Abstract
Background
More than 300 million people carry a diagnosis of asthma, with data to suggest that they are at a higher risk for infection or adverse outcomes from severe acute respiratory syndrome coronavirus 2. Asthma is remarkably heterogeneous, and it is currently unclear how patient-intrinsic factors may relate to coronavirus disease 2019.
Objective
We sought to identify and characterize subsets of patients with asthma at increased risk for severe acute respiratory syndrome coronavirus 2 infection.
Methods
Participants from 2 large asthma cohorts were stratified using clinically relevant parameters to identify factors related to angiotensin-converting enzyme-2 (ACE2) expression within bronchial epithelium. ACE-2–correlated gene signatures were used to interrogate publicly available databases to identify upstream signaling events and novel therapeutic targets.
Results
Stratifying by type 2 inflammatory biomarkers, we identified subjects who demonstrated low peripheral blood eosinophils accompanied by increased expression of the severe acute respiratory syndrome coronavirus 2 receptor ACE2 in bronchial epithelium. Genes highly correlated with ACE2 overlapped with type 1 and 2 IFN signatures, normally induced by viral infections. T-cell recruitment and activation within bronchoalveolar lavage cells of ACE2-high subjects was reciprocally increased. These patients demonstrated characteristics corresponding to risk factors for severe coronavirus disease 2019, including male sex, history of hypertension, low peripheral blood, and elevated bronchoalveolar lavage lymphocytes.
Conclusions
ACE2 expression is linked to upregulation of viral response genes in a subset of type 2–low patients with asthma with characteristics resembling known risk factors for severe coronavirus disease 2019. Therapies targeting the IFN family and T-cell–activating factors may therefore be of benefit in a subset of patients.
Key words: COVID-19, ACE2, coronavirus, SARS-CoV-2, asthma, interferons, viral response, Type-2 low
Abbreviations used: ACE2, Angiotensin-converting enzyme-2; BAL, Bronchoalveolar lavage; BEC, Bronchial epithelial cell; COVID-19, Coronavirus disease 2019; IMSA, Immune Modulation in Severe Asthma; PC1, Patient cluster 1; PC4, Patient cluster 4; SARP, Severe Asthma Research Program; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; STAT, Signal transducer and activator of transcription; T1, Type 1; T2, Type 2
Evolving epidemiological data during the coronavirus disease 2019 (COVID-19) pandemic have shed light on populations at risk for severe infection. Chronic lung disease is consistently listed as a risk factor, though relationship to specific underlying conditions is unclear.1 The impact of asthma remains particularly controversial,2 ranging anywhere from protective effect to 3-fold risk for hospitalization in young adults.1 Furthermore, asthma is a remarkably heterogeneous disease with differing underlying immunobiology, severity, and response to treatment.3 Thus, a more granular understanding of asthma and its subtypes is required to address the controversies related to the health impact of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Entry of SARS-CoV-2 is mediated by fusion of the viral spike protein and cellular membranes through interaction with cell surface angiotensin-converting enzyme-2 (ACE2).4 , 5 Previous work confirms ACE2 expression by bronchial epithelial cells (BECs) and supports a link to type 1 (T1) immunity.6 , 7 In contrast, asthma, for most, is thought to be a type 2 (T2) (IL-4, IL-5, and IL-13)-driven immune process. Whether this skewing toward T2 immunity and away from T1 could alter COVID-19 clinical outcomes is not yet clear.
Despite this relationship to T2 immunity, the BEC transcriptome of patients with asthma, particularly those with severe disease, is markedly heterogeneous and includes both T2 and T1 signature genes.8, 9, 10 This understanding contributed to the current paradigm for defining asthma by the presence or absence of T2 biomarkers, such as fractional exhaled nitric oxide, blood, and sputum eosinophils.5 While T2-high asthma has been associated with more frequent exacerbations, a subset of severe patients exhibit a T1-polarized immune response typified by increased IFN-γ.11 , 12 As a known viral response gene, IFN-γ could critically impact COVID-19. Thus, we hypothesized that T1 polarization of the immune response in BECs would link to a high-risk COVID-19 phenotype that could be identified in relation to ACE2 gene expression.
To assess the impact of immune polarization on ACE2 expression, we stratified patients by biomarkers of T2 inflammation. We next looked for genes correlated with ACE2 expression to assemble a signature for patients with elevated expression. Ontological term enrichment was performed to ascribe function to these genes and comparison to in vitro stimulation and knockdown condition data sets was done to confirm relationship to pathways and upstream ligands. Patient clustering with our ACE2 gene signature then informed a supervised classification model in which commonly available clinical data were used to predict epithelial gene expression.
Methods
Lead contact and materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sally Wenzel (swenzel@pitt.edu).
Human subjects
All subjects provided informed consent in accordance with an institutional review board protocol approved by the University of Pittsburgh. Male and female nonsmoking (<10 pack-year and no smoking in the previous year) subjects meeting the European Respiratory Society/American Thoracic Society definition of severe asthma were recruited.13 Also recruited were subjects with milder asthma on no or lower doses of inhaled corticosteroid (FEV1 >60% predicted with no history of frequent or severe exacerbations of asthma in previous year) and healthy controls. Subjects comprised all racial/ethnic backgrounds and were between the ages of 18 and 60 years. Subjects underwent extensive baseline characterization, including physiologic (spirometry, diffusing capacity, and PC20), allergic, and clinical characterization.14 Subjects also underwent bronchoscopy per published protocols and consistent with procedures established at the University of Pittsburgh consistent with the Severe Asthma Research Program (SARP).
Epithelial cell transcriptional profiling
Microarray expression profiling was performed on isolated epithelial cells from subjects of the SARP cohort as previously described.9 Participants enrolled in the Immune Modulation in Severe Asthma (IMSA) cohort underwent research bronchoscopy with endobronchial brushing. RNA isolated from endobronchial brushings was used to generate cDNA libraries using the Illumina TruSeq RNA Library Prep Kit (Illumina, San Diego, Calif). The libraries were sequenced using the Illumina NextSeq500 System. Quality control for raw FASTQ files was performed using FastQC, and the low-quality reads and 3’ adapters were trimmed with Trim Galore! (Babraham Bioinformatics, Cambridge, United Kingdom). The RNA sequence aligner STAR was used to align the trimmed reads to the reference human genome (hg19).15 Gene expression was subsequently quantified by counting the number of read fragments uniquely mapped to genes using featureCounts.16 Gene expression data were normalized using the rLog package from the DESeq2 suite.17 Samples were controlled for batch effect using the COMBAT algorithm.18 Duplicate gene identities were resolved using maximum values.
