See the article by Mouraviev et al in this issue, pp. 797–805.
The study by Mouraviev et al1 brings to light the important concept of assimilating radiomics predictions with other clinical information to augment diagnostic performance. In the most basic terms, radiomics models integrate non-invasive imaging (typically MRI) with specific clinical parameters—such as genetic status from corresponding biopsies—as matched data inputs to train machine-learning (ML) algorithms. These ML algorithms identify the statistical correlations within the training data to non-invasively predict the clinical parameter of interest (eg, epidermal growth factor receptor [EGFR] amplification) on new unseen cases using the MR images alone. This has potentially transformative implications for many aggressive CNS tumors that may not be amenable to complete surgical resection, like glioblastoma (GBM) or multiple brain metastases. Namely, one of the fundamental challenges for individualized oncology (ie, precision medicine) lies in the inability to resolve the internal genetic heterogeneity of tumors like GBM using surgical biopsies alone, because the genetic drug targets from one biopsy location may not accurately reflect those from other parts of the same tumor.2 Similarly, the genetic alterations within a surgically biopsied brain metastasis may differ from other surgically unsampled metastases elsewhere in the brain.3 Ironically, these unbiopsied/unresected tumor populations harbor the most clinically relevant sensitivities to targeted drug therapy, despite remaining “unknown” even after gross total resection. In light of these challenges, recent use of image-localized biopsies and spatially resolved radiomics maps of intratumoral genetic heterogeneity have introduced clinically viable solutions for diagnosing the potentially unique drug targets within the residual unresected tumor segment.4 These maps also offer insight into the dynamics between neighboring genetic subpopulations, which could be leveraged for therapeutic benefit.5 This positions radiomics at the forefront of decision support for the still evolving paradigm of individualized oncology.
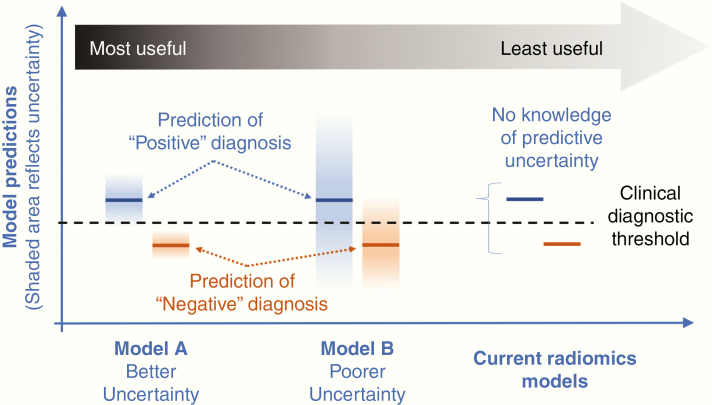
With that said, there are challenges that must be addressed before radiomics can reliably integrate with clinical decision making. For instance, existing radiomics models lack the ability to quantify or report the uncertainty associated with each prediction. Past studies have focused on accuracy (eg, sensitivity/specificity) and covariance (eg, standard error, 95% confidence intervals) in group analyses but have not yet addressed the uncertainty for individual predictions. This uncertainty represents a key feature of all modeling approaches, because it informs how each patient’s data coincide with the model’s ability to make a prediction on those data.6 In the context of ML, predictive uncertainty generally relates to the abundance or sparsity of the model’s training data. The scope of this training data (whether sparse or abundant) establishes the upper and lower bounds of the model domain, which guides the predictions for all new unseen test cases (ie, new prospective patients). Ideally, the new patient’s test data fall within the distribution of the training domain, which allows the radiomics model to interpolate predictions with the lowest uncertainty. However, if the patient’s test data fall outside of the training domain, predictions must be extrapolated, at the cost of greater model uncertainty. Essentially all clinical models suffer from the universal problem of sparse training data, because patient biopsy and imaging data are usually limited. This is particularly true for heterogeneous tumors like GBM, which require image-localized biopsies (and spatially matched MRI measurements) to resolve the regional genetic variability within each tumor. As such, radiomics models invariably run the risk of extrapolation, which underscores the importance of quantifying the uncertainty of each model prediction. As an example, reporting whether a patient’s tumor has a 55% likelihood of EGFR amplification may present different clinical implications than reporting a 99% likelihood of EGFR amplification. But both scenarios are more informative than simply stating that the amplification is present (Fig. 1).7 Predictive uncertainty allows clinicians to understand how each patient’s data relate to the training domain of the model, which can inform clinical confidence and decisions on how to stratify the radiomics outputs relative to other available clinical information and the overall management considerations as a whole.
Fig. 1.
