, Liao P, et al. . CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017;19(suppl_5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kline C, Felton E, Allen IE, Tahir P, Mueller S. Survival outcomes in pediatric recurrent high-grade glioma: results of a 20-year systematic review and meta-analysis. J Neurooncol. 2018;137(1):103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jones C, Karajannis MA, Jones DTW, et al. . Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro Oncol. 2017;19(2):153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sturm D, Pfister SM, Jones DTW. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J Clin Oncol. 2017;35(21):2370–2377. [DOI] [PubMed] [Google Scholar]
- 7. Gröbner SN, Worst BC, Weischenfeldt J, et al. ; ICGC PedBrain-Seq Project; ICGC MMML-Seq Project The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321–327. [DOI] [PubMed] [Google Scholar]
- 8. Zhang J, Walsh MF, Wu G, et al. . Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373(24): 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bouffet E, Larouche V, Campbell BB, et al. . Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 10. Shlien A, Campbell BB, de Borja R, et al. ; Biallelic Mismatch Repair Deficiency Consortium Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–262. [DOI] [PubMed] [Google Scholar]
- 11. Bougeard G, Renaux-Petel M, Flaman JM, et al. . Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol. 2015;33(21):2345–2352. [DOI] [PubMed] [Google Scholar]
- 12. Jones DT, Hutter B, Jäger N, et al. ; International Cancer Genome Consortium PedBrain Tumor Project Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Helfferich J, Nijmeijer R, Brouwer OF, et al. . Neurofibromatosis type 1 associated low grade gliomas: a comparison with sporadic low grade gliomas. Crit Rev Oncol Hematol. 2016;104:30–41. [DOI] [PubMed] [Google Scholar]
- 14. Listernick R, Ferner RE, Liu GT, Gutmann DH. Optic pathway gliomas in neurofibromatosis-1: controversies and recommendations. Ann Neurol. 2007;61(3):189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yap YS, McPherson JR, Ong CK, et al. . The NF1 gene revisited—from bench to bedside. Oncotarget. 2014;5(15):5873–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Listernick R, Charrow J, Greenwald M, Mets M. Natural history of optic pathway tumors in children with neurofibromatosis type 1: a longitudinal study. J Pediatr. 1994;125(1):63–66. [DOI] [PubMed] [Google Scholar]
- 17. Wiemels JL, Walsh KM, de Smith AJ, et al. . GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nat Commun. 2018;9(1):286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van der Auwera GA, Carneiro MO, Hartl C, et al. . From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poplin R, Ruano-Rubio V, DePristo MA, et al. . Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2018:201178. [Google Scholar]
- 20. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carson AR, Smith EN, Matsui H, et al. . Effective filtering strategies to improve data quality from population-based whole exome sequencing studies. BMC Bioinformatics. 2014;15:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang C, Zhan X, Liang L, Abecasis GR, Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet. 2015;96(6):926–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;1(5):2008. [Google Scholar]
- 25. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frankish A, Diekhans M, Ferreira AM, et al. . GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Landrum MJ, Lee JM, Riley GR, et al. . ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. BRAVO variant browser: University of Michigan and NHLBI. The NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program 2018; https://bravo.sph.umich.edu/freeze5/hg38/. Accessed December, 12, 2019.
- 31. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF. Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet. 2018;103(4):522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karczewski KJ, Francioli LC, Tiao G, et al. . Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019:531210. [Google Scholar]
- 35. Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kobayashi Y, Yang S, Nykamp K, Garcia J, Lincoln SE, Topper SE. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bainbridge MN, Armstrong GN, Gramatges MM, et al. ; Gliogene Consortium Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst. 2015;107(1):384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwartzentruber J, Korshunov A, Liu XY, et al. . Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 39. Yan H, Parsons DW, Jin G, et al. . IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fiddes IT, Lodewijk GA, Mooring M, et al. . Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 2018;173(6):1356–1369 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ronellenfitsch MW, Oh JE, Satomi K, et al. . CASP9 germline mutation in a family with multiple brain tumors. Brain Pathol. 2018;28(1):94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bouaoun L, Sonkin D, Ardin M, et al. . TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat. 2016;37(9):865–876. [DOI] [PubMed] [Google Scholar]
- 43. Poley JW, Wagner A, Hoogmans MM, et al. ; Rotterdam Initiative on Gastrointestinal Hereditary Tumors Biallelic germline mutations of mismatch-repair genes: a possible cause for multiple pediatric malignancies. Cancer. 2007;109(11):2349–2356. [DOI] [PubMed] [Google Scholar]
- 44. Aslan A, Borcek AO, Pamukcuoglu S, Baykaner MK. Intracranial undifferentiated malign neuroglial tumor in Smith-Lemli-Opitz syndrome: a theory of a possible predisposing factor for primary brain tumors via a case report. Childs Nerv Syst. 2017;33(1):171–177. [DOI] [PubMed] [Google Scholar]
- 45. Li FP, Fraumeni JF Jr, Mulvihill JJ, et al. . A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48(18):5358–5362. [PubMed] [Google Scholar]
- 46. de Andrade KC, Frone MN, Wegman-Ostrosky T, et al. . Variable population prevalence estimates of germline TP53 variants: a gnomAD-based analysis. Hum Mutat. 2019;40(1):97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Druker H, Zelley K, McGee RB, et al. . Genetic counselor recommendations for cancer predisposition evaluation and surveillance in the pediatric oncology patient. Clin Cancer Res. 2017;23(13):e91–e97. [DOI] [PubMed] [Google Scholar]
- 48. Villani A, Shore A, Wasserman JD, et al. . Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol. 2016;17(9):1295–1305. [DOI] [PubMed] [Google Scholar]
- 49. Barrett JC, Buxbaum J, Cutler D, et al. . New mutations, old statistical challenges. bioRxiv. 2017:115964. [Google Scholar]
- 50. Sudmant PH, Rausch T, Gardner EJ, et al. ; 1000 Genomes Project Consortium An integrated map of structural variation in 2504 human genomes. Nature. 2015;526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study used biospecimens from the California Biobank Program. Any uploading of genomic data and/or sharing of these biospecimens or individual data derived from these biospecimens has been determined to violate the statutory scheme of the California Health and Safety Code Sections 124980(j), 124991(b), (g), (h), and 103850 (a) and (d), which protect the confidential nature of biospecimens and individual data derived from biospecimens. Certain aggregate results may be available from the authors by request.