Whole transcriptome correlation analysis
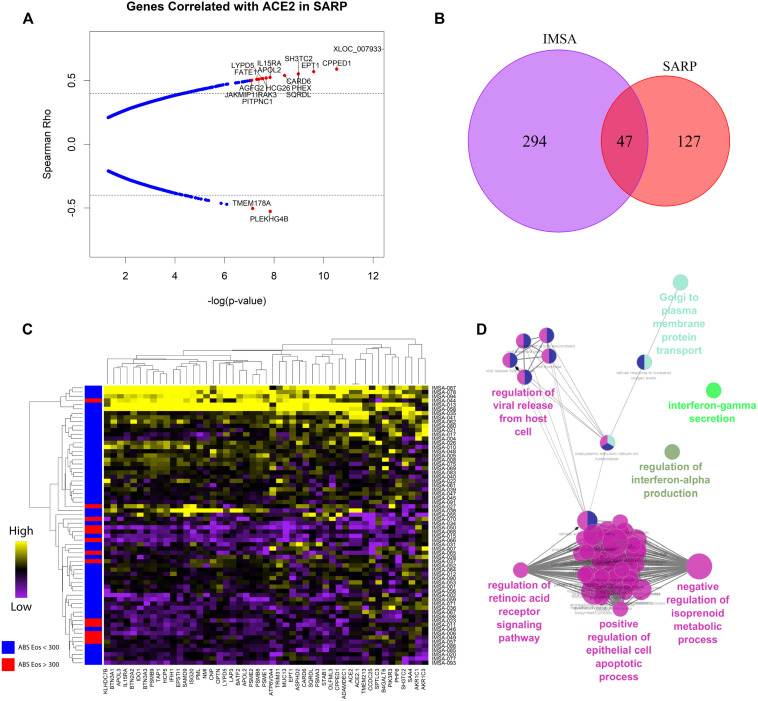
Spearman rank correlation was calculated between ACE2 and all other genes in both the IMSA and SARP epithelial cell data sets, followed by P-value adjustment for a false-discovery rate of less than 5%. For assembly of ACE2-correlated signatures, an arbitrary cutoff set at the 99th percentile of all positive rho values for a given data set was selected. This corresponded to 122 genes in IMSA and 189 in SARP. Hashed lines correlation plots indicate these respective cutoffs. Negative correlations were not included in overlap analysis.
Epithelial phenotype scoring
Patients in the SARP and IMSA cohorts were scored for strength of expression for curated lists of genes related to asthma phenotype. T1 asthma scoring was performed using the geometric mean of 8 genes identified as upregulated by IFN family members and increased in patients with asthma compared with healthy controls.19 The T2 score was similarly calculated using 3 genes identified as upregulated by IL-13 stimulation of cells and increased in asthma.10
Overlap with Library of Integrated Network-Based Cellular Signatures gene sets
Hypergeometric overlap of the ACE2-correlated signature was performed via tools available through the National Institutes of Health Library of Integrated Network-Based Cellular Signatures.20 Comparison with gene lists related to viral infection was accomplished using Enrichr21 to interrogate the “Virus Perturbations from GEO” data set. Metadata from gene lists that demonstrated significant overlap with P value less than .05 after correction for multiple testing were mined for semantic similarity using hierarchical clustering based on cosine similarity measurement, identifying SARS-CoV as the most common term. CommunityMap was used to interrogate overexpressed gene signatures and ligand stimulation conditions for similarity with ACE2-correlated genes.
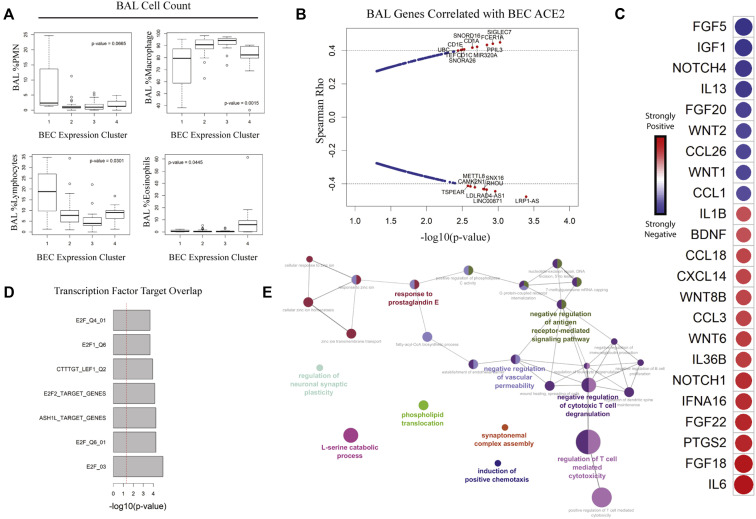
Transcription factor target analysis
Using Enrichr, the ACE2-correlated gene signature was used to interrogate the ChEA,22 TTRUST,23 and ENCODE24 databases for overlapping target sets. Metadata from target lists that demonstrated significant overlap with P value less than .05 after correction for multiple testing were mined for semantic similarity using hierarchical clustering based on cosine similarity measurement, allowing for quantification of term frequency for pie chart assembly.
Graph-based transcription factor activity prediction
Genes identified as transcription factor targets from interrogation of databases as detailed above were used for network assembly using the R package linkcomm (version 1.0-12).25 Connectivity between network neighborhoods was used to identify transcription factors most likely to account for ACE2-correlated genes via greatest connectedness scoring.
Transcription factor binding site prediction
The National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq NM_021804, chrX:15620192) of the ACE2 gene was used for transcription factor binding site prediction with the Web-based Contra (version 3) platform.26 The top-ranking transcription factor predictions from semantic similarity and graph-based analysis were used to interrogate the genomic region 500 bp upstream of the ACE2 locus through the promoter region.
Gene ontology map construction
Gene lists were submitted to the Cytoscape27 (version 3.8.0) application ClueGo.28 Network complexity was set to medium, with filtering of gene ontology terms that met a P value of overlap less than .001. Redundant term collapse was enabled.
Estimation of number of patient clusters
The determination of patient group number used to cut the dendrogram of epithelial expression data for both IMSA and SARP cohorts was accomplished via gap statistic calculation using the R package “NbClust” (version 3.0).29
Clustering of patients based on BEC gene expression
Following filtering for the cohort-specific ACE2-correlated gene signature, dendrograms were assembled using Euclidean distance and the “ward.D2” method of hierarchical clustering. The dendrograms were then cut using the number of patient groups determined by gap statistic as described above.
BEC cluster prediction modeling
To build a predication model for BEC expression cluster using cell count and clinical data, we used sparse partial least squares discriminant analysis within the “mixOmics” package in R (version 6.10.8).30 Model training was performed using the IMSA data set using all patients including healthy controls, patients with mild to moderate asthma, and patients with severe asthma. Initial input for model parameters included age, FEV1% predicted, exhaled nitric oxide, systolic blood pressure, diastolic blood pressure, sex, and absolute blood cell counts. The number of weighted vectors and variables included in our model was chosen on the basis of tuning and cross-validation. MixOmics offers leave-one-out or fold cross-validation as options for these steps, and models assembled using both protocols performed similarly in area under the receiver operating characteristic curve generation and model error rate measurement. A model featuring absolute lymphocyte count, absolute eosinophil count, diastolic blood pressure, and sex was determined to have optimal performance. Because blood pressure measurements were not available for SARP, external cross-validation could not be performed. For the area under the receiver operating characteristic curve, 5-fold cross-validation was used. Receiver operating characteristic curves were calculated as one class versus the others using 5-fold validation. Reported area under the curve values (right of plot) are based on comparison of predicted scores of one class versus the others using a 2-component model. Wilcoxon test for one class versus the others met a significance threshold of P less than .05 for all groups.