Predictive uncertainty of radiomics models. Many nonmedical ML applications (eg, weather mapping, hydrologic forecasting) employ a probabilistic approach to quantifying predictive uncertainty.7 Such approaches can be implemented in radiomics models to inform the clinical confidence in predicted diagnoses. As an example, in the scenario of high predictive certainty (ie, low uncertainty) (Model A, far left), individual predictions are determined from the mean values (dark blue and orange lines) of relatively narrow probability distributions (blue and orange bars). These narrow distributions represent relatively low predictive uncertainty, and in this case, are well separated from the clinical diagnostic threshold (dotted line) guiding prospective diagnosis. This type of model provides the greatest utility when it achieves both high classification accuracy (ie, positive vs negative diagnosis) as well as high predictive certainty (ie, narrow probability distributions and low uncertainty) in the diagnosis provided by the model. An example of a less useful model (Model B, middle) would exhibit lower predictive certainty (ie, higher uncertainty) as represented by broader probability distributions (blue and orange bars) that cross the diagnostic threshold (dotted line). Such overlap would convey lower clinical confidence in the model-generated predictions of positive vs negative diagnosis (dark blue and orange lines, respectively). Regardless, having knowledge of predictive uncertainty—whether high or low—provides valuable information and inherent confidence for clinical teams to stratify model outputs relative to other clinical diagnostic information. Both Models A and B are preferred to current radiomics models (far right) that lack the ability to quantify predictive uncertainty.
The clinical integration of radiomics models will also demand continued improvements in predictive performance and biological interpretability, particularly when applied to individual patients in prospective fashion. It has been shown that interpatient biological variations can impact the strength of correlation between a patient’s imaging data and their underlying tumor biology.8 Interpatient differences are evident even when stratifying by the most fundamental of biological features, such as patient sex (ie, male vs female), which influences not only tumor behavior and gene expression, but also how these events phenotypically manifest on imaging.9 As such, rather than trying to generalize models for a broad patient population through a “one-model-fits-all” approach, a more promising avenue may be to tailor the model to fit the individual patient, or subgroups of patients, much like treatments are tailored in the context of personalized medicine. This individualized approach to radiomics has already shown substantial gains in model performance for predicting tumor cell density and extent.8 Similar approaches should be translated to other radiomics applications in the future. It is also incumbent upon us, as physicians and clinical scientists, to continue to impose our biological understanding and expertise in the development and application of radiomics models. We should not accept a purely “black-box” approach to diagnosing and managing our patients. While ML offers obvious benefits, it is a data-driven approach with inherent limitations. For instance, ML generally suffers in the ability to generalize correlations beyond the training data (eg, extrapolate) or integrate the spatial relationships of neighboring and potentially correlated predictions/biopsies within the same patient tumor. ML outputs also lack constraints from grounded biological principles, which can affect the feasibility of some predicted outputs (eg, tumor growth/invasion along brain edges, prediction of genetic aberrations in normal brain). As an example, initial efforts in hybrid modeling—through the integration of biological inferences from mechanistic models—have shown improvements in biological interpretability and predictive performance over pure ML data-driven models in specific clinical contexts.10 Continued work is needed to expand such approaches to broader radiomics applications.
Acknowledgment
This editorial is the sole product of the author(s). No third party had input or gave support to its writing.
Funding
This work was supported by NIH NS082609 (L.S.H.), NIH CA221938 (L.S.H.), NIH CA220378 (L.S.H., K.R.S.), NIH CA210180 (L.S.H., K.R.S.), James S. McDonnell Foundation (K.R.S.), Ben and Catherine Ivy Foundation (K.R.S.), and the Arizona Biomedical Research Commission (L.S.H., K.R.S.).
Conflict of interest statement
US Patents: US8571844B2 (K.R.S.); US Patent Applications: 15/290,963 (L.S.H.); PCT/US2018/061887 (L.S.H., K.R.S.); PCT/US2019/019687 (L.S.H., K.R.S.).
References
- 1. Mouraviev A, Detsky J, Sahgal A, et al. . Use of radiomics for the prediction of local control of brain metastases after stereotactic radiosurgery. Neuro Oncol. 2020;22(6):797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Snuderl M, Fazlollahi L, Le LP, et al. . Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. [DOI] [PubMed] [Google Scholar]
- 3. Brastianos PK, Carter SL, Santagata S, et al. . Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5(11):1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu LS, Ning S, Eschbacher JM, et al. . Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol. 2017;19(1):128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69(11):4894–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hawkins-Daarud A, Johnston SK, Swanson KR. Quantifying uncertainty and robustness in a biomathematical model-based patient-specific response metric for glioblastoma. JCO Clin Cancer Inform. 2019;3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quiñonero-Candela J, Rasmussen CE, Sinz F, Bousquet O, Schölkopf B.. Evaluating Predictive Uncertainty Challenge. Machine Learning Challenges. Evaluating Predictive Uncertainty, Visual Object Classification, and Recognising Tectual Entailment 1–27. 2006. doi: 10.1007/11736790_1. [DOI] [Google Scholar]
- 8. Hu LS, Yoon H, Eschbacher JM, et al. . Accurate patient-specific machine learning models of glioblastoma invasion using transfer learning. AJNR Am J Neuroradiol. 2019;40(3):418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang W, Warrington NM, Taylor SJ, et al. . Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci Transl Med. 2019;11(473):pii: eaao5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaw N, Hawkins-Daarud A, Hu LS, et al. . Integration of machine learning and mechanistic models accurately predicts variation in cell density of glioblastoma using multiparametric MRI. Sci Rep. 2019;9(1):10063. [DOI] [PMC free article] [PubMed] [Google Scholar]