Ligand response estimation
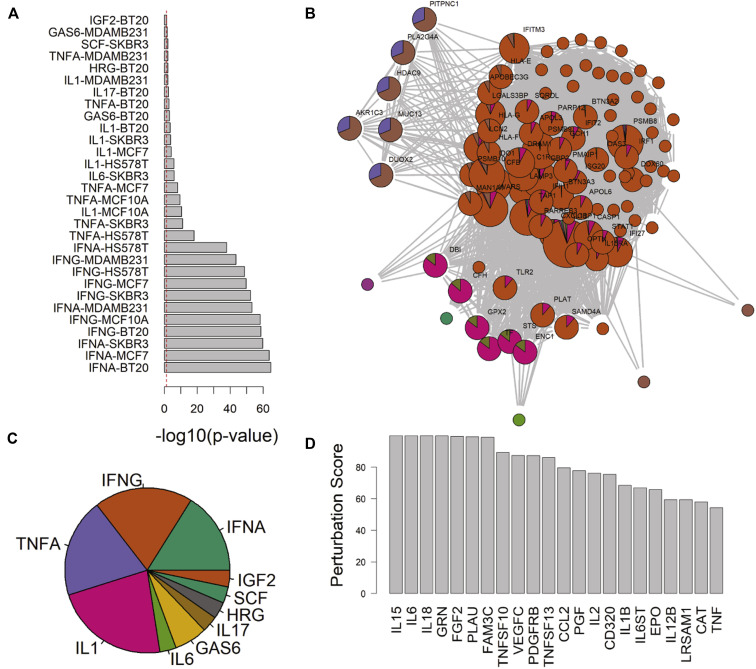
Ligand response data sets were aggregated for pie chart construction using semantic similarity as described above. A graph-based approach was taken to identify genes that belonged to at least 5 response conditions to form a list of “hub” genes. This list was then used for CommunityMap interrogation of knockdown perturbations that opposed their expression. The resultant conditions were filtered for cytokines and soluble factors.
Statistical analysis
All statistical analysis was performed using the R computing environment (version 3.5.3 or 3.6.1 contingent on package dependencies) unless otherwise noted. Statistical testing and methodology are described within figure legends or above within context-specific methodologies. For presentation of boxplots, bars represent median values, with bounds of boxes representing interquartile range and whiskers representing 1.5 times the upper or lower interquartile range. For comparison of categorical variables across BEC expression clusters, independence of distribution was calculated via likelihood-ratio chi-square test.
Data and software availability
BEC and BAL gene expression datasets are available through the NCBI Gene Expression Omnibus (GEO). Software and custom code will be available upon request.
Results
T2-low asthma expresses higher levels of ACE2 and viral response genes
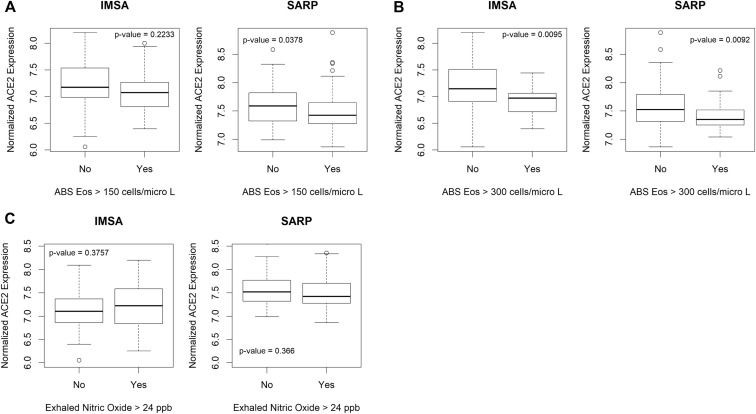
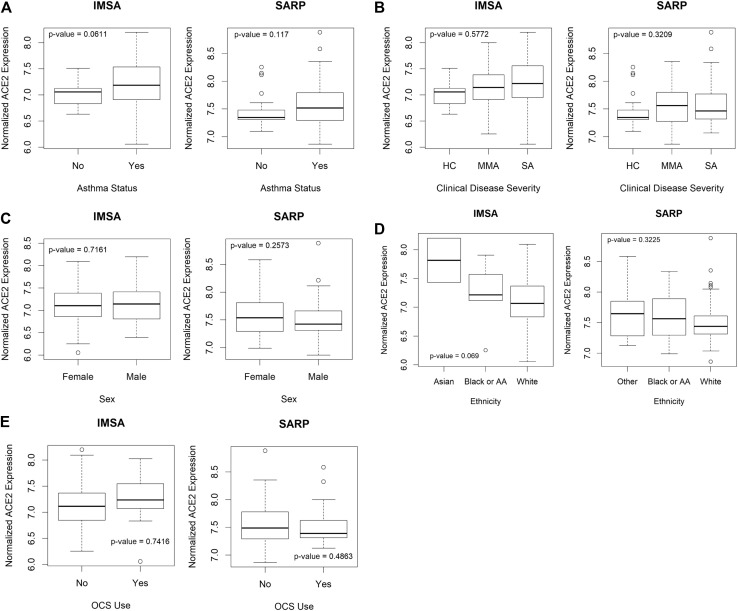
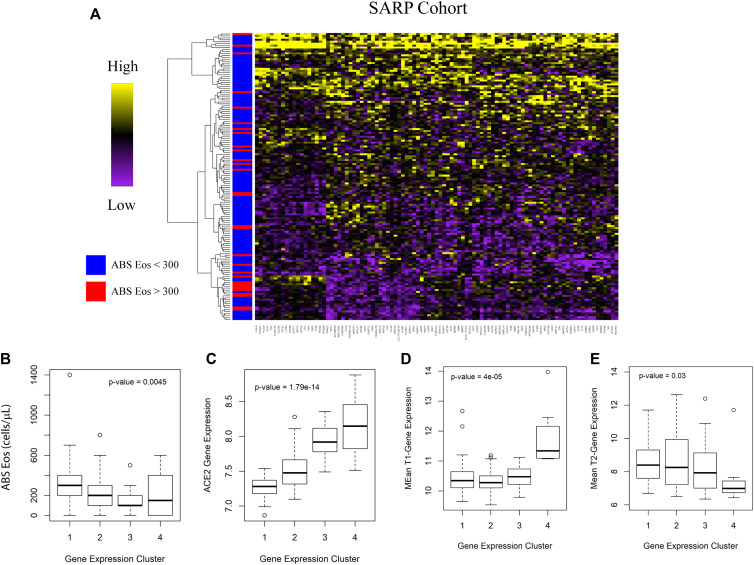
To assess whether expression of ACE2 may be linked to parameters used in the evaluation of patients with asthma, we interrogated 2 large cohorts: our local IMSA cohort (demographic characteristics listed in Table E1 in this article’s Online Repository at www.jacionline.org) as well as BEC gene expression data from phases 1 and 2 of the SARP (demographic characteristics previously reported in Modena et al9).8 , 9 We found that absolute blood eosinophil count, a commonly used biomarker of T2 inflammation, effectively identified patients with differential expression of ACE2 using clinically relevant cutoffs of 150 and 300 cells/μL (see Fig E1 in this article’s Online Repository at www.jacionline.org and Fig 1 , A). Increased ACE2 expression in participants with low blood eosinophils suggests increased capacity for viral binding. No other parameter, including sex, ethnicity, age, or oral corticosteroid use, showed a relationship (see Figs E2 and E3 in this article’s Online Repository at www.jacionline.org).
Fig E1.
Biomarkers for T2 inflammation identify patients with differential ACE2 expression. Evaluation of participants in asthma cohorts for differential expression of ACE2 based on clinically relevant biomarkers of T2 inflammation. A, A cutoff of more than 300 eosinophils/μL of peripheral blood identifies patients with differential expression of ACE2 by bronchial epithelium in the IMSA cohort. Boxplot represents normalized ACE2 expression broken down by blood absolute eosinophil cutoff. Bars represent median values, with bounds of boxes representing interquartile range (IQR) and whiskers representing 1.5 times the upper or lower IQR. Testing was performed using Student t test. B, Differential expression of ACE2 can be identified using a cutoff of either more than 150 or more than 300 eosinophils/μL of peripheral blood in the SARP cohort. C, Comparison of ACE2 level for patients with measured exhaled nitric oxide over the commonly used threshold of more than 24 parts per billion showed no difference in expression in either the IMSA cohort or the SARP cohort. ABS, Abslolute; Eos, eosinophils.
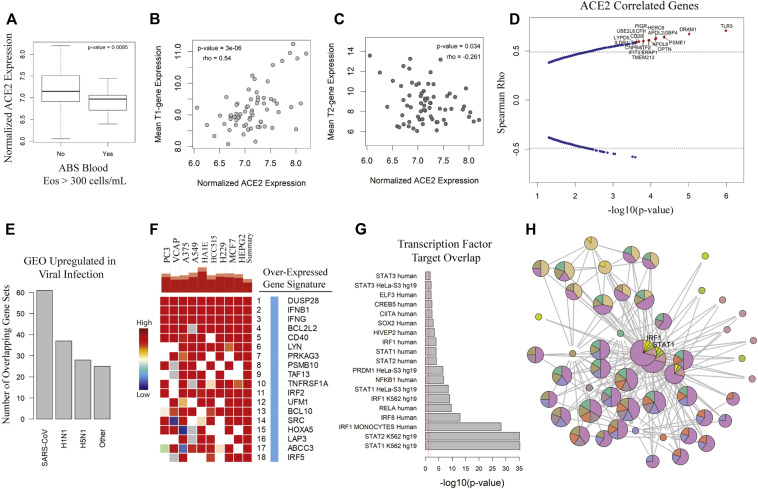
Fig 1.
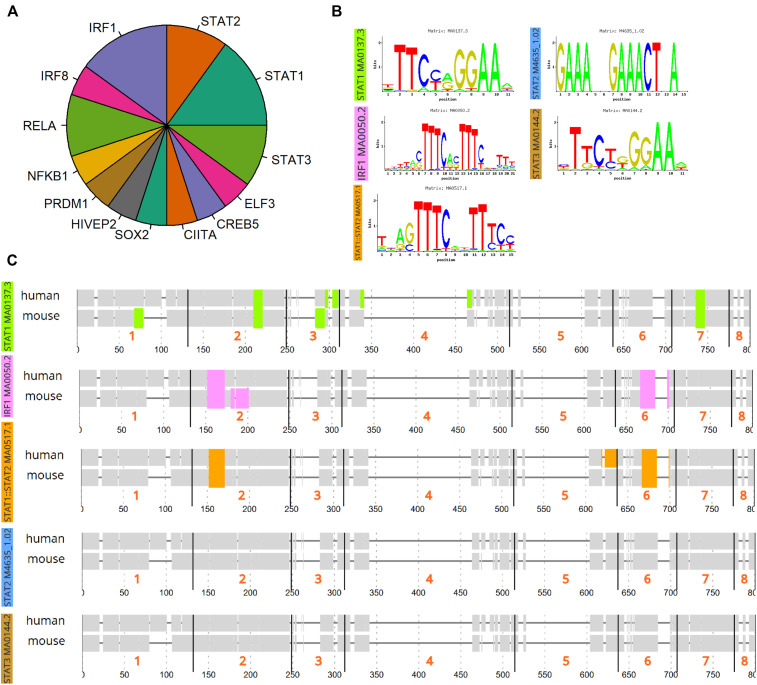
ACE2 is increased in T2-low patients with asthma and is correlated with viral response genes. A, Boxplot of ACE2 by blood absolute (ABS) eosinophil (Eos) cutoff. Comparison made using Student t test. Spearman rank correlation for composite T1 (B) or T2 (C) gene expression score vs ACE2 transcript level with rho and P value as indicated. D, Spearman rho vs −log10(P value) for genes associated with ACE2. Hashed lines indicate 99th percentile of rho values. Genes of interest are highlighted in red. E, Count of GEO data sets upregulated in viral infections overlapping with ACE2-correlated genes. F, Heatmap indicating strength of overlap between in vitro overexpression conditions from the LINCS database vs ACE2-correlated signature from the IMSA cohort. G, Plotting of TFT sets overlapped with BEC ACE2-correlated signature from the CHEA, TTRUST, and ENCODE libraries. H, Graphical resolution of membership between TFT sets identifying STAT1 and IRF1 as having greatest enrichment in the ACE2-correlated gene list. LINCS, Library of Integrated Network-Based Cellular Signatures; TFT, transcription factor target.
Fig E2.
Relationship between clinical parameters and ACE2 expression. Testing for differential expression of ACE2 using clinical parameters including (A) asthma status, (B) clinical disease severity, (C) sex, (D) ethnicity, or (E) oral corticosteroid (OCS) use did not reveal significant variation in either the IMSA cohort or the SARP cohort. Boxplots of normalized ACE2 expression broken down clinical parameters are plotted as indicated. Bars represent median values, with bounds of boxes representing IQR and whiskers representing 1.5 times the upper or lower IQR. Testing was performed using Student t test. AA, African American; HC, healthy control; IQR, interquartile range; MMA, mild to moderate asthma; SA, severe asthma.
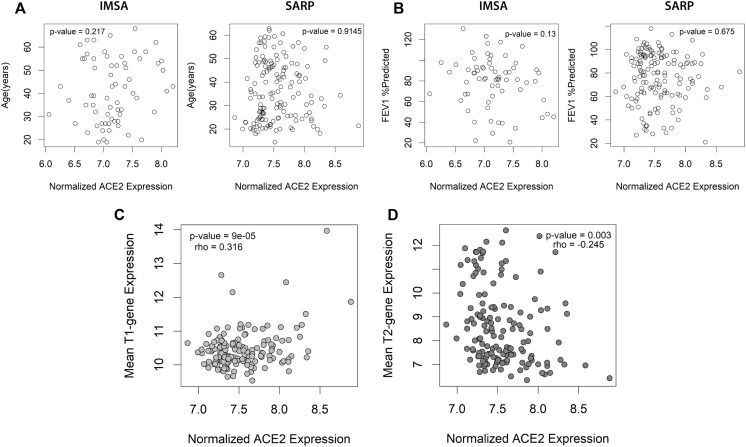
Fig E3.
Correlation between other clinical parameters and ACE2 expression. Spearman rank correlation between (A) age or (B) FEV1% predicted and normalized ACE2 expression using Spearman rank was performed in both IMSA and SARP cohorts, demonstrating no significant relationship. Spearman rank correlation for composite T1 (C) or T2 (D) gene expression score vs ACE2 transcript level for participants in the SARP cohort with rho and P value as indicated.
Patients with asthma with lower blood eosinophils and higher ACE2 expression are also likely to have low levels of T2 inflammation as suggested by poor historic response to therapies targeted at T2 cytokines such as IL-4 or IL-5.3 To assess for relationship to clinical asthma phenotype, we compared ACE2 expression to T1 or T2 signatures in BECs (see Fig E3 and Fig 1, B and C), identifying positive correlation between ACE2 and T1 score and negative correlation between ACE2 and T2 score. These data suggest that high levels of ACE2 expression are more likely in T2-low patients with asthma. On examining genes correlated with ACE2 in BECs of the IMSA cohort, we found striking correlation to transcripts identified as upregulated in viral infection31 (Fig 1, B and C). This included genes known to be upregulated by influenza family members H1N1 and H5N1, as well as the highly related SARS-CoV, which also uses ACE2 for cell entry.5 , 6 Interrogation of publicly available gene signatures through the Library of Integrated Network-Based Cellular Signatures showed overlap between ACE2-correlated genes from BECs and those induced by in vitro overexpression of known viral response genes IFNB1, IFNG, and CD40.32
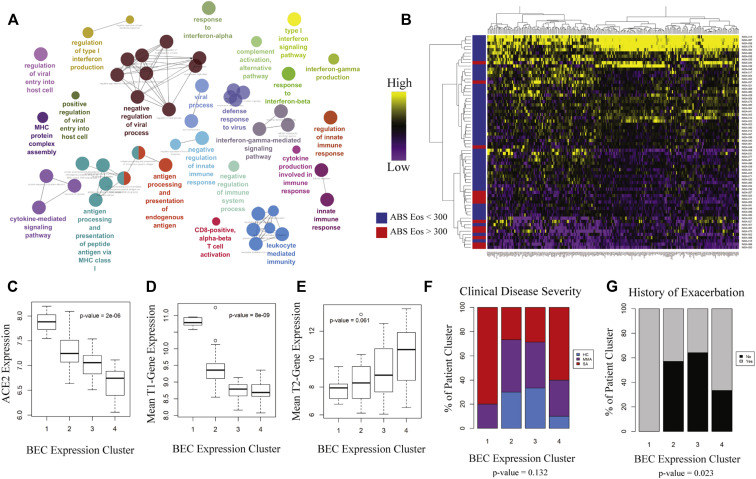
To corroborate activation of downstream pathways, we searched transcription factor target databases ChEA, TTRUST, and ENCODE using our ACE-2 correlated gene signature (Fig 1, E).22, 23, 24 Aggregating these results by semantic similarity, we found strong evidence supporting activation of multiple transcription factors including signal transducer and activator of transcription (STAT) 1, IFN regulatory factor 1, nuclear factor Kappa B subunit 1, and STAT3 (see Fig E4 in this article's Online Repository at www.jacionline.org). Implementing a graph-based approach to resolve redundancy between enrichment sets, we identified STAT1 and IRF1 as having the highest predicted activity in relation to expression of ACE2-correlated genes (Fig 1, F). Supporting a role for STAT1 and IRF1 activation, putative binding sites were identified at the ACE2 locus on the human X chromosome using genomic regulatory factor databases26 , 33 (see Fig E3). Importantly, the correlation between ACE2 and viral response genes was independently recapitulated in the SARP cohort (see Fig E5 in this article’s Online Repository at www.jacionline.org). Transcripts associated with ACE2 overlapped between cohorts and shared enrichment for ontological terms such as regulation of IFN-α and IFN-γ as well as viral release from host cell.34 , 35 These gene ontology terms, as well as those relating to T-cell activation, innate immune regulation, and antigen processing, were significantly enriched in an expanded ACE2-correlated signature (Fig 2, A ).
Fig E4.
Transcription factor binding predictions at the ACE2 genomic locus. A, Results of transcription factor target analysis from the CHEA, ENCODE, and TTRUST data sets were aggregated using semantic similarity and relative count plotted as fractions in pie chart. B, ConTra analysis of predicted transcription factor binding sites at the ACE2 genomic locus on the X chromosome was performed using the relevant JASPAR binding sequences indicated in the figure. C, Predicted sites for STAT1, IRF1, STAT1:STAT2, STAT2, and STAT3 within the ACE2 promoter region through 500 bp upstream.
Fig E5.
ACE2-correlated genes are consistent between independent asthma cohorts. A, Plotting of Spearman correlation rho vs −log10(P value) for genes associated with ACE2 in BECs in the SARP cohort with P value less than .05 after correction for multiple testing for FDR less than 5%. Hashed lines indicate the 99th percentile of rho values. Genes of interest are highlighted in red as indicated in the figure. B, Venn diagram indicating overlap of genes in the IMSA and SARP cohorts that had a Spearman rho value of more than 0.4, with P value less than .05 after correction for multiple testing for FDR less than 5%. C, Heatmap of expression of genes identified in overlap analysis across patients in the IMSA cohort. Row side bar indicates those participants with absolute blood eosinophil counts less than (blue) or greater than (red) 300 cells/μL. Patients (rows) and genes (columns) are ordered according to similarity. D, Gene ontology enrichment term mapping for transcripts correlated with ACE2 in both IMSA and SARP cohorts using the Cytoscape plug-in ClueGo. FDR, False-discovery rate.
Fig 2.
Patient clustering using ACE2-correlated genes identifies 2 groups of patients with severe asthma with distinct immune profiles. Following identification of an ACE2-correlated gene signature within the IMSA cohort, (A) gene ontology enrichment mapping was performed using the Cytoscape plug-in ClueGo. Only terms that met a significance threshold of P value less than .001 are included in the graph. B, Heatmap of expression of ACE2-correlated genes within BECs across patients in the IMSA cohort. Row side bar indicates those participants with absolute blood eosinophil counts less than (blue) or greater than (red) 300 cells/μL. C, Boxplot of normalized ACE2 expression across BEC expression clusters. Kruskal-Wallis testing for variation in expression was performed, with P value indicated on the graph. Boxplot of mean T1 (D) or T2 (E) gene expression across BEC expression clusters. Breakdown of (F) clinical disease severity and (G) history of exacerbation in preceding year across BEC expression clusters. ABS, Absolute; Eos, eosinophils.
Clustering of patients using an ACE2-correlated gene signature identifies differing asthma risk phenotypes
Having found relationship between T1 immune response and ACE2 expression within BECs, we next identified subjects within the IMSA cohort who demonstrated upregulation of this axis. Using genes most highly correlated with ACE2, we clustered participants on the basis of BEC gene expression (Fig 2, B and C). As anticipated, the 4 clusters differed in levels of blood eosinophils (Fig 3 , D), which varied significantly along with their ACE2 expression (Fig 2, C). Mean T1 gene expression score was highest in patient cluster 1 (PC1), which also exhibited the highest ACE2 transcript level (Fig 2, D). PC1 scored lowest in mean T2 gene expression despite being composed entirely of patients with severe asthma (Fig 2, E and F). Although the clusters did not differ by disease severity (Fig 2, E), asthma exacerbation history was highest in PC1 and patient cluster 4 (PC4) (Fig 2, G), which corresponded to opposite ends of the spectrum for blood eosinophils, T1/T2 scoring, and ACE2 expression. Importantly, patient clusters with similar molecular and clinical characteristics could be independently identified within the SARP cohort (see Fig E6 in this article’s Online Repository at www.jacionline.org), suggesting generalizability to these findings.
Fig 3.
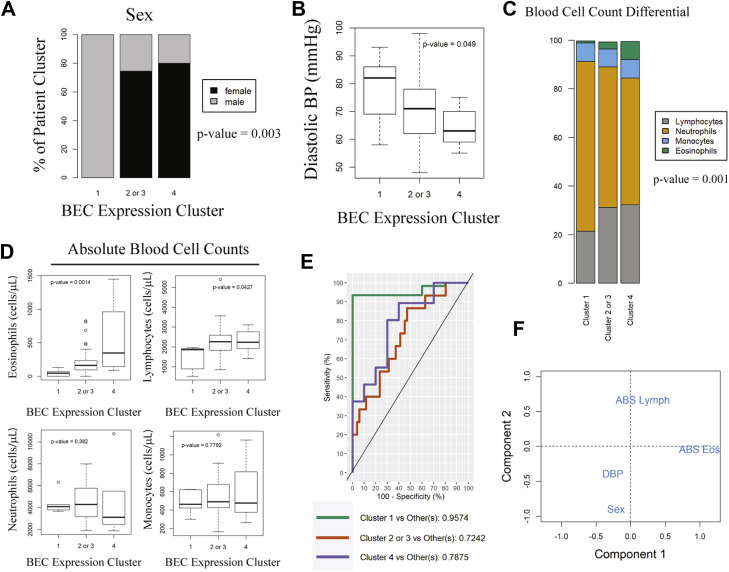
Noninvasive clinical and peripheral blood factors identify the potentially high COVID-19 risk in PC1. A, Distribution of sex across BEC expression clusters. Independence of distribution was calculated via likelihood-ratio chi-square test. B, Boxplot of diastolic BP across BEC expression clusters. Variation was tested by Kruskal-Wallis. C, Stacked bar chart of peripheral blood differential cell counts across BEC expression clusters in the IMSA cohort. Height of bars represents mean cell percentages across patients in the BEC cluster. P value of difference in proportion for neutrophils and lymphocytes is reported in the figure. D, Absolute blood cell counts plotted across BEC expression clusters. Variation was tested using Kruskal-Wallis. E, ROC curve of prediction model for BEC expression cluster using differential blood cell count, sex, and diastolic BP. Wilcoxon test for one class vs the others met a significance threshold of P less than .05 for all groups. F, Plot of variance for included prediction parameters across components used in model. ABS, Absolute; BP, blood pressure; DBP, diastolic blood pressure; Eos, eosinophils; Lymph, lymphocytes; ROC, receiver operating characteristic.
Fig E6.
Clustering of the SARP cohort by ACE2-correlated genes recapitulates groupings similar to IMSA. Ability to cluster patients into clinically meaningful groups was validated in the SARP cohort using ACE2-correlated genes. A, Heatmap of expression of ACE2-correlated genes across patients in the SARP cohort. Row side bar indicates those participants with absolute blood eosinophil counts less than (blue) or greater than (red) 300 cells/μL. Patients are clustered according to similarity in gene expression. B, Boxplot of peripheral blood absolute eosinophil count across BEC clusters in the SARP cohort. Bars represent median values, with bounds of boxes representing IQR and whiskers representing 1.5 times the upper or lower IQR. Kruskal-Wallis testing was performed, with P value indicated on the graph. C, Boxplot of normalized ACE2 expression across BEC expression clusters. Variation in expression was tested by Kruskal-Wallis. ABS, Absolute; Eos, eosinophils; IQR, interquartile range.
Noninvasive biomarkers predict a high-risk ACE2 patient cluster
Several risk factors have been associated with more severe COVID-19 outcomes including male sex, hypertension, and high neutrophil-to-lymphocyte ratios in peripheral blood.2 , 36 , 37 The ACE2-high PC1, which was exclusively male (Fig 3, A), also exhibited higher resting diastolic blood pressure and trended toward a greater history of physician-diagnosed hypertension (χ2 P value of .113) (Fig 3, B). Consistent with increased risk for severe COVID-19, PC1 demonstrated a higher neutrophil-to-lymphocyte ratio36 (Fig 3, C). Absolute blood cell counts also varied between BEC gene expression clusters; PC1 demonstrated significantly lower absolute lymphocytes and eosinophils (Fig 3, D), similarly reflecting reports of lymphopenia and eosinopenia in COVID-19.2 , 36 , 38 , 39 Given these strong relationships with reported COVID-19 risk factors, we developed a predictive model trained on the IMSA data set for patient classification using blood differential cell count, sex, and resting diastolic blood pressure, which achieved an area under the receiver operating characteristic curve of 0.96 for PC1 (Fig 3, E). This model did not require information on asthma severity for classification, making it broadly applicable for BEC gene expression prediction. Examination of model variables showed that readily available parameters could identify ACE2-high patients who are potentially at risk for severe COVID-19 (Fig 3, F).
Reciprocal T-cell activation from a separate lung compartment associates with high-risk PC1
Study of asthma has underscored reciprocal signaling between epithelial and immune cell compartments as crucial for propagation of inflammation.3 , 40 We found variation in the relative composition of compartmentally distinct bronchoalveolar lavage (BAL) immune cells between our BEC patient clusters, with PC1 demonstrating the highest percentages of lymphocytes and PC4 the highest percentages of eosinophils (Fig 4 , A).
Fig 4.
Intercompartmental crosstalk links high epithelial ACE2 with activated BAL lymphocytes. A, Boxplot of BAL immune cell composition across BEC clusters. Variation was tested using Kruskal-Wallis. B, Spearman rho vs −log10(P value) for association of BEC ACE2 with genes in BAL cells. Hashed lines indicate 99th percentile of rho values. Genes of interest are highlighted in red. C, Correlogram of cytokine expression by BAL cells vs epithelial ACE2. Colors of circles indicate directionality: red for positive and blue for negative Spearman rho values. Circle sizes are inversely proportional to P values. D, TFT sets overlapped with BAL ACE2-correlated signature. E, Gene ontology enrichment mapping of BAL ACE2-correlated signature. PMN, Polymorphonuclear cell; TFT, transcription factor target.
To gain more granular insight into how BAL immune cells may affect epithelium, we assessed bulk BAL (RNAsequencing) transcriptional data for genes correlated with BEC ACE2 expression (Fig 4, B). Consistent with the observed increase in T lymphocytes within ACE2-high PC1, BAL genes associated with antigen-presenting cells such as the CD1 family were highly correlated with BEC ACE2.41 Looking at expression of cytokines and soluble factors, we found multiple potential links between immune and epithelial cells (Fig 4, C). Among BAL genes positively correlated with BEC ACE2 were IL6, CCL3, IL36B, and IL1B. CCL3 was shown to regulate the function and migration of CD8+ T cells, a source of IFN-γ, following viral infection.42 T1 immune response genes showed a positive relationship with BEC ACE2 expression, whereas those associated with T2 response such as IL-13, CCL1, and CCL26 showed a negative relationship. These data suggest that BECs from ACE2-high patients interact with BAL immune cells to induce expression of cytokines and chemokines associated with viral infection leading to T-cell activation and T1 polarization.42, 43, 44, 45, 46, 47
To further investigate this relationship, we performed transcription factor target analysis on BAL transcription data and found enrichment of E2F family-related genes in association with BEC ACE2 (Fig 4, D). The transcriptional repressor E2F2 may cooperate with cAMP response element-binding protein (CREB) in the silencing of genes responsible for DNA metabolism and cell cycle regulation,48 suggesting that loss of T-cell quiescence is associated with higher levels of BEC ACE2. Gene ontology term enrichment confirms this relationship between T-cell cytotoxicity and expression of ACE2 in bronchial epithelium (Fig 4, E).
ACE2-correlated genes predict novel therapeutic targets
Targeted treatments for T2-low asthma have remained elusive despite widespread recognition of this phenotype.49 To identify candidates for intervention that may counteract changes seen in ACE2-high epithelium, we interrogated the Library of Integrated Network-Based Cellular Signatures database to find ligand stimulation conditions that induce similar patterns in vitro (Fig 5 , A). In aggregate, this list included multiple hits for type I and II IFNs, TNF-α, and IL-1 (Fig 5, C). Because many of these genes were listed in multiple sets, we used a graph-based approach to identify hub genes shared between them (Fig 5, B). Querying the database for knockdown conditions shown to decrease expression of these hub genes, we pinpointed cytokines that may be reasonable targets for abrogating ACE2-correlated gene expression (Fig 5, D). This included T-cell stimulatory factors IL-15 and IL-12 as well as inflammasome-activated proteins IL-18 and IL-1β. Intriguingly, IL-6 knockdown was tied for the highest predicted impact on ACE2-correlated expression. Anecdotally, treatment of patients with COVID-19 with immunosuppressive drugs, including those specifically targeted at IL-6, has demonstrated some efficacy in severe disease.50 Although these data have not yet been confirmed by randomized controlled trials, pathological data from autopsy have supported increased IL-6 and an overabundance of T cells in fatal cases.51 Thus, it is tempting to hypothesize that an essential element of SARS-CoV2 response may be captured by gene expression changes noted in our cohort studies.
Fig 5.
Novel targets for antagonizing ACE2-correlated gene expression are identified using curated ligand stimulation models. A, Plot of −log10(P value) overlap between ligand stimulation conditions in cultured cell lines and ACE2-correlated BEC signature. B, Graphical approach to resolving overlapping membership of genes between ligand stimulation conditions identified hub genes shared between data sets. C, Pie chart of count for aggregated results from ligand stimulation and ACE2-signature overlap. D, Arbitrary perturbation score plots from the LINCS database for cytokines that negatively impact the expression of ACE2 hub genes upon knockdown in vitro. LINCS, Library of Integrated Network-Based Cellular Signatures.
Discussion
Given the known heterogeneity in asthma, it is not surprising that reports on COVID-19 outcomes in relation to the disease have reported conflicting results. We found that a subset of patients with asthma display a distinctive gene expression signature in their bronchial epithelium that includes co-expression of ACE2 with a viral response network related to IFN signaling among others. These patients exhibit a high T1 and low T2 epithelial gene expression signature, in addition to low blood eosinophils, consistent with a T2-low asthma phenotype. Separately, BAL cells from these individuals show reciprocal increase in their expression of upstream signaling molecules, including CCL3, IL36B, and IFNA16 and IL1B, consistent with compartmental crosstalk. These participants were exclusively male and had higher diastolic blood pressure. Consistent with a higher risk phenotype, they also had eosinopenia, lymphopenia, a higher neutrophil-to-lymphocyte ratio in peripheral blood, and increased BAL lymphocytes.2 , 36 , 38 , 51 Transcription factor target analysis of ACE2-correlated genes identifies potential binding sites for STAT1 and IRF1 at the ACE2 genomic locus, with growing evidence to suggest that ACE2 expression is indeed positively regulated by upstream IFN stimulation.7 These data suggest that activation of host response pathways in some patients with asthma may in fact be deleterious during the COVID-19 pandemic. Further work is needed to confirm the ability to mitigate SARS-CoV-2 infectivity via decrease of cell surface ACE2 through manipulation of specific signaling pathways.
As part of these analyses, another PC was identified (PC4) with shared severity and exacerbation measures but had high blood eosinophils and low ACE2. This cluster is consistent with a recent report that the T2 cytokine IL-13 is suppressive of ACE2 expression in BECs.52 This suppression may explain the associated lower nasal epithelial ACE2 expression in children with atopic asthma. Interestingly, low ACE in children was not associated with high blood eosinophils, suggesting differences between children and adults, and perhaps contributing to the largely low risk of children for severe COVID-19.53
It is important to note that patients in both the IMSA and SARP cohorts were not actively exacerbating and were on stable doses of controller medication at the time of bronchoscopy. Although further validation is required to conclusively determine whether the antiviral gene signature is related to upregulation or downregulation of cell surface ACE2, the concurrent association of ACE2 mRNA with clinical risk factors for COVID-19 gives some reassurance that transcript dose reflects protein. Whether level of ACE2 alone mediates susceptibility to SARS-CoV-2 or severity of COVID-19 remains to be determined.
Conclusions
We provide evidence that easily available noninvasively obtained clinical and inflammatory biomarkers can identify patients with a BEC profile concerning for an overwhelming inflammatory response to SARS-CoV-2. In addition, the substantial association of ACE2-correlated genes with immune pathways for which targeted immunosuppressive therapies are currently available supports future work studying the impact of targeting IFNs, IL-1β, and IL-6 pathways in severe cases.
Clinical implications.
T2-low patients with asthma may be at increased risk for adverse outcome from COVID-19 and deserve increased vigilance upon developing symptoms.
Acknowledgments
We thank Renee Wunderley for human subject recruitment and for obtaining informed consent, and Kathryn Scholl and John Trudeau for processing of the BAL samples. We thank the SARP 1&2 investigators, Eugene Bleecker, William Busse, Serpil C. Erzurum, and Deborah Meyers. We thank Jay Kolls, Wei Chen, Qi Yan, and Mike Gorry for their assistance with RNA sequencing.
Footnotes
This study was supported by the National Institutes of Health (NIH) (grant no. P01AI106684 to A.R. and S.E.W., grant no. R01HL113956 to A.R., and grant no. R01AI048927 to A.R.), NIH grant number U10HL109152 (to S.E.W.), and NIH grant number F32HL14741501 (to M.C.).
Disclosure of potential conflict of interest: S. E. Wenzel is a consultant for AstraZeneca, GlaxoSmithKline, and Sanofi and is also involved in clinical trials being run by Knopp, Sanofi, and AstraZeneca. S. E. Wenzel and A. Ray have research support from Pieris Pharmaceuticals. The rest of the authors declare that they have no relevant conflicts of interest.
Appendix
Table E1.
Demographic characteristics of the IMSA cohort
| Characteristic | Healthy control |
Mild to moderate asthma (MMA) |
Severe asthma (SA) |
P value |
|---|---|---|---|---|
| (n = 17) | (n = 25) | (n = 24) | ||
| Mean age (y) | 40.4 ± 12.9 | 36.6 ± 14.6 | 46.7 ± 11.6 | .025 |
| Sex: male/female | 5/12 | 6/19 | 9/15 | .587 |
| Race (Asian/black and AA/white) | 0/1/16 | 1/4/20 | 1/5/18 | .6192216 |
| Mean BMI (kg/m2) | 26.6 ± 4.5 | 29 ± 6.1 | 32.7 ± 8 | .015 |
| Median Feno (ppb) | 14 ± 6.1 | 30 ± 37.1 | 21 ± 50.2 | .002 |
| Mean ABS blood eosinophils (cells/μL) | 169 ± 86 | 272 ± 233 | 273 ± 386 | .216 |
| Mean ABS blood lymphocytes (cell/μL) | 2077 ± 521 | 2389 ± 514 | 2165 ± 972 | .1274207 |
| Mean FEV1 (%predicted) | 99 ± 13.7 | 84.6 ± 17.3 | 58 ± 22.1 | <.0001 |
| Mean diastolic blood pressure (mm Hg) | 70 ± 9.3 | 69 ± 12.2 | 72.7 ± 11.2 | .4970527 |
ABS, Absolute; BMI, body mass index; Feno, fractional exhaled nitric oxide; ppb, parts per billion.
Mean or median values are as reported in the table. Age, BMI, Feno, ABS blood eosinophils, ABS blood lymphocytes, and diastolic blood pressure are presented as ±1 SD. FEV1% predicted is presented as ±1 SEM. P value of differential testing for age, BMI, Feno, ABS blood eosinophils, ABS blood lymphocytes, diastolic blood pressure, and FEV1 was calculated using Kruskal-Wallis. P value reported for race and sex was calculated using Pearson χ2.
References
- 1.Garg S., Kim L., Whitaker M., O’Halloran A., Cummings C., Holstein R. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019—COVID-NET, 14 states, March 1–30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69 doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan [April 12, 2020]. J Allergy Clin Immunol. https://doi.org/10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed]
- 3.Ray A., Camiolo M., Fitzpatrick A., Gauthier M., Wenzel S.E. Are we meeting the promise of endotypes and precision medicine in asthma? Physiol Rev. 2020;100:983–1017. doi: 10.1152/physrev.00023.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sajuthi SP, DeFord P, Jackson ND, Montgomery MT, Everman JL, Rios CL, et al. Type 2 and interferon inflammation strongly regulate SARS-CoV-2 related gene expression in the airway epithelium [published online ahead of print April 10, 2020]. bioRxiv. https://doi.org/10.1101/2020.04.09.034454. [DOI] [PMC free article] [PubMed]
- 8.Modena B.D., Bleecker E.R., Busse W.W., Erzurum S.C., Gaston B.M., Jarjour N.N. Gene expression correlated with severe asthma characteristics reveals heterogeneous mechanisms of severe disease. Am J Respir Crit Care Med. 2017;195:1449–1463. doi: 10.1164/rccm.201607-1407OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modena B.D., Tedrow J.R., Milosevic J., Bleecker E.R., Meyers D.A., Wu W. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190:1363–1372. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodruff P.G., Modrek B., Choy D.F., Jia G., Abbas A.R., Ellwanger A. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauthier M., Chakraborty K., Oriss T.B., Raundhal M., Das S., Chen J. Severe asthma in humans and mouse model suggests a CXCL10 signature underlies corticosteroid-resistant Th1 bias. JCI Insight. 2017;2 doi: 10.1172/jci.insight.94580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raundhal M., Morse C., Khare A., Oriss T.B., Milosevic J., Trudeau J. High IFN-γ and low SLPI mark severe asthma in mice and humans. J Clin Investig. 2015;125:3037–3050. doi: 10.1172/JCI80911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung K.F., Wenzel S.E., Brozek J.L., Bush A., Castro M., Sterk P.J. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 14.Moore W.C., Bleecker E.R., Curran-Everett D., Erzurum S.C., Ameredes B.T., Bacharier L. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–413. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 17.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhakta N.R., Christenson S.A., Nerella S., Solberg O.D., Nguyen C.P., Choy D.F. IFN-stimulated gene expression, type 2 inflammation, and endoplasmic reticulum stress in asthma. Am J Respir Crit Care Med. 2018;197:313–324. doi: 10.1164/rccm.201706-1070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stathias V., Turner J., Koleti A., Vidovic D., Cooper D., Fazel-Najafabadi M. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 2020 Jan 8;48:D431–D439. doi: 10.1093/nar/gkz1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han H., Cho J.W., Lee S., Yun A., Kim H., Bae D. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalinka A.T., Tomancak P. linkcomm: an R package for the generation, visualization, and analysis of link communities in networks of arbitrary size and type. Bioinformatics. 2011;27:2011–2012. doi: 10.1093/bioinformatics/btr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreft Ł., Soete A., Hulpiau P., Botzki A., Saeys Y., De Bleser P. ConTra v3: a tool to identify transcription factor binding sites across species, update 2017. Nucleic Acids Res. 2017;45:W490–W494. doi: 10.1093/nar/gkx376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charrad M., Ghazzali N., Boiteau V., Niknafs A. NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Softw. 2014;61:1–36. [Google Scholar]
- 30.Rohart F., Gautier B., Singh A., Lê Cao K.-A. mixOmics: an R package for 'omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouillard A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016;2016 doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd Golub AS. L1000 Dataset -small molecule perturbagens- LINCS Trans-Center Project.
- 33.Lesurf R., Cotto K.C., Wang G., Griffith M., Kasaian K., Jones S.J.M. ORegAnno 3.0: a community-driven resource for curated regulatory annotation. Nucleic Acids Res. 2016;44:D126–D132. doi: 10.1093/nar/gkv1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, et al. Neutrophil-to-lymphocyte ratio predicts severe illness patients with 2019 novel coronavirus in the early stage [published online ahead of print February 12, 2020]. medRxiv. 10.1101/2020.02.10.20021584. [DOI]
- 37.Wang B., Li R., Lu Z., Huang Y. Does comorbidity increase the risk of patients with COVID-19: evidence from meta-analysis. Aging (Albany NY) 2020;12:6049–6057. doi: 10.18632/aging.103000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China [published online ahead of print February 19, 2020]. Allergy. https://doi.org/10.1111/all.14238. [DOI] [PubMed]
- 39.Li Q., Ding X., Xia G., Chen H.G., Chen F., Geng Z. Eosinopenia and elevated C-reactive protein facilitate triage of COVID-19 patients in fever clinic: a retrospective case-control study. EClinicalMedicine. 2020 doi: 10.1016/j.eclinm.2020.100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roan F., Obata-Ninomiya K., Ziegler S.F. Epithelial cell-derived cytokines: more than just signaling the alarm. J Clin Investig. 2019;129:1441–1451. doi: 10.1172/JCI124606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porcelli S., Brenner M.B., Greenstein J.L., Terhorst C., Balk S.P., Bleicher P.A. Recognition of cluster of differentiation 1 antigens by human CD4−CD8>−cytolytic T lymphocyte. Nature. 1989;341:447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 42.Trifilo M.J., Bergmann C.C., Kuziel W.A., Lane T.E. CC chemokine ligand 3 (CCL3) regulates CD8(+)-T-cell effector function and migration following viral infection. J Virol. 2003;77:4004. doi: 10.1128/JVI.77.7.4004-4014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Queen D., Ediriweera C., Liu L. Function and regulation of IL-36 signaling in inflammatory diseases and cancer development. Front Cell Dev Biol. 2019;7:317. doi: 10.3389/fcell.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buhl A.L., Wenzel J. Interleukin-36 in infectious and inflammatory skin diseases. Front Immunol. 2019;10:1162. doi: 10.3389/fimmu.2019.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanneganti T.-D. The inflammasome starts rolling. Nat Rev Immunol. 2018;18:483. doi: 10.1038/s41577-018-0027-1. [DOI] [PubMed] [Google Scholar]
- 47.Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 48.Laresgoiti U., Apraiz A., Olea M., Mitxelena J., Osinalde N., Rodriguez J.A. E2F2 and CREB cooperatively regulate transcriptional activity of cell cycle genes. Nucleic Acids Res. 2013;41:10185–10198. doi: 10.1093/nar/gkt821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wenzel S.E. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W., Zhao Y., Zhang F., Wang Q., Li T., Liu Z. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): the perspectives of clinical immunologists from China. Clin Immunol. 2020;214:108393. doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu B., Xu X., Wei H. Why tocilizumab could be an effective treatment for severe COVID-19? J Transl Med. 2020;18:164. doi: 10.1186/s12967-020-02339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson DJ, Busse WW, Bacharier LB, Kattan M, O’Connor GT, Wood RA, et al. Association of respiratory allergy, asthma and expression of the SARS-CoV-2 receptor, ACE2 [published online ahead of print April 22, 2020]. J Allergy Clin Immunol. https://doi.org/10.1016/j.jaci.2020.04.009. [DOI] [PMC free article] [PubMed]
- 53.Li L.Q., Huang T., Wang Y.Q., Wang Z.P., Liang Y., Huang T.B. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol. 2020;92:577–583. doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
BEC and BAL gene expression datasets are available through the NCBI Gene Expression Omnibus (GEO). Software and custom code will be available upon request.